Abstract
Hox genes play a pivotal role in the determination of anteroposterior axis specificity during bilaterian animal development. They do so by acting as a master control and regulating the expression of genes important for development. Recently, however, we showed that Hox genes can also function in terminally differentiated tissue of the lepidopteran Bombyx mori. In this species, Antennapedia (Antp) regulates expression of sericin-1, a major silk protein gene, in the silk gland. Here, we investigated whether Antp can regulate expression of multiple genes in this tissue. By means of proteomic, RT-PCR, and in situ hybridization analyses, we demonstrate that misexpression of Antp in the posterior silk gland induced ectopic expression of major silk protein genes such as sericin-3, fhxh4, and fhxh5. These genes are normally expressed specifically in the middle silk gland as is Antp. Therefore, the evidence strongly suggests that Antp activates these silk protein genes in the middle silk gland. The putative sericin-1 activator complex (middle silk gland-intermolt-specific complex) can bind to the upstream regions of these genes, suggesting that Antp directly activates their expression. We also found that the pattern of gene expression was well conserved between B. mori and the wild species Bombyx mandarina, indicating that the gene regulation mechanism identified here is an evolutionarily conserved mechanism and not an artifact of the domestication of B. mori. We suggest that Hox genes have a role as a master control in terminally differentiated tissues, possibly acting as a primary regulator for a range of physiological processes.
Keywords: Bombyx mori, development, gene regulation, proteomics, silkworm, transcription regulation, transgenic, Antennapedia, Hox, silk gland
Introduction
Hox genes have a critical role in specifying identity along the anteroposterior axis during bilaterian animal development. In the fruit fly Drosophila melanogaster, Hox gene mutations can cause transformation of one segment into another and occasionally produce very striking phenotypes such as antenna-to-leg or haltere-to-wing transformation (1, 2). Each Hox gene encodes a protein with a homeodomain and acts as a transcription factor. Hox genes regulate a number of downstream genes as a master control gene during segment determination in arthropods or tissue development in vertebrates (3). This process is well illustrated by haltere and wing development in Drosophila. The fruit fly hind wing (haltere) is smaller and has a different shape from the forewing. These morphological differences are controlled by the Hox gene Ultrabithorax (Ubx),3 a member of the Bithorax Complex group of genes that determine the identity of the metathoracic segment (2, 4). During haltere development, Ubx represses expression of wing-patterning genes at multiple points in development and does not simply act as an upstream activator of the haltere developmental cascade (5). Genes under the control of Ubx include transcriptional factors such as vestigial, spalt-related, and achaete-scute, and signaling molecules such as wingless, decapentaplegic, and egfr/ras, which all play key roles in wing development (5–10). Many Ubx downstream genes have recently been identified using technologies such as microarrays and chromatin immunoprecipitation; these studies revealed that hundreds of genes are potentially targets of Ubx (11–13). A number of Hox target genes have also been identified in Drosophila and vertebrates (14–16).
Although there has been extensive study of Hox functions during development, comparatively little is known about its functions in other biological processes. In mice, the homeobox transcription factor genes Engrailed-1/2 and Otx2 are known to be involved in the patterning and compartmentalization of the developing nervous system as well as in physiological regulation in adults (17). Recently, we identified a unique Hox gene function in the silkworm Bombyx mori, a lepidopteran species. Lepidopteran larvae produce silk to form cocoons and/or support larval molt. Silk genes are expressed at a very high level in the larval silk gland, and a number of studies have explored the transcriptional regulation mechanisms of these genes, mainly using biochemical approaches. The studies using B. mori revealed that the expression level and the spatial expression pattern of silk genes are determined by transcription factors such as forkhead, Pit-1/Oct/Unc-86 (POU)-M1, and Arrowhead (18–22). We recently found that a Hox gene, Antennapedia (Antp), plays a key role in the expression of sericin-1 (ser1), one of the major silk protein genes (23). Our analysis demonstrated that Antp protein is a component of a putative ser1 activator called middle silk gland (MSG)-intermolt-specific complex (MIC); moreover, induced misexpression of Antp results in the induction of ser1 expression in the posterior silk gland (PSG) where there is no expression in normal individuals (23). The silk gland is a terminally differentiated tissue, and we therefore speculated that Antp can regulate physiological as well as developmental processes.
The main question raised by our previous observations was whether Hox genes could play a fundamental role in physiological regulation. To answer this question here, we sought to identify novel Antp-regulated genes in the Bombyx silk gland. Using proteomic, RT-PCR, and in situ hybridization analyses, we found that Antp could induce expression of multiple major silk protein genes such as ser3, fhxh4, and fhxh5 in the PSG. These genes are normally expressed only in the MSG. Moreover, MIC binds to the upstream regions of these genes, suggesting that Antp directly regulates their expressions. We also found that this pattern of gene expression is well conserved between Bombyx mori and the wild species Bombyx mandarina, indicating that this transcriptional regulation mechanism is an evolutionarily conserved process and not an artifact of silkworm domestication. Our results here support the speculation that Hox genes have a role as a key regulator in physiological regulation as well as in developmental processes. This finding provides further understanding of the functional evolution of Hox genes.
Experimental Procedures
Silkworm Strains
The silkworms were reared on an artificial diet (Nihon Nosan Kogyo, Yokohama, Japan) at 25 °C under a photoperiod of 12-h light:12-h dark for daizo and transgenic strains and 16-h light:8-h dark for the Kinshu × Showa strain. daizo was used as the wild type strain for gene expression analysis, and Kinshu × Showa was utilized for protein extraction for the electrophoretic mobility shift assay (EMSA). The Ayfib-431a (Ayfib-GAL4) transgenic strain was used for the PSG-specific GAL4 driver. This strain harbors the Antheraea yamamai fibroin promoter-GAL4 cassette and can induce PSG-specific gene expression in first instar larva or earlier stages (24, 25). The hs-GAL4 strain (modified from Bmhsp70-GAL4 (26)) was used for heat shock-induced gene expression in this or previous studies (21–23). The heat shock was a treatment of 42 °C for 2 h on the 1st day of fifth instar larvae (L5D1) following the method described previously (23). The UAS-Antp strain is described elsewhere (23). For misexpression analysis, UAS strains were crossed with GAL4 strains, and the genotype of the progeny was determined by screening for the transgenic marker (DsRed for GAL4 and AmCyan for UAS-Antp). The wild silkmoth B. mandarina was sampled from the field in the Shimonita and Maebashi areas of Japan, and their hybrid has been maintained in our laboratory for 19 generations.
EMSA
EMSA was carried out as described previously (23, 27). The protein extract was prepared from the posterior portion of the MSG of Kinshu × Showa larvae at L5D2. The sequences of the oligonucleotides and competitors are shown in Table 1. The protein-probe complexes were separated on 7% polyacrylamide gels. A 100-fold molar excess of unlabeled oligonucleotides was added to the reaction for the competition experiments. The antibodies used for the supershift assay are as described previously (23).
TABLE 1.
Oligonucleotide sequences of probes and competitors used for the EMSA
Lower case letters show extra sequences for labeling.
| Name | Position | Sequence |
|---|---|---|
| ser3 −90 | −114/−69 | aattCAAGTGTATTAAACAAATAATTAATTATTTATTTTATTGGTAACTG |
| Sr-70 | −81/−52 | GAAGCGAAAATTTATTACTCTCTACGTAAG |
| Sr-70M2 | −81/−52 | GAAGCGAAAATTTGGTACTCTCTACGTAAG |
| fhxh4 −1660 | −1682/−1637 | aattGAAAAACGTAAATAAATTGATTTATGACAGAGTATATTTTTAACCG |
| fhxh5 −300 | −324/−279 | aattTTAATCTCTTGTCATCAATATTATAAATCTCATTTTTTGGCCCTTA |
Sample Preparation and Two-dimensional Electrophoresis
Larvae of the Ayfib-GAL4/+ and Ayfib-GAL4/UAS-Antp strains were dissected at L5D3 to isolate PSGs. Sample preparation and electrophoresis were conducted as described previously (28). Briefly, the PSG was dissociated in lysis buffer (10 mg of tissue/300 μl of buffer) containing 7 m urea, 2 m thiourea, 4% CHAPS, 2% Nonidet P-40, 5% 2-mercaptoethanol, 0.2% Ampholine (pH 3.5–10), and EDTA-free protease inhibitor. The dissociated tissue was subjected to repeated freeze/thaw cycles in liquid nitrogen and then stored at −80 °C until used for electrophoresis. The two-dimensional PAGE gels were stained with Coomassie Brilliant Blue.
Mass Spectrometry and Data Analysis
Mass spectrometry and data analysis were performed largely as described previously (28). Spots were excised from five or six gels, and amaZon SL (Bruker Daltonics, Bremen, Germany) was used for the MS analysis. The MS data were analyzed with Mascot software (29) using the amino acid sequence data in the National Center for Biotechnology Information data bank (www.ncbi.nlm.nih.gov/) and silkworm genome database (30).
RNA Extraction, cDNA Synthesis, and RT-PCR
RNA was extracted from each part of the silk gland of B. mori daizo strain (L5D3) and B. mandarina (just before spinning stage); additionally, RNA was obtained from the PSGs of B. mori Ayfib-GAL4/+ (L5D3), Ayfib-GAL4/UAS-Antp (L5D3), hsp70-GAL4/+ (L5D2), and hsp70-GAL4/UAS-Antp (L5D2) strains. The Illustra RNAspin MINI RNA Isolation kit (GE Healthcare) was used for RNA extraction from hsp70-GAL4/+ and hsp70-GAL4/UAS-Antp individuals and was used for cDNA synthesis using a PrimeScript RT-PCR kit (Takara, Otsu, Japan) (23). For other samples, ISOGEN (Nippongene, Tokyo, Japan) and the SV Total RNA Isolation System (Promega, Madison, WI) were used for RNA extraction, and Superscript III (Life Technologies) was used for cDNA synthesis (25). KOD-FX polymerase (Toyobo, Osaka, Japan) was used for RT-PCR. Primer sequences are listed in Table 2.
TABLE 2.
Primers used for gene expression analysis
| Gene name | Forward | Reverse |
|---|---|---|
| rp49 | 5′-CAGGCGGTTCAAGGGTCAATAC-3′ | 5′-TGCTGGGCTCTTTCCACGA-3′ |
| ser1 | 5′-CAAAGACCGCCAACATGCGT-3′ | 5′-CAGCGTTCCAATTGGCCTGA-3′ |
| ser2 | 5′-CACTTTTCGGGGGCTTAGTT-3′ | 5′-TTCGTATTCGGAACCTTTGC-3′ |
| ser3 | 5′-AGTTGCTCTATTCCTGATAG-3′ | 5′-TGTCGTCGGAATTCTCACCA-3′ |
| Antp | 5′-ATGGATGGCTGCGATCAGCAG-3′ | 5′-TAAGGTTTACTGTGGCGAGGT-3′ |
| fhxh4 | 5′-ACTGTGGCGGATGAGGTAGA-3′ | 5′-TCTGGGATTAGAGTCTTCGA-3′ |
| fhxh5 | 5′-ACGAGGCGTGTCTTGCCCTT-3′ | 5′-GGTCCGTAATCACACGTGGC-3′ |
| BGIBMGA010891 | 5′-GCATTTCCATTATTGCGTTG-3′ | 5′-ATGCAAAAACCTTGATGTCT-3′ |
| BGIBMGA011308 | 5′-TCACAACACCTTAGTCAACT-3′ | 5′-CCTTCGTTCAAGAAACCAGC-3′ |
| BGIBMGA007061 | 5′-CACTCTCCGATGAATTCATC-3′ | 5′-TCCGTTAGAATAGGTACATA-3′ |
| h-fib | 5′-TGCGCTCTGCAGTATGTCGC-3′ | 5′-CACTGTTTGATACGTATGGC-3′ |
| l-fib | 5′-TCGCCATCCTCAACGTTCAA-3′ | 5′-GACGATGCAGTACTCTTCAT-3′ |
| fhx | 5′-GATACCGGTGTTCCAGTTTG-3′ | 5′-GTGCAGCAGGGTCAGATCTT-3′ |
Probe Synthesis for in Situ Hybridization
For ser1 probe synthesis, a plasmid in which a partial fragment (106–775 bp; J01040) was inserted into the pSPT18 vector (Roche Diagnostics) was PCR-amplified using T7 and SP6 primers. The PCR product was utilized as the template for probe labeling. For fhxh4 and fhxh5, L5D3 MSG-P cDNA from the daizo strain was amplified using primers 5′-ACTGTGGCGGATGAGGTAGA-3′ and 5′-GTAATACGACTCACTATAGGGCTCTGGGATTAGAGTCTTCGA-3′ (fhxh4) or 5′-ACGAGGCGTGTCTTGCCCTT-3′ and 5′-GTAATACGACTCACTATAGGGCGGTCCGTAATCACACGTGGC-3′ (fhxh5) and used for probe labeling. The labeling reaction was carried out with a digoxigenin RNA labeling kit (SP6/T7) following the manufacturer's protocol (Roche Diagnostics).
In Situ Hybridization Analysis
Silk glands were dissected from individuals of Ayfib-GAL4/+ and Ayfib-GAL4/UAS-Antp L5D3 and sequentially fixed with 100% methanol, 100% ethanol, and 4% paraformaldehyde. They were washed with PBS, treated with 0.2 n HCl for 20 min, and washed again with PBS. They were treated with 10 μg/ml proteinase K (Roche Diagnostics) at room temperature for 20 min and then washed with PBS. Prehybridization was carried out in hybridization buffer (5× SSC, 50% formamide, 50 μg/ml heparin, and 50 μg/ml salmon sperm DNA) at 50 °C for 1 h. The probe was then added at a final concentration of 100 ng/ml and incubated at 50 °C overnight. The silk glands were washed with 2× SSC and 0.1% Tween 20 at 50 °C for 20 min and then washed twice with 0.2× SSC and 0.1% Tween 20 at room temperature. For the antibody reaction, a blocking step was performed using 1% goat normal serum, the anti-digoxigenin antibody (Roche Diagnostics) was applied at a 1:4000 dilution, and the tissue was incubated at 37 °C for 1 h. The silk gland was washed with PBS, stained with nitro blue tetrazolium/5-bromo-4-chloro-3-indolyl phosphate (Roche Diagnostics), and counterstained with a graded series of ethanol. The image was captured using Nikon Ds-Vi1 (Nikon Corp., Tokyo, Japan) and processed using Photoshop CS5 (Adobe Systems Inc., San Jose, CA).
Results
Expression of sericin Genes following Antp Misexpression
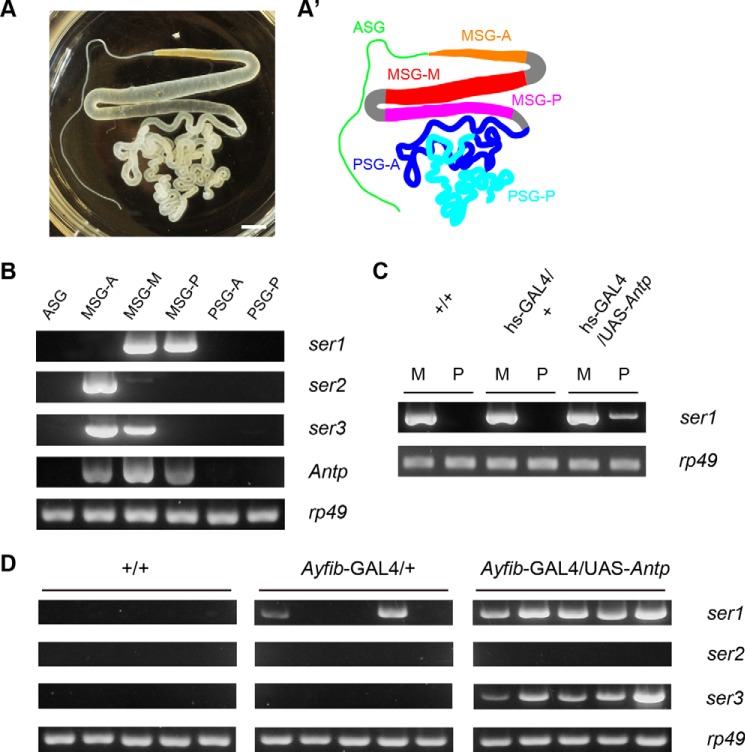
Antp is a Hox transcription factor gene that shows MSG-specific expression in the silk gland (23, 31). The MSG is separated into three territories termed MSG-A, MSG-M, and MSG-P (Fig. 1, A and A′), all of which express Antp (Fig. 1B) (23). Ectopic expression of Antp in the PSG using a heat shock promoter and the PSG-specific Antheraea yamamai fibroin promoter resulted in the induction of ser1 expression in the PSG (Fig. 1, C and D) (23). In the latter case, ectopic ser1 was observed in some GAL4/+ PSGs (Fig. 1D), which might result from the transcriptional perturbation; in the silkworm, GAL4 frequently exhibits a toxic effect (32–34). We conclude that ser1 is regulated by Antp because it was expressed in all Ayfib-GAL4/UAS-Antp PSGs and was also induced by hs-GAL4 (Fig. 1, C and D). ser1 is normally expressed in the MSG-M and MSG-P but not in MSG-A (Fig. 1B). The absence of ser1 expression in MSG-A may be associated with a lack of Antp protein expression or failure of MIC formation in this territory (23). Induction of ser1 expression by ectopic Antp suggested that it was acting as a ser1 activator in the MSG-M/MSG-P of normal individuals. In addition to ser1, B. mori has two sericin genes (ser2 and ser3) that are expressed specifically in MSG-A and/or MSG-M (Fig. 1B) (35, 36). As a first step to identify novel Antp target genes, we investigated whether ser2 and/or ser3 could be induced by Antp. RT-PCR analyses showed that ser3 but not ser2 was induced in the PSG of individuals misexpressing Antp (Fig. 1D). ser3 is expressed normally in the MSG-A and MSG-M (Fig. 1B); we suggest that Antp functions for the activation of ser3 expression in MSG-M.
FIGURE 1.
Induction of sericin genes by Antp. A photograph (A) and schematic drawing (A′) of the silk gland are shown. The MSG can be subdivided into three regions along the anteroposterior axis (MSG-A, MSG-M, and MSG-P). PSG can be also subdivided into two regions, although there is no apparent morphological difference (PSG-A and PSG-P). Scale bar, 0.3 cm. B–D, silk gland gene expression investigated by RT-PCR. B, expression of three sericin genes in daizo (wild type) strain. These genes are expressed specifically in MSG with different patterns of spatial expression. C, induction of ser1 by heat shock-induced Antp misexpression. M and P indicate MSG and PSG, respectively. D, expression of sericin genes in the PSGs of +/+, Ayfib-GAL4/+, and Ayfib-GAL4/UAS-Antp individuals. Five individuals were investigated for each genotype. ser1 and ser3 are induced strongly by Antp misexpression. ser1 expression is also apparent in two individuals of Ayfib-GAL4/+, possibly due to the transcriptional perturbation by GAL4. ASG, anterior silk gland.
Antp Binds to the Promoter of ser3
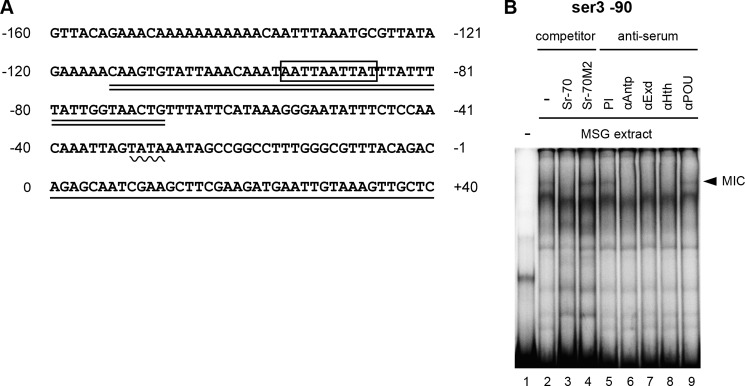
Antp protein can bind to the promoter of ser1 and directly activate its expression (23). We asked whether this was also the case for ser3. A search of the ser3 upstream region identified a putative MIC binding site ((G/A)ATT(T/A)ATNA(T/C)) (27) at the −90 position (Fig. 2A). We carried out an EMSA using an oligonucleotide that included this element (ser3 −90) and found that the MSG extract showed significant binding to this probe (Fig. 2B, lane 2). This binding is likely mediated by MIC because the ser1 −70 oligonucleotide (Sr-70) interfered with the binding, whereas the mutated probe (Sr-70M2) did not (Fig. 2B, lanes 3 and 4). The MIC is composed of Antp and its cofactors Extradenticle and Homothorax (23). The addition of antiserum against these proteins clearly inhibited binding, but the preimmune serum or antiserum against POU-M1 did not (Fig. 2B, lanes 5–9). This further suggests that the binding complex is certainly the MIC. We therefore conclude that Antp directly activates ser3 expression by binding to its promoter sequence.
FIGURE 2.
A, sequence of the putative promoter region for ser3. The boxed region indicates the hypothetical MIC binding site, the wavy line indicates the putative TATA box, the underlined sequence indicates the transcribed region, and the double underlined sequence indicates the oligonucleotide utilized for the EMSA analysis (ser3 −90). B, binding of MIC to the ser3 promoter. The band representing the presumptive MIC binding is indicated with the arrowhead. The addition of the non-labeled Sr-70 oligonucleotide (lane 3) abolished the binding, whereas Sr-70M2 did not (lane 4). Antiserum against Antp (lane 6), Extradenticle (Exd) (lane 7), and Homothorax (Hth) (lane 8) also interfered with binding, but the preimmune serum (lane 5) or antiserum against POU-M1 (lane 9) did not show an effect.
Identification of Novel Antp Target Genes
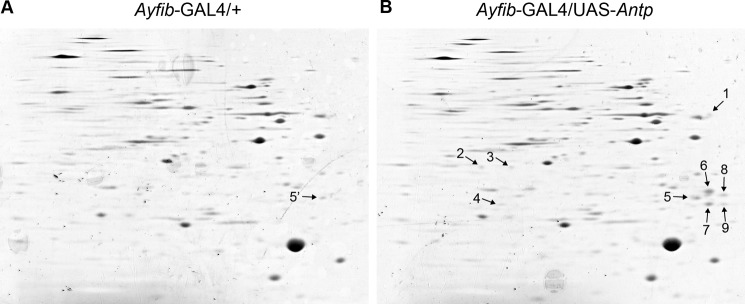
The fact that ser3 was induced via ectopic Antp expression raises the possibility that genes other than sericin might also be regulated by Antp. To identify such genes, we carried out a proteomic analysis. PSG proteins were extracted from normal and Antp-misexpressing larvae at the 3rd day of the fifth instar and used for two-dimensional electrophoresis. Comparison of the gel images showed that the overall spot patterns were similar between the two experimental groups (Fig. 3). Nevertheless, we did identify nine spots that were apparently present only in the Antp-misexpressing PSG or showed stronger staining compared with the normal PSG (Fig. 3B, arrows). These spots were isolated and analyzed by mass spectrometry. The analysis showed that spot 1 was a protease inhibitor (BGIBMGA010891), spot 3 was acyl-coenzyme A dehydrogenase (BGIBMGA011308) and cathepsin B (BGIBMGA007061), and spots 5–9 were fibrohexamerin-like proteins Fhxh4 and Fhxh5 (Table 3). Spots 5–9 showed relatively strong staining in the gel image (Fig. 3). Thus, Fhxh4 and Fhxh5 were apparently strongly induced by Antp misexpression. Spot 5 also appeared to be present in normal individuals (Fig. 3, designated as spot 5′), but we suspect that this spot was some other protein(s) as Fhxh4/Fhxh5 transcripts were not detected in normal PSGs (see the description later). Fhxh4/Fhxh5 proteins are abundant in the cocoon of normal individuals (37) and are predicted to be major silk proteins similar to sericins.
FIGURE 3.
Gel images of PSG proteins after two-dimensional electrophoresis. The gels were stained with Coomassie Brilliant Blue. A, Ayfib-GAL4/+. B, Ayfib-GAL4/UAS-Antp. Spots detected specifically in Ayfib-GAL4/UAS-Antp are marked with arrows and numbers. Spot 5′ in the Ayfib-GAL4/+ gel indicates a spot that appears to be located at the identical position to spot 5 of Ayfib-GAL4/UAS-Antp.
TABLE 3.
Profile of PSG proteins expressed specifically in Antp-misexpressing individuals
| Spot number | Gene numbera | Gene name | Accession number | Annotation | Mascot score |
|---|---|---|---|---|---|
| 1 | BGIBMGA010891 | AK383125 | Protease inhibitor I8 | 62 | |
| 2 | ND | ||||
| 3b | BGIBMGA011308 | AK383321 | Acyl-coenzyme A dehydrogenase | 119 | |
| BGIBMGA007061 | AK383399 | Cathepsin B | 99 | ||
| 4 | ND | ||||
| 5b | fhxh4 | XP_004922636 | Fibrohexamerin-like | 173 | |
| fhxh5 | XP_004922696 | Fibrohexamerin-like | 38 | ||
| 6 | fhxh4 | XP_004922636 | Fibrohexamerin-like | 83 | |
| 7 | fhxh4 | XP_004922636 | Fibrohexamerin-like | 73 | |
| 8 | fhxh4 | XP_004922636 | Fibrohexamerin-like | 83 | |
| 9 | fhxh4 | XP_004922636 | Fibrohexamerin-like | 80 |
a Gene number in the silkworm genome database is shown. ND, not determined.
b These spots showed partial homology against two distinct proteins.
Induction of the Identified Proteins Is Mediated by Transcriptional Activation
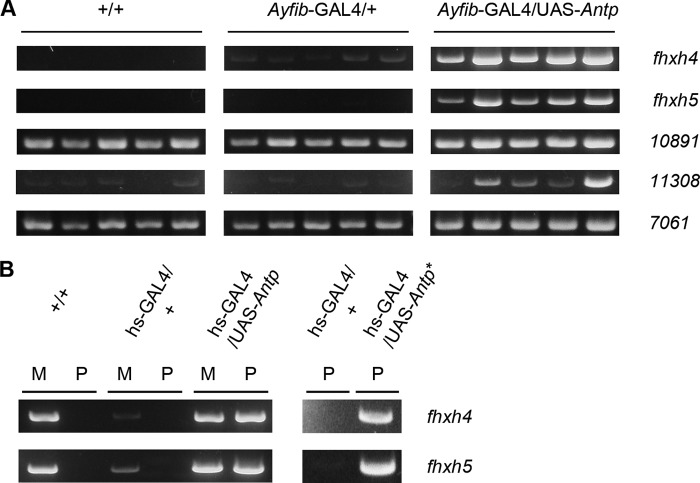
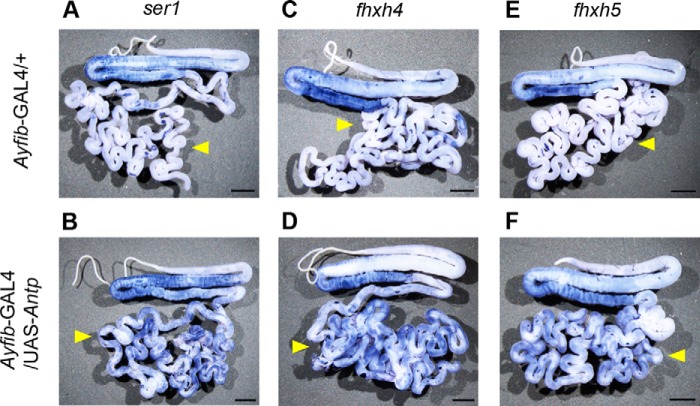
Antp encodes a transcription factor and is presumed to induce the identified proteins by transcriptional activation. An RT-PCR analysis confirmed this was the case for all the identified genes. mRNAs of fhxh4 and fhxh5 were induced strongly when Antp was misexpressed using the PSG promoter or the heat shock promoter (Fig. 4, A and B). Induction of the mRNAs was also observed for other genes when Antp was induced using the PSG promoter (Fig. 4A). We also examined fhxh4 and fhxh5 expression by in situ hybridization analysis and found that induction occurred in a widespread region of the PSG (Fig. 5).
FIGURE 4.
Expression analysis of genes identified by proteomic analysis. Expression was analyzed by RT-PCR. A, expression in the PSGs of +/+, Ayfib-GAL4/+, and Ayfib-GAL4/UAS-Antp individuals. 10891 indicates gene BGIBMGA010891, 11308 indicates BGIBMGA011308, and 7061 indicates BGIBMGA007061. B, induction of fhxh4 and fhxh5 by heat shock-induced Antp misexpression. M and P indicate MSG and PSG, respectively. Induction was also observed when Antp was misexpressed using an independent UAS-Antp strain (UAS-Antp*). Gene expression was analyzed in fifth instar larvae.
FIGURE 5.
In situ hybridization of ser1 (A and B), fhxh4 (C and D), and fhxh5 (E and F). A, C, and E are the silk glands of Ayfib-GAL4/+, and B, D, and F are those of Ayfib-GAL4/UAS-Antp individuals. Yellow arrowheads indicate the PSG. Scale bar, 0.3 cm.
In the silkworm, it is known that transgene expression is affected by position of insertion of the transposon (33, 38). The insertion can also cause disruption of endogenous genes (25) and this might possibly give rise to an artifactual effect that is irrespective of the transgene expression. It is therefore important to verify whether two or more independent transgenic strains can give identical results. We used another independent UAS-Antp strain (23) for this purpose and examined fhxh4 and fhxh5 expression in the PSG after Antp misexpression. A clear induction effect was also observed in this strain (Fig. 4B), indicating that induction was undoubtedly provided by Antp activity.
Gene Expression in the Normal Silk Gland
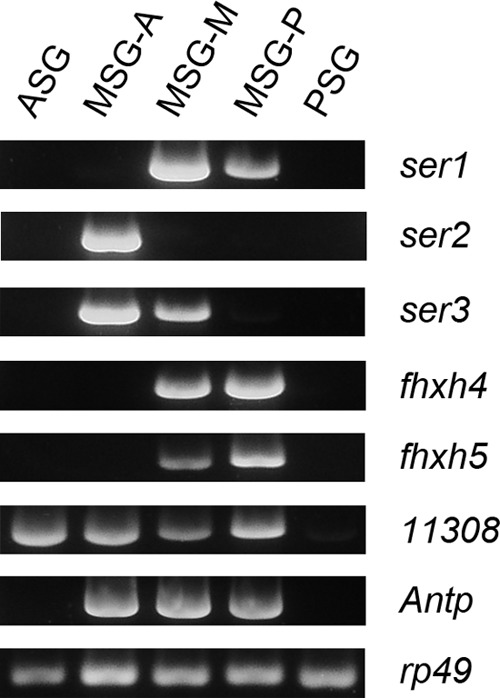
We next examined the expression of the genes described above in the normal silk gland. Antp is expressed specifically in the MSG (Fig. 1B) (23); it seems reasonable to presume that genes positively regulated by Antp also show this expression pattern. Our RT-PCR analysis showed that fhxh4 and fhxh5 are expressed strongly in the MSG-P and weakly in the MSG-M; they could not be detected in other regions of the silk gland (Fig. 6). This expression pattern correlates well with that of ser1 (Fig. 1B). For other Antp-induced genes, we found that they were expressed not only in the MSG but also in other silk gland regions (Fig. 6).
FIGURE 6.
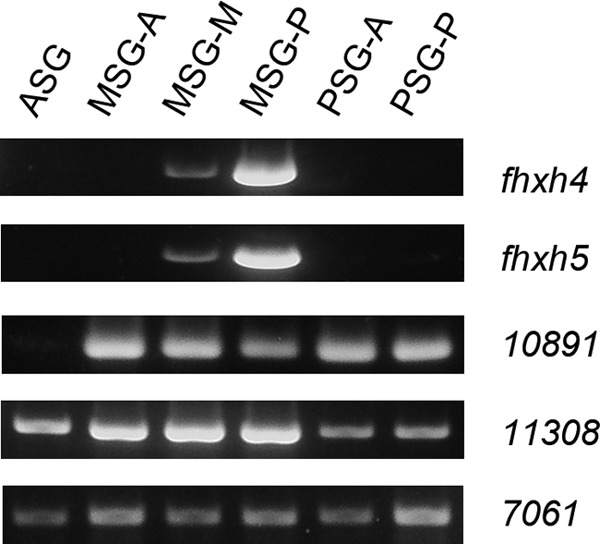
RT-PCR analysis of gene expression in the silk gland of the daizo strain. ASG, anterior silk gland.
Antp Binds to the Upstream Region of fhxh4 and fhxh5
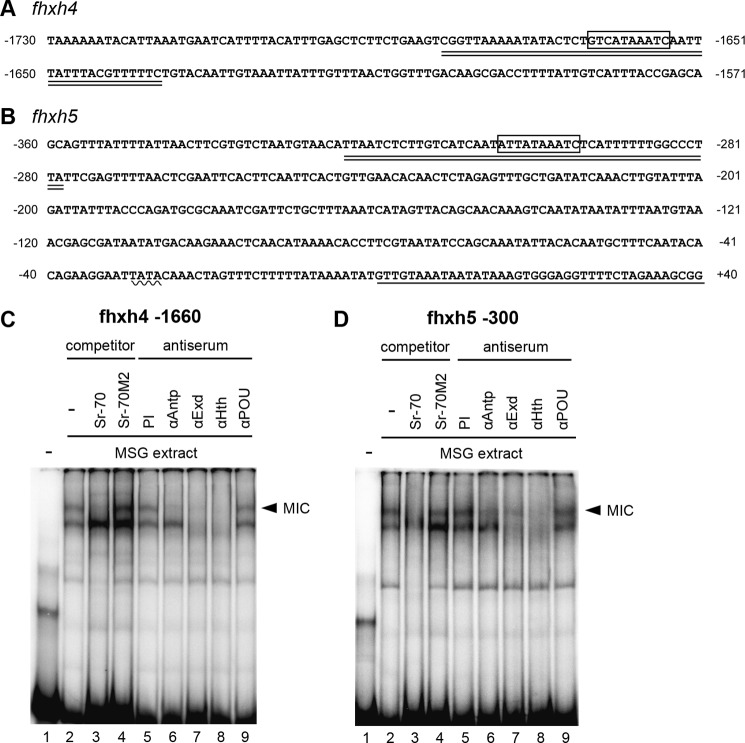
The expression of fhxh4 and fhxh5 shows good agreement with the production of Antp protein. This suggests that Antp might directly activate their expressions. A search of upstream genome sequences revealed that a putative MIC binding site was present at around −1660 for fhxh4 and −300 for fhxh5 (Fig. 7, A and B). We carried out an EMSA using an oligonucleotide probe including these elements and found that the MSG extract could bind to both of these sequences (Fig. 7, C and D). A competition analysis and a supershift assay further supported the interpretation that this binding was mediated by the MIC (Fig. 7, C and D). Therefore, we conclude that Antp directly regulates fhxh4 and fhxh5 expression by binding to upstream sequences of these genes.
FIGURE 7.
A and B, sequence of the upstream region of fhxh4 (A) and fhxh5 (B). The boxed region indicates the hypothetical MIC binding site, the wavy line indicates the putative TATA box, the underlined sequence indicates the transcribed region, and the double underlined sequence indicates the oligonucleotide utilized for the EMSA analysis (fhxh4 −1660 and fhxh5 −300). C and D, binding of MIC to the upstream region of fhxh4 (C) and fhxh5 (D). The band that represents the presumptive MIC binding is indicated with the arrowhead. The binding was abolished by the non-labeled Sr-70 probe but not by the Sr-70M2 probe (lanes 3 and 4). Interference with the binding was also apparent after addition of antisera against Antp, Extradenticle (Exd), and Homothorax (Hth) but not after addition of anti-POU-M1 (POU) serum or preimmune (PI) serum (lanes 5–9).
Effect of Antp Misexpression on PSG-specific Genes
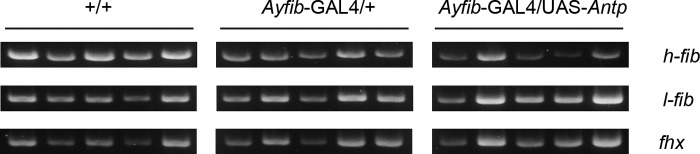
In addition to their role as a transcriptional activator, Hox genes can also act as a repressor of expression (39). This suggests that Antp may function in the repression of PSG-specific genes. Fibroin component genes such as h-fibroin (h-fib), l-fibroin (l-fib), and fibrohexamerin (fhx) are known to be expressed strongly and specifically in the PSG (22, 40, 41). We examined the expression of these genes following misexpression of Antp. Previously, we showed that the heat shock promoter-induced misexpression of Antp did not affect h-fib expression (23). Here, we overexpressed Antp using the fibroin promoter and obtained a similar result (Fig. 8). In addition, this overexpression did not affect the expression of l-fib and fhx genes (Fig. 8). In a proteomic analysis, we found that none of the proteins that are expressed in the PSG showed a reduced expression level in individuals misexpressing Antp (Fig. 3). We conclude from these results that Antp has little or no effect on the expression of PSG-specific genes.
FIGURE 8.
RT-PCR analysis of expression of h-fib, l-fib, and fhx in the PSG of Antp-misexpressing individuals.
Gene Expression in the Silk Gland of a Wild Bombyx Species
B. mori is a domesticated species that has been selected for cocoon yield for more than 5000 years. It is possible that this long term artificial selection might have influenced the patterns of gene expression in this species such that the results here might not necessarily reflect events in a wild population. To determine whether the gene regulation mechanism identified here is also present in the natural environment, we examined the pattern of gene expression in B. mandarina, a wild relative of B. mori. We conducted an RT-PCR analysis of the B. mandarina silk gland and found that the gene expression patterns were similar in the two species (Fig. 9). Importantly, Antp showed MSG-specific expression in both species (Fig. 9; compare with Fig. 1B). Thus, we conclude that the gene regulation pattern in B. mori is not an artifact of domestication but reflects the natural situation.
FIGURE 9.
RT-PCR analysis of gene expression in the silk gland of B. mandarina. 11308 indicates the BGIBMGA011308 gene. ASG, anterior silk gland.
Discussion
In this study, we confirmed that Hox genes regulate expression of multiple genes in the silk gland. Our analyses demonstrated that Antp induced the expression of a number of major silk protein genes such as ser3, fhxh4, and fhxh5 in addition to ser1 (Figs. 1D, 4, and 5). This finding is a significant contribution to our understanding of Hox functions because the roles of these regulators in terminally differentiated tissue had not been elucidated previously. The silk gland is a good model for studying additional Hox functions because Antp-mediated regulation of ser1 expression has already been studied in detail (23). Lepidopteran insects produce cocoons to protect the larvae and/or pupae from environmental dangers, and we speculate that the function of Antp in the silk gland indicates its role as a regulator of physiological processes. We found that individuals misexpressing Antp in the silk gland exhibited little or no morphological changes despite the expression of the gene from an early larval stage (see Fig. 5, B, D, and F) (25). This further suggests that Antp acts in regulatory pathways that are distinct from those required for development. Our finding that Antp controls expression of multiple genes in the silk gland indicates its role as a master control gene in this tissue. This result also implies that acquisition of Antp expression in the MSG could have been one of the critical events that enabled the silkworm to be able to produce a cocoon. We investigated Antp expression in another lepidopteran species and, interestingly, found that MSG-specific expression was also present in B. mandarina (Fig. 9). This finding suggests that Antp function might be conserved in the genus Bombyx. In contrast, Antp is expressed strongly in the anterior silk gland/MSG and the PSG of the eri silkworm (Samia ricini), a Saturniidae species (data not shown), indicating that it might have distinct biological functions in this family. One possibility is that Antp regulates sericin expression in the MSG and fibroin in the PSG in a similar manner as B. mori forkhead (19, 43).
Our misexpression analysis identified seven Antp-induced genes (ser1, ser3, fhxh4, fhxh5, BGIBMGA010891, BGIBMGA011308, and BGIBMGA007061; Figs. 1D and 4A). Among these genes, expression of ser1, fhxh4, and fhxh5 was specific to MSG-M and MSG-P in the normal silk gland (Figs. 1B and 6). We found that MIC could bind to the −1660 sequence of fhxh4 and −300 sequence of fhxh5 (Fig. 7, C and D), and we presume that Antp directly activates their expression by binding to these elements. The MIC binding consensus sequence was also present at −190 of fhxh4, but our analysis failed to detect binding to this region (data not shown). We therefore hypothesize that not all consensus sequences are recognized by the MIC. fhxh4/fhxh5 expression was confined to the middle and posterior portions of the MSG, and this expression shows a good correlation with the territories in which the MIC is formed (23). In addition to transcriptional activation by MIC, a POU-type transcription factor, POU-M1, might act to repress fhxh4/fhxh5 in the MSG-A and/or in the MSG-M; its role in ser1 repression has already been demonstrated (20). ser3 is normally expressed in the MSG-A/MSG-M (Fig. 1B), and our finding that MIC can bind to position −90 (Fig. 2A) indicates that Antp directly activated its transcription in the MSG-M. The MIC is not formed in the MSG-A (23), and we speculate that there is probably another activation mechanism in this region of the gland. ser3 transcription is absent in the MSG-P; this may be due to expression of a factor such as invected, a putative transcriptional repressor (22, 23). Other Antp-induced genes showed more widespread expression (Fig. 6), and it is unclear whether Antp is indeed involved in the transcriptional regulation of these genes. There are two possible scenarios. First, Antp activates their expression in MSG-M/MSG-P, whereas other factor(s) act in other locations. Second, Antp regulates these genes only in particular biological contexts such as embryogenesis. It should be feasible to discriminate between these possibilities using Antp knockdown and/or knock-out in the MSG.
The fhxh genes were recently identified and found to show weak homology to fhx (30). In total, eight fhxh genes have been identified in the silkworm, and all are aligned tandemly in the genome. Of these genes, only fhxh4 and fhxh5 are expressed in the silk gland and produce major silk proteins. At present, the biological roles of Fhxh4/5 are unknown. Fhx is involved in the appropriate organization and secretion of the fibroin complex (44), and the presence of a putative glycosylation site that is essential for Fhx function in Fhxh4/5 (data not shown) (45) indicates that they might act in a similar manner. In a two-dimensional gel analysis, the Fhxh4 protein was detected as a multiple cluster of spots (Fig. 3 and Table 3), supporting the interpretation that it undergoes post-translational modifications in the silk gland. Recently fhxh genes were also found in S. ricini, and interestingly some are expressed strongly in the anterior silk gland/MSG (42). These proteins retain the glycosylation sites (42), and it is possible that they have conserved biological functions.
Here we succeeded in identifying a number of novel Hox target genes in the silk gland using a proteomics approach. However, this method provides limited information due to limitation in sensitivity, and it is possible that Antp might regulate a larger numbers of genes. Currently, we are in the process of trying to identify further candidate genes using RNA sequencing analysis; this approach is expected to produce more information on the number of genes regulated by Hox genes in biological pathways unrelated to the developmental process.
Author Contributions
T. T. designed and carried out all of the experiments with the exceptions described below. K. U. generated transgenic silkworm strains. M. K. conducted experiments concerned with heat shock-induced Antp expression, RNA sampling, and cDNA synthesis. Shi. T. carried out the EMSA experiment. H. K. performed proteomic and mass spectrometry analysis. Shu. T. and T. Y. provided instructions. T. T. wrote the paper with support from all authors. H. S. supervised the work.
Acknowledgments
We are grateful to Kaoru Nakamura, Koji Hashimoto, and Toshihiko Misawa for maintaining the silkworms and Dr. Natuo Kômoto for providing B. mandarina. We also thank Satoko Kawamoto for technical assistances and Dr. Takao K. Suzuki for helpful discussion. This work was supported by the National Institute of Agrobiological Sciences technical support system.
This work was supported by grants-in-aid from the Ministry of Agriculture, Forestry, and Fisheries. The authors declare that they have no conflicts of interest with the contents of the article.
- Ubx
- Ultrabithorax
- POU
- Pit-1/Oct/Unc-86
- Antp
- Antennapedia
- ser
- sericin
- MIC
- MSG-intermolt-specific complex
- PSG
- posterior silk gland
- MSG
- middle silk gland
- h-fib
- h-fibroin
- l-fib
- l-fibroin
- fhx
- fibrohexamerin
- fhxh
- fibrohexamerin-like.
References
- 1. Lewis E. B. (1978) A gene complex controlling segmentation in Drosophila. Nature 276, 565–570 [DOI] [PubMed] [Google Scholar]
- 2. Kaufman T. C., Lewis R., and Wakimoto B. (1980) Cytogenetic analysis of chromosome 3 in Drosophila melanogaster: the homeotic gene complex in polytene chromosome interval 84 A-B. Genetics 94, 115–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hueber S. D., and Lohmann I. (2008) Shaping segments: Hox gene function in the genomic age. BioEssays 30, 965–979 [DOI] [PubMed] [Google Scholar]
- 4. Robertson L. K., and Mahaffey J. W. (2009) in Insect Development (Gilbert L. I., ed) pp. 1–57, Elsevier, San Diego, CA [Google Scholar]
- 5. Weatherbee S. D., Halder G., Kim J., Hudson A., and Carroll S. (1998) Ultrabithorax regulates genes at several levels of the wing-patterning hierarchy to shape the development of the Drosophila haltere. Genes Dev. 12, 1474–1482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Prasad M., Bajpai R., and Shashidhara L. S. (2003) Regulation of Wingless and Vestigial expression in wing and haltere discs of Drosophila. Development 130, 1537–1547 [DOI] [PubMed] [Google Scholar]
- 7. Crickmore M. A., and Mann R. S. (2006) Hox control of organ size by regulation of morphogen production and mobility. Science 313, 63–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. de Navas L. F., Garaulet D. L., and Sánchez-Herrero E. (2006) The Ultrabithorax Hox gene of Drosophila controls haltere size by regulating the Dpp pathway. Development 133, 4495–4506 [DOI] [PubMed] [Google Scholar]
- 9. Pallavi S. K., Kannan R., and Shashidhara L. S. (2006) Negative regulation of Egfr/Ras pathway by Ultrabithorax during haltere development in Drosophila. Dev. Biol. 296, 340–352 [DOI] [PubMed] [Google Scholar]
- 10. Makhijani K., Kalyani C., Srividya T., and Shashidhara L. S. (2007) Modulation of Decapentaplegic gradient during haltere specification in Drosophila. Dev. Biol. 302, 243–255 [DOI] [PubMed] [Google Scholar]
- 11. Hersh B. M., Nelson C. E., Stoll S. J., Norton J. E., Albert T. J., and Carroll S. B. (2007) The UBX-regulated network in the haltere imaginal disc of D. melanogaster. Dev. Biol. 302, 717–727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Agrawal P., Habib F., Yelagandula R., and Shashidhara L. S. (2011) Genome-level identification of targets of Hox protein Ultrabithorax in Drosophila: novel mechanisms for target selection. Sci. Rep. 1, 205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Slattery M., Ma L., Négre N., White K. P., and Mann R. S. (2011) Genome-wide tissue-specific occupancy of the Hox protein Ultrabithorax and Hox cofactor Homothorax in Drosophila. PLoS One 6, e14686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Schwab K., Hartman H. A., Liang H.-C., Aronow B. J., Patterson L. T., and Potter S. S. (2006) Comprehensive microarray analysis of Hoxa11/Hoxd11 mutant kidney development. Dev. Biol. 293, 540–554 [DOI] [PubMed] [Google Scholar]
- 15. Hueber S. D., Bezdan D., Henz S. R., Blank M., Wu H., and Lohmann I. (2007) Comparative analysis of Hox downstream genes in Drosophila. Development 134, 381–392 [DOI] [PubMed] [Google Scholar]
- 16. Rohrschneider M. R., Elsen G. E., and Prince V. E. (2007) Zebrafish Hoxb1a regulates multiple downstream genes including prickle1b. Dev. Biol. 309, 358–372 [DOI] [PubMed] [Google Scholar]
- 17. Prochiantz A., and Di Nardo A. A. (2015) Homeoprotein signaling in the developing and adult nervous system. Neuron 85, 911–925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Fukuta M., Matsuno K., Hui C. C., Nagata T., Takiya S., Xu P.-X., Ueno K., and Suzuki Y. (1993) Molecular cloning of a POU domain-containing factor involved in the regulation of the Bombyx sericin-1gene. J. Biol. Chem. 268, 19471–19475 [PubMed] [Google Scholar]
- 19. Mach V., Takiya S., Ohno K., Handa H., Imai T., and Suzuki Y. (1995) Silk gland factor-1 involved in the regulation of Bombyx sericin-1 gene contains fork head motif. J. Biol. Chem. 270, 9340–9346 [DOI] [PubMed] [Google Scholar]
- 20. Kimoto M., Kitagawa T., Kobayashi I., Nakata T., Kuroiwa A., and Takiya S. (2012) Inhibition of the binding of MSG-intermolt-specific complex, MIC, to the sericin-1 gene promoter and sericin-1 gene expression by POU-M1/SGF-3. Dev. Genes Evol. 222, 351–359 [DOI] [PubMed] [Google Scholar]
- 21. Ohno K., Sawada J., Takiya S., Kimoto M., Matsumoto A., Tsubota T., Uchino K., Hui C.-C., Sezutsu H., Handa H., and Suzuki Y. (2013) Silk gland factor-2, involved in fibroin gene transcription, consists of LIM homeodomain, LIM-interacting, and single-stranded DNA-binding proteins. J. Biol. Chem. 288, 31581–31591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kimoto M., Tsubota T., Uchino K., Sezutsu H., and Takiya S. (2015) LIM-homeodomain transcription factor Awh is a key component activating all three fibroin genes, fibH, fibL and fhx, in the silk gland of the silkworm, Bombyx mori. Insect. Biochem. Mol. Biol. 56, 29–35 [DOI] [PubMed] [Google Scholar]
- 23. Kimoto M., Tsubota T., Uchino K., Sezutsu H., and Takiya S. (2014) Hox transcription factor Antp regulates sericin-1 gene expression in the terminal differentiated silk gland of Bombyx mori. Dev. Biol. 386, 64–71 [DOI] [PubMed] [Google Scholar]
- 24. Sezutsu H., Uchino K., Kobayashi I., Tatematsu K., Iizuka T., Yonemura N., and Tamura T. (2009) Conservation of fibroin gene promoter function between the domesticated silkworm Bombyx mori and the wild silkmoth Antheraea yamamai. J. Insect Biotechnol. Sericol. 78, 1–10 [Google Scholar]
- 25. Tsubota T., Uchino K., Suzuki T. K., Tanaka H., Kayukawa T., Shinoda T., and Sezutsu H. (2014) Identification of a novel strong and ubiquitous promoter/enhancer in the silkworm Bombyx mori. G3 4, 1347–1357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Uchino K., Imamura M., Sezutsu H., Kobayashi I., Kojima K., Kanda T., and Tamura T. (2006) Evaluating promoter sequences for trapping an enhancer activity in the silkworm Bombyx mori. J. Insect. Biotechnol. Sericol. 75, 89–97 [Google Scholar]
- 27. Takiya S., Inoue H., and Kimoto M. (2011) Novel enhancer and promoter elements indispensable for the tissue-specific expression of the sericin-1 gene of the silkworm Bombyx mori. Insect. Biochem. Mol. Biol. 41, 592–601 [DOI] [PubMed] [Google Scholar]
- 28. Kajiwara H., Imamaki A., Nakamura M., Mita K., Xia Q., and Ishizaka M. (2009) Proteome analysis of silkworm. 1. Fat body. J. Electrophor. 53, 19–26 [Google Scholar]
- 29. Perkins D. N., Pappin D. J., Creasy D. M., and Cottrell J. S. (1999) Probability-based protein identification by searching database using mass spectrometry data. Electrophoresis 20, 3551–3567 [DOI] [PubMed] [Google Scholar]
- 30. International Silkworm Genome Consortium (2008) The genome of a lepidopteran model insect, the silkworm Bombyx mori. Insect. Biochem. Mol. Biol. 38, 1036–1045 [DOI] [PubMed] [Google Scholar]
- 31. Dhawan S., and Gopinathan K. P. (2003) Expression profiling of homeobox genes in silk gland development in the mulberry silkworm Bombyx mori. Dev. Genes Evol. 213, 523–533 [DOI] [PubMed] [Google Scholar]
- 32. Imamura M., Nakai J., Inoue S., Quan G.-X., Kanda T., and Tamura T. (2003) Targeted gene expression using the GAL4/UAS system in the silkworm Bombyx mori. Genetics 165, 1329–1340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Uchino K., Sezutsu H., Imamura M., Kobayashi I., Tatematsu K., Iizuka T., Yonemura N., Mita K., and Tamura T. (2008) Construction of a piggyBac-based enhancer trap system for the analysis of gene function in silkworm Bombyx mori. Insect. Biochem. Mol. Biol. 38, 1165–1173 [DOI] [PubMed] [Google Scholar]
- 34. Kobayashi I., Kojima K., Uchino K., Sezutsu H., Iizuka T., Tatematsu K., Yonemura N., Tanaka H., Yamakawa M., Ogura E., Kamachi Y., and Tamura T. (2011) An efficient binary system for gene expression in the silkworm, Bombyx mori, using GAL4 variants. Arch. Insect Biochem. Physiol. 76, 195–210 [DOI] [PubMed] [Google Scholar]
- 35. Takasu Y., Yamada H., Tamura T., Sezutsu H., Mita K., and Tsubouchi K. (2007) Identification and characterization of a novel sericin gene expressed in the anterior middle silk gland of the silkworm Bombyx mori. Insect Biochem. Mol. Biol. 37, 1234–1240 [DOI] [PubMed] [Google Scholar]
- 36. Takasu Y., Hata T., Uchino K., and Zhang Q. (2010) Identification of Ser2 proteins as major sericin components in the non-cocoon silk of Bombyx mori. Insect Biochem. Mol. Biol. 40, 339–344 [DOI] [PubMed] [Google Scholar]
- 37. Zhang Y., Zhao P., Dong Z., Wang D., Guo P., Guo X., Song Q., Zhang W., and Xia Q. (2015) Comparative proteome analysis of multi-layer cocoon of the silkworm, Bombyx mori. PLoS One 10, e0123403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Tatematsu K., Kobayashi I., Uchino K., Sezutsu H., Iizuka T., Yonemura N., and Tamura T. (2010) Construction of a binary transgenic gene expression system for recombinant protein production in the middle silk gland of the silkworm Bombyx mori. Transgenic Res. 19, 473–487 [DOI] [PubMed] [Google Scholar]
- 39. Krasnow M. A., Saffman E. E., Kornfeld K., and Hogness D. S. (1989) Transcriptional activation and repression by Ultrabithorax proteins in cultured Drosophila cells. Cell 57, 1031–1043 [DOI] [PubMed] [Google Scholar]
- 40. Bello B., Horard B., and Couble P. (1994) The selective expression of silk-protein-encoding genes in Bombyx mori silk gland. Bull. Inst. Pasteur 92, 81–100 [Google Scholar]
- 41. Inoue S., Kanda T., Imamura M., Quan G.-X., Kojima K., Tanaka H., Tomita M., Hino R., Yoshizato K., Mizuno S., and Tamura T. (2005) A fibroin secretion-deficient silkworm mutant, Nd-sD, provides an efficient system for producing recombinant proteins. Insect Biochem. Mol. Biol. 35, 51–59 [DOI] [PubMed] [Google Scholar]
- 42. Tsubota T., Yamamoto K., Mita K., and Sezutsu H. (2016) Gene expression analysis in the larval silk gland of the eri silkworm Samia ricini. Insect Sci., in press [DOI] [PubMed] [Google Scholar]
- 43. Takiya S., Kokubo H., and Suzuki Y. (1997) Transcriptional regulatory elements in the upstream and intron of the fibroin gene bind three specific factors POU-M1, Bm Fkh and FMBP-1. Biochem. J. 321, 645–653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Inoue S., Tanaka K., Arisaka F., Kimura S., Ohtomo K., and Mizuno S. (2000) Silk fibroin of Bombyx mori is secreted, assembling a high molecular mass elementary unit consisting of H-chain, L-chain, and P25, with a 6:6:1 molar ratio. J. Biol. Chem. 275, 40517–40528 [DOI] [PubMed] [Google Scholar]
- 45. Tanaka K., Inoue S., and Mizuno S. (1999) Hydrophobic interaction of P25, containing Asn-linked oligosaccharide chains, with the H-L complex of silk fibroin produced by Bombyx mori. Insect Biochem. Mol. Biol. 29, 269–276 [DOI] [PubMed] [Google Scholar]