Abstract
Whole-genome duplication (WGD) results in new genomic resources that can be exploited by evolution for rewiring genetic regulatory networks in organisms. In metazoans, WGD occurred before the last common ancestor of vertebrates, and has been postulated as a major evolutionary force that contributed to their speciation and diversification of morphological structures. Here, we have sequenced genomes from three of the four extant species of horseshoe crabs—Carcinoscorpius rotundicauda, Limulus polyphemus and Tachypleus tridentatus. Phylogenetic and sequence analyses of their Hox and other homeobox genes, which encode crucial transcription factors and have been used as indicators of WGD in animals, strongly suggests that WGD happened before the last common ancestor of these marine chelicerates >135 million years ago. Signatures of subfunctionalisation of paralogues of Hox genes are revealed in the appendages of two species of horseshoe crabs. Further, residual homeobox pseudogenes are observed in the three lineages. The existence of WGD in the horseshoe crabs, noted for relative morphological stasis over geological time, suggests that genomic diversity need not always be reflected phenotypically, in contrast to the suggested situation in vertebrates. This study provides evidence of ancient WGD in the ecdysozoan lineage, and reveals new opportunities for studying genomic and regulatory evolution after WGD in the Metazoa.
Introduction
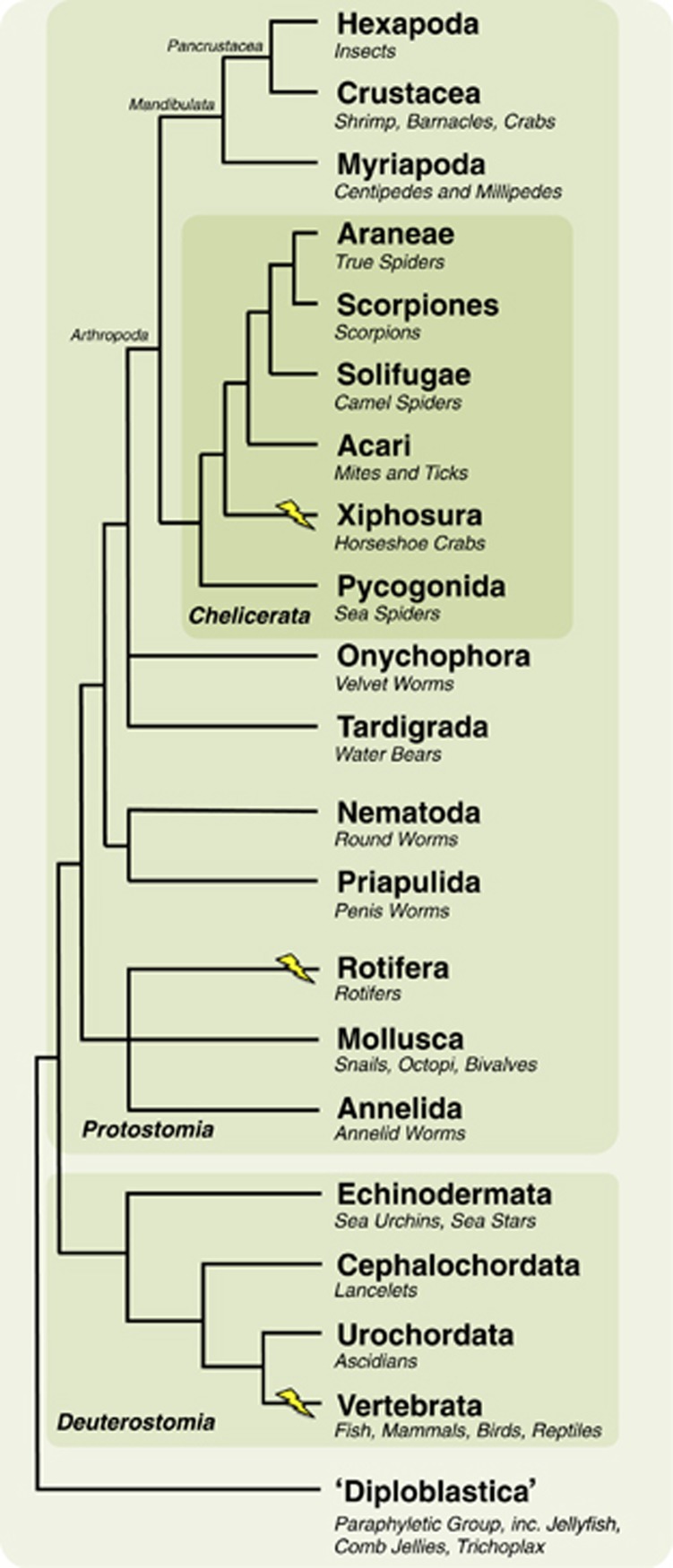
Whole-genome duplication (WGD) events provide organisms with additional copies of their entire genomes, and have long been postulated as the major source of evolutionary innovations that contribute to the diversification of anatomical structures and rapid speciation seen in some lineages (Ohno, 1970; Holland, 2003; Sémon and Wolfe, 2007; Jaillon et al., 2009; Van de Peer et al., 2009). WGD events are distributed unequally across eukaryote phylogeny. They are common in plants (Adams and Wendel, 2005) and several have been reported in fungi (Albertin and Marullo, 2012), but the repeated rounds of WGD seen in the Vertebrata are the most prominent examples yet reported in metazoans, as can be seen in Figure 1. Outside the Vertebrata, only the fast-evolving parthenogenetic bdelloid rotifers (Flot et al., 2013) have been shown to exhibit WGD in the metazoan clade.
Figure 1.
Whole-genome duplication events across metazoan phylogeny. Xiphosuran, chelicerate and metazoan consensus phylogeny, with arthropod relationships based on Regier et al. (2010) and wider metazoan relationships based on Dunn et al. (2008). Note that some inter-relationships, notably in the Chelicerata, are controversial. Specific clades noted in the text, such as the Chelicerata, Protostomia and Deuterostomia, are indicated by differential shading. Known WGD events are marked with a lightning bolt. Note multiple rounds of WGD have occurred in all lineages thus marked.
The presence of multiple paralogous copies of different developmental gene families, especially the homeobox Hox cluster genes, which are arranged in large syntenic genomic regions, are essential for the discovery and study of WGDs in vertebrates (Amores et al., 1998). Homeobox genes usually contain a 180-nucleotide long motif that encodes a DNA-binding domain, the homeodomain. Most homeodomain-containing proteins act as transcription factors, with roles in a wide range of developmental processes. High degrees of homeobox gene conservation, and in some cases functional conservation, are found across different phyla from fungi, plants and animals/metazoans (Gehring, 1984, 1994; De Robertis, 1994; Holland and Hogan, 1988; Galliot et al., 1999; Bürglin, 2005, 2011; Ryan et al., 2006; Holland et al., 2007, 2012; Larroux et al., 2008; Zhong et al., 2008; Zhong and Holland, 2011a), with the encoded homeodomain generally being the most highly conserved region (Bürglin, 2005, 2011). Owing to their roles as gene regulators and their highly conserved sequences, homeobox genes have become ideal candidates to follow large-scale changes to genomes in evolution (Holland, 1999; 2012).
The homeobox gene superclass is extremely diverse, with over 200 genes in vertebrate genomes and ~100 genes in many invertebrate genomes. Based on the homeodomain sequence, metazoan homeobox genes can be classified into 11 classes—the ANTP (including the Hox genes), CERS (ceramide synthase), CUT, HNF, LIM, PRD, PROS, POU, SINE, TALE (three amino-acid loop extension) and ZF (zinc finger) (Bürglin, 2005, 2011; Holland et al., 2007; Zhong et al., 2008; Holland, 2012). According to phylogenetic analyses and surveys of many genomes, two major classes of homeobox genes, the ANTP and PRD are thought to be confined to the metazoans (Galliot et al., 1999; Bürglin, 2005, 2011; Holland and Takahashi, 2005).
The ANTP-class homeobox genes are named after the Antennapedia gene of Drosophila melanogaster. This family of genes can be recognised by their distinctive homeodomain, and in some cases, also five or six amino acids constituting the hexapeptide (or pentapeptide) motif upstream of the homeodomain (Bürglin, 2005, 2011; Holland et al., 2007; Holland, 2012). Nomenclature within the ANTP-class homeobox genes is complex, and includes Hox, Hox-like (or Hox-linked), Hox/ParaHox related, extended Hox, NK and NK-like or NK-linked genes (Holland et al., 2007; Ferrier, 2008; Hui et al., 2012). Comparing the chromosomal positions of the ANTP-class homeobox genes in human and mouse, Pollard and Holland (2000) first postulated that the whole ANTP-class of homeobox genes originated from a single ancestral gene, via an ancient mega-ANTP-class homeobox cluster (megacluster) that existed deep in metazoan ancestry (for details, see Pollard and Holland, 2000; review by Garcia-Fernàndez, 2005). Mapping of ANTP class homeobox genes in the cephalochordate, Branchiostoma floridae and polychaete Platynereis dumerilii has provided further support for the hypothesis (Castro and Holland, 2003; Luke et al., 2003; Hui et al., 2009, 2012).
The PRD class homeobox genes derive their name from the paired gene of D. melanogaster (Bürglin, 2005, 2011; Holland et al., 2007), and are further subdivided into the PAX and PAXL subclasses found throughout the animal kingdom (Galliot et al., 1999; Ryan et al., 2006; Holland et al., 2007; Hoshiyama et al., 2007; Larroux et al., 2008). Unlike ANTP class genes, PRD genes are usually dispersed; however, two PRD-homeobox clusters have been proposed—the mammalian Rhox homeobox gene cluster (MacLean et al., 2005) and a conserved cluster containing homeobrain, rx and orthopedia (Mazza et al., 2010).
Recently, however, a number of investigations in the Chelicerata have provided circumstantial evidence for the possibility of one or more WGD events in that clade. These include the discovery of multiple copies of homeobox-containing genes in the spider Cupiennius salei (Schwager et al., 2007), the scorpion Centruroides sculpturatus (Sharma et al., 2014a) and the horseshoe crab Limulus polyphemus (Nossa et al., 2014). The latter paper showed by a variety of methods, including linkage mapping, comparison of age distribution of paralogous genes and gene cluster comparison, that WGD occurred in L. polyphemus. However, whether WGD is limited to L. polyphemus alone, or is more commonly found in the Xiphosura, is uninvestigated, and would have a range of implications for our understanding of the prevalence of WGD in the Chelicerata. The Chelicerata are a diverse clade, found both on land and in aquatic habitats and although many chelicerate lineages are strikingly diverse both in terms of number of species and morphological and ecological adaptations, other groups exhibit a remarkable morphological stasis. The marine chelicerate horseshoe crabs, of the Order Xiphosura, are a prominent example of the latter situation.
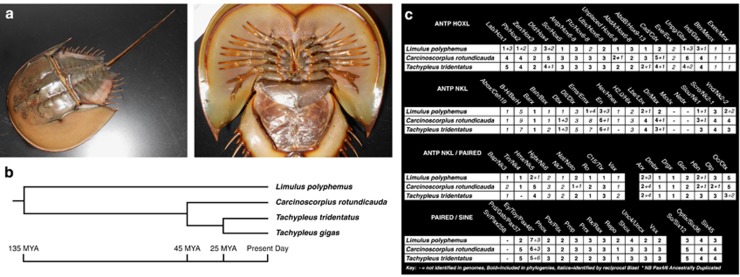
With the exception of details in abdominal segment and biramous limb morphology (Briggs et al., 2012) living horseshoe crabs, such as Carcinoscorpius rotundicauda (Figure 2a), are remarkably similar in appearance to fossil species 400 million years in age, and almost indistinguishable to some found in the Jurassic (Briggs et al., 2005). In addition, very limited diversity has been catalogued in the Xiphosura across geological time (Störmer 1952; Störmer 1955; Anderson and Selden, 1997). Present xiphosuran diversity is limited to just four species, L. polyphemus found in the Atlantic, and C. rotundicauda, Tachypleus gigas and Tachypleus tridentatus found in Southeast Asia. These species separated from one another relatively recently, ~135 million years before the present day (Figure 2b, Obst et al., 2012).
Figure 2.
Xiphosuran biology and homeodomain-containing gene number. (a) Adult Carcinoscorpinus rotundicauda, oblique and ventral view. (b) Xiphosuran phylogeny and divergence time after Obst et al. (2012). MYA=millions of years ago. (c) Homeobox genes extracted from sequenced horseshoe crab species with phylogenetic evidence of identity shown in bold text. Genes identified but not included in phylogeny are shown in italics, and can be seen in Supplementary File 1.
Here we analyze the homeobox gene complements in the genomes of the three extant horseshoe crab genera (in the species C. rotundicauda, L. polyphemus and Tachypleus tridentatus) and provide strong evidence for the presence of at least one WGD event in the xiphosuran lineage. The orthologous relationships of the multiple copies of homeobox genes in all horseshoe crab genomes indicate that the duplication predates the last common ancestor of extant xiphosurans. We present reverse transcription (RT)-PCR and RNAseq-based assays providing evidence of the diversification of Hox gene functionality after this event, with sub- and neo-functionalisation along the anterior–posterior axis of these species. This represents the first evidence and study of the consequences of a WGD event in an ecdysozoan lineage. Such evidence has important implications for how we consider genomic evolution and phenotypic diversification after such events, representing a key comparison point for the vertebrate lineage when assessing the causes and consequences of genomic and phenotypic changes in metazoan evolution.
Materials and methods
Genomic and transcriptomic sequencing
Horseshoe crab samples were acquired commercially from local suppliers in Hong Kong (C. rotundicauda and T. tridentatus) and a collaborator (S Smith) in the USA (L. polyphemus). Genomic DNA samples were extracted from muscle tissue using a PureLink Genomic DNA Mini Kit and the manufacturer's protocol, and sequenced by BGI (Hong Kong) on Illumina HiSeq2000. A variety of assembly software platforms and settings were trialled empirically before selection of Velvet for generation of a final assembly. Fragment size distributions (nominally 170 bp) were found using Bowtie (Langmead and Salzberg, 2012) on a preliminary assembly (k-mer 51, Velvet 1.2.09 (Zerbino and Birney, 2008)) of genomes. These preliminary assemblies were also used to determine expected genome coverage. Using this information, genome assembly was performed using Velvet 1.2.09 (Zerbino and Birney, 2008), with a k-mer size of 51, minimum fragment length of 200 bp, fragment sizes as noted in Supplementary File 1 and expected coverage as noted in text. The raw read data from the three horseshoe crabs examined in this study have been uploaded to NCBI's short read archive, and are available under Bioproject Accession PRJNA243016, SRP040718, and assemblies can be downloaded from webpage (tinyurl.com/HSCgenomes) and the Dryad repository linked to this article (url: http://dx.doi.org/10.5061/dryad.81fv1).
mRNA samples for sequencing were extracted from whole chelicerae samples of adult C. rotundicauda and T. tridentatus using Trizol according to the manufacturer's instructions, followed by RNeasy (Qiagen, Venlo, Netherlands) purification. Samples were sequenced by BGI (Hong Kong) on an Illumina HiSeq2000. Trinity r2013_08_14 (Grabherr et al., 2011) was used to assemble transcriptomes with all default settings.
Gene identification and phylogenetic analysis
Genes were identified in our data set, and in the sequenced genomes of other chelicerates by tblastn with ncbi-blast-2.2.23+ (Altschul et al., 1990) using homeodomain sequences of known identity downloaded from HomeoDB (Zhong and Holland, 2011b). All contig sequences hit with an E-value <10−9 were reciprocally blasted (blastx) to the NCBI nr database to putatively confirm identity. Identified contigs with exonic regions of clear homology with known homeodomain genes were converted to amino-acid sequence using the EMBOSS Transeq server (Rice et al., 2000) and standard codon table. Sequences were aligned using MAFFT (Katoh et al., 2002) under the L-INS-i model to known chelicerate sequences downloaded from GenBank and relevant genome resources, along with B. floridae, Homo sapiens and T. castaneum sequences downloaded from HomeoDB (Zhong and Holland, 2011b). Aligned regions had gaps removed using MEGA (Kumar et al., 2008). Phylogenetic analysis was performed using PhyML (Guindon et al., 2009) and MrBayes (Ronquist & Huelsenbeck, 2003) using models as described in figure legends. Bayesian phylogenies were then displayed in FigTree for visualisation (tree.bio.ed.ac.uk/software/figtree/).
Expression analysis
Walking legs, book gills and the telson of two adult individuals of each of C. rotundicauda and T. tridentatus were excised and samples taken from the most proximal portion of each appendage to the main body mass. Primers were designed to conserve exonic regions using Primer3Plus for each gene, and aligned to other paralogues to ensure specificity. RNA was extracted using Trizol followed by two rounds of phenol/chloroform extraction and RNA precipitation, and reverse transcription performed using Takara Reverse Polymerase. PCR master mixes were prepared for each gene examined, which were split into 18 μl reaction mixes containing 0.5 μM of primer, 200 μM of each dNTP (Takara Bio Inc., Shiga, Japan), 1 × PCR buffer, 3 mM MgCl2 and 2 units Taq (Roche, Penzberg, Germany), to which 2 μl of 10–30 ng cDNA from each appendage was added to individual tubes. PCR was performed using a 3 min 95 °C initial denaturation, 38 × 95 °C/60 s, 60 °C/60 s, 72 °C/60 s steps and a final 72 °C/600 s elongation. Electrophoresis was performed on a 1% agarose gel and visualized under UV with SYBR Safe (Life Technologies, Carlsbad, CA, USA). All primer sequences and gels can be seen in Supplementary File 2.
Results and Discussion
Genome sequencing and assembly
Genome and transcriptome sequencing/assembly results are summarised in Supplementary File 1. Paired end read quality was high, with lower quartile Phred scores above 30 through to the 100th base. The data from the three horseshoe crabs examined in this study have been uploaded to NCBI's short read archive, and are available under Bioproject Accession PRJNA243016, SRP040718. Using tblastn (Altschul et al., 1990), comparing the 458 gene family CEGMA data set (Parra et al., 2007) to the horseshoe crab genome assemblies with an E-value cutoff of 10−6; 98% recovery of identifiable homologues of the 458 Core Eukaryotic Genes were found in the L. polyphemus, T. tridentatus and C. rotundicauda genomes (448, 450 and 448 Core Eukaryotic Genes, respectively), confirming excellent recovery of the coding areas of these genomes. Five common Core Eukaryotic Genes were absent from all three genomes, and may be particularly divergent or absent in the xiphosuran lineage (Supplementary File 1). The genome sequence depth for all three species is around sixfold, recovering ~1.5 Gbp per species. This is lower than the previously estimated genome sizes of these species of ~2.8 Gbp (Goldberg et al., 1975), which together with CEGMA data suggest the assembled genome data sets are biased towards coding sequence to the exclusion of longer repeat sequences, or perhaps that previous calculations were overestimates. A L. polyphemus genome has been noted online (Nossa et al., 2014), but the data presented here represents a large increase in publically available sequence data for the Xiphosura clade, particularly by including Southeast Asian species.
Multiple copies of Hox genes in all three horseshoe crab genomes
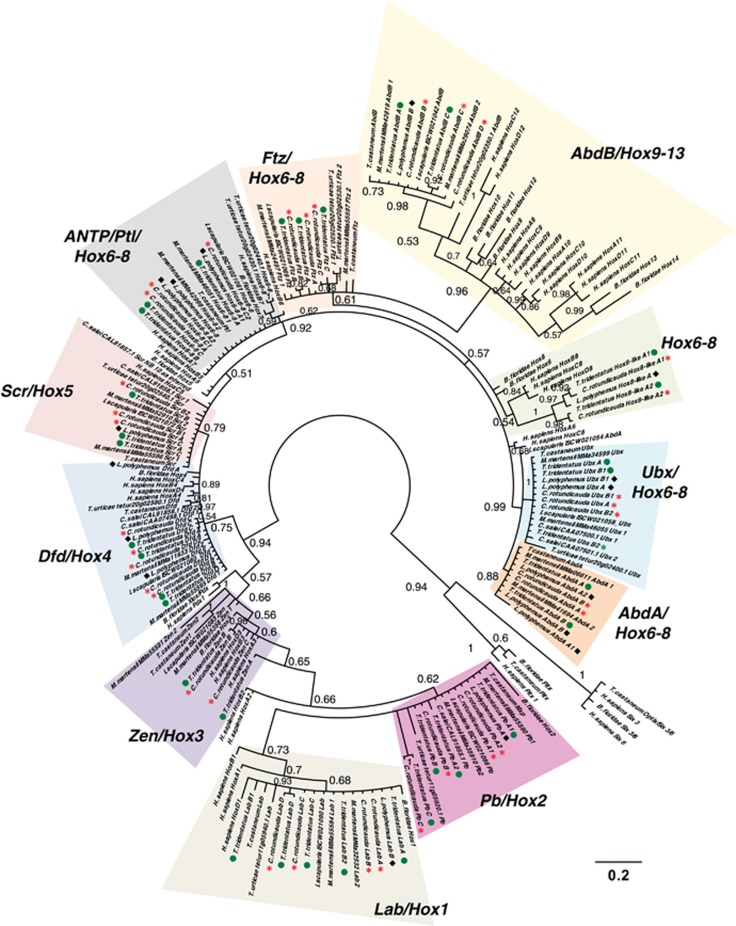
Homeobox genes encode transcription factors deployed in animal development and contain several well-conserved classes such as the ANTP, PRD and SINE genes (Zhong and Holland, 2011b; Holland, 2012; Hui et al., 2012). Genes of the ANTP class, notably the Hox-linked genes, including the well-known Hox cluster genes and their chromosomal neighbors, such as Evx and Mox genes (also known as the extended Hox cluster), are very stable in animal evolution. Owing to their characteristic arrangement into linked gene clusters occupying large chromosomal regions, they have been widely used to identify WGD events and other major genomic changes in animals (Amores et al., 1998; Castro et al., 2006; Tsai et al., 2013; Smith et al., 2013). To date, fully duplicated Hox cluster genes have only been found in species that have undergone WGDs. Examination of Hox and extended Hox cluster genes in the three horseshoe crab assemblies revealed between two and four representatives of almost every gene in each species (Figure 2c, row 1). These numbers could result from a single round of WGD followed by independent duplication or two rounds of WGD followed by loss, the latter being arguably the more likely scenario, given the number of independent duplications that would be necessary across widely separated genomic regions. Our data provide a minimum estimate for these homeobox gene groups in the Xiphosura; there is a possibility that more genes may exist in the genomes than recovered in our analyses. Orthology was confirmed by examining multiple sequence alignments and phylogenetic trees made using Bayesian phylogenetic methods (MrBayes, Ronquist & Huelsenbeck, 2003), including sequences from horseshoe crabs, other chelicerates, humans, the amphioxus B. floridae and beetle Tribolium castaneum (Figure 3 and in Supplementary Files 1, 3 and 4, where nodes have not been collapsed). Where precise prediction of exonic sequence was impaired due to truncated contig size or divergent gene sequence, genes were excluded from phylogenetic consideration, and are shown in italics in Figure 2c. The contig sequences of all genes are listed in Supplementary File 1. Owing to the low level of nucleotide sequence divergence between the three horseshoe crabs species, orthologous relationships between paralogues in the three horseshoe crabs are clear in sequence alignments, such as all investigated Hox genes (Supplementary File 5). This contrasts with the local tandem duplications seen in the ftz and AbdA genes of the Hox cluster in mite (Grbić et al., 2011) and the zen/Hox3 genes of Drosophila and T. castaneum (Falciani et al., 1996; Stauber et al., 1999). Thus, as observed in vertebrate and teleost genomes (Hoegg and Meyer, 2005), all three xiphosuran genomes investigated here also contain multiple copies of all Hox and extended Hox cluster genes paralogous groups, strongly suggesting WGDs in this lineage.
Figure 3.
ANTP HoxL class gene inter-relationships. Bayesian phylogenetic tree of ANTP HoxL class genes from sequenced xiphosuran species, along with the previously described complements of other chelicerates, amphioxus Branchiostoma floridae, human Homo sapiens and beetle Tribolium castaneum. Phylogenies inferred with the Jones model and 55 informative sites. Numbers at base of nodes represent posterior probability to two significant figures. Trees shown are the result of Bayesian analysis, run for 10 000 000 generations, and analyses run until the s.d. of split frequencies was below 0.01, with the first 25% of sampled trees discarded as ‘burn-in'. All sequences and alignments detailed in Supplementary File 1. Shading shows major gene families. Coloured dots aid in identification of species, with green circles for T. tridentatus, black diamonds for L. polyphemus and red asterisks for C. rotundicauda. Scale bar represents substitutions per site at given distances. Rooted using Pitx and Six 3/6 sequence outgroups.
Multiple copies of other homeobox and homeobox cluster genes in all three horseshoe crab genomes
Evidence for an ancestral WGD in the Xiphosura does not only come from the Hox and extended Hox cluster genes. Multiple gene copies were also found in the three sequenced horseshoe crab genomes for other dispersed groups of homeobox genes that ordinarily exist as single copy in invertebrate genomes and are dispersed over different chromosomal regions in all animals surveyed to date (Supplementary Files 3 and 4, Zhong and Holland, 2011a; Hui et al., 2012). This finding makes it highly unlikely that the Hox gene duplications are the result of local segmental duplication.
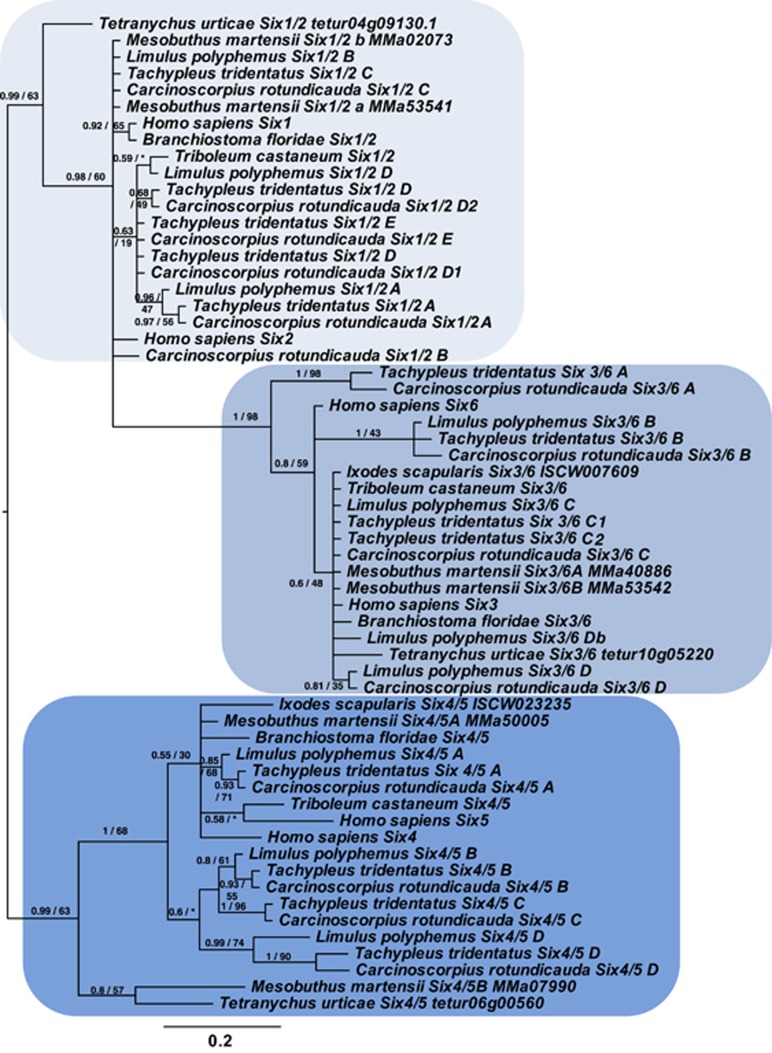
For example, multiple paralogs shared between horseshoe crab species are also found in the SINE class tree (Figure 2c, Figure 4). In metazoans, there are three families of SINE class gene—Six 1/2, Six 3/6 and Six 4/5, and most invertebrates possess a single gene copy in each Six family (Zhong and Holland, 2011a; Holland, 2012). In our three sequenced species of the horseshoe crab genomes, multiple copies of members of each of the three SINE gene families are found. Each family has clear posterior probability/bootstrap support for their internal relationships with relatively short branches (for example, 1.0/98 for monophyly of Six 3/6; 0.99/63 monophyly of Six 4/5; 0.99/63 monophyly of Six 1/2). Clear orthologous relationships between some xiphosuran SINE paralogues can be observed (for example, the subfamily A of the Six 1/2 clade has a single representative in every horseshoe crab examined (Figure 4 with single-letter suffix).
Figure 4.
Phylogenetic tree of SINE class homeobox genes. Phylogenetic tree of SINE class homeobox genes from three xiphosuran species, along with the amphioxus Branchiostoma floridae, human Homo sapiens and beetle Tribolium castaneum. Bayesian/ML phylogenies inferred using the Jones and LG models, respectively, on the basis of 51 informative sites. Numbers at base of nodes represent posterior probability/bootstrap proportions (1000 replicates, expressed as %age) to two significant figures. Trees shown are the result of Bayesian analysis, run for 1 500 000 generations, until the s.d. of split frequencies was below 0.01, with the first 25% of sampled trees discarded as ‘burn-in'. Where ML analysis collapses to a polytomy at the previous node and does not match topology of Bayesian tree, bootstrap proportions are replaced with a ‘*'. All sequences and alignments detailed in Supplementary File 1. Blue-shaded areas indicate SINE gene families. Scale bar represents substitutions per site at given distances. Rooted at midpoint.
Multiple gene copies in the three sequenced horseshoe crab genomes were also found for representatives from other homeobox clusters found in other bilaterians, such as the ParaHox, Hbn/Otp/Rx and NK clusters (additional file 1, Zhong and Holland, 2011a; Hui et al., 2008, 2012; Mazza et al., 2010). Although much less studied than the Hox, ParaHox and NK clusters, the trio of hbn, rx and otp constitutes a well-conserved PRD-homeobox gene cluster whose organization has been maintained in many protostome genomes (Mazza et al., 2010). Together, these results reveal that xiphosuran genomes contain paralogous arrays of genes that are well known for being single copy in other animal species. In a recent study analyzing the hormonal pathway genes in arthropods, it has also shown that the three species of horseshoe crabs contain multiple copies for some of the analyzed genes (Qu et al., 2015). Even if one assumes a particularly high rate of tandem duplication in the horseshoe crab lineage, the presence of multiple copies of genes that are typically arranged into large and complex syntenic and regulatory genomic regions (and thus less prone to tandem duplication) suggests a WGD scenario is more likely than independent duplication of all loci independently. Although gene order evidence would provide even more proof for WGD rather than independent duplication in all species in this lineage, Nossa et al. (2014) provide a strong basis for this claim. Further syntenic data from Asiatic horseshoe crab species would provide a conclusive test for WGD ancestrally in this clade.
It is necessary to ask whether the deduced WGD is a shared ancestral event, or whether WGD occurred independently in three horseshoe crab species. This cannot be easily distinguished using gene numbers. Considering the gene trees as shown above, the presence of orthologous relationships between multiple paralogues of homeobox gene families in the xiphosuran lineage strongly implies that the WGD events that are seen in these three species are shared ancestrally, and not the product of several independent WGD events in the species sequenced here. Although the Chelicerata phylogeny is controversial (Figure 1, Regier et al., 2010; Rota-Stabelli et al., 2013; Sharma et al., 2014b), the diversification of the crown group Xiphosura is well established (Figure 2b). Therefore, the ancestral WGD that we found here would date back to at least 135 million years ago.
WGD may therefore have occurred multiple times in the Xiphosura, but assessing the extent of this is not trivial. Using gene numbers to assess gene duplication is sometimes complicated by confusion between allelic variants and paralogous (duplicate) loci. In the present case, the fact that each species has multiple variants of a gene, and the demonstration that these group into orthologous groups when compared between species (Supplementary File 5), strongly supports the conclusion that these variants are duplicate genes dating to before divergence of currently extant species, not alleles differing within the sequenced individuals. As genome sequences were determined from individual animals, cases with greater than two individual sequences cannot be due to allelic variation alone. Given the presence of four or more paralogues for many genes in all three horseshoe crab species presented here, the occurrence of at least one round of WGD in their common ancestors is more parsimonious than independent duplication of all loci in question.
Subfunctionalisation of Hox gene expression
Using both RNAseq of the chelicerae and RT-PCR on the appendages along the anterior–posterior axis, we have also investigated the impact that WGD had on the expression of Hox genes in adult C. rotundicauda and T. tridentatus, whose lineages diverged approximately 45 million years ago (Figure 2b). We excluded chilaria (rudimentary appendages posterior to last walking legs) from our analysis due to their small size.
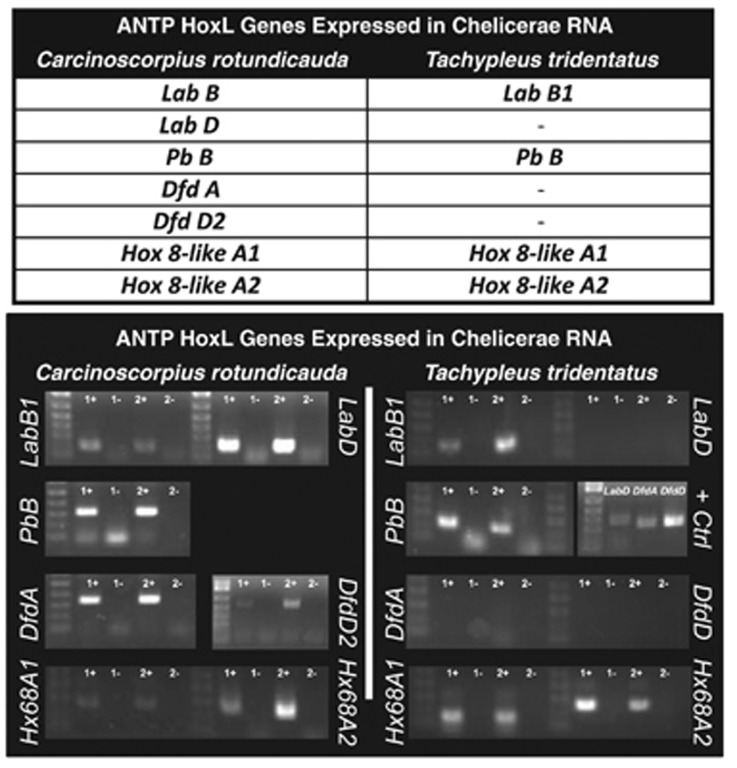
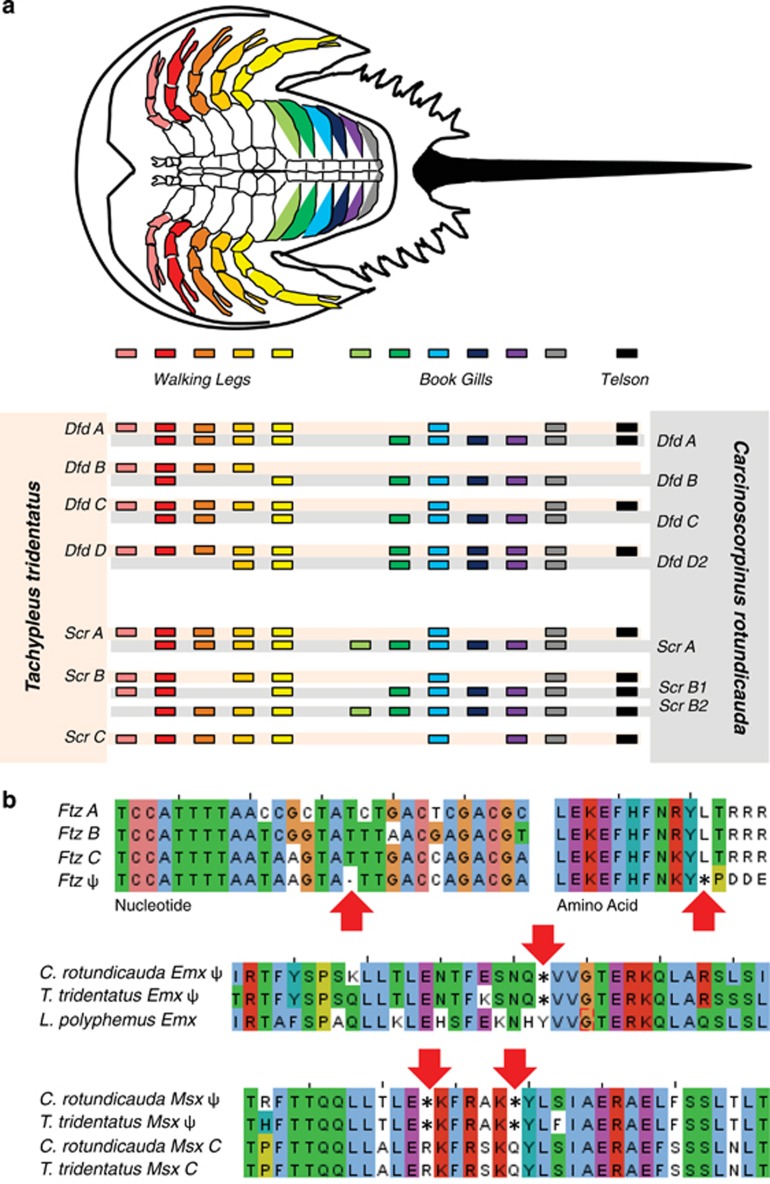
Only seven (C. rotundicauda) and four (T. tridentatus) Hox genes were expressed in adult chelicerae tissue (Figure 5). The expression of these genes in the chelicerae of these species was tested by RT-PCR in independent adult samples. In general, T. tridentatus expresses fewer of these genes than C. rotundicauda, such as the lab and Dfd duplicates. In the case Dfd families, no orthologues are expressed in the former species. To further test whether duplicated Hox genes have diverged in function anterior to the chelicerae, expression patterns of the Hox genes Dfd and Scr were examined on appendages along the anterior–posterior axis of adult C. rotundicauda and T. tridentatus by RT-PCR. These two genes were chosen based on their key roles in patterning and maintaining the identity of limbs in arthropods (Telford and Thomas, 1998). In Figure 6a, a variety of contrasting patterns of orthologous genes are shown, reflecting flexibility in remodeling transcriptional regulation, possibly facilitated by WGD (evidence of orthology in Supplementary File 4). For example, in T. tridentatus, expression of Dfd D is consistent between the two species, whereas Dfd A, DfdB and DfdC are expressed in different domains in the two species (Dfd A, DfdB and DfdC are confined to predominantly anterior, walking limb domains in T. tridentatus but are found expressed in the appendages along much of the anterior–posterior axis of C. rotundicauda). The expression patterns of Hox genes shown above confirmed that following WGD events in horseshoe crabs, paralogous genes have undergone subfunctionalisation in these lineages.
Figure 5.
RNA expression evidence for subfunctionalisation of ANTP HoxL class genes. ANTP HoxL class genes found in chelicerae RNAseq data in two species of horseshoe crab, along with RT-PCR confirmation of expression in two independent adult samples. No RT controls, also shown, confirm that expression derives from RNA and not gDNA contamination.
Figure 6.
Sub- and pseudofunctionalisation of ANTP HoxL class genes. (a) Subfunctionalisation of ANTP class homeobox genes: figure showing the expression of a number of ANTP class homeodomain genes along the AP axis of adults of two species of horseshoe crabs, Carcinoscorpinus rotundicauda and Tachypleus tridentatus, as determined using RT-PCR as described in methods. Significant subfunctionalisation is seen in the expression of these paralogous genes. (b) Nucleotide (left) and amino-acid (right) alignments of portion of T. tridentatus Ftz genes and transcribed Ftz pseudogene. Note indel, leading to frameshift error in translation. Amino-acid sequences of Emx and Msx genes and pseudogenes identified in genomic sequence. Asterisks indicate where single-nucleotide substitutions have resulted in missense mutations, coding for a premature stop codon. Red arrows indicate location of such signals.
Pseudogenization
A number of homeobox pseudogenes, with clear missense and indel mutations in the homeodomain, were also identified. The sequences of these homeobox pseudogenes have also been independently confirmed in separate individuals to those originally sequenced (data not shown). The process of pseudogenisation of these homeobox pseudogenes most likely occurred at different times in the three species. For example, Msx and Emx genes in T. tridentatus and C. rotundicauda share signatures of pseudogenization (Figure 6b), but the other pseudogenes are not detected in common between these two species. Transcription of a pseudogenised representative of the Hox gene ftz in T. tridentatus can be observed in the RT-PCR-based assays of several independent individuals (Figure 6b, Supplementary File 1). Whether other xiphosuran Hox pseudogenes are transcribed is unknown, but this example suggests the complete ‘silencing' of pseudogenes may take millions of years after a WGD event, allowing sufficient time for extensive remodelling of regulatory mechanisms (Shakhnovich & Koonin, 2006). Birth and death rates for pseudogenes vary from species to species (Podlaha and Zhang, 2010) and the rates can be heightened by high neutral mutation rate, as observed in mouse (Waterston et al., 2002), or by high genomic deletion rates as seen in the fly (Petrov et al., 1996, 1998).
WGDs in horseshoe crabs, chelicerates and vertebrates
The Xiphosura is generally accepted as the sister group of the Arachnida (Figure 1, Regier et al., 2010, Rota-Stabelli et al., 2013). As shown in Figure 3, Figure 4 and Supplementary Files 3 and 4, the genome of the scorpion Mesobuthus martensii (Cao et al., 2013) possesses multiple copies of many of the homeobox gene families studied here. Furthermore, the spider C. salei and the scorpion C. sculpturatus also possess multiple copies of several Hox cluster genes (Schwager et al., 2007; Sharma et al., 2014a). The presence of WGD in the horseshoe crabs led us to consider whether WGD might have occurred in the chelicerate stem lineage, rather than being limited to the Xiphosura. Our phylogenetic analyses (Figures 3 and 4,Supplementary Files 3 and 4), however, do not find unambiguous orthologous relationships between duplicated homeobox genes in scorpion and spider lineages, or between these animals and horseshoe crabs. With the present data, we cannot draw firm conclusions about the number of WGD events, but suggest that independent WGD events may have occurred in different chelicerate lineages.
In the case of M. martensii the presence of duplicates of a wide range of homeobox gene families strongly suggests that WGD may also be present in the scorpion lineage. In some cases, very weak support (with posterior probability >0.5) is found for orthologous relationships between scorpion M. martensii and Xiphosuran paralogues (for example, Supplementary File 3 Abd B clade). This could be the result of shared WGD, independent WGD or convergence, or more local gene duplication prior to the divergence of these lineages. At present, given contig lengths in our sequence and publically released scaffold lengths available from the M. martensii genome project, it is not possible to resolve by syntenic analysis whether duplication is shared. With present data, we hypothesize that either independent WGD events account for the patterns seen here, or (less likely given present evidence) an ancient chelicerate WGD is shared by scorpions and horseshoe crabs. Previous studies have noted morphological links between xiphosurans and scorpions (Wheeler & Hayashi, 1998) although not normally to the exclusion of other chelicerates; hence, any inference of shared WGD between scorpions and xiphosurans would have profound phylogenetic implications. Our reasons for favouring the first scenario (independent WGD) include the general lack of support found in our trees for shared WGD, and the fact that the spider mite T. urticae, which is generally thought more closely related to scorpions than xiphosurans, lacks any clear sign of WGD (it has a single Hox cluster in its genome, and although there are some expanded highly expanded homeobox genes, these are limited to just a few families (Grbić et al., 2011)).
Spider C. salei Hox paralogues also mirror the pattern of in-paralogue clustering (see Supplementary File 3, C. salei Ubx, Scr, Dfd) suggesting further that duplication of homeobox genes occurred in multiple chelicerate lineages independently. Whether the duplicated spider Hox genes could be indicative of a possible WGD in that lineage is an open question. Whole-genome sequencing in C. salei or further analyses of the two recently published spider genomes (Sanggaard et al., 2014), specifically looking for traces of WGD, would help to solve this question. Overall, these comparative data indicate that WGDs, particularly in chelicerates, are more common phenomena than previously appreciated.
WGD events have been suggested to be a potent source of innovation in genetic regulatory networks, a driver of genomic evolution, and a contributing factor in animal diversification. However, WGD events have a patchy phylogenetic distribution in the Metazoa (Figure 1), and to date the evolutionary implications have only been inferred from vertebrates. In addition to this well-known case, WGD has recently been detected in the bdelloid rotifer Adenita vaga (Flot et al., 2013). However, the obligately asexual reproduction of this species could limit any conclusions drawn about genome evolution when using it as a comparison point with the vertebrates. The discovery of WGD in sexually reproducing horseshoe crabs will thus be useful for wider comparison of patterns of functional gain and loss with other animals, and in particular, with vertebrate WGD.
The presence of multiple rounds of WGD in the Xiphosura mirrors may be in both vertebrates and rotifer. We suggest, therefore, that WGD may be prone to occur in succession in animal lineages that survive genome duplication. Whether this is the result of how WGDs occur in the first instance, or how likely they are to be evolutionarily successful is a question for further consideration.
The contrast between how the Xiphosura (which has been phenotypically consistent since the Cambrian (Rota-Stabelli et al., 2013) and the Vertebrata, (which exhibit extreme phenotypic diversity) have utilised WGD events is stark. Even during the Carboniferous period, when the xiphosuran group experienced its highest diversity peak, the lineage consisted of not >50 species, as evidenced by the fossil record, indicating a very low radiation potential (Anderson and Selden, 1997) although it is possible that the recovered fossil record is an imperfect reflection of actual diversity. Nevertheless, our data are consistent with the idea that WGD events need not result in morphological changes (at least in horseshoe crabs), and could allow the buffering of transcriptional networks to external insult through complex overlapping patterns of expression, as suggested in Chapman et al., 2006, and further reinforced by the fact that transcriptional regulators are disproportionately retained after WGD events (Maere et al., 2005; Blomme et al., 2006). It is important to note that a range of other factors, particularly increased network-interaction complexity, may have contributed to the diversification in the Vertebrata, and this issue will be an interesting topic for future study in cases of invertebrate WGD.
The relative morphological stasis of horseshoe crabs implies that novelty at a genomic level has not been mirrored phenotypically in this clade. Does this suggest a general difference between invertebrates and vertebrates in the evolutionary significance of WGD? Before drawing such a conclusion, it is important to note that the extent to which vertebrate WGD affected the later evolution of this clade has been a subject of debate (Furlong and Holland, 2002; Dehal and Boore, 2005; Donoghue and Purnell, 2005). Additional data will be of much aid in discerning what influence WGD has on subsequent genomic and phenotypic evolution. Further examination of patterns of gene gain/loss, expression and functional divergence of paralogues, and regulatory network changes (such as post-transcriptional gene regulation by microRNAs) following WGD in the Xiphosura and in vertebrates are warranted to disentangle this complex issue.
This work provides clear evidence of an ancient WGD event in the horseshoe crab lineage through the genomic sequencing of three of the four extant species. This is the first WGD event to be described in the ecdysozoans, and is one of very few such events to be known from the animal kingdom. The data presented here urge reexamination of long-standing hypothesis regarding the evolutionary outcomes of WGDs in animals. The three xiphosuran cases identified here will provide evolutionary comparison points for the inference of the effect of polyploidy on animals.
Data archiving
All data related to this work are available for download from the NCBI short read archive and our own website (tinyurl.com/HSCgenomes) and the Dryad repository linked to this article (url: http://dx.doi.org/10.5061/dryad.81fv1).
Acknowledgments
This study was supported by a Direct Grant (4053034) of The Chinese University of Hong Kong (JHLH), and Collaborative Research Fund of the Research Grants Council, HKSAR Government (C4042-14G) (TFC, HSK, KHC, JHLH). PWHH was supported by the ERC. We thank Stephen Smith of Virginia Tech for help supplying L. polyphemus samples.
Author contributions
JHLH conceived and designed this work. NJK assembled genomes, mined genomic data and performed phylogenetic analysis. KWC acquired, kept and extracted gDNA and RNA from animals, performed PCR, cloning and plasmid sequencing. WN assembled transcriptomes and performed bioinformatics analyses. KWC, HYY performed RT-PCR assay. JHLH and ZQ aided animal husbandry and experimental work. NJK, KWC, WN, ZQ, IM, HYY, TFC, HSK PWHH, KHC and JHLH analysed the data presented here, and wrote and approved the final manuscript.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Heredity website (http://www.nature.com/hdy)
Supplementary Material
References
- Adams KL, Wendel JF. (2005). Polyploidy and genome evolution in plants. Curr Opin Plant Biol 8: 135–141. [DOI] [PubMed] [Google Scholar]
- Albertin W, Marullo P. (2012). Polyploidy in fungi: evolution after whole-genome duplication. Proc Biol Sci 279: 2497–2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. (1990). Basic local alignment search tool. J Mol Biol 215: 403–410. [DOI] [PubMed] [Google Scholar]
- Amores A, Force A, Yan YL, Joly L, Amemiya C, Fritz A et al. (1998). Zebrafish hox clusters and vertebrate genome evolution. Science 282: 1711–1714. [DOI] [PubMed] [Google Scholar]
- Anderson LI, Selden PA. (1997). Opisthosomal fusion and phylogeny of Palaeozoic Xiphosura. Lethaia 30: 19–31. [Google Scholar]
- Blomme T, Vandepoele K, De Bodt S, Simillion C, Maere S, Van De Peer Y. (2006). The gain and loss of genes during 600 million years of vertebrate evolution. Genome Biol 7: R43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briggs DE, Siveter DJ, Siveter DJ, Sutton MD, Garwood RJ, Legg D. (2012). Silurian horseshoe crab illuminates the evolution of arthropod limbs. Proc Natl Acad Sci USA 109: 15702–15705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briggs DE, Moore RA, Shultz JW, Schweigert G. (2005). Mineralization of soft-part anatomy and invading microbes in the horseshoe crab Mesolimulus from the Upper Jurassic Lagerstatte of Nusplingen, Germany. Proc Biol Sci 272: 627–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bürglin TR. (2005) Homeodomain proteins. In: Myers RA (ed) Encyclopedia of molecular cell biology and molecular medicine. John Wiley & Sons: New Jersey, NJ, USA, pp, 179–222. [Google Scholar]
- Bürglin TR. (2011) Homeodomain subtypes and functional diversity. In: Hughes TR (ed) A handbook of transcription factors. Springer: The Netherland, pp, 95–122. [DOI] [PubMed] [Google Scholar]
- Cao Z, Yu Y, Wu Y, Hao P, Di Z, He Y, Chen Z et al. (2013). The genome of Mesobuthus martensii reveals a unique adaptation model of arthropods. Nat Commun 4: 2602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro LFC, Holland PWH. (2003). Chromosomal mapping of ANTP class homeobox genes in amphioxus: piecing together ancestral genomes. Evol Dev 5: 459–465. [DOI] [PubMed] [Google Scholar]
- Castro LFC, Rasmussen SL, Holland PW, Holland ND, Holland LZ. (2006). A Gbx homeobox gene in amphioxus: insights into ancestry of the ANTP class and evolution of the midbrain/hindbrain boundary. Dev Biol 295: 40–51. [DOI] [PubMed] [Google Scholar]
- Chapman BA, Bowers JE, Feltus FA, Paterson AH. (2006). Buffering of crucial functions by paleologous duplicated genes may contribute cyclicality to angiosperm genome duplication. Proc Natl Acad Sci USA 103: 2730–2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehal P, Boore JL. (2005). Two rounds of whole genome duplication in the ancestral vertebrate. PLoS Biol 3: e314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Robertis EM. (1994) The homeobox in cell differentiation and evolution. In: Duboule D (Ed). Guidebook to the homeobox genes. Oxford University Press: New York, NY, USA, pp, 13–23. [Google Scholar]
- Donoghue PCJ, Purnell MA. (2005). Genome duplication, extinction, and vertebrate evolution. Trends Ecol Evol 20: 312–319. [DOI] [PubMed] [Google Scholar]
- Dunn CW, Hejnol A, Matus DQ, Pang K, Browne WE, Smith SA et al. (2008). Broad phylogenomic sampling improves resolution of the animal tree of life. Nature 452: 745–749. [DOI] [PubMed] [Google Scholar]
- Falciani F, Hausdorf B, Schröder R, Akam M, Tautz D, Denell R, Brown S. (1996). Class 3 Hox genes in insects and the origin of zen. Proc Natl Acad Sci USA 93: 8479–8484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrier DEK. (2008) When is a Hox gene not a Hox gene? The importance of gene nomenclature. In: Minelli A, Fusco G (eds). Evolving Pathways: key themes in Evolutionary Developmental Biology,. Cambridge University Press: Cambridge, UK, 175–193. [Google Scholar]
- Flot JF, Hespeels B, Li X, Noel B, Arkhipova I, Danchin EG, Hejnol A et al. (2013). Genomic evidence for ameiotic evolution in the bdelloid rotifer Adineta vaga. Nature 500: 453–457. [DOI] [PubMed] [Google Scholar]
- Furlong RF, Holland PW. (2002). Were vertebrates octoploid? Philos Trans R Soc Lond B Biol Sci 357: 531–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galliot B, de Vargas C, Miller D. (1999). Evolution of homeobox genes: Q50 paired-like genes founded the paired class. Dev Genes Evol 209: 186–197. [DOI] [PubMed] [Google Scholar]
- Gehring WJ. (1984). Homeotic genes, the homeobox, and the spatial organization of the embryo. Harvey Lect 81: 153–172. [PubMed] [Google Scholar]
- Gehring WJ. (1994) A history of the homeobox. In: Duboule D (ed). Guidebook to the homeobox genes. Oxford University Press: New York, NY, USA, pp, 3–10. [Google Scholar]
- Garcia-Fernàndez J. (2005). The genesis and evolution of homeobox gene clusters. Nat Rev Genet 6: 881–892. [DOI] [PubMed] [Google Scholar]
- Goldberg RB, Crain WR, Ruderman JV, Moore GP, Barnett TR, Higgins RC et al. (1975). DNA sequence organization in the genomes of five marine invertebrates. Chromosoma 51: 225–251. [DOI] [PubMed] [Google Scholar]
- Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I et al. (2011). Full-length transcriptome assembly from RNA-seq data without a reference genome. Nat Biotechnol 29: 644–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grbić M, Van Leeuwen T, Clark RM, Rombauts S, Rouzé P, Grbić V et al. (2011). The genome of Tetranychus urticae reveals herbivorous pest adaptations. Nature 479: 487–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S, Delsuc F, Dufayard JF, Gascuel O. (2009) Estimating maximum likelihood phylogenies with PhyML. In: Posada D (ed). Bioinformatics for DNA Sequence Analysis. Humana Press: New York, NY, USA, pp, 113–137. [DOI] [PubMed] [Google Scholar]
- Hoegg S, Meyer A. (2005). Hox clusters as models for vertebrate genome evolution. Trends Genet 21: 421–424. [DOI] [PubMed] [Google Scholar]
- Holland PW. (1999). Gene duplication: past, present and future. Semin Cell Dev Bio 10: 541–547. [DOI] [PubMed] [Google Scholar]
- Holland PWH, Hogan BL. (1988). Expression of homeobox genes during mouse development: a review. Genes Dev 2: 773–782. [DOI] [PubMed] [Google Scholar]
- Holland PWH. (2003). More genes in vertebrates? J Struct Funct Genomics 3: 75–84. [PubMed] [Google Scholar]
- Holland PWH, Takahashi T. (2005). The evolution of homeobox genes: Implications for the study of brain development. Brain Res Bull 66: 484–490. [DOI] [PubMed] [Google Scholar]
- Holland PWH, Booth HA, Bruford EA. (2007). Classification and nomenclature of all human homeobox genes. BMC Biol 5: 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland PWH. (2012). Advanced review: evolution of homeobox genes. Wiley Interdiscip Rev Dev Biol 2: 31–45. [DOI] [PubMed] [Google Scholar]
- Hoshiyama D, Iwabe N, Miyata T. (2007). Evolution of the gene families forming the Pax/Six regulatory network: Isolation of genes from primitive animals and molecular phylogenetic analyses. FEBS Lett 581: 1639–1643. [DOI] [PubMed] [Google Scholar]
- Hui JHL, Holland PWH, Ferrier DEK. (2008). Do cnidarians have a ParaHox cluster? Analysis of synteny around a Nematostella homeobox gene cluster. Evol Dev 10: 725–730. [DOI] [PubMed] [Google Scholar]
- Hui JHL, Raible F, Korchagina N, Dray N, Samain S, Magdelenat G et al. (2009). Features of the ancestral bilaterian inferred from Platynereis dumerilii ParaHox genes. BMC Biol 7: 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hui JHL, McDougall C, Monteiro AS, Holland PWH, Arendt D, Balavoine G, Ferrier DEK. (2012). Extensive chordate and annelid macrosynteny reveals ancestral homeobox gene organization. Mol Biol Evol 29: 157–165. [DOI] [PubMed] [Google Scholar]
- Jaillon O, Aury J-M, Wincker P. (2009). ‘Changing by doubling', the impact of whole genome duplications in the evolution of eukaryotes. C R Biol 332: 241–253. [DOI] [PubMed] [Google Scholar]
- Katoh K, Misawa K, Kuma KI, Miyata T. (2002). MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res 30: 3059–3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Nei M, Dudley J, Tamura K. (2008). MEGA: a biologist-centric software for evolutionary analysis of DNA and protein sequences. Brief Bioinform 9: 299–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. (2012). Fast gapped-read alignment with Bowtie 2. Nat Methods 9: 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larroux C, Luke GN, Koopman P, Rokhsar DS, Shimeld SM, Degnan BM. (2008). Genesis and expansion of metazoan transcription factor gene classes. Mol Biol Evol 25: 980–996. [DOI] [PubMed] [Google Scholar]
- Luke GN, Castro LFC, McLay K, Bird C, Coulson A, Holland PW. (2003). Dispersal of NK homeobox gene clusters in amphioxus and humans. Proc Natl Acad Sci USA 100: 5292–5295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maere S, De Bodt S, Raes J, Casneuf T, Van Montagu M, Kuiper M, Van De Peer Y. (2005). Modeling gene and genome duplications in eukaryotes. Proc Natl Acad Sci USA 102: 5454–5459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazza ME, Pang K, Reitzel AM, Martindale MQ, Finnerty JR. (2010). A conserved cluster of three PRD-class homeobox genes (homeobrainrx and orthopedia in the Cnidaria and Protostomia. EvoDevo 1: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLean JA, Chen MA, Wayne CM, Bruce SR, Rao M, Meistrich ML et al. (2005). Rhox: a new homeobox gene cluster. Cell 120: 369–382. [DOI] [PubMed] [Google Scholar]
- Nossa C, Havlak P, Yue JX, Lv J, Vincent K, Brockmann HJ, Putnam NH. (2014). Joint assembly and genetic mapping of the Atlantic horseshoe crab genome reveals ancient whole genome duplication. Gigascience 3: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obst M, Faurby S, Bussarawit S, Funch P. (2012). Molecular phylogeny of extant horseshoe crabs (Xiphosura, Limulidae) indicates Paleogene diversification of Asian species. Mol Phylogenet Evol 62: 21–26. [DOI] [PubMed] [Google Scholar]
- Ohno S. (1970) Evolution by gene duplication. Springer-Verlag: Berlin, Germany.. [Google Scholar]
- Parra G, Bradnam K, Korf I. (2007). CEGMA: a pipeline to accurately annotate core genes in eukaryotic genomes. Bioinformatics 23: 1061–1067. [DOI] [PubMed] [Google Scholar]
- Petrov DA, Lozovskaya ER, Hartl DL. (1996). High intrinsic rate of DNA loss in Drosophila. Nature 384: 346–349. [DOI] [PubMed] [Google Scholar]
- Petrov DA, Chao YC, Stephenson EC, Hartl DL. (1998). Pseudogene evolution in Drosophila suggests a high rate of DNA loss. Mol Biol Evol 15: 1562–1567. [DOI] [PubMed] [Google Scholar]
- Podlaha O, Zhang J. (2010) Pseudogenes and their evolution. In: Encyclopedia of Life Sciences (eLS). John Wiley & Sons, Ltd: Chichester, England. [Google Scholar]
- Pollard SL, Holland PWH. (2000). Evidence for 14 homeobox gene clusters in human genome ancestry. Curr Biol 10: 1059–1062. [DOI] [PubMed] [Google Scholar]
- Regier JC, Shultz JW, Zwick A, Hussey A, Ball B, Wetzer R et al. (2010). Arthropod relationships revealed by phylogenomic analysis of nuclear protein-coding sequences. Nature 463: 1079–1083. [DOI] [PubMed] [Google Scholar]
- Qu Z, Kenny NJ, Lam HM, Chan TF, Chu KH, Bendena WG, Tobe SS, Hui JH. (2015). How did arthropod sesquiterpenoids and ecdysteroids arise? comparison of hormonal pathway genes in noninsect arthropod genomes. Genome Biol Evol 7: 1951–1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice P, Longden I, Bleasby A. (2000). EMBOSS: the European molecular biology open software suite. Trends Genet 16: 276–277. [DOI] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP. (2003). MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19: 1572–1574. [DOI] [PubMed] [Google Scholar]
- Rota-Stabelli O, Daley AC, Pisani D. (2013). Molecular timetrees reveal a Cambrian colonization of land and a new scenario for ecdysozoan evolution. Curr Biol 23: 392–398. [DOI] [PubMed] [Google Scholar]
- Ryan JF, Burton PM, Mazza ME, Kwong GK, Mullikin JC, Finnerty JR. (2006). The cnidarian-bilaterian ancestor possessed at least 56 homeoboxes: evidence from the starlet sea anemone. Nematostella vectensis Genome Biol 7: R64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanggaard KW, Bechsgaard JS, Fang X, Duan J, Dyrlund TF, Gupta V et al. (2014). Spider genomes provide insight into composition and evolution of venom and silk. Nat Commun 5: 3765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwager EE, Schoppmeier M, Pechmann M, Damen WG. (2007). Duplicated Hox genes in the spider Cupiennius salei. Front Zool 4: 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sémon M, Wolfe KH. (2007). Consequences of genome duplication. Curr Opin Genet Dev 17: 505–512. [DOI] [PubMed] [Google Scholar]
- Shakhnovich BE, Koonin EV. (2006). Origins and impact of constraints in evolution of gene families. Genome Res 16: 1529–1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma PP, Schwager EE, Extavour CG, Wheeler WC. (2014. a). Hox gene duplications correlate with posterior heteronomy in scorpions. Proc Biol Sci 281: 20140661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma PP, Kaluziak ST, Pérez-Porro AR, González VL, Hormiga G, Wheeler WC, Giribet G. (2014. b). Phylogenomic interrogation of Arachnida reveals systemic conflicts in phylogenetic signal. Mol Biol Evol 31: 2963–2984. [DOI] [PubMed] [Google Scholar]
- Smith JJ, Kuraku S, Holt C, Sauka-Spengler T, Jiang N, Campbell MS et al. (2013). Sequencing of the sea lamprey (Petromyzon marinus genome provides insights into vertebrate evolution. Nat Genet 45: 415–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stauber M, Jäckle H, Schmidt-Ott U. (1999). The anterior determinant bicoid of Drosophila is a derived Hox class 3 gene. Proc Natl Acad Sci USA 96: 3786–3789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Störmer L. (1952). Phylogeny and taxonomy of fossil horseshoe crabs. J Paleontol 26: 630–639. [Google Scholar]
- Störmer L. (1955) Merostomata. In: Kaesler RL (ed). Treatise of Invertebrate Paleontology – Part P: Arthropoda 2. University of Kansas and Geological Society of America: Lawrence, Kansas, USA, pp 4–41. [Google Scholar]
- Telford MJ, Thomas RH. (1998). Expression of homeobox genes shows chelicerate arthropods retain their deutocerebral segment. Proc Natl Acad Sci USA 95: 10671–10675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai IJ, Zarowiecki M, Holroyd N, Garciarrubio A, Sanchez-Flores A, Brooks KL et al. (2013). The genomes of four tapeworm species reveal adaptations to parasitism. Nature 496: 57–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van de Peer Y, Maere S, Meyer A. (2009). The evolutionary significance of ancient genome duplications. Nat Rev Genet 10: 725–732. [DOI] [PubMed] [Google Scholar]
- Waterston RH, Lindblad-Toh K, Birney E et al. (2002). Initial sequencing and comparative analysis of the mouse genome. Nature 420: 520–562. [DOI] [PubMed] [Google Scholar]
- Wheeler WC, Hayashi CY. (1998). The phylogeny of the extant chelicerate orders. Cladistics 14: 173–192. [DOI] [PubMed] [Google Scholar]
- Zerbino DR, Birney E. (2008). Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res 18: 821–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong YF, Butts T, Holland PWH. (2008). HomeoDB: a database of homeobox gene diversity. Evol Dev 10: 516–518. [DOI] [PubMed] [Google Scholar]
- Zhong YF, Holland PWH. (2011. a). The dynamics of vertebrate homeobox gene evolution: gain and loss of genes in mouse and human lineages. BMC Evol Biol 11: 169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong YF, Holland PWH. (2011. b). HomeoDB2: functional expansion of a comparative homeobox gene database for evolutionary developmental biology. Evol Dev 13: 567–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.