Abstract
Serum amyloid A is a major acute-phase plasma protein that modulates innate immunity and cholesterol homeostasis. We combine sequence analysis with x-ray crystal structures to postulate that SAA acts as an intrinsically disordered hub mediating interactions among proteins, lipids and proteoglycans. A structural model of lipoprotein-bound SAA monomer is proposed wherein two α-helices from the N-domain form a concave hydrophobic surface that binds lipoproteins. A C-domain, connected to the N-domain via a flexible linker, binds polar/charged ligands including cell receptors, bridging them with lipoproteins and re-routing cholesterol transport. Our model is supported by the SAA cleavage in the inter-domain linker to generate the 1–76 fragment deposited in reactive amyloidosis. This model sheds new light on functions of this enigmatic protein.
Keywords: Immune response and reactive AA amyloidosis, Reverse cholesterol transport and atherosclerosis, Acute-phase high-density lipoprotein, Amino acid sequence and structural analyses, Intrinsically disordered protein hub
1. INTRODUCTION
1.1 Overview of SAA function and structure
Serum amyloid A (SAA, 12–14 kDa) is a family of homologous proteins that has been conserved for half a billion years, from sea cucumbers and oysters to humans [1–3]. Humans have four SAA genes; saa1 and saa2 encode for acute-phase isoforms, saa3 is an apparent pseudogene, and saa4 encodes for a constitutively expressed isoform [4, 5]. SAA is synthesized under the cytokine control by hepatocytes and other cells [6] and is secreted into plasma where it associates with lipoproteins. Most circulating SAA is bound to the surface of high-density lipoproteins (HDL) [7–11]. In acute injury, infection or inflammation, plasma concentration of SAA can increase more than a thousand-fold reaching over 1 mg/ml [2, 12]. The biological advantage conferred by this dramatic increase is unclear but is thought to result, in part, from the ability of SAA to mobilize cholesterol for cell repair [2, 13]. In acute phase response, SAA partially displaces the main HDL protein, apolipoprotein (apo) A-I, and thereby re-directs HDL metabolism from the classic pathway of cholesterol removal (termed reverse cholesterol transport) to cholesterol recycling [2, 13]. SAA is also implicated strongly in retinol transport [14], in modulating innate immunity, and in various diseases including atherosclerosis and certain cancers [2, 6, 9, 15–18]. Plasma levels of SAA provide a clinical marker of inflammation, and elevated plasma SAA has emerged as a causal risk factor for cardiovascular disease [9, 17, 18].
Prolonged elevation of SAA can lead to reactive amyloidosis wherein SAA and its N-terminal fragments termed amyloid A (AA), mainly residues 1–76, deposit in kidneys and other organs and damage them [19, 20]. AA amyloidosis is a life-threatening complication of chronic inflammation and a major form of human systemic amyloidosis worldwide [20, 21]. Although animal model studies and in vitro studies show that proteolytic processing of SAA critically influences AA amyloidosis [22], many aspects of AA generation and deposition in vivo remain unclear. For example, it is not known whether the direct protein precursor of AA fragments is the largely unfolded lipid-free SAA, the HDL-bound partially α-helical SAA, or the misfolded β-sheet-rich aggregated SAA [20, 21].
Functions of SAA in health and disease are subject of debate and involve SAA binding to a wide range of ligands. These include hydrophobic ligands such as HDL and other plasma lipoproteins [7–11], cell membranes, model liposomes [23–25], cholesterol [26] and retinol [14]; charged ligands such as metal ions [27] and heparan sulfate proteoglycans [28–30]; and numerous proteins such as cystatin C [31], extracellular matrix proteins [32, 33] and cell receptors. The latter include several G-protein coupled receptors (such as formyl peptide receptor-like 1 and toll-like receptors 2 and 4) and scavenger receptors (SR-BI, CD36, and LOX-1). Although SAA-induced signaling via the formyl peptide and toll-like receptors is apparent only for lipid-free SAA, scavenger receptors interact productively with SAA-containing HDL (SAA-HDL). Examples include SR-BI that interacts productively with both normal HDL and SAA-HDL [34, 35], or CD36 and LOX-1 that preferentially bind and internalize modified lipoproteins including SAA-HDL ([15, 36, 37] and references therein). The ability of SAA to bind these and other diverse ligands and to readily form amyloid is facilitated by the dynamic conformation of this small protein.
Under near-physiologic conditions in solution without bound ligands, SAA spans multiple conformations and has little or no ordered secondary structure [21, 38, 39], which qualifies it as an intrinsically disordered protein (IDP). Like other IDPs, SAA readily self-associates in vitro to form various partially folded oligomers (dimer, tetramer, hexamer, octamer) that can interconvert [38]. Although such oligomers have not been directly observed in vivo, they have been attributed distinct biological properties [14, 38–40]. Quaternary and secondary structures of SAA depend upon environmental conditions such as temperature. For example, under near-physiologic solvent conditions, lipid-free murine SAA isoform 1.1 (mSAA1.1) is ~17% α-helical and self-associated at 4 °C, but this helical stricture unfolds and oligomers are disrupted upon heating to 37 °C [25]. Other ligand-free SAA isoforms are also self-associated and partially α-helical at 4 °C, but their secondary and quaternary structures are progressively disrupted upon heating to 37 °C [38]. Although such unfolded ligand-free forms of SAA have been observed in many in vitro studies, these forms may be only transiently present in vivo as they are highly labile to proteolysis and misfolding as compared to the SAA-HDL, which is the major circulating form.
Ligand binding and folding are usually coupled in IDPs [41, 42]. In SAA, binding to phospholipid surface and lipoprotein formation are coupled to α-helical folding. For example, in model HDL reconstituted from mSAA1 and phosphatidylcholine, α-helical content at 37 °C increases from near-zero to ~35%, as compared to 70–80% observed in similar complexes containing other HDL apolipoproteins such as apoA-I or apoA-II [21, 25]. The SAA conformation on HDL is unknown and is proposed, for the first time, in the current study.
1.2 Atomic structures of lipid-free apolipoproteins: Insights into lipid-bound conformations
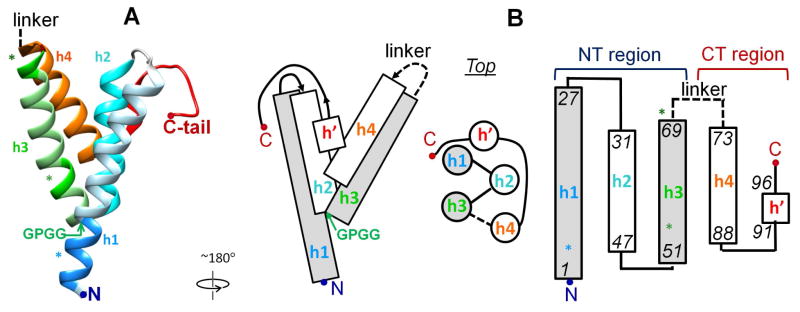
Remarkably for an IDP, high-resolution x-ray crystal structures of lipid-free human and murine SAA have been determined in 2014 in three different forms: hSAA1.1 hexamer (PDB ID 4IP9), hSAA1.1 dimer of dimers (PDB ID 4IP8) [40], and mSAA3 tetramer (PDB ID 4Q5G) [14]. All structures revealed a similar monomer fold, indicating strongly that this fold reflects an intrinsic propensity of the SAA molecule to form a well-ordered structure and is not an artifact of intermolecular packing (Fig. 1A). Residues 1–88 formed a unique Y-shaped 4-helix bundle comprised of helices h1 (residues 1–27), h2 (32–47), h3 (50–69), and h4 (73–88), with the polar C-terminal (CT) tail (residues 89–104) containing a short 3/10 helix h′ (91–96) interacting with h1 and h2 via multiple hydrogen bonds and salt bridges (Fig. 1) [14, 40].
Figure 1. Helix bundle structure of SAA determined by X-ray crystallography [14, 40].
A. Atomic structure of hSAA1.1 (PDB ID 4IP8) [40]. Secondary structural elements are color-coded: h1 (residues 1–27) – blue; h2 (32–47) – teal; h3 (50–69) – green; h4 (73–88) – orange; CT tail (89–104) including a 3/10 helix h′ (90–96) – red. The disordered h3/h4 linker that starts at residue 70 is indicated. Asterisks mark predicted amyloidogenic segments in residues 2–9 (blue), 51–53 (light green) and 69–71 (dark green) [54]. The highly conserved GPGG motif in residues 48–51 located at the N-terminal end of h3, which is tightly packed against h1, is indicated.
B. Cartoon showing SAA helices and their packing and connectivity in the helix bundle. Rectangles (side view) or circles (top view) show four α-helices, h1 to h4. Shapes filled in gray represent amphipathic α-helices (see Fig. 3 for detail). Lines show non-helical segments. Dashed line shows the flexible h3/h4 linker (see Fig. 2 and text for detail). N-terminal (NT, residues 1–69) and C-terminal domains (CT, residues 70–104 in hSAA1) are indicated. The two-domain structure of SAA was first proposed by Fred Stevens [64] a decade before the crystal structure determination.
The hexamer observed in one crystal structure of hSAA1.1 (PDB ID 4IP9) was proposed to represent the HDL-bound form [40]. However, this idea has several flaws: (i) SAA hexamer lacks a large contiguous apolar surface that is prerequisite for HDL binding; (ii) HDL surface, which is comprised mainly of phosphatidylcholine and cholesterol, is uncharged yet the proposed HDL binding site on the hexamer is charged; (iii) this site is not selective for HDL; (iv) cross-linking studies showed that SAA hexamer must dissociate into monomers to bind HDL [55]. Moreover, the crystal structures had ~85% α-helical content, more than twice as high as that observed in any solution studies, including lipid-bound or oligomeric SAA at 4 °C. This suggests that in the absence of additional ligands, only a fraction of the helical structure observed by crystallography is folded in SAA-HDL; the nature of this folded structure is addressed below in part 2.2.
Other HDL apolipoproteins, such apoA-I and apoE whose high-resolution structures have been determined for lipid-free mutant forms, also fold in solution into NT 4-helix bundles, with the unstructured CT tails wrapped around the bundle. However, there are several important differences between these apolipoproteins and SAA.
First, CT tails in apoA-I and apoE are highly hydrophobic and dynamic in the lipid-free state in solution and form the primary lipid binding sites [43, 44]. Upon binding to the lipid surface, these tails fold into amphipathic α-helices that form the major lipid surface-binding motif in the apolipoprotein family [45]. In contrast, the CT tail in SAA is largely polar and is well-ordered albeit mostly non-helical in the crystal structure (Fig. 1) [14, 40]. Notably, the CT tail of SAA does not form an amphipathic secondary structure and, hence, is not expected to directly bind to lipoprotein surface (Fig. 3B discussed below).
Second, helix bundles in apoE, apoA-I and apoA-IV are comprised of amphipathic α-helices whose apolar faces form the interior of the bundle. In contrast, the SAA monomer is stabilized by extensive hydrogen bonding among conserved polar residues and water molecules in the interior of the bundle [14]. Such tertiary interactions in monomeric SAA are expected to be highly labile in aqueous solution.
Third, the shape of the 4-helix bundle, which is elongated and relatively straight in the available atomic structures of apoE and apoA-IV or semi-circular in apoA-I, differs from the cone-shaped 4-helix bundle in SAA (supplemental Fig. S1). The apolipoprotein shape is important for conferring 2D curvature to the lipoprotein surface whose constituent lipids have near-zero spontaneous curvature. For example, the highly curved dimer seen in the crystal structures of apoA-I [46, 47] has a diameter d~11 nm that complements the convex surface of HDL (d=8–12 nm) (Fig. S1). This explains the strong preference of apoA-I for binding to HDL as compared to larger less curved low- or very low-density lipoproteins, LDL (d=20–24 nm) or VLDL (d=40–100 nm). Compared to apoA-I, 4-helix bundles of apoE and apoA-IV are relatively straight, which explains the relatively high affinity of these proteins for VLDL [48]. Notably, amino acid sequences of these and other apolipoproteins contain Pro-punctuated 11/22-mer tandem helical repeats that help confer the overall molecular curvature (Fig. S1) [45–48]. In contrast, the SAA sequence lacks Pro punctuation, and the molecular basis for preferential binding of SAA to HDL as compared to LDL or VLDL is unknown.
Fourth, extensive biophysical studies, including protein cross-linking followed by mass spectrometry and distance measurements using FRET or EPR, indicate that the 4-helix bundles in apoA-I, apoA-IV and apoE open up on the lipid to form two pairs of helices that wrap around the lipoprotein perimeter in an antiparallel “double-belt” conformation ([46–50] and references therein). Such a helix bundle opening exposes the apolar faces of the amphipathic apolipoprotein α-helices to the lipid surface (Fig. S1). It is unknown whether the SAA helix bundle opens upon HDL binding.
In summary, despite major recent advances in high-resolution structural studies [14, 40], it remains unclear how the well-ordered helix bundle structure observed by x-ray crystallography in lipid-free oligomeric SAA relates to the much less helical highly dynamic conformations of HDL-bound or lipid-free SAA, facilitating SAA binding to a wide range of diverse ligands in vivo.
Here we integrate the results of our amino acid sequence analyses with the existing biophysical, structural and functional data on SAA, which were obtained by many teams over nearly four decades. We identify a novel lipoprotein-binding site in the SAA monomer, which explains SAA selectivity for HDL, and propose a molecular basis for its dynamic ligand binding and functions.
2. RESULTS AND DISCUSSION
2.1 Properties of individual α-helices in SAA from various species
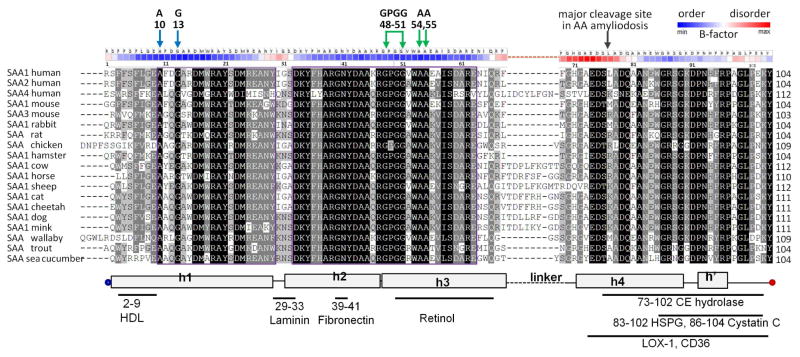
Figure 2 shows amino acid sequences of representative SAA proteins. Physicochemical and structural properties of these proteins were assessed by using sequence-based prediction methods. For all proteins explored, secondary structural predictions agreed with the crystal structures of hSAA1.1 and mSAA3, indicating α-helices h1, h2, h3 and h4 and the turns between them, along with the largely non-helical CT tail (Fig. S2). This result supports the idea that the 4-helix bundle in SAA is evolutionally conserved [40]. Surprisingly, h4 consistently showed high helical propensity, even though its N-terminal half was disordered in all crystal structures (Fig. 2); the functional implications of this observation are discussed in part 2.5 below.
Figure 2.
Amino-acid sequence alignment in the SAA family. The residue numbers correspond to human SAA1 and SAA2. The conservation degree is shown by the text highlight, with more conserved residues in darker highlight. The alignment was carried out by using Clustal Omega server [65]. Secondary structural elements observed by x-ray crystallography are shown at the bottom: rectangles – helices, lines – non-helical structure [14, 40]. Known locations of ligand binding sites in SAA are indicated at the bottom. Colored arrows at the top show key residues from h1 and h3 (A, G and the GPGG motif) that, we propose, lock the relative orientation of these helices. Black arrow shows the cleavage site that generates the major AA fragment 1–76 found in human amyloid deposits. Structural disorder is illustrated by B-factors obtained from the x-ray crystal structure of hSAA1.1 (PDB ID 4IP9). Atomic B-factors are color coded as shown. Similar B-factor distribution is seen in all six SAA molecules from the three available crystal structures (PDB ID 4IP8, 4IP9, 4Q5G).
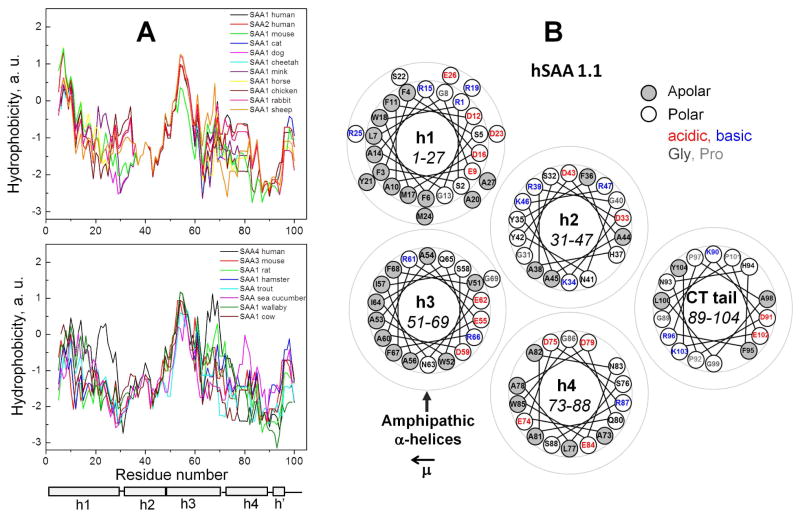
Hydrophobicity profiles of all proteins showed a local maximum at residues 50–56 often followed by a smaller peak near residue 68 (Fig. 3A). These peaks delineated the NT and CT ends of the highly conserved h3 that showed high hydrophobicity in all proteins. In addition, several SAA proteins (human SAA1 and SAA2, mouse SAA1, cat SAA1, etc.) showed the major hydrophobicity maximum near the NT end of h1 (Fig. 3A, top panel). This is not surprising since residues 1–11 in hSAA have been implicated strongly in forming the major HDL binding site and to contain the major amyloidogenic segment 2–9 [40, 51] (Fig. 1A, bright blue). In several SAA proteins (e. g. human SAA1 and SAA2, mouse SAA1, etc.) the hydrophobicity was high at the NT end of h1 but progressively declined towards its CT end (Fig. 3A top), suggesting that h1 may penetrate the lipid surface at an angle, with deeper insertion of the NT end. In contrast, in other proteins (such as human SAA4, mouse SAA3, rat SAA, trout SAA), the NT end of h1 was substantially charged or polar (Fig. 2, Fig. 3A bottom), suggesting reduced protein affinity for HDL. Consistent with this idea, in acute phase in rat and trout, the levels of circulating HDL-bound SAA do not increase; only local increase in SAA, which is probably not bound to HDL, was reported in trout [52, 53]. In summary, h3 in all species and h1 in some species have high hydrophobicity, while the rest of the SAA molecule is relatively polar.
Figure 3. Hydrophobicity properties of SAA proteins.
A. Hydropathy profiles of representative SAA family members obtained by using ExPASy server [66] with Kyte-Doolittle scale. Residue numbering follows hSAA1; additional residues that are present at the NT end or in the h3–h4 linker of other proteins (Fig. 2) are not shown. Some proteins show the major hydrophobicity maximum near the N-terminus of h1 (top panel); others show relatively low hydrophobicity in h1 (bottom panel). All proteins show a hydrophobicity maximum in h3 that is a part of a highly conserved region.
B. Helix wheel diagrams of hSAA1.1 depict five segments observed in the crystal structure, including α-helices h1 to h4 and the largely non-helical C-terminal tail. Apolar and polar residues are color coded as indicated. In each segment the direction of the hydrophobic moment μ is right to left (left arrow). In hSAA1.1, μ(h1)=11.9, μ(h2)=6.7, with much lower values of μ<3 for h2, h4 and the CT tail. Similarly, in all other SAA proteins, only h1 and h3 form amphipathic helices with a large well-demarcated apolar face and a high hydrophobic moment. The values of μ were obtained by using a publically available on-line server created by D. Armstrong and R. Zidovetzki (http://rzlab.ucr.edu/scripts/wheel/wheel.cgi?sequence=ABCDEFGHIJLKMNOP&submit=Submit). The results are consistent with the previous secondary structure prediction for SAA [64].
Importantly, helix wheel diagrams of all SAA proteins revealed that only h1 and h3 can form amphipathic α-helices with large relatively straight apolar faces (Fig. 3B). Similar helices form the major lipid surface-binding motif in apolipoproteins [45]. However, the non-random radial charge distribution characteristic of apolipoprotein α-helices, which is thought to augment their affinity for the lipid surface [45], is not observed in SAA (Fig. 3B). In all SAA proteins, h1 showed the highest hydrophobic moment (μ =8–12) followed by h3 (μ =5–8); in contrast, h2 and h4 consistently showed low hydrophobic moments (μ =0–3) and low overall hydrophobicity (Fig. 3). Notably, h1 and h3 are located on the same side of the SAA monomer (Fig. 1). Together, our results indicate that possible lipid surface binding sites in SAA are located in h1 and h3, and not just in the NT end of h1 as previously thought. This is in stark contrast with apoA-I, apoE and other apolipoproteins whose entire molecules can bind lipid surface via contiguous apolar faces formed by tandem amphipathic α-helical repeats [45, 46].
Although further discussion focuses on SAA from mice and men, particularly hSAA1.1 and mSAA3 whose atomic structures have been determined experimentally, it sheds light on the structure-function relationship in other members of this protein family.
2.2 h1 and h3 form a concave apolar surface that contains amyloid hot spots and binds HDL
In hSAA1, the predicted amyloid-forming sequence propensity is confined to residues 1–72, consistent with the finding that the 1–76 fragment is the main constituent of human AA deposits [40, 54]. One major and two minor amyloidogenic segments, or hot spots, have been predicted in residues 2–9 (SFFSFLG) from h1 and residues 53–55 (WAA) and 67–70 (RFFG) from h3, respectively [54]. In the crystal structure of hSAA1.1, these segments are largely exposed in the monomer (Fig. 1A, asterisks). In vivo, amyloid hot spots are normally protected from misfolding by the native packing, which in apolipoproteins includes protein-protein and protein-lipid interactions [54]. Therefore, amyloid hot spots in SAA are expected to be sequestered in vivo upon lipid binding or self-association. In fact, the major hot spot segment 2–9 of hSAA binds HDL [51] and is partially sequestered in the crystal structure of the hexamer [40]. Deletion of this segment or its point mutations greatly decrease (by up to 95%) but do not completely abolish hSAA binding to HDL [51], suggesting that the NT end forms the major but not the sole HDL binding site.
We posit that both h1 and h3 in SAA contribute to HDL binding. This idea stems from the observation that only h1 and h3 in SAA form amphipathic α-helices with large apolar faces (Fig. 3B). Also, hydrophobicity (Fig. 3A) and transmembrane tendency profiles (not shown) suggest that h1 and h3 are the only segments in SAA with significant affinity for lipid. Moreover, structural studies of mSAA3 tetramer have implicated h3 as a binding site for a lipophilic ligand retinol [14].
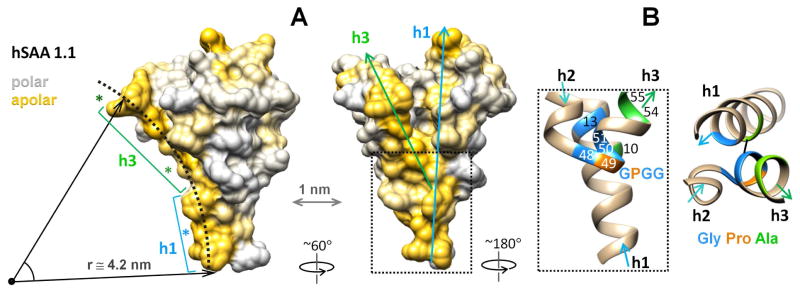
Although the direct role of h3 in HDL binding remains to be established experimentally, strong evidence for such binding comes from the crystal structures of SAA. A striking feature in the surface hydrophobicity distribution in the SAA monomer is an elongated concave hydrophobic surface formed by h1 and h3 (Fig. 4A). The curvature of this surface (radius r ≅ 4.2 nm) is complementary to that of HDL (d = 8–11.5 nm), making it immediately clear how SAA monomer binds HDL via this site, with a particular preference for the smaller particles termed HDL3 [8], and why such binding is favored over larger lipoproteins [5, 10, 11]. The predicted amyloidogenic segments in SAA are located at the ends (residues 2–9 and 67–70) or in the middle (53–33) of this surface (Fig. 4A, asterisks) and are solvent-exposed in the SAA monomer. By binding to HDL via this concave apolar surface, SAA can protect these sensitive segments from initiating the misfolding as well as maintain the lipoprotein surface curvature.
Figure 4. Proposed HDL binding site in the SAA monomer and the helical packing that determines its curvature.
A - Space-filling model of hSAA1 monomer illustrates its surface hydrophobicity. Two projections show the concave surface formed by apolar faces of h1 and h3 (in gold). Dotted arc with a radius of r~4.2 nm indicates the putative HDL binding site. Asterisks show amyloidogenic segments in h1 (blue) and h3 (green), which are exposed in monomeric SAA. Arrows show the orientation of h1 and h3. Boxed region contains junction of h1 and h3 that determines their relative packing at ~45°.
B - Zoomed-in view of the boxed region shows h1 and h3 packing via the GPGG motif in residues 48–51 and other conserved residues (G13, A10, A54, A55) that are numbered and color-coded. Black line in the right panel indicates the shortest distance of 3.6Å between main chains of A10 and G50. Additional details of the main and side chain interactions in this region are shown in Figure S3.
What are the molecular determinants for the SAA binding affinity and specificity for HDL? The curvature of the apolar surface formed by h1 and h3 is defined by the ~45° angle between these helices. This angle is critically hinged upon the strictly conserved GPGG motif in residues 48–51. In the crystal structures, this well-ordered motif forms a tight turn and the NT end of h3 that is packed against h1 (Fig. 1, left panels). G50, G51 and A54, A55 from h3 pack against A10 and G13 from h1, locking h1 in a groove formed by the GPGG motif at the NT end of h3 and facilitating an unusually close spacing between these helices (Fig. 4B), with only 3.6Å separation between the main chain atoms Cα of A10 and C of G50 and between N of A10 and C of G50. Additional main and side chain interactions involved in h1–h3 packing in this region are shown in the supplementary Figure S3. To our knowledge, such helical packing via the GPGG motif has not been reported previously.
Notably, the structure in this region, including the h2–h3 linker, is well ordered, as evident from low atomic B-factors; in contract, other interhelical linkers, such as h1–h2 and h3–h4, are relatively disordered (Fig. 2 top). Therefore, despite high potential flexibility of the Gly-rich GPGG motif, folding of this region freezes out local flexibility and locks h1 against h3. The high entropic cost of such folding must be counterbalanced by water exclusion from the apolar surface formed by h1 and h3 and by the favorable enthalpy of HDL binding at this surface. We propose that HDL surface forms a template that augments the folding of h1 and h3 accompanied by the insertion of their apolar faces into the lipid surface.
Notably, the residues critical for the packing of h1 against h3, including A10, G13, G48, P49, G50, G51, A54 and A55, are strictly conserved (Fig. 2 and Fig. S3), suggesting that the angle between h1 and h3 (and hence, the curvature of the hypothetical lipoprotein-binding surface) is also conserved throughout evolution. In fact, the crystal structures of hSAA1.1 (Figs. 1, 3) and mSAA3 (Fig. S4) show a similar angle between h1 and h3 and a similarly curved hydrophobic surface formed by these helices. We propose that the GPGG motif and other conserved residues involved in h1–h3 packing determine the binding selectivity of SAA for HDL-size lipoproteins, while variable residues 1–11 at the NT end of h1 modulate the species- and isoform-specific affinity of SAA for HDL. Still, one cannot exclude that the angle between h1 and h3 may be adjusted upon SAA binding to less curved surfaces of larger lipoproteins and lipid vesicles.
Apart from lipid binding, the large apolar surface formed by h1 and h3, which is exposed in the SAA monomer, is expected to drive protein self-association in solution. In fact, this surface is directly involved in self-association in all three oligomeric forms of SAA that have been crystallized [14, 40]. To make this surface available for HDL binding, such oligomers must dissociate. In fact, biophysical and cross-linking studies of mSAA2.2 by an expert team reported that the hexamer has little affinity for HDL and dissociates into monomers during binding to HDL [55]. Hence, we propose that the HDL-bound SAA is monomeric, although self-association via alternative surfaces cannot be excluded.
2.3 The helix bundle of SAA does not open on the lipid surface
In contrast to h1 and h3, SAA helices h2 and h4 lack large apolar faces (Fig. 3) and are unlikely to directly bind to lipid surface. The latter is consistent with the SAA crystal structures wherein h2 is packed against h1 and h3 opposite to the putative HDL binding site. Therefore, we propose that in SAA-HDL, h1 and h3 are sandwiched between h2 and HDL and partially retain their conformation seen in the crystal structures, particularly the packing of h1 against h3 via the GPGG motif (Fig. 1A, Fig. 5). Hence, we posit that the 4-helix bundle of SAA does not open into two helical pairs on the lipid surface. Rather, SAA oligomers dissociate to expose the curved apolar surface in the monomer that binds to HDL via h1 and h3; the conformation of these helices resembles that seen in the crystal structures of lipid-free proteins. This lipid binding mode is unique to SAA and differs from the helix bundle opening into two pairs of antiparallel helices, h1–h2 and h3–h4, which was inferred for apoE, apoA-I, and apoA-IV on the lipid surface (Fig. S1).
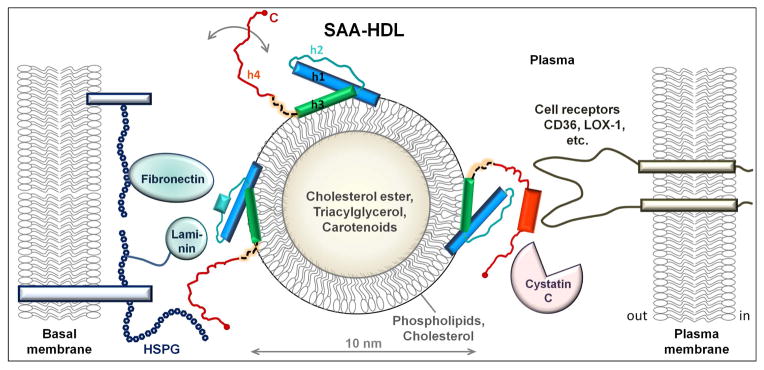
Figure 5.
Hypothetical model of SAA on HDL surface. SAA and HDL are drawn to scale. HDL is shown in cross-section to illustrate its polar surface containing amphipathic proteins, phospholipids (mainly phosphatidylcholine) and cholesterol, and the apolar core containing cholesterol esters, triacylglycerides and small amounts of fat-soluble vitamins such as carotenoids. Other HDL proteins such as apoA-I or apoA-II that are also present on SAA-HDL [25] are not shown. In the NT domain of SAA (residues 1–69 in blue, teal and green), amphipathic α-helices h1 and h3 form the putative HDL binding site. We posit that h1 and h3 on HDL retain their relative packing via the GPGG motif observed by x-ray crystallography. Parts of h1 and h3 that bind HDL are folded, accounting for an average α-helical content of ~35% observed in lipid-bound SAA by circular dichroism [25]. In the absence of additional ligands, the rest of the SAA molecule spans multiple conformations (circular arrow). The dynamic behavior of the CT domain (red) is facilitated by the flexible interdomain linker (dashed line). Apart from HDL, the NT domain can also bind other ligands, such as laminin and fibronectin that bind via h2 and h2/h3 linker [32, 33]. Alternatively, the NT domain of SAA can bind retinol via h3 (not shown) [14] and/or self-associate via h1 and h3 [14, 39]. CT domain can bind diverse ligands [64] including various cell receptors (CD36, LOX-1, etc.) [37], heparan sulfate proteoglycans (HSPG) [28], cystatin C [31], cholesterol ester hydrolase [58] and others. Ligand binding is probably augmented by the high intrinsic helical propensity of h4 (Fig. S2) that is expected to undergo coupled binding and folding. Thus, in the presence of additional bound ligands, the helical content in SAA on HDL may ultimately approach that seen in the crystal structure. SAA brings together ligands that are bound to NT- and CT-domains of the same SAA molecule, as well as those bound to different SAA molecules on the same HDL particle. Such bridging of diverse ligands, which may bind with positive or negative cooperativity depending on the environment, can modulate their functional interactions. We propose that SAA acts as a molecular hub modulating a wide range of functional interactions among diverse proteins, lipids, and proteoglycans.
Although experimental evidence for the idea that the packing of h1 and h3 observed by x-ray crystallography is maintained on SAA-HDL remains to be established, several lines of evidence support this idea. First, the most conserved regions in the SAA family (>85% sequence identity [2]) encompass residues 13–29 from h1 and 33–63 from h2–h3 (Fig. 2, boxed). In the crystal structure, these regions are packed against each other in the helix bundle, while more variable residues 1–12 (NT end of h1) and 30–32 (h1/h2 linker) are exposed (Fig. 1A, Fig. 2). Such a direct correlation between the sequence conservation and the tertiary packing of h1–h3 helices in the bundle suggests that this packing is conserved for function. Since a major function of SAA is lipid transport via HDL, it is likely that the packing of h1–h3 helices in the crystal structures of lipid-free SAA represents, in part, their HDL-bound conformation.
Second, our model of SAA-HDL is consistent with studies of SAA effects in cell adhesion, which identified a laminin-binding motif in residues 29–33 and a fibronectin-binding motif in residues 39–41 of SAA [32, 33] and showed that these motifs are solvent-exposed both in lipid-free and in HDL-bound SAA [33]. In the crystal structure, these motifs are located in h1/h2 linker and in the middle of h2 (Fig. 2, bottom), and are exposed both in lipid-free SAA and in our model of SAA-HDL (Fig. 5, in teal).
Third, additional support for our model comes from epitope mapping studies reporting that: i) accessible epitopes in SAA comprise residues 31–39 (part of h2), 64–78 (h3/h4 linker and adjacent ends of h3 and h4 helices), and 95–104 (CT tail); ii) inaccessible epitopes include most of h1 and h3; iii) the same epitopes are exposed in HDL-associated and in lipid-free SAA that is presumably self-associated [56]. All these findings are consistent with our model of SAA-HDL in which h1 and h3 are largely helical and are directly involved in lipid binding and/or self-association, while the rest of the molecule, which is largely unfolded, is accessible for binding to other ligands and probably becomes folded upon such binding (Fig. 5).
2.4 NT and CT domains are connected via a flexible linker
Apart from the NT end of h1, the most variable sequence region in SAA starts at residue 70 of hSAA and encompasses the h3/h4 linker and the NT half of h4. This region can vary in the amino acid composition and in length, and can contain a 7- to 9-residue insert found in several proteins including hSAA4 (Fig. 2). The enzymatic N-glucosylation observed in the insert of hSAA4 indicates that this region is disordered in vivo [57]. Moreover, crystal structures of hSAA1.1 and mSAA3, which lack the insert, also show that the h3/h4 linker forms the most disordered region in all six molecules in three available crystal structures of SAA (two molecules per asymmetric unit), with high atomic B-factors (Fig. 2, top). Consequently, the disorder in this linker is an inherent property of the SAA molecule and not an artifact of the crystal packing. We posit that this flexible linker between NT and the CT domains of SAA is important for function.
2.5 Helix h4 and the CT tail can dissociate to bind other ligands
Our analysis suggests that SAA contains two domains, one encompassing NT two-thirds of the protein (residues 1–69) and the other its CT one-third (residues 70–104 in hSAA1). The NT domain contains highly conserved helical segments from h1–h3 that can bind hydrophobic ligands, such as lipoproteins, cholesterol and retinol, via the extended apolar surface formed by h1 and h3; this surface is also central to protein self-association in solution [14, 26, 40]. The NT domain is connected by a flexible linker to the dynamic CT domain that encompasses h4 and the largely non-helical C-terminal tail. We posit that the relatively polar CT domain can dissociate from the h1–h3 helices and recruit additional interacting partners, bridging them with the NT-bound ligands (Fig. 5). Such dissociation is probably augmented by the unusually polar water-containing interior of the SAA monomer [14]. Consistent with this idea, several polar/charged ligands of SAA reportedly bind at the CT residues 70–104. These ligands include heparan sulfate that binds to the basic residues from the 83–102 region in mSAA [28]; cystatin C that forms electrostatic interactions with SAA residues 86–104 [31]); and cell receptors such as LOX-1 and CD36 that bind modified lipoproteins [37] (Figs. 2, 5). By bridging HDL to LOX-1 or CD36, SAA can facilitate HDL internalization by many cell types expressing these receptors, and thereby re-direct cholesterol transport to these cells. Similarly, SAA can probably re-direct the transport of other HDL lipids, including phospholipids, triglycerides and a small amount of fat-soluble vitamins such as carotenoids and retinol esters present in HDL core (Fig. 5). Thus, SAA can modulate transport of these vitamins not only in HDL-dissociated [14] but also in HDL-bound form.
Synergistic action of NT and CT domains of SAA is also exemplified by its effects on cholesterol esterification. Via its CT residues 74–103, SAA stimulates cholesterol ester hydrolase that de-esterifies cellular cholesterol. Via its lipid-binding NT residues 1–20, SAA diminishes the activity of acetyl: and lecithin:cholesterol acyltransferase that esterify cellular and plasma cholesterol, respectively [58]. Through these combined effects, SAA shifts cholesterol distribution from the esterified (storage) to the unesterified (transport) form, and thereby augments efflux of excess cholesterol from cells to HDL.
In a small protein such as SAA, spatial proximity of multiple ligand binding sites is expected to result in binding cooperativity. In fact, SAA binding to heparan sulfate was reported to dissociate HDL from SAA [29], suggesting negative cooperativity between HDL binding at the NT domain of SAA and heparan sulfate binding at the CT domain. SAA dissociation from HDL exposes amyloid hot spots in h1 and h3 (Fig. 4A), which is expected to promote amyloid formation. Consequently, blocking SAA binding to heparan sulfate chains in vivo provides a potential therapeutic strategy to combat AA amyloidosis. Furthermore, interactions of HDL-bound SAA with heparan sulfate were reported to interfere with SAA-HDL binding to cell receptors [30], suggesting negative cooperativity in the binding of heparan sulfate and the receptor to the CT domain of SAA.
Ligand binding by IDPs typically involves α-helical folding. Both affinity and specificity of binding can benefit from the high α-helical propensity of the ligand binding sites [41]. We posit that, due to its polar character and high intrinsic α-helical propensity (Fig. 3, Fig. S2), h4 can readily dissociate from h1–h3, recruit polar/charged ligands and undergo coupled folding and binding.
In other IDPs, ligand binding at various sites may have positive or negative cooperativity depending upon the relative location of the binding sites and environmental conditions such as ligand concentration or pH [41, 59]. An environment-dependent allosteric regulation was proposed to provide a common mechanism for the functional modulation in IDPs that act as molecular hubs in protein interaction networks [41]. We speculate that tunable environment-dependent allosteric regulation of ligand binding to NT and CT domains of SAA can modulate complex functions of this protein in signaling, immune response and lipid homeostasis. If so, such context-dependent effects may help reconcile the conflicting data regarding biological properties of SAA, such as its reported ability to either promote or prevent inflammation ([2, 13, 16] and references therein).
2.6 SAA as a protein hub in interaction networks modulating lipid homeostasis
Our analysis suggests that, like many other IDPs that are promiscuous binders, SAA acts a molecular hub facilitating a wide range of functional interactions among diverse ligands, including various proteins, lipids and proteoglycans. One example is interactions between HDL, which binds to the NT domain of SAA, and cell receptors CD36 or LOX-1, which bind to its CT domain. SAA-mediated HDL binding and internalization via these receptors likely re-routs transport of cholesterol and other HDL lipids and lipid-soluble vitamins towards cell repair, which is thought to benefit immediate survival in acute injury or inflammation [13, 14]. However, internalization of SAA-HDL by arterial macrophages, which express CD36 and LOX-1, is also expected to augment the accumulation of macrophage cholesterol, ultimately leading to foam cell formation and cholesterol deposition in arterial plaques. Therefore, preferential internalization of SAA-HDL by arterial macrophages [13, 60] likely promotes atherosclerosis. This effect is partially alleviated by the ability of SAA to augment cholesterol efflux from cells by i) shifting the equilibrium towards the unesterified form of cholesterol, and ii) supplying extracellular acceptors for cholesterol in a process mediated by the lipid transporter ABCA1 ([13] and references therein). The net effect is that SAA promotes efficient efflux of most but not all cholesterol that is taken up by macrophages upon internalization of acute-phase lipoproteins. Thus, elevated plasma SAA probably augments cholesterol accumulation in the arteries, which contributes to the established causal link between inflammation and atherosclerosis [17, 18].
Since each HDL particle can carry several copies of SAA ([25] and references therein), ligand binding to one copy can probably influence the binding of a similar or a different ligand to another copy, adding another layer of the tunable allosteric regulation via SAA. In this scenario, each SAA-HDL particle acts as a multivalent macromolecular hub in interaction networks involving plasma and membrane-associated proteins and proteoglycans (Fig. 5), which amplifies the ability of SAA to bridge a wide range of ligands and modulate their functional interactions.
A similar concept has been proposed by several teams to describe various functions of HDL in cardiovascular disease, inflammation, and immune response. For example, HDL was proposed to provide a platform for the assembly of multiple proteins whose synergistic action is important for the evolution of innate immunity to bacterial infections [61]. These ideas are supported by ongoing proteomic studies of lipoproteins using mass spectrometry, which are aimed to elucidate key functional determinants of these heterogeneous particles [60–63]. Such proteomic studies are particularly useful to identify proteins associated with circulating lipoproteins. The current study extends these ideas towards membrane-associated moieties, such as cell receptors and proteoglycans that preferentially bind SAA-HDL (Fig. 5).
3. SUMMARY
We propose the first structural model of HDL-bound SAA monomer, which is distinct from any other lipoprotein models. In contrast to the helix bundle opening into two pairs of antiparallel helices that bind to lipid surface in apoE, apoA-I and apoA-IV, SAA probably binds HDL via the concave apolar surface formed by helices h1 and h3 that are packed at an angle of ~45°. Relative orientation of these helices in lipid-bound and in free SAA is determined by several conserved residues including the GPGG motif at the NT end of h3 that locks h1 like a socket wrench. Our model explains, for the first time, why SAA preferentially binds to HDL over larger lipoproteins. Further, we postulate that the NT domain of SAA, which binds HDL, and the CT domain, which binds cell receptors and other ligands and bridges them with HDL, act in synergy to modulate lipid homeostasis. This synergy potentially extends to other less well-understood functions of SAA in immune response and cell signaling.
Importantly, the proposed two-domain model of SAA, with its flexible interdomain linker starting at residue 70, is consistent with the observation that the 1–76 fragment is the major protein constituent of AA deposits in vivo. The proteolytic cleavage of this linker produces the amyloidogenic NT fragment that forms AA deposits in vivo, and the non-amyloidogenic CT fragment [22]. Our model suggests that the h3/h4 linker is flexible and can be susceptible to cleavage in lipid-free as well as in HDL-bound SAA. Our model of SAA-HDL is also consistent with the location of amyloid hot spots in h1 and h3, which are protected on HDL but are labile in free SAA monomer [54]. Future experiments will directly probe the proposed structural model of lipid-bound SAA and its functional ramifications.
Supplementary Material
Highlights.
Serum amyloid A is a multifunctional intrinsically disordered acute-phase protein
Bioinformatics, structural and functional studies suggest two domains in SAA
Amphipathic α-helices in residues 1–69 form a concave surface that binds lipoproteins
Polar segment 70–104 recruits receptors and other functional ligands for lipoproteins
SAA acts as a hub in interaction networks in lipid homeostasis and immune response
Acknowledgments
We are grateful to Dr. Shobini Jayaraman for many useful discussions and helpful comments on the manuscript prior to publication and to Drs. Elena S. Klimtchuk and Isabel Morgado for helpful suggestions. We thank Dr. Assen Marintchev for helpful comments on the manuscript and Dr. Barbara Schreiber for extremely useful discussions. This work was supported by the National Institutes of Health grant GM067260 to OG; NF was supported, in part, by the T32 training grant HL007969.
ABBREVIATIONS
- SAA
serum amyloid A
- hSAA1.1
human SAA
- mSAA
murine SAA
- AA
amyloid A
- IDP
intrinsically disordered protein
- HDL
high-density lipoprotein
- SAA-HDL
SAA-containing HDL
- LDL
low-density lipoprotein
- VLDL
very low-density lipoprotein
- apo
apolipoprotein
- NT
N-terminal
- CT
C-terminal
- CD36
cluster differentiation 36 scavenger receptor
- LOX-1
oxidized low-density lipoprotein receptor 1
- SR B1
scavenger receptor class B member 1
Footnotes
Author Contribution
NF carried out bioinformatics studies, prepared the graphics and contributed to writing. OG conceived the project, prepared the graphics and wrote the paper.
References cited
- 1.Uhlar CM, Burgess CJ, Sharp PM, Whitehead AS. Evolution of the serum amyloid A (SAA) protein superfamily. Genomics. 1994;19(2):228–235. doi: 10.1006/geno.1994.1052. [DOI] [PubMed] [Google Scholar]
- 2.Kisilevsky R, Manley PN. Acute-phase serum amyloid A: perspectives on its physiological and pathological roles. Amyloid. 2012;19:5–14. doi: 10.3109/13506129.2011.654294. [DOI] [PubMed] [Google Scholar]
- 3.Qu F, Xiang Z, Yu Z. The first molluscan acute phase serum amyloid A (A-SAA) identified from oyster Crassostrea hongkongensis: molecular cloning and functional characterization. Fish Shellfish Immunol. 2014;39(2):145–151. doi: 10.1016/j.fsi.2014.05.013. [DOI] [PubMed] [Google Scholar]
- 4.Uhlar CM, Whitehead AS. Serum amyloid A, the major vertebrate acute-phase reactant. Eur J Biochem. 1999;265(2):501–523. doi: 10.1046/j.1432-1327.1999.00657.x. [DOI] [PubMed] [Google Scholar]
- 5.De Beer MC, Yuan T, Kindy MS, Asztalos BF, Roheim PS, de Beer FC. Characterization of constitutive human serum amyloid A protein (SAA4) as an apolipoprotein. J Lipid Res. 1995;36(3):526–534. [PubMed] [Google Scholar]
- 6.Urieli-Shoval S, Linke RP, Matzner Y. Expression and function of serum amyloid A, a major acute-phase protein, in normal and disease states. Curr Opin Hematol. 2000;7(1):64–79. doi: 10.1097/00062752-200001000-00012. [DOI] [PubMed] [Google Scholar]
- 7.Benditt EP, Eriksen N. Amyloid protein SAA is associated with high density lipoprotein from human serum. Proc Natl Acad Sci USA. 1977;74:4025–4028. doi: 10.1073/pnas.74.9.4025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coetzee GA, Strachan AF, van der Westhuyzen DR, Hoppe HC, Jeenah MS, de Beer FC. Serum amyloid A-containing human high density lipoprotein 3. Density, size, and apolipoprotein composition. J Biol Chem. 1986;261:9644–9651. [PubMed] [Google Scholar]
- 9.O’Brien KD, McDonald TO, Kunjathoor V, Eng K, Knopp EA, Lewis K, Lopez R, Kirk EA, Chait A, Wight TN, de Beer FC, LeBoeuf RC. Serum amyloid A and lipoprotein retention in murine models of atherosclerosis. Arterioscler Thromb Vasc Biol. 2005;25(4):785–790. doi: 10.1161/01.ATV.0000158383.65277.2b. [DOI] [PubMed] [Google Scholar]
- 10.Jahangiri A, Wilson PG, Hou T, Brown A, King VL, Tannock LR. Serum amyloid A is found on apoB-containing lipoproteins in obese humans with diabetes. Obesity. 2013;21(5):993–996. doi: 10.1002/oby.20126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lepedda AJ, Nieddu G, Zinellu E, De Muro P, Piredda F, Guarino A, Spirito R, Carta F, Turrini F, Formatom M. Proteomic analysis of plasma-purified VLDL, LDL, and HDL fractions from atherosclerotic patients undergoing carotid endarterectomy: identification of serum amyloid A as a potential marker. Oxid Med Cell Longev. 2013;2013:385214. doi: 10.1155/2013/385214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gabay C, Kushner I. Acute-phase proteins and other systemic responses to inflammation. N Engl J Med. 1999;340(6):448–454. doi: 10.1056/NEJM199902113400607. [DOI] [PubMed] [Google Scholar]
- 13.Manley PN, Ancsin JB, Kisilevsky R. Rapid recycling of cholesterol: the joint biologic role of C-reactive protein and serum amyloid A. Med Hypotheses. 2006;66(4):784–792. doi: 10.1016/j.mehy.2005.10.018. [DOI] [PubMed] [Google Scholar]
- 14.Derebe MG, Zlatkov CM, Gattu S, Ruhn KA, Vaishnava S, Diehl GE, MacMillan JB, Williams NS, Hooper LV. Serum amyloid A is a retinol binding protein that transports retinol during bacterial infection. eLife. 2014;3:e03206. doi: 10.7554/eLife.03206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eklund KK, Niemi K, Kovanen PT. Immune functions of serum amyloid A. Crit Rev Immunol. 2012;32(4):335–348. doi: 10.1615/critrevimmunol.v32.i4.40. [DOI] [PubMed] [Google Scholar]
- 16.Ye RD, Sun L. Emerging functions of serum amyloid A in inflammation. J Leukoc Biol. 2015;98(6):923–929. doi: 10.1189/jlb.3VMR0315-080R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dong Z, Wu T, Qin W, An C, Wang Z, Zhang M, Zhang Y, Zhang C, An F. Serum amyloid A directly accelerates the progression of atherosclerosis in apolipoprotein E–deficient mice. Mol Med. 2011;17(11–12):1357–1364. doi: 10.2119/molmed.2011.00186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thompson JC, Jayne C, Thompson J, Wilson PG, Yoder MH, Webb N, Tannock LR. A brief elevation of serum amyloid A is sufficient to increase atherosclerosis. J Lipid Res. 2015;56:286–293. doi: 10.1194/jlr.M054015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Simons JP, Al-Shawi R, Ellmerich S, Speck I, Aslam S, Hutchinson WL, Mangione PP, Disterer P, Gilbertson JA, Hunt T, Millar DJ, Minogue S, Bodin K, Pepys MB, Hawkins PN. Pathogenetic mechanisms of amyloid A amyloidosis. Proc Natl Acad Sci USA. 2013;110(40):16115–16120. doi: 10.1073/pnas.1306621110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Westermark GT, Fändrich M, Westermark P. AA amyloidosis: pathogenesis and targeted therapy. Annu Rev Pathol. 2015;10:321–344. doi: 10.1146/annurev-pathol-020712-163913. [DOI] [PubMed] [Google Scholar]
- 21.Takase H, Tanaka M, Miyagawa S, Yamada T, Mukai T. Effect of amino acid variations in the central region of human serum amyloid A on the amyloidogenic properties. Biochem Biophys Res Commun. 2014;444(1):492–497. doi: 10.1016/j.bbrc.2014.01.029. [DOI] [PubMed] [Google Scholar]
- 22.Röcken C, Fändrich M, Stix B, Tannert A, Hortschansky P, Reinheckel T, Saftig P, Kähne T, Menard R, Ancsin JB, Bühling F. Cathepsin protease activity modulates amyloid load in extracerebral amyloidosis. J Pathol. 2006;210(4):478–487. doi: 10.1002/path.2076. [DOI] [PubMed] [Google Scholar]
- 23.Stonik JA, Remaley AT, Demosky SJ, Neufeld EB, Bocharov A, Brewer HB. Serum amyloid A promotes ABCA1-dependent and ABCA1-independent lipid efflux from cells. Biochem Biophys Res Commun. 2004;321:936–941. doi: 10.1016/j.bbrc.2004.07.052. [DOI] [PubMed] [Google Scholar]
- 24.Takase H, Furuchi H, Tanaka M, Yamada T, Matoba K, Iwasaki K, Kawakami T, Mukai T. Characterization of reconstituted high-density lipoprotein particles formed by lipid interactions with human serum amyloid A. Biochim Biophys Acta. 2014;1842:1467–1474. doi: 10.1016/j.bbalip.2014.07.012. [DOI] [PubMed] [Google Scholar]
- 25.Jayaraman S, Haupt C, Gursky O. Thermal transitions in serum amyloid A in solution and on the lipid: implications for structure and stability of acute-phase HDL. J Lipid Res. 2015;56(8):1531–1542. doi: 10.1194/jlr.M059162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liang JS, Schreiber BM, Salmona M, Phillip G, Gonnerman WA, de Beer FC, Sipe JD. Amino terminal region of acute phase, but not constitutive, serum amyloid A (apoSAA) specifically binds and transports cholesterol into aortic smooth muscle and HepG2 cells. J Lipid Res. 1996;37(10):2109–2116. [PubMed] [Google Scholar]
- 27.Wang L, Colón W. Effect of zinc, copper, and calcium on the structure and stability of serum amyloid A. Biochemistry. 2007;46(18):5562–5569. doi: 10.1021/bi602629y. [DOI] [PubMed] [Google Scholar]
- 28.Ancsin JB, Kisilevsky R. The heparin/heparan sulfate-binding site on apo-serum amyloid A. Implications for the therapeutic intervention of amyloidosis. J Biol Chem. 1999;274(11):7172–7181. doi: 10.1074/jbc.274.11.7172. [DOI] [PubMed] [Google Scholar]
- 29.Noborn F, Ancsin JB, Ubhayasekera W, Kisilevsky R, Li JP. Heparan sulfate dissociates serum amyloid A (SAA) from acute-phase high-density lipoprotein, promoting SAA aggregation. J Biol Chem. 2012;287(30):25669–25677. doi: 10.1074/jbc.M112.363895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Han CY, Tang C, Guevara ME, Wei H, Wietecha T, Shao B, Subramanian S, Omer M, Wang S, O’Brien KD, Marcovina SM, Wight TN, Vaisar T, de Beer MC, de Beer FC, Osborne WR, Elkon KB, Chait A. Serum amyloid A impairs the anti-inflammatory properties of HDL. J Clin Invest. 2016;126(1):266–281. doi: 10.1172/JCI83475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Spodzieja M, Rafalik M, Szymańska A, Kołodziejczyk AS, Czaplewska P. Interaction of serum amyloid A with human cystatin C--assessment of amino acid residues crucial for hCC-SAA formation (part II) J Mol Recognit. 2013;26(9):415–425. doi: 10.1002/jmr.2283. [DOI] [PubMed] [Google Scholar]
- 32.Preciado-Patt L, Levartowsky D, Prass M, Hershkoviz R, Lider O, Fridkin M. Inhibition of cell adhesion to glycoproteins of the extracellular matrix by peptides corresponding to serum amyloid A. Toward understanding the physiological role of an enigmatic protein. Eur J Biochem. 1994;223(1):35–42. doi: 10.1111/j.1432-1033.1994.tb18963.x. [DOI] [PubMed] [Google Scholar]
- 33.Urieli-Shoval S, Shubinsky G, Linke RP, Fridkin M, Tabi I, Matzner Y. Adhesion of human platelets to serum amyloid A. Blood. 2002;99(4):1224–1229. doi: 10.1182/blood.v99.4.1224. [DOI] [PubMed] [Google Scholar]
- 34.Cai L, de Beer MC, de Beer FC, van der Westhuyzen DR. Serum amyloid A is a ligand for scavenger receptor class B type I and inhibits high density lipoprotein binding and selective lipid uptake. J Biol Chem. 2005;280(4):2954–2961. doi: 10.1074/jbc.M411555200. [DOI] [PubMed] [Google Scholar]
- 35.van der Westhuyzen DR, Cai L, de Beer MC, de Beer FC. Serum amyloid A promotes cholesterol efflux mediated by scavenger receptor B-I. J Biol Chem. 2005;280:35890–35895. doi: 10.1074/jbc.M505685200. [DOI] [PubMed] [Google Scholar]
- 36.Baranova IN, Bocharov AV, Vishnyakova TG, Kurlander R, Chen Z, Fu D, Arias IM, Csako G, Patterson AP, Eggerman TL. CD36 is a novel serum amyloid A (SAA) receptor mediating SAA binding and SAA-induced signaling in human and rodent cells. J Biol Chem. 2010;285(11):8492–8506. doi: 10.1074/jbc.M109.007526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tomita T, Ieguchi K, Sawamura T, Maru Y. Human serum amyloid A3 (SAA3) protein, expressed as a fusion protein with SAA2, binds the oxidized low density lipoprotein receptor. PLoS One. 2015;10(3):e0118835. doi: 10.1371/journal.pone.0118835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang Y, Srinivasan S, Ye Z, Javier Aguilera J, Lopez MM, Colón W. Serum amyloid A 2.2 refolds into a octameric oligomer that slowly converts to a more stable hexamer. Biochem Biophys Res Commun. 2011;407(4):725–729. doi: 10.1016/j.bbrc.2011.03.090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Colón W, Aguilera JJ, Srinivasan S. Intrinsic stability, oligomerization, and amyloidogenicity of HDL-free serum amyloid A. Adv Exp Med Biol. 2015;855:117–134. doi: 10.1007/978-3-319-17344-3_5. [DOI] [PubMed] [Google Scholar]
- 40.Lu J, Yu Y, Zhu I, Cheng Y, Sun PD. Structural mechanism of serum amyloid A-mediated inflammatory amyloidosis. Proc Natl Acad Sci USA. 2014;111(14):5189–5194. doi: 10.1073/pnas.1322357111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ferreon AC, Ferreon JC, Wright PE, Deniz AA. Modulation of allostery by protein intrinsic disorder. Nature. 2013;498(7454):390–394. doi: 10.1038/nature12294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Uversky VN. Protein misfolding in lipid-mimetic environments. Adv Exp Med Biol. 2015;855:33–66. doi: 10.1007/978-3-319-17344-3_2. [DOI] [PubMed] [Google Scholar]
- 43.Chetty PS, Mayne L, Lund-Katz S, Stranz D, Englander SW, Phillips MC. Helical structure and stability in human apolipoprotein A-I by hydrogen exchange and mass spectrometry. Proc Natl Acad Sci USA. 2009;106(45):19005–19010. doi: 10.1073/pnas.0909708106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen J, Li Q, Wang J. Topology of human apolipoprotein E3 uniquely regulates its diverse biological functions. Proc Natl Acad Sci USA. 2011;108(36):14813–14818. doi: 10.1073/pnas.1106420108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Segrest JP, Jones MK, De Loof H, Brouillette CG, Venkatachalapathi YV, Anantharamaiah GM. The amphipathic helix in the exchangeable apolipoproteins: a review of secondary structure and function. J Lipid Res. 1992;33(2):141–166. [PubMed] [Google Scholar]
- 46.Borhani DW, Rogers DP, Engler JA, Brouillette CG. Crystal structure of truncated human apolipoprotein A-I suggests a lipid-bound conformation. Proc Natl Acad Sci USA. 1997;94(23):12291–12296. doi: 10.1073/pnas.94.23.12291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mei X, Atkinson D. Crystal structure of C-terminal truncated apolipoprotein A-I reveals the assembly of high density lipoprotein (HDL) by dimerization. J Biol Chem. 2011;286(44):38570–38582. doi: 10.1074/jbc.M111.260422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Deng X, Walker RG, Morris J, Davidson WS, Thompson TB. Role of conserved proline residues in human apolipoprotein A-IV structure and function. J Biol Chem. 2015;290(17):10689–10702. doi: 10.1074/jbc.M115.637058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gursky O. Crystal structure of Δ(185–243)ApoA-I suggests a mechanistic framework for the protein adaptation to the changing lipid load in good cholesterol: from flatland to sphereland via double belt, belt buckle, double hairpin and trefoil/tetrafoil. J Mol Biol. 2013;425:1–16. doi: 10.1016/j.jmb.2012.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Phillips MC. New insights into the determination of HDL structure by apolipoproteins: Thematic review series: high density lipoprotein structure, function, and metabolism. J Lipid Res. 2013;54(8):2034–2048. doi: 10.1194/jlr.R034025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Patel H, Bramall J, Waters H, De Beer MC, Wo P. Expression of recombinant human serum amyloid A in mammalian cells and demonstration of the region necessary for high-density lipoprotein binding and amyloid fibril formation by site directed mutagenesis. Biochem J. 1996;318:1041–1049. doi: 10.1042/bj3181041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Villarroel F, Casado A, Vásquez J, Matamala E, Araneda B, Amthauer R, Enriquez R, Concha MI. Serum amyloid A: a typical acute-phase reactant in rainbow trout? Dev Comp Immunol. 2008;32(10):1160–1169. doi: 10.1016/j.dci.2008.03.004. [DOI] [PubMed] [Google Scholar]
- 53.Rossmann C, Windpassinger C, Brunner D, Kovacevic A, Schweighofer N, Malli R, Schuligoi R, Prokesch A, Kluve-Beckerman B, Graier WF, Kratky D, Sattler W, Malle E. Characterization of rat serum amyloid A4 (SAA4): a novel member of the SAA superfamily. Biochem Biophys Res Commun. 2014;450(4):1643–1649. doi: 10.1016/j.bbrc.2014.07.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Das M, Gursky O. Amyloid-forming properties of human apolipoproteins: Sequence analyses and structural insights. Adv Exp Med Biol. 2015;855:175–211. doi: 10.1007/978-3-319-17344-3_8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang L, Colón W. The interaction between apolipoprotein serum amyloid A and high-density lipoprotein. Biochem Biophys Res Commun. 2004;317(1):157–161. doi: 10.1016/j.bbrc.2004.03.027. [DOI] [PubMed] [Google Scholar]
- 56.Malle E, Herz R, Artl A, Ibovnik A, Andreae F, Sattler W. Mapping of antigenic determinants of purified, lipid-free human serum amyloid A proteins. Scand J Immunol. 1998;48(5):557–561. doi: 10.1046/j.1365-3083.1998.00439.x. [DOI] [PubMed] [Google Scholar]
- 57.Yamada T, Sato J, Kotani K, Tanaka M. Influence of polymorphism on glycosylation of serum amyloid a4 protein. Biochem Res Int. 2014;2014:527254. doi: 10.1155/2014/527254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kisilevsky R, Tam SP. Macrophage cholesterol efflux and the active domains of serum amyloid A 2.1. J Lipid Res. 2003;44(12):2257–2269. doi: 10.1194/jlr.M300133-JLR200. [DOI] [PubMed] [Google Scholar]
- 59.Motlagh HN, Wrabl JO, Li J, Hilser VJ. The ensemble nature of allostery. Nature. 2014;508(7496):331–339. doi: 10.1038/nature13001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vaisar T, Tang C, Babenko I, Hutchins P, Wimberger J, Suffredini AF, Heinecke JW. Inflammatory remodeling of the HDL proteome impairs cholesterol efflux capacity. J Lipid Res. 2015;56(8):1519–1530. doi: 10.1194/jlr.M059089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shiflett AM, Bishop JR, Pahwa A, Hajduk SL. Human high density lipoproteins are platforms for the assembly of multi-component innate immune complexes. J Biol Chem. 2005;280(38):32578–3285. doi: 10.1074/jbc.M503510200. [DOI] [PubMed] [Google Scholar]
- 62.Heinecke JW. The protein cargo of HDL: implications for vascular wall biology and therapeutics. J Clin Lipidol. 2010;4(5):371–375. doi: 10.1016/j.jacl.2010.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shah AS, Tan L, Long JL, Davidson WS. Proteomic diversity of high density lipoproteins: our emerging understanding of its importance in lipid transport and beyond. J Lipid Res. 2013;54(10):2575–2585. doi: 10.1194/jlr.R035725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Stevens FJ. Hypothetical structure of human serum amyloid A protein. Amyloid. 2004;11(2):71–80. doi: 10.1080/13506120412331272296. [DOI] [PubMed] [Google Scholar]
- 65.Sievers F, Wilm A, Dineen DG, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Söding J, Thompson JD, Higgins D. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Systems Bio. 2011;7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gasteiger E, Hoogland C, Gattiker A, Duvaud S, Wilkins MR, Appel RD, Bairoch A. Protein identification and analysis tools on the ExPASy server. In: Walker JM, editor. The Proteomics Protocols Handbook. Humana Press; 2005. pp. 571–607. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.