Abstract
Metabolic modeling of dynamic 13C labeling curves during infusion of 13C-labeled substrates allows quantitative measurements of metabolic rates in vivo. However metabolic modeling studies performed in the brain to date have only modeled time courses of total isotopic enrichment at individual carbon positions (positional enrichments), not taking advantage of the additional dynamic 13C isotopomer information available from fine-structure multiplets in 13C spectra. Here we introduce a new 13C metabolic modeling approach using the concept of bonded cumulative isotopomers, or bonded cumomers. The direct relationship between bonded cumomers and 13C multiplets enables fitting of the dynamic multiplet data. The potential of this new approach is demonstrated using Monte-Carlo simulations with a brain two-compartment neuronal-glial model. The precision of positional and cumomer approaches are compared for two different metabolic models (with and without glutamine dilution) and for different infusion protocols ([1,6-13C2]glucose, [1,2-13C2]acetate, and double infusion [1,6-13C2]glucose + [1,2-13C2]acetate). In all cases, the bonded cumomer approach gives better precision than the positional approach. In addition, of the three different infusion protocols considered here, the double infusion protocol combined with dynamic bonded cumomer modeling appears the most robust for precise determination of all fluxes in the model. The concepts and simulations introduced in the present study set the foundation for taking full advantage of the available dynamic 13C multiplet data in metabolic modeling.
Keywords: bonded cumomer, isotopomer, metabolic modeling, 13C magnetic resonance spectroscopy, brain, neuronal-glial metabolism, Monte-Carlo simulation
Introduction
Carbon-13 NMR spectroscopy is a unique tool to quantitatively assess metabolic fluxes in vivo [1–3]. Measurements rely on infusion of a 13C-labeled substrate (e.g. glucose, acetate and other substrates) and on subsequent detection of 13C incorporation into metabolites using NMR spectroscopy. Analysis of these experimentally measured 13C labeling curves using metabolic models ultimately yields quantitative estimates of metabolic rates.
Two different families of metabolic models can be distinguished depending on the temporal dynamics of the input data: isotopic steady-state models versus isotopic dynamic models. In the first case, metabolic modeling using 13C enrichments at isotopic steady-state allows determination of metabolic flux ratios, but does not provide absolute flux values [4,5]. In contrast, dynamic metabolic models, which are based on the measurement of progressive 13C incorporation into metabolites (i.e. before isotopic steady state has been reached), can yield absolute values of metabolic rates.
Independently of the temporal dynamics, metabolic models also differ depending on the chemical specificity of the input data. Here again two main families can be distinguished: positional models versus isotopomer models. While positional models are based only on the total amount of label accumulated at each carbon position (yielding at most N independent isotopic time-courses or steady-state values for a metabolite with N different carbons), isotopomer models take full advantage of the biochemical information that can be assessed with NMR spectroscopy by considering all different isotopomers, i.e. all the different possible combinations of labeled and unlabeled carbons (yielding at most 2N−1 independent variables for a metabolite with N different carbons).
In principle, dynamic modeling of isotopomer time-courses should allow the derivation of metabolic fluxes with the highest achievable precision and accuracy by making optimal use of all the available data. However, such dynamic modeling of isotopomer time courses is still in its infancy. Although isotopomer modeling at isotopic steady-state is commonly used on microorganisms [6,7], it has only been rarely used to analyze in vivo data. Conversely, dynamic modeling in the brain in vivo has been performed using time courses of total positional enrichments (for review [1–3]) but not using time courses of 13C multiplets arising from isotopomers. To the best of our knowledge, dynamic analysis of 13C multiplet time courses has only been used to study heart metabolism ex vivo and indeed led to increased precision on the determination of metabolic fluxes [8,9]. Early seminal work in perfused heart with a one-compartment model also used time courses of isotopomer concentrations as variables in the model in order to simulate time courses of positional enrichment, but only positional enrichment curves were fitted [10].
The fact that so few in vivo metabolic modeling studies have taken advantage of the additional dynamic 13C isotopomer information available from 13C multiplets can be attributed to two factors. First, dynamic measurement of fine-structure multiplets in 13C spectra is difficult in vivo due to broader linewidth and limited sensitivity. However, it has been shown in the recent years that using high field NMR systems allowed the dynamic detection of 13C multiplets in the rat brain during an infusion of [1,6-13C]glucose [11] or [U-13C]glucose [12]. The second reason is that no metabolic model is currently readily available to take this additional dynamic isotopomer information into account in the modeling.
In this context, the goal of the present paper is to introduce and assess a new metabolic modeling approach that allows analysis of time courses of individual 13C multiplets measured in the brain from 13C spectra, thereby taking full advantage of the information available in 13C spectra. Because not all possible isotopomers are detectable by NMR, a model including all possible isotopomers would be unnecessarily cumbersome. The concept of bonded cumulative isotopomers or bonded cumomers introduced in the present work leads to a reduced number of equations as well as a more simple derivation of equations compared to a model including all possible isotopomers, while retaining all the NMR-measurable isotopomer information. As explained in the theory section below, there is a straightforward relationship between bonded cumomers and 13C multiplets observed in 13C NMR spectra. The concept of cumomer was used in early studies by Muzykantov and Shestov [13] and the term cumomer was first proposed by Wiechert et al. in 1999 [14]. After exposing the theory and the practical construction of a bonded cumomer model, we investigate the potential of the new approach for metabolic modeling of brain compartmentalized neuronal-glial metabolism. Monte-Carlo simulations are performed to compare the precision of the new approach using bonded cumomers with the precision of the standard positional modeling approach for different labeling strategies, namely for the infusion of either [1,6-13C2]glucose or [1,2-13C2]acetate or of both substrates simultaneously.
Theory
Definition and properties
Let us denote M{i} the isotopomer of a metabolite M, where {i} is a set of integers i1, i2…, in indicating the positions of all the labeled carbons. The isotopomer fraction [M{i}]/[M] of molecules M labeled exactly at positions i1, i2…, in will be referred to as m{i}. By definition, a cumulative isotopomer fraction (or cumomer fraction), noted πM{i}, is the sum of isotopomer fractions for all isotopomers labeled at least at positions i1, i2…, in, whatever the label at other positions. The size n of the set {i} will be referred to as the order of the π-function. For example, assuming that M has a total of four carbons, the second order π-function πM{1,3} can be expressed as:
| (1) |
The cumomer fraction πM{1,3} for cumomer M{1,3} can be interpreted as the probability that both first and third carbons are labeled.
By definition, cumomer fractions are always comprised between 0 and 1. It is also clear that, if set {i} is included in set {j}, then πM{i}≥πM{j}. For example set {1,2} is included in set {1,2,3} and consequently πM{1,2}≥πM{1,2,3} because the probability of M being labeled on carbons 1 and 2 (irrespective of carbon 3) is greater than the probability of M being labeled on carbons 1, 2 and 3 simultaneously. Moreover, if the label input in the system is not decreasing, cumomers fractions are never decreasing (this is not the case with isotopomers). Finally, first-order cumomers fractions correspond to the positional enrichments as used in positional models.
By definition, bonded cumomers are cumomers whose indexes refer to adjacent carbons. For example, M{1,2} and M{1,2,3} are bonded cumomers, whereas M{1,3} and M{1,2,4} are not.
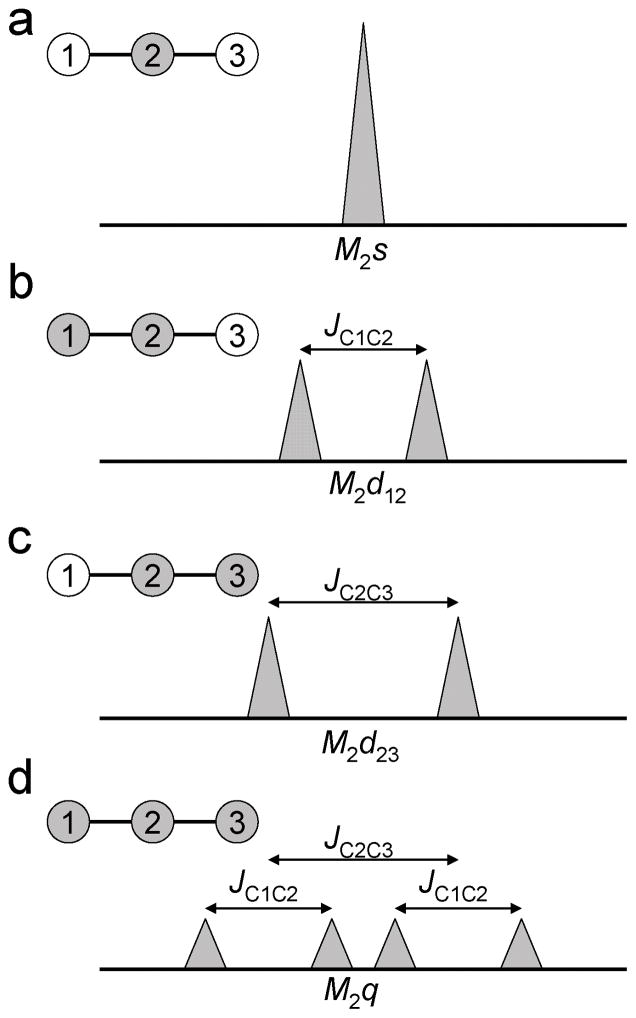
Bonded cumomers and fine structure of 13C NMR spectra (13C multiplets)
The fine structure of 1H-decoupled 13C NMR spectra exhibits spectral patterns characteristic of different isotopomers. Let us for example consider the 2nd carbon C2 of metabolite M, with two adjacent carbons C1 and C3 so that C2 is directly bound to C1 and C3. If C2 is a 13C isotope while C1 and C3 are 12C, the C2 resonance will appear as a singlet on 13C spectrum, whose intensity will be referred to as M2s (fig. 1A). If, in addition to C2, C1 is labeled but not C3, the carbon-carbon one-bond scalar coupling between C1 and C2 will result in the splitting of the C2 resonance into a doublet, which will be referred to as M2d12 (fig. 1B). Symmetrically, if C3 is labeled but not C1, the C2 resonance is split into a doublet M2d23 (fig. 1C). Finally, if both C1 and C3 are labeled in addition to C2, the C2 resonance is split into a doublet of doublet (quartet), referred to as M2q (fig. 1D). At this point it is important to realize that the C2 NMR spectral pattern is only sensitive to the labeling of its two direct neighbors, and is not significantly affected by the labeling of further carbons such a C4, C5…, due to the fact that long-range scalar coupling is too small (a few Hz) to yield any further visible splitting in vivo [15]. Therefore, the 4 different splitting patterns possible for any carbon with two direct neighbors are very naturally expressed in terms of the “bonded” cumomers of order n≤3, i.e. π-functions whose indexes refer to 3 or less adjacent carbons, using the following transformation:
| (2) |
Figure 1.
NMR spectra fine structure and isotopomers. For a given 13C labeled carbon in the carbon chain of metabolite M (C2 in this example), the resonance fine structure depends on the labeling of the direct neighbors C1 and C3. (a) Unlabeled C1 and C3 correspond to the singlet M2s. (b) Labeled C1 and unlabeled C3 correspond to the doublet M2d12, the frequency gap between the two peaks being the scalar coupling constant between C1 and C2, JC1C2. (c) Unlabeled C1 and labeled C3 correspond to the doublet M2d23, the frequency gap being JC2C3. (d) Simultaneously labeled C1 and C3 correspond to the doublet of doublet M2q.
In the equation above, intensities are normalized so that the total signal M2total (which is the sum of all multiplet components M2s + M2d23 + M2d12 + M2q) is equal to the isotopic enrichment of the carbon at the C2 position. Furthermore, if the coupling is identical between C1 and C2 on one hand, and C2 and C3 and the other hand, then M2d12 and M2d23 are undistinguishable (M2d), and the quartet M2q is actually a triplet (M2t). In this context, the above equation simplifies to:
| (3) |
It should be mentioned that a labeled carbon can in principle be bound to more than 2 labeled carbons, provided the carbon chain presents some branching. This would lead to some additional carbon-carbon coupling, and the corresponding additional spectral patterns would require π-functions of order n=4 for adequate representation. In the case of the two-compartment neuronal-glial model considered in the present paper, the carbon chains of relevant metabolites have no branching, so that the above connection matrix between spectral pattern and bonded cumomers with n≤3 describes exhaustively the NMR fine structure discernable in vivo.
Writing the differential equations for bonded cumomers
Using the probabilistic interpretation of cumomers, writing the differential equations describing time-evolution of each bonded cumomer of order n≤3 in the model is achieved by tracing the provenance of carbon atoms in chemical reactions involving this particular cumomer. This is most conveniently done by using atom distribution matrices (ADM) [13,16]. An ADM describes how carbons are transferred between reactants and products for a given biochemical reaction, and is closely related to the concepts of atom mapping matrices [17], isotopomer mapping matrices [18] and transition matrices [19]. For example, let’s consider the reaction A+B↔C+D occurring at chemical equilibrium at the rate V (in moles per time unit) in both directions, as illustrated in figure 2a, with the transfer of carbon atoms as indicated by the arrows. The corresponding ADM (fig. 2b) for the forward reaction lists the carbon positions for the reactants A and B along its first dimension (rows), while it lists the carbon positions for products C and D along its second dimension (columns), with coefficients 1 in the ADM indicating how the corresponding source carbon is transferred to the products. From this ADM, writing the equations for the cumomers is straightforward, since the provenance of the label in the products is readily identified in the matrix. Let’s for example consider the formation of πC{2,3}. From the ADM of fig. 2b, one can trace the provenance of the 2nd carbon of C from the 2nd carbon of A, while the 3rd carbon of C comes from 1st carbon of B. Hence, for each individual reaction, the probability for the formed C molecule to be labeled at least at its 2nd and 3rd carbons is the probability that the substrate molecule A is labeled at its 2nd carbon (whatever the label for other carbons) and the substrate molecule B is labeled at least at its 1st carbon (whatever the label for other carbons), which by definition is the product πA{2}×πB{1}. For πC{1,2}, the ADM shows that the 1st and 2nd carbons of C come from the 1st and 2nd carbons of a single molecule A, so that the probability for the formed C molecule to be labeled at least at 1st and 2nd carbons is πA{1,2}.
Figure 2.

(a) Example of a bi-molecular reaction A+B↔C+D. The gray arrows represent the way carbons are transferred during the reaction. (b) The corresponding Atom Distribution Matrix (ADM).
Assuming that C is an NMR-observed metabolite, one can then derive the complete set of isotope balance equations for the bonded cumomer fractions of order n≤3. The variation of concentration of labeled molecule over time is equal to the amount of labeled molecule that enters the metabolic pool minus the amount of labeled molecule that leaves the metabolic pool. For the first order:
| (4) |
For the second order:
| (5) |
For the third order:
| (6) |
Starting from the set of bonded π-functions for NMR-observed metabolites (e.g. first, second and third order π-functions for glutamate and glutamine), the system of equations is recursively extended, writing differential equations for all new π-functions that appear at each stage in the metabolic model. For example, in Eq (4), (5) and (6), writing the equation for C leads to new π-functions appearing for A and B; the next step would then be to write equations for each of these new π-functions, and so on. Note that some non-bonded π-functions may be involved in the generation of bonded π-functions associated with NMR-observed metabolites. For example in Eq (5) above πB{1,3} is a source for πC{3,4}, so that the differential equation for πB{1,3} has to be written. When the differential equations have been written for all intermediate π-functions appearing as input terms (except for metabolites corresponding to input of carbon inside the network), the time-evolution of the “observed” π-functions can be computed for any label input into the biochemical network by numerically solving the set of coupled differential equations. A particular case needs to be mentioned. In the case of a metabolite invariant by rotation (such as succinate HOOC-CH2-CH2-COOH), the overall symmetry of the molecule implies that symmetric carbons are biochemically indistinguishable. This results in label scrambling between symmetric carbons, as it is traditionally recognized in 13C positional models. For example, the molecule succinate is left invariant by a 180° rotation, so that C1 is equivalent to C4, and C2 is equivalent to C3 (provided the consecutive carbons of the chain are consecutively labeled). In terms of cumomers, the following equalities can therefore be written for succinate:
| (7) |
In this case, although the ADM can be expressed in an arbitrary numeration for the succinate carbon chain, care must be taken when writing the differential equations to consider not only the input cumomers for the considered cumomer, but also for all its equivalents. Let’s assume that the ADM representation yields input fluxes of the following kind:
| (8) |
Here FSuc{1,2,3}+ and FSuc{2,3,4}+ are input functions depending on multiple input cumomers as derived from the ADM representation, and hence are likely to be different. The equivalence πSuc{1,2,3}=πSuc{2,3,4} is taken into account by summing the two previous equations, which immediately yields:
| (9) |
Conversely, if πSuc{2,3,4} appears as a source term in some differential equation (i.e. on the right side), it can simply be replaced by πSuc{1,2,3}, for which the differential equation is written.
Bonded cumomer versus isotopomer metabolic modeling
In most cases, the mathematical expression of a bonded cumomer model requires significantly less differential equations than a full isotopomer model that takes into account every possible isotopomer in the system.
A first reduction of the system arises from the fact that, in practice, π-functions of order n>3 have no effect on the observable NMR fine structures and can therefore be eliminated from the system. Furthermore, since only bonded π-functions are associated to observable NMR fine structures, all π-functions of order n≤3 that are not implied in the production of bonded π-functions for the observed metabolites can also be discarded. In contrast, when using isotopomers as the basic “building block” to construct the model, equations have to be included for every possible isotopomer, which quickly leads to a very large number of equations.
Another simplification occurs for bonded cumomers in the case of a cleavage reaction. In this case, the system is reduced because the labeling state of carbons exiting the network has no effect on the labeling of metabolites belonging to the biochemical network. For example, let us consider the reaction A↔B+C at chemical equilibrium, where the 3-carbon molecule A loses its third carbon to yield the 2-carbon molecule B, at the rate V in both directions. The following system is obtained for the isotopomers of B:
| (10) |
In contrast, the differential equations for the cumomers of B are:
| (11) |
The last set of equations only implies three π-functions associated to A (πA{1}, πA{2}, πA{1,2}), while the corresponding set of isotopomer equations implies six different isotopomer fractions of A: a{1}, a{2}, a{1,2}, a{1,3}, a{2,3}, a{1,2,3}.
In summary, a metabolic model based on bonded cumomers allows the reduction of the number of equations to the minimal number of variables required to describe the temporal evolution of NMR-detectable 13C-multiplets. Although the burden of solving the system is left to a computer, two advantages can be identified for reducing the number of equations: i) from a computational point of view, solving a reduced set of differential equations is faster, which becomes particularly advantageous when the system is extremely large, or when it has to be solved repeatedly thousands of times (for example in the case of Monte-Carlo simulations); ii) for a human operator, a reduced system of equations is much easier to assess, verify and understand.
Methods
Brain two-compartment neuronal-glial model: biochemical network and system of equations
The oxidative metabolism in neurons and astrocytes (glia), as well as the glutamate-glutamine cycle between these two kinds of cells, associated to glutamatergic neurotransmission, play a central role in brain metabolism, and non-invasive measurement of these metabolic rates using neuronal-glial models has been the subject of continuous research efforts in the past 10 years. Here we derive the differential equations for bonded cumomers in the case of a neuronal-glial compartmentalized metabolic network similar to those recently published [4,20,21]. The specific model used in the present work is shown in Figure 3.
Figure 3.

A two-compartment metabolic model for brain energy metabolism and glu/gln neurotransmission. Metabolic fluxes: CMRglc-cerebral metabolic rate of glucose, VTCA(N), VTCA(A)- neuronal and glial TCA fluxes; VPC glial pyruvate carboxylase flux; VNT- glu/gln neurotransmission; VX- exchange flux between 2-OG and Glu and between OAA and Asp; VOUT - dilution of labeled lactate by unlabeled lactate; VDILGLN – dilution of labeled Gln by unlabeled glutamine.
The model was constructed as follows. First, resonances of interest for metabolic modeling were identified, in this case the resonances most often detected in the brain in vivo (glutamate C4, C3, C2 and glutamine C4, C3, C2). Second, all bonded cumomers of order n≤3 corresponding to these detected resonances were identified. Third, the system of equations was recursively built by writing differential equations for all intermediates implied in the generation of these cumomers (using ADM as described earlier in the theory section). Finally, the system was then simplified by eliminating unnecessary equations as described before. The final system describing the incorporation of 13C from labeled-glucose towards the bonded cumomers for glutamate and glutamine consisted in a total of 129 differential equations, after suppressing unnecessary equations. The biochemical network and the corresponding set of cumomer equations will be referred to as two-compartment dynamic bonded cumomer model. Note that the same biochemical network described in terms of isotopomers would consist in over 300 differential equations (2N−1 for each metabolite of length N, except for succinate which has 9 independent isotopomers).
Precision of positional versus cumomer models for different labeling strategies: Monte-Carlo simulations
Three different infusion protocols were investigated by numerical simulation. In the first situation, the 13C time-courses were simulated for an infusion of [1,6-13C2]glucose enriched at 70%, which in terms of plasma glucose cumomers correspond to πGlc_pl{1}=πGlc_pl{6}=πGlc_pl{1,6}=0.7, the other plasma glucose cumomer corresponding to the 1.1% 13C natural abundance, where any carbon has a 1.1% chance to be labeled, independently from the labeling of its neighbors: πGlc_pl{k}=0.011 and πGlc_pl{1,k}=πGlc_pl{k,6}=πGlc_pl{1,k,6}=0.7×0.011 for 1<k<6; πGlc_pl{j,k}=0.0112 and πGlc_pl{1,j,k}=πGlc_pl{j,k,6}=0.7×0.0112 for 1<j<k<6; πGlc_pl{i,j,k}=0.0113 for 1<i<j<k<6. The second infusion protocol consisted in an infusion of 100% enriched [1,2-13C2]acetate (πAce_pl{1}=πAce_pl{2}=πAce_pl{1,2}=1, and here again the other cumomers corresponding to the natural 13C abundance). The third infusion protocol consisted in the simultaneous infusion of [1,6-13C2]glucose enriched at 70% and [1,2-13C2]acetate enriched at 100%. For the acetate infusion protocol and the double infusion protocols, acetate was assumed to be metabolized in glia only [22], with an acetate/glucose partition coefficient ϕ = 0.8 (i.e. 80% of astrocytic acetyl-CoA is synthesized from acetate and 20% from glucose) based on studies of acetate transport with similar acetate infusion rates [23].
For each infusion protocol, the system was solved in order to generate time-courses of bonded cumomers for glutamate and glutamine, using Matlab (The MathWorks Inc., Natick, MA) and the built-in function ode15s. As an illustration, the solution for an infusion of [1,6-13C2]glucose (modeled here as a step function being 0.011 for t<0 and 0.7 for t>0) is displayed on fig. 4 for glutamate C4 and C3. The values of the fluxes used for the simulation were based on metabolic rates representative of awake human brain metabolism or anesthetized rat brain metabolism: VTCA(N) = 1 μmol/g/min, VTCA(A) = 0.1 μmol/g/min, VNT = 0.3 μmol/g/min, VPC = 0.1 μmol/g/min, VX = 1.0 μmol/g/min, VOUT = 0.3 μmol/g/min, and VDILGLN = 0.1 μmol/g/min (if included). Using the connection matrices introduced in Theory section, the cumomer time-courses of fig. 4A and 4B can be converted to the time-courses of NMR fine structures (fig. 4C and 4D). For example, eq. (2) was used to convert time courses of bonded cumomers for glutamate and glutamine C2 into time courses of the corresponding NMR multiplets.
Figure 4.
Example of 13C labeling time courses for glutamate C4 (top raw) and glutamate C3 (bottom row) simulated using the metabolic model shown in Figure 3 during an infusion of 70%-enriched [1,6-13C2]glucose. (Left) Time evolution of π functions corresponding to bonded cumomers for glutamate C4 (A) and glutamate C3 (B). (Right) Time evolution of the associated spectral fine structure (13C multiplets) for glutamate C4 (C) and glutamate C3 (D). There is a direct correspondence between the π functions computed by the model and the measured 13C multiplets signals measured by 13C NMR. All time courses were simulated with the metabolic model shown in Figure 3 and the following metabolic fluxes: VTCA(N) = 1 μmol/g/min, VTCA(A) = 0.1 μmol/g/min, VNT = 0.3 μmol/g/min, VPC = 0.1 μmol/g/min, VX = 1.0 μmol/g/min, VOUT = 0.3 μmol/g/min, VDILGLN = 0. The glucose isotopic enrichment was simulated as a “square” function, increasing from 1.1% to 70% at t = 0.
The reliability of the flux values was first derived for the positional model (i.e. the cumomer model reduced to first order cumomers) using Monte-Carlo simulations. Briefly, for each Monte-Carlo draw (at least 500 draws total), random Gaussian noise (standard deviation σ = 0.2 μmol/g) was added to the time courses of total 13C concentration of glutamate and glutamine C4, C3, and C2 (six curves in total). Total 13C concentration was obtained by multiplying positional enrichments curves (i.e. first order cumomers) by total glutamate and glutamine concentrations ([Glu] = 10 mM, [Gln] = 4 mM). Then, for each draw, the noised time-courses (with a time-resolution of 7.5 min over 150 minutes, corresponding to 20 time points per curve) were fitted with the metabolic model using Simplex and/or BFGS algorithms. Proper convergence was confirmed by verifying that goodness-of-fit values were close to expected theoretical values. Stability of the simulation was further verified by ensuring that results did not depend on initial conditions for the fit. The six following fluxes were determined: neuronal TCA cycle VTCA(N), astroglial TCA cycle VTCA(A), pyruvate carboxylase VPC, exchange between glutamate and 2OG VX, glutamate-glutamine cycle VNT, and the lactate dilution flux VOUT. The effect of including an additional flux for dilution of glutamine (VDILGLN) was also investigated. Note that VOUT was not fitted for the acetate infusion, since acetate enters the TCA cycle after the pyruvate/lactate pool. From the flux distribution obtained during the Monte-Carlo simulation, the probability distribution and the standard deviation could be calculated for each flux.
The precision of the bonded cumomer model was assessed with an equivalent Monte-Carlo simulation, but here the noised time-courses (noise standard deviation σ = 0.2 μmol/g) were generated for every NMR-observable multiplet. These noised time-courses were then fitted using the bonded cumomer model. Positional labeling curves and all labeling curves for measurable multiplets at the glutamate and glutamine C4, C3 and C2 positions were included in the fit, with the exception of time courses of multiplets whose concentration remained low, because they may not be detectable experimentally with sufficient precision. Specifically, labeling curves that did not reach a 13C concentration of at least 1 mM at their maximum point were excluded from the fit. Because of this criterion, 13C labeling curves included in the fit were different for each infusion protocol. For example, with [1,6-13C2]glucose, all glutamate carbons (C4, C3, C2) reach high enrichment, so that Glu-C3S intensity is low (most signal is in Glu-C3D and Glu-C3T). In contrast, with acetate infusion, glutamate enrichment is much smaller, so that Glu-C3S is proportionally higher. Therefore Glu-C3S was included for acetate infusion and not for glucose infusion.
For glucose infusion, the fitted time-courses were: GluC4total, GluC4S, GluC4D43, GluC3total, GluC3D, GluC3T, GluC2total, GluC2S, GluC2D23, GluC2Q, GlnC4total, GlnC4S, GlnC4D43, GlnC3total, GlnC3D, GlnC3T, GlnC2total, GlnC2S, GlnC2D21, GlnC2D23 (total 20 curves). For acetate infusion, the fitted time-courses were: GluC4total, GluC4D45, GluC4Q, GluC3total, GluC3S, GluC3D, GluC2total, GluC2S, GluC2D12, GlnC4total, GlnC4D45, GlnC4Q, GlnC3total, GlnC3S, GlnC2total, GlnC2D12 (total 16 curves). For double infusion, fitted time courses were: GluC4total, GluC4S, GluC4D43, GluC4D45, GluC3total, GluC3D, GluC3T, GluC2total, GluC2S, GluC2D23, GluC2D21, GlnC4total, GlnC4S, GlnC4D43, GlnC4D45, GlnC3total, GlnC3D, GlnC3T, GlnC2total, GlnC2S, GlnC2D23, GlnC2D21 (total 22 curves).
Results
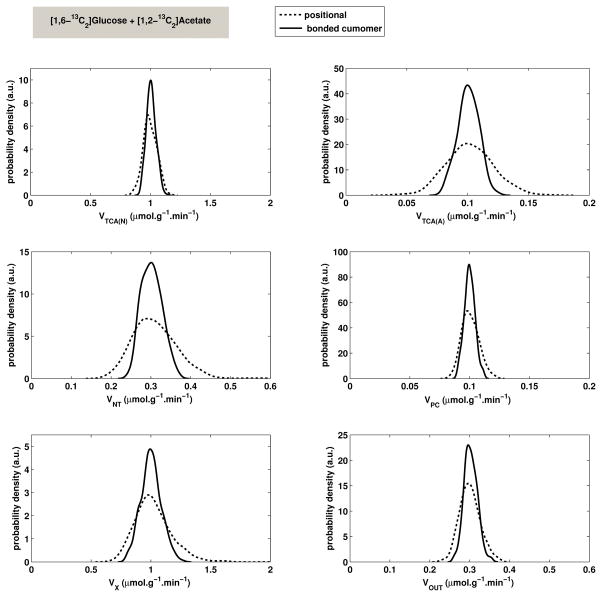
Figure 5 shows an example of probability density functions in the particular case of double infusion [1,6-13C2]glucose + [1,2-13C2]acetate and with no glutamine dilution included in the metabolic model. From these probability density functions, the relative standard deviations (s.d.) were determined for each metabolic flux in the model.
Figure 5.
Example of probability density functions for metabolic fluxes estimated during a double infusion of 70% [1,6-13C2]glucose and 100% [1,2-13C2]acetate by MC simulation (500 draws), with random noise (s.d.=0.2 μmol/g), human synthetic data set, 20 experimental time points per curve (150 min duration, with a 7.5 min time interval between experimental points). The bonded cumomer model improves the precision of all metabolic fluxes compared to the positional model (solid line). The specific model used for this figure did not include glutamine dilution (VDILGLN = 0). Bonded cumomers give a similar improvement in precision when glutamine dilution is included.
Results for the different simulated conditions are regrouped in Table 1. Three different infusion protocols where simulated: (i) ([1,6-13C2]glucose alone,. (ii) [2-13C]acetate alone, and (iii) double infusion of [1,6-13C2]glucose and [1,2-13C2]acetate. Each infusion protocol was simulated in the case without glutamine dilution (VDILGLN = 0, not iterated) and in the case with glutamine dilution (nominal VDILGLN = 0.1, iterated), and in each case the positional modeling approach was compared to the cumomer modeling approach.
Table 1.
Relative standard deviation for fitted metabolic rates obtained by Monte-Carlo simulation with the positional and the bonded cumomer models, for the three infusion protocols: [1,6-13C]glucose only, [1,2-13C]acetate only, and [1,6-13C]glucose and [1,2-13C]acetate simultaneously, without glutamine dilution (VDILGLN = 0) and with glutamine dilution (VDILGLN = 0.1). Monte-Carlo simulations were performed with 20 experimental points per curve, tmax =150 min, noise σ = 0.2 μmol.g−1.
| [1,6-13C2]Glucose | [U-13C]acetate | [1,6-13C2]Glucose + [U-13C]acetate | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| without gln dilution | with gln dilution | without gln dilution | with gln dilution | without gln dilution | with gln dilution | |||||||
| positional | cumomer | positional | cumomer | positional | cumomer | positional | cumomer | positional | cumomer | positional | cumomer | |
| VTCA(N) | 7% | 5% | 7% | 6% | 22% | 14% | 46% | 45% | 6% | 4% | 6% | 5% |
| VTCA(A) | 52% | 43% | 61% | 49% | 16% | 9% | 25% | 13% | 20% | 9% | 36% | 12% |
| VNT | 77% | 17% | 22% | 15% | 11% | 9% | 39% | 30% | 19% | 8% | 20% | 9% |
| VPC | 12% | 9% | 19% | 12% | 14% | 9% | 36% | 27% | 7% | 4% | 12% | 6% |
| VX | 19% | 14% | 20% | 13% | 24% | 12% | 71% | 18% | 18% | 9% | 18% | 9% |
| VOUT | 7% | 6% | 17% | 10% | n/a | n/a | n/a | n/a | 9% | 6% | 13% | 9% |
| VDILGLN | n/a | n/a | 19% | 11% | n/a | n/a | 98% | 70% | n/a | n/a | 15% | 9% |
Bonded cumomer vs. positional model
The main result emerging from Table 1 is that, regardless of the infusion protocol and whether glutamine dilution is present in the metabolic model or not, the cumomer model provided significantly improved precision on the determination of metabolic fluxes. The improvement in precision ranged from 3% to 77% depending on the metabolic flux and the conditions of the simulation (infusion protocol and inclusion or not of glutamine dilution). In the case without glutamine dilution, cumomer modeling led to an average 30% improvement in the precision of metabolic fluxes with [1,6-13C2]glucose, 39% improvement with [2-13C]acetate and 44% improvement with double infusion. Similarly, in the case with glutamine dilution, the average improvement was 32% with [1,6-13C2]glucose, 33% improvement with [2-13C]acetate and 45% improvement with double infusion.
Impact of glutamine dilution
Including glutamine dilution had a surprisingly large impact on the results of Monte-Carlo simulations. For example, the s.d. on VNT with [1,6-13C2]glucose and positional model was markedly improved when glutamine dilution was included in the model (going from 77% without glutamine dilution to 17% with glutamine dilution). In contrast, determination of metabolic fluxes with [2-13C]acetate and positional models degraded considerably when glutamine dilution was included in the model, presumably because the glutamine dilution flux itself was poorly determined in that case. Interestingly, for the double infusion protocol, the estimated precision of metabolic rates was similar whether glutamine dilution was included or not. This suggests that the cumulative metabolic information coming from two different substrates renders the precision on fitted metabolic fluxes less sensitive to changes in the metabolic model.
Overall, the impact of glutamine dilution on the results of MC simulations was much less significant when using the cumomer model than when using the positional model.
Comparison of infusion protocols. Positional model
The infusion protocol had a large impact on the estimated precision of metabolic fluxes as can be seen from Table 1. Using [1,6-13C2]glucose allowed excellent determination of VTCA(N) even with positional models (with s.d. comprised between 5% and 7%). In contrast, the determination of VNT was less precise with positional models (s.d. ranging from 77% without glutamine dilution to 22% with glutamine dilution). Determination of VPC and VX was not as good as VTCA(N) (precision ranging from 12% to 20%). The determination of VTCA(A) was poor with [1,6-13C2]glucose (precision ranging from 52% to 61%. With 13C-acetate, precision was improved most notably for VTCA(A) (which is not surprising because acetate is a glial-specific substrate) but the precision on other fluxes remained unsatisfactory, especially with glutamine dilution included in the model. Finally, the double infusion protocol with positional model broadly yielded similar precision to the [1,6-13C2]glucose protocol, except for VTCA(A) which was markedly more precise with the double infusion protocol (s.d. ranging from 52% to 61% with glucose only and 20% to 36% with double infusion).
Comparison of infusion protocols. Bonded cumomer model
As noted above, using the cumomer model led to an improvement in the precision of metabolic fluxes in all cases. However, there were still notable differences in precision depending on the metabolic fluxes of interest and on the infusion protocol used. Using [1,6-13C2]glucose alone with the cumomer model yielded satisfactory precision (5% to 17%) on all fluxes except VTCA(A) (which had a precision of 43% or 49% without and with glutamine dilution respectively). Using 13C-acetate led to an improvement in VTCA(A), but a degradation in the precision of most other fluxes in the model. With the double infusion model, however, the precision was excellent for all metabolic fluxes, ranging from 4% to 9% (depending on the metabolic flux) when glutamine dilution was not included, and ranging from 5% to 12% when glutamine dilution was included. Of all the cases considered in this study, the best results were obtained with the double infusion protocol and the cumomer model.
Discussion
Dynamic 13C metabolic modeling using bonded cumomer
In the present study, we propose a new metabolic modeling approach for dynamic analysis of time courses of 13C multiplets measured by in vivo 13C MRS. The new framework, which uses bonded cumomers, opens the possibility to include dynamic information from individual 13C multiplets measured by 13C MRS into the metabolic modeling procedure. So far, nearly all dynamic metabolic modeling studies have focused on the analysis of time courses of positional enrichment. The new method sets the foundation for taking full advantage of the additional information available from 13C multiplets.
It is important to note that the use of cumomers by itself does not lead to a reduced number of equations compared to a complete isotopomer or cumomer model that includes every possible isotopomer or cumomer [14]. In order to achieve the desired reduction in the number of equations, the new concept of bonded cumomers is critical. Furthermore, in contrast to positional models, a bonded cumomer model retains all the information potentially available from 13C spectra. Therefore, the use of bonded cumomers results in a metabolic model that is optimal for both compactness and completeness.
Our study builds on a large body of work in the field of chemical engineering. In that field of research, most studies are performed at isotopic steady-state, whereas our focus is decidedly on dynamic analysis of time courses, because one of the strengths of in vivo 13C NMR is the ability to measure dynamic data in intact tissue.
A closely related concept is the elementary metabolite unit (EMU) approach recently introduced by Antoniewicz et al. [24]. It consists in considering some sub-groups of atoms in molecular atomic chains, only a small fraction of which is sufficient to describe NMR-detectable isotopic labeling. Therefore, the EMU approach, like the bonded cumomer approach that we propose here, leads to a considerable reduction in the number of equations. However, in the paper by Antoniewicz et al., in contrast to our approach, relevant EMUs are transformed to isotopomers, which are subsequently converted to NMR multiplets. In contrast, with bonded cumomers, it is possible to make a direct correspondence between bonded cumomers and 13C NMR multiplets. Although there are clearly several possible approaches to construct a model adapted to the analysis of dynamic 13C isotopomer data, the present work suggests that bonded cumomers are particularly well-suited for this purpose.
Superiority of bonded cumomer modeling compared to positional modeling
Monte-Carlo simulations show that metabolic modeling of dynamic time courses of 13C multiplets using the bonded cumomers model yields standard deviations on fitted metabolic rates which are systematically smaller than for positional modeling, whatever the infusion protocol considered. Two main reasons may be advanced to explain this increased precision. First, due to the larger number of measured time-courses, more information is used as input in the model (even though some of this information is obviously redundant). Second, the intensity of multiplets is in some cases more sensitive to flux variations than the intensity of total positional enrichment. This can be easily assessed by simulating multiplet time-courses for different flux values (not shown). In summary, metabolic modeling using bonded cumomers gives increased precision on fitted metabolic rates by making the best possible use of the metabolic information available in 13C spectra.
One possible limitation is that, depending on the infused substrate and the level of isotopic enrichment, some of the multiplets may not be measurable in vivo (due to lower signal-to-noise). For example, when using [1-13C]glucose (as in most humans studies), 13C multiplets have proportionally much lower intensity than when using [1,6-13C2]glucose. However, measurement of time courses for at least some of the 13C multiplets should still be possible (perhaps with a lower temporal resolution). Fortunately, it is not necessary to include all possible multiplets in order to obtain improved precision on metabolic fluxes. For example, Monte-Carlo simulations with the double infusion protocol performed when fitting only 15 dynamic curves of 13C multiplets in glutamate and glutamine leads to nearly the same precision as when fitting the entire set of 22 curves (data not shown). This can be explained by the fact that some of the information in 13C spectra is redundant. In this context, Monte-Carlo simulations are the tool of choice to determine the expected precision on metabolic fluxes based on the available or anticipated experimental data that will be used for the metabolic modeling.
Glutamine dilution
One of the surprising results of the present study is the large impact of the glutamine dilution flux on the determination of VNT, as pointed out in a recent study [25]. We previously reported that the determination of VNT was not very precise when using a positional two-compartment model with no glutamine dilution and using [1-13C]glucose as the infused substrate [26]. In that study, the standard deviation on VNT was very large (670%) when using [1-13C]glucose (noise level 0.2 μmol.g−1, 20 data points per curve, 6 free parameters in the model, 7 curves fitted). In the present study, under similar simulated experimental conditions but with [1,6-13C2]glucose, the standard deviation on VNT was improved due to the increased isotopic enrichment in amino acids when using [1,6-13C2]glucose instead of [1-13C]glucose, but remained relatively high (77%). The precision of VNT was markedly improved (from 77% to 22%) with when glutamine dilution was included in the positional model (Table 1). In contrast, some fluxes were less well determined when glutamine dilution was included. For example, when using 13C-acetate as the infused substrate, the s.d. on metabolic fluxes was increased for most fluxes with glutamine dilution compared to without glutamine dilution (Table 1).
Therefore, the precision on specific metabolic fluxes is very dependent not only on the particular experimental conditions of the study (infused substrate(s), signal-to-noise of 13C data, number of 13C time courses measured, etc) but also on the specific metabolic network used. This underscores the necessity to carefully evaluate the precision of metabolic modeling as part of every study. In particular, the precision of a given two-compartment model cannot be assumed based on Monte-Carlo simulations performed with a different two-compartment model, even if the two models are very similar (eg differing only by the addition of a dilution flux). This is an important point because two-compartment models have been evolving constantly in the past 10 years. For example, initial two-compartment models did not include astrocytic TCA cycle or pyruvate carboxylase [27,28]. Similarly, glutamine dilution has been included in some studies (e.g. [29,30]) but not included in other studies (e.g. [20,31]). Although such changes have been made in two-compartment brain metabolic models over the years, the impact of these changes on the precision of fitted metabolic rates has not been systematically evaluated.
Interestingly, glutamine dilution had much less impact on the precision of metabolic fluxes when using the bonded cumomer model than when using the positional model. This suggests that taking the additional 13C multiplet data into account in the modeling makes the modeling less sensitive to assumptions or modifications in the metabolic model.
Usefulness of Monte-Carlo simulations
The current study illustrates the usefulness of Monte-Carlo simulations to evaluate the precision of metabolic models and determine the best experimental conditions for adequate precision on the metabolic fluxes of interest.
This precision on metabolic fluxes depends on a large number of factors. Experimental factors that can affect the precision of fitted fluxes include: signal-to-noise of 13C data, duration of 13C time courses, number of data points per curve, infused substrate, infusion protocol, etc. Other factors affecting the precision depend on how the metabolic modeling is performed: number of independent time courses fitted simultaneously, numbers of degrees of freedom (free parameters in the model), actual metabolic model used (with assumptions and constraints).
Finally, it is worth emphasizing that Monte-Carlo simulations provide only a lower bound for the precision on metabolic fluxes. Furthermore, they do not ensure that the determination of metabolic fluxes is accurate, since the data, the fitting procedure, or the model itself may be biased. In spite of these limitations, Monte-Carlo simulations are an irreplaceable tool to predict the best precision that can be achieved with a specific metabolic model and under specific experimental conditions.
Toward improved precision in the measurement of glutamatergic neurotransmission flux in vivo
Monte-Carlo simulations in the present study clearly show that cumomer dynamic metabolic modeling should lead to a large improvement in the precision of metabolic fluxes compared to position metabolic modeling.
Bonded cumomer metabolic modeling with [1,6-13C2]glucose only gives excellent precision on all metabolic fluxes except VTCA(A). The addition of [1,2-13C2]acetate allows simultaneous determination of VTCA(A) precisely in the same experiment.
The simulations in the present paper were performed assuming an acetate/glucose partition coefficient ϕ = 0.8 (i.e. 80% of astrocytic acetyl-CoA is synthesized from acetate and 20% from glucose), as estimated from rat studies. In humans, however, such a high partition coefficient is not realistic, because the infusion rate of acetate in humans is typically much lower than in rats for safety reasons. In addition, [1-13C]glucose is often used in humans rather than the much more expensive [1,6-13C2]glucose. Using [1-13C]glucose rather than [1,6-13C2]glucose greatly reduces the intensity of 13C-13C multiplets. Therefore, the results presented here may not be directly transposable to human studies.
The next step is to apply this new metabolic modeling approach to actual in vivo data. We previously reported measurement of dynamic 13C multiplet data during [1,6-13C2]glucose infusion [11] and during simultaneous infusion of [1,6-13C2]glucose and [1,2-13C2]acetate [32]. In a separate study, we characterized acetate transport in the brain for metabolic modeling of acetate [33]. The double infusion approach is attractive because, when infusing [1,6-13C2]glucose and [1,2-13C2]acetate simultaneously, glucose and acetate give rise to different isotopomer patterns that reflect predominantly neuronal and glial metabolism respectively, as demonstrated earlier on brain extracts [34]. Glucose is taken up primarily by neurons and gives rise to a singlet at Glu-C4S and Gln-C4S position, whereas acetate is taken by astrocytes and gives rise to doublets in GluC4-D45 and GlnC4-D45. The cumomer model is ideally suited to exploit this exquisite isotopomer information that directly reflects brain cellular compartmentation.
Conclusion
In this work, a novel approach was proposed for 13C dynamic metabolic modeling. The concept of bonded cumomer was presented, which allows a complete description of NMR spectra fine structures, while retaining a minimal amount of variables and equations compared to isotopomer models. A dynamic bonded cumomer model was then derived for the neuron-glia two-compartment model. The reliability of the model was investigated using Monte-Carlo simulation, showing the systematic superiority of bonded cumomer modeling compared to positional modeling as classically performed for in vivo studies, irrespective the specific substrate(s) and infusion protocol used. In all cases, the precision obtained with cumomer modeling was much better than with positional modeling. Bonded cumomer modeling with [1,6-13C2]glucose led to a good precision on all fluxes except VTCA(A). Excellent precision was reached on all fluxes when using double infusion of [1,6-13C2]glucose and [1,2-13C2]acetate combined with bonded cumomer modeling. This raises the prospect of greatly increased precision for future metabolic modeling studies using multi-compartment models in the brain.
Acknowledgments
This work was supported by NIH grants P41RR008079, P41EB015894, P30NS057091 and R01NS038672 (P.G.H.)
Abbreviations used
- MRS
Magnetic Resonance Spectroscopy
- ADM
Atom Distribution Matrix
- TCA cycle
Tricarboxylic acid cycle
- VNT
Rate of glutamate-glutamine cycle
- VTCA(N)
Rate of neuronal TCA cycle
- VTCA(A)
Rate of astrocytic TCA cycle
- VPC
Rate of pyruvate carboxylase
- VX
rate of exchange between 2-oxoglutarate and glutamate
- VOUT
Rate of lactate dilution
- VDILGLN
Rate of glutamine dilution
- Glu
Glutamate
- Gln
Glutamine
Indices
- N
Neuronal
- A
Astrocytic
References
- 1.de Graaf RA, Mason GF, Patel AB, Behar KL, Rothman DL. In vivo 1H-[13C]-NMR spectroscopy of cerebral metabolism. NMR Biomed. 2003;16(6–7):339–357. doi: 10.1002/nbm.847. [DOI] [PubMed] [Google Scholar]
- 2.Gruetter R, Adriany G, Choi I-Y, Henry P-G, Lei H-X, Oz G. Localized in vivo 13C NMR spectroscopy of the brain. NMR Biomed. 2003;16:313–338. doi: 10.1002/nbm.841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Henry PG, Adriany G, Deelchand D, Gruetter R, Marjanska M, Oz G, Seaquist ER, Shestov A, Ugurbil K. In vivo 13C NMR spectroscopy and metabolic modeling in the brain: a practical perspective. Magn Reson Imaging. 2006;24(4):527–539. doi: 10.1016/j.mri.2006.01.003. [DOI] [PubMed] [Google Scholar]
- 4.Lebon V, Petersen KF, Cline GW, Shen J, Mason GF, Dufour S, Behar KL, Shulman GI, Rothman DL. Astroglial contribution to brain energy metabolism in humans revealed by 13C nuclear magnetic resonance spectroscopy: elucidation of the dominant pathway for neurotransmitter glutamate repletion and measurement of astrocytic oxidative metabolism. J Neurosci. 2002;22(5):1523–1531. doi: 10.1523/JNEUROSCI.22-05-01523.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Malloy CR, Sherry AD, Jeffrey FM. Analysis of tricarboxylic acid cycle of the heart using 13C isotope isomers. Am J Physiol. 1990;259(3 Pt 2):H987–995. doi: 10.1152/ajpheart.1990.259.3.H987. [DOI] [PubMed] [Google Scholar]
- 6.Klapa MI, Park SM, Sinskey AJ, Stephanopoulos G. Metabolite and isotopomer balancing in the analysis of metabolic cycles: I. Theory. Biotechnol Bioeng. 1999;62(4):375–391. doi: 10.1002/(sici)1097-0290(19990220)62:4<375::aid-bit1>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- 7.Wiechert W. 13C metabolic flux analysis. Metab Eng. 2001;3(3):195–206. doi: 10.1006/mben.2001.0187. [DOI] [PubMed] [Google Scholar]
- 8.Carvalho RA, Rodrigues TB, Zhao P, Jeffrey FM, Malloy CR, Sherry AD. A (13)C isotopomer kinetic analysis of cardiac metabolism: influence of altered cytosolic redox and [Ca(2+)](o) Am J Physiol Heart Circ Physiol. 2004;287(2):H889–895. doi: 10.1152/ajpheart.00976.2003. [DOI] [PubMed] [Google Scholar]
- 9.Jeffrey FM, Reshetov A, Storey CJ, Carvalho RA, Sherry AD, Malloy CR. Use of a single (13)C NMR resonance of glutamate for measuring oxygen consumption in tissue. Am J Physiol. 1999;277(6 Pt 1):E1111–1121. doi: 10.1152/ajpendo.1999.277.6.E1111. [DOI] [PubMed] [Google Scholar]
- 10.Chance MC, Seeholzer SH, Kobayashi K, Williamson JR. Mathematical analysis of isotope labeling in the citric acid cycle with applications to 13C NMR studies in perfused rat hearts. J Biol Chem. 1983;258:13785–13794. [PubMed] [Google Scholar]
- 11.Henry P-G, Oz G, Provencher S, Gruetter R. Toward dynamic isotopomer analysis in the rat brain in vivo: automatic quantitation of 13C NMR spectra using LCModel. NMR Biomed. 2003;16:400–412. doi: 10.1002/nbm.840. [DOI] [PubMed] [Google Scholar]
- 12.Xu S, Shen J. In vivo dynamic turnover of cerebral 13C isotopomers from [U-13C]glucose. J Magn Reson. 2006;182(2):221–228. doi: 10.1016/j.jmr.2006.07.003. [DOI] [PubMed] [Google Scholar]
- 13.Muzykantov VS, Shestov AA. Kinetic equations for the redistribution of isotopic molecules due to reversible dissociation. Homoexchange of methane. React Kinet Catal Lett. 1986;32(2):307–312. [Google Scholar]
- 14.Wiechert W, Mollney M, Isermann N, Wurzel M, de Graaf AA. Bidirectional reaction steps in metabolic networks: III. Explicit solution and analysis of isotopomer labeling systems. Biotechnol Bioeng. 1999;66(2):69–85. [PubMed] [Google Scholar]
- 15.Deelchand DK, Ugurbil K, Henry PG. Investigating brain metabolism at high fields using localized 13C NMR spectroscopy without 1H decoupling. Magn Reson Med. 2006;55(2):279–286. doi: 10.1002/mrm.20756. [DOI] [PubMed] [Google Scholar]
- 16.Muzykantov VS. Distribution and transfer of atoms by elementary reactions. React Kinet Catal Lett. 1980;13(4):419–424. [Google Scholar]
- 17.Zupke C, Stephanopoulos G. Modeling of Isotope Distributions and Intracellular Fluxes in Metabolic Networks Using Atom Mapping Matrices. Biotechnology Progress. 1994;10(5):489–498. [Google Scholar]
- 18.Schmidt K, Carlsen M, Nielsen J, Villadsen J. Modeling Isotopomer Distributions in Biochemical Networks Using Isotopomer Mapping Matrices. Biotechnol Bioeng. 1997;55:831–840. doi: 10.1002/(SICI)1097-0290(19970920)55:6<831::AID-BIT2>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 19.Wiechert W, De Graaf AA. Bidirectional Reaction Steps in Metabolic Networks: I. Modeling and Simulation of Carbon Isotope Labeling Experiments. Biotechnol Bioeng. 1997;55(1):101–117. doi: 10.1002/(SICI)1097-0290(19970705)55:1<101::AID-BIT12>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 20.Gruetter R, Seaquist ER, Ugurbil K. A mathematical model of compartmentalized neurotransmitter metabolism in the human brain. Am J Physiol. 2001;281:E100–E112. doi: 10.1152/ajpendo.2001.281.1.E100. [DOI] [PubMed] [Google Scholar]
- 21.Shen J, Petersen KF, Behar KL, Brown P, Nixon TW, Mason GF, Petroff OAC, Shulman GI, Shulman RG, Rothman DL. Determination of the rate of the glutamate/glutamine cycle in the human brain by in vivo 13C NMR. Proc Natl Acad Sci USA. 1999;96:8235–8240. doi: 10.1073/pnas.96.14.8235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Waniewski RA, Martin DL. Preferential utilization of acetate by astrocytes is attributable to transport. J Neurosci. 1998;18(14):5225–5233. doi: 10.1523/JNEUROSCI.18-14-05225.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Deelchand DK, Koski DM, Shestov AA, Ugurbil K, Henry P-G. Acetate Utilization is Rate-Limited in the Rat Brain. 15th ISMRM; Berlin, Germany. 2007. p. 717. [Google Scholar]
- 24.Antoniewicz MR, Kelleher JK, Stephanopoulos G. Elementary metabolite units (EMU): a novel framework for modeling isotopic distributions. Metab Eng. 2007;9(1):68–86. doi: 10.1016/j.ymben.2006.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shen J, Rothman DL, Behar KL, Xu S. Determination of the glutamate-glutamine cycling flux using two-compartment dynamic metabolic modeling is sensitive to astroglial dilution. J Cereb Blood Flow Metab. 2009;29(1):108–118. doi: 10.1038/jcbfm.2008.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shestov AA, Valette J, Ugurbil K, Henry PG. On the reliability of (13)C metabolic modeling with two-compartment neuronal-glial models. J Neurosci Res. 2007 doi: 10.1002/jnr.21269. [DOI] [PubMed] [Google Scholar]
- 27.Sibson NR, Dhankhar A, Mason GF, Behar KL, Rothman DL, Shulman RG. In vivo 13C NMR measurements of cerebral glutamine synthesis as evidence for glutamate-glutamine cycling. Proc Natl Acad Sci USA. 1997;94:2699–2704. doi: 10.1073/pnas.94.6.2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sibson NR, Dhankhar A, Mason GF, Rothman DL, Behar KL, Shulman RG. Stoichiometric coupling of brain metabolism and glutamatergic neuronal activity. Proc Natl Acad Sci USA. 1998;95:316–321. doi: 10.1073/pnas.95.1.316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.de Graaf RA, Mason GF, Patel AB, Rothman DL, Behar KL. Regional glucose metabolism and glutamatergic neurotransmission in rat brain in vivo. Proc Natl Acad Sci USA. 2004;101(34):12700–12705. doi: 10.1073/pnas.0405065101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Oz G, Berkich DA, Henry PG, Xu Y, LaNoue K, Hutson SM, Gruetter R. Neuroglial metabolism in the awake rat brain: CO2 fixation increases with brain activity. J Neurosci. 2004;24(50):11273–11279. doi: 10.1523/JNEUROSCI.3564-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mason GF, Petersen KF, de Graaf RA, Shulman GI, Rothman DL. Measurements of the anaplerotic rate in the human cerebral cortex using 13C magnetic resonance spectroscopy and [1-13C] and [2-13C] glucose. J Neurochem. 2007;100(1):73–86. doi: 10.1111/j.1471-4159.2006.04200.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Deelchand DK, Nelson C, Shestov AA, Ugurbil K, Henry PG. Simultaneous measurement of neuronal and glial metabolism in rat brain in vivo using co-infusion of [1,6-13C2]glucose and [1,2-13C2]acetate. J Magn Reson. 2009;196(2):157–163. doi: 10.1016/j.jmr.2008.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Deelchand DK, Shestov AA, Koski DM, Ugurbil K, Henry PG. Acetate transport and utilization in the rat brain. J Neurochem. 2009;109(Suppl 1):46–54. doi: 10.1111/j.1471-4159.2009.05895.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Taylor A, McLean M, Morris P, Bachelard H. Approaches to studies on neuronal/glial relationships by 13C-MRS analysis. Dev Neurosci. 1996;18(5–6):434–442. doi: 10.1159/000111438. [DOI] [PubMed] [Google Scholar]