Abstract
Rationale
Cocaine addiction is a major public health problem with a substantial genetic basis for which the biological mechanisms remain largely unknown. Systems genetics is a powerful method for discovering novel mechanisms underlying complex traits, and intravenous drug self-administration (IVSA) is the gold standard for assessing volitional drug use in preclinical studies. We have integrated these approaches to identify novel genes and networks underling cocaine use in mice.
Methods
Mice from 39 BXD strains acquired cocaine IVSA (0.56 mg/kg/infusion). Mice from 29 BXD strains completed a full dose-response curve (0.032 – 1.8 mg/kg/infusion).
Results
We identified independent genetic correlations between cocaine IVSA and measures of environmental exploration and cocaine sensitization. We identified genome-wide significant QTL on chromosomes 7 and 11 associated with shifts in the dose-response curve and on chromosome 16 associated with sessions to acquire cocaine IVSA. Using publicly available gene expression data from the nucleus accumbens, midbrain, and prefrontal cortex of drug-naïve mice, we identified Aplp1 and Cyfip2 as positional candidates underlying the behavioral QTL on chromosomes 7 and 11, respectively. A genome-wide significant trans-eQTL linking Fam53b (a GWAS candidate for human cocaine dependence) on chromosome 7 to the cocaine IVSA behavioral QTL on chromosome 11 was identified in the midbrain; Fam53b and Cyfip2 were co-expressed genome-wide significantly in the midbrain. This finding indicates that cocaine IVSA studies using mice can identify genes involved in human cocaine use.
Conclusions
These data provide novel candidate genes underlying cocaine IVSA in mice, and suggest mechanisms driving human cocaine use.
Keywords: Cyfip2, Fam53b, Aplp1, addiction, QTL, eQTL, genetic correlation, cocaine sensitization, open field, light dark box
INTRODUCTION
After more than 40 years of sustained efforts to control the production, distribution, and consumption of cocaine (Global Commission on Drug Policy 2011), cocaine addiction persists as a widespread public health problem. Over 17 million people use cocaine worldwide (UNODC 2013). In the United States there are over 1.6 million cocaine users, and 69% of these individuals meet DSM-IV criteria for substance dependence or abuse (SAMHSA 2013). Cocaine has the highest abuse potential of all illicit drugs (Goldstein and Kalant 1990), and addiction to drugs including cocaine, alcohol, nicotine, and opiates is highly heritable (Goldman et al. 2005). Twin studies reveal that heritability of heavy cocaine use and abuse is greater than 70%, making cocaine addiction the most heritable of all addiction disorders (Goldman et al. 2005). The combination of high addiction liability and high heritability makes discovery of the genetic underpinnings of cocaine addiction a public health priority. Nevertheless, the genetic factors driving cocaine use and addiction remain largely unknown.
Although a variety of assays have been used to evaluate the reinforcing properties and behavioral effects of cocaine (Sanchis-Segura and Spanagel 2006), intravenous self-administration (IVSA) is widely considered to be the gold standard for assessing volitional drug use in preclinical studies (Mello and Negus 1996; Sanchis-Segura and Spanagel 2006; Thomsen and Caine 2007). The voluntary nature of drug use in IVSA mirrors that of human drug use and confers a high level of construct validity to the paradigm. To date, investigators utilizing the IVSA paradigm to probe the genetic mechanisms underlying cocaine use have primarily used a reverse genetics approach in which individual genes that are hypothesized to contribute to cocaine use are perturbed (e.g., knocked out), and the effect of these perturbations on the phenotype are assessed (Sora et al. 2010). While this approach provides rigorous genetic and environmental control, it relies on preexisting assumptions about genes underlying drug use (Hall et al. 2013), and this precludes discovery of novel and perhaps unexpected genes and mechanisms driving drug use. Furthermore, reverse genetics studies typically test the effect of gene perturbation on a single genetic background, hampering the identification of polygenic networks underlying behavioral traits. In contrast to the reverse genetics approach, systems genetics utilizes an experimental population with known trait variation to perform a global analysis of all biological molecules underlying that trait (Civelek and Lusis 2014). In this way, the genes and networks underlying a trait can be identified in the absence of a priori hypotheses.
In the present study, we used a systems genetics approach in the BXD recombinant inbred (RI) strains (Peirce et al. 2004) to discover novel genes and networks underlying cocaine IVSA. We assessed the acquisition of cocaine IVSA in mice from 39 BXD strains, obtained a complete dose-response curve in mice from 29 strains, and assessed heritability of these traits. We assessed genetic relationships between cocaine IVSA and commonly used assays indexing effects of experimenter-administered cocaine (sensitization, conditioned place preference) as well as phenotypes related to cocaine use in mice and rats (exploration in a novel open field and light/dark box). We identified genome-wide significant quantitative trait loci (QTL) associated with cocaine IVSA phenotypes. We used publicly available gene expression data from the nucleus accumbens (NAc), midbrain, and prefrontal cortex (PFC) of drug-naïve mice to prioritize candidate genes and identify genome-wide significant expression QTL (eQTL), genes, and networks associated with cocaine IVSA phenotypes.
METHODS AND MATERIALS
Subjects
Cocaine IVSA was assessed in male and female mice (N = 99) from 44 BXD RI strains: BXD1/TyJ, BXD2/TyJ, BXD9/TyJ, BXD11/TyJ, BXD13/TyJ, BXD14/TyJ, BXD15/TyJ, BXD16/TyJ, BXD19/TyJ, BXD20/TyJ, BXD21/TyJ, BXD27/TyJ, BXD28/TyJ, BXD29/Ty, BXD31/TyJ, BXD32/TyJ, BXD33/TyJ, BXD34/TyJ, BXD36/TyJ, BXD39/TyJ, BXD40/TyJ, BXD43/RwwJ, BXD50/RwwJ, BXD55/RwwJ, BXD56/RwwJ, BXD61/RwwJ, BXD62/RwwJ, BXD63/RwwJ, BXD65/RwwJ, BXD65b/RwwJ (originally 92), BXD69/RwwJ, BXD70/RwwJ, BXD71/RwwJ, BXD73a/RwwJ (originally 80), BXD75/RwwJ, BXD77/RwwJ, BXD83/RwwJ, BXD84/RwwJ, BXD86/RwwJ, BXD87/RwwJ, BXD90/RwwJ, BXD98/RwwJ, BXD99/RwwJ, BXD100/RwwJ. BXD strains 1 - 40 were obtained from the Jackson Laboratory (Bar Harbor, ME), and BXD strains 43 - 100 were provided by Drs. Lu Lu and Robert W. Williams (University of Tennessee Health Science Center, Memphis, TN). Mice were bred in the Russell Vivarium at Oak Ridge National Laboratory (ORNL). At three weeks of age litters were weaned and shipped from ORNL to the vivarium in the Department of Psychology at the University of Memphis for cocaine IVSA testing. Throughout the experiment, mice had free access to food and water and were maintained in a temperature controlled environment (70 ± 2°F) on a 14:10 light:dark cycle (lights on at 06:00).
Behavioral Testing
The cocaine IVSA testing procedure and chambers have been described in detail previously (Dickson et al. 2011). Prior to catheter implantation, mice were housed in same-sex and same-strain groups of 3 to 5. At 12 weeks of age an indwelling catheter was implanted into the right external jugular vein under oxygen/isoflurane anesthesia. Mice were individually housed following catheter implantation. Behavioral testing began following 3 days of post-surgical recovery. IVSA data were collected using Med Associates operant conditioning chambers. Manipulanda were two retractable response levers (active and inactive). Following three days of post-surgical recovery, mice began cocaine IVSA testing on a fixed-ratio 1 schedule at a dose of 0.56 mg/kg/infusion. Mice were tested in two-hour sessions at the same time daily seven days per week throughout the experiment. Acquisition of cocaine IVSA was defined as two consecutive sessions during which ≥ 10 infusions occurred, infusions did not vary by more than 20%, and at least 80% of lever presses were on the active lever. Following acquisition of cocaine IVSA, the self-administration response was allowed to stabilize at 7 additional doses to establish a dose-response curve. Doses were presented in the following order: 0.56, 1.8, 1.0, 0.32, 0.18, 0.1, 0.056, 0.032 mg/kg/infusion. Stabilization was defined as two consecutive sessions during which infusions did not vary by more than 20% and at least 70% of lever presses were on the active lever.
To maintain patency, catheters were flushed before and after each daily testing session with 25 μl of a heparin lock solution (Hospira; Lake Forest, IL; 100 U/ml heparin/saline). To forestall bacterial infection, mice were infused (2 μl/g) with an enrofloxacin/saline solution (10 mg/kg) immediately before the heparin flush at the end of each session. Catheters were tested for patency with an infusion (2 μl/g) of a methohexital/saline solution (5 mg/kg) between each cocaine dose. Rapid loss of muscle tone was interpreted as an indication of patency. Cocaine hydrochloride was obtained from Sigma-Aldrich Chemical Co. (St Louis, MO, USA). Cocaine doses were calculated as the salt. Methohexital (Brevital®) and enrofloxacin (Baytril®) were obtained from Henry Schein (Melville, NY). Methohexital and cocaine hydrochloride were dissolved in 0.9% saline. All solutions were filtered through 0.22 μm syringe filters.
Genetic Correlations
Using GeneNetwork (www.genenetwork.org), we assessed the genetic correlations between cocaine IVSA infusions and four other measures: cocaine conditioned place preference (CPP), cocaine sensitization, distance traveled in a novel open field, and time on the light side of a light/dark box. We have published these four measures previously (Philip et al. 2010) and they are publicly available on GeneNetwork (GeneNetwork IDs 12051, 12013, 12068, 11904). BXD mice tested on cocaine IVSA, cocaine CPP, cocaine sensitization, open field, and light/dark assays were generated at ORNL; testing on these five assays was performed at the University of Memphis. Consequently, all mice were exposed to the same environmental variables including light cycle, food, water, bedding, and animal handlers. Subjects tested on cocaine IVSA were not the same mice as those tested on other assays.
QTL Mapping
We performed QTL mapping of cocaine IVSA data using GeneNetwork. Whole genome interval mapping was performed using 5000 permutations. When a genome-wide significant QTL (p < .05) was identified, we attempted to identify additional QTLs with composite interval mapping using the peak marker from the initial significant QTL as a control locus. One-way or factorial analysis of variance (ANOVA) was used to assess effects of the C57BL/6J (B6) and DBA/2J (D2) alleles at the peak of identified QTL on IVSA phenotypes. Potential effects of sex were also examined. When repeated measures ANOVA was used, the assumption of homogeneity of variance across groups was assessed using Mauchly’s test of sphericity. If this assumption was violated, the Huynh–Feldt correction was used.
When performing interval mapping using BXD RI strains, the allele at a locus (B6 or D2), not BXD strain, is the between-subjects factor. Consequently, only a single animal of each genotype (i.e., BXD strain) is required in a mapping study, and it is the total number of strains tested, not the number of replicates within a strain, that has the greatest effect on power to detect a QTL (Belknap 1998). Moreover, because allele at a locus is the between subjects factor as opposed to strain, interactions of sex and allele can be assessed without phenotyping male and female mice from all strains.
Gene Candidate Identification and Prioritization
Following the identification of significant QTL, we used QTL Miner (Alberts and Schughart 2010) on GeneNetwork to identify all genes located within the 2 logarithm of odds (LOD) confidence interval (CI) of a behavioral QTL. Genes from this set that exhibited genome-wide significant (p < .05) cis-acting eQTL (cis-eQTL) were considered positional candidates for behavioral QTL. To prioritize positional candidates, we examined co-variation of IVSA phenotypes and expression of candidate genes in the NAc, midbrain, and PFC using publicly available gene expression data sets (GeneNetwork study IDs: 44, 141, 36) (Wolen et al. 2012; Ye et al. 2014). These brain regions are critically involved in cocaine IVSA (Bocklisch et al. 2013; Ito et al. 2004; Marinelli et al. 2003; Pettit et al. 1984; Roberts and Koob 1982; Vassoler et al. 2013; Weissenborn et al. 1997; Wise 2009). For each QTL, candidate genes exhibiting expression levels that covaried significantly with the IVSA phenotype associated with the QTL were considered priority positional candidates. For covariation analysis, the family-wise error rate for each QTL was adjusted using the Bonferonni correction (.05 / number of gene candidates).
Trans-acting eQTL Associated with Cocaine IVSA Behavioral QTL
We reasoned that genes exhibiting expression levels that map to cocaine IVSA behavioral QTL in a brain region with known relevance to cocaine IVSA may be functionally related to the gene or genes underlying those same behavioral QTL. Therefore, we identified genes that exhibited genome-wide significant trans-acting eQTL (trans-eQTL) (p < .05) with a peak located within the 2 LOD confidence interval of a cocaine IVSA behavioral QTL.
Genome-wide Covariation of Cocaine IVSA Phenotypes and Gene Expression
We reasoned that genes exhibiting expression levels that covary with a cocaine IVSA phenotype in a brain region with known relevance to cocaine IVSA may be part of a larger network involved in driving cocaine IVSA behaviors. Therefore, we identified all genes across the genome exhibiting expression levels that covaried genome-wide significantly with cocaine IVSA phenotypes associated with a behavioral QTL. To calculate the genome-wide error rate for each brain region, we used the Bonferroni correction (.05 / number of probe sets). Probe sets not associated with a gene were excluded. Bonferroni-adjusted genome-wide significance levels for the NAc, midbrain, and PFC were 1.1 × 10−6, 1.5 × 10−6, and 1.1 × 10−6, respectively.
RESULTS
Acquisition of Cocaine IVSA
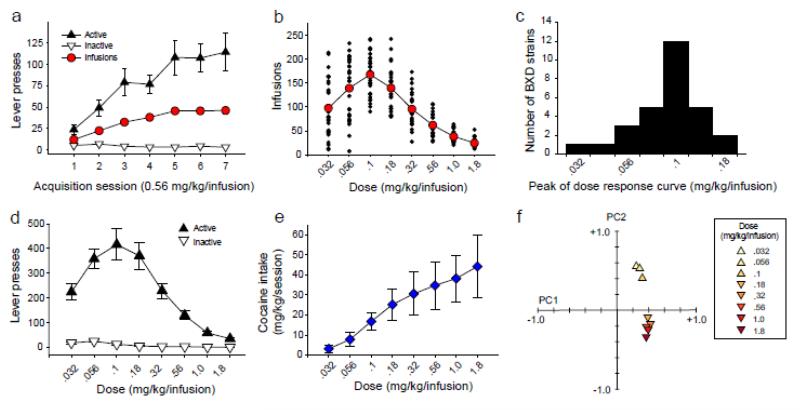
Mice from 39 strains acquired cocaine IVSA (Table S1). The number of days to meet acquisition criteria ranged from 2 to 11 (M = 4.62, SD = 2.12) across the 39 BXD strains. Heritability of this measure was .28 (Table S2).To assess acquisition of cocaine IVSA, we performed a repeated measures ANOVA using number of lever presses as the dependent measure, session (1 - 7) as a within-subjects factor, and lever (active or inactive) as a within-subjects factor. For mice that reached criterion prior to day 7, the mean of active and inactive lever presses on the final two acquisition sessions was carried forward. As a group, BXD strains rapidly learned to lever press for cocaine (Figure 1a) as indicated by a significant session × lever interaction [F (6, 228) = 9.43, p = 3.70 × 10−5]. Main effects of lever [F (1, 38) = 44.79, p = 6.42 × 10−8] and session [F (6, 228) = 8.51, p = 8.30 × 10−5] were also significant. Post hoc tests indicated that the number of active presses was significantly greater than the number of inactive presses on all session (p < .01 for all comparisons). The number of active presses increased significantly (p = 1.14 × 10−4) from session 1 (M = 23.79; SD = 33.00) to session 7 (M = 114.48; SD = 137.75). In contrast, inactive presses did not change significantly from session 1 (M = 5.16; SD = 8.25) to session 7 (M = 3.10; SD = 3.27).
Figure 1. Intravenous cocaine self-administration in the BXD recombinant inbred panel.
(a) Active presses, inactive presses, and cocaine infusions (mean ± SE) during the first seven 120-min sessions of cocaine IVSA testing in 39 BXD strains. Number of active presses was significantly greater than number of inactive presses on all sessions (p < .01). (b) Number of cocaine infusions at each dose (mean of final two 120 min sessions) along the dose-response curve in 29 BXD strains. Red circles represent the mean of all BXD strains at each dose and black diamonds represent individual strain means. (c) Frequency distribution of the peak of the dose-response curve for individual BXD strains. (d, e) Active and inactive lever presses (mean ± SE) and cocaine intake (mean ± SD) across the dose-response curve. (f) Principal components associated with cocaine IVSA infusions at all doses along the dose-response curve.
Cocaine IVSA Dose-response Curve
Mice from 29 strains completed a dose-response curve (Table S1). Heritability of infusions at each of the 8 doses ranged from .46 to .60 (Table S2). The dose-response curve (Figure 1b) exhibited a characteristic ascending limb and descending limb, with a peak at 0.1 mg/kg/infusion. The peak of the dose-response curve varied across individual BXD strains and ranged from .032 to .18 mg/kg/infusion (Figure 1c, S1). To assess performance across the dose-response curve, we conducted a repeated measures ANOVA using number of infusions as the dependent measure and dose as a within-subjects factor. Number of infusions varied significantly between doses as indicated by a significant main effect of dose [F (7, 196) = 77.74, p = 5.09 × 10−23]. Post hoc comparisons indicated that (1) the number of infusions at all doses on the ascending limb, including the peak dose, differed significantly, and (2) the number of infusions at all doses on the descending limb, including the peak dose, differed significantly (p < .01 for all comparisons). Analysis of lever press data (Figure 1d) revealed the same pattern. Cocaine intake across doses is provided in Figure 1e.
Genetic Correlations
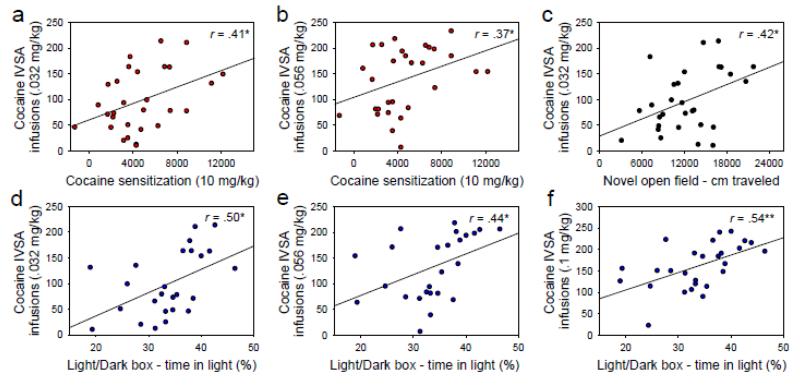
Using GeneNetwork, we assessed genetic correlations of cocaine IVSA and two commonly used assays relevant to the behavioral effects of cocaine: cocaine CPP and cocaine sensitization. Cocaine CPP (3.2 mg/kg i.p.) was not genetically correlated with cocaine IVSA at any dose. Cocaine sensitization (10 mg/kg i.p.) was positively genetically correlated with cocaine IVSA infusions at the two lowest cocaine doses (Figure 2a, 2b). We also assessed genetic correlations among cocaine IVSA and (1) distance traveled in a novel open field and (2) time spent in the light side of a light/dark box. Distance traveled (cm) during 60 minutes in a novel open field was positively genetically correlated with cocaine IVSA at the lowest cocaine dose (Figure 2c); this relationship approached significance at the 0.1 mg/kg/infusion dose (r = .34, p < .06) and 0.18 mg/kg/infusion dose (r = .307, p < .07). Percentage of time spent in the light side of a light/dark box during a 10 min testing session was positively genetically correlated with cocaine IVSA at the three lowest cocaine doses (Figure 2d, 2e, 2f).
Figure 2. Genetic correlations.
(a, b) Cocaine sensitization was positively genetically correlated with cocaine IVSA infusions at the two lowest cocaine doses. (c) Distance traveled over 60 min in a novel open field was positively genetically correlated with cocaine IVSA infusions at the lowest dose. (d, e, f) Percentage of time in the light side of a light/dark box during a single 10 min session was positively genetically correlated with cocaine infusions at the three lowest cocaine doses. After adjusting for the effects of the other two phenotypes, each of these measures explained unique genetic variance on cocaine IVSA infusions at the lowest cocaine dose: cocaine sensitization (26.2%, p < .01), light/dark box (29.8%, p < .001), and open field (11.7%, p < .05). Data points in all panels represent BXD strain means.
Notably, the three traits which were genetically correlated with cocaine IVSA infusions (cocaine sensitization, distance traveled in a novel open field, and time on the light side of a light/dark box) were not genetically intercorrelated. This suggests that dissociable underlying mechanisms drive the genetic relationship between cocaine IVSA and each of these behaviorsin the BXD RI panel. To assess the hypothesis that these three traits explained unique genetic variance on cocaine IVSA, we performed a simultaneous multiple regression using cocaine IVSA infusions at the .032 mg/kg/infusion dose as the dependent measure and cocaine sensitization, distance traveled in a novel open field, and time on the light side of a light/dark box as predictors. The overall regression was statistically significant [F (3, 21) = 12.26, p = 7.50 × 10−5], and the three predictor variables collectively accounted for 63.7% of the genetic variance around IVSA infusions at the lowest cocaine dose (R = .798). After controlling for the effects of the other two predictors, each predictor explained statistically significant unique genetic variance on cocaine IVSA infusions at the lowest cocaine dose: cocaine sensitization (26.2%, p < .01), time on the light side of a light/dark box (29.8%, p < .001), and distance traveled in a novel open field (11.7%, p < .05).
QTL Mapping of Cocaine IVSA Phenotypes
Indexing dose-response curve characteristics
To facilitate QTL mapping of traits affecting the shape of the cocaine IVSA dose-response curve, we used the Correlation Matrix and PCA tool in GeneNetwork to generate principal components associated with the number of infusions at each of the 8 cocaine doses. This procedure generated 2 principal components (Figure 1f): All cocaine doses loaded on the first principal component, and the highest 5 doses loaded strongly. The 3 cocaine doses comprising the ascending limb and peak of the dose-response curve loaded strongly on the second principal component. From this point we refer to these principal components as (1) infusions on the descending limb of the dose-response curve (IDL) and (2) infusions on the ascending limb of the dose-response curve (IAL), respectively.
At low cocaine doses it can be challenging to determine if lever pressing reflects titration of cocaine intake or responding in extinction. To dissociate these two types of responding, we created an index termed consistency of cocaine intake across the ascending limb of the dose-response curve (CAL) by calculating the relative standard deviation of mean cocaine intake of the three lowest doses. Mice with low CAL values maintained relatively consistent cocaine intake across these doses, whereas mice with high CAL values exhibited variable intake across doses. Thus, low CAL values indicate relatively precise titration of cocaine intake across the three lowest doses and suggest that lever presses on the ascending limb of the dose-response curve reflect volitional cocaine intake. In contrast, high CAL values reflect a failure to titrate cocaine intake and may indicate that mice were in extinction.
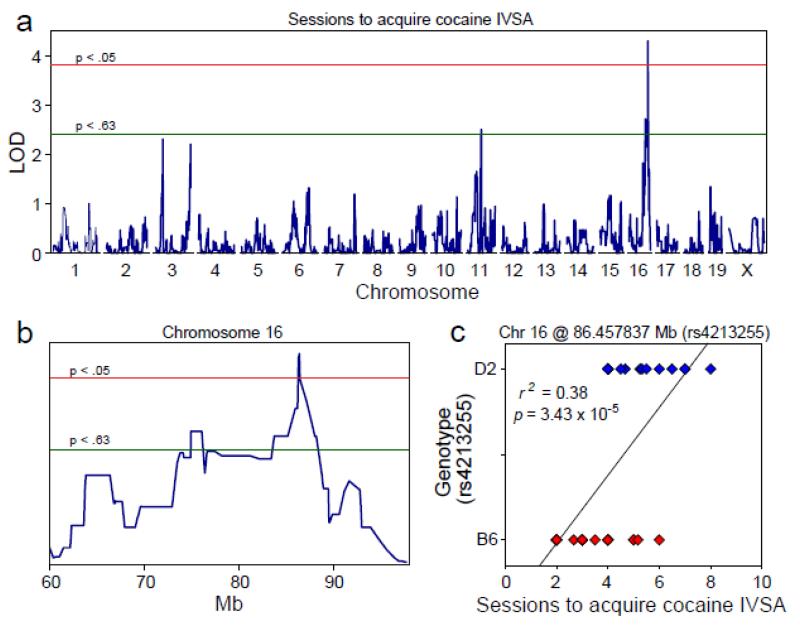
Genome-wide significant QTL associated with sessions to acquire cocaine IVSA
Three BXD strains were identified by GeneNetwork as outliers (13, 27, 33) and were excluded from the QTL analysis. We identified a significant QTL (Figures 3a, 3b) on chromosome (Chr) 16 with a peak locus of 86.393 Mb (LOD = 4.28, p = 1.80 × 10−2). The 2 LOD CI was 83.407 Mb – 88.783 Mb and the closest listed marker to the QTL peak was rs4213255 at 86.458 Mb. To assess the effect of genotype at this locus on sessions to acquire cocaine IVSA, we grouped the 36 BXD strains by the B6 (n = 21) or D2 (n = 15) genotype at the rs4213255 locus (B6- and D2-allele groups, respectively). The D2-allele group required significantly more sessions to reach acquisition criteria (M = 5.32, SD = 1.32) relative to the B6-allele group (M = 3.40, SD = 1.18) [F (1, 34) = 20.97, p = 6.00 × 10−5], and this effect accounted for 38.19% of the variance on the sessions to acquire cocaine IVSA measure (Figures 3c). Including sex as a between-subjects factor in the analysis (i.e., calculating strain means in males and females separately) resulted in the same main effect of genotype [F (1, 43) = 14.76, p = 3.97 × 10−4] but no main effect of sex or interaction of genotype and sex.
Figure 3. QTL associated with the number of sessions to meet cocaine IVSA acquisition criteria.
(a, b) Genome-wide significant QTL on chromosome 16 associated with sessions to reach cocaine IVSA acquisition criteria (2 LOD CI: 83.407 - 88.783 Mb). (c) BXD strains carrying the D2 allele at the QTL peak required significantly more sessions to reach acquisition criteria (M = 5.32, SD = 1.32) relative to BXD strains carrying the B6 allele (M = 3.40, SD = 1.18). This effect accounted for 38% of the variance on the sessions to acquire cocaine IVSA measure.
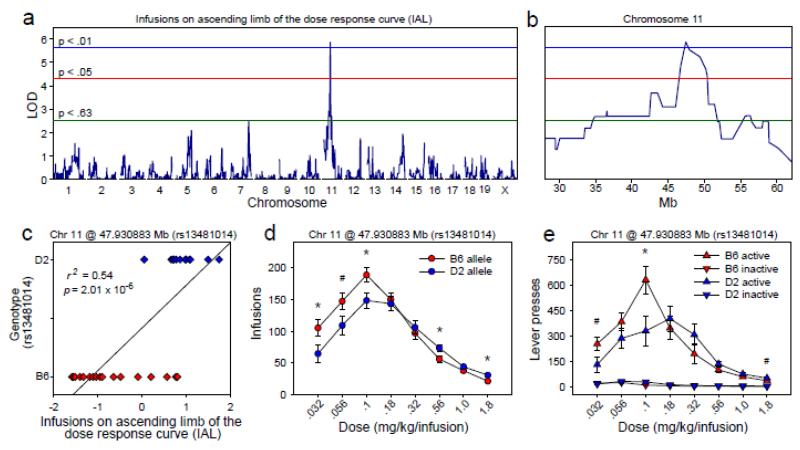
Genome-wide significant QTL associated with infusions on the ascending limb (IAL) of the dose-response curve
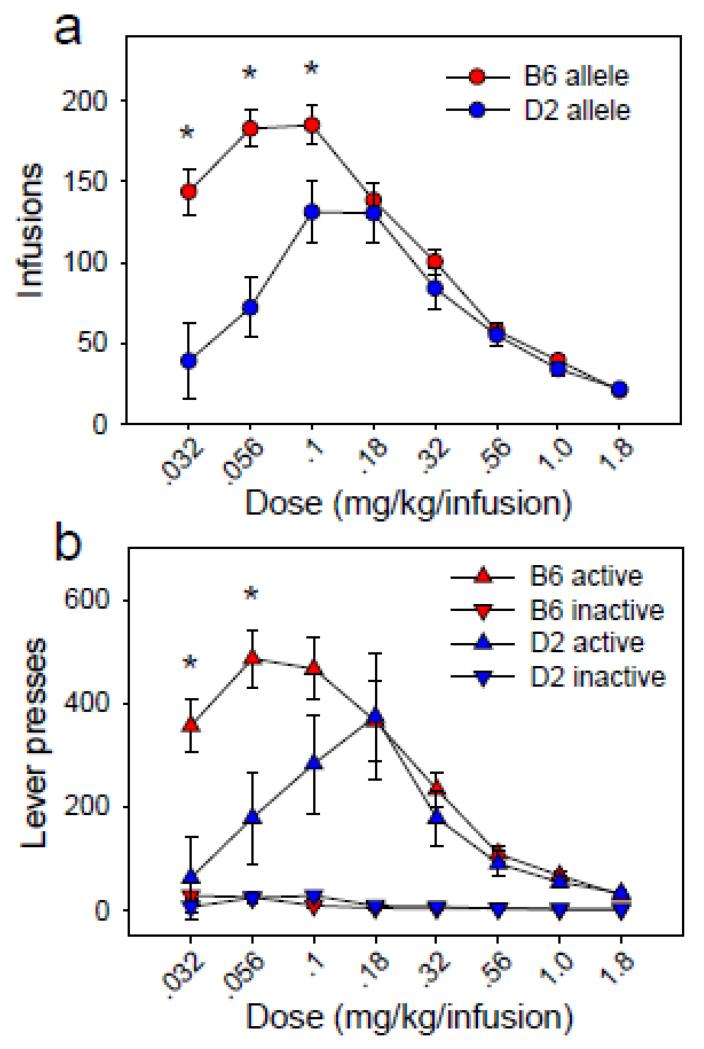
We identified a significant QTL (Figures 4a, 4b) on Chr 11 with a peak locus of 47.436 Mb (LOD = 5.87, p = 7.00 × 10−3). The 2 LOD CI was 46.00 Mb – 50.39 Mb and the closest listed marker to the QTL peak was rs13481014 at 47.931 Mb. To assess the effect of genotype at this locus on the IAL factor, we grouped the 29 BXD strains by the B6 (n = 17) or D2 (n = 12) genotype at the rs13481014 locus. IAL factor scores of the B6-allele group (M = −0.76, SD = 1.02) differed significantly from those of the D2-allele group (M = 1.08, SD = 0.60) [F (1, 27) = 31.08, p = 7.00 ×10−6], and this effect accounted for 53.52% of the variance on the IAL factor (Figure 4c).
Figure 4. QTL associated with infusions on the ascending limb of the dose response curve.
(a, b) Genome-wide significant QTL on chromosome 11 associated with infusions on the ascending limb of the dose response curve (IAL) (2 LOD CI: 46.00 - 50.39 Mb). (c) IAL factor scores of BXD mice carrying the B6 allele at the QTL peak differed significantly from scores of BXD mice carrying the D2 allele at the QTL peak, and this effect accounted for 54% of the variance on the IAL factor. (d, e) BXD strains carrying the B6 allele at the QTL peak infused significantly more cocaine on the ascending limb of the dose response curve relative to strains carrying the D2 allele at the QTL peak. This effect was significantly reversed on the descending limb, indicating a leftward shift of the dose response curve in BXD strains carrying the B6 allele at the QTL peak. Data points represent group means ± SE.
Genome-wide significant QTL associated with consistency of cocaine intake across the ascending limb of the dose-response curve (CAL)
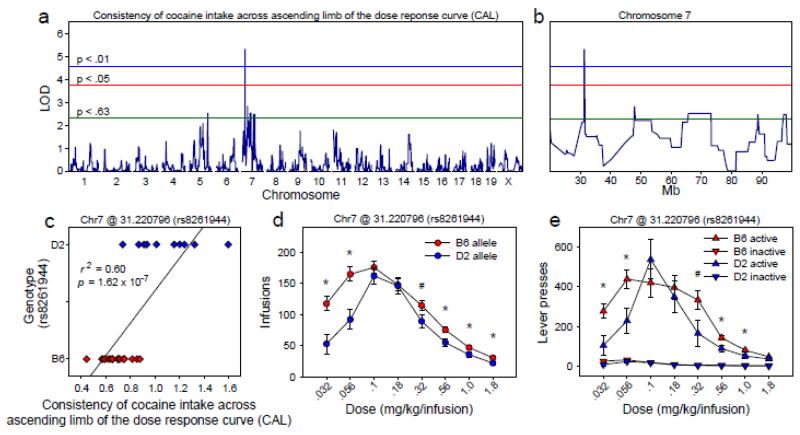
We identified a significant QTL (Figures 5a, 5b) on Chr 7 with a peak locus of 31.221 Mb (LOD = 5.73, p = 2.00 × 10−3). The 2 LOD CI was 31.206 Mb – 31.467 Mb, and the marker at the QTL peak was rs8261944. To assess the effect of genotype at this locus on CAL, we grouped the 29 BXD strains by the B6 (n = 19) or D2 (n = 10) genotype at the rs8261944 locus. CAL scores of the B6-allele group (M = 0.68, SD= 0.11) were significantly lower than those of the D2-allele group (M = 1.10, SD = 0.25) [F (1, 27) = 40.04, p = 8.96 × 10−7], and this effect accounted for 59.73% of the variance on the CAL index (Figure 5c).
Figure 5. QTL associated with consistency of cocaine intake across the ascending limb of the dose response curve.
(a, b) Genome-wide significant QTL on chromosome 7 associated with consistency of cocaine intake across the ascending limb of the dose response curve (CAL) (2 LOD CI: 31.206 - 31.467 Mb). (c) CAL index scores of BXD strains carrying the B6 allele at the QTL peak differed significantly from scores of BXD strains carrying the D2 allele at the QTL peak, and this effect accounted for 60% of the variance on the CAL index. (d, e) BXD strains carrying the B6 allele at the QTL peak infused significantly more cocaine on the ascending and descending limbs of the dose response curve relative to strains carrying the D2 allele at the QTL peak. Collectively, these observations indicate an upward shift of the dose response curve in BXD strains carrying the B6 allele at the QTL peak. Data points represent group means ± SE.
Effect of genotype at the IAL and CAL QTL on the cocaine IVSA dose-response curve
To assess the effect of genotype at the IAL and CAL QTL on infusions across the dose-response curve, we performed a repeated measures ANOVA using number of cocaine infusions as the dependent measure and dose as the within-subjects factor (.032 – 1.8 mg/kg/infusion). Between-subjects factors were sex, genotype at the IAL QTL peak (B6 or D2 allele), and genotype at the CAL QTL peak (B6 or D2 allele). Infusions varied as a function of cocaine dose [F (7, 161) = 84.36, p = 5.06 × 10−42], and this effect was moderated by genotype at the IAL QTL [dose × IAL genotype: F (7, 161) = 5.19, p = 1.01 × 10−4] (Figures 4d, 4e) and genotype at the CAL QTL [dose × CAL genotype: F (7, 161) = 5.84, p = 2.60 × 10−5] (Figures 5d, 5e). Genotypes at the IAL and CAL QTL did not interact. There was no main effect of sex and sex did not interact with other factors.
BXD strains in the B6-allele group at the QTL associated with IAL (i.e., B6 allele at rs13481014) infused significantly more cocaine on the ascending limb of the dose-response curve relative to BXD strains in the IAL D2-allele group (Figures 4d, 4e). Conversely, the IAL B6-allele group infused significantly less cocaine on the descending limb of the dose-response curve relative to the IAL D2-allele group. Collectively, these data reveal a leftward shift of the dose-response curve in BXD strains carrying the B6 allele at Chr 11 @ 47.93 Mb.
BXD strains in the B6-allele group at the QTL associated with CAL (i.e., B6 allele at rs8261944) infused significantly more cocaine on both the ascending and descending limbs of the dose-response curve relative to BXD strains in the CAL D2-allele group (Figures 5d, 5e). These data reveal an upward shift of the dose-response curve in BXD strains carrying the B6 allele at Chr 7 @ 31.22 Mb.
BXD strains carrying the B6 allele at both Chr 11 @ 47.93 Mb (IAL QTL peak) and Chr 7 @ 31.22 Mb (CAL QTL peak) infused significantly more cocaine and made significantly more active lever presses on the ascending limb of the dose-response curve relative to BXD strains carrying the D2 allele at both QTL (Figures 6a, 6b). The combined allelic effects were significantly greater than the independent effects on the two lowest cocaine doses (p < .05). Dose-response curves of individual BXD strains grouped by allele at the IAL and CAL QTL are provided in Figure S1.
Figure 6. Combined allelic effects on the cocaine IVSA dose response curve of the behavioral QTL on chromosome 7 and 11.
BXD strains carrying the B6 allele at both Chr 11 @ 47.93 Mb (IAL QTL peak) and Chr 7 @ 31.22 Mb (CAL QTL peak) infused significantly more cocaine (a) and made significantly more active lever presses (b) on the ascending limb of the dose response curve relative to BXD strains carrying the D2 allele at both QTL. The combined allelic effects were significantly greater than the independent effects on the two lowest cocaine doses (p < .05). Data points represent group means ± SE.
QTL Positional Candidates
Positional candidates were those genes within the 2 LOD confidence interval of a behavioral QTL that exhibited significant cis-eQTL in the NAc, midbrain, or PFC (Table 1). Candidates for the QTL on Chr 11 @ 47.43 Mb associated with the IAL phenotype were Cyfip2 and Itk. Candidates for the QTL on Chr 7 @ 31.221 Mb associated with the CAL phenotype were Aplp1, Nfkbid, Hspb6, Kirrel2, Lin37, Nphs1, Psenen, and Tmem149. Candidates for the QTL on Chr 16 @ 86.393 Mb associated with sessions to acquire cocaine IVSA were Adamts1, App, Bach1, Cyyr1, Grik1, and Mrpl39. App exhibited a suggestive cis-eQTL in the PFC and was included as a candidate because of its functional relevance to Aplp1 (Aydin et al. 2012; Cousins et al. 2015; Korte et al. 2012). With the exception of Mrpl39, all candidates contained non-synonymous SNPs. With the exception of Psenen, all candidates contained indels.
Table 1.
Priority positional candidates for behavioral QTL associated with cocaine IVSA phenotypes
| Gene |
cis-eQTL |
Covariation2 |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Phenotype | Region | Symbol | Chr | Mb | n | Probe set | Chr | Mb | LRS | p 1 | r | p 3 | References |
| IAL | Midbrain | Cyfip2 | 11 | 46.007 | 15 | A_55_P1962771 | 11 | 42.844 | 62.7 | < 1.00E-03 | 0.79 | 2.16E-04 | Kumar et al. (2013) |
| Midbrain | Itk | 11 | 46.138 | 15 | A_55_P1953788 | 11 | 42.631 | 19.4 | 2.80E-02 | 0.77 | 4.48E-04 | ||
| CAL | PFC | Lin37 | 7 | 31.341 | 12 | 1426577_a_at | 7 | 31.220 | 34.9 | 1.00E-03 | 0.74 | 4.01 E-03 | |
| Midbrain | Kirrel2 | 7 | 31.233 | 15 | A_51_P512306 | 7 | 31.220 | 35.6 | < 1.00E-03 | 0.74 | 9.36E-04 | ||
| Midbrain | Aplpl | 7 | 31.220 | 15 | A_55_P1991605 | 7 | 31.220 | 21.2 | 9.00E-03 | 0.66 | 5.56E-03 | ||
p-values < .05 indicate genome-wide significant cis-eQTL
covariation of phenotype and gene expression in the listed brain region
all p-values are statistically significant (family-wise error rate = .05) following Bonferroni adjustment for the total number of positional candidates within the QTL confidence interval
QTL positional candidates exhibiting expression levels that covaried significantly with their cocaine IVSA phenotype in one of the three brain regions were elevated to priority candidates (Table 2). All priority candidates exhibited genome-wide significant cis-eQTL in the same brain region that covariation of phenotype and expression was observed. Cyfip2 and Aplp1 also exhibited genome-wide significant trans-eQTL in those regions. Interval mapping of Cyfip2 expression in the midbrain revealed a genome-wide significant trans-eQTL on Chr 3 with a peak locus of 28.53 Mb (LOD = 4.09, p = 4.40 × 10−2) and a 2 LOD CI of 24.78 – 36.34. Notably, the fragile X related gene Fxr1 (Chr 3 @ 33.96 Mb) resides within this CI and is functionally related to Cyfip2 (Schenck et al. 2001).
Table 2.
Genes exhibiting midbrain expression levels that map genome-wide significantly to the behavioral QTL associated with infusions on the ascending limb of the dose-response curve (IAL phenotype)
| Gene |
trans-eQTL |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Symbol | Chr | Mb | n | Probe set | Chr | Mb | LRS | p 1 | References |
| Fam53b | 7 | 139.951 | 37 | A_55_P2178496 | 11 | 49.367 | 23.51 | 2.00E-03 | Gelernter et al. (2014) |
| Mapk12 | 15 | 88.961 | 37 | A_52_P252258 | 11 | 47.930 | 23.58 | 2.00E-03 | |
| Lppr5 | 3 | 117.392 | 37 | A_55_P1987291 | 11 | 50.386 | 20.50 | 8.00E-03 | |
| Chchd7 | 4 | 3.866 | 37 | A_51_P510939 | 11 | 50.386 | 17.77 | 1.30E-02 | |
| Serpini1 | 3 | 75.421 | 37 | A_66_P100165 | 11 | 50.197 | 20.18 | 1.80E-02 | |
| Rps6ka6 | X | 108.502 | 37 | A_55_P2020911 | 11 | 50.386 | 19.92 | 2.40E-02 | |
p-values < .05 indicate genome-wide significant trans-eQTL
Interval mapping of Aplp1 expression in the midbrain revealed a genome-wide significant trans-eQTL on Chr 11 with a peak locus of 57.65 Mb (LOD = 3.94, p = 3.90 × 10−2) and a 2 LOD CI of 50.19 – 61.28. The location of this trans-eQTL is notable because the genes exhibiting expression levels that covaried most strongly with the IAL phenotype were located within this CI. These relationships reached genome-wide significance for Hist3h2a and Skp1a (Table 3). These associations suggest a functional relationship between a priority gene candidate for the CAL phenotype (Aplp1) and genes strongly associated with the IAL phenotype.
Table 3.
Genome-wide significant covariation of IVSA phenotypes and gene expression in the nucleus accumbens (NAc), midbrain, and prefrontal cortex (PFC).
| Gene |
eQTL |
Covariation2 |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Phenotyp e |
Region | Symbol | Ch r |
Mb | n | Probe set | Ch r |
Mb | LR S |
p 1 | r | p 3 |
| Sessions to acquisitio n |
NAc Midbrai n |
Nudt8
Gm579 7 |
19 | 4.001 | 1 7 |
1450111_a_at | - | - | - | - | 0.87 3 |
4.99e -07 |
| 14 | 8.156 | 1 9 |
A_55_P203608 3 |
- | - | - | - | 0.87 2 |
8.22e -08 |
|||
| PFC | Rpl35 | 2 | 38.85 7 |
1 3 |
1436840_x_at | - | - | - | - | 0.93 4 |
9.13e -08 |
|
| PFC | Rpl35 | 2 | 38.85 7 |
1 3 |
1455950_x_at | - | - | - | - | 0.93 3 |
1.07e -07 |
|
| IAL | NAc |
Hist3h2
a |
11 | 58.76 9 |
1 6 |
1455712_at | 11 | 56.51 5 |
35. 5 |
< 1.00E -03 |
0.92 2 |
7.75e -09 |
| NAc | Skp1a | 11 | 52.05 7 |
1 6 |
1423148_at | 11 | 50.19 6 |
72. 0 |
1.00E -03 |
0.88 1 |
6.63e -07 |
|
| PFC | Rpl31 | 1 | 39.42 7 |
1 2 |
1460008_x_at | - | - | - | - | 0.92 8 |
7.85e -07 |
|
| CAL | Midbrai n |
Pate4 | 9 | 35.41 5 |
1 5 |
A_51_P194293 | - | - | - | - | 0.88 6 |
1.15e -06 |
| PFC | Fin14 | X | 12.86 9 |
1 2 |
1423043_s_at | 7 | 31.21 2 |
18. 9 |
3.40E -02 |
0.93 0 |
6.25e -07 |
|
p-values < .05 indicate genome-wide significant eQTL
covariation of phenotype and gene expression in the listed brain region
all p-values are statistically significant (family-wise error rate = .05) following Bonferroni adjustment for number of probe sets in the brain region
Trans-eQTL Associated with Cocaine IVSA Behavioral QTL
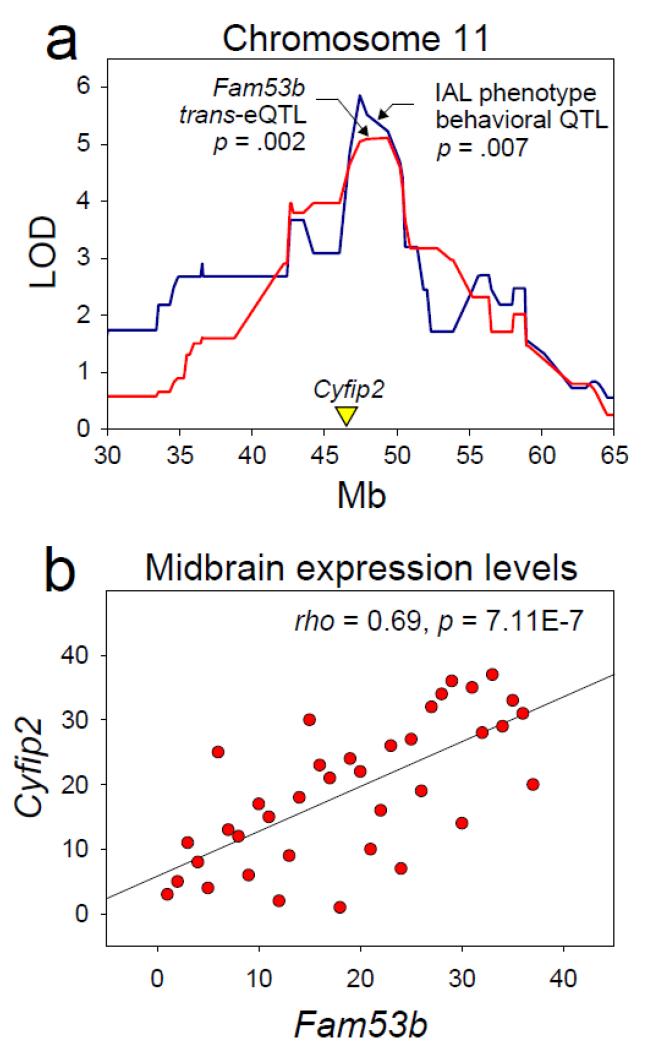
We identified genes exhibiting genome-wide significant trans-eQTL that mapped to a cocaine IVSA behavioral QTL. We performed this analysis using the midbrain gene expression data set because four of the five priority positional candidates covaried significantly with their QTL phenotype in the midbrain (Table 1). We identified six genes, Fam53b, Mapk12, Lppr5, Chchd7, Serpini1, and Rps6ka6 that exhibited genome-wide significant trans-eQTL located within the CI of the IAL behavioral QTL on Chr 11 @ 47.43 Mb (Table 2). Notably, the only genome-wide association study of cocaine dependence in humans to date identified Fam53b as the sole genome-wide significant gene candidate (Gelernter et al. 2014). Figure 7a shows the genome-wide significant trans-eQTL associated with expression of Fam53b (Chr 7 @ 139.95 Mb) superimposed on the behavioral QTL associated with the IAL phenotype. Fam53b and the highest priority gene candidate associated with the IAL QTL (Cyfip2) were co-expressed genome-wide significantly in the midbrain (Figure 7b). There were no genome-wide significant trans-eQTL within the CI of the behavioral QTL on Chr 7 @ 31.221.
Figure 7. Fam53b maps to a cocaine IVSA behavioral QTL on chromosome 11.
(a) Fam53b is a human GWAS candidate for cocaine dependence (Gelernter et al. 2014). Midbrain expression of Fam53b (Chr 7 @ 139.95 Mb) maps genome-wide significantly to the IAL behavioral QTL located on Chr 11. (b) Fam53b and the top gene candidate for the IAL behavioral QTL (Cyfip2) were co-expressed genome-wide significantly in the midbrain. Collectively, these data suggest a functional relationship between Cyfip2 and Fam53b in the context of cocaine use.
Covariation of Cocaine IVSA Phenotypes and Gene Expression Across the Genome
We identified genes that covaried genome-wide significantly in the NAc, midbrain, or PFC with one of the three cocaine IVSA phenotypes associated with a behavioral QTL (Table 3). Covariation of phenotype and expression of all genes listed in Table 3 reached statistical significance in the listed brain region after Bonferroni adjustment (.05 / number of probe sets). Three of these genes (Hist3h2a, Skp1a, Fin14) exhibited genome-wide significant eQTL.
DISCUSSION
Systems genetics is a powerful method for identifying the biological mechanisms underlying complex traits, and cocaine IVSA is the gold standard for assessing volitional drug use in animals. In the present study, we integrated these two approaches to identify novel genes and networks underlying cocaine IVSA in the BXD recombinant inbred mouse panel. To our knowledge, this is the first report of a systems genetics analysis of cocaine IVSA phenotypes. Mice from 39 BXD strains acquired cocaine IVSA, and mice from 29 BXD strains completed a full dose-response curve (Figure 1). We identified independent genetic associations between cocaine IVSA and measures of environmental exploration and cocaine sensitization (Figure 2). We identified a genome-wide significant QTL for sessions to acquire cocaine IVSA (Figure 3) and two genome-wide significant QTL associated with cocaine IVSA on the ascending limb of the dose-response curve (Figures 4, 5, 6). Using publicly available gene expression data from the NAc, midbrain, and PFC, we prioritized gene candidates within behavioral QTL (Table 1) and identified genome-wide significant trans-eQTL (Table 2, Figure 7) and genes (Table 3) associated with cocaine IVSA.
Genetic Correlations
We identified positive genetic correlations between cocaine IVSA and cocaine sensitization, distance traveled in a novel open field, and time in the light side of a light/dark box (Figure 2). Importantly, each of these measures explained unique genetic variance on low-dose cocaine IVSA. Time in the light side of a light/dark box explained the most genetic variance of the three phenotypes (29.8%). Elevated time in the dark compartment of a light/dark box may reflect an anxious phenotype, whereas elevated time in the light compartment may reflect a risk taking or impulsive phenotype (Arrant et al. 2013). Regarding this interpretation of light/dark box behavior, the genetic associations reported here are consistent with previous findings of positive associations between psychostimulant IVSA and operant measures of impulsive action and impulsive choice (Dalley et al. 2007; Marusich and Bardo 2009). In humans, trait-impulsivity is reported to be a risk factor for the development of psychostimulant dependence (Ersche et al. 2013; Ersche et al. 2012; Ersche et al. 2010).
Distance traveled in a novel open field explained 11.7% of the genetic variance on low-dose cocaine IVSA. High levels of exploration in a novel open field may be viewed as reflecting a sensation seeking phenotype, and sensation seeking may be a risk factor for drug use or abuse (Blanchard et al. 2009). Previous studies in outbred rats have shown a phenotypic relationship between exploration in a novel open field and methamphetamine IVSA (Cain et al. 2005; Gancarz et al. 2011; Klebaur et al. 2001; Piazza et al. 1990), but the genetic component of this relationship has been unclear. The present study illustrates that shared genetic mechanisms drive open field exploration and low-dose cocaine IVSA in the BXD population.
Cocaine sensitization explained 26.2% of the genetic variance on low-dose cocaine IVSA, whereas cocaine CPP did not explain statistically significant genetic variance on cocaine IVSA. Collectively, these data suggest that the genetic mechanisms driving cocaine IVSA are largely, though not completely, dissociable from those driving the most commonly used indexes of the behavioral effects of experimenter-administered cocaine. Moreover, the pattern of genetic associations among all examined phenotypes indicates that the observed genetic relationship between cocaine and the open field and light/dark exploratory traits is captured by the cocaine IVSA paradigm, but not by the cocaine CPP or sensitization paradigms. It should be noted, however, that expanded genetic diversity in mouse populations such as the Collaborative Cross RI lines or Diversity Outbred mice may allow identification of relationships not detectable in the BXD lines (Chesler 2014). The use of IVSA methods such as a progressive ratio schedule (Richardson and Roberts 1996), a second order schedule (Everitt and Robbins 2000), or extended-access sessions (Mantsch et al. 2004) may enable identification of relationships not observed in this study.
Genome-Wide Significant QTL on Chromosomes 7 and 11 Associated with Shifts in the Cocaine IVSA Dose-response Curve
We detected a QTL on Chr 11 @ 47.43 Mb associated with infusions on the ascending limb of the dose-response curve (IAL QTL) and a QTL on Chr 7 @ 31.22 Mb associated with consistency of cocaine intake across the ascending limb of the dose-response curve (CAL QTL). Presence of the B6 allele at the peak of the IAL QTL (Figures 4d, 4e) and CAL QTL (Figures 5d, 5e) was strongly associated with elevated cocaine intake on the ascending limb of the dose-response curve, and the effect of carrying the B6 allele at both loci was striking (Figure 6a, 6b).
Some lever pressing at low doses was likely due to extinction responding, and we attempted to quantify this phenomenon with the CAL phenotype. Specifically, mice with a high CAL score exhibited relatively variable cocaine intake across the lowest cocaine doses, suggesting that these mice did not adjust active lever pressing across doses to titrate cocaine intake. The effect of the D2 allele at the CAL QTL was a precipitous drop in responding on the lowest two doses relative to the peak of the dose-response curve (Figure 5d). In contrast, the effect of the D2 allele at the peak of the IAL QTL was an ascending limb that was shifted down equivalently at the three lowest doses (Figure 4d). These differential effects suggest that the genes underlying the IAL and CAL QTL drive dissociable aspects of cocaine IVSA. In support of this hypothesis, the alleles at the peak of the IAL and CAL QTLs differentially affected the overall dose-response curve. Specifically, we observed a statistically significant leftward shift in the dose-response curve in BXD strains carrying the B6 allele at the IAL QTL, whereas a statistically significant upward shift was observed in BXD strains carrying the B6 allele at the CAL QTL. Studies in knockout mice will be required to confirm the effect of gene candidates proposed to underlie these QTL (Table 1) and to dissociate the effects of these genes on dose-response curve shifts, responding at low doses, and responding during extinction.
QTL Positional Candidates
Cyfip2 and Itk were the sole positional candidates for the IAL QTL on Chr 11. Expression of both genes was significantly correlated with the IAL phenotype in the midbrain (Table 1). Two recent studies suggest that Cyfip2 may be the driver of this QTL. First, Cyfip2 plays a critical role in the behavioral sensitization to cocaine (Kumar et al. 2013), and the observation of a genetic correlation between cocaine sensitization and low-dose cocaine IVSA (Fig 2a, b) indicates shared genetic underpinnings of these phenotypes. Second, expression of Fam53b in the midbrain maps genome-wide significantly to the IAL QTL (Figure 7a), and Fam53b and Cyfip2 (but not Itk) are co-expressed genome-wide significantly in the midbrain (Figure 7b). This association is relevant because Fam53b was the sole genome-wide significant candidate in the only human genome-wide association study of cocaine dependence to date (Gelernter et al. 2014). Notably, all relationships involving the IAL phenotype, Cyfip2 expression, and Fam53b expression were observed in the midbrain; this suggests that the putative effect of Cyfip2 and Fam53b on cocaine use may occur in this region.
The priority candidates for the CAL QTL on Chr 7 were Aplp1, Lin37, and Kirrel2 (Table 1). Two observations suggest that Aplp1 may be the driver of this QTL. First, expression of App, which is closely related to Aplp1 (Aydin et al. 2012; Cousins et al. 2015; Korte et al. 2012), is significantly associated with chronic cocaine and heroin abuse in humans (Albertson et al. 2004; Albertson et al. 2006). Notably, App is a gene candidate for the QTL on Chr 16 associated with sessions to IVSA acquisition. Second, gene expression data suggest a functional relationship between Aplp1 and the genetic mechanisms underlying the IAL phenotype. Specifically, Aplp1 exhibits a genome-wide significant trans-eQTL on Chr 11 @ 57.65 Mb, and two of the three genes exhibiting genome-wide significant covariation with the IAL phenotype reside within the CI of this trans-eQTL (Table 3). Moreover, Wolen et al. (2012) observed an association between these regions on Chr 7 and 11 in the context of ethanol response and proposed involvement of Aplp1 and Gria1 (Chr 11 @ 57.14). Considering this putative association and that the CAL phenotype may index extinction responding, it is notable that Gria1 knockouts over-respond during extinction in a cocaine IVSA paradigm (Mead et al. 2007).
The positional candidates for the QTL on Chr 16 associated with sessions to acquire cocaine IVSA were Adamts1, App, Bach1, Cyyr1, Grik1, and Mrpl39. We did not identify a priority candidate because covariation of expression and phenotype did not reach statistical significance for any gene candidate. However, other sources of evidence suggest App and Grik1 as genes of interest. App is significantly associated with chronic cocaine and heroin abuse in humans (Albertson et al. 2004; Albertson et al. 2006) and is a member of the same gene family as Aplp1, a positional candidate for the CAL QTL on Chr 7. Grik1 encodes the GluR5 kainate receptor subunit (Contractor et al. 2011), and deletion of this subunit affects cocaine sensitization in B6 mice (Gregus et al. 2010).
The positional candidates described here putatively underlie their associated cocaine IVSA phenotypes. Future studies using knockout mice will be necessary to confirm involvement of these gene candidates in cocaine IVSA and uncover the mechanisms driving observed involvement.
Relevance to human cocaine addiction
Addiction is a complex phenomenon, the core of which is persistent drug use in the face of harmful consequences (American Psychiatric Association 2000). Although addiction-like qualities have been demonstrated in rats (Deroche-Gamonet et al. 2004), the addiction construct is difficult to measure in a laboratory setting. In the present study, we tested mice under time-limited access sessions to identify candidate genes underlying characteristics of the cocaine IVSA dose-response curve and latency to acquire cocaine IVSA. Although not direct indexes of addiction, these measures may serve as addiction endophenotypes and, as such, enable identification of genes and mechanisms underlying human addiction. A critical observation from the present study supports this hypothesis: The only genome-wide association study of cocaine dependence in humans to date identified Fam53b as the sole genome-wide significant gene candidate (Gelernter et al. 2014). Remarkably, in the present study we found that midbrain expression of Fam53b, which is located on chromosome 7, mapped genome-wide significantly to a behavioral QTL on chromosome 11 (Figure 7a). This behavioral QTL was associated with a lateral shift in the cocaine IVSA dose-response curve (Figure 4). Moreover, expression of Fam53b and the top gene candidate for this behavioral QTL (Cyfip2) were co-expressed genome-wide significantly in the midbrain (Figure 7b). These data suggest that the gene underlying the behavioral QTL on chromosome 11, putatively Cyfip2, mediates expression of Fam53b on chromosome 7. Collectively, these data illustrate that variation in classical pharmacological measures such as dose-response curves may serve as addiction endophenotypes which can reveal putative genes and mechanisms associated with human addiction.
CONCLUSIONS
We conducted a systems genetics study using the BXD RI mouse strains to identify novel genes and networks underlying cocaine IVSA. To our knowledge, this is the first report of a systems genetics analysis of cocaine IVSA. We identified novel genome-wide significant behavioral QTL, expression QTL, and genes associated with cocaine IVSA phenotypes. A genome-wide significant trans-eQTL linking Fam53b (a GWAS candidate for human cocaine dependence) on chromosome 7 to a cocaine IVSA behavioral QTL on chromosome 11 suggests the ability of mouse preclinical studies using the cocaine IVSA paradigm to identify genes and networks involved in human cocaine dependence. Collectively, these data provide novel candidate genes driving cocaine IVSA in mice and suggest novel mechanisms underlying cocaine use in human populations.
Supplementary Material
Acknowledgements
This project was made possible by NIDA grant 1R01DA020677. The authors gratefully acknowledge Drs. Lu Lu and Robert W. Williams for providing new BXD strains, Erin Clardy and Tom Schneider for assistance with behavioral data collection, Lei Yan for technical assistance with GeneNetwork, and Darla Miller.
References
- Alberts R, Schughart K. QTLminer: identifying genes regulating quantitative traits. BMC Bioinformatics. 2010;11:516. doi: 10.1186/1471-2105-11-516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albertson DN, Pruetz B, Schmidt CJ, Kuhn DM, Kapatos G, Bannon MJ. Gene expression profile of the nucleus accumbens of human cocaine abusers: evidence for dysregulation of myelin. J Neurochem. 2004;88:1211–9. doi: 10.1046/j.1471-4159.2003.02247.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albertson DN, Schmidt CJ, Kapatos G, Bannon MJ. Distinctive profiles of gene expression in the human nucleus accumbens associated with cocaine and heroin abuse. Neuropsychopharmacology. 2006;31:2304–12. doi: 10.1038/sj.npp.1301089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- American Psychiatric Association . Diagnostic and statistical manual of mental disorders. Revised 4th ed. Author; Washington, DC: 2000. [Google Scholar]
- Arrant AE, Schramm-Sapyta NL, Kuhn CM. Use of the light/dark test for anxiety in adult and adolescent male rats. Behav Brain Res. 2013;256:119–27. doi: 10.1016/j.bbr.2013.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aydin D, Weyer SW, Muller UC. Functions of the APP gene family in the nervous system: insights from mouse models. Exp Brain Res. 2012;217:423–34. doi: 10.1007/s00221-011-2861-2. [DOI] [PubMed] [Google Scholar]
- Belknap JK. Effect of within-strain sample size on QTL detection and mapping using recombinant inbred mouse strains. Behav Genet. 1998;28:29–38. doi: 10.1023/a:1021404714631. [DOI] [PubMed] [Google Scholar]
- Blanchard MM, Mendelsohn D, Stamp JA. The HR/LR model: Further evidence as an animal model of sensation seeking. Neurosci Biobehav Rev. 2009;33:1145–54. doi: 10.1016/j.neubiorev.2009.05.009. [DOI] [PubMed] [Google Scholar]
- Bocklisch C, Pascoli V, Wong JC, House DR, Yvon C, de Roo M, Tan KR, Luscher C. Cocaine disinhibits dopamine neurons by potentiation of GABA transmission in the ventral tegmental area. Science. 2013;341:1521–5. doi: 10.1126/science.1237059. [DOI] [PubMed] [Google Scholar]
- Cain ME, Saucier DA, Bardo MT. Novelty seeking and drug use: contribution of an animal model. Exp Clin Psychopharmacol. 2005;13:367–75. doi: 10.1037/1064-1297.13.4.367. [DOI] [PubMed] [Google Scholar]
- Chesler EJ. Out of the Bottleneck: The Diversity Outcross and Collaborative Cross Mouse Populations in Behavioral Genetics Research. Mamm Genome. 2014;25:3–11. doi: 10.1007/s00335-013-9492-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Civelek M, Lusis AJ. Systems genetics approaches to understand complex traits. Nat Rev Genet. 2014;15:34–48. doi: 10.1038/nrg3575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contractor A, Mulle C, Swanson GT. Kainate receptors coming of age: milestones of two decades of research. Trends Neurosci. 2011;34:154–63. doi: 10.1016/j.tins.2010.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cousins SL, Dai W, Stephenson FA. APLP1 and APLP2, members of the APP family of proteins, behave similarly to APP in that they associate with NMDA receptors and enhance NMDA receptor surface expression. J Neurochem. 2015;133:879–85. doi: 10.1111/jnc.13063. [DOI] [PubMed] [Google Scholar]
- Dalley JW, Fryer TD, Brichard L, Robinson ES, Theobald DE, Laane K, Pena Y, Murphy ER, Shah Y, Probst K, Abakumova I, Aigbirhio FI, Richards HK, Hong Y, Baron JC, Everitt BJ, Robbins TW. Nucleus accumbens D2/3 receptors predict trait impulsivity and cocaine reinforcement. Science. 2007;315:1267–70. doi: 10.1126/science.1137073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deroche-Gamonet V, Belin D, Piazza PV. Evidence for addiction-like behavior in the rat. Science. 2004;305:1014–7. doi: 10.1126/science.1099020. [DOI] [PubMed] [Google Scholar]
- Dickson PE, Rogers TD, Lester DB, Miller MM, Matta SG, Chesler EJ, Goldowitz D, Blaha CD, Mittleman G. Genotype-dependent effects of adolescent nicotine exposure on dopamine functional dynamics in the nucleus accumbens shell in male and female mice: a potential mechanism underlying the gateway effect of nicotine. Psychopharmacology (Berl) 2011;215:631–42. doi: 10.1007/s00213-010-2159-2. [DOI] [PubMed] [Google Scholar]
- Ersche KD, Jones PS, Williams GB, Smith DG, Bullmore ET, Robbins TW. Distinctive personality traits and neural correlates associated with stimulant drug use versus familial risk of stimulant dependence. Biol Psychiatry. 2013;74:137–44. doi: 10.1016/j.biopsych.2012.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ersche KD, Turton AJ, Chamberlain SR, Muller U, Bullmore ET, Robbins TW. Cognitive dysfunction and anxious-impulsive personality traits are endophenotypes for drug dependence. Am J Psychiatry. 2012;169:926–36. doi: 10.1176/appi.ajp.2012.11091421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ersche KD, Turton AJ, Pradhan S, Bullmore ET, Robbins TW. Drug addiction endophenotypes: impulsive versus sensation-seeking personality traits. Biol Psychiatry. 2010;68:770–3. doi: 10.1016/j.biopsych.2010.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everitt BJ, Robbins TW. Second-order schedules of drug reinforcement in rats and monkeys: measurement of reinforcing efficacy and drug-seeking behaviour. Psychopharmacology (Berl) 2000;153:17–30. doi: 10.1007/s002130000566. [DOI] [PubMed] [Google Scholar]
- Gancarz AM, San George MA, Ashrafioun L, Richards JB. Locomotor activity in a novel environment predicts both responding for a visual stimulus and self-administration of a low dose of methamphetamine in rats. Behav Processes. 2011;86:295–304. doi: 10.1016/j.beproc.2010.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelernter J, Sherva R, Koesterer R, Almasy L, Zhao H, Kranzler HR, Farrer L. Genome-wide association study of cocaine dependence and related traits: FAM53B identified as a risk gene. Mol Psychiatry. 2014;19:717–23. doi: 10.1038/mp.2013.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Global Commission on Drug Policy War on Drugs: Report of the Global Commission on Drug Policy. 2011 Availableat http://www.globalcommissionondrugs.org/wp-content/themes/gcdp_v1/pdf/Global_Commission_Report_English.pdf.
- Goldman D, Oroszi G, Ducci F. The genetics of addictions: uncovering the genes. Nat Rev Genet. 2005;6:521–32. doi: 10.1038/nrg1635. [DOI] [PubMed] [Google Scholar]
- Goldstein A, Kalant H. Drug policy: striking the right balance. Science. 1990;249:1513–21. doi: 10.1126/science.2218493. [DOI] [PubMed] [Google Scholar]
- Gregus AM, Tropea TF, Wang Y, Hauck SC, Costa AC, Rajadhyaksha AM, Inturrisi CE. Deletion of the GluR5 subunit of kainate receptors affects cocaine sensitivity and preference. Neurosci Lett. 2010;468:186–9. doi: 10.1016/j.neulet.2009.10.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall FS, Drgonova J, Jain S, Uhl GR. Implications of genome wide association studies for addiction: are our a priori assumptions all wrong? Pharmacol Ther. 2013;140:267–79. doi: 10.1016/j.pharmthera.2013.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito R, Robbins TW, Everitt BJ. Differential control over cocaine-seeking behavior by nucleus accumbens core and shell. Nat Neurosci. 2004;7:389–97. doi: 10.1038/nn1217. [DOI] [PubMed] [Google Scholar]
- Klebaur JE, Bevins RA, Segar TM, Bardo MT. Individual differences in behavioral responses to novelty and amphetamine self-administration in male and female rats. Behav Pharmacol. 2001;12:267–75. doi: 10.1097/00008877-200107000-00005. [DOI] [PubMed] [Google Scholar]
- Korte M, Herrmann U, Zhang X, Draguhn A. The role of APP and APLP for synaptic transmission, plasticity, and network function: lessons from genetic mouse models. Exp Brain Res. 2012;217:435–40. doi: 10.1007/s00221-011-2894-6. [DOI] [PubMed] [Google Scholar]
- Kumar V, Kim K, Joseph C, Kourrich S, Yoo SH, Huang HC, Vitaterna MH, de Villena FP, Churchill G, Bonci A, Takahashi JS. C57BL/6N mutation in cytoplasmic FMRP interacting protein 2 regulates cocaine response. Science. 2013;342:1508–12. doi: 10.1126/science.1245503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantsch JR, Yuferov V, Mathieu-Kia AM, Ho A, Kreek MJ. Effects of extended access to high versus low cocaine doses on self-administration, cocaine-induced reinstatement and brain mRNA levels in rats. Psychopharmacology (Berl) 2004;175:26–36. doi: 10.1007/s00213-004-1778-x. [DOI] [PubMed] [Google Scholar]
- Marinelli M, Cooper DC, Baker LK, White FJ. Impulse activity of midbrain dopamine neurons modulates drug-seeking behavior. Psychopharmacology (Berl) 2003;168:84–98. doi: 10.1007/s00213-003-1491-1. [DOI] [PubMed] [Google Scholar]
- Marusich JA, Bardo MT. Differences in impulsivity on a delay-discounting task predict self-administration of a low unit dose of methylphenidate in rats. Behav Pharmacol. 2009;20:447–54. doi: 10.1097/FBP.0b013e328330ad6d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mead AN, Zamanillo D, Becker N, Stephens DN. AMPA-receptor GluR1 subunits are involved in the control over behavior by cocaine-paired cues. Neuropsychopharmacology. 2007;32:343–53. doi: 10.1038/sj.npp.1301045. [DOI] [PubMed] [Google Scholar]
- Mello NK, Negus SS. Preclinical evaluation of pharmacotherapies for treatment of cocaine and opioid abuse using drug self-administration procedures. Neuropsychopharmacology. 1996;14:375–424. doi: 10.1016/0893-133X(95)00274-H. [DOI] [PubMed] [Google Scholar]
- Peirce JL, Lu L, Gu J, Silver LM, Williams RW. A new set of BXD recombinant inbred lines from advanced intercross populations in mice. BMC Genet. 2004;5:7. doi: 10.1186/1471-2156-5-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettit HO, Ettenberg A, Bloom FE, Koob GF. Destruction of dopamine in the nucleus accumbens selectively attenuates cocaine but not heroin self-administration in rats. Psychopharmacology (Berl) 1984;84:167–73. doi: 10.1007/BF00427441. [DOI] [PubMed] [Google Scholar]
- Philip VM, Duvvuru S, Gomero B, Ansah TA, Blaha CD, Cook MN, Hamre KM, Lariviere WR, Matthews DB, Mittleman G, Goldowitz D, Chesler EJ. High-throughput behavioral phenotyping in the expanded panel of BXD recombinant inbred strains. Genes Brain Behav. 2010;9:129–59. doi: 10.1111/j.1601-183X.2009.00540.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piazza PV, Deminiere JM, Maccari S, Mormede P, Le Moal M, Simon H. Individual reactivity to novelty predicts probability of amphetamine self-administration. Behav Pharmacol. 1990;1:339–345. doi: 10.1097/00008877-199000140-00007. [DOI] [PubMed] [Google Scholar]
- Richardson NR, Roberts DC. Progressive ratio schedules in drug self-administration studies in rats: a method to evaluate reinforcing efficacy. J Neurosci Methods. 1996;66:1–11. doi: 10.1016/0165-0270(95)00153-0. [DOI] [PubMed] [Google Scholar]
- Roberts DC, Koob GF. Disruption of cocaine self-administration following 6-hydroxydopamine lesions of the ventral tegmental area in rats. Pharmacol Biochem Behav. 1982;17:901–4. doi: 10.1016/0091-3057(82)90469-5. [DOI] [PubMed] [Google Scholar]
- SAMHSA Results from the 2012 National Survey on Drug Use and Health: Summary of National Findings. 2013 Availableathttp://www.samhsa.gov/data/NSDUH/2012SummNatFindDetTables/NationalFindings/NSDUHresults2012.pdf.
- Sanchis-Segura C, Spanagel R. Behavioural assessment of drug reinforcement and addictive features in rodents: an overview. Addict Biol. 2006;11:2–38. doi: 10.1111/j.1369-1600.2006.00012.x. [DOI] [PubMed] [Google Scholar]
- Schenck A, Bardoni B, Moro A, Bagni C, Mandel JL. A highly conserved protein family interacting with the fragile X mental retardation protein (FMRP) and displaying selective interactions with FMRP-related proteins FXR1P and FXR2P. Proc Natl Acad Sci U S A. 2001;98:8844–9. doi: 10.1073/pnas.151231598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sora I, Li B, Igari M, Hall FS, Ikeda K. Transgenic mice in the study of drug addiction and the effects of psychostimulant drugs. Ann N Y Acad Sci. 2010;1187:218–46. doi: 10.1111/j.1749-6632.2009.05276.x. [DOI] [PubMed] [Google Scholar]
- Thomsen M, Caine SB. Intravenous drug self-administration in mice: practical considerations. Behav Genet. 2007;37:101–18. doi: 10.1007/s10519-006-9097-0. [DOI] [PubMed] [Google Scholar]
- UNODC World Drug Report. 2013 Available at http://www.unodc.org/unodc/secured/wdr/wdr2013/World_Drug_Report_2013.pdf.
- Vassoler FM, White SL, Schmidt HD, Sadri-Vakili G, Pierce RC. Epigenetic inheritance of a cocaine-resistance phenotype. Nat Neurosci. 2013;16:42–7. doi: 10.1038/nn.3280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissenborn R, Robbins TW, Everitt BJ. Effects of medial prefrontal or anterior cingulate cortex lesions on responding for cocaine under fixed-ratio and second-order schedules of reinforcement in rats. Psychopharmacology (Berl) 1997;134:242–57. doi: 10.1007/s002130050447. [DOI] [PubMed] [Google Scholar]
- Wise RA. Roles for nigrostriatal--not just mesocorticolimbic--dopamine in reward and addiction. Trends Neurosci. 2009;32:517–24. doi: 10.1016/j.tins.2009.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolen AR, Phillips CA, Langston MA, Putman AH, Vorster PJ, Bruce NA, York TP, Williams RW, Miles MF. Genetic dissection of acute ethanol responsive gene networks in prefrontal cortex: functional and mechanistic implications. PLoS One. 2012;7:e33575. doi: 10.1371/journal.pone.0033575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye R, Carneiro AM, Airey D, Sanders-Bush E, Williams RW, Lu L, Wang J, Zhang B, Blakely RD. Evaluation of heritable determinants of blood and brain serotonin homeostasis using recombinant inbred mice. Genes Brain Behav. 2014;13:247–60. doi: 10.1111/gbb.12092. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.