Abstract
A new class of broadly neutralizing antibodies (bNAbs) from HIV donors has been reported to target the glycans on gp120, thus renewing hope of developing carbohydrate-based HIV vaccines. However, the version of gp120 used in previous studies was not from human T cells and so the glycosylation pattern could be somewhat different to that found in the native system. Moreover, some antibodies recognized two different glycans simultaneously and this cannot be detected with the commonly used glycan microarrays on glass slides. Here, we have developed a glycan microarray on an aluminium oxide-coated glass slide containing a diverse set of glycans, including homo- and mixed N-glycans (high-mannose, hybrid and complex types) that were prepared by modular chemo-enzymatic methods to detect the presence of hetero-glycan binding behaviours. This new approach allows rapid screening and identification of optimal glycans recognized by neutralizing antibodies, and could speed up the development of HIV-1 vaccines targeting cell surface glycans.
Graphical Abstract
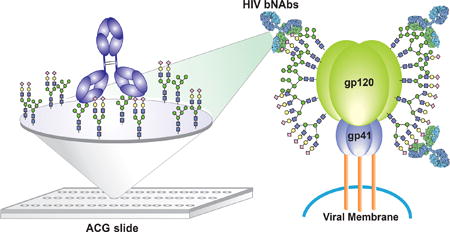
The extraordinary genetic diversity of HIV-1 and its capacity to evade host immune responses that elicit neutralizing anti- bodies are among the greatest hurdles in the development of an effective and safe HIV-1 vaccine1–4. One mechanism by which HIV escapes the host immune response is to coat the envelope glycoprotein gp120 with a glycan shield composed of diverse N-linked oligosaccharides derived from the host glycosylation machinery, so called immunologically ‘self’ glycans5–7. Furthermore, during transport of the nascent glycoprotein gp160 polypeptide through the Endoplasmic reticulum and Golgi complex, the glycans acquire complex and highly diverse structures by terminal glycosylation, leading to tremendous glycan heterogeneity on gp1208–13. Although the antibody 2G12 isolated from HIV carriers has been shown to have both heavy and light chains intertwined to recognize the oligomannose epitopes on gp12014, and is capable of neutralizing about 30% of the existing HIV variants, the antibodies induced by the oligomannose-based vaccine(s) unfortunately failed to neutralize HIV-1 virions, despite the presence of high-mannose glycans on the expressed gp12015–19. One possible reason for this failure is that the synthetic epitope did not represent the native 2G12 epitope. Recently, a series of new monoclonal broadly neutralizing antibodies (bNAbs) isolated from HIV-1-infected patients were found to neutralize a broad spec-trum of HIV-1 strains20–29. Among this pool of antibodies, some recognize peptide epitopes located at the CD4 binding site23 or glycan epitopes at the variable loops (antibodies PG9/PG16)24–26. The excellent neutralization potency exhibited by these bNAbs, especially those targeting the N-glycans, suggests that these epitopes may be used for vaccine development. This hypothesis was validated by a recent structural study of antibodies PG9 and PG16, which recognize two adjacent heteroglycans30, and the observation was further supported by a binding study using synthetic glycopeptides31. Unfortunately, the gp120 used in these structural studies was from GlcNAc transferase-deficient (GnTI−/−) human embryonic kidney (HEK) 293S or 293F cells26,30, which mainly produce the high-mannose-type N-glycans, which may not be the true ligands for these antibodies. Similarly, the gp120 from different expression systems such as insect cells32,33, HEK 293T cells10,34,35, 37 CHO cells36,37 and swainsonine-treated HEK 293F cells30 also resulted in specific glycosylation profiles8, most of which were also high-mannose types, and therefore any functional study of gp120 glycans using these systems could be problematic. A microarray approach with diverse glycan structures may be used to elucidate the binding specificities of bNAbs, but to our knowledge, no glycan microarray-based study has given a complete understanding of specificity, especially when such antibodies target different glycans simultaneously on gp120.
The development of glycan microarrays has allowed an unprecedented high-throughput exploration of the specificities of a diverse range of glycan-binding proteins38–46, including the most comprehensive one available, from the Consortium of Functional Glycomics (CFG), which contains more than 600 oligosaccharides on an N-hydroxysuccinimide (NHS)-activated glass slide41. However, the spacer group and immobilization chemistry used in different array formats clearly result in differences in the density, distribution and orientation of glycan presentation, which may dramatically affect the binding affinity and even specificity in glycan protein interactions. Therefore, a cross-comparison among different array platforms and development of new glycan arrays to improve the sensitivity of detection of hetero-ligand bindings are particularly important. Our group has recently introduced a glycan array on an aluminium-oxide-coated glass (ACG) slide using phosphonic acid- ended glycans for immobilization. Preliminary studies showed that the new array produced superior results in terms of signal intensity, homogeneity and sensitivity when compared with the commonly used arrays on glass slides47,48. Access to gp120-related N-glycans is a formidable task, because such structures are species- and cell-specific33,37 and difficult to obtain due to their structural diversity and micro-heterogeneity, as well as synthetic challenges. However, a major advance in N-glycan synthesis was achieved recently49–51 with the stepwise enzymatic extension of a chemically synthesized tri-antennary acceptor by Boons49 and a similar strategy by Wang51. Despite these advances, the development of a more efficient strategy for the synthesis of diverse N-glycans of high-mannose, hybrid- and complex-type structures (estimated to be around 20,000) remains a major challenge.
Results and discussion
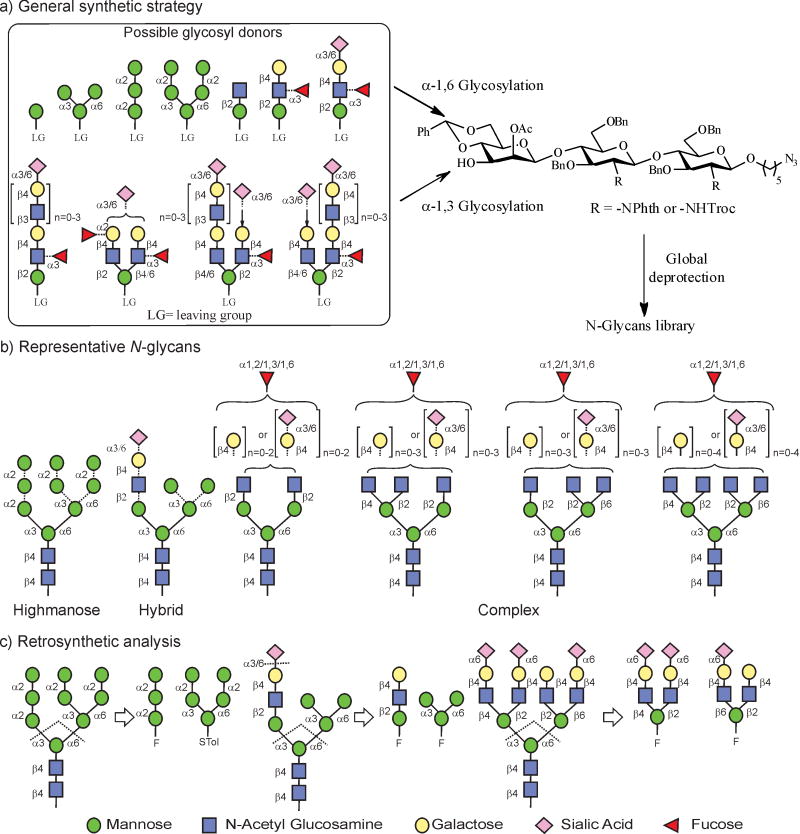
Modular synthesis of high-mannose, hybrid- and complex-type N-glycans. Recent analysis of a recombinant monomeric HIV-1gp120JRCSF from HEK 293T cells suggested the presence of extensive structural diversity with a characteristic cell-specific pattern32–37. The study of the glycan specificities of HIV-1 bNAbs thus requires pure, diverse and well-defined glycan samples in sufficient amounts. For this reason we developed a modular approach to the synthesis of a diverse array of N-glycans, as depicted in Fig. 1a. The strategy was designed on the basis that diversity can be created by assembly of the so-called ‘D1 and D2/D3 arm modules’, followed by the α-specific mannosylation at the O3 and/or O6 position of the mannose residue of the common core trisaccharide. Previously, the versatility of the glycosyl fluoride strategy was combined with enzymatic sialylation to build a library of symmetric bi-, tri- and tetra-antennary complex-type glycans50. Here, we extend the scope of this modular strategy to prepare high-mannose, hybrid and, more importantly, asymmetrically sialylated multi-antennary glycans (Fig. 1b).
Figure 1. A general strategy for the modular synthesis of gp120-related N-glycans.
Due to the large number of possibilities in glycosidic linkages generating a huge diversity of structures (around 20,000), especially from the GlcNAc residues to the non-reducing end, a modular approach is necessary to minimize the reaction steps and create enough diversity to reflect the nature of N-glycosylation. a, Synthesis of high-mannose-, hybrid- and complex-type N-glycans through regio- and stereoselective glycosidation of orthogonally protected core trisaccharide at the O3 and O6 positions with a modular set of diverse glycosyl donors. b, Representative N-glycans that can be generated by this strategy. c, Retrosynthetic disconnections of high-mannose-, hybrid- and complex-type glycans, showing the building blocks required for assembly.
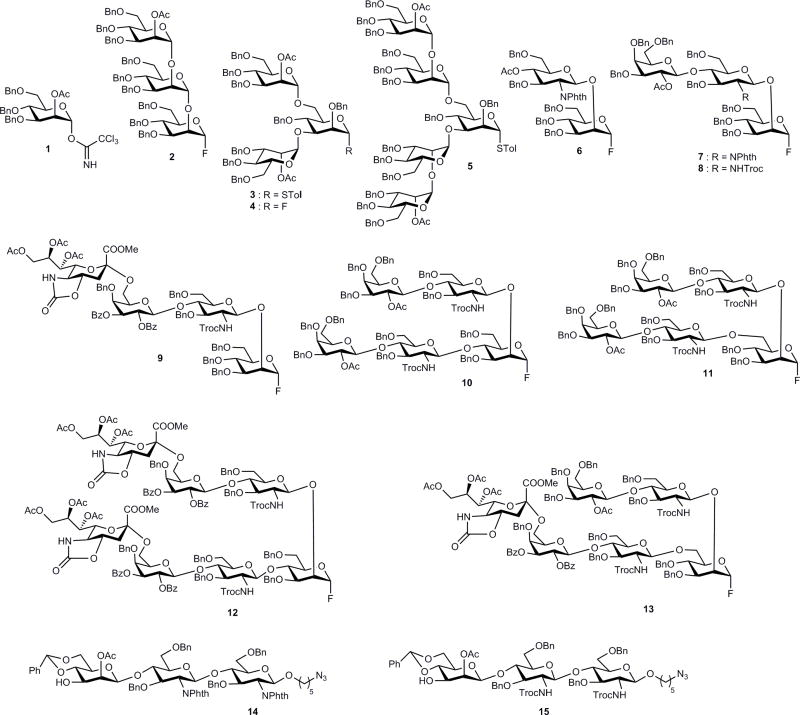
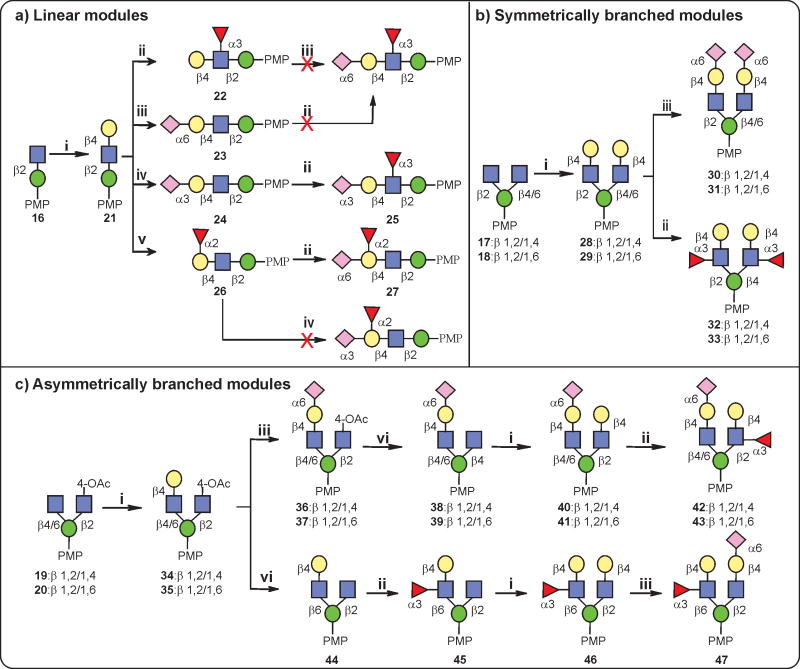
Based on the retrosynthetic disconnection of N-glycan structures (Fig. 1c), we envisioned that a modular set of building blocks 1–13 and core trisaccharides 14,15 with crucial β-mannoside linkage (Fig. 2) could be used as starting materials for the preparation of various N-glycans (G1–33, Supplementary Fig. 11). To illustrate this strategy, oligomannose-type (mono- to pentasaccharides, 1–5), complex-type (di- to heptasaccharides, 6–13) and core trisaccharide (14, 15) building blocks were first chemically synthesized on multigram scales (Supplementary Schemes 1–8), with temporary anomeric protecting groups installed before transformation into fluorides. For the high-mannose series (Man3/Man4/Man5/Man9 GlcNAc2) glycans, donors 1 and 2, and for the hybrid series glycans, donors 6 and 7 were stereoselectively linked to the O3 position of 1450. The benzylidene ring was then removed to obtain 4,6-diol, and finally a regioselective glycosylation was achieved at the O6 position with donors 1–7. In the course of glycosylation reactions, various promoters were employed depending on the choice of glycosyl donors. The phthalimide protections at all glucosamine residues were modified to acetamides, and deacetylation and finally debenzylation were performed to obtain free glycans Supplementary Schemes 9–11 and Supplementary Fig. 1: glycans 1, 2, 4–9, 12). Taking advantage of their remarkable specificity, the complex-type D1 arm of the hybrid glycan was enzymatically sialylated52,53 to obtain α-2,3/6-Neu5Ac isoforms (Supplementary Schemes 12 and 13 and Supplementary Fig. 2, glycans 10, 11, 13, 14). Having established a rapid route to the complex-type glycans (glycans 15–17, 20–23, 26–28, 32 and 33, Supplementary Fig. 3), we turned our attention to more diverse asymmetric glycans and focused on the α-2,6-sialylated antigens recognized by PG9 and PG1630,31 using building blocks 9–13 prepared in gram quantities (Supplementary Schemes 4–8). In this process we observed that the removal of N-phthalimide protection from glycans with preinstalled sialic acid under strong heating conditions often provided undesirable side products, so the more versatile NH-Troc protection was installed at all GlcNAc residues. Fluoride donors 8–13 were then used for glycosidation of core 1550,54 under the promotion of AgOTf/Cp2HfCl2. Surprisingly, all these complex conjugations were found to be very clean, albeit in moderate yields (Supplementary Schemes 14–19), and the stereoselectivity was excellent, except for glycan G18 (Supplementary Scheme 14), where the glycosylation of 8 resulted in a mixture of isomers, suggesting that non-participating mannosylations are complex in terms of selectivity in some cases. Finally, a global deprotection afforded naturally occurring positional isomers of bi-, tri- and tetra-antennary asymmetric N-glycans (Supplementary Fig. 3, glycans 18, 24, 25, 29–31). To study the role of core fucose, examples 3 and 19 were also prepared (Supplementary Scheme 20). With this convergent synthesis strategy in place, a chemo-enzymatic approach to the synthesis of D1 and D2/D3 arm donors was explored to allow a rapid assembly of diverse N-glycans55,56. To this end, various glycosyl transferases were used, including β-1,4-galactosyltransferases51,57, α-2,3/2,6-sialyltransferases52,53 and α-1,3/1,2-fucosyltransferases49,51,58, for the preparation of linear and branched modules by enzymatic extension of chemically synthesized acceptors 16–20 (Fig. 3 and Supplementary Scheme 22). The GlcNAc moiety of acceptor 16 was transformed into LacNAc using β-1,4-galactosyltransferase and uridine 5′-diphosphogalactose (UDP-Gal) to form 21, which was further extended by α-2,6/2,3-sialyltransferase in the presence of cytidine-5′-monophospho-N-acetylneuraminic acid (CMP-Neu5Ac) to provide targets 23 and 24, respectively. Next, treatment of 21, 23 and 24 with the α-1,3-fucosyltransferase from Helicobacter pylori (Hpα1,3FT) resulted in the modification of LacNAc and α-2,3-sialyl LacNAc but not 2,6-sialyl LacNAc to afford 22 and 25. In addition, the α-1,3-fucosylated LacNAc was found to restrict the access of enzymatic α-2,3/6-sialylation to the terminal galactose. Acceptor 21 was modified with the α-1,2-fucosyltransferase from HEK293 cells in the presence of guanosine 5′-diphospho-β-L-fucose (GDP-fucose) to provide 26 (Fig. 3a). It was observed that the α-1,2-fucosylated module 26 was accepted by the α-2,6-sialyltransferase to give 27, but the α-2,3-sialyltransferase failed to accept this substrate (Fig. 3a and Supplementary Scheme 23). Next, symmetric modules 28–33 were prepared from acceptors 17 and 18 (Fig. 3b and Supplementary Scheme 24). In the case of asymmetric modules, selective incorporation of sialic acid or fucose to one of the antennas is necessary. Therefore, acceptors 19 and 20 were designed in such a way that the GlcNAc at the mannose O2 position was differentiated from the GlcNAc at the O4 or O6 position by masking the 4-hydroxy group through acetylation to prevent enzymatic galactosylation while retaining its water solubility. This strategy allowed a selective extension of one arm while keeping the other intact. As depicted in Fig. 3c, a Gal residue was added by β-1,4-GalT to the GlcNAc residue at the β-1,4/1,6-mannose branch, whereas the GlcNAc residue at the β-1,2 branch remained intact. By taking advantage of their specificity, α-1,3-FucT and α-2,6-SiaT were used for the preparation of asymmetric modules 36–43 and 44–47 (Fig. 3c and Supplementary Scheme 25), which were purified and fully characterized (Supplementary Section III).
Figure 2. Structures of D1 and D2/D3 arm building blocks.
A modular set of building blocks prepared by total chemical synthesis and used for oligosaccharide assembly.
Figure 3. Chemo-enzymatic synthesis of modules.
a–c, Representative chemo-enzymatic approach to the synthesis of linear (a), symmetric (b) and asymmetrically branched (c) modules essential for N-glycan assembly. Reagents and conditions: (i), UDP-galactose, β-1,4-GalT; (ii), GDP-fucose, α-1,3-FucT;(iii), CMP-Neu5Ac, α-2,6-SiaT; (iv), CMP-Neu5Ac, α-2,3-SiaT; (v), GDP-fucose, α-1,2-FucT; (vi), NaOH.
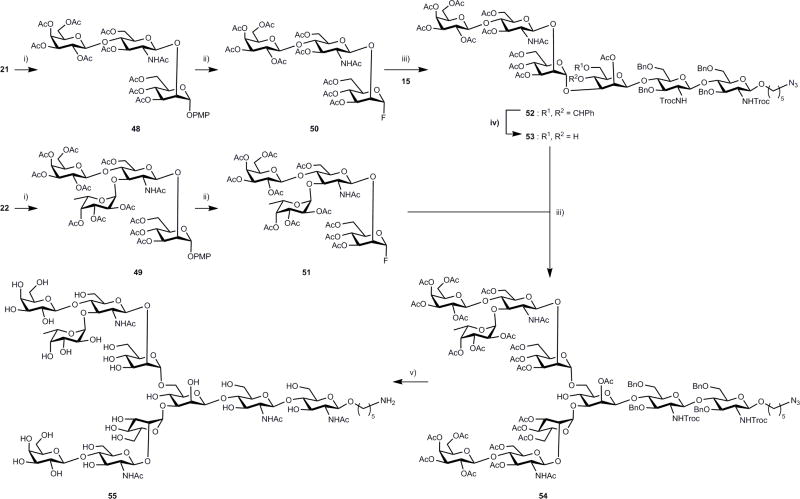
To illustrate the use of modules prepared by the chemo-enzymatic method for further glycosylation, modules 21 and 22 were selected for a proof-of-concept experiment (Fig. 4). Peracetylation of modules 21 and 22, followed by transformation into the glycosyl fluoride, provided donors 50 and 51, respectively. Glycosylation with fluoride 50, in the presence of AgOTf/Cp2HfCl2, indeed provided the expected hexasaccharide 52 in 70% yield. Next, the benzylidene was cleaved in the presence of p-toluene sulfonic acid catalyst, and donor 51 was stereospecifically installed at the 6-position to give decasaccharide 54, which was deprotected to afford glycan 55 (Supplementary Schemes 26 and 27).
Figure 4. Proof-of-concept demonstration of chemo-enzymatic strategy to N-glycan synthesis.
Reagents and conditions: (i), acetic anhydride, pyridine;(ii), (1) CAN, toluene:ACN:H2O:toluene; (2) DAST, CH2Cl2, −30 °C; (iii), AgOTf, Cp2HfCl2, toluene, 4 Å MS, 0 °C to rt; (iv), p-TSA, acetonitrile, rt; (v), (1) LiOH, 1,4-dioxane:H2O; 90 °C, overnight; (2) Ac2O, pyridine, overnight; (3) NaOMe, MeOH, overnight; (4) Pd(OH)2, MeOH:H2O:HCOOH (5:3:2), H2.CAN, cerium ammonium nitrate; DAST, diethylaminosulfur trifluoride; AgOTf, silver trifluromethanesulfonate; Cp2HfCl2, bis(cyclopentadienyl)hafnium dichloride; MS, molecular sieves.
Taken together, we have demonstrated an efficient way to prepare the complex-type N-glycans of interest through a proper selection of a defined set of modules that are generated chemically as well as chemo-enzymatically. The versatility of oligosaccharyl fluoride donors has allowed a clean conjugation of highly branched modules to the core with excellent stereo- and regio-selectivity. The oligosaccharides with a pre-installed alkyl amine linker at the reducing end can be used directly for reaction with NHS slides through amide bond formation, further modified for other array formats, or conjugated to proteins for structural and functional studies.
Glycan microarray on NHS-activated and aluminium-oxide coated glass slides
It has been reported that PG9, PG16 and PGTs 128 and 141–145 are able to neutralize 70–80% of circulating HIV-1 isolates with potent activity20, suggesting that the targeted epitopes are highly conserved among the HIV-1 variants and could guide the design of immunogens. To gain insights into the glycan specificities of these antibodies, we used our newly developed array to profile the ligands of HIV-1 bNAbs. The synthetic N-glycan ligands were printed on NHS-activated glass slides through amide bond formation, with 100 μM each of glycans 1–33 (Supplementary Fig. 11). Each sample was printed with five replicates, and slide images were obtained from a fluorescence scan after incubation with DyLight649-conjugated donkey anti-human IgG antibody. Our results revealed that PG16 binds to the α-2,6-sialylated complex-type oligosaccharides, consistent with our previous results50, and the binding affinity is proportional to the number of terminal sialic acid residues (Supplementary Fig. 12). In addition, we found that the PG16 binding was not affected by the presence of the core fucose (glycan 19 versus 16). Interestingly, the binding of PG16 to asymmetric glycans 29–33 suggests the importance of sialic acid on the D1 arm. Finally, we could not observe binding to the high-mannose-type glycan Man5GlcNAc2 (Supplementary Fig. 12). PG9 and PGTs 141–145 were not observed to bind any of the glycans on the NHS array, probably due to their extremely weak binding (Supplementary Figs 13–15). In our binding studies, we observed a strong fluorescent signal against glycan 5 (Man4GlcAc2), which was later confirmed to be from the non- specific binding of secondary antibody (Supplementary Fig. 16).
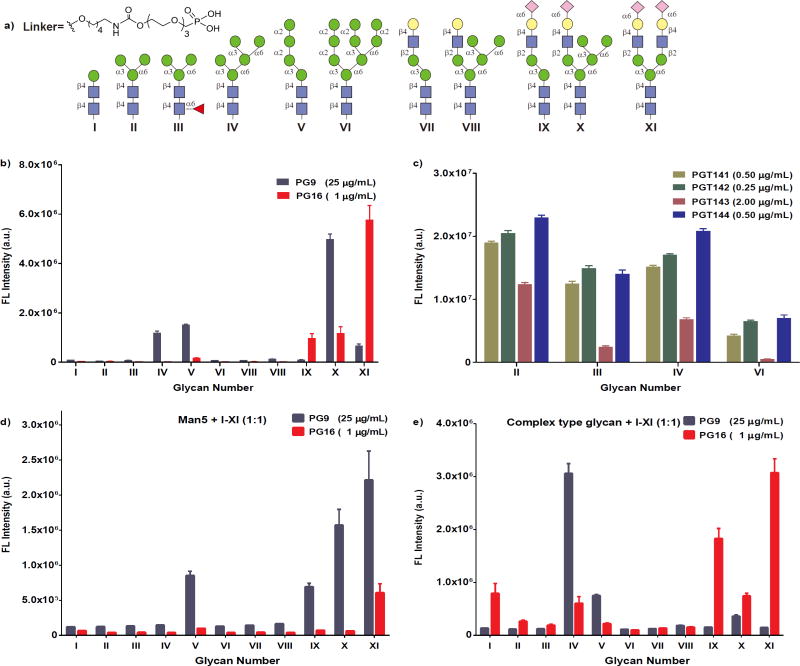
To further understand why glycan binding to these bNAbs was not readily observed on the NHS-coated glass slides, we conducted a specificity test with the ACG array. A comparison of homogeneity between the ACG- and NHS-coated glass slides showed that the ACG slide provided a more homogeneous glycan distribution on its surface (Supplementary Figs 4–9). Also, based on atomic force microscopy (AFM), the structural orientation of the glycans on the ACG slide was in a more extended conformation (Supplementary Fig. 10). We can therefore simply adjust the concentration of glycan to control the density and distance of glycans on the ACG surface. To prepare a representative ACG array, glycans I–XI (Fig. 5a) were linked to a phosphonic acid tail for spontaneous covalent immobilization on the ACG slide (Supplementary Scheme 21). After incubation with a secondary antibody, we determined the dissociation constant (KD.surf.)59 of the glycans interacting with the antibody of interest. On using the ACG slide the signal intensity was enhanced, as shown by a comparison of the use of PG16 at 1 μg ml−1 on the ACG slide and 5 μg ml−1 on the NHS-coated glass slide (Fig. 5b and Supplementary Fig. 12). The binding affinity of PG16 to the bi- antennary complex-type N-glycan (XI) (KD = 0.320 μM) was higher than that to the hybrid-type glycan (X, KD = 0.935 μM; Supplementary Fig. 19 and Supplementary Table 2), supporting the proposed existence of these glycans at Asn173, as suggested by a structural study of PG16 in complex with gp12030. To evaluate whether the ACG array format could enhance the detection sensitivity we performed ligand specificity profiling at various concentrations of PG9. Interestingly, on the ACG array, PG9 showed an apparent specificity for the hybrid-type glycan (X, Fig. 5b) and detectable binding to Man5GlcNAc2 (IV) and the α-2,6-sialylated bi-antennary complex-type oligosaccharide (XI). Previously, it was shown that PG9 required Man5GlcNAc2 at primary (Asn160) and secondary (Asn156 or Asn173) binding sites together with a short peptide strand for gp120 recognition26, but the composition of glycan at Asn156 or Asn173 was defined as a complex-type glycan in later studies30,31. In the present study, the strong PG9 interaction with the hybrid-type structure compared with both Man5GlcNAc2 and complex-type glycans indicated the presence of a hybrid-type glycan or an oligomannose and a complex-type glycan in close proximity as ligand(s). Nonetheless, to our knowledge, these results represent the first evidence of PG9 binding to carbohydrates without protein or peptide domains.
Figure 5. Glycan specificities of HIV-1 bNAbs on ACG array.
a, Synthetic N-glycans are chemically modified with a phosphonic acid tail for covalent attachment to the ACG slide through phosphonate chemistry. b,c, Binding of PG9, PG16 and PGTs 141–144 to structures I–XI printed on an ACG slide. d,e, Binding of PG9 and PG16 to each of the glycan mixtures was evaluated to determine the effect of adjacent glycans on binding affinity. Arrays were printed by mixing 100 μM of Man5GlcNAc2 or the complex-type glycan with every structure from I to XI in a 1:1 ratio. Molar concentrations (in μM) for antibodies are given in the legend. The mean signal intensities and standard errors calculated for five independent replicates on the array are shown.
To understand the exact glycan epitopes recognized by antibodies PGTs 141–145 (ref. 20), a panel of glycans I–XI on an ACG slide was prepared for analysis. The result revealed that PGTs 141–144 could recognize the oligomannose glycans Man3/5GlcNAc2, and the observed trend in binding affinity was PGT142 > PGT144 > PGT141 > PGT143 (Fig. 5c and Supplementary Fig. 18). However, PGT145, the most potent of the group, failed to show detectable binding. The significant decrease in the affinity for PGTs 141–144 towards Man9GlcNAc2 was probably due to shielding of the inner core (Man3/5GlcNAc2) by terminal mannose residues. Taken together, these results demonstrate the efficiency of the ACG array format in detecting the low-affinity interactions of recently isolated HIV-1 bNAbs.
Heteroglycan binding of PG9 and PG16
Due to the absence of a co-crystal structure of PG9 in complex with a hybrid-type glycan, it is difficult to determine the molecular details of the interaction. The structural features suggest that PG9 could accommodate the high- mannose-type D2/D3 arm and the complex-type D1 arm present in a hybrid-type glycan, or has binding sites that can accommodate Man5GlcNAc2 at Asn160 and complex-type glycans at Asn156 or 17330,31. However, both complex- and hybrid-type glycans contain the α-2,6-NeuAc-Gal-GlcNAc arm.
To evaluate the glycan combination at Asn160 and Asn156/Asn173 of gp120 that best fits into the PG9 binding pocket, we printed two different mixed-glycan arrays. In one array, Man5GlcNAc2 (IV) was mixed with every glycan from I to XI in a 1:1 mole ratio (Fig. 5d), while in the other, the bi-antennary complex-type structure (XI) was mixed with every glycan from I to XI (Fig. 5e). The binding profile of PG9 to various mixtures suggests that a mixture containing Man5GlcNAc2 and a bi-antennary glycan [(IV+XI) or (XI+IV)] interacted more strongly with PG9 than IV or XI alone. Furthermore, we also observed a comparable binding to Man5 combined with X and XI, suggesting that Man5GlcNAc2 at Asn160 was the primary binding site, while structures IX, X and XI used the complex-type D1 arm for interaction with the secondary binding site. Based on the homogeneous array results of PG9 (Fig. 5b), Man5GlcNAc2 IV or complex-type glycan XI alone did not seem to provide sufficient binding affinity. On the other hand, the hybrid-type glycan X showed a significant enhancement in binding. In the mixed glycans study (Fig. 5c), however, a combination of IV and XI was found to achieve the strongest binding to PG9, followed by a combination of Man5 and hybrid type. In a similar manner, we studied the binding specificity of PG16 (Fig. 5e) and it was found that the combination of Man5GlcNAc2 and complex-type N-glycan (IV+XI) (KD = 0.827 μM) or the combination of hybrid and complex glycans (X+XI) (KD = 0.988 μM) was weaker than the complex-type glycan alone (KD = 0.320 μM) (Supplementary Fig. 19 and Supplementary Table 2). These results indicate the importance of sialylated antennas in the PG16 binding site, including possibly the tri- and tetra-antennary complex-type N-glycans reported previously50.
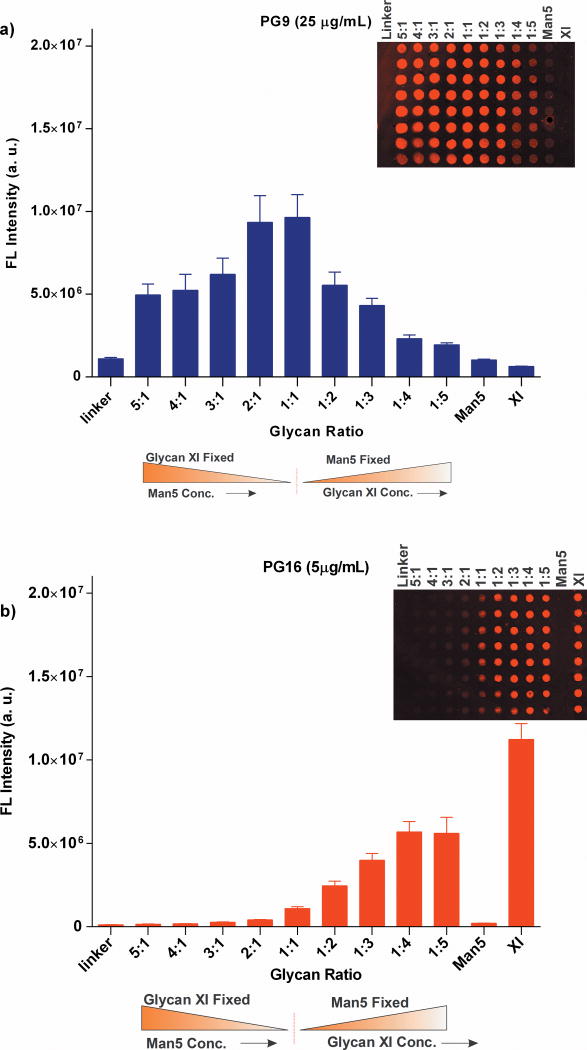
To further understand the exact ratio of IV and XI in the mixture we performed a serial dilution experiment. IV was mixed with XI in various ratios (1:1/2/3/4/5) and vice versa. A 100 μM solution of each of these mixtures was printed on the ACG surface together with linker and glycans alone as control. Interestingly, at fixed IV, the PG9 binding was gradually decreased by increasing the XI ratio in the mixture. By changing the IV ratio at fixed XI, PG9 achieved the strongest binding at ratios of 1:1 and 2:1 of IV to XI (Fig. 6a). These results suggest that a glycan ratio of 1:1 provides the best ligands for PG9. However, PG16 responded in a different manner to each of these mixtures, and the interactions were greatly enhanced by the presence of complex-type glycan in the mixtures (Fig. 6b). However, we were unable to detect PG16 binding to Man5GlcNAc2. We conclude that PG9 recognizes a mixture of Man5 and complex-type glycan, whereas the complex-type glycan alone is enough to elicit a PG16 response.
Figure 6. Glycan specificities of PG9 and PG16 to mixtures of Man5 and XI in various ratios.
Arrays were printed with 100 μM of linker, Man5GlcNAc2 (IV), complex-type glycan (XI) and each of the mixtures of (IV+XI) or (XI+IV) in 1:1/2/3/4/5 ratios. Molar concentrations (in μM) for antibodies are given in the legend. Mean signal intensities and standard errors calculated for eight independent replicates on the array are shown.
Conclusions
We have successfully developed a modular synthetic strategy for the rapid production of a diverse array of high-mannose-, hybrid- and complex-type N-linked oligosaccharides in highly pure and sufficient amounts, making possible the study of various N-glycans and the development of new glycan array platforms to determine the glycan specificities of newly discovered HIV-1 bNAbs. The ACG array and the binding measurements obtained in a high throughput manner together provide an effective means for detecting the extremely weak binding of HIV-1 bNAbs to glycans and enable the discovery and understanding of essential epitopes and hetero-ligands recognized by antibodies. These findings may aid the speedy design of effective carbohydrate-based vaccines against HIV-1.
METHODS
All reactions were performed under an inert atmosphere using dry solvents in anhydrous conditions, unless otherwise noted. Full experimental details, glycan microarray analysis and characterization data (1H and 13C NMR, high-resolution mass spectrometry and Rf values) for all new compounds are provided in Supplementary Sections I–III.
Supplementary Material
Acknowledgments
The authors thank D. Chiang and C.-N. Hsiao for their internal coordination within the Thin Film Technology Division, Instrument Technology Research Center (ITRC) and National Applied Research Laboratories, Hsinchu Science Park, Taiwan. The authors also Thank W.-H. Cho of ITRC for the vapor deposition polymerization (VDP) coating of aluminium-coated glass slides as well as the ellipsometry characterization work, N.-N. Chu and C.-Y. Su of ITRC for their work on SEM and AFM instrumental measurements, and L.-W. Lo of GRC for help with the use of various microscopy instruments, especially the confocal microscope (Leica SP5). Special thanks go to N.-H. Lin, L.-M. Wang and C.-C. Huang of CHO Pharma Inc. This work was supported by the National Science Council (grant no. NSC101-2321-B-001-024), Academia Sinica, the National Institutes of Health and the International Aids Vaccine Initiative (IAVI).
Footnotes
Author contributions
S.S.S., C.-Y.W. and C.-H.W. conceived and designed the experiments. S.S.S., S.-H.C., T.-I.T., S.Y.T, V.S.S., Y.-S.L., Y.-Y.C., C.-T.R., C.-C.L., S.P., C.-S.T., H.-W.S., Y.-F.Z. and C.-H.L. performed the experiments. S.S.S., C.-Y.W. and C.-H.W. analysed the data. D.R.B. and P.D.K. contributed materials/analysis tools. S.S.S., C.-Y.W. and C.-H.W. wrote the paper.
Supplementary information and chemical compound information are available in the online version of the paper.
Competing financial interests. The authors declare no competing financial interests.
References
- 1.Barouch DH. Challenges in the development of an HIV-1 vaccine. Nature. 2008;455:613–619. doi: 10.1038/nature07352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gaschen B, et al. AIDS - Diversity considerations in HIV-1 vaccine selection. Science. 2002;296:2354–2360. doi: 10.1126/science.1070441. [DOI] [PubMed] [Google Scholar]
- 3.Astronomo RD, Burton DR. Carbohydrate vaccines: developing sweet solutions to sticky situations? Nat Rev Drug Discov. 2010;9:308–324. doi: 10.1038/nrd3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Craigo JK, Montelaro RC. Lessons in AIDS vaccine development learned from studies of equine infectious, anemia virus infection and immunity. Viruses. 2013;5:2963–76. doi: 10.3390/v5122963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Scanlan CN, Offer J, Zitzmann N, Dwek RA. Exploiting the defensive sugars of HIV-1 for drug and vaccine design. Nature. 2007;446:1038–1045. doi: 10.1038/nature05818. [DOI] [PubMed] [Google Scholar]
- 6.Kwong PD, Mascola JR, Nabel GJ. Rational Design of Vaccines to Elicit Broadly Neutralizing Antibodies to HIV-1. Cold Spring Harb Perspect Med. 2011;1:a007278. doi: 10.1101/cshperspect.a007278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burton DR, Mascola JR. Antibody responses to envelope glycoproteins in HIV-1 infection. Nature Immunol. 2015;16:571–6. doi: 10.1038/ni.3158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pritchard LK, Harvey DJ, Bonomelli C, Crispin M, Doores KJ. Cell- and Protein-Directed Glycosylation of Native Cleaved HIV-1 Envelope. J Virol. 2015;89:8932–44. doi: 10.1128/JVI.01190-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pritchard LK, et al. Structural Constraints Determine the Glycosylation of HIV-1 Envelope Trimers. Cell Rep. 2015;11:1604–13. doi: 10.1016/j.celrep.2015.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Doores KJ, et al. Envelope glycans of immunodeficiency virions are almost entirely oligomannose antigens. Proc Natl Acad Sci U S A. 2010;107:13800–13805. doi: 10.1073/pnas.1006498107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pabst M, Chang M, Stadlmann J, Altmann F. Glycan profiles of the 27 N-glycosylation sites of the HIV envelope protein CN54gp140. Biol Chem. 2012;393:719–730. doi: 10.1515/hsz-2012-0148. [DOI] [PubMed] [Google Scholar]
- 12.Go EP, et al. Comparative Analysis of the Glycosylation Profiles of Membrane-Anchored HIV-1 Envelope Glycoprotein Trimers and Soluble gp140. J Virol. 2015;89:8245–57. doi: 10.1128/JVI.00628-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang W, et al. A systematic study of the N-glycosylation sites of HIV-1 envelope protein on infectivity and antibody-mediated neutralization. Retrovirology. 2014;10:14. doi: 10.1186/1742-4690-10-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Calarese DA, et al. Antibody domain exchange is an immunological solution to carbohydrate cluster recognition. Science. 2003;300:2065–2071. doi: 10.1126/science.1083182. [DOI] [PubMed] [Google Scholar]
- 15.Wang LX. Carbohydrate-based vaccines against HIV/AIDS. Acs Sym Ser. 2006;932:133–160. [Google Scholar]
- 16.Joyce JG, et al. An oligosaccharide-based HIV-1 2G12 mimotope vaccine induces carbohydrate-specific antibodies that fail to neutralize HIV-1 virions. Proc Natl Acad Sci U S A. 2008;105:15684–15689. doi: 10.1073/pnas.0807837105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang LX. Synthetic carbohydrate antigens for HIV vaccine design. Curr Opin Chem Biol. 2013;17:997–1005. doi: 10.1016/j.cbpa.2013.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Horiya S, MacPherson IS, Krauss IJ. Recent strategies targeting HIV glycans in vaccine design. Nat Chem Biol. 2014;10:990–999. doi: 10.1038/nchembio.1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fernandez-Tejada A, Haynes BF, Danishefsky SJ. Designing synthetic vaccines for HIV. Expert Rev Vaccines. 2015;14:815–31. doi: 10.1586/14760584.2015.1027690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Walker LM, et al. Broad neutralization coverage of HIV by multiple highly potent antibodies. Nature. 2011;477:466–470. doi: 10.1038/nature10373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sok D, Moldt B, Burton DR. SnapShot: broadly neutralizing antibodies. Cell. 2013;155:728–728. doi: 10.1016/j.cell.2013.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Garces F, et al. Structural evolution of glycan recognition by a family of potent HIV antibodies. Cell. 2014;159:69–79. doi: 10.1016/j.cell.2014.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu X, et al. Rational design of envelope identifies broadly neutralizing human monoclonal antibodies to HIV-1. Science. 2010;329:856–861. doi: 10.1126/science.1187659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pancera M, et al. Crystal structure of PG16 and chimeric dissection with somatically related PG9: structure-function analysis of two quaternary-specific antibodies that effectively neutralize HIV-1. J Virol. 2010;84:8098–8110. doi: 10.1128/JVI.00966-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Doores KJ, Burton DR. Variable Loop Glycan Dependency of the Broad and Potent HIV-1-Neutralizing Antibodies PG9 and PG16. J Virol. 2010;84:10510–10521. doi: 10.1128/JVI.00552-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McLellan JS, et al. Structure of HIV-1 gp120 V1/V2 domain with broadly neutralizing antibody PG9. Nature. 2011;480:336–343. doi: 10.1038/nature10696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pejchal R, et al. A potent and broad neutralizing antibody recognizes and penetrates the HIV glycan shield. Science. 2011;334:1097–1103. doi: 10.1126/science.1213256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mouquet H, et al. Complex-type N-glycan recognition by potent broadly neutralizing HIV antibodies. Proc Natl Acad Sci U S A. 2012;109:E3268–E3277. doi: 10.1073/pnas.1217207109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Falkowska E, et al. Broadly neutralizing HIV antibodies define a glycan-dependent epitope on the prefusion conformation of gp41 on cleaved envelope trimers. Immunity. 2014;40:657–68. doi: 10.1016/j.immuni.2014.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pancera M, et al. Structural basis for diverse N-glycan recognition by HIV-1-neutralizing V1–V2-directed antibody PG16. Nat Struct Mol Biol. 2013;20:804–813. doi: 10.1038/nsmb.2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Amin MN, et al. Synthetic glycopeptides reveal the glycan specificity of HIV-neutralizing antibodies. Nat Chem Biol. 2013;9:521–526. doi: 10.1038/nchembio.1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Murphy CI, et al. Enhanced expression, secretion, and large-scale purification of recombinant HIV-1 gp120 in insect cell using the baculovirus egt and p67 signal peptides. Protein Expres Purif. 1993;4:349–357. doi: 10.1006/prep.1993.1046. [DOI] [PubMed] [Google Scholar]
- 33.Kong L, et al. Expression-system-dependent modulation of HIV-1 envelope glycoprotein antigenicity and immunogenicity. J Mol Biol. 2010;403:131–147. doi: 10.1016/j.jmb.2010.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Go EP, et al. Characterization of glycosylation profiles of HIV-1 transmitted/founder envelopes by mass spectrometry. J Virol. 2011;85:8270–8284. doi: 10.1128/JVI.05053-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Eggink D, et al. Lack of complex N-glycans on HIV-1 envelope glycoproteins preserves protein conformation and entry function. Virology. 2010;401:236–247. doi: 10.1016/j.virol.2010.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhu X, Borchers C, Bienstock RJ, Tomer KB. Mass spectrometric characterization of the glycosylation pattern of HIV-gp120 expressed in CHO cells. Biochemistry. 2000;39:11194–11204. doi: 10.1021/bi000432m. [DOI] [PubMed] [Google Scholar]
- 37.Raska M, et al. Glycosylation patterns of HIV-1 gp120 depend on the type of expressing cells and affect antibody recognition. J Biol Chem. 2010;285:20860–20869. doi: 10.1074/jbc.M109.085472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.De Paz JL, Horlacher T, Seeberger PH. Oligosaccharide microarrays to map interactions of carbohydrates in biological systems. Methods Enzymol. 2006;415:269–292. doi: 10.1016/S0076-6879(06)15017-X. [DOI] [PubMed] [Google Scholar]
- 39.Oyelaran O, Gildersleeve JC. Glycan arrays: recent advances and future challenges. Curr Opin Chem Biol. 2009;13:406–413. doi: 10.1016/j.cbpa.2009.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rillahan CD, Paulson JC. Glycan microarrays for decoding the glycome. Annu Rev Biochem. 2011;80:797–823. doi: 10.1146/annurev-biochem-061809-152236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Blixt O, et al. Printed covalent glycan array for ligand profiling of diverse glycan binding proteins. Proc Natl Acad Sci U S A. 2004;101:17033–17038. doi: 10.1073/pnas.0407902101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fukui S, Feizi T, Galustian C, Lawson AM, Chai W. Oligosaccharide microarrays for high-throughput detection and specificity assignments of carbohydrate-protein interactions. Nat Biotechnol. 2002;20:1011–1017. doi: 10.1038/nbt735. [DOI] [PubMed] [Google Scholar]
- 43.Wang D, Liu S, Trummer BJ, Deng C, Wang A. Carbohydrate microarrays for the recognition of cross-reactive molecular markers of microbes and host cells. Nat Biotechnol. 2002;20:275–281. doi: 10.1038/nbt0302-275. [DOI] [PubMed] [Google Scholar]
- 44.Fazio F, Bryan MC, Blixt O, Paulson JC, Wong C-H. Synthesis of sugar arrays in microtiter plate. J Am Chem Soc. 2002;124:14397–14402. doi: 10.1021/ja020887u. [DOI] [PubMed] [Google Scholar]
- 45.Paulson JC, Blixt O, Collins BE. Sweet spots in functional glycomics. Nat Chem Biol. 2006;2:238–248. doi: 10.1038/nchembio785. [DOI] [PubMed] [Google Scholar]
- 46.Scurr DJ, et al. Surface characterization of carbohydrate microarrays. Langmuir. 2010;26:17143–17155. doi: 10.1021/la1029933. [DOI] [PubMed] [Google Scholar]
- 47.Chang SH, et al. Glycan array on aluminum oxide-coated glass slides through phosphonate chemistry. J Am Chem Soc. 2010;132:13371–13380. doi: 10.1021/ja1046523. [DOI] [PubMed] [Google Scholar]
- 48.Tseng SY, et al. Glycan arrays on aluminum-coated glass slides. Chem-Asian J. 2008;3:1395–1405. doi: 10.1002/asia.200800154. [DOI] [PubMed] [Google Scholar]
- 49.Wang Z, et al. A general strategy for the chemoenzymatic synthesis of asymmetrically branched N-glycans. Science. 2013;341:379–383. doi: 10.1126/science.1236231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shivatare SS, et al. Efficient convergent synthesis of bi-, tri-, and tetra-antennary complex type N-glycans and their HIV-1 antigenicity. J Am Chem Soc. 2013;135:15382–15391. doi: 10.1021/ja409097c. [DOI] [PubMed] [Google Scholar]
- 51.Li L, et al. Efficient chemoenzymatic synthesis of an N-glycan isomer library. Chem Sci. 2015;6:5652–5661. doi: 10.1039/c5sc02025e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Takakura Y, Tsukamoto H, Yamamoto T. Molecular cloning, expression and properties of an alpha/beta-Galactoside alpha 2,3-sialyltransferase from Vibrio sp JT-FAJ-16. J Biochem. 2007;142:403–412. doi: 10.1093/jb/mvm147. [DOI] [PubMed] [Google Scholar]
- 53.Tsukamoto H, Takakura Y, Mine T, Yamamoto T. Photobacterium sp JT-ISH-224 produces two sialyltransferases, alpha-/beta-galactoside alpha2,3-sialyltransferase and beta-galactoside alpha2,6-sialyltransferase. J Biochem. 2008;143:187–197. doi: 10.1093/jb/mvm208. [DOI] [PubMed] [Google Scholar]
- 54.Toshima K. Glycosyl fluorides in glycosidations. Carbohydr Res. 2000;327:15–26. doi: 10.1016/s0008-6215(99)00325-0. [DOI] [PubMed] [Google Scholar]
- 55.Rabbani S, Schwardt O, Ernst B. Glycosyltransferases: An efficient tool for the enzymatic synthesis of oligosaccharides and derivatives as well as mimetics thereof. Chimia. 2006;60:23–27. [Google Scholar]
- 56.Muthana S, Yu H, Huang S, Chen X. Chemoenzymatic synthesis of size-defined polysaccharides by sialyltransferase-catalyzed block transfer of oligosaccharides. J Am Chem Soc. 2007;129:11918–11919. doi: 10.1021/ja075736b. [DOI] [PubMed] [Google Scholar]
- 57.Lau K, et al. Highly efficient chemoenzymatic synthesis of β1–4-linked galactosides with promiscuous bacterial β1–4-galactosyltransferases. Chem Commun. 2010;46:6066–6068. doi: 10.1039/c0cc01381a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Soriano del Amo D, et al. Chemoenzymatic synthesis of the sialyl Lewis X glycan and its derivatives. Carbohydr Res. 2010;345:1107–13. doi: 10.1016/j.carres.2010.03.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liang PH, Wang SK, Wong C-H. Quantitative analysis of carbohydrate-protein interactions using glycan microarrays: Determination of surface and solution dissociation constants. J Am Chem Soc. 2007;129:11177–11184. doi: 10.1021/ja072931h. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.