CHAPTER SUMMARY
Bacterial pathogens utilize a multitude of methods to invade mammalian hosts, damage tissue sites, and thwart the immune system from responding. One essential component of these strategies for many bacterial pathogens is the secretion of proteins across phospholipid membranes. Secreted proteins can play many roles in promoting bacterial virulence, from enhancing attachment to eukaryotic cells, to scavenging resources in an environmental niche, to directly intoxicating target cells and disrupting their functions. Many pathogens use dedicated protein secretion systems to secrete virulence factors from the cytosol of the bacteria into host cells or the host environment. In general, bacterial protein secretion apparatuses can be divided into different classes, based on their structures, functions, and specificity. Some systems are conserved in all classes of bacteria and secrete a broad array of substrates, while others are only found in a small number of bacterial species and/or are specific to only one or a few proteins. In this chapter, we review the canonical features of several common bacterial protein secretion systems, as well as their roles in promoting the virulence of bacterial pathogens. Additionally, we address recent findings that indicate that the innate immune system of the host can detect and respond to the presence of protein secretion systems during mammalian infection.
INTRODUCTION
One essential prokaryotic cell function is the transport of proteins from the cytoplasm into other compartments of the cell, the environment, and/or other bacteria or eukaryotic cells — a process known as protein secretion. Prokaryotes have developed numerous ways of transporting protein cargo between locations, which largely involve the assistance of dedicated protein secretion systems. Protein secretion systems are essential for the growth of bacteria and are used in an array of processes. Some secretion systems are found in almost all bacteria and secrete a wide variety of substrates, while others have been identified in only a small number of bacterial species or are dedicated to secreting only one or a few proteins. In certain cases, these dedicated secretions systems are used by bacterial pathogens to manipulate the host and establish a replicative niche. Other times, they are required to take advantage of an environmental niche, perhaps by secreting proteins that help bacteria to compete with nearby microorganisms. There are several different classes of bacterial secretion systems, and their designs can differ based on whether their protein substrates cross a single phospholipid membrane, two membranes, or even three membranes, where two are bacterial and one is a host membrane. Due to the specificity of expression of some of these secretion systems in bacterial pathogens, antimicrobials are being developed against these systems to augment our current repertoire of antibiotics. This topic will be discussed in Section VII of this textbook
Five secretion systems will be discussed in depth in subsequent chapters in this section: the Type III Secretion System (T3SS), T4SS, T5SS, T6SS, and T7SS. In this overview, we provide a brief introduction to a number of protein secretion systems, including those that are not discussed in depth in succeeding chapters, in order to highlight the structural and functional similarities and differences between these systems. Our discussions will focus on the canonical features of each system and not the multitude of variations in each one (Table 1). In addition, we briefly review recent findings that indicate that the innate immune system of the host can detect and respond to the presence of protein secretion systems during mammalian infection.
Table 1.
Classes of bacterial protein secretion systems
| Secretion Apparatus | Secretion Signal | Steps in Secretion | Folded Substrates? | Number of Membranes | Gram (+) or Gram (−) |
|---|---|---|---|---|---|
| Sec | N-terminus | 1 | No | 1 | Both |
| Tat | N-terminus | 1 | Yes | 1 | Both |
| T1SS | C-terminus | 1 | No | 2 | Gram (−) |
| T2SS | N-terminus | 2 | Yes | 1 | Gram (−) |
| T3SS | N-terminus | 1–2 | No | 2–3 | Gram (−) |
| T4SS | C-terminus | 1 | No | 2–3 | Gram (−) |
| T5SS | N-terminus | 2 | No | 1 | Gram (−) |
| T6SS | No known secretion signal | 1 | Unknown | 2–3 | Gram (−) |
| SecA2 | N-terminus | 1 | No | 1 | Gram (+) |
| Sortase | N-terminus (Sec) C-terimnus (cws) |
2 | Yes | 1 | Gram (+) |
| Injectosome | N-terminus | 2 | Yes | 1 | Gram (+) |
| T7SS | C-terminus | 1 | Yes | 1–3 | Gram (+) |
SECRETION ACROSS THE CYTOPLASMIC MEMBRANE
A major focus of this chapter is the use of dedicated secretion systems to transport proteins out of the bacterial cell and into the environment or into a recipient cell. However, protein secretion from the bacterial cytoplasmic compartment into other compartments of the cell, particularly into or across the cytoplasmic membrane, also occurs. The general secretion (Sec) and twin arginine translocation (Tat) pathways are the bacterial secretion systems most commonly used to transport proteins across the cytoplasmic membrane (1). The Sec and Tat pathways are the most highly conserved mechanisms of protein secretion, and have been identified in all domains of life (bacteria, archaea, and eukarya) (2, 14). Most proteins transported by the Sec and Tat pathways remain inside of the cell, either in the periplasm or the inner membrane. However, in Gram-negative bacteria, proteins delivered to the cytoplasmic membrane or periplasm of the cell by the Sec or Tat pathways can either stay in those compartments, or may be transported outside of the cell with the help of another secretion system. While the Sec and Tat systems have several common elements, they transport proteins by fundamentally different mechanisms.
The Sec Secretion Pathway
The Sec pathway primarily translocates proteins in their unfolded state. This system consists of three parts: a protein targeting component, a motor protein, and a membrane integrated conducting channel, called the SecYEG translocase (2). Additionally, a number of Gram-positive bacteria produce Sec accessory proteins that serve important roles in the secretion of specific proteins. While proteins secreted by the Sec apparatus can serve many roles, a number of proteins that promote virulence of bacterial pathogens are transported through this pathway. Pathogens that use Sec-dependent secretion to transport virulence factors across the cytoplasmic membrane include the Gram-negative bacteria Vibrio cholerae, Klebsiella pneumoniae, and Yersinia enterocolitica (3). Examples of Gram-positive pathogens that employ Sec accessory systems include Staphylococcus aureus and Listeria monocytogenes (4–7). The use of Sec accessory proteins in secretion by Gram-positive bacteria will be covered in more detail later in this chapter.
Export by the Sec pathway relies on a hydrophobic signal sequence at the N-terminus of the secreted protein, which is typically 20 amino acids in length and contains 3 regions: a positively charged amino terminal, a hydrophobic core, and a polar carboxyl-terminal (2). Proteins that will be secreted into the periplasm or outside of the cell by the Sec pathway contain SecB-specific signal sequences, while proteins meant to remain in the inner membrane contain a signal recognition particle (SRP)-specific signal sequence. The differences between these two pathways are outlined below (2).
The SecB pathway
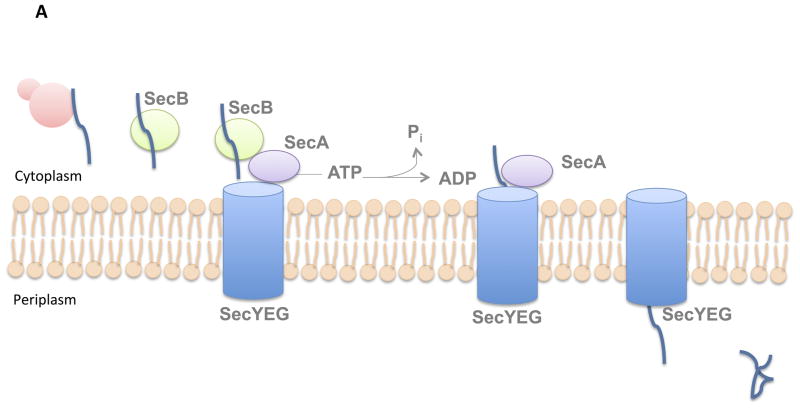
In many Gram-negative bacteria, proteins destined for transport to the periplasm or outside of the cell contain a removable signal sequence recognized by the SecB protein (Fig 1A). This protein serves as a chaperone, binding to pre-secretory proteins and preventing them from folding (8). SecB then delivers its substrates to SecA, a multi-functional protein that both guides proteins to the SecYEG channel, and also serves as the ATPase that provides the energy for protein translocation (9). Prior to transport through the channel, a protease protein cleaves off the SecB signal sequence from the protein, and the secreted protein is then is folded upon delivery to the periplasm (10). While many proteins delivered by the SecB system remain in the periplasm, some will ultimately become extracellular. Once these proteins are delivered to the periplasm, they can be transported across the outer membrane with the help of the Type II and Type V secretion systems. We will discuss these Sec-dependent mechanisms of protein secretion in detail later on in this chapter.
Fig. 1. Export through the Sec pathway.
In bacteria, the Sec pathway transports unfolded proteins across the cytoplasmic membrane. Proteins secreted by this pathway may either become embedded in the inner membrane, or will be released into the periplasm. In Gram-negative organisms, these periplasmic proteins may be released extracellularly with the help of an additional secretion system. (A) Proteins destined for the periplasm (or extracellular release) are translocated by a post-translational mechanism and contain a removable signal sequence recognized by the SecB protein. SecB binds pre-secretory proteins and prevents them from folding, while also delivering its substrates to SecA. SecA both guides proteins to the SecYEG channel, and also serves as the ATPase that provides the energy for protein translocation. Following transport through the SecYEG channel, proteins are folded in the periplasm. (B) The Sec pathway utilizes a co-translational mechanism of export to secrete proteins destined for the inner membrane. These proteins contain a signal sequence recognized by the SRP particle. During translation, SRP binds target proteins as they emerge from the ribosome, and recruits the docking protein FtsY. FtsY delivers the ribosome-protein complex to the SecYEG channel, which translocates the nascent protein across the cytoplasmic membrane. During translocation across the channel, the transmembrane domain is able to escape through the side of the channel into the membrane, where the protein remains attached.
The SRP pathway
The Sec system can also transport proteins that are meant to remain in the inner membrane by way of the SRP pathway (Fig 1B). Transmembrane proteins often contain hydrophobic domains, and thus are unstable when cytoplasmic. For this reason, secretion by the SRP pathway utilizes a co-translational mechanism of export that couples translation of the protein by the ribosome with secretion through the SecYEG channel (11). The SRP pathway relies on the SRP particle, which contains a small, 4.5S RNA bound to a protein called Ffh (11). During protein secretion by this pathway, SRP first binds the transmembrane domain of proteins as they emerge from the ribosome (12). SRP then binds the docking protein FtsY, which delivers the ribosome-protein complex to the SecYEG channel (2). Translation of the protein then drives the secretion of the nascent protein through the channel. During this process, the transmembrane domain of the protein escapes through the side of the channel into the membrane, where it remains attached (13). The mechanism of this final step is not yet known.
The Tat Secretion Pathway
In contrast to the Sec pathway, the Tat pathway primarily secretes folded proteins (14) (Fig. 2). This pathway is critical because not all proteins can be secreted in their unfolded state, as certain proteins that contain post-translational modifications, such as redox factors, are synthesized in the cytoplasm (15). The materials required for these modifications would not be available extracellularly or in the periplasm and, thus, these proteins must be folded and modified in the cytoplasm prior to secretion in their 3-dimensional state.
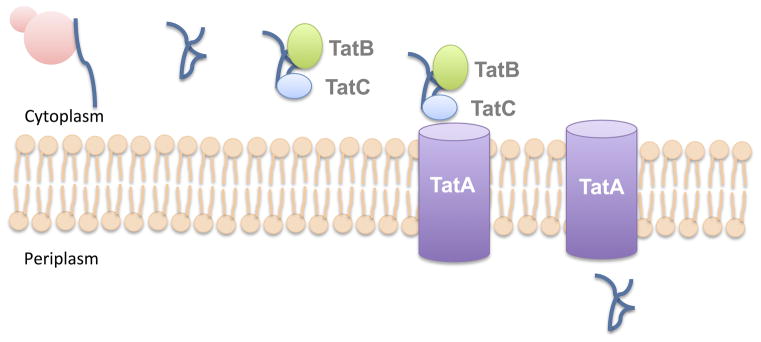
Fig. 2. Secretion through the Tat pathway.
Bacteria secrete folded proteins across the cytoplasmic membrane using the Tat secretion pathway. This pathway consists of 2–3 components (TatA, TatB, and TatC). In Gram-negative bacteria, TatB and TatC bind a specific N-terminal signal peptide containing a “twin” arginine motif on folded Tat secretion substrates. TatB and TatC then recruit TatA to the cytoplasmic membrane, where it forms a channel. Folded proteins are then translocated across the channel and into the periplasm. In Gram-negative bacteria, these proteins may remain in the periplasm, or can be exported out of the cell by the T2SS.
The Tat pathway of protein secretion consists of 2–3 subunits: TatA, TatB, and TatC (in Gram-positive bacteria, TatA and TatB are combined into one multi-functional protein) (16, 17). In Escherichia coli, TatB and TatC bind the signal peptide of Tat-secreted proteins and then recruit TatA, which forms the membrane-spanning channel (18). The Tat signal sequence contains a pair of “twin” arginines in the motif S-R-R at the N-terminus of the folded protein (18). Whereas most proteins secreted by the Tat apparatus in Gram-positive bacteria are released extracellularly, Tat-secreted proteins in Gram-negative bacteria can either remain periplasmic or are transported out of the cell by the Type II Secretion System through a mechanism that will be reviewed later in this chapter.
While the Tat pathway is important for the physiology and survival of both pathogenic and non-pathogenic bacteria, a number of pathogenic bacteria, including Pseudomonas aeruginosa (19), Yersinia pseudotuberculosis (20), and E. coli O157:H7 (21) require a functional Tat pathway for full virulence in animal infection models. Phospholipase C enzymes are a notable example of Tat-secreted proteins that serve as virulence factors for a number of pathogens, including P. aeruginosa (19), Legionella pneumophila (22), and Mycobacterium tuberculosis (23). These enzymes, which cleave phospholipids immediately before their phosphate group, can serve a variety of functions during infection, including suppression of immune cell activity and promotion of intracellular survival (24).
PROTEIN SECRETION BY GRAM-NEGATIVE BACTERIA
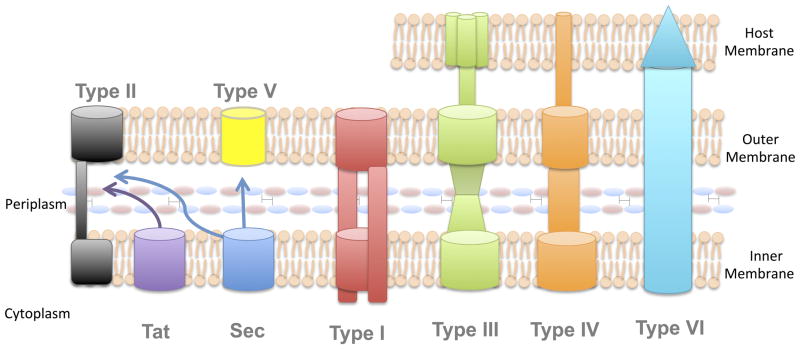
A number of Gram-negative bacteria rely on dedicated secretion systems to transport virulence proteins outside of the cell and, in some cases, directly into the cytoplasm of a eukaryotic or prokaryotic target cell. Extracellular protein secretion can be a challenge for Gram-negative bacteria, because these secreted proteins must cross two (and, in some cases, three) phospholipid membranes in order to reach their final destination (Fig 3). Some secreted proteins in Gram-negative bacteria traverse these membranes in two separate steps, where they are first delivered to the periplasm through the Sec or Tat secretion systems, as discussed in the preceding section, and are then transferred across the outer membrane by a second transport system. This process is known as Sec- or Tat-dependent protein secretion. Additionally, many other proteins are secreted through channels that span both the inner and outer bacterial membranes through a process known as Sec- or Tat-independent protein secretion. The dedicated secretion systems in Gram-negative bacteria are numbered Type I through Type VI, with each system transporting a specific subset of proteins. These systems all rely on β-barrel channels that form a ring in the outer membrane of the bacterial cell, but otherwise exhibit a fair amount of diversity in their structures and mechanistic functions, as will be outlined below.
Fig. 3. Secretion systems in Gram-negative bacteria.
Gram-negative bacteria utilize a number of dedicated protein secretion systems to transport proteins across 1, 2, or 3 phospholipid membranes. Some proteins are secreted in a two-step, Sec- or Tat-dependent mechanism. These proteins cross the inner membrane with the help of either the Sec or Tat secretion pathways and are then transported across the outer membrane using a second secretion system. The T2SSs and T5SSs secrete proteins in this manner. Because it secretes folded substrates, the T2SS translocates proteins initially transported by either the Tat or Sec pathway (where Sec substrates are folded in the periplasm). In contrast, autotransporters of the T5SS must be unfolded prior to outer membrane transport and thus must be secreted across the inner membrane by the Sec pathway. Additionally, several Gram-negative protein secretion systems transport their substrates across both bacterial membranes in a onestep, Sec- or Tat-independent process. These include the T1SSs, T3SSs, T4SSs, and T6SSs. All of these pathways contain periplasm-spanning channels and secrete proteins from the cytoplasm outside the cell, however, their mechanisms of protein secretion are quite different. Three of these secretion systems, the T3SS, T4SS, and T6SS can also transport proteins across an additional host cell membrane, delivering secreted proteins directly to the cytosol of a target cell.
The Type I Secretion System
Type I secretion systems (T1SSs) have been found in a large number of Gram-negative bacteria, including pathogens of plants and animals, where they transport their substrates in a one-step process (as demonstrated in Fig. 3) across both the inner and outer bacterial membranes (recently reviewed in (25)). Unlike other protein transport systems found in Gram-negative bacteria, T1SSs closely resemble a large family of ATP-binding cassette (ABC) transporters, which export small molecules such as antibiotics and toxins out of the cell (26). Some bacteria may have several T1SSs, each of which is dedicated to transporting one or a few unfolded substrates (27). These substrates range in function and include digestive enzymes, such as proteases and lipases, as well as adhesins, heme-binding proteins, and proteins with repeats-in-toxins (RTX) motifs. T1SS substrates are generally Sec-independent and typically, but don’t always, contain a C-terminal signal sequence that is recognized by the T1SS and remains uncleaved (see below).
T1SSs have three essential structural components: an ABC transporter protein in the inner membrane, a membrane fusion protein (MFP) that crosses the inner membrane and bridges it to the outer membrane factor (OMF) in the outer membrane (25). The ABC transporter component associated with the T1SS has several critical functions – it catalyzes ATP to provide the energy to transport the substrate, interacts with the MFP, and participates in substrate recognition (28). The MFP associates with the ABC transporter in the inner membrane and spans the periplasm to associate with the OMF (29–31). In addition, the cytoplasmically located N-terminus of the MFP is believed to play a role in substrate selection (30, 32). The OMF generates a pore in the outer membrane, through which the substrate passes in an unfolded state. Interestingly, T1SSs often use the multi-purpose protein TolC as their OMF (27). This pore-forming protein is also used to export molecules and other compounds, and is recruited to the MFP after the ABC transporter and MFP have contacted a substrate(32).
The T1SS ABC transporters have been further divided into three groups based on their N-terminal sequences (reviewed in (28)). One class of ABC transporters contains a C39 peptidase domain, which belongs to the papain superfamily structural motif. The C39-peptidase-containing ABC-transporters are critical for recognizing and cleaving the N-termini of substrates. An example of a T1SS substrate with a C39 peptidase domain is Colicin V of E. coli (33). A second class of ABC transporters contains a C39-like peptidase domain (CLD) that lacks proteolytic activity and, therefore, does not cleave its designated substrates (34). Substrates of CLD-containing ABC transporters generally contain RTX motifs and are much larger than those secreted by a C39-containing peptidase ABC transporter. Interestingly, RTX motifs bind to calcium at extracellular, but not intracellular levels. Because calcium binding promotes the folding of these proteins, these large substrates are able to remain unfolded inside the cell (35). Finally, a third class of T1SS ABC transporters lacks any additional sequences in the N-terminal domain. Their substrates may or may not contain RTX motifs but are smaller in size than substrates transported by CLD-containing ABC transporters and contain secretion signals at their C-termini (27).
T1SS substrates contribute to virulence in a variety of bacterial pathogens, including V. cholerae, which uses its T1SS to secrete the MARTX toxin (36), and Serratia marcescens, which secretes the hemophore HasA via the T1SS pathway (29). One of the best-studied T1SS substrates is the HlyA hemolysin protein of uropathogenic E. coli (37–39). This RTX-family toxin inserts into the membranes of both erythrocytes and nucleated eukaryotic cells, causing them to rupture (37). Rupture of host cells by HlyA can help the bacteria to cross mucosal barriers, and additionally, can damage effector immune cells, which prevents clearance of the infection.
The Type II Secretion System
Type II secretion systems (T2SSs) are conserved in most Gram-negative bacteria, where they transport folded proteins from the periplasm into the extracellular environment. Because the T2SS channel is only found in the outer membrane (Fig. 3), proteins secreted through this apparatus must first be delivered to the periplasm via the Sec or Tat secretion pathways, which transfer protein substrates across the inner membrane, as described earlier in this chapter. This secretion system was originally called the main terminal branch of the Sec secretion pathway due to its ability to export proteins transported across the inner membrane by the Sec secretion system (3). However, this nomenclature has since been updated to the T2SS to reflect the ability of these secretion systems to transport Tat-secreted proteins as well (40). Because proteins destined for secretion by the T2SS apparatus must first pass through the Sec or Tat inner membrane transporters, T2SS substrates must have a Sec- or Tat-type cleavable signal sequence at their N termini (3). Additionally, because the T2SS secretes folded substrates, proteins transported across the cytoplasmic membrane by the Sec pathway must be folded in the periplasm prior to export through the T2SS.
T2SSs have a broad specificity and are capable of secreting a diverse array of substrates outside of the bacterial cell, some of which contribute to the virulence of bacterial pathogens (3). In some bacterial species, the T2SS is required for the secretion of multiple substrates, while in others, it is only used to transport a single protein (41). These secreted proteins have a range of biological functions, but are generally enzymes, such as proteases, lipases, and phosphatases, as well as several proteins that process carbohydrates (3).
T2SSs are complex and consist of as many as 15 different proteins, which can be broken into four subassemblies: the outer-membrane complex, the inner-membrane platform, the secretion ATPase, and the pseudopilus (3). As its name suggests, the outer-membrane complex resides in the outer membrane, where it serves as the channel through which folded periplasmic T2SS substrates are translocated (42). This channel is composed of a multimeric protein called the secretin. The secretin has a long N terminus, which is believed to extend all the way to the periplasm to make contact with other T2SS proteins in the inner membrane (42). The inner membrane platform, which is composed of multiple copies of at least 4 proteins, is embedded in the inner membrane and extends into the periplasm, contacting the secretin. This platform plays a crucial role in the secretion process, by communicating with the secretin, pseudopilus, and the ATPase to coordinate export of substrates (3). The ATPase is located in the cytoplasm and provides the energy to power the system. As its name implies, the T2SS pseudopilus is evolutionarily related and structurally similar to proteins that comprise type IV pili on bacterial cell surfaces, as well as some bacterial competence systems (43). Therefore, one model for secretion through the T2SS channel proposes that these pseudopili retract in order to push the folded T2SS substrate through the outer membrane channel. In this “piston” model, “secretion-competent” proteins in the periplasm contact the periplasmic domain of the secretin. This interaction is believed to stimulate the cytoplasmic ATPase to drive retraction of the T2SS pseudopili, which push proteins through the secretin channel (3, 44, 45).
A number of bacterial pathogens employ T2SSs to transport virulence factors outside of the cell. Examples of T2SS substrates that are important for virulence in a mammalian host include the cholera toxin of V. cholerae (46), which causes the watery diarrhea associated with the disease cholera, and exotoxin A of P. aeruginosa (47), which blocks protein synthesis in host cells, leading to lethal infection by this bacterium. Still, other pathogens use their T2SSs to secrete enzymes that help them adapt to their environment, which can include plant and animal hosts. These pathogens include Legionella pneumophila (48), enterotoxigenic and enterohemorrhagic E. coli (ETEC and EHEC) (49–51), K. pneumoniae (52), Aeromonas hydrophila (53), and Dickeya dadantii (54).
The Type III Secretion System
Type III secretions systems (T3SSs) are found in a large number of Gram-negative bacterial pathogens and symbionts (reviewed in (55)). T3SSs have been described as “injectisomes” and “needle and syringe”-like apparatuses because of their structure (see Fig. 3). They secrete a wide variety of proteinaceous substrates across both the inner and outer bacterial membranes. In addition, most T3SSs also transport substrates into a target eukaryotic cell membrane in the same step and, therefore, actually transport proteins across three membranes. Secretion of T3SS substrates is generally thought to be a one-step process, although recently this notion has been challenged in Yersinia (discussed below). T3SS substrates are generically called effector proteins. Pathogens may secrete only a few effectors proteins, as in the cases of Pseudomonas and Yersinia, or several dozen, as in the cases of Shigella and EHEC. Secretion signals are embedded within the N-termini of T3SS substrates and are not cleaved. Many, but not all T3SS effectors have chaperones that guide them to the T3SS base, where they are secreted in an ATP-dependent, unfolded state.
The T3SS has a core of 9 proteins that are highly conserved among all known systems (reviewed in (56, 57)). They share 8 of these proteins with the flagellar apparatus found in many bacteria and are evolutionarily related to flagellin (58). In addition to these 9 core proteins, T3SSs have an additional 10 to 20 proteins that play either essential or important roles in their function. The structural components of T3SSs are typically encoded in a few operons, which can be found either in pathogenicity islands in the bacterial chromosome or on plasmids. Because T3SSs are typically horizontally acquired, bacteria that are evolutionary distinct may have closely related systems and vice versa (58). For example, the genomes of Shigella and E. coli are highly homologous, yet the Shigella T3SS is more similar to the Salmonella T3SS than it is to systems found in the E. coli pathogens EHEC and EPEC. Seven families of T3SSs have been proposed primarily based on the homology of their extracellularly elaborated needles, tips, and translocons (58).
The T3SS can be broken down into three main components: a base complex or basal body, the needle component, and the translocon (56). The base complex contains cytoplasmic components and spans the inner and outer membrane, forming a socket-like structure consisting of several rings with a center rod (59). In most systems it is comprised of at least 15 proteins (56, 57). Encased by and emanating from this socket and rod-like structure is a filament called the needle, which extends through the secretin and into the extracellular space (59). The T3SS needle has an inner hollow core that is wide enough to permit an unfolded effector to traverse (60, 61). Excitingly, recent work has visualized a ‘trapped’ effector protein by cryo-EM and single particle analysis, supporting the model that substrates can traverse through the needle (62, 63).
The T3SS tip complex, which resides on the outer end of the needle, is critical for sensing contact with host cells and regulating secretion of effectors (64, 65). It is also necessary for insertion of the translocon into host cell membranes (65, 66). The T3SS translocon is essential for passage of effectors through host cell membranes, but not for secretion of effectors outside of the bacterium (67, 68). Translocons are assembled upon contact with host cells and form a pore that is essential for effector delivery (66). Recently, however, an alternative two-step model of translocation of Type 3 effectors has been proposed, where effectors and translocon components are secreted prior to host cell contact and remain associated with the bacteria, perhaps in lipid vesicles (69, 70). After contact with host cells, perhaps sensed through the needle, the translocon and tip proteins form a pore through which the effectors pass. Additional experiments are needed to determine the mechanism by which translocation occurs.
Translocation of T3SS effectors into host cells is essential for the virulence of many pathogens, including pathogenic species of Yersinia, Salmonella, and Shigella (55). Over the last 25 years, much work has focused on understanding the functions of T3SS effector proteins. Their functions vary widely among different pathogens and how they jointly orchestrate their effects on host cells is still being elucidated (71–74). Many of these effectors remodel normal cellular functions to enable the pathogen to establish an infectious niche either within the host cell or in mammalian tissue sites. Impressively, the study of these effectors has provided fundamental insights into a number of different facets of eukaryotic cell biology.
The Type IV Secretion System
Type IV secretion systems (T4SSs) are ancestrally related to bacterial DNA conjugation systems and can secrete a variety of substrates, including single proteins and protein-protein and DNA-protein complexes (75). T4SSs secrete substrates into a wide range of target cells, including other bacteria (of the same or different species) and eukaryotic cells. These macromolecular complexes are largely found in Gram-negative bacteria, where they transport substrates across both the inner and outer membranes (Fig. 3). Like T3SSs, T4SSs can span an additional, host cell membrane, allowing for direct transfer of substrates into the cytoplasm of the recipient cell. Because T4SSs are capable of transferring both DNA and proteins, they can serve a variety of functions, including conjugative transfer of DNA, DNA uptake and release, and translocation of effector proteins or DNA/protein complexes directly into recipient cells.
Despite the diversity in their substrates and functions, all T4SSs are evolutionarily related, sharing common components and operating in a similar manner (76). For that reason, the remainder of this section will focus on the VirB/D system of Agrobacterium tumeficans as a model of Type IV Secretion. A. tumeficans uses its T4SS to transport oncogenic T-DNA into plant cells, and has served as the paradigm for studying T4SS assembly and function (77). The VirB/D T4SS contains 12 proteins, named VirB1-VirB11 and VirD4 (78). Most of these proteins are membrane associated and multi-copy, interacting with themselves and each other. The VirB6-10 proteins are found in the periplasm, inner and outer membranes, and form the secretion channel as well as its accessory proteins. VirB4, VirB11, and VirD4 localize to the inner membrane and serve as the ATPases that power the system. VirD4 also functions as a coupling protein, binding proteins prior to secretion through the channel. Generally, T4SSs also include an extracellular pilus, composed of a major (VirB2) and minor (VirB5) subunit.
The process of substrate secretion through the T4SS apparatus is still an active area of investigation. However, it is believed that substrate DNA or protein first makes contact with VirD4, which functions as a molecular “gate” at the base of the secretion apparatus (79). VirD4 then transfers the substrate to VirB11, which delivers the substrate to the inner membrane channel complex. Finally, the substrate is transferred across the periplasm to the outer membrane protein complex. It is not currently known what role the T4SS pilus plays in the secretion process. Some believe that the pilus may serve merely as an attachment device, allowing bacteria to come into tight contact with target cells (78). Still, others have predicted that the pilus may actually serve as the conduit for substrate translocation, particularly into target cells (80). Work to determine which of these two models is correct is currently ongoing.
T4SSs play pivotal roles in the pathogenesis of a wide range of bacteria. Notable examples of bacterial pathogens that employ T4SSs for virulence are Neisseria gonorrhoeae, which uses its T4SS to mediate DNA uptake (which promotes virulence gene acquisition)(81), and L. pneumophila, Brucella suis, and Helicobacter pylori, which use their T4SSs to translocate effector proteins into host cells during infection to disrupt their defense strategies (82). These effector proteins have a wide range of functions. For example, the intracellular pathogen L. pneumophila uses its T4SS to translocate more than 200 effector proteins into the host cell, where they play important roles in remodeling the host cell architecture in order to create a vacuole suitable for bacterial replication (83). A major focus in the T4SS field is now on understanding the roles these effector proteins affect host cell functions. In addition to enhancing our understanding of host-pathogen interactions, these studies have also led to novel insights into eukaryotic cellular biology.
The Type V Secretion System
Type V secretion system (T5SS) substrates are unique in that, unlike other secreted substrates, which cross the bacterial membrane with the help of a dedicated secretion apparatus or membrane channel, they secrete themselves. These proteins or groups of proteins carry their own β-barrel domain, which inserts into the outer membrane and forms a channel that either the remainder of the protein or a separate protein is transported through (84, 85). Because protein secretion by T5SSs only occurs in the outer membrane, these proteins must first be translocated across the inner membrane and into the periplasm in an unfolded state by the Sec apparatus (Fig. 3). Therefore, T5SS proteins carry an N-terminal Sec signal sequence that is cleaved off as they pass into the periplasm (86).
Most well known T5SS substrates are virulence proteins, serving as toxins and receptor-binding proteins. Some examples of T5SS substrates that play important roles in pathogenesis include the immunoglobulin A protease of N. gonorrhoeae, which cleaves host antibodies (87), the IcsA protein of Shigella flexneri (88), which promotes actin-based intracellular motility and also serves as an adhesin (89), and YadA of Y. enterocolitica (90), which helps to promote translocation of T3SS substrates into host cells, and assists in mediating resistance to attack by the host complement system (91). T5SSs can be separated into three classes, depending on the number of proteins involved in the secretion process. These classes include autotransporter secretion, two-partner secretion, and chaperone-usher secretion (92).
Autotransporter secretion
The most simplistic form of Type V secretion is known as the autotransporter system. As its name implies, autotransporters contain components that allow them to secrete themselves (92). More specifically, autotransporters contain 3–4 domains: a translocator domain at the C-terminus that forms the outer membrane channel, a linker domain, a passenger domain that contains the functional part of the autotransporter protein, and sometimes, a protease domain that cleaves off the passenger domain once it passes through the channel (85).
Following secretion of the unfolded autotransporter protein through the inner membrane, the translocator domain assembles in the outer membrane, forming a 12-stranded β-barrel, usually with the help of a number of accessory factors, including the periplasmic chaperone Skp and the Bam complex (93, 94). The flexible linker domain then leads the passenger domain through the channel to the outside of the cell. Once the transporter domain has reached the outside of the cell, it is either released by its own protease domain or remains attached to the translocator domain and protrudes outside the cell (85).
Two-partner secretion
While the majority of T5SS substrates are secreted via the autotransporter mechanism, a few rely on different polypeptides for transport outside of the cells. In a process called two-partner secretion, a pair of proteins participates in the secretion process, in which one partner carries the β-barrel domain, while the other partner serves as the secreted protein (95). Two-partner secretion has been observed in a large variety of Gram-negative bacteria and is primarily responsible for transporting large virulence proteins, such as the filamentous haemagglutinin of Bordetella pertussis and the high-molecular weight adhesins HWM1 and HWM2 of Haemophilus influenzae (96, 97).
Chaperone-usher secretion
A third subcategory of T5SSs involves proteins secreted with the help of two other proteins: the usher protein, which forms the β-barrel channel in the outer membrane, and the chaperone, a periplasmic protein that facilitates folding of the secreted protein prior to delivery to the channel (98). Chaperone-usher systems are commonly used to assemble pilins on the surface of Gram-negative bacteria, such as the P pilus of uropathogenic E. coli (98).
The Type VI Secretion System
Type VI secretion systems (T6SSs) are the most recent bacterial secretion systems to be discovered (99) and, therefore, there is still much to learn about their structure and functions. T6SSs translocate proteins into a variety of recipient cells, including eukaryotic cell targets and, more commonly, other bacteria (100). These systems are fairly well conserved in a wide-range of Gram-negative bacterial species, with nearly a quarter of sequenced genomes containing genes for T6SS components (101). Unlike many of the other characterized Gram-negative secretion systems, T6SSs are capable of transporting effector proteins from one bacterium to another in a contact-dependent manner, which is believed to play a role in bacterial communication and interactions in the environment (100).
T6SSs are very large, with up to 21 proteins encoded within a contiguous gene cluster (100). Thirteen of these proteins appear to be conserved in all T6SSs and are thought to play a structural role in the secretion apparatus. Intriguingly, T6SSs share structural homology to phage tails, and it has been hypothesized that T6SSs may have arisen from inverted phage tails that eject proteins outside of the bacterial cell rather than injecting them inside the cell (Fig. 3) (102). It has been proposed that some structural components of the T6SS apparatus may also serve as effector proteins, though other T6SS effector proteins have also been identified. These effectors have many forms and functions, with many directed against the bacterial cell wall and membrane, which supports a role for this secretion apparatus in promoting interspecies bacterial competition (100, 101). Lending further credence to this hypothesis, many T6SS effectors are encoded alongside a gene that provides immunity to the effector, thereby preventing self-intoxication (100).
T6SSs are hypothesized to contribute to the virulence of some bacterial pathogens, both through delivery of protein substrates to host cells, and by secreting substrates into neighboring bacteria that may be competing to exploit a specific host niche. While we know that many bacterial pathogens, including P. aeruginosa, V. cholerae, and S. marcescens are able to use their T6SSs under laboratory conditions (101–103), the mechanisms of how these T6SSs contribute to survival in the environment (and in mammalian infection) have not been determined.
PROTEIN SECRETION BY GRAM-POSITIVE BACTERIA
Thus far, our discussion of protein secretion has been primarily focused on Gram-negative bacteria, which possess two phospholipid membranes separated by a periplasm compartment containing a thin layer of peptidoglycan. In contrast, as demonstrated in Fig. 4, Gram-positive bacteria contain only one lipid bilayer and are surrounded by a very thick cell wall (considerably thicker than that of Gram-negative bacteria). Additionally, some species of Gram-positive bacteria, most notably the Mycobacteria, contain a cell wall heavily modified by lipids called a mycomembrane (Fig. 5). Because of these differences in basic cell structure, it is not surprising that Gram-positive bacteria differ from Gram-negative organisms in their mechanisms of extracellular protein secretion. Like Gram-negative organisms, Gram-positive bacteria employ both the Tat and Sec pathways to transport proteins across the cytoplasmic membrane. However, in many cases, this transport is not sufficient to deliver proteins to their final destinations. In the following section, we review conserved mechanisms of protein secretion in Gram-positive and Mycobacteria, many of which are used by pathogens to transport important virulence factors out of the cell during mammalian infection.
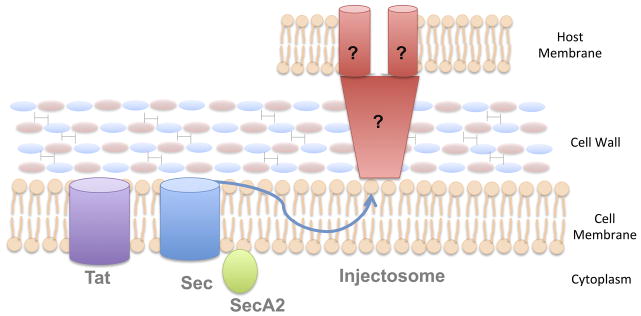
Fig. 4. Secretion systems in Gram-positive bacteria.
Gram-positive bacteria contain a single cytoplasmic membrane surrounded by a very thick cell wall. These organisms can secrete proteins across the membrane using the Tat and Sec secretion systems. In contrast to Gram-negative organisms, many Gram-positive bacteria use an additional factor for Sec secretion of a smaller subset of proteins, called SecA2. Additionally, there is evidence that some Gram-positive bacteria may use dedicated secretion apparatuses, called “injectosomes” to transport proteins from the bacterial cytoplasm into the cytoplasm of a host cell in a 2-step process. The specific mechanism of this process has not been determined, though it has been proposed that the injectosome may utilize a protected channel to transport proteins across the cell wall during export.
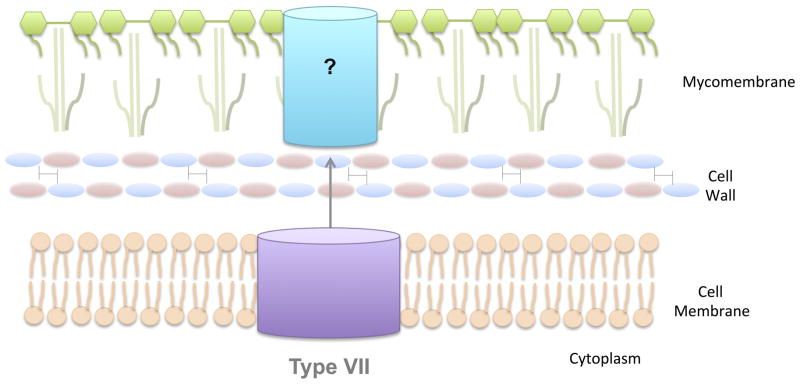
Fig. 5. The Type VII Secretion System.
Certain Gram-positive organisms, including members of the genus Mycobacteria, contain a cell wall layer that is heavily modified by lipids, called a mycomembrane. These organisms contain a distinct protein secretion apparatus called a T7SS. T7SSs contain several core inner membrane proteins that interact with cytosolic chaperones and form a channel through which proteins are secreted. Additionally, it has been proposed that T7SSs may contain an additional, mycomembrane-spanning channel that aids in extracellular secretion of substrates, though this model has not been experimentally proven.
SecA2 Secretion
As we discussed earlier in this chapter, the Sec secretion pathway is one of the most conserved mechanisms of protein export and is found in all classes of bacteria. As mentioned above, an essential component of post-translational Sec export (through the SecB pathway) is the targeting and ATPase protein SecA. For years, it was thought that all bacteria contained a single SecA protein; however, recent discoveries have shown that many Gram-positive organisms, including L. monocytogenes, Bacillus subtilis, Clostridium difficile, M. tuberculosis, and Corynbacteria glutamicum, actually contain two SecA homologues, called SecA1 and SecA2 (reviewed in (104, 105)) (Fig. 4). In these organisms, SecA1 is essential, and aids in the secretion of proteins via the canonical Sec pathway, as described earlier in this chapter. In contrast, SecA2 is seldom required for growth under standard laboratory conditions, and is used to export a smaller set of proteins. Generally, SecA2 substrates are involved in stress responses and/or cell wall modifications, repair and metabolism. While SecA2 is often not essential for growth under normal laboratory conditions, it is required under specific stress conditions and has also been linked to virulence.
It is currently thought that the SecYEG core transporter transports SecA2 substrates and that SecA2 provides an additional means of regulation of specific substrates. The SecA2 protein, like SecA, contains two nucleotide-binding domains, a pre-protein cross-linking domain, a helical wing and helical scaffold domain, and a C- terminal domain (106). However, most SecA2 proteins are smaller than their corresponding SecA homologs, because they have small deletions in one of these domains (107). These deletions may alter the rate of ATP hydrolysis, the interaction with the rest of the Sec apparatus, and/or the location of SecA2 molecules (108, 109). For these reasons, secretion of the substrates associated with SecA2 molecules may be regulated differently compared to secretion of SecA substrates.
Although some information has been gleaned from studies of SecA2 in various bacteria, there are a number of unanswered questions about SecA2 systems. While it appears most likely that SecA2 substrates are secreted through the normal SecYEG secretion system, SecA2 interactions with other transporters cannot be ruled out. Likewise, it is unclear how SecA2 interacts with SecYEG and how SecA versus SecA2 substrates are selected for secretion, if indeed both are directed to the same core apparatus. Finally, how SecA2 discriminates among potential substrates is poorly understood.
A number of Streptococci and Staphylococci express a second Sec secretion system called aSec or SecA2-SecY2. These systems not only contain SecA2, but also have other proteins that serve to transport SecA2 substrates, including SecY2 and at least three accessory Sec transport proteins. aSec systems typically transport large, highly glycosylated cell-wall anchor proteins with serine-rich repeats (SRR) (110, 111). These SRR glycosylated proteins function as adhesins in a number of Streptococcus and Staphylococcus species, and can contribute to virulence in these pathogens.
In contrast to bacteria that only express SecA1, bacteria expressing aSec are thought to transport substrates through a channel called SecY2. SecY2 lacks the cytoplasmic loops in SecY that normally interact with SecA1, and therefore SecA1 bound substrates are unlikely to interact with SecY2 transporters (107). In addition, there are three accessory proteins in this system, whose roles are not yet understood, although they are essential for secretion of these glycoproteins (112). All of these proteins localize to the membrane and cytosol and may help deliver the SecA2-substrate complex to SecY2, open the pore, assist in transport of the substrate, and participate in complete glycosylation of these substrates.
Sortases
Like their Gram-negative counterparts, many Gram-positive pathogens express proteins on their outer surfaces that assist in survival during infection of a mammalian host. Because Gram-positive bacteria lack an outer membrane, these proteins must embed themselves into the Gram-positive cell wall in order to be retained on the outer surface of the bacterium. In order to fulfill this function, Gram-positive bacteria encode a class of enzymes, called sortases, which covalently attach proteins to the cell wall following secretion across the cytoplasmic membrane (reviewed in (113, 114)).
Most Gram-positive bacteria express a variety of sortases, which can range in specificity. For example, the general, “housekeeping” sortases, SrtA, can attach as many as 40 proteins to the cell wall, while other sortases are specific for only one or two proteins (114). Sortases carry out covalent linkages of proteins to the cell wall in a catalytic reaction called transpeptidation. Targets of SrtA typically contain an N-terminal signal peptide, as well as a 30–40 residue C-terminal cell wall sorting (cws) signal, which is composed of a pentapeptide cleave site, LPXTG, and a hydrophobic domain(114, 115). Proteins destined for cell wall attachment by SrtA are first targeted to the Sec translocase by their N-terminal signal sequences and are translocated across the membrane through the canonical Sec pathway. Next, the sorting signals of these proteins are processed by SrtA, which cuts between the threonine and glycine residues of their LPXTG sites (114, 115). The carboxyl group of the threonine is then covalently linked to a cysteine residue on the C terminus of the sortase, effectively cleaving off the C-terminus of the original protein (114, 115). Finally, this intermediate is covalently attached to the amino group on the cell wall precursor lipid II. This modified lipid II is then incorporated into peptidoglycan during cell wall synthesis, effectively embedding the SrtA substrate into the cell wall. Other sortases attach their substrates via a similar catalytic reaction to that used by SrtA, but they have specificities for different LPXTG motifs and amino groups.
Sortases are found in nearly all Gram-positive bacteria and, as mentioned above, process some proteins that contribute to the virulence of many pathogens. Pili are a classical example of cell surface proteins in Gram-positive bacteria (113, 114). These filamentous protein structures are often involved in adhering to and invading host tissue sites during infection. Pili are typically composed of a major subunit, called pilin, and one or more minor subunits, which are usually assembled sequentially using specialized sortases (113, 114). Pathogens that are known to utilize pili during infection include Streptococcus pneumoniae, Streptococcus pyogenes, and S. aureus (113, 114).
Extracellular Protein Secretion In Gram-positive Bacteria
Not all secreted proteins in Gram-positive bacteria will remain embedded in the cell wall. Rather, many proteins exported across the cytoplasmic membrane by the Sec or Tat pathways will eventually be released into the extracellular environment, often through passive diffusion through the peptidoglycan layer. Additionally, some Gram-positive organisms use a separate secretion apparatus (named the T7SS) to export certain proteins across the cytoplasmic membrane and, potentially, through the cell wall.
Like the T3SS and T4SS effectors of Gram-negative bacteria, some proteins secreted by Gram-positive pathogens will be delivered to the cytoplasmic compartments of eukaryotic host cells. The mechanisms by which Gram-positive effector proteins reach the cytoplasm of eukaryotic cells are varied, and are in large part exemplified by the self-translocating AB toxin model of delivery. However, recent evidence suggests that some Gram-positive organisms may actually use a protein secretion apparatus to directly deliver certain effector proteins to eukaryotic cells (116).
Gram-positive “injectosomes”: a model for effector transport into host cells
One secretion apparatus in Gram-positive organisms, coined the “injectosome” (note the difference in spelling from the Gram-negative T3SS injectisome), is proposed to be functionally analogous (but structurally unrelated) to the T4SSs and T3SSs of Gram-negative bacteria. This model of protein secretion has been observed in the Gram-positive pathogen Streptococcus pyogenes, which injects at least one virulence factor, NAD glycohydrolase (SPN), into the cytoplasm of keratinocytes by this mechanism (116, 117). In order to create the pore required for SPN translocation, another protein, SLO, is first secreted via the Sec pathway. SLO is a member of a class of toxins called cholesterol-dependent cytolysins (CDCs), which bind cholesterol on the surface of eukaryotic cells and insert into their membranes, creating pores (118). Following pore formation by SLO, SPN is translocated across the plasma membrane by Sec, and into the eukaryotic cell through the pore (119). Once it reaches the cytosol of host cells, SPN cleaves the glycosidic bond of β-NAD+ to produce nicotinamide and ADP-ribose, a potent second messenger — thereby disrupting normal cell functions (120). For this reason, SPN serves as a major virulence factor for Streptococcus pyogenes, particularly during severe infections with this bacterium.
Interestingly, there is some evidence that SPN translocation is not simply the result of diffusion of effectors through the pore (116, 121). Rather, some have speculated that a protected channel is formed between the bacterium and the translocation apparatus, similar to the T3SS (see Fig. 4). However, much more work still needs to be done to determine whether this model is accurate.
The Type VII Secretion System
As mentioned above, certain Gram-positive organisms, including species of Mycobacteria and Corynebacteria, contain a heavily lipidated cell wall layer called a mycomembrane. These lipids form a very dense, waxy, hydrophobic layer on the outer surface of the bacteria, which serves as an effective barrier against a number of environmental stresses and anti-microbial therapies. However, as one might imagine, this layer makes extracellular protein transport more difficult for these species of bacteria. Therefore, these bacteria utilize specialized mechanisms for protein transport across their inner and mycomembranes, called Type VII secretion systems (T7SSs) (reviewed in (105, 122, 123)) (see Fig. 5). T7SSs were originally identified in M. tuberculosis in 2003 (124), where they are called ESX systems. These systems have now been identified in a number of bacteria in the order Corynebacteriales. In addition, analogous substrates and some components of these systems have also been identified in a number of Gram-positive organisms that lack mycomembranes, including the pathogens S. aureus, Bacillus anthracis, and L. monocytogenes ut
The core structural components of T7SSs, and frequently their substrates, are typically encoded in linked gene clusters (123). In general, five core structural proteins appear in most of these gene clusters. These core components, called EccB, EccC, EccD, EccE and MycP in the ESX systems, are all membrane proteins (128). All except EccD have hydrophobic domains and thus all may interact with various other accessory components in the cytoplasm or peptidoglycan layer, such as chaperones or the cytosolic protein, EccA, which may supply the source of energy for substrate transport (129). The four membrane associated Ecc proteins from ESX-5 form a large inner membrane complex, which presumably contains a channel through which the substrates traverse (128). The fifth component, MycP, is a mycosin, or subtilisin-like protease. The role of MycP in protein translocation through the T7SS is not completely understood, however, it is believed to play an important role in the regulation of secretion (130). T7SSs also have a variable number of additional proteins that are important for the function of specific systems. For instance, some systems may derive energy from the EccA cytosolic protein, an ATPase associated with diverse cellular activities (129) Other T7SSs may use energy generated by EccCb1, a protein that forms a SpoIIIE-type ATPase with the integral membrane component EccCa1 (131). Additionally, in Mycobacteria, chaperones are used to recognize specific substrates and target them to specific ESX systems (132).
While there has been tremendous amount of work done to understand the mechanism of protein translocation through the T7SS since its discovery 12 years ago, we do not yet understand how substrates navigate through the mycomembrane. The mycomembrane has been likened to the outer membrane of Gram-negative bacteria based on electron microscopy studies, so presumably there should be factors required for passage through this layer (133, 134). It remains to be seen whether some of the already recognized components of the T7SS, or other not yet identified factors, are critical for passage through this barrier.
T7SSs play a variety of roles in bacterial physiology and pathogenesis. The first system identified, ESX-1, is a major virulence factor in M. tuberculosis (135, 136). To date, five T7SS have been identified in Mycobacteria, named ESX-1 thru ESX-5, although not all Mycobacteria harbor all five systems (137). In fact, only ESX-3 and ESX-4 are found in all Mycobacterial species. Curiously, although ESX-4 appears to be the most ancient system, secretion from ESX-4 has not yet been demonstrated and it lacks several genes found in most other systems. Secretion from ESX-2 has also not been demonstrated.
While some T7SSs can contribute to virulence, as outlined above, not all of these systems function in virulence or even in secretion of substrates. For example, the T7SS in Listeria does not appear essential for virulence (126). Likewise the ESX-1, which is essential for the virulence of M. tuberculosis, is also present in the non-pathogenic M. smegmatis, where it functions in conjugation of DNA substrates (reminiscent of some T4SS) (138). In M. tuberculosis, ESX-3 secreted substrates are critical in iron acquisition and growth in vitro (139), whereas ESX-1 and ESX-5 are essential for virulence but not for growth in medium (124, 140). Thus, these systems apparently play diverse roles depending on the species of Mycobacteria. This diversity of function is likely to be also true in their roles in other bacteria.
HOST IMMUNE RECOGNITION OF BACTERIAL SECRETION SYSTEMS
A recent area of active investigation in the fields of bacterial pathogenesis and innate immunity has been the study of how the mammalian immune system discriminates between pathogenic and commensal bacteria. One way in which innate immune systems recognize pathogens is through the utilization of specific receptors and immune cell proteins that sense mechanisms, or patterns, of bacterial pathogenesis (141). This is in contrast to simply sensing molecules carried by both pathogens and commensals, such as lipopolysaccharide (LPS) or peptidoglycan. As discussed in this chapter, one hallmark of bacterial infection for many pathogens is the use of dedicated protein secretion systems to directly deposit effector proteins or toxins into mammalian tissue sites and/or host cells. For this reason, the mammalian innate immune system has developed strategies to detect the use of bacterial secretion systems and/or their secreted substrates during infection (141). Because bacterial secretion of virulence proteins can occur by many different mechanisms, the host immune system has developed multiple methods of sensing these processes (Fig. 6).
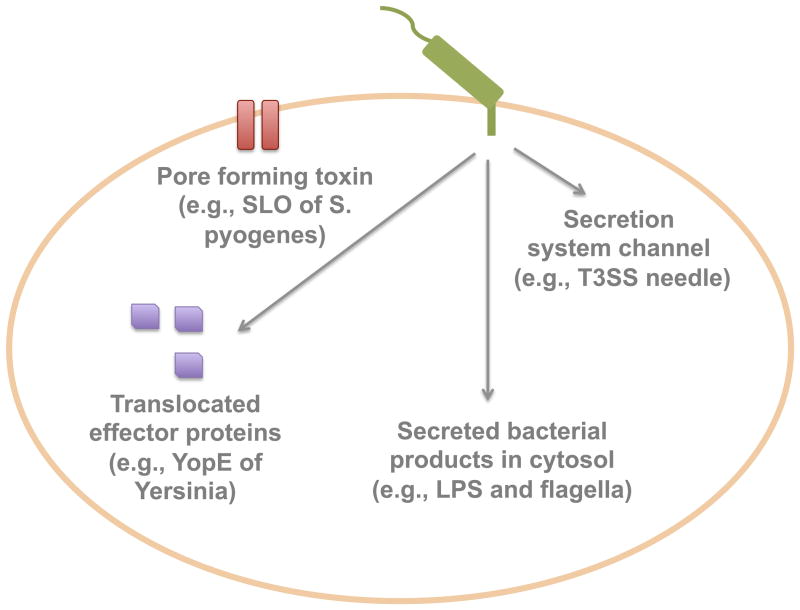
Fig. 6. Mechanisms of innate immune recognition of bacterial secretion systems.
In order to distinguish between pathogenic and commensal bacteria, the mammalian innate immune system has developed methods to directly recognize patterns unique to bacterial pathogens, such as the use of protein secretion apparatuses. The immune system can sense several facets of bacterial protein secretion. These include the pore formation by secretion systems or secreted proteins, aberrant translocation of bacterial molecules into the cytosol, presence of effector proteins and/or their activities, as well as the components of the secretion systems themselves.
One mechanism by which the innate immune response can detect bacterial secretion systems is by sensing the cytosolic access of bacterial products. These products are often small molecules, such as peptidoglycan, flagellin, and LPS, which can be aberrantly translocated into the bacterial cytoplasm through bacterial secretion systems. Because these receptors are limited to the bacterial cytosol, their activation would be indicative of the specific use of a secretion system by an invading pathogen. One such example is the family of cytoplasmic receptors called Nod-like receptors (NLRs), which can directly sense cytosolic molecules, including LPS and flagellin (141). Activation of these receptors leads to a cascade of signaling that ultimately induces the production of inflammatory cytokines. Additionally, the immune system has developed methods to directly sense the translocation of secreted effectors. For example, recent findings have shown that macrophages can sense manipulation of Rho GTPases by the Yersinia T3SS effector YopE (142). Following detection of this signal, macrophages mount (through an unknown mechanism) a response that ultimately results in clearance of intracellular bacteria.
The mammalian innate immune system can also detect the disruption of membranes by pore-forming proteins, such as the translocons of T3SSs or the CDCs of Gram-positive pathogens, which are often secreted or inserted by bacterial secretion systems that directly deliver effector proteins to target cells. For example, pore formation by SLO of Streptococcus pyogenes has been shown to activate the NLR family receptor NLRP3 (143). This activation eventually stimulates the production of inflammatory cytokines that help the host to respond to and clear the infection.
Finally, there is some evidence that the host immune system can detect components of secretion channels that protrude out of the bacterial membrane, such as the T3SS needle and components of the translocon. Recently, it has been shown that recognition of several T3SS needle proteins, including YscF of Yersinia and MxiH from Shigella, can induce proinflammatory cytokine production in host cells (144).
CONCLUSIONS
Bacterial pathogens utilize a multitude of methods to invade mammalian hosts, damage tissue sites, and thwart the immune system from responding. One essential component of these strategies for many bacterial pathogens is the secretion of proteins across phospholipid membranes. Secreted proteins can play many roles in promoting bacterial virulence, from enhancing attachment to eukaryotic cells, to scavenging resources in an environmental niche, to directly intoxicating target cells and disrupting their functions. As we discussed in this chapter, these proteins may be transferred out of the bacterial cytoplasm through a variety of mechanisms, usually involving the use of dedicated protein secretion systems. For this reason, the study of protein secretion systems has been an important focus in the field of bacterial pathogenesis. The remaining chapters in this section will offer a more detailed focus on the molecular and functional characteristics of some of these secretion systems.
References
- 1.Natale P, Bruser T, Driessen AJ. Sec- and Tat-mediated protein secretion across the bacterial cytoplasmic membrane--distinct translocases and mechanisms. Biochim Biophys Acta. 2008;1778(9):1735–56. doi: 10.1016/j.bbamem.2007.07.015. [DOI] [PubMed] [Google Scholar]
- 2.Papanikou E, Karamanou S, Economou A. Bacterial protein secretion through the translocase nanomachine. Nature Reviews Microbiology. 2007;5:839–851. doi: 10.1038/nrmicro1771. [DOI] [PubMed] [Google Scholar]
- 3.Korotkov KV, Sandkvist M, Hol WGJ. The type II secretion system: biogenesis, molecular architecture and mechanism. Nature Reviews Microbiology. 2012;10:336–351. doi: 10.1038/nrmicro2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lenz LL, Mohammadi S, Geissler A, Portnoy DA. SecA2-dependent secretion of autolytic enzymes promotes Listeria monocytogenes pathogenesis. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:12432–12437. doi: 10.1073/pnas.2133653100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Braunstein M, Espinosa BJ, Chan J, Belisle JT, Jacobs WR. SecA2 functions in the secretion of superoxide dismutase A and in the virulence of Mycobacterium tuberculosis. Molecular Microbiology. 2003;48:453–464. doi: 10.1046/j.1365-2958.2003.03438.x. [DOI] [PubMed] [Google Scholar]
- 6.Lenz LL, Portnoy DA. Identification of a second Listeria secA gene associated with protein secretion and the rough phenotype. Molecular Microbiology. 2002;45:1043–1056. doi: 10.1046/j.1365-2958.2002.03072.x. [DOI] [PubMed] [Google Scholar]
- 7.Bensing BA, Sullam PM. An accessory sec locus of Streptococcus gordonii is required for export of the surface protein GspB and for normal levels of binding to human platelets. Molecular Microbiology. 2002;44:1081–1094. doi: 10.1046/j.1365-2958.2002.02949.x. [DOI] [PubMed] [Google Scholar]
- 8.Randall LL, Hardy SJ. SecB, one small chaperone in the complex milieu of the cell. Cell Mol Life Sci. 2002;59(10):1617–23. doi: 10.1007/PL00012488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hartl FU, Lecker S, Schiebel E, Hendrick JP, Wickner W. The binding cascade of SecB to SecA to SecY/E mediates preprotein targeting to the E. coli plasma membrane. Cell. 1990;63(2):269–79. doi: 10.1016/0092-8674(90)90160-g. [DOI] [PubMed] [Google Scholar]
- 10.Mogensen JE, Otzen DE. Interactions between folding factors and bacterial outer membrane proteins. Molecular Microbiology. 2005;57(2):326–346. doi: 10.1111/j.1365-2958.2005.04674.x. [DOI] [PubMed] [Google Scholar]
- 11.Luirink J, Sinning I. SRP-mediated protein targeting: structure and function revisited. Biochimica Et Biophysica Acta-Molecular Cell Research. 2004;1694(1–3):17–35. doi: 10.1016/j.bbamcr.2004.03.013. [DOI] [PubMed] [Google Scholar]
- 12.Sijbrandi R, Urbanus ML, ten Hagen-Jongman CM, Bernstein HD, Oudega B, Otto BR, Luirink J. Signal recognition particle (SRP)-mediated targeting and sec-dependent translocation of an extracellular Escherichia coli protein. Journal of Biological Chemistry. 2003;278(7):4654–4659. doi: 10.1074/jbc.M211630200. [DOI] [PubMed] [Google Scholar]
- 13.Paetzel M, Karla A, Strynadka NCJ, Dalbey RE. Signal peptidases. Chemical Reviews. 2002;102(12):4549–4579. doi: 10.1021/cr010166y. [DOI] [PubMed] [Google Scholar]
- 14.Robinson C, Bolhuis A. Tat-dependent protein targeting in prokaryotes and chloroplasts. Biochimica Et Biophysica Acta-Molecular Cell Research. 2004;1694(1–3):135–147. doi: 10.1016/j.bbamcr.2004.03.010. [DOI] [PubMed] [Google Scholar]
- 15.Berks BC, Palmer T, Sargent F. Protein targeting by the bacterial twin-arginine translocation (Tat) pathway. Current Opinion in Microbiology. 2005;8(2):174–181. doi: 10.1016/j.mib.2005.02.010. [DOI] [PubMed] [Google Scholar]
- 16.Sargent F, Stanley NR, Berks BC, Palmer T. Sec-independent protein translocation in Escherichia coli - A distinct and pivotal role for the TatB protein. Journal of Biological Chemistry. 1999;274(51):36073–36082. doi: 10.1074/jbc.274.51.36073. [DOI] [PubMed] [Google Scholar]
- 17.Pop O, Martin U, Abel C, Muller JP. The twin-arginine signal peptide of PhoD and the TatA(d)/C-d proteins of Bacillus subtilis form an autonomous tat translocation system. Journal of Biological Chemistry. 2002;277(5):3268–3273. doi: 10.1074/jbc.M110829200. [DOI] [PubMed] [Google Scholar]
- 18.Müller M. Twin-arginine-specific protein export in Escherichia coli. Research in Microbiology. 2005;156:131–136. doi: 10.1016/j.resmic.2004.09.016. [DOI] [PubMed] [Google Scholar]
- 19.Ochsner UA, Snyder A, Vasil AI, Vasil ML. Effects of the twin-arginine translocase on secretion of virulence factors, stress response, and pathogenesis. Proceedings of the National Academy of Sciences of the United States of America. 2002;99(12):8312–8317. doi: 10.1073/pnas.082238299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lavander M, Ericsson SK, Broms JE, Forsberg A. The twin arginine translocation system is essential for virulence of Yersinia pseudotuberculosis. Infection and Immunity. 2006;74(3):1768–1776. doi: 10.1128/IAI.74.3.1768-1776.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pradel N, Ye CY, Livrelli V, Xu HG, Joly B, Wu LF. Contribution of the twin arginine translocation system to the virulence of enterohemorrhagic Escherichia coli O157 : H7. Infection and Immunity. 2003;71(9):4908–4916. doi: 10.1128/IAI.71.9.4908-4916.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rossier O, Cianciotto NP. The Legionella pneumophila tatB gene facilitates secretion of phospholipase C, growth under iron-limiting conditions, and intracellular infection. Infection and Immunity. 2005;73(4):2020–2032. doi: 10.1128/IAI.73.4.2020-2032.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McDonough JA, McCann JR, Tekippe EM, Silverman JS, Rigel NW, Braunstein M. Identification of functional Tat signal sequences in Mycobacterium tuberculosis proteins. Journal of Bacteriology. 2008;190(19):6428–6438. doi: 10.1128/JB.00749-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Songer JG. Bacterial phospholipases and their role in virulence. Trends in Microbiology. 1997;5(4):156–161. doi: 10.1016/S0966-842X(97)01005-6. [DOI] [PubMed] [Google Scholar]
- 25.Thomas S, Holland IB, Schmitt L. The Type 1 secretion pathway - the hemolysin system and beyond. Biochim Biophys Acta. 2014;1843(8):1629–41. doi: 10.1016/j.bbamcr.2013.09.017. [DOI] [PubMed] [Google Scholar]
- 26.Symmons MF, Bokma E, Koronakis E, Hughes C, Koronakis V. The assembled structure of a complete tripartite bacterial multidrug efflux pump. Proc Natl Acad Sci U S A. 2009;106(17):7173–8. doi: 10.1073/pnas.0900693106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Delepelaire P. Type I secretion in gram-negative bacteria. Biochim Biophys Acta. 2004;1694(1–3):149–61. doi: 10.1016/j.bbamcr.2004.05.001. [DOI] [PubMed] [Google Scholar]
- 28.Kanonenberg K, Schwarz CK, Schmitt L. Type I secretion systems - a story of appendices. Res Microbiol. 2013;164(6):596–604. doi: 10.1016/j.resmic.2013.03.011. [DOI] [PubMed] [Google Scholar]
- 29.Letoffe S, Delepelaire P, Wandersman C. Protein secretion in gram-negative bacteria: assembly of the three components of ABC protein-mediated exporters is ordered and promoted by substrate binding. EMBO J. 1996;15(21):5804–11. [PMC free article] [PubMed] [Google Scholar]
- 30.Pimenta AL, Young J, Holland IB, Blight MA. Antibody analysis of the localisation, expression and stability of HlyD, the MFP component of the E. coli haemolysin translocator. Mol Gen Genet. 1999;261(1):122–32. doi: 10.1007/s004380050949. [DOI] [PubMed] [Google Scholar]
- 31.Lee M, Jun SY, Yoon BY, Song S, Lee K, Ha NC. Membrane fusion proteins of type I secretion system and tripartite efflux pumps share a binding motif for TolC in gram-negative bacteria. PLoS One. 2012;7(7):e40460. doi: 10.1371/journal.pone.0040460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Balakrishnan L, Hughes C, Koronakis V. Substrate-triggered recruitment of the TolC channel-tunnel during type I export of hemolysin by Escherichia coli. J Mol Biol. 2001;313(3):501–10. doi: 10.1006/jmbi.2001.5038. [DOI] [PubMed] [Google Scholar]
- 33.Wu KH, Tai PC. Cys32 and His105 are the critical residues for the calcium-dependent cysteine proteolytic activity of CvaB, an ATP-binding cassette transporter. J Biol Chem. 2004;279(2):901–9. doi: 10.1074/jbc.M308296200. [DOI] [PubMed] [Google Scholar]
- 34.Lecher J, Schwarz CK, Stoldt M, Smits SH, Willbold D, Schmitt L. An RTX transporter tethers its unfolded substrate during secretion via a unique N-terminal domain. Structure. 2012;20(10):1778–87. doi: 10.1016/j.str.2012.08.005. [DOI] [PubMed] [Google Scholar]
- 35.Linhartova I, Bumba L, Masin J, Basler M, Osicka R, Kamanova J, Prochazkova K, Adkins I, Hejnova-Holubova J, Sadilkova L, Morova J, Sebo P. RTX proteins: a highly diverse family secreted by a common mechanism. FEMS Microbiol Rev. 2010;34(6):1076–112. doi: 10.1111/j.1574-6976.2010.00231.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dolores JS, Agarwal S, Egerer M, Satchell KJ. Vibrio cholerae MARTX toxin heterologous translocation of beta-lactamase and roles of individual effector domains on cytoskeleton dynamics. Mol Microbiol. 2015;95(4):590–604. doi: 10.1111/mmi.12879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Welch RA, Dellinger EP, Minshew B, Falkow S. Haemolysin contributes to virulence of extra-intestinal E. coli infections. Nature. 1981;294(5842):665–7. doi: 10.1038/294665a0. [DOI] [PubMed] [Google Scholar]
- 38.Hughes C, Muller D, Hacker J, Goebel W. Genetics and pathogenic role of Escherichia coli haemolysin. Toxicon. 1982;20(1):247–52. doi: 10.1016/0041-0101(82)90210-0. [DOI] [PubMed] [Google Scholar]
- 39.Mackman N, Holland IB. Functional characterization of a cloned haemolysin determinant from E. coli of human origin, encoding information for the secretion of a 107K polypeptide. Mol Gen Genet. 1984;196(1):129–34. doi: 10.1007/BF00334104. [DOI] [PubMed] [Google Scholar]
- 40.Voulhoux R, Ball G, Ize B, Vasil ML, Lazdunski A, Wu LF, Filloux A. Involvement of the twin-arginine translocation system in protein secretion via the type II pathway. Embo Journal. 2001;20(23):6735–6741. doi: 10.1093/emboj/20.23.6735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cianciotto NP. Type II secretion: a protein secretion system for all seasons. Trends in Microbiology. 2005;13:581–588. doi: 10.1016/j.tim.2005.09.005. [DOI] [PubMed] [Google Scholar]
- 42.Korotkov KV, Gonen T, Hol WGJ. Secretins: dynamic channels for protein transport across membranes. Trends in Biochemical Sciences. 2011;36(8):433–443. doi: 10.1016/j.tibs.2011.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sauvonnet N, Vignon G, Pugsley AP, Gounon P. Pilus formation and protein secretion by the same machinery in Escherichia coli. Embo Journal. 2000;19(10):2221–2228. doi: 10.1093/emboj/19.10.2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hobbs M, Mattick JS. Common Components in the Assembly of Type-4 Fimbriae, DNA Transfer Systems, Filamentous Phage and Protein-Secretion Apparatus - a General System for the Formation of Surface-Associated Protein Complexes. Molecular Microbiology. 1993;10(2):233–243. doi: 10.1111/j.1365-2958.1993.tb01949.x. [DOI] [PubMed] [Google Scholar]
- 45.Shevchik VE, RobertBaudouy J, Condemine G. Specific interaction between OutD, an Erwinia chrysanthemi outer membrane protein of the general secretory pathway, and secreted proteins. Embo Journal. 1997;16(11):3007–3016. doi: 10.1093/emboj/16.11.3007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sandkvist M, Michel LO, Hough LP, Morales VM, Bagdasarian M, Koomey M, DiRita VJ, Bagdasarian M. General secretion pathway (eps) genes required for toxin secretion and outer membrane biogenesis in Vibrio cholerae. Journal of Bacteriology. 1997;179(22):6994–7003. doi: 10.1128/jb.179.22.6994-7003.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lu HM, Lory S. A specific targeting domain in mature exotoxin A is required for its extracellular secretion from Pseudomonas aeruginosa. Embo Journal. 1996;15(2):429–436. [PMC free article] [PubMed] [Google Scholar]
- 48.Cianciotto NP. Type II secretion and Legionella virulence. Current Topics in Microbiology and Immunology. 2013;376:81–102. doi: 10.1007/82_2013_339. [DOI] [PubMed] [Google Scholar]
- 49.Kulkarni R, Dhakal BK, Slechta ES, Kurtz Z, Mulvey MA, Thanassi DG. Roles of Putative Type II Secretion and Type IV Pilus Systems in the Virulence of Uropathogenic Escherichia coli. Plos One. 2009;4(3) doi: 10.1371/journal.pone.0004752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tauschek M, Gorrell RJ, Strugnell RA, Robins-Browne RM. Identification of a protein secretory pathway for the secretion of heat-labile enterotoxin by an enterotoxigenic strain of Escherichia coli. Proceedings of the National Academy of Sciences of the United States of America. 2002;99(10):7066–7071. doi: 10.1073/pnas.092152899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lathem WW, Grys TE, Witowski SE, Torres AG, Kaper JB, Tarr PI, Welch RA. StcE, a metalloprotease secreted by Escherichia coli O157 : H7, specifically cleaves C1 esterase inhibitor. Molecular Microbiology. 2002;45(2):277–288. doi: 10.1046/j.1365-2958.2002.02997.x. [DOI] [PubMed] [Google Scholar]
- 52.Pugsley AP, Chapon C, Schwartz M. Extracellular Pullulanase of Klebsiella-Pneumoniae Is a Lipoprotein. Journal of Bacteriology. 1986;166(3):1083–1088. doi: 10.1128/jb.166.3.1083-1088.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jiang B, Howard SP. The Aeromonas-Hydrophila Exee Gene, Required Both for Protein Secretion and Normal Outer-Membrane Biogenesis, Is a Member of a General Secretion Pathway. Molecular Microbiology. 1992;6(10):1351–1361. doi: 10.1111/j.1365-2958.1992.tb00856.x. [DOI] [PubMed] [Google Scholar]
- 54.He SY, Lindeberg M, Chatterjee AK, Collmer A. Cloned Erwinia-Chrysanthemi out Genes Enable Escherichia-Coli to Selectively Secrete a Diverse Family of Heterologous Proteins to Its Milieu. Proceedings of the National Academy of Sciences of the United States of America. 1991;88(3):1079–1083. doi: 10.1073/pnas.88.3.1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Buttner D. Protein export according to schedule: architecture, assembly, and regulation of type III secretion systems from plant- and animal-pathogenic bacteria. Microbiol Mol Biol Rev. 2012;76(2):262–310. doi: 10.1128/MMBR.05017-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Abrusci P, McDowell MA, Lea SM, Johnson S. Building a secreting nanomachine: a structural overview of the T3SS. Curr Opin Struct Biol. 2014;25:111–7. doi: 10.1016/j.sbi.2013.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Burkinshaw BJ, Strynadka NC. Assembly and structure of the T3SS. Biochim Biophys Acta. 2014;1843(8):1649–63. doi: 10.1016/j.bbamcr.2014.01.035. [DOI] [PubMed] [Google Scholar]
- 58.Troisfontaines P, Cornelis GR. Type III secretion: more systems than you think. Physiology (Bethesda) 2005;20:326–39. doi: 10.1152/physiol.00011.2005. [DOI] [PubMed] [Google Scholar]
- 59.Kubori T, Matsushima Y, Nakamura D, Uralil J, Lara-Tejero M, Sukhan A, Galan JE, Aizawa SI. Supramolecular structure of the Salmonella typhimurium type III protein secretion system. Science. 1998;280(5363):602–5. doi: 10.1126/science.280.5363.602. [DOI] [PubMed] [Google Scholar]
- 60.Deane JE, Cordes FS, Roversi P, Johnson S, Kenjale R, Picking WD, Picking WL, Lea SM, Blocker A. Expression, purification, crystallization and preliminary crystallographic analysis of MxiH, a subunit of the Shigella flexneri type III secretion system needle. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2006;62(Pt 3):302–5. doi: 10.1107/S1744309106006555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Demers JP, Habenstein B, Loquet A, Kumar Vasa S, Giller K, Becker S, Baker D, Lange A, Sgourakis NG. High-resolution structure of the Shigella type-III secretion needle by solid-state NMR and cryo-electron microscopy. Nat Commun. 2014;5:4976. doi: 10.1038/ncomms5976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dohlich K, Zumsteg AB, Goosmann C, Kolbe M. A substrate-fusion protein is trapped inside the Type III Secretion System channel in Shigella flexneri. PLoS Pathog. 2014;10(1):e1003881. doi: 10.1371/journal.ppat.1003881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Radics J, Konigsmaier L, Marlovits TC. Structure of a pathogenic type 3 secretion system in action. Nat Struct Mol Biol. 2014;21(1):82–7. doi: 10.1038/nsmb.2722. [DOI] [PubMed] [Google Scholar]
- 64.Price SB, Cowan C, Perry RD, Straley SC. The Yersinia pestis V antigen is a regulatory protein necessary for Ca2(+)-dependent growth and maximal expression of low-Ca2+ response virulence genes. J Bacteriol. 1991;173(8):2649–57. doi: 10.1128/jb.173.8.2649-2657.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Picking WL, Nishioka H, Hearn PD, Baxter MA, Harrington AT, Blocker A, Picking WD. IpaD of Shigella flexneri is independently required for regulation of Ipa protein secretion and efficient insertion of IpaB and IpaC into host membranes. Infect Immun. 2005;73(3):1432–40. doi: 10.1128/IAI.73.3.1432-1440.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Holmstrom A, Olsson J, Cherepanov P, Maier E, Nordfelth R, Pettersson J, Benz R, Wolf-Watz H, Forsberg A. LcrV is a channel size-determining component of the Yop effector translocon of Yersinia. Mol Microbiol. 2001;39(3):620–32. doi: 10.1046/j.1365-2958.2001.02259.x. [DOI] [PubMed] [Google Scholar]
- 67.Hakansson S, Bergman T, Vanooteghem JC, Cornelis G, Wolf-Watz H. YopB and YopD constitute a novel class of Yersinia Yop proteins. Infect Immun. 1993;61(1):71–80. doi: 10.1128/iai.61.1.71-80.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hakansson S, Schesser K, Persson C, Galyov EE, Rosqvist R, Homble F, Wolf-Watz H. The YopB protein of Yersinia pseudotuberculosis is essential for the translocation of Yop effector proteins across the target cell plasma membrane and displays a contact-dependent membrane disrupting activity. EMBO J. 1996;15(21):5812–23. [PMC free article] [PubMed] [Google Scholar]
- 69.Akopyan K, Edgren T, Wang-Edgren H, Rosqvist R, Fahlgren A, Wolf-Watz H, Fallman M. Translocation of surface-localized effectors in type III secretion. Proc Natl Acad Sci U S A. 2011;108(4):1639–44. doi: 10.1073/pnas.1013888108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Edgren T, Forsberg A, Rosqvist R, Wolf-Watz H. Type III secretion in Yersinia: injectisome or not? PLoS Pathog. 2012;8(5):e1002669. doi: 10.1371/journal.ppat.1002669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Angot A, Vergunst A, Genin S, Peeters N. Exploitation of eukaryotic ubiquitin signaling pathways by effectors translocated by bacterial type III and type IV secretion systems. PLoS Pathog. 2007;3(1):e3. doi: 10.1371/journal.ppat.0030003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ham H, Sreelatha A, Orth K. Manipulation of host membranes by bacterial effectors. Nature Reviews. Microbiology. 2011;9:635–646. doi: 10.1038/nrmicro2602. [DOI] [PubMed] [Google Scholar]
- 73.Spano S, Galan JE. A novel anti-microbial function for a familiar Rab GTPase. Small GTPases. 2013;4(4):252–4. doi: 10.4161/sgtp.27282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tosi T, Pflug A, Discola KF, Neves D, Dessen A. Structural basis of eukaryotic cell targeting by type III secretion system (T3SS) effectors. Res Microbiol. 2013;164(6):605–19. doi: 10.1016/j.resmic.2013.03.019. [DOI] [PubMed] [Google Scholar]
- 75.Cascales E, Christie PJ. The versatile bacterial type IV secretion systems. Nature Reviews. Microbiology. 2003;1:137–149. doi: 10.1038/nrmicro753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lessl M, Lanka E. Common mechanisms in bacterial conjugation and Ti-mediated T-DNA transfer to plant cells. Cell. 1994;77(3):321–4. doi: 10.1016/0092-8674(94)90146-5. [DOI] [PubMed] [Google Scholar]
- 77.Bundock P, den Dulk-Ras A, Beijersbergen A, Hooykaas PJ. Trans-kingdom T-DNA transfer from Agrobacterium tumefaciens to Saccharomyces cerevisiae. EMBO J. 1995;14(13):3206–14. doi: 10.1002/j.1460-2075.1995.tb07323.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Fronzes R, Christie PJ, Waksman G. The structural biology of type IV secretion systems. Nature Reviews. Microbiology. 2009;7:703–714. doi: 10.1038/nrmicro2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Atmakuri K, Cascales E, Christie PJ. Energetic components VirD4, VirB11 and VirB4 mediate early DNA transfer reactions required for bacterial type IV secretion. Mol Microbiol. 2004;54(5):1199–211. doi: 10.1111/j.1365-2958.2004.04345.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Babic A, Lindner AB, Vulic M, Stewart EJ, Radman M. Direct visualization of horizontal gene transfer. Science. 2008;319(5869):1533–6. doi: 10.1126/science.1153498. [DOI] [PubMed] [Google Scholar]
- 81.Hamilton HL, Dillard JP. Natural transformation of Neisseria gonorrhoeae: from DNA donation to homologous recombination. Mol Microbiol. 2006;59(2):376–85. doi: 10.1111/j.1365-2958.2005.04964.x. [DOI] [PubMed] [Google Scholar]
- 82.Backert S, Meyer TF. Type IV secretion systems and their effectors in bacterial pathogenesis. Curr Opin Microbiol. 2006;9(2):207–17. doi: 10.1016/j.mib.2006.02.008. [DOI] [PubMed] [Google Scholar]
- 83.Isberg RR, O’Connor TJ, Heidtman M. The Legionella pneumophila replication vacuole: making a cosy niche inside host cells. Nat Rev Microbiol. 2009;7(1):13–24. doi: 10.1038/nrmicro1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Pohlner J, Halter R, Beyreuther K, Meyer TF. Gene structure and extracellular secretion of Neisseria gonorrhoeae IgA protease. Nature. 1987;325(6103):458–62. doi: 10.1038/325458a0. [DOI] [PubMed] [Google Scholar]
- 85.Leyton DL, Rossiter AE, Henderson IR. From self sufficiency to dependence: mechanisms and factors important for autotransporter biogenesis. Nat Rev Microbiol. 2012;10(3):213–25. doi: 10.1038/nrmicro2733. [DOI] [PubMed] [Google Scholar]
- 86.van Ulsen P, Rahman Su, Jong WSP, Daleke-Schermerhorn MH, Luirink J. Type V secretion: From biogenesis to biotechnology. Biochimica et Biophysica Acta (BBA) - Molecular Cell Research. 2014;1843:1592–1611. doi: 10.1016/j.bbamcr.2013.11.006. [DOI] [PubMed] [Google Scholar]
- 87.Pohlner J, Halter R, Meyer TF. Neisseria gonorrhoeae IgA protease. Secretion and implications for pathogenesis. Antonie Van Leeuwenhoek. 1987;53(6):479–84. doi: 10.1007/BF00415506. [DOI] [PubMed] [Google Scholar]
- 88.Brandon LD, Goehring N, Janakiraman A, Yan AW, Wu T, Beckwith J, Goldberg MB. IcsA, a polarly localized autotransporter with an atypical signal peptide, uses the Sec apparatus for secretion, although the Sec apparatus is circumferentially distributed. Molecular Microbiology. 2003;50(1):45–60. doi: 10.1046/j.1365-2958.2003.03674.x. [DOI] [PubMed] [Google Scholar]
- 89.Zumsteg AB, Goosmann C, Brinkmann V, Morona R, Zychlinsky A. IcsA Is a Shigella flexneri Adhesion Regulated by the Type III Secretion System and Required for Pathogenesis. Cell Host & Microbe. 2014;15(4):435–445. doi: 10.1016/j.chom.2014.03.001. [DOI] [PubMed] [Google Scholar]
- 90.Roggenkamp A, Ackermann N, Jacobi CA, Truelzsch K, Hoffmann H, Heesemann H. Molecular analysis of transport and oligomerization of the Yersinia enterocolitica adhesin YadA. Journal of Bacteriology. 2003;185(13):3735–3744. doi: 10.1128/JB.185.13.3735-3744.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Mikula KM, Kolodziejczyk R, Goldman A. Yersinia infection tools-characterization of structure and function of adhesins. Frontiers in Cellular and Infection Microbiology. 2013;3 doi: 10.3389/fcimb.2012.00169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Leyton DL, Rossiter AE, Henderson IR. From self sufficiency to dependence: mechanisms and factors important for autotransporter biogenesis. Nature Reviews Microbiology. 2012;10(3):213–225. doi: 10.1038/nrmicro2733. [DOI] [PubMed] [Google Scholar]
- 93.Wagner JK, Heindl JE, Gray AN, Jain S, Goldberg MB. Contribution of the periplasmic chaperone Skp to efficient presentation of the autotransporter IcsA on the surface of Shigella flexneri. J Bacteriol. 2009;191(3):815–21. doi: 10.1128/JB.00989-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ruiz-Perez F, Henderson IR, Leyton DL, Rossiter AE, Zhang Y, Nataro JP. Roles of periplasmic chaperone proteins in the biogenesis of serine protease autotransporters of Enterobacteriaceae. J Bacteriol. 2009;191(21):6571–83. doi: 10.1128/JB.00754-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Henderson IR, Navarro-Garcia F, Desvaux M, Fernandez RC, Ala’Aldeen D. Type V protein secretion pathway: the autotransporter story. Microbiology and molecular biology reviews: MMBR. 2004;68:692–744. doi: 10.1128/MMBR.68.4.692-744.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Lambert-Buisine C, Willery E, Locht C, Jacob-Dubuisson F. N-terminal characterization of the Bordetella pertussis filamentous haemagglutinin. Mol Microbiol. 1998;28(6):1283–93. doi: 10.1046/j.1365-2958.1998.00892.x. [DOI] [PubMed] [Google Scholar]
- 97.McCann JR, St Geme JW., 3rd The HMW1C-like glycosyltransferases--an enzyme family with a sweet tooth for simple sugars. PLoS Pathog. 2014;10(4):e1003977. doi: 10.1371/journal.ppat.1003977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Waksman G, Hultgren SJ. Structural biology of the chaperone-usher pathway of pilus biogenesis. Nat Rev Microbiol. 2009;7(11):765–74. doi: 10.1038/nrmicro2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Mougous JD, Cuff ME, Raunser S, Shen A, Zhou M, Gifford CA, Goodman AL, Joachimiak G, Ordoñez CL, Lory S, Walz T, Joachimiak A, Mekalanos JJ. A virulence locus of Pseudomonas aeruginosa encodes a protein secretion apparatus. Science (New York, NY) 2006;312:1526–1530. doi: 10.1126/science.1128393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Russell AB, Peterson SB, Mougous JD. Type VI secretion system effectors: poisons with a purpose. Nature Reviews. Microbiology. 2014;12:137–148. doi: 10.1038/nrmicro3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Russell AB, Hood RD, Bui NK, LeRoux M, Vollmer W, Mougous JD. Type VI secretion delivers bacteriolytic effectors to target cells. Nature. 2011;475(7356):343–7. doi: 10.1038/nature10244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Pukatzki S, Ma AT, Revel AT, Sturtevant D, Mekalanos JJ. Type VI secretion system translocates a phage tail spike-like protein into target cells where it cross-links actin. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:15508–15513. doi: 10.1073/pnas.0706532104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.English G, Trunk K, Rao VA, Srikannathasan V, Hunter WN, Coulthurst SJ. New secreted toxins and immunity proteins encoded within the Type VI secretion system gene cluster of Serratia marcescens. Mol Microbiol. 2012;86(4):921–36. doi: 10.1111/mmi.12028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Feltcher ME, Braunstein M. Emerging themes in SecA2-mediated protein export. Nat Rev Microbiol. 2012;10(11):779–89. doi: 10.1038/nrmicro2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Freudl R. Leaving home ain’t easy: protein export systems in Gram-positive bacteria. Res Microbiol. 2013;164(6):664–74. doi: 10.1016/j.resmic.2013.03.014. [DOI] [PubMed] [Google Scholar]
- 106.Rigel NW, Braunstein M. A new twist on an old pathway--accessory Sec [corrected] systems. Mol Microbiol. 2008;69(2):291–302. doi: 10.1111/j.1365-2958.2008.06294.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Bensing BA, Seepersaud R, Yen YT, Sullam PM. Selective transport by SecA2: an expanding family of customized motor proteins. Biochim Biophys Acta. 2014;1843(8):1674–86. doi: 10.1016/j.bbamcr.2013.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Hou JM, D’Lima NG, Rigel NW, Gibbons HS, McCann JR, Braunstein M, Teschke CM. ATPase activity of Mycobacterium tuberculosis SecA1 and SecA2 proteins and its importance for SecA2 function in macrophages. J Bacteriol. 2008;190(14):4880–7. doi: 10.1128/JB.00412-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Fagan RP, Fairweather NF. Clostridium difficile has two parallel and essential Sec secretion systems. J Biol Chem. 2011;286(31):27483–93. doi: 10.1074/jbc.M111.263889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Siboo IR, Chaffin DO, Rubens CE, Sullam PM. Characterization of the accessory Sec system of Staphylococcus aureus. J Bacteriol. 2008;190(18):6188–96. doi: 10.1128/JB.00300-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Mistou MY, Dramsi S, Brega S, Poyart C, Trieu-Cuot P. Molecular dissection of the secA2 locus of group B Streptococcus reveals that glycosylation of the Srr1 LPXTG protein is required for full virulence. J Bacteriol. 2009;191(13):4195–206. doi: 10.1128/JB.01673-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Seepersaud R, Bensing BA, Yen YT, Sullam PM. Asp3 mediates multiple protein-protein interactions within the accessory Sec system of Streptococcus gordonii. Mol Microbiol. 2010;78(2):490–505. doi: 10.1111/j.1365-2958.2010.07346.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Telford JL, Barocchi MA, Margarit I, Rappuoli R, Grandi G. Pili in gram-positive pathogens. Nat Rev Microbiol. 2006;4(7):509–19. doi: 10.1038/nrmicro1443. [DOI] [PubMed] [Google Scholar]
- 114.Hendrickx AP, Budzik JM, Oh SY, Schneewind O. Architects at the bacterial surface - sortases and the assembly of pili with isopeptide bonds. Nat Rev Microbiol. 2011;9(3):166–76. doi: 10.1038/nrmicro2520. [DOI] [PubMed] [Google Scholar]
- 115.Mazmanian SK, Liu G, Hung TT, Schneewind O. Staphylococcus aureus sortase, an enzyme that anchors surface proteins to the cell wall. Science. 1999;285(5428):760–763. doi: 10.1126/science.285.5428.760. [DOI] [PubMed] [Google Scholar]
- 116.Madden JC, Ruiz N, Caparon M. Cytolysin-mediated translocation (CMT): A functional equivalent of type III secretion in Gram-positive bacteria. Cell. 2001;104(1):143–152. doi: 10.1016/s0092-8674(01)00198-2. [DOI] [PubMed] [Google Scholar]
- 117.Ghosh J, Caparon MG. Specificity of Streptococcus pyogenes NAD(+) glycohydrolase in cytolysin-mediated translocation. Molecular Microbiology. 2006;62(4):1203–1214. doi: 10.1111/j.1365-2958.2006.05430.x. [DOI] [PubMed] [Google Scholar]
- 118.Tweten RK. Cholesterol-dependent cytolysins, a family of versatile pore-forming toxins. Infection and Immunity. 2005;73:6199–6209. doi: 10.1128/IAI.73.10.6199-6209.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Madden JC, Ruiz N, Caparon M. Cytolysin-mediated translocation (CMT): a functional equivalent of type III secretion in gram-positive bacteria. Cell. 2001;104:143–152. doi: 10.1016/s0092-8674(01)00198-2. [DOI] [PubMed] [Google Scholar]
- 120.Ghosh J, Anderson PJ, Chandrasekaran S, Caparon MG. Characterization of Streptococcus pyogenes beta-NAD(+) Glycohydrolase REEVALUATION OF ENZYMATIC PROPERTIES ASSOCIATED WITH PATHOGENESIS. Journal of Biological Chemistry. 2010;285(8):5683–5694. doi: 10.1074/jbc.M109.070300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Magassa NG, Chandrasekaran S, Caparon MG. Streptococcus pyogenes cytolysin-mediated translocation does not require pore formation by streptolysin O. EMBO reports. 2010;11:400–405. doi: 10.1038/embor.2010.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Simeone R, Bottai D, Brosch R. ESX/type VII secretion systems and their role in host-pathogen interaction. Curr Opin Microbiol. 2009;12(1):4–10. doi: 10.1016/j.mib.2008.11.003. [DOI] [PubMed] [Google Scholar]
- 123.Houben EN, Korotkov KV, Bitter W. Take five - Type VII secretion systems of Mycobacteria. Biochim Biophys Acta. 2014;1843(8):1707–16. doi: 10.1016/j.bbamcr.2013.11.003. [DOI] [PubMed] [Google Scholar]
- 124.Stanley SA, Raghavan S, Hwang WW, Cox JS. Acute infection and macrophage subversion by Mycobacterium tuberculosis require a specialized secretion system. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:13001–13006. doi: 10.1073/pnas.2235593100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Burts ML, Williams WA, DeBord K, Missiakas DM. EsxA and EsxB are secreted by an ESAT-6-like system that is required for the pathogenesis of Staphylococcus aureus infections. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:1169–1174. doi: 10.1073/pnas.0405620102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Way SS, Wilson CB. The Mycobacterium tuberculosis ESAT-6 homologue in Listeria monocytogenes is dispensable for growth in vitro and in vivo. Infect Immun. 2005;73(9):6151–3. doi: 10.1128/IAI.73.9.6151-6153.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Baptista C, Barreto HC, Sao-Jose C. High levels of DegU-P activate an Esat-6-like secretion system in Bacillus subtilis. PLoS One. 2013;8(7):e67840. doi: 10.1371/journal.pone.0067840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Houben EN, Bestebroer J, Ummels R, Wilson L, Piersma SR, Jimenez CR, Ottenhoff TH, Luirink J, Bitter W. Composition of the type VII secretion system membrane complex. Mol Microbiol. 2012;86(2):472–84. doi: 10.1111/j.1365-2958.2012.08206.x. [DOI] [PubMed] [Google Scholar]
- 129.Luthra A, Mahmood A, Arora A, Ramachandran R. Characterization of Rv3868, an essential hypothetical protein of the ESX-1 secretion system in Mycobacterium tuberculosis. J Biol Chem. 2008;283(52):36532–41. doi: 10.1074/jbc.M807144200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Ohol YM, Goetz DH, Chan K, Shiloh MU, Craik CS, Cox JS. Mycobacterium tuberculosis MycP1 protease plays a dual role in regulation of ESX-1 secretion and virulence. Cell Host Microbe. 2010;7(3):210–20. doi: 10.1016/j.chom.2010.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Champion PAD, Stanley SA, Champion MM, Brown EJ, Cox JS. C-terminal signal sequence promotes virulence factor secretion in Mycobacterium tuberculosis. Science (New York, NY) 2006;313:1632–1636. doi: 10.1126/science.1131167. [DOI] [PubMed] [Google Scholar]
- 132.Daleke MH, van der Woude AD, Parret AH, Ummels R, de Groot AM, Watson D, Piersma SR, Jimenez CR, Luirink J, Bitter W, Houben EN. Specific chaperones for the type VII protein secretion pathway. J Biol Chem. 2012;287(38):31939–47. doi: 10.1074/jbc.M112.397596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Hoffmann C, Leis A, Niederweis M, Plitzko JM, Engelhardt H. Disclosure of the mycobacterial outer membrane: cryo-electron tomography and vitreous sections reveal the lipid bilayer structure. Proc Natl Acad Sci U S A. 2008;105(10):3963–7. doi: 10.1073/pnas.0709530105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Zuber B, Chami M, Houssin C, Dubochet J, Griffiths G, Daffe M. Direct visualization of the outer membrane of mycobacteria and corynebacteria in their native state. J Bacteriol. 2008;190(16):5672–80. doi: 10.1128/JB.01919-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Pym AS, Brodin P, Brosch R, Huerre M, Cole ST. Loss of RD1 contributed to the attenuation of the live tuberculosis vaccines Mycobacterium bovis BCG and Mycobacterium microti. Mol Microbiol. 2002;46(3):709–17. doi: 10.1046/j.1365-2958.2002.03237.x. [DOI] [PubMed] [Google Scholar]
- 136.Lewis KN, Liao R, Guinn KM, Hickey MJ, Smith S, Behr MA, Sherman DR. Deletion of RD1 from Mycobacterium tuberculosis mimics bacille Calmette-Guerin attenuation. J Infect Dis. 2003;187(1):117–23. doi: 10.1086/345862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Gey Van Pittius NC, Gamieldien J, Hide W, Brown GD, Siezen RJ, Beyers AD. The ESAT-6 gene cluster of Mycobacterium tuberculosis and other high G+C Gram-positive bacteria. Genome Biol. 2001;2(10):RESEARCH0044. doi: 10.1186/gb-2001-2-10-research0044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Coros A, Callahan B, Battaglioli E, Derbyshire KM. The specialized secretory apparatus ESX-1 is essential for DNA transfer in Mycobacterium smegmatis. Mol Microbiol. 2008;69(4):794–808. doi: 10.1111/j.1365-2958.2008.06299.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Siegrist MS, Unnikrishnan M, McConnell MJ, Borowsky M, Cheng TY, Siddiqi N, Fortune SM, Moody DB, Rubin EJ. Mycobacterial Esx-3 is required for mycobactin-mediated iron acquisition. Proc Natl Acad Sci U S A. 2009;106(44):18792–7. doi: 10.1073/pnas.0900589106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Ekiert DC, Cox JS. Structure of a PE-PPE-EspG complex from Mycobacterium tuberculosis reveals molecular specificity of ESX protein secretion. Proc Natl Acad Sci U S A. 2014;111(41):14758–63. doi: 10.1073/pnas.1409345111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Vance RE, Isberg RR, Portnoy DA. Patterns of pathogenesis: discrimination of pathogenic and nonpathogenic microbes by the innate immune system. Cell Host & Microbe. 2009;6:10–21. doi: 10.1016/j.chom.2009.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Wang X, Parashar K, Sitaram A, Bliska JB. The GAP activity of type III effector YopE triggers killing of Yersinia in macrophages. PLoS Pathog. 2014;10(8):e1004346. doi: 10.1371/journal.ppat.1004346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Harder J, Franchi L, Munoz-Planillo R, Park JH, Reimer T, Nunez G. Activation of the Nlrp3 inflammasome by Streptococcus pyogenes requires streptolysin O and NF-kappa B activation but proceeds independently of TLR signaling and P2X7 receptor. J Immunol. 2009;183(9):5823–9. doi: 10.4049/jimmunol.0900444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Osei-Owusu P, Jessen DL, Toosky M, Roughead W, Bradley DS, Nilles ML. The N-terminus of the type III secretion needle protein YscF from Yersinia pestis functions to modulate innate immune responses. Infect Immun. 2015 doi: 10.1128/IAI.02687-14. [DOI] [PMC free article] [PubMed] [Google Scholar]