Abstract
Background
Dietary exposure to cytotoxic and carcinogenic aristolochic acid (AA) causes severe nephropathy typically associated with urological cancers. Monitoring of AA exposure uses biomarkers such as aristolactam-DNA adducts, detected by mass spectrometry in the kidney cortex, or the somatic A>T transversion pattern characteristic of exposure to AA, as revealed by previous DNA sequencing studies using fresh frozen tumors.
Methods
Here we report a low-coverage whole-exome sequencing method (LC-WES) optimized for multi-sample detection of the AA mutational signature, and demonstrate its utility in 17 formalin-fixed paraffin-embedded urothelial tumors obtained from 15 patients with endemic nephropathy, an environmental form of aristolochic acid nephropathy.
Results
LC-WES identified the AA signature, alongside signatures of age and APOBEC enzyme activity, in 15 samples sequenced at the average per-base coverage of ~10x. Analysis at 3–9x coverage revealed the signature in 91% of the positive samples. The exome-wide distribution of the predominant A>T transversions exhibited a stochastic pattern whereas 83 cancer driver genes were enriched for recurrent non-synonymous A>T mutations. In two patients, pairs of tumors from different parts of the urinary tract, including the bladder, harbored overlapping mutation patterns, suggesting tumor dissemination via cell seeding.
Conclusion
LC-WES analysis of archived tumor tissues is a reliable method applicable to investigations of both the exposure to AA and its biologic effects in human carcinomas.
Impact
By detecting cancers associated with AA exposure in high-risk populations, LC-WES can support future molecular epidemiology studies and provide evidence-base for relevant preventive measures.
Keywords: aristolochic acid, urothelial carcinoma, formalin-fixed paraffin-embedded tumors, whole-exome sequencing, mutational signature
INTRODUCTION
The International Agency for Research on Cancer (IARC) classified aristolochic acid (AA) as a Group 1 carcinogen (1). Exposure to AA, following intake of Aristolochia herbaceous plants as traditional medicines or due to consumption of bread from flour contaminated by Aristolochia seeds, can lead to aristolochic acid nephropathy (AAN). AAN is a progressive tubulo-interstitial nephropathy with high risk of developing upper tract urothelial carcinoma (UTUC) (2–5). Additionally, recent studies proposed AA as a factor contributing to the development of hepatocellular (6–8), renal cell (9, 10) and urinary bladder carcinomas (11) and intrahepatic cholangiocarcinoma (12). Given this growing spectrum of AA-associated tumor types, AA exposure detection methods for screening of disease-risk populations are of key importance.
Following metabolic activation of AA, aristolactam (AL)-DNA adducts accumulate in the proximal tubules of the renal cortex and are used as biomarker of exposure (4, 13, 14). AL-DNA adducts may persist for over 20 years after the exposure had ceased (15, 16) and can be measured by 32P-postlabelling (14, 17) or by ultra-performance-liquid chromatography-electrospray ionization-multistage scan mass spectrometry (UPLC-ESI-MS/MSn), both applicable to formalin-fixed paraffin-embedded (FFPE) tissues (16, 18, 19). However, the 32P-postlabeling method lacks specificity, and access to the UPLC-ESI-MS/MSn methodology and its optimization for biomaterial of low quantity are limiting factors.
DNA sequencing established a characteristic AA mutational signature marked by accumulation of A>T transversions within the 5’-Pyr-A-Pur-3’ sequence context (enriched for 5’-CpApG-3’), preferentially located on the non-transcribed strand (8–10, 20, 21). In cancers not associated with AA, such A>T transversions are infrequent (22, 23).
We exploited the unique features of the AA mutational signature to devise a sensitive method for AA exposure detection, based on low-coverage whole-exome sequencing (LC-WES, at approximately 10x in contrast with the conventional 100x coverage), optimized for analysis of tumor-specific DNA of limited quantity and integrity extracted from archived FFPE tissues. The studied urothelial tumor samples originated from a well-characterized population residing in the endemic nephropathy (EN) regions of Croatia and Bosnia and Herzegovina (13), with EN being thus far the only recognized environmental form of AAN (4, 24). For the first time, we report in the urothelial tumors of EN patients the genome-wide signatures of AA, age and APOBEC cytidine deaminase activity, thereby extending previous mutational analyses of this population based solely on the mutations of the TP53 tumor suppressor gene (4, 13, 25). In addition, we demonstrate the ability of LC-WES to elucidate the impact of the AA mutation spectra on key homeostatic biological pathways and to reveal possible mechanisms of tumor dissemination along the urinary tract.
MATERIALS AND METHODS
Patients and tumor samples
Exposure to AA was investigated in 15 patients with urothelial tumors, diagnosed with EN following established criteria (13, 26). As controls, UTUC samples were obtained from 4 patients from a metropolitan area of the United States, unlikely exposed to AA. All specimens were FFPE-converted in the histopathologic laboratories of the participating centers. The involved anatomical sites were renal pelvis, ureter and bladder (ICD-10 codes C65, C66 and C67, respectively). Clinicopathological features and Aristolochia exposure history are listed in Supplementary Table S1. The study protocols included patients’ informed consent and were approved by the IARC Ethics Committee and the Institutional Review Boards of the participating institutions.
DNA isolation from paraffin sections
Hematoxylin-eosin preparations of the paraffin block sections were used to identify tumor tissue free of necrotic areas. The tumor cell areas were measured by ImageJ software (27). Ten (10) µm sections, cut with the Leica RM 2145 microtome (Leica Microsystems), were used to macrodissect the tumor-enriched areas and isolate genomic DNA yielding 1–2 µg (5–10 ng/mm2) per sample. Prior to DNA isolation, slides were deparaffinized for 5 minutes (min) in 100% xylene, kept for 5 min in absolute ethanol, 5 min in 85% ethanol, 5 min in 75% ethanol and stored in milliQ water. DNA isolation was done using QIAamp DNA FFPE Tissue kit (Qiagen) following the manufacturer’s protocol. DNA yields and concentrations were measured using the Picogreen assay (LifeTechnologies) and Fluoroskan Ascent FL microplate fluorometer (Thermo Fisher Scientific). DNA purity was evaluated by the NanoDrop 8000 spectrophotometer (Thermo Fisher Scientific), and DNA integrity assessed by 0.8% agarose gel electrophoresis.
AL-DNA adduct analysis and TP53 resequencing
DNA was isolated from the renal cortex and tumor tissues by standard phenol-chloroform extraction techniques. The level of AL-DNA adducts in the renal cortex DNA (10–20 µg) was determined using 32P-postlabeling polyacrylamide gel electrophoresis, as previously described (13). The TP53-specific mutations were identified using the AmpliChip p53 Research Test (Roche Molecular Diagnostics, Pleasanton, CA), sensitively detecting all single base-pair substitutions and single-base deletions (13).
WES library preparation, exome capture and sequencing
Two hundred-fifty (250) ng of genomic DNA were sheared by adaptive focused acoustics™ method (Covaris, Inc.) to obtain ~300 bp fragments, with water temperature of 4°C, one cycle at 175 Watt peak power, duty factor 10 and 200 cycles per burst. Resulting fragment size was assessed using the 2100 Bioanalyzer and High Sensitivity DNA kit (Agilent Technologies). The sheared DNA was converted into libraries using the Kapa LTP Library Preparation Kit (Kapa Biosystems). Briefly, the fragmented DNA was subjected to end repair reaction followed by poly-A-tailing and adapter ligation, excess adapters removed by Agencourt AMPure XP beads (Beckman Coulter). Eight cycles of PCR were performed to amplify the libraries with correct adapters on both ends. Four libraries (250 ng each) were pooled per exome capture with the Nimblegen SeqCap EZ Exome reagent. Exome-enriched mixes were PCR-amplified in 10 cycles, post-enrichment libraries pooled in 420 µl of water to a final concentration of 6 pM. This volume was divided and loaded in two lanes of the rapid run mode flow cell for cluster generation and sequencing on the HiSeq2500, in a paired-end 50 bp cycle run. Multiplexing 16 samples per run resulted in the target coverage of ~10x.
Four additional EN UTUC samples and two UTUC samples from the metropolitan United States were analyzed in a validation assay using the SOLiD 5500XL sequencer (Life Technologies). See Supplementary Methods for details.
Raw HiSeq2500 sequencing data were deposited to the Sequence Read Archive (SRA) of the National Center for Biotechnology Information (NCBI) repository (ID SRP042035) to become available from the NCBI’s dbGaP database. The annotated list of single-base substitutions (HiSeq2500 data) is provided in Supplementary Table S2.
Sequencing data analysis
FastQ reads were aligned to the human genome (hg19) using Burrows-Wheeler Aligner. Realignment and base quality score recalibration was done by the Genome Analysis Toolkit (GATK) and the duplicate-read removal by Picard. GATK HaplotypeCaller was used to call variants subsequently annotated on the RefSeq Gene transcript contents by ANNOVAR (28). Polymorphisms present in normal population and removed from our data originated from these collections: 1000 genomes (1000g, http://www.1000genomes.org/), Exome Sequencing Project (ESP, http://exome.gs.washington.edu/) and the SNP database build 137 (dbSNP, http://www.ncbi.nlm.nih.gov/SNP/). We removed variants with frequency above 0.1% in either the 1000g or ESP databases, or annotated in the dbSNP database, or present in a custom germline variant catalog built from 560 cases from The Cancer Genome Atlas (TCGA, http://cancergenome.nih.gov/). Variants mapping to repetitive sequences contained in the genomic segmental duplication database (29) alongside variants with ≥90% homology with multiple regions were excluded. R functions were developed to compute the mutation type distributions and strand bias. The strand bias significance was determined by Pearson χ2 test. These tertiary analysis parameters were computed in two separate coverage ranges, ≥3x with no defined maximum, and between 3–9x to emulate ultra-low coverage.
Mutational signature analysis using non-negative matrix factorization (NMF)
NMF decomposes mutational patterns based on factorization of one matrix (n×m) in two matrices W (n×r) and H (r×m) with the constraint that all three matrices must be composed of non-negative elements (30). The r is the rank of factors to be extracted from the input matrix, corresponding to the number of signatures. The input matrix contained one column per patient (only HiSeq2500 data considered) and in rows the frequency of mutations types in 96 possible two-base sequence contexts. The R package NMF (31) was used to extract mutational signatures. The correlation between the extracted signatures and previously published ones (7, 21, 22, 32) and/or available in COSMIC (23) was computed as the inner product of the two signatures (vectors) divided by the product of their norms.
Functional analysis of tumor-specific non-synonymous mutations
To examine the biological impact of the gene mutants in the AA signature-positive samples, analysis was performed using the DAVID tool (33), with two input gene lists: 1) genes harboring non-synonymous (missense, stop-gain or stop-loss) SBS and 2) genes non-synonymously mutated in the EN data set and in AA signature-positive samples from at least one of the two published datasets on UTUC in Taiwanese patients (8, 21). The list was further narrowed by classifying the mutated genes as established oncogenes or tumor suppressors listed by the Gene Set Enrichment Analysis (GSEA) database (34) and/or a cancer driver genes defined by recent seminal studies (35–39).
RESULTS AND DISCUSSION
Low-coverage detection of AA exposure signature in urothelial tumors of EN patients
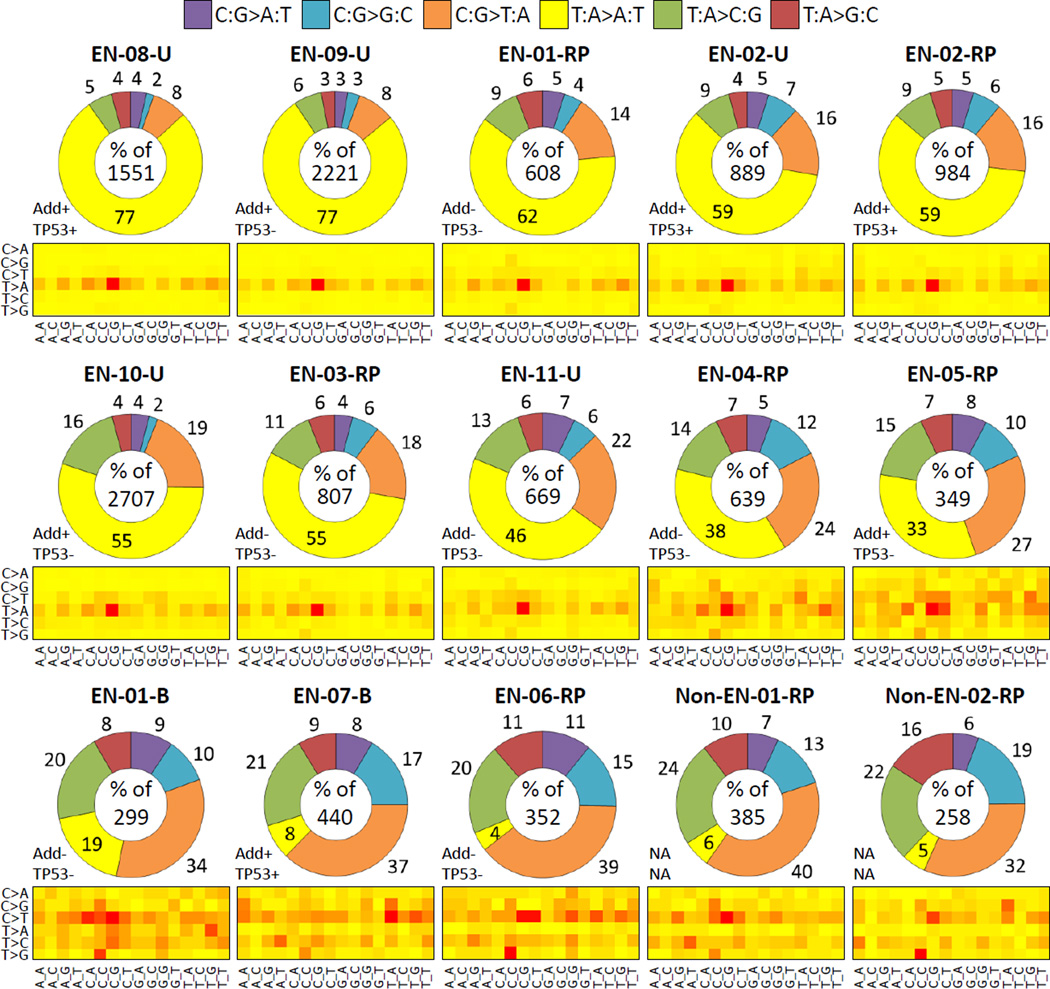
We applied HiSeq2500 LC-WES to genomic DNAs isolated from FFPE urothelial tumors from 11 EN cases, two of whom had concurrent UTUC and bladder carcinoma, and from two US patients providing non-EN control samples (13 patients and 15 tumors in total, see Supplementary Table S1). Features of AA signature had been described earlier, as follows: mutational load of ≥40 SBS or ≥10 A>T in exonic positions, high proportion of A>T (>35% of all SBS types or as the predominant type) with a strand bias of ≥1.25, and ≥33% enrichment of A>T in the 5’-(C/T)pApG-3’ sequence context (8, 21). We used analogous criteria for the AA signature (≥50 SBS per sample of which ≥15% are A>T SBS, of which ≥20% are in the 5’-CpApG-3’ context), applying more stringent statistical analysis of the strand bias ratio combined with a cut-off of ≥1.5 (9, 11). Under these criteria the AA signature was readily observed in 10 of the 13 analyzed EN tumor samples, with 33–77% of A>T transversions per sample (Fig. 1), a non-transcribed strand bias of 2.0–3.3 and the 5’-C_G-3’ context enrichment above 19% (mean 24.6%, SD=4.9, range of 19.1–27.4%) (Table 1). In contrast, A>T mutations and their enrichment in the 5’-CpApG-3’context are generally low in cancers of non-AA etiology, based on our analysis of 7,160 tumors of 52 cancer types in the COSMIC database (average 5.8% A>T, range 0–12.1%, of which 10% are in the 5’-CpApG-3’ context). Similarly, the average percentage of A>T in the 5’-CpApG-3’ context in TCGA urothelial carcinoma data (only bladder data available) is 10.8% (0–50%), while the mean percentage of all A>T mutations is low (average of 3.9%, range 0.8–8.3%).
Figure 1. SBS alterations in urothelial tumors analyzed by LC-WES.
The distribution of six SBS types and their trinucleotide context are shown for variants detected at ≥3x per-base coverage. The doughnut charts correspond to individual samples (sample ID on top), ordered from high to low percentage of A>T. Total SBS counts per sample are provided in the center of each graph. The numbers outside the chart sections denote each mutation type percentage. The suffix -B, -RP and -U stands for bladder, renal pelvis and ureter, respectively. Add+/− = sample positive or negative for aristolactam-DNA adducts; TP53+/− = mutated (+) or wild-type (−) TP53 gene. The heat-maps summarize relative frequencies of the six mutation types (C>A stands for C:G>A:T etc.) across the 16 possible trinucleotide contexts listed at the bottom. Red=high frequency, yellow=low frequency.
Table 1.
Summary of the SBS detected at ≥3x per-base coverage and of the AA-signature analysis results
| Case ID | Total SBS |
SBS per Mbp |
A>T SBS |
A>T per Mbp |
A>T (%) |
CAG context (%) |
SB A>T ratio (NTr/Tr) |
aSB P value |
SB FDR Q value |
Contribution to Sig 22 (AA) |
AA signature |
|---|---|---|---|---|---|---|---|---|---|---|---|
| EN-01-RP | 608 | 9.1 | 376 | 5.6 | 61.8 | 26.1 | 3.3 (272/82) | 0 | 0 | 380 (63%) | Yes |
| EN-01-B | 299 | 4.5 | 56 | 0.8 | 18.7 | 12.5 | 2.1 (37/18) | 0.015 | 1 | 53 (18%) | Yesb |
| EN-02-RP | 984 | 14.7 | 585 | 8.7 | 59.5 | 26.7 | 3.1(424/136) | 0 | 0 | 514 (52%) | Yes |
| EN-02-U | 889 | 13.3 | 529 | 7.9 | 59.5 | 26.1 | 3.3 (384/118) | 0 | 0 | 457 (51%) | Yes |
| EN-03-RP | 807 | 12.0 | 443 | 6.6 | 54.9 | 25.0 | 2.0 (278/140) | 2.1E-11 | 1.7E-09 | 402 (50%) | Yes |
| EN-04-RP | 639 | 9.5 | 241 | 3.6 | 37.7 | 19.5 | 2.3 (157/68) | 4.4E-09 | 3.6E-07 | 164 (26%) | Yes |
| EN-05-RP | 349 | 5.2 | 115 | 1.7 | 33.0 | 19.1 | 2.8 (79/28) | 1.3E-06 | 1.1E-04 | 87 (25%) | Yes |
| EN-06-RP | 352 | 5.3 | 15 | 0.2 | 4.3 | 0.0 | 5.5 (11/2) | 0.027 | 1 | 0 (0%) | -- |
| EN-07-B | 440 | 6.6 | 35 | 0.5 | 8.0 | 11.4 | 1.2 (15/13) | 0.850 | 1 | 0 (0%) | -- |
| EN-08-U | 1551 | 23.1 | 1195 | 17.8 | 77.0 | 23.4 | 2.8 (840/303) | 0 | 0 | 1391 (90%) | Yes |
| EN-09-U | 2221 | 33.1 | 1701 | 25.4 | 76.6 | 27.4 | 2.7 (1185/431) | 0 | 0 | 1931 (87%) | Yes |
| EN-10-U | 2707 | 40.4 | 1486 | 22.2 | 54.9 | 26.2 | 2.8 (1052/377) | 0 | 0 | 2706 (100%) | Yes |
| EN-11-U | 669 | 10.0 | 309 | 4.6 | 46.2 | 26.9 | 2.5 (211/85) | 3.7E-13 | 0 | 297 (44%) | Yes |
| Non-EN-01-RP | 385 | 5.7 | 24 | 0.4 | 6.2 | 0.0 | 0.6 (9/14) | 0.400 | 1 | 0 (0%) | -- |
| Non-EN-02-RP | 258 | 3.9 | 14 | 0.2 | 5.4 | 14.3 | 5.0 (10/2) | 0.043 | 1 | 0 (0%) | -- |
NOTE: Suffices -B, -RP and -U indicate bladder, renal pelvis and ureter, respectively.
Abbreviations: SBS, single base substitution. Mbp, megabase pair. A>T; A>T transversion(s); SB A>T, strand bias, the ratio of A>T transversions on the non-transcribed versus transcribed strand (the number of respective transversions is shown in brackets); CAG context (%), percentage of A>T transversions in the most frequent context reported for the AA signature; NTr/Tr, ratio of A>T variants on non-transcribed versus transcribed strand. SB P value and FDR q value, measures of significance of the strand bias, see Materials and Methods. Contribution to Sig 22 (AA) (Sig = NMF-determined mutational signature) shown as the number of SBS and the corresponding percentage (in parentheses) of the total SBS per sample. AA signature, positivity for AA signature considering the co-occurrence of ≥50 SBS and ≥15% A>T, ≥20% CAG context, strand bias (SB) and its significance (SB P and/or q value) and mutation load contribution to Sig 22 (AA), as described previously (9).
Zero values correspond to a P value below 2×10−16.
Lower percentage in the 5’-CpApG-3’ context and non-significant q value; supported by the NMF analysis.
A weaker signature marked by 18.7% A>T, strand bias of 2.1 and the 5’-CpApG-3’ context proportion of 12.5% was observed in the bladder tumor sample (EN-01-B) of a patient with a concurrent AA signature-positive UTUC (EN-01-RP, see Table 1 and Supplementary Table S1). Two EN samples (EN-06 and EN-07) and the two non-EN controls were found negative for the AA signature, with A>T transversions present at 4–8%. In the case of EN-07 (bladder carcinoma with no history of UTUC), despite the presence of AL-DNA adducts in the patient’s renal cortex, the mutation profile (Fig. 1) suggested AA-unrelated etiology. Among the AA signature-positive samples, we detected an average of 1,142 (range 349–2,707) mutations per tumor (~18 SBS per sample per exome megabase [Mb]) whereas the mutation rate in the control and negative samples (including the weaker AA-signature bladder cancer) was on average 357 (range 258–440) mutations per sample (~6 per sample per exome Mb). As shown in Table 1, the predominant A>T transversions substantially contributed to the high SBS counts.
LC-WES analysis of the EN UTUC thus generates results consistent with previous reports on the highly mutagenic potential of AA (8, 21, 40) and our results justify the use of exome sequencing for reliable detection of exposure to AA in archived FFPE material.
LC-WES identifies AA signature at ultra-low coverage
We next investigated whether the AA signature can be identified at ultra-low coverage. Upon considering 3–9 non-duplicate per-base reads, mutation counts in the AA-associated samples decreased to 233 per tumor on average (~4 per sample per exome Mb) and to average 67 per tumor (1 per sample per exome Mb) in the negative samples and the weakly positive bladder tumor (EN-01-B). The 10 tumors shown in Fig. 1 (two top rows) still exhibited the AA signature at ultra-low coverage (Supplementary Fig. S1), with the strand bias ratios between 1.7–4.7, and retained prominent enrichment of the 5’-CpApG-3’context (>25%). Thus, the specific and unique features of the AA signature can be reliably detected in FFPE tumor samples by superficial coverage sequencing.
These results open an attractive opportunity for retrospective analyses of archived pathological specimens from the regions of AA exposure risk. In comparison with the 32P-post-labelling and mass spectrometry adduct detection techniques, the LC-WES approach is based on a commodity technology that generates genome-wide information. LC-WES is also very sensitive, using low input DNA amounts (250 ng compared to 5–10 µg required for adduct analysis). Finally, it can indicate exposure to AA when neither AL-DNA adducts nor mutations in TP53 are detected, as we demonstrate for the AA signature-positive cases EN-01, EN-03, EN-04 and EN-11 (Fig. 1 and Supplementary Table S1).
AA-associated urothelial tumors harbor three major mutational signatures
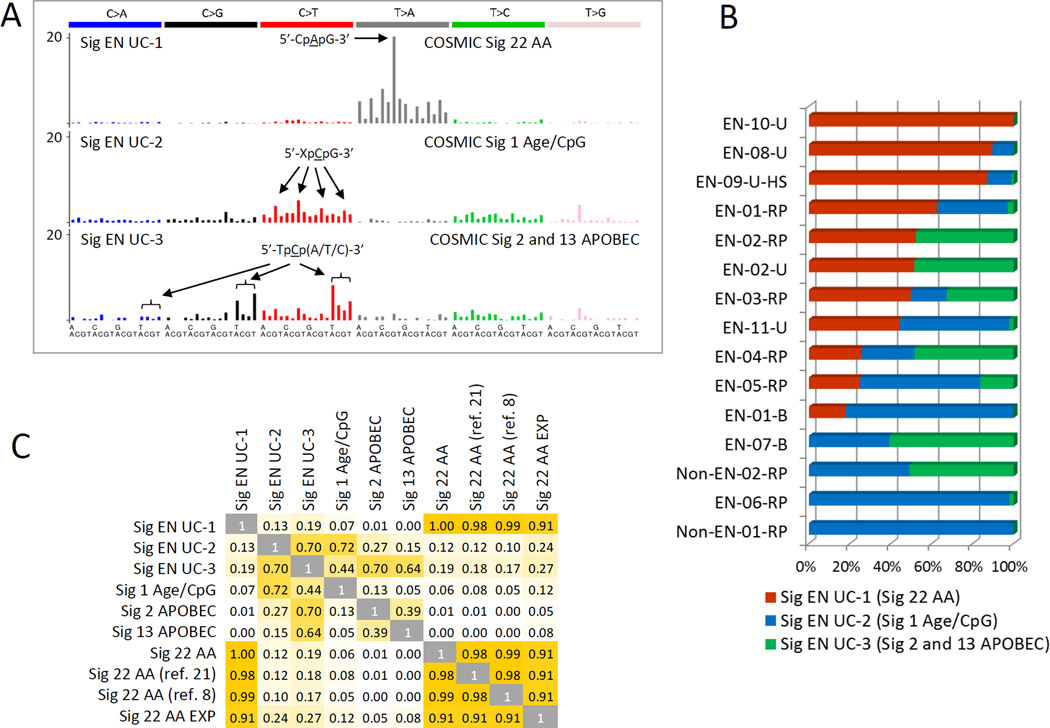
NMF extracts individual mutational signatures from complex alteration patterns observed in primary tumors, reflecting thus the specific effects of etiological factors (7, 9, 22, 41). NMF was used to describe the AA signature in human UTUC, bladder, liver and renal carcinomas (7, 9, 11) and in experimental in vitro system designed to model mutational signatures of carcinogens (32). Here, in the EN urothelial tumors, the NMF approach identified three distinct signatures, the AA-specific signature (Signature 22) (42), the signature related to age (C:G>T:A in the 5’-XpCpG-3’ context, Signature 1) (22), and the Signatures 2 and/or 13 associated with the cytidine deaminase activity of the APOBEC enzymes (22) (Fig. 2). All three signatures are currently listed in the COSMIC database (23). Furthermore, NMF aided in classifying the EN-01-B bladder tumor as positive due to non-negligible sample contribution to the AA signature (18%), in contrast with the negative samples (EN-06-RP and EN-07-B, with contributions of 0% for both) and non-EN controls (each 0% contribution, see Table 1 and Fig. 2B). The identified EN UTUC AA signature correlated highly (>90%) with the COSMIC Signature 22 (23), derived from AA-associated primary UTUC tumors from Taiwanese patients (8, 21), and with the AA signature modeled in vitro (32) (Fig. 2C). The other EN tumor signatures matched their COSMIC counterparts with 72% similarity (Age) and 70% and 64% similarity (APOBEC, Signature 2 and 13, respectively) (Fig. 2C).
Figure 2. Mutational signatures determined by NMF.
Results are shown for urothelial carcinoma samples sequenced on HiSeq2500. A) Contribution of each mutation type to signatures of AA, age/CpG and APOBEC. The x-axis represents the trinucleotide sequence contexts, with the 5’-flank base in the first row and the 3’-flank in the second. B) Contributions of the studied urothelial tumors to the individual signatures shown in A. C) Correlation between NMF-identified EN sample (EN UC) signatures and previously described COSMIC Signatures 1, 2/13 and 22 (22), signature 22 identified in Taiwan UTUC samples (8, 21), and signature 22 AA Exp, modeled experimentally in vitro (32).
Validation of the LC-WES performance on a distinct sequencing platform
To validate the LC-WES performance using another sequencing chemistry and platform, we analyzed four additional EN UTUCs positive for AL-DNA adducts and p53 A>T mutations (EN-12, EN-13, EN-14, EN-15), and two control UTUCs from US patients (Non-EN-03 and Non-EN-04), on the SOLiD 5500×l sequencer. At the average 14.5x coverage we observed the AA signature in all EN samples, although in samples EN-13-RP and EN-14-RP the A>T transversion was the second most abundant mutation type following C>T (see Supplementary Fig. S2A). The signature remained detectable at ultra-low coverage (~4.6x), when considering only the 3–9 read interval (Supplementary Fig. S2B).
Chromosomal distribution of the AA-specific mutations and recurrently mutated cancer driver genes
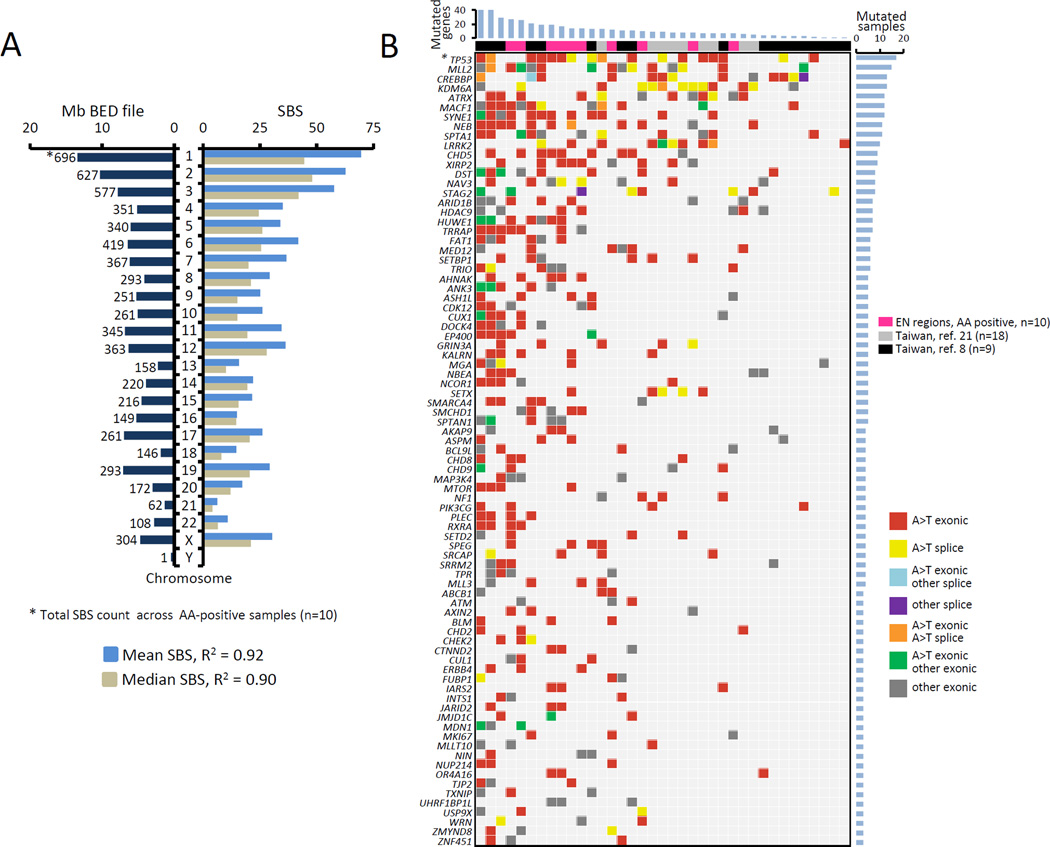
In the A>T enriched samples, A>T transversions were randomly distributed along the sequenced regions, with linear correlation between A>T SBS counts and chromosome size (R2=0.9) (Fig. 3A). Similar correlation was maintained in the minimum coverage interval of 3–9x (data not shown). This result was confirmed by the analysis of the Taiwan UTUCs (8, 21) in which a similar although less linear trend was observed (R2=0.61–0.64). These findings suggest a stochastic A>T mutation distribution within the gene/transcription units represented by the exome.
Figure 3. Distribution of A>T mutations and their impact on cancer driver genes.
A) Correlation (squared Pearson product-moment correlation coefficient R2) between the mean (light blue) and median (gray) values of A>T SBS counts per chromosome and the chromosome size in Mb (dark blue, left side). Variants based on ≥3 unique reads were considered. B) Meta-analysis of recurrently mutated genes in AA-associated UTUC. Genes with non-synonymous SBS variants identified in this study were compared with gene mutants found by two previously published AAN-UTUC data sets from Taiwan (8, 21). * = TP53 mutations combine results from the AmpliChip and LC-WES analyses. The list of recurrently mutated genes was narrowed down to cancer driver genes only, as described in Materials and Methods. See also Supplementary Table S4 for detailed annotation of these mutations.
Despite this apparently random pattern, we identified 83 cancer driver genes carrying protein sequence-altering A>T SBS, that were recurrently mutated across the three datasets of the AA signature-positive tumor samples (this study, n=10, and the two previously reported Taiwanese sets of n=18 (21) and n=9 (8)). These findings are summarized in Fig. 3B and in Supplementary Table S4. The recurrently mutated genes included numerous known drivers and chromatin-associated factors such as TP53, ARID1B, ATRX, CREBBP, CHD2, CHD5, CHD8, FAT1, KDM6A, MLL2 (KMT2D), SETBP1, TRRAP. TP53 was the most frequently mutated gene (17 of 37 [46%] samples) with all its mutations being A>T transversions. Fifteen samples exhibited mutations in the histone methyl-transferase KMT2D (MLL2), with varying SBS types, suggesting secondary mutation processes possibly linked due to high mutational loads and increased genomic instability. Further systematic investigations should be undertaken to establish possible recurrent alterations in particular genes and pathways in UTUC across studies of different populations/geographical areas. For instance, data in Fig. 3 and in Supplementary Table S3 indicate that TP53, CREBBP and LRRK2 are mutated mostly in the Taiwanese samples whereas mutations in the AHNAK, ATRX, SMCHD1 and XIRP2 genes are enriched in the EN UTUC samples. Other factors contributing to these differences merit further investigations, including varying modes of AA exposure (low-dose chronic intake in the EN regions as compared to higher-dose, (sub)acute exposures resulting from the use of traditional herbal medicines in Asia) and disease susceptibility due to the patients’ genetic background.
Biological impact of the AA-signature
Using NIH DAVID, we performed Gene Ontology (GO) and KEGG pathway analyses of the genes harboring non-synonymous A>T mutations in the AA-signature positive samples analyzed by HiSeq2500 (n=10). We identified gene targets from the functional classes of cell adhesion, cell-matrix contact, cell migration, cell cycle, cell signaling (MAPKKK/RAS and PI3K cascades, mTOR pathway), pathways of WNT, insulin and ERBB signaling, nucleotide excision repair, and the DNA-dependent ATPase and helicase activity, chromatin modification and histone binding related to gene expression regulation, with dozens to hundreds of mutated genes per category (Supplementary Table S3). This observation suggests massive deregulation and/or destabilization of key homeostatic pathways by the high A>T mutation loads.
Next, for the 83 recurrently mutated cancer genes (Fig. 3B and Supplementary Table S4) we observed enrichment of GO and KEGG categories related to regulation of transcription, chromatin/histone modification, and categories of DNA damage response and DNA repair (Supplementary Table S5). These included numerous previously established cancer driver genes (TP53, AHNAK, ARID1B, ATRX, BLM, CHD2, CHD5, CHD8, CHD9, CHEK2, CLTC, ERBB4, FN1, HUWE1 IARS2, KALRN, LRRK2, MLL2, NEB, RXRA, SMCHD1, SPEG, STAG2, SYNE1, TRIO (35–39)). Thus, the LC-WES analysis of AA-exposed urothelial tumors and associated data mining can reveal biological information contents, particularly upon meta-analysis with data from different populations characterized by identical etiology and tumor types.
Overlapping mutation patterns in distinct tumors from same patients
Two EN patients had synchronous urothelial tumors in distinct anatomical sites (renal pelvis and bladder, samples EN-01-RP and EN-01-B; and renal pelvis and ureter, samples EN-02-RP and EN-02-U). By using LC-WES, we investigated the common genetic origins of these synchronous tumor pairs. In patient EN-01, the overlapping SBS were enriched for C>T mutations (42%) followed by A>G (20%), and only 7.7% of the overlapping SBS were A>T transversions affecting the coding sequence of mere 3 non-cancer genes (VWA3B, KDM3B and ACIN1). However, the A>T SBS were enriched among the mutations unique to the renal pelvis and to the bladder tumor (77% and 28%, respectively, Supplementary Fig. S3A), suggestive of a common precursor carrying mainly non-A>T driver mutations, giving rise to two tumor progenies subsequently accumulating distinct patterns of A>T alterations in either anatomical site. The distinct AA signature in the bladder tumor is in keeping with a recent study of Asian bladder cancer patients in whose tumors the AA signature manifested without the involvement of upper tract or a history of renal disease (11). In contrast, the tumors in the renal pelvis and ureter of patient EN-02 shared the majority of mutations contributing to a prominent AA signature, suggesting a common precursor carrying mostly A>T alterations (Supplementary Fig. S3B). This genetic relationship between same-patient tumors suggests cell seeding along the tract as the basis for tumor dissemination. However, further investigations of a larger multiple-tumor case series and with the use of deep sequencing is needed to further elucidate the exact mechanisms of multifocal and recurrent tumorigenesis in the urinary tract of AA-exposed patients.
In summary, we report successful detection of the genome-wide AA signature in urothelial tumors of EN patients, using archived FFPE specimens and a customized low-coverage exome sequencing. The described technique is a cost-effective screening tool potentially applicable to molecular epidemiology studies aiming at identifying cancers associated with AA exposure. This ability of the LC-WES and its applicability to archived biomaterial may be exploited in future systematic studies on AAN and associated cancers, in support of established or future disease prevention programs.
Supplementary Material
ACKNOWLEDGMENTS
We thank the Genetic Platform, IARC, and the Genome Technology Center, NYU Langone Medical Center for expert assistance with exome sequencing; we thank Ms Christine Carreira, Dr Behnoush Abedi-Ardekani and Dr Elisabetta Kuhn for assistance with the biospecimen evaluation and processing. We thank the staff of the General Hospital Slavonski Brod, the University Hospital Centre Zagreb and of the Weill Medical College of Cornell University, New York, for expert assistance and support. We are grateful to Drs Monica Hollstein and Michael Korenjak for critical comments on the manuscript.
FINANCIAL SUPPORT
IARC Regular Budget; Grant P01 ES04068 to A.P. Grollman from the NIH/NIEHS; R03-TW007042 grant to A.P. Grollman from the NIH Fogarty International Center; grant to A.P. Grollman from Henry and Marsha Laufer; Grant no. 108-0000000-0329 to B. Jelaković from the Croatian Ministry of Science, and Grant 04/38 to B. Jelaković from the Croatian Foundation for Science; and Grant NIH/NCI P30 CA016087-33, which partially supports the NYU Genome Technology Center.
Footnotes
DISCLOSURE OF POTENTIAL CONFLICTS OF INTEREST
S.F. Shariat has received honoraria from speakers bureau from Astellas, Takeda, Ipsen, Janssen, Sanofi, Olympus, Wolff, Pierre Fabre, and Sanochenia, has ownership interest (including patents) in prostate and bladder cancer biomarkers, and is a consultant/advisory board member for Astellas, Ipsen, Sanochemia, Olympus, Wolff, and Janssen. No potential conflicts of interest were disclosed by the other authors.
REFERENCES
- 1.Lyon: International Agency for Research on Cancer; 2012. IARC Monographs on the Evaluation of Carcinogenic Risks to Humans Volume 100A; A Review of Human Carcinogens: Pharmaceuticals. [Google Scholar]
- 2.Debelle FD, Vanherweghem JL, Nortier JL. Aristolochic acid nephropathy: a worldwide problem. Kidney International. 2008;74:158–169. doi: 10.1038/ki.2008.129. [DOI] [PubMed] [Google Scholar]
- 3.Grollman AP. Aristolochic acid nephropathy: Harbinger of a global iatrogenic disease. Environmental and Molecular Mutagenesis. 2013;54:1–7. doi: 10.1002/em.21756. [DOI] [PubMed] [Google Scholar]
- 4.Grollman AP, Shibutani S, Moriya M, Miller F, Wu L, Moll U, et al. Aristolochic acid and the etiology of endemic (Balkan) nephropathy. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:12129–12134. doi: 10.1073/pnas.0701248104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nortier JL, Martinez MC, Schmeiser HH, Arlt VM, Bieler CA, Petein M, et al. Urothelial carcinoma associated with the use of a Chinese herb (Aristolochia fangchi) The New England Journal of Medicine. 2000;342:1686–1692. doi: 10.1056/NEJM200006083422301. [DOI] [PubMed] [Google Scholar]
- 6.Huang J, Deng Q, Wang Q, Li KY, Dai JH, Li N, et al. Exome sequencing of hepatitis B virus-associated hepatocellular carcinoma. Nature Genetics. 2012;44:1117–1121. doi: 10.1038/ng.2391. [DOI] [PubMed] [Google Scholar]
- 7.Poon SL, McPherson JR, Tan P, Teh BT, Rozen SG. Mutation signatures of carcinogen exposure: genome-wide detection and new opportunities for cancer prevention. Genome Medicine. 2014;6:24. doi: 10.1186/gm541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Poon SL, Pang ST, McPherson JR, Yu W, Huang KK, Guan P, et al. Genome-wide mutational signatures of aristolochic acid and its application as a screening tool. Science Translational Medicine. 2013;5:197ra01. doi: 10.1126/scitranslmed.3006086. [DOI] [PubMed] [Google Scholar]
- 9.Jelakovic B, Castells X, Tomic K, Ardin M, Karanovic S, Zavadil J. Renal cell carcinomas of chronic kidney disease patients harbor the mutational signature of carcinogenic aristolochic acid. International Journal of Cancer. 2015;136:2967–2972. doi: 10.1002/ijc.29338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Scelo G, Riazalhosseini Y, Greger L, Letourneau L, Gonzalez-Porta M, Wozniak MB, et al. Variation in genomic landscape of clear cell renal cell carcinoma across Europe. Nature Communications. 2014;5:5135. doi: 10.1038/ncomms6135. [DOI] [PubMed] [Google Scholar]
- 11.Poon S, Huang M, Choo Y, McPherson J, Yu W, Heng H, et al. Mutation signatures implicate aristolochic acid in bladder cancer development. Genome Medicine. 2015;7:38. doi: 10.1186/s13073-015-0161-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zou S, Li J, Zhou H, Frech C, Jiang X, Chu JS, et al. Mutational landscape of intrahepatic cholangiocarcinoma. Nature Communications. 2014;5:5696. doi: 10.1038/ncomms6696. [DOI] [PubMed] [Google Scholar]
- 13.Jelakovic B, Karanovic S, Vukovic-Lela I, Miller F, Edwards KL, Nikolic J, et al. Aristolactam-DNA adducts are a biomarker of environmental exposure to aristolochic acid. Kidney International. 2012;81:559–567. doi: 10.1038/ki.2011.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schmeiser HH, Bieler CA, Wiessler M, van Ypersele de Strihou C, Cosyns JP. Detection of DNA adducts formed by aristolochic acid in renal tissue from patients with Chinese herbs nephropathy. Cancer Research. 1996;56:2025–2028. [PubMed] [Google Scholar]
- 15.Schmeiser HH, Nortier JL, Singh R, da Costa GG, Sennesael J, Cassuto-Viguier E, et al. Exceptionally long-term persistence of DNA adducts formed by carcinogenic aristolochic acid I in renal tissue from patients with aristolochic acid nephropathy. International Journal of Cancer. 2014;135:502–507. doi: 10.1002/ijc.28681. [DOI] [PubMed] [Google Scholar]
- 16.Yun BH, Yao L, Jelakovic B, Nikolic J, Dickman KG, Grollman AP, et al. Formalin-fixed paraffin-embedded tissue as a source for quantitation of carcinogen DNA adducts: aristolochic acid as a prototype carcinogen. Carcinogenesis. 2014;35:2055–2061. doi: 10.1093/carcin/bgu101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dong H, Suzuki N, Torres MC, Bonala RR, Johnson F, Grollman AP, et al. Quantitative determination of aristolochic acid-derived DNA adducts in rats using 32P-postlabeling/polyacrylamide gel electrophoresis analysis. Drug metabolism and disposition: the biological fate of chemicals. 2006;34:1122–1127. doi: 10.1124/dmd.105.008706. [DOI] [PubMed] [Google Scholar]
- 18.Yun BH, Rosenquist TA, Sidorenko V, Iden CR, Chen CH, Pu YS, et al. Biomonitoring of aristolactam-DNA adducts in human tissues using ultra-performance liquid chromatography/ion-trap mass spectrometry. Chemical Research in Toxicology. 2012;25:1119–1131. doi: 10.1021/tx3000889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yun BH, Rosenquist TA, Nikolic J, Dragicevic D, Tomic K, Jelakovic B, et al. Human formalin-fixed paraffin-embedded tissues: an untapped specimen for biomonitoring of carcinogen DNA adducts by mass spectrometry. Analytical Chemistry. 2013;85:4251–4258. doi: 10.1021/ac400612x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hollstein M, Moriya M, Grollman AP, Olivier M. Analysis of TP53 mutation spectra reveals the fingerprint of the potent environmental carcinogen, aristolochic acid. Mutation Research. 2013;753:41–49. doi: 10.1016/j.mrrev.2013.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hoang ML, Chen CH, Sidorenko VS, He J, Dickman KG, Yun BH, et al. Mutational signature of aristolochic acid exposure as revealed by whole-exome sequencing. Science Translational Medicine. 2013;5:197ra02. doi: 10.1126/scitranslmed.3006200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SA, Behjati S, Biankin AV, et al. Signatures of mutational processes in human cancer. Nature. 2013;500:415–421. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Forbes SA, Beare D, Gunasekaran P, Leung K, Bindal N, Boutselakis H, et al. COSMIC: exploring the world's knowledge of somatic mutations in human cancer. Nucleic Acids Research. 2015;43:D805–D811. doi: 10.1093/nar/gku1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hranjec T, Kovac A, Kos J, Mao W, Chen JJ, Grollman AP, et al. Endemic nephropathy: the case for chronic poisoning by aristolochia. Croatian Medical Journal. 2005;46:116–125. [PubMed] [Google Scholar]
- 25.Moriya M, Slade N, Brdar B, Medverec Z, Tomic K, Jelakovic B, et al. TP53 Mutational signature for aristolochic acid: an environmental carcinogen. International Journal of Cancer. 2011;129:1532–1536. doi: 10.1002/ijc.26077. [DOI] [PubMed] [Google Scholar]
- 26.Jelakovic B, Nikolic J, Radovanovic Z, Nortier J, Cosyns JP, Grollman AP, et al. Consensus statement on screening, diagnosis, classification and treatment of endemic (Balkan) nephropathy. Nephrology, Dialysis, Transplantation. 2014;29:2020–2027. doi: 10.1093/ndt/gft384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nature Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Research. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bailey JA, Gu Z, Clark RA, Reinert K, Samonte RV, Schwartz S, et al. Recent segmental duplications in the human genome. Science. 2002;297:1003–1007. doi: 10.1126/science.1072047. [DOI] [PubMed] [Google Scholar]
- 30.Lee DD, Seung HS. Learning the parts of objects by non-negative matrix factorization. Nature. 1999;401:788–791. doi: 10.1038/44565. [DOI] [PubMed] [Google Scholar]
- 31.Gaujoux R, Seoighe C. A flexible R package for nonnegative matrix factorization. BMC Bioinformatics. 2010;11:367. doi: 10.1186/1471-2105-11-367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Olivier M, Weninger A, Ardin M, Huskova H, Castells X, Vallee MP, et al. Modelling mutational landscapes of human cancers in vitro. Scientific Reports. 2014;4:4482. doi: 10.1038/srep04482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature Protocols. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 34.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lawrence MS, Stojanov P, Mermel CH, Robinson JT, Garraway LA, Golub TR, et al. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature. 2014;505:495–501. doi: 10.1038/nature12912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Plass C, Pfister SM, Lindroth AM, Bogatyrova O, Claus R, Lichter P. Mutations in regulators of the epigenome and their connections to global chromatin patterns in cancer. Nature Reviews Genetics. 2013;14:765–780. doi: 10.1038/nrg3554. [DOI] [PubMed] [Google Scholar]
- 37.Shen H, Laird PW. Interplay between the cancer genome and epigenome. Cell. 2013;153:38–55. doi: 10.1016/j.cell.2013.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tamborero D, Gonzalez-Perez A, Perez-Llamas C, Deu-Pons J, Kandoth C, Reimand J, et al. Comprehensive identification of mutational cancer driver genes across 12 tumor types. Scientific Reports. 2013;3:2650. doi: 10.1038/srep02650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA, Jr, Kinzler KW. Cancer genome landscapes. Science. 2013;339:1546–1558. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Clyne M. Bladder cancer: aristolochic acid--one of the most potent carcinogens known to man. Nature Reviews Urology. 2013;10:552. doi: 10.1038/nrurol.2013.186. [DOI] [PubMed] [Google Scholar]
- 41.Alexandrov LB, Nik-Zainal S, Wedge DC, Campbell PJ, Stratton MR. Deciphering signatures of mutational processes operative in human cancer. Cell Reports. 2013;3:246–259. doi: 10.1016/j.celrep.2012.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Helleday T, Eshtad S, Nik-Zainal S. Mechanisms underlying mutational signatures in human cancers. Nature Reviews Genetics. 2014;15:585–598. doi: 10.1038/nrg3729. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.