Abstract
An understanding of how the conformational behavior of single-stranded DNAs and RNAs depend on sequence is likely to be important for attempts to rationalize the thermodynamics of nucleic acid folding. In an attempt to further our understanding of such sequence dependences we report here the results of 192 (1 μs) explicit-solvent molecular dynamics (MD) simulations of 48 DNA and 48 RNA tetranucleotide sequences performed using recently reported modifications to the AMBER force field. Each tetranucleotide was simulated starting from two different conformations – a fully natively-stacked, and a completely unstacked conformation – and populations of the various possible base stacking arrangements were analyzed. The simulations indicate that, for both DNA and RNA, the populations of fully natively stacked conformations increase linearly with increasing numbers of purines in the sequence, while the conformational entropies, computed by two complementary methods, decrease. Despite the comparatively short simulation times, the computed free energies of stacking of the 16 possible combinations of bases in the middle of the sequences are found to be in good correspondence with values reported recently from simulations of dinucleoside monophosphates using the same force field. Finally, consistent with recent reports from other groups, non-native stacking interactions, i.e. between bases that are not adjacent in sequence, are shown to be a recurring feature of the simulations; in particular, stacking interactions of bases in a i:i+2 relationship are shown to occur significantly more frequently when the intervening base is a pyrimidine. Given that the high prevalence of non-native stacking interactions is thought to be unrealistic, it appears that further parameterization work will be required before accurate conformational descriptions of single-stranded nucleic acids can be obtained with current force fields.
Introduction
While molecular dynamics (MD) simulations have been used to simulate the conformational dynamics of nucleic acids for many years,1, 2 recent times have seen substantial effort focused on the reparameterization of nucleic acid force fields to improve agreement with experimental data. This has been especially true for the widely-used AMBER force fields, for which the parm99 parameter set has proven to be an established jumping-off point.3 One relatively early modification was proposed by the Orozco group to improve the description of the α and γ backbone dihedrals; these modifications were implemented in the now very widely-used bsc0 parameter set.4 Since then, work by a number of groups has identified the need to reparameterize the terms that describe the glycosidic bond dihedrals in order to overcome poor reproduction of NMR observables for nucleosides,5 and to improve the modeling of A-RNA regions,6, 7 for which the formation of artefactual “ladder-like” structures had been reported.8, 9 Thanks to these efforts, there are now several alternative parameter sets for the glycosidic bond dihedrals for RNA that are available5, 7, 10 as well as at least one such parameter set for DNA.11 More recently, reparameterizations of other backbone dihedrals have been proposed.12, 13 It is not yet clear, however, that these newer parameter sets always offer a very significant improvement in behavior, e.g. capturing the conformational behavior of Z-DNA remains challenging.13
An attractive way to test parameterizations of nucleic acids is by comparison with NMR data for oligonucleotides. One important class of test system has been the RNA hairpin tetraloop, for which a number of experimental structures have been solved.14 Maintaining the correct tertiary structure of such tetraloops in MD simulations initially proved to be challenging, but was shown to be achievable6 when some of the proposed improvements to glycosidic dihedrals were incorporated,5, 7 especially when used in combination with the bsc0 parameter set.4 A second important type of test system are single-stranded tetranucleotides: in particular, these offer valuable opportunities to test the abilities of current force fields to describe RNAs that, while generally stacked in A-form conformations (see below), exhibit flexibility of the kind that might be present in the unfolded state of RNAs. Experimental scalar coupling constants and nuclear Overhauser effects (NOEs) have been reported for a number of different tetranucleotide systems by the Turner group.15–18 Studies carried out by the same group comparing these NMR data with long MD simulations for r(GACC)15 and r(CCCC)14 have provided further evidence in favor of the use of reparameterized glycosidic dihedral parameters.5 In addition, however, they have shown that simulations can predict substantial populations of non-A-form, “intercalated” conformations – in which bases become stacked in an order different from that suggested by the linear sequence – for which no experimental data (NOEs) have been observed.
Similar intercalated conformations have been observed in MD simulations performed using enhanced sampling techniques carried out by the Cheatham group.19, 20 Despite their comparatively small size, it is challenging to fully sample the conformational free energy landscape of tetranucleotide systems using conventional, i.e. ‘brute force’, MD simulations. Because of this, these systems provide an excellent vehicle for testing different sampling methodologies, such as variants of replica exchange techniques.19–22 In their most recent study, for example, the Cheatham group showed that a converged view of the conformational behavior of r(GACC) could be achieved using multi-dimensional replica exchange sampling techniques, but that it required a very significant computational investment:20 in that work, 192 system replicas were simulated, each for 300 ns, and replica exchanges were invoked simultaneously in both the temperature and Hamiltonian dimensions, with the latter involving lowered dihedral barriers. Importantly, a number of different intercalated structures were again observed, including the ones identified in previous simulation studies of the same tetranucleotide,15, 19 and in ‘brute force’ 1.5 μs simulations of the r(CCCC) tetranucleotide previously carried out by the Turner group.16 As noted above, these intercalated structures are in apparent conflict with NMR data.15, 16
Comparative studies of simulation and experiment for RNA tetranucleotides have been taken to a new level by very recent work from the Turner group.18 In that work, which was published as this work was being completed, NMR data for four RNA tetranucleotides: r(AAAA), r(CAAU), r(GACC) and r(UUUU), were used as a benchmark for testing four modified versions of the original AMBER parm99 force fields. ‘Brute force’ MD simulations of each tetranucleotide were carried out for 8 to 10 μs and were repeated starting from four or five very different initial conformations. The extraordinary aggregate simulation time of 739 μs of that study, coupled with its thorough testing of a number of different simulation force fields, makes the study itself a new benchmark for MD simulations of RNAs. Experimentally, r(UUUU) is the only one of the four tetranucleotides studied that does not adopt a predominantly A-form, base-stacked conformation. The lower degree of ‘native’ stacking in r(UUUU) was correctly reproduced by each of the four force fields tested, but they also predicted large numbers of NOEs that were not observed experimentally, suggesting that the simulations produced conformations that were more compact and/or structured than is realistic. For the other tetranucleotides, the force fields again predicted substantial populations of conformations in which stacking interactions occur between bases that are not adjacent in the sequence. The overall effect of this repeated sampling of structures that are inconsistent with the NMR data was that none of the tested force fields produced a level of agreement with experimental observables that was above 40%. Given these results, it seems inescapable that substantial changes to the AMBER force fields for RNAs will be needed in the near future.
Given the very considerable computational demands of the simulations, it is not surprising that all of the MD studies described above have focused on only one or a few tetranucleotide sequences. The use of very long simulation times,18 or of enhanced sampling methods,19, 20 has the clear advantage of enabling unambiguous conclusions to be drawn about the simulated behavior of the selected systems. But the exploration of only a few such systems places limits on our ability to determine the extent to which simulation results are sequence or composition dependent. In addition, the focus of such studies on RNA means that corresponding information on the behavior of simulations of similar DNAs is currently lacking, although studies of single-stranded CGCGAATTCGCG have been reported.23, 24 As one way of addressing these gaps in our knowledge we report here the results of 192 1-μs MD simulations of 48 RNA and 48 DNA tetranucleotides, performed using a combination of the parm99 AMBER force field,3 the bsc0 parameter set,4 and the χOL37 and χOL411 parameterizations of the glycosidic bond angles for RNA and DNA, respectively. While each of the individual simulation times is comparatively short at 1 μs, the simulations allow us to identify clear trends due to sequence composition and sugar identity on the conformational behavior of tetranucleotides and reinforce recent conclusions regarding the surprising prevalence of stacking interactions involving bases that are not adjacent in sequence.18
Methods
Initial structures of all nucleic acids simulated here were generated using the Stroud group’s Make-NA server (http://structure.usc.edu/make-na/server.html) and formatted to be recognizable by the MD simulation program GROMACS version 4.6.5.25, 26 For both DNA and RNA we simulated all 36 tetranucleotides of the form CXYG and GXYC, with X and Y being the four standard nucleotides; in addition, for both DNA and RNA we simulated eight purine-only tetranucleotides (GAAA, GAAG, GAGA, GAGG, GGAA, GGAG, GGGA, GGGG) and eight pyrimdine-only tetranucleotides (in the case of DNA these were CTTT, CTTC, CTCT, CTTT, CCTT, CCTC, CCCT, CCCC; in the case of RNA these were CUUU, CUUC, CUCU, CUUU, CCUU, CCUC, CCCU, CCCC). In total, therefore 48 DNA and 48 RNA tetranucleotides were simulated. Since each was simulated for 1 μs twice – once using a fully natively-stacked initial conformation and once using a completely unstacked initial conformation (see below) – the total simulation time reported here is 192 μs.
Each tetranucleotide was simulated in a comparatively large 50 × 50 × 50 Å box to which periodic boundary conditions were applied. Each nucleic acid was modeled using the AMBER parm993 force field supplemented with the bsc0 parameters4 that improve modeling of α and γ torsions, and with improved parameters for the glycosidic torsions for both RNA (χOL3)7 and DNA (χOL4).11 This combination of parameter sets has been shown by the Otyepka and Šponer groups to perform quite well in applications to a variety of challenging systems27, 28 and, at least for RNA, is now the set recommended for use in AMBER;11 hence, in the recent study by the Turner group it was referred to by the short-hand description ‘ff10’.18 As noted by a reviewer, the εζOL1 parameters13 are now also recommended for use in modeling B-DNA by the AMBER developers; we chose not to include those parameters as our focus here is on modeling single-stranded DNAs, for which the parameters appear to be largely untested. We anticipate that none of our qualitative results will be affected by this decision. Water was modeled explicitly using the TIP4P-Ew model.29 As in our recent work,30 this was selected in preference to the more commonly used TIP3P model31 since our eventual goal is to model protein-nucleic acid interactions: when used together with AMBER protein parameters,32–34 TIP4P-Ew has been shown to perform well at describing the conformational behavior of small peptides.35 Na+ and Cl– ions – which were added to 150 mM concentrations in order to crudely mimic physiological concentrations – were modeled using the appropriate set of parameters derived by Joung & Cheatham;36 three additional Na+ ions were added to ensure overall system electroneutrality. Previous work by the Cheatham group has indicated that – at least for the r(GACC) system – there is little difference in conformational behavior between simulations that include only enough ions to ensure electroneutrality and those that incorporate additional salt.20
All systems were first equilibrated for 1.35 ns, with the temperature being raised incrementally from 50 to 298 K over the course of the first 350 ps; following this equilibration period, all MD simulations were carried out for a production period of 1 μs. During MD, pressure and temperature were maintained at their equilibrium values using the Parrinello-Rahman37 barostat and Nosé38-Hoover39 thermostat, respectively. All covalent bonds were constrained to their equilibrium lengths with LINCS,40 allowing a 2.5 fs timestep to be used. Short-range van der Waals and electrostatic interactions were truncated at 10 Å and longer-range electrostatic interactions were computed using the smooth Particle Mesh Ewald method.41 During the production period of the simulations, all solute coordinates were saved at intervals of 0.1 ps giving a total of 10 million snapshots for each simulation for subsequent analysis.
As noted above, one complete set of 96 MD simulations was performed using fully ‘natively’ stacked initial conformations that would be found, for example, in the context of double-stranded DNA or RNA (e.g. Figure S1A). A second set of simulations was performed in which the tetranucleotides were forced to assume unstacked conformations at the beginning of the production period. These simulations were carried out using the same protocol outlined above with the exception that during the equilibration period harmonic restraints were applied to the C1’ atoms of neighboring ribose rings in order to move them ~5 Å further apart from each other than the distance found in A-form (for RNA) and B-form (for DNA) conformations; a similar strategy was one of a number of approaches used recently by the Turner group to generate alternative initial conformations.18 The imposition of these restraints was found to be sufficient to cause all tetranucleotides to adopt fully unstacked conformations (e.g. Figure S1B); during the subsequent 1 μs production period all harmonic restraints were removed so that ‘refolding’ to stacked conformations was possible.
A number of groups have devised geometric criteria to define stacked conformations of nucleic acid bases during MD simulations.18, 42–45 As in our previous work,30 we used a combination of three different criteria: (1) a minimum distance between any pair of heavy atoms in the two bases < 4 Å, (2) a distance between the center of mass of each base of < 5 Å, and (3) a vector angle between the normals to the planes of the two bases between 0 – 45° or between 135 – 180°. The first two of these properties were measured for each snapshot using the standard GROMACS utilities g_mindist, and g_dist respectively, the third was measured using a script written in-house. Pairs of bases simultaneously satisfying all three of these criteria were considered to be stacked. For the central two bases of each tetranucleotide, free energies of stacking, ΔGstack, were computed using ΔGstack = –RT ln (Pstacked/Punstacked), where Pstacked and Punstacked are the respective populations of conformations in which bases 2 and 3 are stacked and unstacked, R is the Gas constant, and T is the temperature.
There has also been significant interest in developing methods to describe19, 46 and cluster nucleic acid conformations.47 Here we devised two simple but complementary approaches to cluster the structures sampled during the simulations in order to gain a quantitative measure of conformational flexibility. In one approach, we clustered snapshots according to the similarity of their backbone conformations. This was achieved using the g_clust utility of Gromacs, selecting only those heavy atoms that are part of the sugar-phosphate backbone, and applying a RMSD cutoff of 1 Å for deciding whether two snapshots are to be included in the same cluster. In a second approach, we clustered snapshots according to the similarity of their base stacking arrangements. For a tetranucleotide of the form 1–2-3–4 there are six possible base stacking interactions that can, in principle, be adopted: these are 1–2, 1–3, 1–4, 2–3, 2–4, and 3–4. Of these, 1–2, 2–3, and 3–4 are the ‘native’ stacking arrangements of bases in adjacent nucleotides that would be found, for example, in double-stranded regions. As noted in Results, however, each of the other three ‘non-native’ stacking arrangements is frequently observed in the simulations described here. To provide a short-hand description of the overall stacking arrangement in a given snapshot, therefore, we use a six-character ‘label’ with each of the six characters representing the status of each possible base stacking interaction in the order 1–2, 1–3, 1–4, 2–3, 2–4, 3–4. For each possible interaction we assign a character of ‘0’ if the bases are not in contact, ‘1’ if they are stacked, and ‘2’ if they are in contact but not considered stacked (i.e. if the minimum distance between any pair of heavy atoms in the two bases is < 4 Å). Defined in this way, the ‘label’ for a fully natively stacked conformation would be 100101 since only the 1–2, 2–3, and 3–4 pairs of bases would be stacked. Since there are six characters, each of which can take on three values, there are in principle 36 = 729 possible labels that could be required to describe the stacking status of a given tetranucleotide. In practice, however, many of these are never sampled, e.g. a label such as 111000 does not occur since it is not structurally possible for the base at the ‘1’ position to be simultaneously stacked with the 2, 3 and 4 bases.
Following the clustering of snapshots – either on the basis of backbone similarity or on the basis of their base-stacking arrangements – we can compute effective conformational entropies using the standard statistical thermodynamic relationship: S = −R Σ pi ln pi, where the summation runs over all identified clusters (or labels) and pi is the fractional population of each cluster/label.
Results
As outlined in Methods, we have performed independent 1 μs MD simulations of 48 DNA and 48 RNA tetranucleotides starting from two extreme initial conformations: fully ‘natively’ stacked (i.e. stacked in residue number order, as would be found in the double-stranded form; Figure S1A) and completely unstacked (Figure S1B). We start with an analysis of the former set of simulations, before verifying that the key trends we observe are recapitulated in the latter set of simulations.
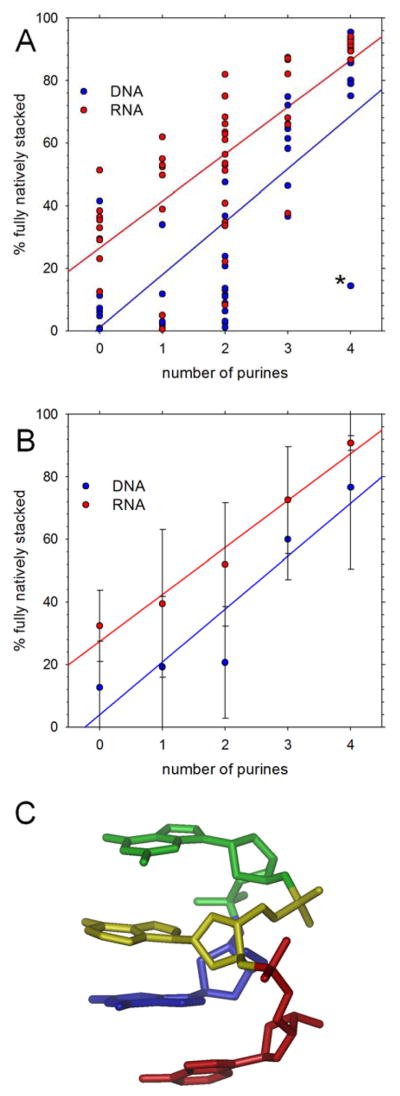
Figure 1A plots the population of simulation snapshots that are found in fully natively stacked conformations as a function of the number of purines for all 48 DNA (blue) and 48 RNA (red) tetranucleotides. With both types of nucleic acid, the fully natively stacked populations tend to increase with the numbers of purines in the sequence (Pearson correlation coefficient, Rcorr, = 0.72 and 0.76 for DNA and RNA respectively), with stacked populations being generally higher in the RNA tetranucleotides than in the DNA tetranucleotides (compare red and blue regression lines). Figure 1B shows the same data grouped and averaged according to the number of purines in the sequence, from which it is clear that the average populations are always higher for RNA than for DNA. The most obvious exception to the general trend shown in Figure 1A is the d(GGAA) DNA tetranucleotide (marked by the asterisk at the lower-right corner of the figure), for which the population of natively stacked conformations is anomalously low. Examination of the trajectory for this system shows that early in the simulation the tetranucleotide adopts and maintains an unusual, fully stacked conformation that is non-native, i.e. in which bases that are not adjacent to each other in sequence are stacked (Figure 1C). For this particular system, the bases are stacked in the order A4-G1-A3-G2 and it therefore represents an example of a conformation in which the two most common intercalation events identified by the Turner group18 are present simultaneously.
Figure 1. Native stacking in simulations starting from natively stacked conformations.
A. Percent of conformations that are fully natively stacked in production simulations; each symbol represents a different tetranucleotide; the asterisk marks the anomalously behaving d(GGAA) tetranucleotide. B. Same as A but with results grouped and averaged by the number of purines in the sequence. C. Snapshot showing the stable, non-native stacking arrangement adopted by d(GGAA); residues are colored using rainbow ordering: G1(blue), G2 (green), A3 (yellow), A4 (red).
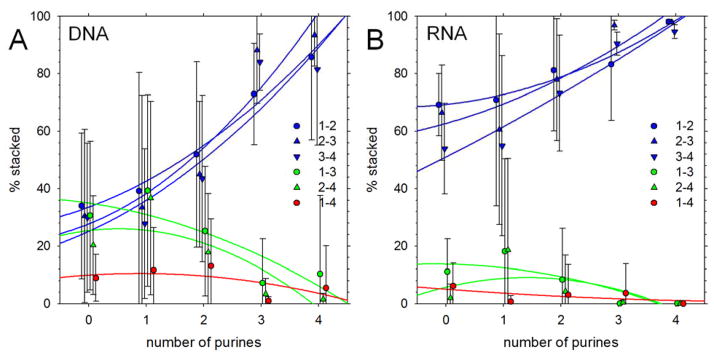
Significant populations of non-native stacking interactions – i.e. interactions involving non-contiguous bases – are found with a number of sequences but their populations vary significantly with sequence composition. Figure 2A shows the populations of the six types of base stacks that can, in principle, form in a tetranucleotide (see Methods) as a function of the number of purines for the 48 DNA sequences; Figure 2B shows a corresponding plot for the 48 RNA sequences. With both types of nucleic acid, the populations of native stacks (i.e. 1–2, 2–3, 3–4 stacks) increase with the number of purines (blue symbols). Non-native 1–3 and 2–4 stacks – which for DNA can reach an average population of nearly 40% (Figure 2A) – occur most frequently in sequences that contain a single purine, and tend to be lower in sequences that contain no purines (hence the negative curvature observed repeatedly in the (green) quadratic regression lines). Stacking interactions involving the most distantly separated bases (1–4 stacks) are, as expected, the least common of the six possible types, but for DNA tetranucleotides average approximately 10% for sequences containing zero, one or two purines.
Figure 2. Populations of base-stacking interactions in simulations starting from natively stacked conformations.
A. Percent of conformations that contain stacking interactions of various types plotted versus number of purines in the DNA sequence. Results shown are averages obtained by grouping sequences by the number of purines they contain; error bars represent standard deviations. Blue symbols represent ‘native’ stacking interactions, i.e. base stacks that involve residues adjacent in the sequence. Lines represent quadratic regression lines. B. Same as A but showing results for RNA tetranucleotides.
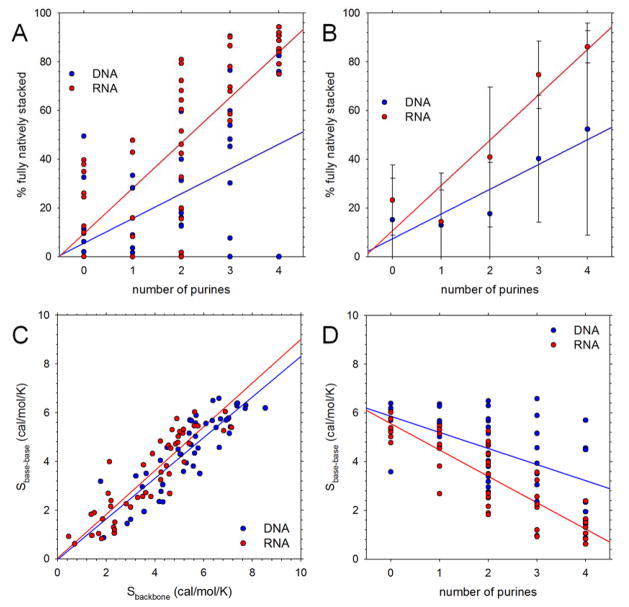
The sampling of non-native stacking arrangements indicates that the tetranucleotides explore a variety of alternative conformations. In an attempt to quantify the degree of conformational flexibility we have used two complementary approaches that cluster conformations sampled during the simulations: one does so according to the root-mean-square deviations of the backbone heavy atoms, and another does so according to the stacking interactions of the bases (see Methods). The two different measures provide qualitatively very similar results: the number of clusters identified on the basis of the base-base stacking interactions is linearly correlated with the number identified using backbone atoms for both DNA and RNA tetranucleotides (Figure 3A). The cluster populations can be converted into effective conformational entropies using standard statistical thermodynamics (see Methods): for both DNA and RNA, the correlation between the two entropy measures Sbase-base and Sbackbone is very high (Figure 3B). As might be expected given the results shown in Figure 1, there is also a clear relationship between these conformational entropy measures and the number of purines in the sequences. Figure 3C, for example, plots Sbase-base versus the number of purines, and shows that the effective conformational entropy decreases considerably with increasing numbers of purines for both DNA (blue) and RNA (red). This trend becomes even clearer when results are grouped and averaged according to the number of purines in the sequence (Figure S2A).
Figure 3. Conformational flexibility in simulations starting from natively stacked conformations.
A. Comparison of number of conformational clusters obtained with two different approaches to clustering (see Methods); each symbol represents a different tetranucleotide. B. Same as A but comparing conformational entropies. C. Plot of conformational entropy, Sbase-base, versus number of purines in the sequence.
Given that it has been shown that simulation times far in excess of 1 μs are likely to be required in order to completely sample the conformational possibilities for tetranucleotides15, 16, 19–21 it is clearly important to determine the extent to which the above trends might be affected by sampling issues. One way of mitigating the effects of running only single-replica simulations is to start from quite different initial conformations, for example, ones in which the glycosidic bond dihedral angles have been flipped.16 Here we have conducted a second set of 96 1 μs simulations of the same tetranucleotides but using a protocol that ensures that all base-base stacking is eliminated during the equilibration period of the simulations (see Methods). If the trends obtained from an analysis of this second set of simulations match those seen from the earlier simulations we can have greater confidence that they are real representations of the simulation force field.
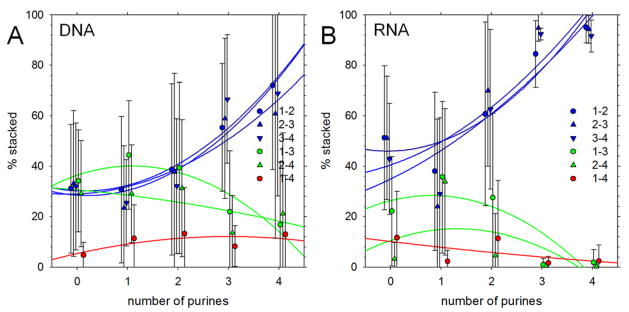
Figure 4A (c.f. Figure 1A) shows that the populations of fully natively stacked conformations seen in this second set of simulations again increase with the number of purines. It can also be seen, however, that there is considerably more scatter in the results than before and that a number of the simulated tetranucleotides do not sample the fully natively stacked conformation at all within 1 μs. These include even sequences containing four purines, such as d(GGGG), d(GAGG) that might be expected to rapidly assume a natively stacked conformation (these are the data points at the bottom right of Figure 4A); interestingly, d(GGAA) – in stark contrast to its behavior in the first set of simulations (see above) – formed and maintained a fully natively stacked conformation. Despite the greater scatter in the results, when grouped and averaged according to the number of purines (Figure 4B) the same overall trend is obtained as was obtained from simulations that started in fully natively stacked conformations (Figure 1B): the average populations of fully natively stacked conformations increase with the numbers of purines for both DNA and RNA. Figure 4C shows that the two entropy measures Sbase-base and Sbackbone computed for this second set of simulations again correlate very highly with each other (c.f. Figure 3C), and Figure 4D shows that again, the conformational entropies of systems decrease significantly as the number of purines increases (c.f. Figure 3D; see Figure S2B for results grouped and averaged by the number of purines). Finally, Figure 5 shows that the relative populations of the different types of stacking interactions follow the same trends as seen in the original set of simulations (c.f. Figure 2): the populations of native stacking interactions increase with the number of purines in the sequence, while the populations of the 1–3 and 2–4 non-native stacking interactions peak (again at ~40%) for sequences that contain only one purine. Interestingly, the average population of 1–3 non-native stacking interactions is, for DNA at least, the highest of any type of stacking interaction for sequences containing one purine (Figure 5A).
Figure 4. Native stacking and conformational flexibility in simulations starting from initially unstacked conformations.
A. Percent of conformations that are fully natively stacked in production simulations; each symbol represents a different tetranucleotide. B. Same as A but with results grouped and averaged by the number of purines in the sequence. D. Plot of conformational entropy, Sbase-base, versus number of purines in the sequence.
Figure 5. Populations of base-stacking interactions in simulations starting from initially unstacked conformations.
A. Percent of conformations that contain stacking interactions of various types plotted versus number of purines in the DNA sequence. Results shown are averages obtained by grouping sequences by the number of purines they contain; error bars represent standard deviations. Blue symbols represent ‘native’ stacking interactions, i.e. base stacks that involve residues adjacent in the sequence. Lines represent quadratic regression lines. B. Same as A but showing results for RNA tetranucleotides.
Direct comparisons of the results obtained from the two sets of simulations are shown in Figures S3 and S4. Figure S3A compares the two Sbase-base measures grouped and averaged according to the number of purines in the sequences. For both DNA and RNA, Sbase-base tends to be somewhat higher in simulations that start from unstacked initial conformations; the differences are small, however, and the correlation coefficients between the two sets of simulation data are accordingly reasonably high (0.84 for DNA and 0.99 for RNA). Figure S3B compares the populations of fully natively stacked conformations, again grouped and averaged according to the number of purines. As expected, the populations are generally higher in simulations that started in natively stacked conformations (i.e. most data points lie below a diagonal running from bottom left to top right of the figure). This is particularly true for DNA, for which a number of initially-unstacked simulations do not sample the fully natively stacked conformation during 1 μs (see above). For RNA, on the other hand, the populations are similar regardless of starting conformation, and this is especially the case for sequences containing three or four purines. Figure S4A compares the average populations of the six types of stacking interactions obtained from the two sets of DNA simulations; Figure S4B shows corresponding results for RNA simulations. As might be anticipated, with both types of nucleic acid, the populations of the non-native 1–3, 2–4 and 1–4 stacking interactions (green and red symbols) tend to be somewhat higher in the simulations that start from initially unstacked conformations, while the native 1–2, 2–3, and 3–4 stacking interactions tend to be somewhat higher in simulations that start from natively stacked conformations. While certainly noticeable, the differences are not drastic, however, which suggests again that the two sets of simulations are capturing similar overall behaviors.
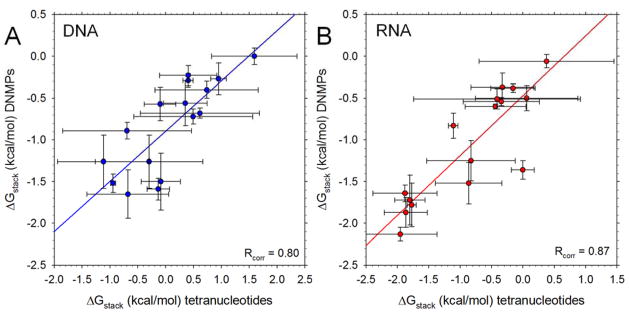
In addition to examining the dependence of native and non-native stacking interactions on the overall purine-pyrimidine composition of the tetranucleotides, we have also explored their dependence upon sequence. We converted the populations of natively stacked interactions into effective free energies of stacking, ΔGstack (see Methods) and compared the results with corresponding values obtained from simulations of dinucleotide monophosphates (DNMPs) that we reported recently.30 Figure 6A compares the ΔGstack values computed here for all 16 types of DNA stack (A-A, A-C, …, T-G, T-T) for the central step of the tetranucleotide (i.e. the 2–3 stacking interaction) with the corresponding ΔGstack values for DNA DNMPs; Figure 6B shows a corresponding set of data for RNAs. In both cases, the correlation is quite good, with Rcorr values of 0.80 and 0.87 for the DNAs and RNAs, respectively. For the DNAs, the ΔGstack values are shifted to more positive values in the tetranucleotides than in the DNMPs, but the statistical reliability of this result is limited given that the ΔGstack values computed from initially-unstacked simulations are poorly correlated with those computed from the initially-stacked simulations (Rcorr = 0.44; Figure S5A). For the RNAs, the ΔGstack values are more similar for the tetranucleotides and DNMPs, and this appears to be more statistically reliable given that the initially-unstacked and initially-stacked data sets are in much better agreement (Rcorr = 0.88; Figure S5B).
Figure 6. Comparison of computed free energies of stacking, ΔGstack, in tetranucleotides and dinucleoside monophosphates (DNMPs).
A. Plot of computed ΔGstack values in DNA DNMPs versus the corresponding ΔGstack values computed here fosr the central bases (i.e. the 2–3 stacking inteaction) of DNA tetranucleotides. Each datapoint represents a different sequence (AA, AC etc); error bars represent standard deviations calculated for all simulations of all tetranucleotides possessing the same 2–3 sequence. Line shows linear regression y = mx + b with m = 0.60 (p-value = 0.0002) and b = −0.89 (p-value < 0.0001). Data for the DNMPs are taken from ref. 25. B. Same as A but showing results for RNAs. Parameters for the linear regression are m = 0.58 (p-value = 0.0008) and b = −0.74 (p-value = 0.0001).
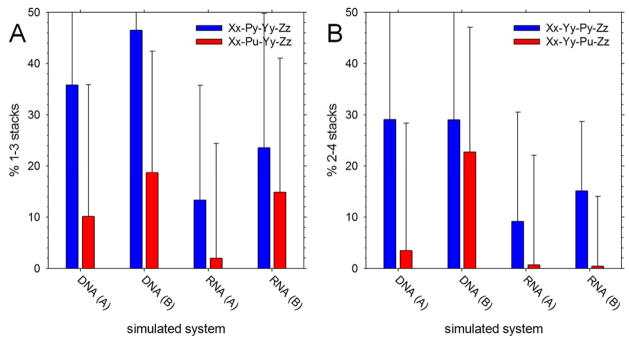
To the extent that we can (see Discussion), we have also explored the sequence dependence of the non-native stacking interactions. Specifically, we have considered the hypothesis that non-native 1–3 and 2–4 stacking interactions might be more likely to occur when the intervening base is a pyrimidine than a purine. To explore this issue for 1–3 stacking we grouped all simulated tetranucleotides according to whether the 2nd base was a pyrimidine or purine and computed the average populations of 1–3 stacks in the two groups. Figure 7A compares these two populations for all types of simulation: results are shown separately for DNA and RNA simulations and according to the simulation’s initial conformation ((A) stacked or (B) unstacked). In all cases, while the error bars are quite large, the population of 1–3 stacks was higher when the intervening (2nd) base was a pyrimidine (compare blue and red bars in Figure 7A). Pursuing a similar strategy for 2–4 stacking interactions (Figure 7B) shows that they also occur more frequently when the intervening (3rd) base is a pyrimidine.
Figure 7. Populations of non-native 1–3 and 2–4 stacks versus identity of the intervening base.
A. Population of 1–3 stacking interactions observed in simulations, averaged and plotted as a function of system type. Blue and red represent 1–3 stacks where the intervening base (2) is a pyrimidine or a purine, respectively; (A) indicates initially natively stacked simulations, (B) indicates initially unstacked simulations. Error bars represent standard deviations. B. Same as A but showing results for 2–4 stacking interactions.
Finally, we have quantified the populations of the various possible intercalated structures that can be formed by the tetranucleotides. The fact that, once formed, intercalated structures can have long lifetimes18 means that there is often a high degree of variability between simulations. Figure 8A, therefore, plots the populations of the 8 possible intercalated structures, averaged over all 48 sequences in each of the four sets of simulations. While there is still considerable scatter in the data, it appears that with both DNA and RNA the most populated structure is the 3-1-4 arrangement, with the 3-2-4 and 1-3-2 arrangements being the second-most populated for DNA and RNA respectively. The generally higher prevalence of 3-1-4 and 1-3-2 arrangements with RNA is consistent with results reported recently using the same force field by the Turner group.18 It is also clear that for RNA, the populations of intercalated structures are higher in the simulations that start in an initially unstacked conformation (compare RNA (B) with RNA (A) in Figure 8A).
Figure 8. Populations of intercalated structures.
A. Population of various possible stacking arrangements observed in simulations, averaged and plotted as a function of system type. B. Number of sequences that sample each type of stacking arrangement at least once during the course of the simulation.
An alternative way of analyzing the data is presented in Figure 8B. Here we plot the number of sequences within each of the four sets of simulations that sample each type of intercalated structure at some point during the 1 μs simulation, regardless of how many times they do so. For DNA, it is the 3-2-4 arrangement that is most likely to be sampled, with 45 of the 48 sequences sampling it in both the A and B sets of simulations; for RNA, the 1-3-2, 2-1-3 and 3-2-4 arrangements are sampled at least once by a similar number of sequences. More interestingly, there appear to be some significant differences between the DNA and RNA. The 2-1-3 arrangement, for example, is sampled at least once by more than 20 RNA sequences (Figure 8A) – even though its overall population remains very low (see Figure 8A) – yet is sampled by only one of the DNA sequences. The 1-4-2 arrangement, on the other hand, is sampled by more than 20 DNA sequences, but fewer than 10 RNA sequences. Together, therefore, Figures 8A and 8B suggests that the relative preferences for forming different types of intercalated structures might differ substantially between DNA and RNA.
Discussion
Before discussing the current simulation results further, it is important to consider whether the strategy adopted here of conducting large numbers of (comparatively) short simulations is justified. As noted in the Introduction, obtaining a completely sampled view of a given tetranucleotide’s conformational behavior using a ‘brute force’ MD approach is likely to require simulation periods far in excess of 1 μs:18, 19 in particular, much longer simulation times would be necessary to exhaustively sample not only the stacking/unstacking equilibria that are of primary interest here but also syn/anti transitions of the glycosidic bonds. While the aggregate simulation time reported here (192 μs) is considerable, we have chosen to expend our computational resources in covering a wide range of different sequences instead of in attempting to obtaining a fully converged view of one or a few sequences. Clearly, we are assuming that the benefits of pursuing such an approach outweigh their disadvantages. We do not claim to have achieved complete sampling for any of the individual sequences modeled here, and it is for this reason that we have refrained from analyzing in detail the conformational behavior of any single sequence. What we do claim, however, is that important general trends can be extracted from the data, especially with regard to what the AMBER force fields predict about the conformational behavior of single-stranded DNAs and RNAs. We think that the fact that we have shown that qualitatively identical results are obtained from simulations that start in fully natively stacked or that start in fully unstacked conformations provides significant support for this idea. In particular, the latter simulations appear to rule out the possibility that the increased stacking observed with more purine-rich sequences in the former simulations (Figure 2) is simply a reflection of slower unstacking kinetics.
The approach that we have followed is clearly complementary to that recently taken by the Turner group, although the aggregate simulation time of the work reported here (192 μs) is considerably lower than that (739 μs) recently reported.18 Importantly, despite their very different goals, the two studies produce very similar results in the areas where they overlap. In particular, both show that repeated sampling of non-native base stacking interactions is a common feature of current state-of-the-art AMBER force fields. The Turner group’s studies go further in demonstrating that this is a consistent feature of a number of variants of the AMBER-based force fields, and most crucially, that it is inconsistent with experiment for those tetranucleotides for which NMR data are available.18 The present study, on the other hand, shows that sampling of non-native base stacking interactions is a clear function of the purine-pyrimidine composition of the tetranucleotides, and occurs with similar frequencies in DNA as well as RNA tetranucleotides (Figures 2 and 5). In addition, it indicates that the Turner group’s substantive conclusions regarding the capabilities of current force fields for describing RNAs are unlikely to be affected by the choice of water model, the treatment of long-range electrostatic interactions, or the inclusion of added salt, all of which differ between the two studies.
The other major results obtained here are the following. The combination of AMBER force fields tested here predict: (a) that native stacking interactions in RNAs are generally stronger than those in the corresponding DNAs (Figures 1 and 4) – as was found in our recent examination of stacking interactions in DNMPs30 – (b) that the sequence dependence of native base stacking interactions in the tetranucleotides mirrors that obtained with DNMPs using the same force field (Figure 6), and (c) that non-native stacking interactions involving bases in a i:i+2 relationship occur significantly more frequently when the intervening base is a pyrimidine (Figure 7). With regard to the first of these results it is difficult to find many experimental studies that directly compare stacking free energies of single stranded RNAs and DNAs. However, the Pollack group has found that the persistence length of (rU)40 is somewhat greater than that of (dT)40 under the same conditions and has partly attributed this difference to the possibility that stacking might occur in (rU)40 but not in (dT)40.48 That interpretation would argue in favor of the present findings, especially so since those systems might be expected a priori to show greater stacking in the DNA form given that, in nucleoside pairs, the presence of the additional methyl group in dT has been shown to promote stacking (see Discussion in ref. 30). The Marszalek group, on the other hand, has shown that in single-molecule stretching experiments performed under identical solution conditions the force required to unstack poly(dA) plateaus at ~23 pN,49 while that for poly(rA) plateaus at 24 +/- 1 pN.50 Since these two estimates are, within error, identical, this might argue that the stronger stacking interactions in RNA predicted by the simulations reported here might be unrealistic.
With regard to the finding that non-native stacking interactions are sensitive to the identity of the intervening nucleotide (see above), it is worth noting that there are likely to be other sequence dependences that could be identified from a more comprehensive sampling of sequences. The present study considers only 48 of the 256 possible sequences that can be formed from the 4 standard nucleotides and, therefore, covers only some of the possible combinations of bases at the 1–3 and 2–4 positions. This precludes us from examining their sequence dependences in the same detail that we have for 2–3 stacking interactions (Figure 6), but future work in this direction might be worth pursuing given the suggestion by the Turner group that the presence of amino groups on the central base might be a factor stabilizing intercalated conformations.18 It might also be of interest to determine whether 1–4 stacking interactions are more likely to occur in sequences of the form Pu-Py-Py-Pu than in sequences of other forms.
On the other hand, the two studies together strongly suggest that the problem of non-native stacking interactions is a persistent one that will need to be resolved if realistic modeling of single-stranded RNA (and probably also DNA) regions is to be achieved. The present finding that the frequency of non-native stacking interactions depends on the purine-pyrimidine composition is consistent with, but does not provide conclusive proof of, problems with the thermodynamic description of stacking interactions, something that has already been suggested as a possible contributing factor by the Turner group.18 Garcia and Chen have recently shown that the simulated stacking interactions of the rG nucleoside are far too favorable using the nonbonded parameters of the current AMBER force field51 and have demonstrated that modifications to the strengths of van der Waals interactions can be made to produce behavior that is more realistic. A similar over-estimation of the favorability of stacking was apparent in our own recent comparisons with experiment of stacking interactions in DNMPs and nucleoside-pairs.30 Other indications that there are significant problems to overcome in the modeling of nucleic acid systems come from the recent studies by the Šponer group showing that sophisticated quantum mechanical methods produce estimates of the relative energies of nucleic acid conformations that can differ qualitatively from those predicted by the Amber force fields both for DNAs52 and RNAs.53 But it is one thing for us to point out that there are problems and quite another to solve them, especially since the Turner group’s recent study shows that modifications that improve a force field’s ability to describe certain situations12, 16 might worsen its behavior in others.18 Given the very complicated inter-dependence of the bonded and nonbonded parameters in current force fields, the current proliferation of alternative parameter sets, and the significant computational demands associated with obtaining converged oligonucleotide simulations, it may be that developing a general solution to the modeling of complex nucleic acid systems might require a concerted, combined effort from the simulation community.
Supplementary Material
Acknowledgments
This work was supported by NIH R01 GM099865 and R01 GM087290 awarded to A.H.E.
Footnotes
System snapshots at the beginning of the simulations; conformational flexibilities averaged according to the number of purines in the sequence; comparison of conformational flexibility obtained from simulations that start with different initial conformations; comparison of base stacking populations obtained from simulations that start with different initial conformations; comparison of base stacking free energies obtained from simulations that start with different initial conformations. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Cheatham TE, 3rd, Case DA. Twenty-five years of nucleic acid simulations. Biopolymers. 2013;99:969–77. doi: 10.1002/bip.22331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Šponer J, Banáš P, Jurečka P, Zgarbová M, Kührová P, Havrila M, Krepl M, Stadlbauer P, Otyepka M. Molecular Dynamics Simulations of Nucleic Acids. From Tetranucleotides to the Ribosome. J Phys Chem Lett. 2014;5:1771–1782. doi: 10.1021/jz500557y. [DOI] [PubMed] [Google Scholar]
- 3.Cheatham TE, Cieplak P, Kollman PA. A Modified Version of the Cornell et al. Force Field with Improved Sugar Pucker Phases and Helical Repeat. J Biomol Struct Dyn. 1999;16:845–862. doi: 10.1080/07391102.1999.10508297. [DOI] [PubMed] [Google Scholar]
- 4.Pérez A, Marchán I, Svozil D, Sponer J, Cheatham TE, III, Laughton CA, Orozco M. Refinement of the AMBER Force Field for Nucleic Acids: Improving the Description of α/γ Conformers. Biophys J. 2007;92:3817–3829. doi: 10.1529/biophysj.106.097782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yildirim I, Stern HA, Kennedy SD, Tubbs JD, Turner DH. Reparameterization of RNA χ Torsion Parameters for the AMBER Force Field and Comparison to NMR Spectra for Cytidine and Uridine. J Chem Theory Comput. 2010;6:1520–1531. doi: 10.1021/ct900604a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Banás P, Hollas D, Zgarbová M, Jurečka P, Orozco M, Cheatham TE, Šponer Ji, Otyepka M. Performance of Molecular Mechanics Force Fields for RNA Simulations: Stability of UUCG and GNRA Hairpins. J Chem Theory Comput. 2010;6:3836–3849. doi: 10.1021/ct100481h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zgarbova M, Otyepka M, Sponer J, Mladek A, Banas P, Cheatham TE, 3rd, Jurecka P. Refinement of the Cornell et al. Nucleic Acids Force Field Based on Reference Quantum Chemical Calculations of Glycosidic Torsion Profiles. J Chem Theory Comput. 2011;7:2886–2902. doi: 10.1021/ct200162x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mlýnský V, Banáš P, Hollas D, Réblová K, Walter NG, Šponer J, Otyepka M. Extensive Molecular Dynamics Simulations Showing That Canonical G8 and Protonated A38H+ Forms Are Most Consistent with Crystal Structures of Hairpin Ribozyme. J Phys Chem B. 2010;114:6642–6652. doi: 10.1021/jp1001258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Banáš P, Sklenovský P, Wedekind JE, Šponer J, Otyepka M. Molecular Mechanism of preQ1 Riboswitch Action: A Molecular Dynamics Study. J Phys Chem B. 2012;116:12721–12734. doi: 10.1021/jp309230v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ode H, Matsuo Y, Neya S, Hoshino T. Force field parameters for rotation around χ torsion axis in nucleic acids. J Comput Chem. 2008;29:2531–2542. doi: 10.1002/jcc.21006. [DOI] [PubMed] [Google Scholar]
- 11.Krepl M, Zgarbova M, Stadlbauer P, Otyepka M, Banas P, Koca J, Cheatham TE, 3rd, Jurecka P, Sponer J. Reference simulations of noncanonical nucleic acids with different chi variants of the AMBER force field: quadruplex DNA, quadruplex RNA and Z-DNA. Journal of chemical theory and computation. 2012;8:2506–2520. doi: 10.1021/ct300275s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yildirim I, Kennedy SD, Stern HA, Hart JM, Kierzek R, Turner DH. Revision of AMBER Torsional Parameters for RNA Improves Free Energy Predictions for Tetramer Duplexes with GC and iGiC Base Pairs. J Chem Theory Comput. 2012;8:172–181. doi: 10.1021/ct200557r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zgarbova M, Luque FJ, Sponer J, Cheatham TE, 3rd, Otyepka M, Jurecka P. Toward Improved Description of DNA Backbone: Revisiting Epsilon and Zeta Torsion Force Field Parameters. J Chem Theory Comput. 2013;9:2339–2354. doi: 10.1021/ct400154j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bevilacqua PC, Blose JM. Structures, Kinetics, Thermodynamics, and Biological Functions of RNA Hairpins. Annu Rev Phys Chem. 2008;59:79–103. doi: 10.1146/annurev.physchem.59.032607.093743. [DOI] [PubMed] [Google Scholar]
- 15.Yildirim I, Stern HA, Tubbs JD, Kennedy SD, Turner DH. Benchmarking AMBER force fields for RNA: comparisons to NMR spectra for single-stranded r(GACC) are improved by revised chi torsions. J Phys Chem B. 2011;115:9261–70. doi: 10.1021/jp2016006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tubbs JD, Condon DE, Kennedy SD, Hauser M, Bevilacqua PC, Turner DH. The nuclear magnetic resonance of CCCC RNA reveals a right-handed helix, and revised parameters for AMBER force field torsions improve structural predictions from molecular dynamics. Biochemistry. 2013;52:996–1010. doi: 10.1021/bi3010347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Condon DE, Yildirim I, Kennedy SD, Mort BC, Kierzek R, Turner DH. Optimization of an AMBER force field for the artificial nucleic acid, LNA, and benchmarking with NMR of L(CAAU) J Phys Chem B. 2014;118:1216–28. doi: 10.1021/jp408909t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Condon DE, Kennedy SD, Mort BC, Kierzek R, Yildirim I, Turner DH. Stacking in RNA: NMR of Four Tetramers Benchmark Molecular Dynamics. J Chem Theory Comput. 2015;11:2729–2742. doi: 10.1021/ct501025q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Henriksen NM, Roe DR, Cheatham TE., 3rd Reliable oligonucleotide conformational ensemble generation in explicit solvent for force field assessment using reservoir replica exchange molecular dynamics simulations. J Phys Chem B. 2013;117:4014–27. doi: 10.1021/jp400530e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bergonzo C, Henriksen NM, Roe DR, Swails JM, Roitberg AE, Cheatham TE., 3rd Multidimensional Replica Exchange Molecular Dynamics Yields a Converged Ensemble of an RNA Tetranucleotide. J Chem Theory Comput. 2014;10:492–499. doi: 10.1021/ct400862k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Roe DR, Bergonzo C, Cheatham TE., 3rd Evaluation of enhanced sampling provided by accelerated molecular dynamics with Hamiltonian replica exchange methods. J Phys Chem B. 2014;118:3543–52. doi: 10.1021/jp4125099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gil-Ley A, Bussi G. Enhanced Conformational Sampling Using Replica Exchange with Collective-Variable Tempering. J Chem Theory Comput. 2015;11:1077–1085. doi: 10.1021/ct5009087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chakraborty K, Mantha S, Bandyopadhyay S. Molecular dynamics simulation of a single-stranded DNA with heterogeneous distribution of nucleobases in aqueous medium. J Chem Phys. 2013;139:075103. doi: 10.1063/1.4818537. [DOI] [PubMed] [Google Scholar]
- 24.Ghosh S, Dixit H, Chakrabarti R. Ion assisted structural collapse of a single stranded DNA: A molecular dynamics approach. Chemical Physics. 2015;459:137–147. [Google Scholar]
- 25.Van Der Spoel D, Lindahl E, Hess B, Groenhof G, Mark AE, Berendsen HJC. GROMACS: Fast, flexible, and free. J Comput Chem. 2005;26:1701–1718. doi: 10.1002/jcc.20291. [DOI] [PubMed] [Google Scholar]
- 26.Hess B, Kutzner C, van der Spoel D, Lindahl E. GROMACS 4: Algorithms for Highly Efficient, Load-Balanced, and Scalable Molecular Simulation. J Chem Theory Comput. 2008;4:435–447. doi: 10.1021/ct700301q. [DOI] [PubMed] [Google Scholar]
- 27.Kührová P, Banáš P, Best RB, Šponer J, Otyepka M. Computer Folding of RNA Tetraloops? Are We There Yet? J Chem Theory Comput. 2013;9:2115–2125. doi: 10.1021/ct301086z. [DOI] [PubMed] [Google Scholar]
- 28.Stadlbauer P, Krepl M, Cheatham TE, 3rd, Koca J, Sponer J. Structural dynamics of possible late-stage intermediates in folding of quadruplex DNA studied by molecular simulations. Nucleic Acids Res. 2013;41:7128–43. doi: 10.1093/nar/gkt412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Horn HW, Swope WC, Pitera JW, Madura JD, Dick TJ, Hura GL, Head-Gordon T. Development of an improved four-site water model for biomolecular simulations: TIP4P-Ew. J Chem Phys. 2004;120:9665–9678. doi: 10.1063/1.1683075. [DOI] [PubMed] [Google Scholar]
- 30.Brown RF, Andrews CT, Elcock AH. Stacking Free Energies of All DNA and RNA Nucleoside Pairs and Dinucleoside-Monophosphates Computed Using Recently Revised AMBER Parameters and Compared with Experiment. J Chem Theory Comput. 2015;11:2315–2328. doi: 10.1021/ct501170h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. Comparison of simple potential functions for simulating liquid water. J Chem Phys. 1983;79:926–935. [Google Scholar]
- 32.Hornak V, Abel R, Okur A, Strockbine B, Roitberg A, Simmerling C. Comparison of multiple Amber force fields and development of improved protein backbone parameters. Proteins: Struct, Funct, Bioinf. 2006;65:712–725. doi: 10.1002/prot.21123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li D-W, Brüschweiler R. NMR-Based Protein Potentials. Angew Chem, Int Ed. 2010;49:6778–6780. doi: 10.1002/anie.201001898. [DOI] [PubMed] [Google Scholar]
- 34.Lindorff-Larsen K, Piana S, Palmo K, Maragakis P, Klepeis JL, Dror RO, Shaw DE. Improved side-chain torsion potentials for the Amber ff99SB protein force field. Proteins: Struct, Funct, Bioinf. 2010;78:1950–1958. doi: 10.1002/prot.22711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Beauchamp KA, Lin Y-S, Das R, Pande VS. Are Protein Force Fields Getting Better? A Systematic Benchmark on 524 Diverse NMR Measurements. J Chem Theory Comput. 2012;8:1409–1414. doi: 10.1021/ct2007814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Joung IS, Cheatham TE. Determination of Alkali and Halide Monovalent Ion Parameters for Use in Explicitly Solvated Biomolecular Simulations. J Phys Chem B. 2008;112:9020–9041. doi: 10.1021/jp8001614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Parrinello M, Rahman A. Polymorphic transitions in single crystals: A new molecular dynamics method. J Appl Phys. 1981;52:7182–7190. [Google Scholar]
- 38.Nosé S. A unified formulation of the constant temperature molecular dynamics methods. The Journal of Chemical Physics. 1984;81:511–519. [Google Scholar]
- 39.Hoover WG. Canonical dynamics: Equilibrium phase-space distributions. Phys Rev A. 1985;31:1695–1697. doi: 10.1103/physreva.31.1695. [DOI] [PubMed] [Google Scholar]
- 40.Hess B, Bekker H, Berendsen HJC, Fraaije JGEM. LINCS: A linear constraint solver for molecular simulations. J Comput Chem. 1997;18:1463–1472. [Google Scholar]
- 41.Essmann U, Perera L, Berkowitz ML, Darden T, Lee H, Pedersen LG. A smooth particle mesh Ewald method. J Chem Phys. 1995;103:8577. [Google Scholar]
- 42.Norberg J, Nilsson L. Potential of mean force calculations of the stacking-unstacking process in single-stranded deoxyribodinucleoside monophosphates. Biophys J. 1995;69:2277–2285. doi: 10.1016/S0006-3495(95)80098-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Norberg J, Nilsson L. Stacking Free Energy Profiles for All 16 Natural Ribodinucleoside Monophosphates in Aqueous Solution. J Am Chem Soc. 1995;117:10832–10840. [Google Scholar]
- 44.Vokáčová Z, Buděšínský M, Rosenberg I, Schneider B, Šponer J, Sychrovský V. Structure and Dynamics of the ApA, ApC, CpA, and CpC RNA Dinucleoside Monophosphates Resolved with NMR Scalar Spin–Spin Couplings. J Phys Chem B. 2009;113:1182–1191. doi: 10.1021/jp809762b. [DOI] [PubMed] [Google Scholar]
- 45.Jafilan S, Klein L, Hyun C, Florián J. Intramolecular Base Stacking of Dinucleoside Monophosphate Anions in Aqueous Solution. J Phys Chem B. 2012;116:3613–3618. doi: 10.1021/jp209986y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schneider B, Morávek Z, Berman HM. RNA conformational classes. Nucleic Acids Res. 2004;32:1666–1677. doi: 10.1093/nar/gkh333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Le Faucheur X, Hershkovits E, Tannenbaum R, Tannenbaum A. Nonparametric clustering for studying RNA conformations. IEEE/ACM Transactions on Computational Biology and Bioinformatics. 2011;8:1604–1619. doi: 10.1109/TCBB.2010.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen H, Meisburger SP, Pabit SA, Sutton JL, Webb WW, Pollack L. Ionic strength-dependent persistence lengths of single-stranded RNA and DNA. Proc Natl Acad Sci USA. 2012;109:799–804. doi: 10.1073/pnas.1119057109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ke C, Humeniuk M, Gracz S-H, Marszalek PE. Direct Measurements of Base Stacking Interactions in DNA by Single-Molecule Atomic-Force Spectroscopy. Phys Rev Lett. 2007;99:018302. doi: 10.1103/PhysRevLett.99.018302. [DOI] [PubMed] [Google Scholar]
- 50.Ke C, Loksztejn A, Jiang Y, Kim M, Humeniuk M, Rabbi M, Marszalek Piotr E. Detecting Solvent-Driven Transitions of poly(A) to Double-Stranded Conformations by Atomic Force Microscopy. Biophys J. 2009;96:2918–2925. doi: 10.1016/j.bpj.2008.12.3939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen AA, García AE. High-resolution reversible folding of hyperstable RNA tetraloops using molecular dynamics simulations. Proc Natl Acad Sci USA. 2013;110:16820–16825. doi: 10.1073/pnas.1309392110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Šponer J, Mládek A, Špačková N, Cang X, Cheatham TE, Grimme S. Relative Stability of Different DNA Guanine Quadruplex Stem Topologies Derived Using Large-Scale Quantum-Chemical Computations. J Am Chem Soc. 2013;135:9785–9796. doi: 10.1021/ja402525c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kruse H, Havrila M, Šponer J. QM Computations on Complete Nucleic Acids Building Blocks: Analysis of the Sarcin–Ricin RNA Motif Using DFT-D3, HF-3c, PM6-D3H, and MM Approaches. J Chem Theory Comput. 2014;10:2615–2629. doi: 10.1021/ct500183w. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.