Abstract
Uptake of copper (Cu) ions into human cells is mediated by the plasma membrane protein Ctr1 and is followed by Cu transfer to cytoplasmic Cu chaperones for delivery to Cu-dependent enzymes. The C-terminal cytoplasmic tail of Ctr1 is a 13-residue peptide harboring an HCH motif that is thought to interact with Cu. We here employ biophysical experiments under anaerobic conditions in peptide models of the Ctr1 C-terminus to deduce Cu-binding residues, Cu affinity, and the ability to release Cu to the cytoplasmic Cu chaperone Atox1. Based on NMR assignments and bicinchoninic acid competition experiments, we demonstrate that Cu interacts in a 1:1 stoichiometry with the HCH motif with an affinity, KD, of ∼10−14 M. Removing either the Cys residue or the two His residues lowers the Cu-peptide affinity, but site specificity is retained. The C-terminal peptide and Atox1 do not interact in solution in the absence of Cu. However, as directly demonstrated at the residue level via NMR spectroscopy, Atox1 readily acquires Cu from the Cu-loaded peptide. We propose that Cu binding to the Ctr1 C-terminal tail regulates Cu transport into the cytoplasm such that the metal ion is only released to high-affinity Cu chaperones.
Introduction
Copper (Cu) ions act as essential cofactors in proteins that facilitate cellular reactions such as respiration, antioxidant defense, neurotransmitter biosynthesis, connective-tissue biosynthesis, and pigment formation (1, 2, 3). Since free Cu is toxic, the intracellular concentration of Cu is regulated via dedicated proteins that facilitate its uptake, efflux, and distribution to target Cu-dependent proteins and enzymes (4, 5, 6). In human cells, Cu is translocated from the plasma to the cytoplasm by the trimeric membrane-spanning protein Ctr1 (5, 7, 8).
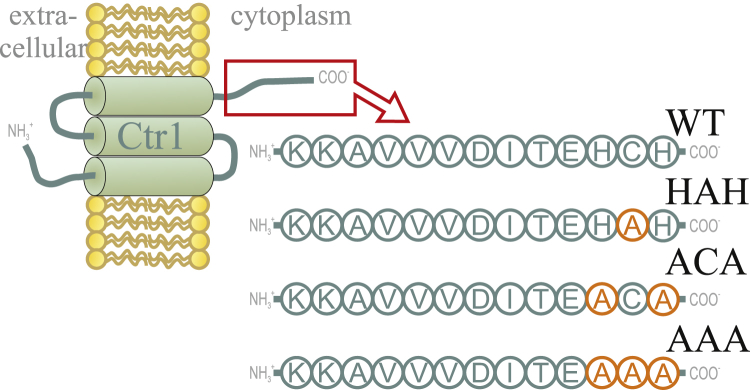
Each Ctr1 polypeptide is 190 residues long. In the assembled trimer, 67 of the amino acids in the N-terminus are extracellular and the last ∼13 residues in the C-terminus are intracellular; the rest of the residues constitute three transmembrane domains (9) (Fig. 1). Based on the 7 Å cryo-electron microscopy structure, the Ctr1 trimer contains a narrow pore at the plasma membrane that widens into a vestibule on the intracellular side (10). Although the N-terminus is rich in Met and His residues, the membrane-spanning pore appears lined with Met triads that transiently interact with Cu (11). It has been hypothesized that once Cu has been transferred through the Ctr1 channel, a recipient protein will bind Cu immediately. Recipient proteins are thought to be the cytoplasmic Cu chaperones Atox1 and CCS, responsible for independent Cu transfer pathways a) to the copper-transporting P1B-type ATPases ATP7A (i.e., Menke disease protein) and ATP7B (i.e., Wilson disease protein) in the trans-Golgi network and b) to cytoplasmic Cu, Zn superoxide dismutase (SOD1), respectively.
Figure 1.
Schematic model of a membrane-spanning Ctr1 monomer with an extracellular N-terminus, three transmembrane domains, and the C-terminal tail on the cytosolic side (red box). The C-terminal WT sequence and the three variants are enlarged. Positions for alanine mutations are highlighted in orange. Peptides are indicated by the abbreviations used here (WT, HAH, ACA, and AAA). To see this figure in color, go online.
Since CCS possesses the capacity to interact with lipid bilayers, membrane scaffolding may be a mechanism that facilitates interactions between Ctr1 and cytoplasmic chaperones (12). As an alternative to direct Ctr1-chaperone interactions, it was proposed that glutathione may act as an intermediate Cu-binding molecule bridging between Ctr1 and the cytoplasmic chaperones (13).
In yeast, Ctr1 has a cytosolic 125-residue C-terminal domain with six cysteine residues that has been shown to interact with the yeast Atox1 homolog, Atx1, and exchange Cu (14, 15). In contrast to yeast, in human Ctr1, the cytoplasmic C-terminus is only a short peptide that nonetheless contains the putative metal-binding motif His-Cys-His (HCH) (9). The HCH motif has been proposed to regulate and/or restrict Ctr1-mediated Cu entry into the cytoplasm, as mutation of this site in the full-length protein (HCH substituted to AAA, or truncation of the C-terminus) substantially increased the rate of Cu transport into human cells (16).
To directly address the Cu(I) binding capacity of the human Ctr1 C-terminus in vitro, we have here applied spectroscopic methods to a set of variant peptides at anaerobic conditions (Fig. 1). The experiments performed demonstrate that Cu(I) binds to the HCH motif, with all three residues contributing to the interaction, and with an intermediate affinity that allows for efficient Cu transfer to the apo-form of Atox1 in solution.
Materials and Methods
Materials, conditions, and sample preparations
Sample buffer was 30 mM MOPS with 50 mM sodium chloride at pH 7.5 in all experiments unless otherwise stated. Four peptides (Fig. 1) were purchased that consisted of the 13 most C-terminal amino acids of the human Ctr1 polypeptide (GenScript USA, Piscataway, NJ). Peptide sequences were KKAVVVDITEHCH (WT), KKAVVVDITEHAH (HAH variant), KKAVVVDITEACA (ACA variant), and KKAVVVDITEAAA (AAA variant) (mutations are underlined). Lyophilized peptides were dissolved in degassed sample buffer and aliquoted under anaerobic conditions in a nitrogen-equilibrated glove box. All further preparation steps were performed under anaerobic conditions as well. The stability of anaerobic conditions was monitored via dithiothreitol (DTT) solutions stored for a couple of days in the glove box. The concentration of nonoxidized thiols was determined by Ellman's assay (see below) as a function of time in the glove box. The concentrations of the peptide stocks were determined as 4.2 mM (for AAA) and 4.6 mM (for HAH) by amino acid analysis at the Amino Acid Analysis Center, Uppsala University. For the cysteine-containing samples (WT and ACA variants), Ellman’s assay provided stock concentrations of 1.2 mM (for WT) and 3.0 mM (for ACA). A stock solution of 1 M CuCl in 37% hydrochloric acid was anaerobically prepared and stored. Further dilution steps with sample buffer were performed directly before experiments. For determination of Cu(I) concentration, the bicinchoninic acid (BCA) assay (see below for details; measuring the absorbance at a wavelength of 562 nm) was used. Solutions of different concentrations of CuCl2 (10–200 μM) were reduced with 300 μM DTT and incubated for 2 h. After addition of 25.7 mM BCA and 1 h incubation time, the absorbance was measured and plotted against Cu(I) concentration. Competition between DTT and BCA was neglected due to the 100-fold higher concentration of BCA. A linear fit of the data points yielded an extinction coefficient of ε562 = 7100 cm−1 M−1 for the BCA2-Cu complex (Fig. S1 C in the Supporting Material).
Using this reference experiment, the concentration of the CuCl stock solution was controlled by mixing with 25.7 mM BCA and measuring absorbance before every single set of experiments. In addition, anaerobic titration of 100 μM BCA with Cu(I) confirmed the assumed Cu(I) concentration. As expected, because of the 2:1 BCA/Cu complex stoichiometry, a linear increase in absorption due to Cu(I) binding to BCA exhibited saturation at half the BCA concentration (Fig. S1 A). Atox1 was expressed and purified as previously reported (17, 18). After the established purification procedure including DTT-containing buffer, the protein sample was transferred to the glove box and a buffer exchange was conducted on a desalting PD10 column (GE Healthcare, Little Chalfont, United Kingdom) with degassed DTT-free sample buffer. The elution profile was analyzed for free thiols with Ellman’s reagent, demonstrating a clear separation of the protein from free DTT (Fig. S1 B). Both Ellman’s assay (determining the concentration of free thiols of Atox1; see details below) and protein absorbance at 280 nm (ε280 = 2980 cm−1 M−1) gave a concentration of 850 μM (±3.5%) for the Atox1 stock solution.
Circular dichroism spectroscopy
A total volume of 1 mL of 100 μM peptide sample was prepared in a 1 cm quartz cuvette (Hellma, Müllheim, Germany) and titrated with 5 mM CuCl solution in the glove box. Between each titration step the cuvette was sealed with a vacuum grease-coated cap and circular dichroism (CD) and absorption spectra were measured outside the glove box.
Ellman’s assay
5,5′-dithiobis-(2-nitrobenzoic acid) (DTNB), or Ellman’s reagent, was purchased from Sigma Aldrich (St. Louis, MO) and prepared as a 10 mM stock solution in degassed sample buffer. For determination of free thiols, each sample was mixed with DTNB and absorbance at 412 nm was measured. For quantitative data analysis, reference absorbance values using known concentrations of l-cysteine were determined. The reference line yielded an extinction coefficient of ε412 = 14,000 cm−1 M−1.
BCA competition assay
BCA was purchased as ready-to-use reagent A of the BCA Protein Assay Kit (Thermo Fisher Scientific, Waltham, MA). This stock solution of 25.7 mM BCA was degassed and transferred to the glove box. Further dilutions in sample buffer were performed anaerobically. For each data point in the peptide titrations to BCA2-Cu, a 100 μL sample was prepared with 100 μM BCA, 44 μM CuCl, and peptide of different concentrations. CuCl was always added as the last step, followed by incubation for 2 h in the glove box before samples were transferred from the glove box to the spectrophotometer. BCA2-Cu absorbance was measured at 562 nm (19).
NMR spectroscopy
For NMR experiments, sealable NMR tubes (Hellma) were used. Total volumes of 600 μL, including 25 μL D2O, were prepared anaerobically, sealed, and transferred to the NMR spectrometer. All NMR experiments were conducted at a Bruker Avance III HD 850 spectrometer equipped with a TCI cryoprobe. The Hα-Cα correlations for the Ctr1 WT peptide were assigned using two-dimensional (2D) homonuclear nuclear Overhauser effect spectroscopy and total correlation spectroscopy experiments at T = 288 K and a concentration of 1 mM. NMR diffusion experiments were performed at T = 298 K and a peptide concentration of 300 μM using a pulse sequence comprising a stimulated echo assisted by bipolar gradients (20), G, employing a diffusion time, Δ, of 50 ms and gradient length, δ, of 6 ms along the z axis. Gradients were calibrated as described in Berger and Braun (21). Integrals for proton signals, I, were determined in the spectral range between 6 and 9 ppm and used for calculation of the diffusion coefficient, D:
where γ is the gyromagnetic ratio. The Cu(I) interaction studies of WT, HAH variant, and AAA variants were performed at T = 288 K at a concentration of 120 μM (WT) and 350 μM (HAH and AAA), respectively. Cu(I) was added from a stock solution of 10 mM CuCl to avoid dilution of the peptide. The one-dimensional proton-observed titration of Cu(I) to HAH variant was performed at T =288 K using an initial peptide concentration of 300 μM and stepwise additions of CuCl aliquots from a 10 mM stock. Two-dimensional 1H-15N HSQC experiments were performed at T = 288 K using Atox1 concentrations of 100 μM (apo condition), 200 μM (when Cu(I) was added), or 350 μM (when apo HAH variant or Cu-loaded HAH variant was added). Additional interaction studies between Atox1 and the WT peptide in the presence or absence of Cu(I) were conducted at T = 288 K at a Bruker Avance III HD 600 spectrometer equipped with a cryogenic BBO probe head. Sample concentrations were 60 μM (apo Atox1) and 120 μM (apo WT peptide) in the absence of Cu(I) and, for Cu transfer analysis, 175 μM (apo Atox1) and 350 μM WT peptide premixed with 175 μM Cu(I).
Results
Ctr1 C-terminal peptides bind Cu(I)
Laboratory experiments with Cu(I) pose the challenge to keep the copper ions in the reduced state throughout the experiments. Whereas many experiments with Cu chaperones such as Atox1 and CCS can be performed under normal atmosphere with DTT as reducing agent (22, 23, 24, 25), this approach only works for proteins with high Cu(I) affinity. As DTT itself has high affinity for Cu(I) (dissociation constant, KD = 10−15 M (26)), it can act as competitor to proteins and peptides with lower or comparable Cu affinities. This was noted for the Ctr1 C-terminal peptide in our first experiments, where we used aerobic DTT conditions (data not shown). In the presence of a fivefold excess of DTT over peptide, there was no Cu binding detected to the WT Ctr1 peptide (by absorbance, CD, or NMR), implying that the peptide had a Cu(I) affinity similar to or weaker than that of DTT.
To overcome this problem, we turned to a cleaner sample preparation, without reducing agents, performed in a glove box with an oxygen-free nitrogen atmosphere and degassed buffers. Cuvettes and sample tubes for spectroscopic analyses were always filled inside the glove box and tightly sealed, keeping oxygen-free conditions also outside the glove box for the duration of the measurements. The latter was confirmed with control experiments using vials of DTT and CuCl sealed in the glove box but then stored outside the glove box and tested for free thiols (DTNB) and reduced Cu (BCA) as a function of time. For at least 2–3 days, there was no change in the spectroscopic values, indicating that the conditions remained anaerobic during this time frame. Nevertheless, as Cu(I) under neutral pH tends to precipitate, copper solutions were always diluted into sample buffer right before experiments.
For the WT Ctr1 peptide, CD spectra measured after anaerobic titration with Cu(I) revealed spectral changes in the 230–280 nm region (reporting on tertiary interactions and Cu-Cys interactions), and absorption data exhibited increased absorbance in the wavelength range 250–350 nm. Both observations implied Cu(I) binding to the peptide. The signal changes appeared to saturate when Cu(I) was added in a 1:1 molar ratio, suggesting a 1:1 binding stoichiometry (Fig. S2). Similar spectral changes were observed upon titrating Cu(I) to the ACA variant (data not shown).
Peptide affinities for Cu(I) through BCA competition
To determine the affinities of the Ctr1 C-terminal peptides (P) for binding of Cu(I) (metal ion, M) we used a competition assay (Eq. 1) with BCA as the competing chromophoric ligand (L) for Cu(I).
| (1) |
As the complex of two BCA molecules with one Cu(I) ion possesses a high absorption band at λ = 562 nm, changes in the BCA2-Cu complex concentration due to competition with another ligand (here, the different peptides) for Cu(I) will be detectable by absorption changes. The principal idea was introduced by Xiao et al. (19) and successfully applied to various Cu(I)-binding proteins (26). Briefly, BCA is loaded with Cu(I) in a BCA/Cu(I) molar ratio of 2.5 at anaerobic conditions. This ensures that all Cu(I) ligand is found in the chromophoric 2:1 complex with BCA, with negligible amounts of the 1:1 complex. The Cu-loaded BCA is then titrated with the peptide of interest (L in Eq. 1) while (through dilutions) keeping the concentrations of Cu(I) and BCA constant. As addition of increasing amounts of peptide will compete for Cu(I), the concentration of the chromophoric BCA2-Cu complex decreases. The experimentally determined decrease in absorbance at λ = 562 nm is plotted against the ratio of peptide to Cu(I). Equation 2 can be used to fit the experimental data:
| (2) |
with [L]total for concentration of BCA and [P]total for the peptide concentration. [MP] and [ML2] are calculated on the basis of absorbance changes normalized to the values without peptide (index 0):
| (3) |
| (4) |
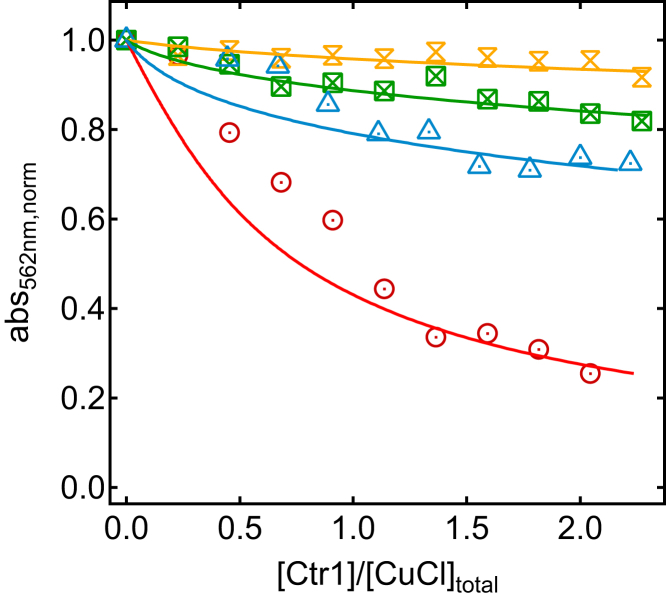
To derive the dissociation constant, KD, for each Cu-peptide complex, we used the carefully determined value of β2 = 1017.2 M−2 as the formation constant for the copper complex with BCA, BCA2-Cu (26). Titration data for the four peptides to BCA2-Cu at anaerobic conditions with accompanying fits to Eq. 2 are shown in Fig. 2, and KD values are reported in Table 1. (We note that there are deviations between the experimental data points and two of the fits at low Ctr1 peptide concentrations.)
Figure 2.
Anaerobic BCA2-Cu competition assay providing Cu(I) affinities for WT peptide (red circles) and peptide variants, HAH (green squares), ACA (blue triangles), and AAA (yellow double triangles). Each sample was prepared anaerobically and Cu(I) was added last, followed by incubation for 2 h before absorption measurements at 562 nm. The whole process was performed at strict anaerobic conditions. Absorbance values were normalized to the value measured for the solution without peptide addition. Results for the best fits of Eq. 2 to the experimental data sets are shown as colored curves (corresponding KD values are reported in Table 1). To see this figure in color, go online.
Table 1.
Cu(I) Dissociation Constants
| Peptide Variant | KD (10−14 M) |
|---|---|
| WT | 2.3 ± 0.2 |
| HAH | 400 ± 30 |
| ACA | 90 ± 10 |
| AAA | ND |
Equation 2 was fitted to the experimental data (Fig. 2) with substitution of Eqs. 3 and 4 using β2 = 1017.2 M−2 as the Cu-BCA2 complex formation constant. ND, not determined.
Whereas the WT peptide competes efficiently with BCA for Cu(I), as visualized by the decreasing absorbance values in the titration, this effect is dramatically attenuated for all three peptide variants with the HCH motif perturbed, and for the AAA peptide, there is in essence no change. For WT, a dissociation constant of KD = (2.3 ± 0.2) × 10−14 M was determined (Table 1). Exchange of the two His to Ala (ACA variant) caused a 45-fold loss in affinity, whereas exchange of the Cys for Ala (HAH variant) decreased the affinity more than two orders of magnitude compared to WT (Table 1). For the AAA variant, because of the small changes in absorption, no affinity value was determined; instead, we concluded that the affinity of this peptide for Cu must be dramatically weaker than for the other peptides.
NMR analysis of Cu(I) binding
The NMR diffusion profile for the WT peptide (Fig. S3) corresponds to a diffusion coefficient of D = (1.96 ± 0.01) × 10−10 m2 s−1, which can be translated to a hydrodynamic radius, rH, of 11.1 Å by using the Stokes-Einstein relationship. In accord with a monomeric state, a rough estimation of rH for an unstructured peptide of 13 residues yields an rH of 9.5 Å (27). For comparison, an unstructured trimer (39 residues) would correspond to an rH of 17.8 Å (27).
One-dimensional NMR was used to confirm Cu(I) interaction with WT and HAH peptides. Cu(I) titration to the HAH variant while monitoring the amide proton signals is shown in Fig. S4. The NMR data demonstrate that there are Cu(I)-peptide interactions, in accord with the data obtained using CD and absorption spectroscopy.
Upon assignments of NMR chemical shifts, residue-specific information on where the copper ion interacts can be obtained. By employing 1H-13C HSQC analysis of Hα-Cα correlations, 12 residues of the WT peptide could be assigned by homonuclear 1H-1H nuclear Overhauser effect spectroscopy, 1H-1H total correlation spectroscopy, and heteronuclear 1H-13C HSQC experiments (with Lys1 missing; see Table S1 for NMR assignments). Comparison of 1H-13C HSQC spectra for WT and the HAH variants revealed that chemical-shift changes occurred only close to the mutation site (Cys12) of the HAH variant as compared to WT (Fig. 3).
Figure 3.
Comparison of 1H-13C HSQC spectra for Hα-Cα correlations for the WT peptide (black) and the HAH variant (red). Assignments of the residues in the WT (black) are shown using the single-letter amino acid code. The Hα-Cα correlation for Ala12 of the HAH variant is shown in red. To see this figure in color, go online.
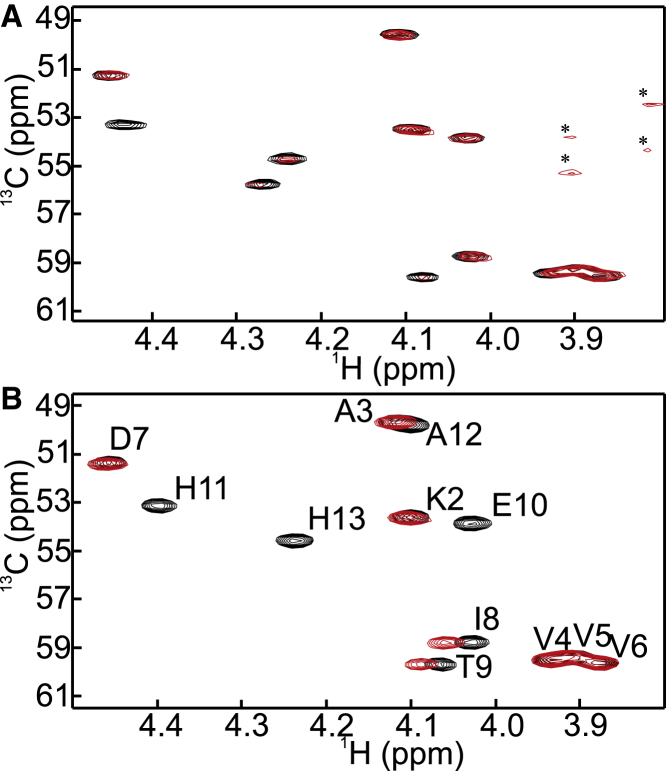
Loss of Hα-Cα signal intensities at the C-terminus of the WT peptide was found upon addition of Cu(I) (Fig. 4 A). Also, loss of signal intensities and changes of chemical shifts were found at the C-terminus of the HAH variant upon addition of Cu(I) (Fig. 4 B). Generally, an interaction may induce 1) a shift of cross signals (fast exchange on the NMR timescale), 2) line broadening of signals (intermediate exchange on the NMR timescale), or 3) a decrease of original peaks and an increase in new peaks (slow exchange on the NMR timescale). Because we observe mostly intensity decreases, along with some broadening, the collected data suggest that Cu(I) interacts with the Ctr1 C-terminus on the slow to intermediate NMR timescale.
Figure 4.
(A) Impact of Cu(I) addition to Hα-Cα correlations of the WT peptide (apo, black; Cu(I)-loaded (holo), red). Cu(I) was added in an equal molar amount, and noise signals of the holo state are indicated by asterisks. (B) Impact of Cu(I) addition to Hα-Cα correlations of the HAH peptide (apo, black; holo, red). Cu(I) was added in an equal molar amount and assignments for the apo state variant are shown. To see this figure in color, go online.
In Fig. 5, the intensity decreases of Hα-Cα correlations for the Cu-loaded WT (Fig. 5 A) and the HAH variant (Fig. 5 B) compared to the apo form are plotted as a function of peptide sequence. The data reveal that Cu(I) binds at the C-terminal residues in both peptide variants. For WT, the highest-intensity attenuation is for Cys12 and His13, whereas upon Cys to Ala mutation, the Cu(I) interaction affects peak intensity all the way from His13 to Glu10, which may imply that Cu(I) utilizes the glutamic acid as a substitute of the removed Cys12. However, since such a side effect is also found for Cu(I) interaction with wild-type Ctr1 (decreased intensity of Thr9 and Glu10), we speculate that this is an indirect effect of nearby Cu(I) coordination. As an important note, amide proton signals of peptides are generally highly sensitive to exchange with the solvent. Consequently, nonexchangeable signals (such as the Hα-Cα correlations used here) should be used for quantitative analysis.
Figure 5.
(A) Intensity decrease of the Hα-Cα correlations of Cu(I)-loaded WT peptide relative to the apo form of WT. Missing assignment of Lys1 is marked by an asterisk. (B) Intensity decrease of Hα-Cα correlations of Cu(I)-loaded HAH variant relative to the apo form of HAH. Missing assignment of Lys1 is marked by an asterisk.
The assignment of Hα-Cα correlations for Ctr1 WT cannot be directly transferred to the AAA variant, as the presence of three mutations strongly changes the chemical shifts in the 2D 1H-13C HSQC spectrum (Ala11-Ala13 chemical shifts are ambiguous; Table S1 and Fig. S5 A). Nonetheless, spectral analysis yields only a weak response upon Cu(I) addition to the AAA variant, showing signal loss of only two chemical shifts (i.e., Lys2 and one of three ambiguous Ala residues; Fig. S5 B). Thus, in agreement with the BCA competition experiments, the AAA variant does not appear to have specific affinity for Cu(I) above that involving weak nonspecific interactions. Because of poor solubility, we could not perform corresponding NMR experiments with the ACA peptide.
Cu-mediated peptide interaction with Atox1
We selected the HAH variant for subsequent 13C-based NMR experiments in the presence of Atox1, because the observed perturbations of Hα-Cα correlations (chemical shifts and signal intensities) in the 2D 1H-13C HSQC upon Cu(I) addition were more pronounced for the HAH variant than for the WT peptide.
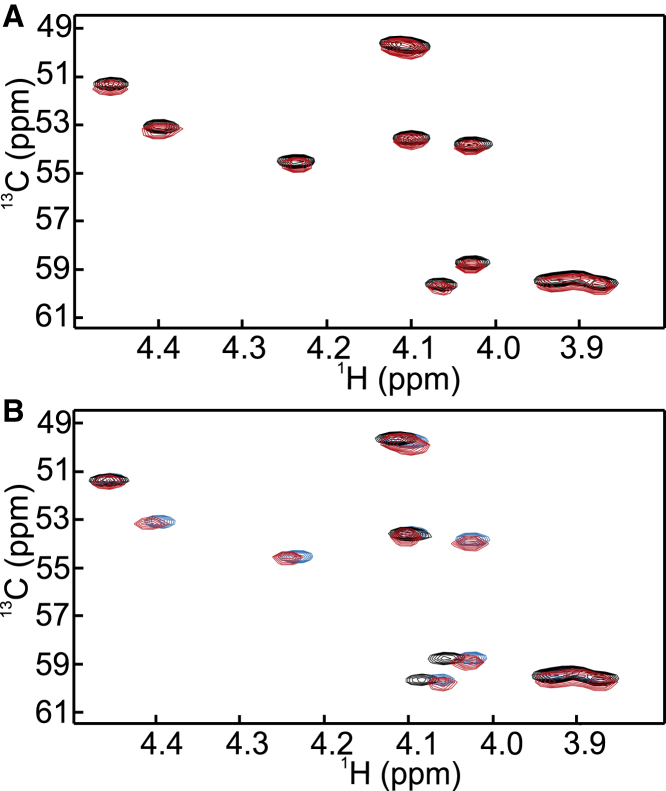
Atox1 does not interact with the HAH variant in the absence of Cu(I) when analyzed via the peptide Hα-Cα chemical shifts (Fig. 6 A). In contrast, addition of Atox1 to the Cu(I)-loaded Ctr1 HAH variant results in Hα-Cα correlations identical to those seen for the apo state of the HAH variant (Fig. 6 B). This implies that Atox1 takes the Cu ion from the metal-loaded peptide, based on observations via the peptide signals.
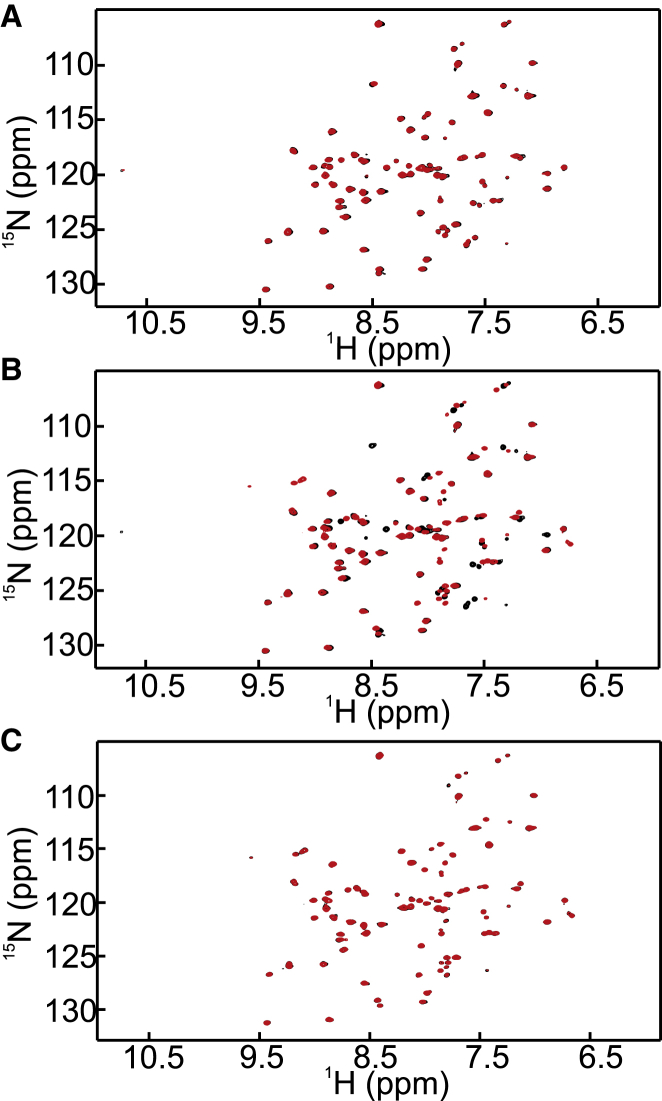
Figure 6.
(A) Comparison of 1H-13C HSQC spectra for Hα-Cα correlations between the HAH variant (black) and the HAH variant mixed with an equal molar amount of Atox1 (red). No Cu(I) is present. (B) Comparison of 1H-13C HSQC spectra for Hα-Cα correlations between the apo form of the HAH variant (blue), the Cu(I) loaded form of the HAH variant (black), and the Cu(I)-loaded form of HAH mixed with one equivalent of the apo form of Atox1 (red). To see this figure in color, go online.
The above reaction was also probed from the other side, i.e., by monitoring the status of Atox1 using 15N-labeled Atox1. Again, the HAH peptide did not interact with Atox1 when probed via the Atox1 chemical shifts (Fig. 7 A). When Atox1 is loaded with Cu(I), there are loss of signals and changes of chemical shifts in the HSQC spectrum of Atox1, as previously reported (28) (Fig. 7 B). Upon addition of Cu(I)-loaded HAH peptide to the apo form of Atox1, the backbone signals alter such that they now match the spectrum for Cu(I)-loaded Atox1 (Fig. 7 C). The data imply that the Cu(I) ion is transferred completely from the peptide to Atox1, which is in accord with the much higher affinity of Cu(I) for Atox1 (26) than for the HAH peptide (Table 1). When we repeated the 1H-15N HSQC experiments with the WT peptide and 15N-labeled Atox1, as expected, the same result of complete Cu(I) transfer from the Ctr1 peptide to Atox1 was found (Fig. S6, A and B).
Figure 7.
(A) Comparison of 1H-15N HSQC spectra between Atox1 (black) and Atox1 titrated with an equal amount of the HAH peptide (red); no Cu(I) is present. (B) Impact of Cu binding on the Atox1 1H-15N HSQC spectrum (apo, black; Cu-loaded (holo) state, red). (C) Comparison of 1H-15N HSQC spectra for Cu(I)-loaded Atox1 (black) and Atox1 after the addition of an equal amount of Cu(I)-loaded HAH peptide (red). To see this figure in color, go online.
Discussion
How Cu is internalized into cells by the trimeric membrane-spanning protein Ctr1 is not fully elucidated. It is believed that oxidized Cu ions are reduced extracellularly, perhaps upon binding to histidine- and methionine-rich Cu-binding sites in the extracellular Ctr1 N-terminal part, followed by shuttling of the reduced metal ion through the internal pore of the Ctr1 trimer, which is lined with methionine triads (5, 7, 8). It is proposed that the last stop for Cu in Ctr1 is the cytoplasmic C-terminus, which contains a putative Cu-binding site in the form of an HCH motif. Because Cu uptake rates by Ctr1 are increased when the C-terminus is mutated (i.e., HCH exchanged to AAA) or truncated (16), it appears that Cu binding to this site regulates Cu uptake into cells, perhaps acting as an open/closed switch of the internal pore. One may speculate that Cu(I) binding to the C-terminal site allosterically blocks further Cu(I) import such that a new Cu ion cannot be channeled through Ctr1 from the extracellular space until the metal ion at the HCH motif has been released.
The subsequent question is what biomolecule(s) obtains Cu(I) from the Ctr1 C-terminal site. There are no free Cu ions in the cytoplasm, and instead, in essence, all Cu ions present in the cytoplasm are bound either to proteins or to small chelating agents (4, 5, 6). The most obvious candidates to receive Cu(I) from Ctr1 are the various cytoplasmic Cu chaperones (CCS and Atox1, as well as the Cu chaperone providing Cu to the mitochondria, Cox17), but the reducing agent glutathione is a possible candidate as well, as its concentration in the cytoplasm is high (13). In the case of CCS, it was demonstrated in vitro that both CCS and its target Cu-dependent enzyme, SOD1, could interact with artificial membranes, and Ctr1 was found capable of delivering Cu to CCS at these membranes, although no mechanistic insight was provided (12).
Accurate estimations of protein affinities for Cu(I) are difficult, and reported values for the same or similar proteins are scattered, likely because of different experimental approaches with different ligand probes and affinity standards. In a thorough study aiming to unify Cu(I)-binding affinities of metal chaperones, the Cu(I) affinity, KD, for Atox1 was determined to be 10−17.4 M at pH 7 (19, 26). We used the same approach and affinity standards here and found a Cu(I) affinity of ∼10−14 M for the HCH-containing peptide at anaerobic conditions. Regardless of error bars, the peptide affinity is clearly much lower (i.e., three orders of magnitude) than the Atox1 affinity for Cu(I), which agrees with the observation of 100% transfer of Cu from the peptide upon mixing with apo-Atox1 at stoichiometric concentrations. Glutathione exists in high concentrations in the cytoplasm (1–10 mM) and may act as a competitor in terms of Cu(I) binding in vivo (26); moreover, parameters such as kinetics and localization may overrule equilibrium thermodynamics in vivo.
We want to emphasize that Cu exchange between the Ctr1 C-terminal peptide and Atox1 cannot proceed via metal release from the peptide followed by free metal uptake by Atox1 with a peptide affinity for Cu(I) of 10−14 M. This implies that even for the fastest on-rate constant of Cu(I) binding (assuming that diffusion controlled, kon ∼ 108 M−1 s−1), Cu(I) dissociation from the peptide would take at least 24 h (1/koff). Therefore, the only way to transport the metal to another partner within the time frame of the experiment is via direct peptide-protein interaction. This mechanism appears common among the copper transport systems (29, 30, 31, 32) and has been elucidated in detail for Atox1 delivering Cu(I) to the fourth metal-binding domain of ATP7B (22).
It appears that both the cysteine and the histidine residues contribute to Cu(I) interaction with the peptide. Cysteine sulfurs are often found as Cu(I) ligands in proteins, as is the case in Atox1 and other Cu chaperones. In contrast, histidine side chains are more often found in metal sites interacting with Cu(II). Nonetheless, linear Cu(I) binding involving two histidine imidazole rings have been reported in a dipeptide system (33). We found that removing either the cysteine or the two histidines in the Ctr1 peptide lowered the affinity by ∼200- and 45-fold, respectively. Thus, the effect is fivefold larger upon removing the cysteine versus the two histidines, implying that the cysteine bond dominates. Although the cysteine or histidine residues can be removed with retention of Cu-binding-site specificity, removing the complete HCH site appeared to abolish specific Cu(I) binding to the peptide. This result agrees with the previous finding of Cu uptake rates into cells being affected by both Ctr1 C-terminal truncation and HCH exchange to AAA (16).
Conclusion
We here show that Cu(I) at anaerobic conditions, mimicking cytoplasmic conditions, interacts with all three residues in the HCH motif in the Ctr1 C-terminus, resulting in a Cu-peptide affinity that is higher than that of Cu-glutathione but low enough to allow for efficient transfer of the metal to the high-affinity cytoplasmic Cu chaperone Atox1. We propose that direct delivery of Cu, via Cu-mediated transient interactions between the C-terminal peptide and a Cu chaperone such as Atox1, is a mechanism that facilitates control of intracellular Cu levels (that matches the need of the cytoplasmic chaperones) and specificity of targets (avoiding unwanted and harmful Cu interactions with other cellular components). Thus, the Cu transfer mechanism from the C-terminal peptide of human Ctr1 to Atox1 appears analogous to the yeast homologs, although the yeast Ctr1 C-terminus is a folded domain (125 residues) and harbors a different metal site (14).
Author Contributions
D.K., M.K., and P.W.S. designed research; D.K. and M.K. performed research and analyzed data; D.K., M.K., and P.W.S. wrote the article.
Acknowledgments
The Swedish Natural Research Council, the Knut and Alice Wallenberg Foundation, Göran Gustafsson Foundation, and Umeå University provided financial support. M.K. was supported by a postdoctoral fellowship from Deutsche Forschungsgemeinschaft (KO-4687/1-1).
Editor: H. Jane Dyson.
Footnotes
This is an open access article under the CC BY-NC-ND license (http://creativecommons.org/licenses/by-nc-nd/4.0/).
Six figures and one table are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(15)01173-X.
Contributor Information
Michael Kovermann, Email: michael.kovermann@uni-konstanz.de.
Pernilla Wittung-Stafshede, Email: pernilla.wittung@chalmers.se.
Supporting Material
References
- 1.Huffman D.L., O’Halloran T.V. Function, structure, and mechanism of intracellular copper trafficking proteins. Annu. Rev. Biochem. 2001;70:677–701. doi: 10.1146/annurev.biochem.70.1.677. [DOI] [PubMed] [Google Scholar]
- 2.Puig S., Thiele D.J. Molecular mechanisms of copper uptake and distribution. Curr. Opin. Chem. Biol. 2002;6:171–180. doi: 10.1016/s1367-5931(02)00298-3. [DOI] [PubMed] [Google Scholar]
- 3.Harris E.D. Basic and clinical aspects of copper. Crit. Rev. Clin. Lab. Sci. 2003;40:547–586. [PubMed] [Google Scholar]
- 4.O’Halloran T.V., Culotta V.C. Metallochaperones, an intracellular shuttle service for metal ions. J. Biol. Chem. 2000;275:25057–25060. doi: 10.1074/jbc.R000006200. [DOI] [PubMed] [Google Scholar]
- 5.Festa R.A., Thiele D.J. Copper: an essential metal in biology. Curr. Biol. 2011;21:R877–R883. doi: 10.1016/j.cub.2011.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Robinson N.J., Winge D.R. Copper metallochaperones. Annu. Rev. Biochem. 2010;79:537–562. doi: 10.1146/annurev-biochem-030409-143539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boal A.K., Rosenzweig A.C. Structural biology of copper trafficking. Chem. Rev. 2009;109:4760–4779. doi: 10.1021/cr900104z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim B.E., Nevitt T., Thiele D.J. Mechanisms for copper acquisition, distribution and regulation. Nat. Chem. Biol. 2008;4:176–185. doi: 10.1038/nchembio.72. [DOI] [PubMed] [Google Scholar]
- 9.Ohrvik H., Thiele D.J. How copper traverses cellular membranes through the mammalian copper transporter 1, Ctr1. Ann. N. Y. Acad. Sci. 2014;1314:32–41. doi: 10.1111/nyas.12371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.De Feo C.J., Aller S.G., Unger V.M. Three-dimensional structure of the human copper transporter hCTR1. Proc. Natl. Acad. Sci. USA. 2009;106:4237–4242. doi: 10.1073/pnas.0810286106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tsigelny I.F., Sharikov Y., Howell S.B. An all-atom model of the structure of human copper transporter 1. Cell Biochem. Biophys. 2012;63:223–234. doi: 10.1007/s12013-012-9358-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pope C.R., De Feo C.J., Unger V.M. Cellular distribution of copper to superoxide dismutase involves scaffolding by membranes. Proc. Natl. Acad. Sci. USA. 2013;110:20491–20496. doi: 10.1073/pnas.1309820110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maryon E.B., Molloy S.A., Kaplan J.H. Cellular glutathione plays a key role in copper uptake mediated by human copper transporter 1. Am. J. Physiol. Cell Physiol. 2013;304:C768–C779. doi: 10.1152/ajpcell.00417.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xiao Z., Wedd A.G. A C-terminal domain of the membrane copper pump Ctr1 exchanges copper(I) with the copper chaperone Atx1. Chem. Commun. (Camb.) 2002;(6):588–589. doi: 10.1039/b111180a. [DOI] [PubMed] [Google Scholar]
- 15.Xiao Z., Loughlin F., Wedd A.G. C-terminal domain of the membrane copper transporter Ctr1 from Saccharomyces cerevisiae binds four Cu(I) ions as a cuprous-thiolate polynuclear cluster: sub-femtomolar Cu(I) affinity of three proteins involved in copper trafficking. J. Am. Chem. Soc. 2004;126:3081–3090. doi: 10.1021/ja0390350. [DOI] [PubMed] [Google Scholar]
- 16.Maryon E.B., Molloy S.A., Kaplan J.H. Rate and regulation of copper transport by human copper transporter 1 (hCTR1) J. Biol. Chem. 2013;288:18035–18046. doi: 10.1074/jbc.M112.442426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Niemiec M.S., Dingeldein A.P., Wittung-Stafshede P. T versus D in the MTCXXC motif of copper transport proteins plays a role in directional metal transport. J. Biol. Inorg. Chem. 2014;19:1037–1047. doi: 10.1007/s00775-014-1147-0. [DOI] [PubMed] [Google Scholar]
- 18.Niemiec M.S., Weise C.F., Wittung-Stafshede P. In vitro thermodynamic dissection of human copper transfer from chaperone to target protein. PLoS One. 2012;7:e36102. doi: 10.1371/journal.pone.0036102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xiao Z., Wedd A.G. The challenges of determining metal-protein affinities. Nat. Prod. Rep. 2010;27:768–789. doi: 10.1039/b906690j. [DOI] [PubMed] [Google Scholar]
- 20.Jones J.A., Wilkins D.K., Dobson C.M. Characterization of protein unfolding by NMR diffusion measurements. J. Biomol. NMR. 1997;10:199–203. [Google Scholar]
- 21.Berger S., Braun S. Wiley-VHC; Weinheim, Germany: 2004. 200 and More NMR Experiments. A Practical Course. [Google Scholar]
- 22.Niemiec M.S., Dingeldein A.P., Wittung-Stafshede P. Enthalpy-entropy compensation at play in human copper ion transfer. Sci. Rep. 2015;5:10518. doi: 10.1038/srep10518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Palm-Espling M.E., Andersson C.D., Wittung-Stafshede P. Determinants for simultaneous binding of copper and platinum to human chaperone Atox1: hitchhiking not hijacking. PLoS One. 2013;8:e70473. doi: 10.1371/journal.pone.0070473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Petzoldt S., Kahra D., Wittung-Stafshede P. Human cytoplasmic copper chaperones Atox1 and CCS exchange copper ions in vitro. Biometals. 2015;28:577–585. doi: 10.1007/s10534-015-9832-1. [DOI] [PubMed] [Google Scholar]
- 25.Palm M.E., Weise C.F., Wittung-Stafshede P. Cisplatin binds human copper chaperone Atox1 and promotes unfolding in vitro. Proc. Natl. Acad. Sci. USA. 2011;108:6951–6956. doi: 10.1073/pnas.1012899108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xiao Z., Brose J., Wedd A.G. Unification of the copper(I) binding affinities of the metallo-chaperones Atx1, Atox1, and related proteins: detection probes and affinity standards. J. Biol. Chem. 2011;286:11047–11055. doi: 10.1074/jbc.M110.213074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wilkins D.K., Grimshaw S.B., Smith L.J. Hydrodynamic radii of native and denatured proteins measured by pulse field gradient NMR techniques. Biochemistry. 1999;38:16424–16431. doi: 10.1021/bi991765q. [DOI] [PubMed] [Google Scholar]
- 28.Anastassopoulou I., Banci L., Rosato A. Solution structure of the apo and copper(I)-loaded human metallochaperone HAH1. Biochemistry. 2004;43:13046–13053. doi: 10.1021/bi0487591. [DOI] [PubMed] [Google Scholar]
- 29.Banci L., Bertini I., Yatsunyk L.A. Metal binding domains 3 and 4 of the Wilson disease protein: solution structure and interaction with the copper(I) chaperone HAH1. Biochemistry. 2008;47:7423–7429. doi: 10.1021/bi8004736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Banci L., Bertini I., Rosato A. An NMR study of the interaction of the N-terminal cytoplasmic tail of the Wilson disease protein with copper(I)-HAH1. J. Biol. Chem. 2009;284:9354–9360. doi: 10.1074/jbc.M805981200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Banci L., Bertini I., Rosato A. Copper(I)-mediated protein-protein interactions result from suboptimal interaction surfaces. Biochem. J. 2009;422:37–42. doi: 10.1042/BJ20090422. [DOI] [PubMed] [Google Scholar]
- 32.Arnesano F., Banci L., O’Halloran T.V. Metallochaperones and metal-transporting ATPases: a comparative analysis of sequences and structures. Genome Res. 2002;12:255–271. doi: 10.1101/gr.196802. [DOI] [PubMed] [Google Scholar]
- 33.Himes R.A., Park G.Y., Karlin K.D. Synthesis and x-ray absorption spectroscopy structural studies of Cu(I) complexes of histidylhistidine peptides: the predominance of linear 2-coordinate geometry. J. Am. Chem. Soc. 2007;129:5352–5353. doi: 10.1021/ja0708013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.