Abstract
Nasopharyngeal colonization by the Gram-positive bacterium Streptococcus pneumoniae is a prerequisite for pneumonia and invasive pneumococcal diseases. Colonization is asymptomatic, involving dynamic and complex interplay between commensals, the host immune system, and environmental factors. The elderly are at an increased risk of developing pneumonia, which might be due to changes in the respiratory microbiota that would impact bacterial colonization and persistence within this niche. We hypothesized that the composition of the upper respiratory tract (URT) microbiota changes with age and subsequently can contribute to sustained colonization and inefficient clearance of S. pneumoniae. To test this, we used a mouse model of pneumococcal colonization to compare the composition of the URT microbiota in young, middle-aged, and old mice in the naive state and during the course of colonization using nasal pharyngeal washes. Sequencing of variable region 3 (V3) of the 16S rRNA gene was used to identify changes occurring with age and throughout the course of S. pneumoniae colonization. We discovered that age affects the composition of the URT microbiota and that colonization with S. pneumoniae is more disruptive of preexisting communities in older mice. We have further shown that host-pathogen interactions following S. pneumoniae colonization can impact the populations of resident microbes, including Staphylococcus and Haemophilus. Together, our findings indicate alterations to the URT microbiota could be detrimental to the elderly, resulting in increased colonization of S. pneumoniae and decreased efficiency in its clearance.
INTRODUCTION
Streptococcus pneumoniae colonizes the mucosal surfaces of the upper respiratory tract (URT), which includes the nose, nasal cavity, pharynx, and larynx (1). Although colonization within the nasal passage often is asymptomatic, access to the airways can result in pneumonia, with further dissemination causing invasive pneumococcal disease (i.e., otitis media, bacteremia, and meningitis) (1, 2). Previous studies analyzing the nasopharyngeal culture of 1,704 samples, including children and adults from the same population, revealed that 53% of children carried S. pneumoniae within the nasopharyngeal tract as opposed to only 4 to 11% that were adult carriers (3–5). Furthermore, S. pneumoniae carriage rates positively correlate with age in young children and then begin to drop in adults (3, 6, 7). These results have been confirmed in epidemiological studies conducted in several locations around the world (7–9).
Despite having significantly lower carriage rates than children (3, 10), colonization within the upper respiratory tract of elderly individuals often leads to the progression and development of pneumonia and invasive pneumococcal disease (11–13). Pneumonia in particular affects elderly individuals approximately four times more often than individuals under the age of 65 (14). The elderly account for approximately 60% of the hospitalizations caused by pneumococcal pneumonia in the United States (15). Since colonization is a prerequisite for infection, the microbe-microbe interactions that contribute to sustaining colonization or promoting expansion must be further studied to understand disease progression in elderly patients.
Using Illumina sequencing of the 16S rRNA gene, we characterized the URT microbiome in young (10 to 14 weeks), middle-aged (12 to 14 months), and old (18 to 22 months) mice in the naive state and throughout the course of nasopharyngeal colonization with Streptococcus pneumoniae. We show that the composition of the URT microbiome differs in the naive state between young, middle-aged, and old mice. Old mice are unable to clear bacterial colonization as effectively as their young counterparts. We observed a number of interspecies interactions between S. pneumoniae and the existing mouse microbiome (e.g., Staphylococcus) that have been reported previously only in experimental models (16–24). In particular, Streptococcus interacted competitively with Staphylococcus and synergistically with Haemophilus. This study begins to characterize how aging impacts bacterial colonization, which will ultimately explain the progression of upper respiratory tract infections in the elderly.
MATERIALS AND METHODS
Mice.
C57BL/6 female mice were from The Jackson Laboratory and were housed under specific-pathogen-free (SPF) conditions at the McMaster Central Animal Facility. The animals were assigned to three groups: 10- to 14-week-old mice (young), 12- to 14-month-old mice (middle-aged), and 18- to 22-month-old mice (old) (n = 72 total). Within each age group, mice were sacrificed at various time points throughout pneumococcal colonization (at day 0, 3, 14, and 21) in order to obtain nasopharyngeal washes (25). Mice that reached the endpoint prematurely were found to have bacteria in the lungs or spleens and were not used in this study. In general, <5% of young mice and 20 to 25% of old mice were euthanized prematurely (26). All procedures were performed in accordance with the McMaster Animal Research Ethics Board guidelines.
Murine model of pneumococcal colonization and nasopharyngeal wash preparation.
Mice were colonized with 107 CFU of a clinical strain of S. pneumoniae, P1547 (serotype 6A), obtained from Jeff Weiser (NYU School of Medicine) as described previously (27, 28). The bacteria were grown in tryptic soy broth medium (Life Technologies) at 37°C and 5% CO2 until cultures reached log phase, with an optical density at 600 nm (OD600) of between 0.45 and 0.55 (27). Anesthetized mice were euthanized by exsanguination. The trachea was expanded and a small incision was made 2 cm above the lungs. Briefly, 1-ml syringes containing 350 μl of sterilized phosphate-buffered saline (PBS) attached to a 26-gauge needle were connected to a 4-cm PE-20 polyethylene tube. The syringe was inserted into the trachea and tied off using a silk suture (Ethicon), and the contents used to wash the nares were collected in 1.5-ml flat-top microtubes (Diamed) (28). Nasal washes then were collected at day 0 (young, n = 5; middle-aged, n = 4; old, n = 7), 3 (young, n = 6; middle-aged, n = 5; old, n = 5), 14 (young, n = 9; middle-aged, n = 6; old, n = 4), and 21 (young, n = 4; middle-aged, n = 3; old, n = 4) after colonization with Streptococcus pneumoniae as previously described (28).
PCR amplification of the 16S rRNA gene.
DNA extraction and 16S variable region 3 (V3) amplification were carried out as described in our recent studies of human nasal swabs (8, 29). The primers were based on the method described in Bartram et al., except the barcodes were incorporated into the forward primer (30).
Briefly, each PCR mixture contained the following in order to amplify V3 of the 16S rRNA gene by PCR: 5 μl of 10× buffer (Life Technologies), 1.5 μl of MgCl2 (50 mM) (Life Technologies), 1 μl of deoxynucleoside triphosphate (dNTP) (10 mM) (Invitrogen), 2 μl of bovine serum albumin (BSA) (10 mg/ml made in pure water and irradiated for 30 min) (Life Technologies), 5 μl of V3F primer (1 μM) (27), 5 μl of V3R primer (1 μM) (27), 0.5 μl of Taq polymerase (Life Technologies), and 200 ng of DNA. The reaction then was run for 30 cycles (94°C for 2 min, 94°C for 30 s, 50°C for 30°C, 72°C for 30 s), with a final polymerization step at 72°C for 10 min (Eppendorf). The products were separated by electrophoresis in a 2% agarose gel and visualized under a UV transilluminator light, and the products corresponding to the amplified V3 (∼300 bp) were excised and purified using standard gel extraction kits (Qiagen).
Illumina sequencing and processing.
Samples were sent to the McMaster DNA Sequencing Facility and sequenced using an Illumina MiSeq per the manufacturer's instructions. The completed run was demultiplexed with Illumina's Casava software. The resulting sequenced data were processed as previously described (8, 29). Briefly, Cutadapt was used to trim the forward and reverse paired-end reads at the opposing primers for input into PANDAseq for assembly (31, 32). Sequences were organized into operational taxonomic units (OTUs) with a clustering threshold of 97% using AbundantOTU+ (33). Single-sequence OTUs (singletons) were removed prior to all analyses using Quantitative Insights into Microbial Ecology (QIIME) (34). All beta diversity plots were generated in R using the Phyloseq package and taxonomic summaries (34). A total of 10,312,391 paired-end reads were observed from the sequencing data (with a minimum of 1,193 reads and a maximum of 347,295 reads). This gave an average of 132,210 counts per sample with a standard deviation of 77,464. These counts corresponded to 7,100 unique OTUs observed within the samples.
Quantitative PCR.
Real-time PCR (quantitative PCR [qPCR]) was used to assess the total bacterial load and lytA abundance using a previously published protocol (8, 35). Briefly, levels of S. pneumoniae in the nasal wash samples using GoTaq qPCR master mix (Promega, WI, USA) and an ABI StepOnePlus (Applied Biosystems, CA, USA) according to the manufacturer's instructions. Forward and reverse primers used for 16S rRNA were the following: 341Fwd, 5′-CCTACGGGAGGCAGCAG-3′; 518Rev, 5′-ATTACCGCGGCTGCTGG-3′ (36). Forward and reverse primers used to measure lytA were the following: Fwd, 5′-AGTACCAGTTGCCGTCTGTG-3′; Rev, 5′-AAATGGGGCATTAGCCGTGA-3′. lytA levels were found to accurately represent CFU, which were previously quantitated and published (26).
Statistics.
Unless otherwise mentioned in the figure legend, statistical significance was determined by unpaired t tests (two-tailed) or a one-way analysis of variance (ANOVA). Data were analyzed with Prism (version 6; GraphPad). Statistical significance of groups by β-diversity (Bray-Curtis) was determined using permutational ANOVA (PERMANOVA) in R phyloseq (37). This was calculated using the ADONIS function, which conducts a permutational multivariate analysis of variance within the samples using distance matrices. Statistical significance was defined as a P value of 0.05.
RESULTS
Upper respiratory microbial communities differ between young, middle-aged, and old mice.
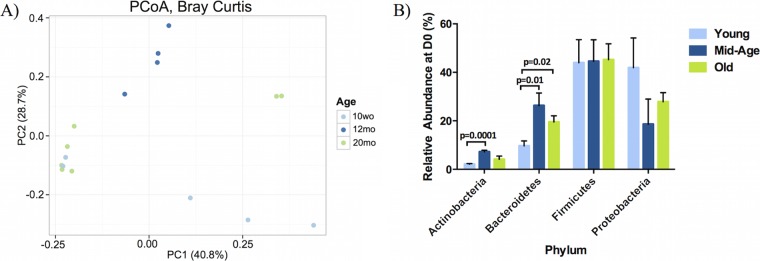
The composition of the microbial community in nasopharyngeal washes from 10-week-old, 12-month-old, and 20-month-old naive mice were compared. There was no detectable difference in total bacterial load between age groups, as measured by qPCR of the 16S rRNA gene (246 ± 106 pg in young mice, 168 ± 38 pg in middle-aged mice, and 379 ± 384 pg in old mice). In order to determine if the composition of the nasopharyngeal tract microbial community changed with age, the β-diversity metric, calculated based on the Bray-Curtis distance, was visualized using a principal coordinate analysis (PCoA) plot (Fig. 1A). Under Bray-Curtis calculations, the samples are statistically different by age (P = 0.04) (Fig. 1A; also see Table S1 in the supplemental material).
FIG 1.
16S rRNA sequencing of nasopharyngeal washes from young, middle-aged, and old mice under naive conditions reveal significant differences within the overall bacterial composition and specific phyla. Bacterial communities were examined under naive conditions to examine the differences between age groups. (A) Communities clustered using principal coordinate analyses (PCoA) of the Bray-Curtis distance matrix. Each point represents one sample and is differentiated by color to indicate the age of the mouse. Plots represent the microbial composition as indicated by the β-diversity between each nasal wash sample. Clustering was observed between the young, middle-aged, and old mice before colonization was statistically significant by PERMANOVA. (B) The four most abundant phyla present in young, middle-aged, and old mice under naive conditions were quantified within the young, middle-aged, and old samples prior to pneumococcal colonization (young, 10 weeks old [10wo], n = 5; middle-aged, 12 months old [12mo], n = 4; old, 20 months old [20mo], n = 7). Values are the means ± standard errors of the means (SEM) of samples within each category. Statistical significance was determined using multiple t tests where appropriate (n = 3 to 9/group). A P value of <0.05 was considered significant. D0, day zero.
Taxonomic summaries prior to colonization indicate that the most abundant phyla are Firmicutes, Proteobacteria, Bacteroidetes, and Actinobacteria (Fig. 1B). The Proteobacteria are present at a relative abundance of 41.9% in young mice but only 27.9% in old mice. In contrast, young mice contain a significantly lower abundance of Bacteroidetes (9.7%) than old mice (19.5%). All three age groups contain comparable levels of Firmicutes (young, 44.0%; middle aged, 44.6%; old, 45.2%), and young and old mice had comparable levels of Actinobacteria (young, 2.1%; middle aged, 7.2%; old, 4.1%).
Old mice do not effectively clear nasopharyngeal colonization.
We next determined whether old mice differed in their ability to clear pneumococcal colonization. Mice were intranasally inoculated with Streptococcus pneumoniae and were monitored up to 21 days (25). The pneumococcal autolysin gene lytA was measured by qPCR to quantitate the levels of S. pneumoniae present in nasal wash samples over the course of the colonization (Fig. 2A). As expected, lytA was not detectable at day 0. Peak levels of S. pneumoniae occurred at day 3 postcolonization in young and middle-aged mice. Levels of S. pneumoniae decreased in young mice by 14 days postcolonization, and most mice had levels of S. pneumoniae that were below the limit of detection by day 21, consistent with their ability to clear pneumococcal colonization. In contrast, lytA levels peak around day 14 in old mice and persist through to day 21 of colonization. At day 21, nasal washes from old mice have considerably higher levels of S. pneumoniae than young mice.
FIG 2.
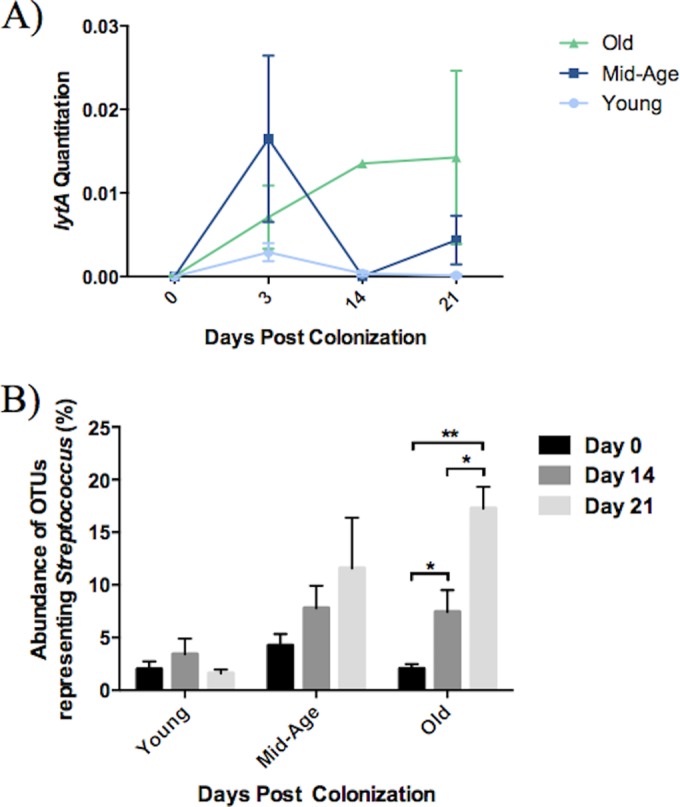
Old mice do not effectively clear nasopharyngeal colonization of S. pneumoniae. S. pneumoniae clearance was examined within young, middle-aged, and old mice using lytA expression (A) and the OTUs representing the Streptococcus genus (B). lytA expression was measured in nasal wash samples of these mice using quantitative PCR. All samples were normalized to the total bacterial load, which was measured by 16S rRNA gene quantitative PCR. Relative abundances of Streptococcus in the nasal microbiome were calculated from all OTUs associated with the Streptococcus genus within each sample. Values are the means ± SEM of samples within each category. Statistical significance was determined using multiple t tests where appropriate (n = 3 to 9/group). A P value of <0.05 was considered significant.
Furthermore, the abundance of OTUs representing the streptococci decreased to very low levels by day 21 in young mice (approximately 1.6% of OTUs). However, this was not seen in the elderly mice. Unlike the young, the elderly mice had a high relative abundance of streptococci at day 21 postcolonization (approximately 17.3% of OTUs) (Fig. 2B). It is important to note that OTUs corresponding to the Streptococcus genus include all streptococci and are not specific to S. pneumoniae. In fact, the OTUs representing the streptococci include a wide array of species, and some of the abundant OTUs assigned to this genus are phylogenetically similar to our bacterium of interest, S. pneumoniae (see Fig. S1 in the supplemental material).
Nasopharynx microbial communities respond differently to pneumococcal colonization with age.
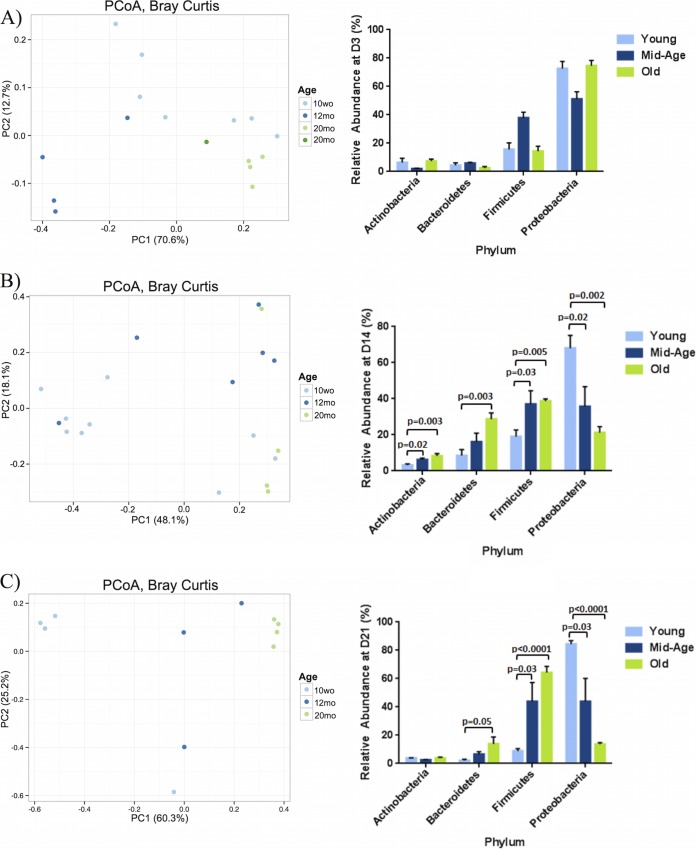
The microbial communities for the nasal wash samples at days 3, 14, and 21 postcolonization were compared to baseline conditions (day 0) (Fig. 1A and 3). Clustering of samples between the age groups was evident at days 3 and 21 postcolonization, suggesting that the microbial communities of mice within the same age group respond similarly to the introduction of S. pneumoniae (Fig. 3A and C). Specifically, 3 days postcolonization, the young, middle-aged, and old microbiota cluster according to their specific age group as visualized using the Bray-Curtis distance metric (Fig. 3A; also see Table S1 in the supplemental material). By day 21, this clustering becomes more distinct between the three age groups, suggesting that the bacterial communities present within young and old mice are distinctly separate after 21 days of colonization (Fig. 3C; also see Fig. S1).
FIG 3.
During colonization, the microbial community changes with age. Principal coordinate analyses (PCoA) plots showing the similarity of the aging URT microbiome postpneumococcal colonization as well as relative abundance plots of the four most abundant phyla were analyzed at day 3 (A), day 14 (B), and day 21 (C) postcolonization. β-Diversity measures were completed using Bray-Curtis calculations and visualized using PCoA. The age groups of the samples are indicated by the designated color. The distance between each of the samples reflects how similar their microbiomes are to one another. Statistical analyses were completed using PERMANOVA. The results are summarized in Table S1 in the supplemental material. The panels on the right represent the mean relative abundances of each phyla within each sample ± SEM of samples within each category. Statistical significance was determined using multiple t test where appropriate (n = 3 to 9/group). P < 0.05 was considered significant.
Taxonomic summaries of microbial communities within the nasopharynx of these mice indicated that the proportion of the four most abundant phyla, Proteobacteria, Firmicutes, Bacteroidetes, and Actinobacteria, changed during the course of pneumococcal colonization (Fig. 3). Proteobacteria comprised the majority of the nasal microbiota within young mice prior to colonization (Fig. 1B), and this was relatively consistent across the course of colonization (Fig. 3). In fact, immediately after colonization at day 3, the OTUs representing Proteobacteria increased from about 40% to 72% relative abundance, and this was maintained throughout colonization until clearance at day 21 (Fig. 3A). The relative abundances of Proteobacteria within young mice were comparable at day 14 postcolonization (68%) and were slightly elevated at day 21 (84.4%) (Fig. 3B). In contrast, although levels of Proteobacteria were elevated in old mice at day 3 postcolonization (74.7%), they continued to decrease throughout colonization, comprising about 21.2% of OTUs by day 14 and only 13.6% by day 21 (Fig. 3C). In fact, by day 21, old mice contained significantly lower levels (P < 0.0001) of OTUs representing the Proteobacteria phylum compared to the levels present at day 3 (Fig. 3C; also see Fig. S2 in the supplemental material).
It was evident that the abundant OTUs within the Firmicutes phylum (which includes S. pneumoniae) were similar between all three age groups at baseline (young, 44%; middle-aged, 44.6%; old, 45.2%) (Fig. 1B). However, within the day 3, 14, and 21 microbiota, there was a decrease in relative abundance of OTUs within the Firmicutes phylum in young mice (day 3, 15.7%; day 14, 19%; day 21, 8.8%) and an increase within the old mice (day 3, 14.3%; day 14, 38.7%; day 21, 64.3%) (Fig. 3C). In fact, by day 21, there is a significantly greater abundance of OTUs (P < 0.0001) representing Firmicutes within the old mice relative to the young and middle-aged samples (see Fig. S2B in the supplemental material).
The abundance of bacterial groups within the URT is altered throughout colonization.
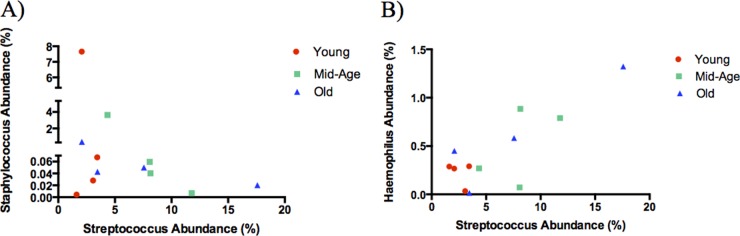
The presence of Streptococcus pneumoniae, Staphylococcus aureus, and Haemophilus influenzae in the nasopharynx previously has been shown to influence subsequent colonization (19). In particular, S. pneumoniae and S. aureus appear to have an antagonistic relationship, while S. pneumoniae and H. influenzae demonstrate a synergistic relationship to promote cocolonization within this microbial niche (17, 18, 20, 21). To further investigate these interactions in a natural community, we examined how existing populations of Staphylococcus and Haemophilus are altered following S. pneumoniae colonization. Prior to colonization (day 0), there is a higher abundance of Staphylococcus than Streptococcus within the URT. However, throughout colonization with S. pneumoniae, the abundance of OTUs representing Staphylococcus appears to decrease as the abundance of Streptococcus increases (Fig. 4A). This trend is particularly evident within the middle-aged and old mice, as the abundance of Streptococcus within their nasopharyngeal washes continues to increase throughout colonization. In contrast, the interaction with Haemophilus appeared to be synergistic, as the relative abundance of Streptococcus throughout colonization was mirrored by the increased abundance of Haemophilus (Fig. 4B). These results support previously published data indicating that colonization with S. pneumoniae can greatly influence the survival and growth of the existing residents within the environment.
FIG 4.
Staphylococcus decreases while Haemophilus increases in proportion to Streptococcus during nasopharyngeal colonization. Relative abundances of OTUs representing Staphylococcus (A) or Haemophilus (B) in correlation to OTUs representing Streptococcus. Each data point within an age group represents the average abundance within nasopharyngeal washes at day 0, 3, 14, or 21 postcolonization (n = 3 to 9/group). The age groups of the samples are indicated by color.
Microbial communities do not return to their baseline composition following S. pneumoniae colonization.
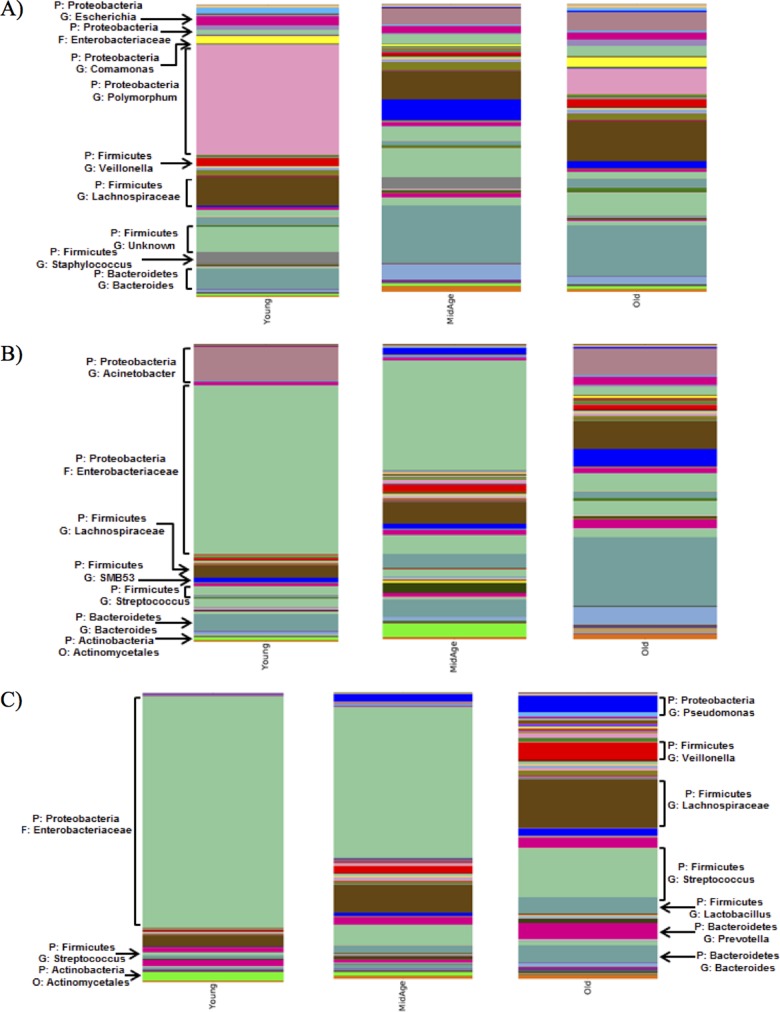
In order to further examine the resolution phase of colonization, the microbiota was examined at days 14 and 21. Taxon summary plots illustrate that the introduction of S. pneumoniae alters the existing bacterial composition, and these changes continue to progress throughout colonization, as examined at the phylum level (see Fig. S3 in the supplemental material) and even more specifically at the genus level (Fig. 5). The microbial communities of mice colonized with S. pneumoniae did not return to baseline composition, as the day 21 microbial communities contained differing abundances of bacterial groups compared to baseline conditions (Fig. 5A and C). Young mice maintained high levels of Proteobacteria throughout colonization with S. pneumoniae. Although the role of Proteobacteria within the URT still remains unclear, some studies have shown its presence to be quite common within this niche (38).
FIG 5.
Bacterial communities within the URT microbiome diverge significantly from precolonization levels. Shown is an averaged taxon summary plot of all samples within a particular group (n = 3 to 9/group). The bacterial groups are labeled according to phylum and then specified to the highest assigned taxonomic group. The height of the bar represents the relative abundance of the associated genus within the day 0 (control) mice (A) and day 14 (B) and day 21 (C) postcolonization mice. The most abundant genera are labeled on the group.
Examining the mice following colonization with S. pneumoniae revealed similar results, as this introduction altered the abundance of existing bacteria within the community (Tables 1 to 3). Indeed, the introduction of S. pneumoniae led to the reduction of certain bacterial groups below the level of detection. The most frequently observed genera showing considerable changes upon colonization were examined in greater detail. These included Streptococcus, Haemophilus, Staphylococcus, Clostridium, and Escherichia. In young mice, Streptococcus OTUs appeared to increase slightly until day 14 and then decreased by day 21 (Table 1). The middle-aged and old mice contained a higher abundance of Streptococcus OTUs that continued to increase until day 21 (Tables 2 and 3). There was a 2-fold increase in the Streptococcus OTU abundance between day 3 and day 21 in the middle-aged mice and an 11-fold increase in the old mice. Bacterial genera such as Haemophilus tended to briefly increase throughout colonization in all three age groups, whereas Escherichia remained constant at low levels postcolonization. Taken together, these data indicated that even if the host is able to clear the pneumococcal infection, their microbiota do not necessarily return to preinfection state, which might ultimately determine future responses to secondary infections from resident or newly acquired infections.
TABLE 1.
Genera most affected by S. pneumoniae within the young URT microbiomea
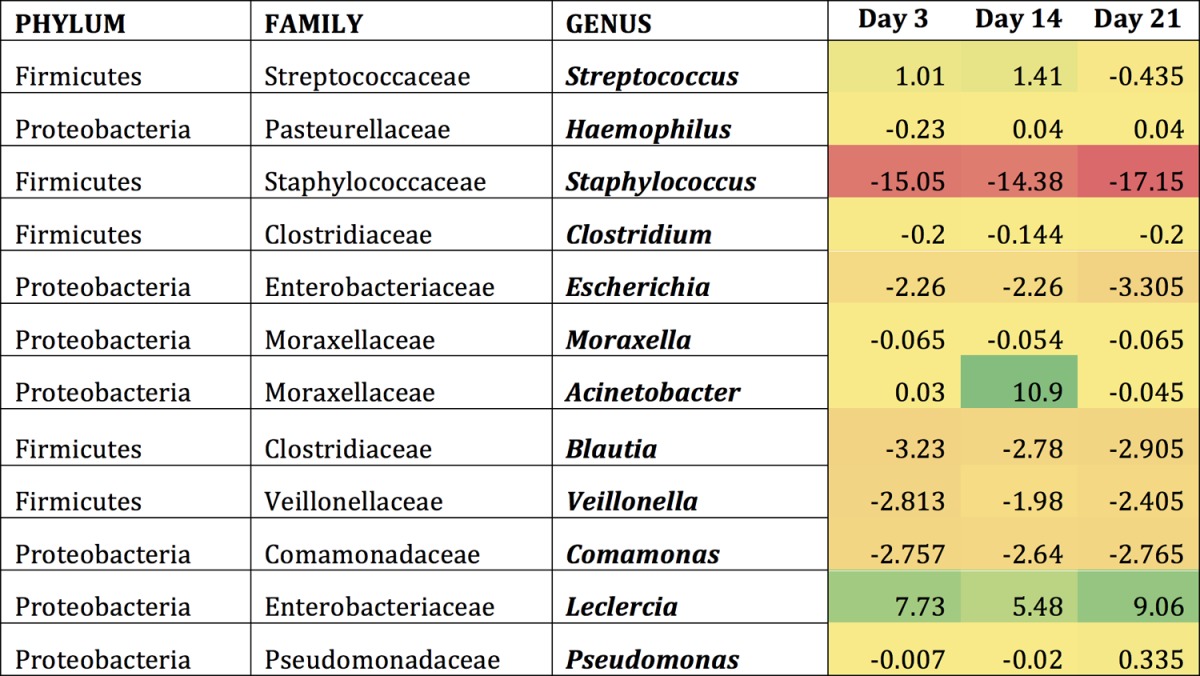
Shown are the differences that occur compared to baseline conditions (day 0) in young mice within each genus. Regions shaded in green indicate an increase from baseline conditions, while regions shaded in red indicate a decrease from baseline conditions.
TABLE 3.
Genera most affected by S. pneumoniae within the elderly URT microbiomea
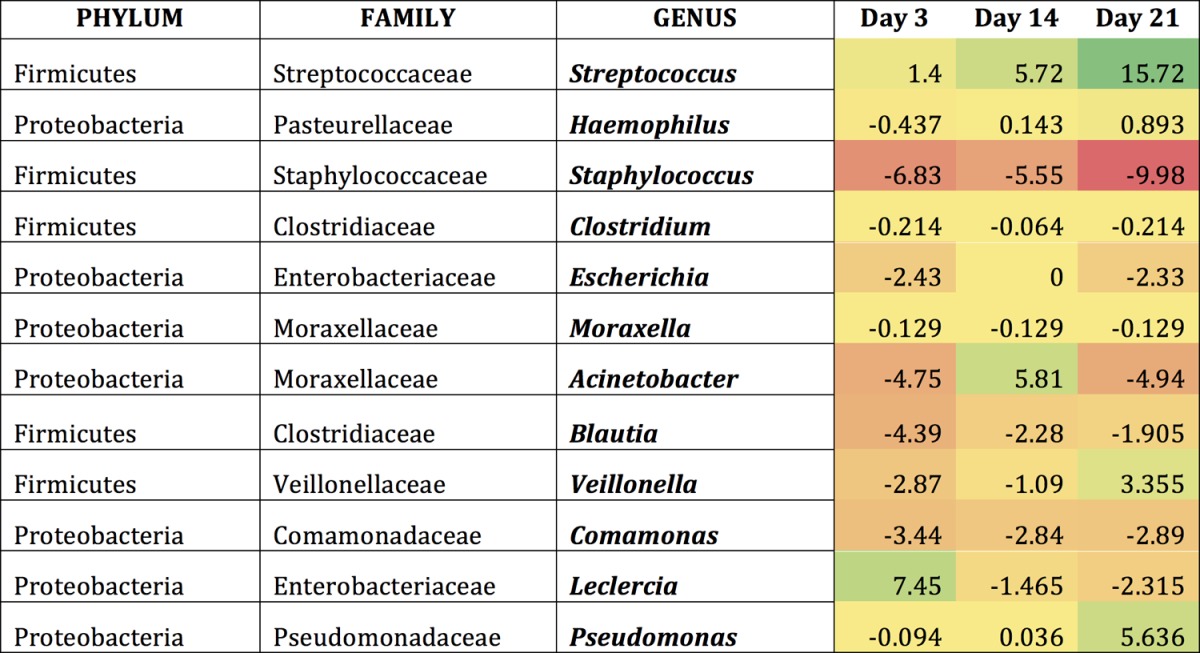
Shown are the differences that occur compared to baseline conditions (day 0) in old mice within each genus. Regions shaded in green indicate an increase from baseline conditions, while regions shaded in red indicate a decrease from baseline conditions.
TABLE 2.
Genera most affected by S. pneumoniae within the middle-aged URT microbiomea
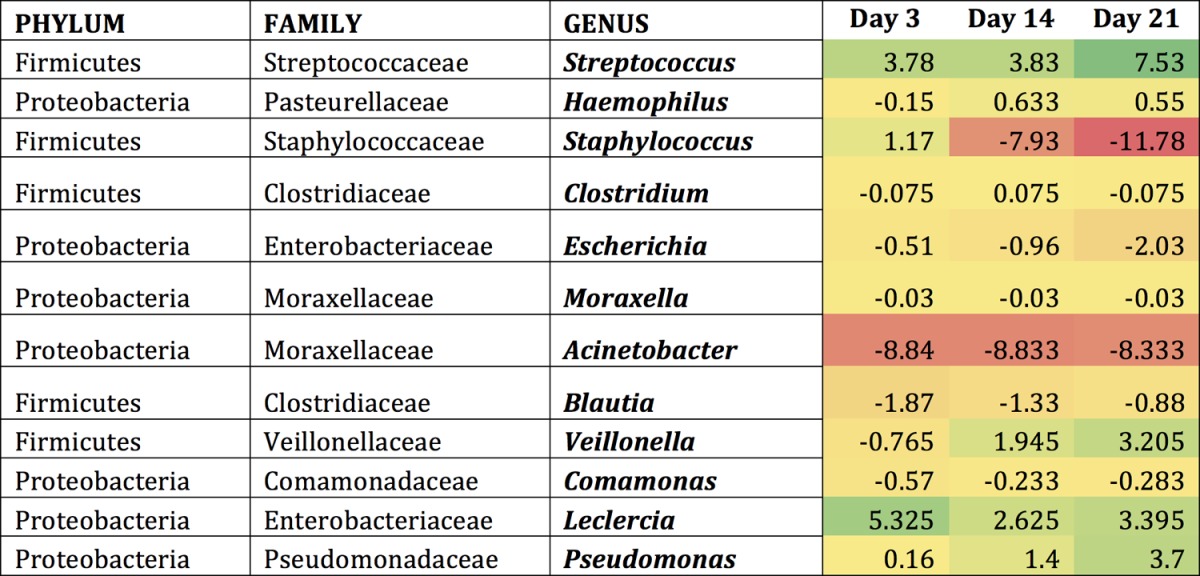
Shown are the differences that occur compared to baseline conditions (day 0) in middle-aged mice within each genus. Regions shaded in green indicate an increase from baseline conditions, while regions shaded in red indicate a decrease from baseline conditions.
DISCUSSION
Nasopharyngeal colonization is a prerequisite for pneumonia or invasive pneumococcal disease (39). In young adults, colonization is cleared within 3 to 6 weeks due to adequate immune control, and the bacteria rarely translocate from the nasopharynx. As a result, disease is rare (3). However, despite the low carriage rates in the elderly (8, 40), Streptococcus pneumoniae is the most common cause of pneumonia in this cohort (41–43). Furthermore, vaccination of older adults does not reduce pneumonia and leads to only a trivial reduction in invasive pneumococcal disease (44, 45). Since S. pneumoniae colonization is influenced by the composition of the nasopharyngeal microbiome, we hypothesized that age-related microbial dysbiosis influences the kinetics of pneumococcal colonization.
Within the upper respiratory tract (URT), Streptococcus pneumoniae can reside as an asymptomatic commensal or a pathogen, resulting in invasive pneumococcal disease (46). As a result of this dual role, a combination of factors within this microbial niche determines how it behaves. Inter- and intraspecies competition between S. pneumoniae and members of the upper respiratory tract microbiome contribute to the ability of S. pneumoniae to establish colonization, the ability of the host to mount a robust antibacterial response, and the expression of virulence factors required to establish disease (47–51). For example, by sensing peptides released by neighboring bacterial species, S. pneumoniae stops replication, initiates a stress response (including the induction of competence), and becomes more adept at maintaining pneumococcal colonization in mouse models (52). Many clinical studies in children demonstrate that pneumococcal carriage is positively associated with Haemophilus influenzae carriage but negatively associated with the carriage of Staphylococcus aureus (17, 23). Furthermore, adults who had more diverse nasal microbiota with a lower number of dominant species, such as Corynebacterium, were more likely to be natural S. pneumoniae carriers and more likely to become experimentally colonized by the pneumococcus (53). Our previous study of nursing home elderly found that the nasal microbiome had more diverse species and a lower percentage of protective Corynebacterium (8), which mimics the colonization-permissive phenotype of the previous study (53). Thus, the composition of the URT microbiome in youth and young adulthood contributes to the ability of S. pneumoniae to establish colonization and, ultimately, infection.
A previous study examining the bacterial profiles of the adult nostril and oropharynx niches in humans revealed a few dominant phyla within each region. Specifically, the nostril bacteria were comprised of Firmicutes and Actinobacteria, while the oropharynx bacteria were comprised of Firmicutes, Proteobacteria, and Bacteroidetes (54–58). Despite the genus- and species-level differences that exist between humans and mice, we observed similar characteristics in our murine URT microbiome at the phylum level. Furthermore, healthy adults are reported to have an inverse correlation between Firmicutes and Actinobacteria within the nasal microbiota (55). This trend was observed within our murine model as well, further revealing the similarities within the two communities.
It is well documented that the gut microbiota changes with age (59–61), and it appears that this may correlate with changes in health (60); however, there are fewer studies on how the airway microbiota changes with age (8) and whether these changes influence the ability of S. pneumoniae to establish colonization. We have shown that the microbial communities of the URT are significantly different with age under steady-state conditions, consistent with a previous report (62). Thus, we sought to further examine the impact of these differences within the old mice on their ability to allow S. pneumoniae colonization and persistence. S. pneumoniae colonization is reported to be influenced by the presence of its community members, including Haemophilus influenzae and Staphylococcus aureus. Since both of these genera are present within the murine URT microbiome, this model allows us to study the natural interactions between S. pneumoniae and the nasopharyngeal microbiota community members.
We examined the changes in the microbial community that occur within the URT following colonization with S. pneumoniae, at least some of which are due to immunosenescence. Antibacterial immunity is impaired in the elderly due to complex changes in innate and adaptive immunity called immunosenescence (63–65). As an example, we have identified that age-related changes in monocytes impair the nasopharyngeal clearance of S. pneumoniae (26). Others have demonstrated that immunity in the lung is impaired because Toll-like receptor (TLR) expression and signaling decreases with age, and this impairs macrophage responses to S. pneumoniae (66). Similarly, the quality of the adaptive antipneumococcal antibodies also have been demonstrated to decrease with age, demonstrating that the adaptive immune response also is impaired (67, 68). The extent to which immunosenescence impairs the maintenance of commensal and microbial communities is not known.
In our mouse model, young mice were able to return to precolonization levels of the streptococci. However, this was not mirrored in the middle-aged and old mice. PCoA plots reveal that young, middle-aged, and old mice cluster within their own age groups following colonization, as shown in a previous publication (62). This trend becomes more distinct by day 21, suggesting that the bacterial community present in each age group responds similarly to colonization. Also, throughout colonization, old mice have an increased abundance of OTUs representing the Firmicutes phylum, which includes S. pneumoniae. Tolerance to commensal bacteria is a key contributor in initiating an immune response against pathogenic bacteria. It has been proposed that changes occurring at the upper respiratory tract barrier within the elderly result in the inability of commensal residents to differentiate between commensal and pathogenic species, ultimately resulting in alterations within innate host defense mechanisms and ineffective bacterial clearance with age (1). Specifically, inhibitors of the NF-κB pathway have been shown to be elevated at the respiratory epithelium in old mice. This increase might be inhibiting the initiation of proinflammatory signaling pathways during bacterial infection and could result in either a more tolerogenic response or a delayed immune response in old mice (1).
Host-pathogen interactions within the nasopharyngeal niche can impact resident microbes. Our data recapitulate experimental models of S. pneumoniae competition or cooperation with other members of the microbiome. We observe that the streptococci appeared to have a competitive interaction with resident Staphylococcus and a synergistic relationship with Haemophilus, as has been described previously (16–18, 21). It is proposed that the inverse relationship with Staphylococcus is a form of bacterial interference (16, 17, 19). It has been suggested that the hydrogen peroxide produced by S. pneumoniae inhibits a variety of competing organisms in the aerobic environment of the URT, including Staphylococcus (16, 17, 20). Contrary to S. pneumoniae's interaction with Staphylococcus, Haemophilus facilitates S. pneumoniae's survival, as shown in previous studies using nasopharyngeal samples of children between 0 and 35 months of age (23). Although the exact mechanisms are unclear, it has been suggested that this phenomenon is related to the downregulation of pneumococcal autolysis and fratricide genes, as well as an increase in pneumococcal biofilm formation (19, 23, 24). As a result, there is decreased production of pneumococcal cell wall hydrolase and a subsequent decrease in pneumococcal lysis during the presence of Haemophilus influenzae. Our observations suggest that the composition of the natural mouse microbiome influences the effectiveness of experimental colonization. Whether this is a factor contributing to differences in susceptibility between mouse strains is not known.
This study examined how age may affect the composition of the microbial community during S. pneumoniae colonization and, in turn, how this phenomenon can play a role in either the clearance or proliferation of a pathogenic species. The overall trends from this study revealed that the elderly mice were not able to clear S. pneumoniae as effectively as the young mice, as they maintained high levels of bacteria even 21 days postcolonization. Characterizing the URT and determining factors driving pneumococcal colonization are important, as pneumonia is a frequently occurring and costly disease in the elderly. Using these data, we can understand the relationship between the aging system and its impact on bacterial colonization to help create alternatives to protect the elderly from age-associated infections.
Supplementary Material
ACKNOWLEDGMENTS
We thank Mark McDermott for critical reading of the manuscript and Jenn Stearns for helpful discussions and suggestions on the analyses. We also thank Michelle Shah and Christine King for their contributions and helpful suggestions during the Illumina library preparations and sequencing.
This work was funded by grants from the Canadian Institutes of Health Research (CIHR) to D.M.E.B. A.P. was supported by an Ontario Graduate Scholarship. N.T. was supported by an Early Researcher Award from the Ontario Ministry of Research and Innovation. Work in the Bowdish laboratory is supported by the McMaster Immunology Research Centre (MIRC) and the M. G. DeGroote Institute for Infectious Disease Research (IIDR). M.G.S. and D.M.E.B. are supported by the CIHR and hold Canada Research Chairs.
Funding Statement
This work was funded by grants from the Canadian Institutes of Health Research (CIHR) to D.M.E.B. A.P. was supported by an Ontario Graduate Scholarship. N.T. was supported by an Early Researcher Award from the Ontario Ministry of Research and Innovation. Work in the Bowdish laboratory is supported by the McMaster Immunology Research Centre (MIRC) and the M. G. DeGroote Institute for Infectious Disease Research (IIDR). M.G.S. and D.M.E.B. are supported by the CIHR and hold Canada Research Chairs.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/IAI.01275-15.
REFERENCES
- 1.Krone CL, Trzciński K, Zborowski T, Sanders EAM, Bogaert D. 2013. Impaired innate mucosal immunity in aged mice permits prolonged Streptococcus pneumoniae colonization. Infect Immun 81:4615–4625. doi: 10.1128/IAI.00618-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Diavatopoulos DA, Short KR, Price JT, Wilksch JJ, Brown LE, Briles DE, Strugnell RA, Wijburg OL. 2010. Influenza A virus facilitates Streptococcus pneumoniae transmission and disease. FASEB J 24:1789–1798. doi: 10.1096/fj.09-146779. [DOI] [PubMed] [Google Scholar]
- 3.Regev-Yochay G, Raz M, Dagan R, Porat N, Shainberg B, Pinco E, Keller N, Rubinstein E. 2004. Nasopharyngeal carriage of Streptococcus pneumoniae by adults and children in community and family settings. Clin Infect Dis 38:632–639. doi: 10.1086/381547. [DOI] [PubMed] [Google Scholar]
- 4.Hamaluba M, Kandasamy R, Ndimah S, Morton R, Caccamo M, Robinson H, Kelly S, Field A, Norman L, Plested E, Thompson BAV, Zafar A, Kerridge SA, Lazarus R, John T, Holmes J, Fenlon SN, Gould KA, Waight P, Hinds J, Crook D, Snape MD, Pollard AJ. 2015. A cross-sectional observational study of pneumococcal carriage in children, their parents, and older adults following the introduction of the 7-valent pneumococcal conjugate vaccine. Medicine (Baltimore, MD) 94:e335. doi: 10.1097/MD.0000000000000335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Watt JP, O'Brien KL, Katz S, Bronsdon MA, Elliott J, Dallas J, Perilla MJ, Reid R, Murrow L, Facklam R, Santosham M, Whitney CG. 2004. Nasopharyngeal versus oropharyngeal sampling for detection of pneumococcal carriage in adults. J Clin Microbiol 42:4974–4976. doi: 10.1128/JCM.42.11.4974-4976.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Le Polain de Waroux O, Flasche S, Prieto-Merino D, Edmunds WJ. 2014. Age-dependent prevalence of nasopharyngeal carriage of Streptococcus pneumoniae before conjugate vaccine introduction: a prediction model based on a meta-analysis. PLoS One 9:e86136. doi: 10.1371/journal.pone.0086136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Abdullahi O, Nyiro J, Lewa P, Slack M, Scott JAG. 2008. The descriptive epidemiology of Streptococcus pneumoniae and Haemophilus influenzae nasopharyngeal carriage in children and adults in Kilifi district, Kenya. Pediatr Infect Dis J 27:59–64. doi: 10.1097/INF.0b013e31814da70c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Whelan FJ, Verschoor CP, Stearns JC, Rossi L, Luinstra K, Loeb M, Smieja M, Johnstone J, Surette MG, Bowdish DME. 2014. The loss of topography in the microbial communities of the upper respiratory tract in the elderly. Ann Am Thorac Soc 11:513–521. doi: 10.1513/AnnalsATS.201310-351OC. [DOI] [PubMed] [Google Scholar]
- 9.Almeida ST, Nunes S, Santos Paulo AC, Valadares I, Martins S, Breia F, Brito-Avô A, Morais A, de Lencastre H, Sá-Leão R. 2014. Low prevalence of pneumococcal carriage and high serotype and genotype diversity among adults over 60 years of age living in Portugal. PLoS One 9:e90974. doi: 10.1371/journal.pone.0090974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nuorti JP, Butler JC, Crutcher JM, Guevara R, Welch D, Holder P, Elliott JA. 1998. An outbreak of multidrug-resistant pneumococcal pneumonia and bacteremia among unvaccinated nursing home residents. N Engl J Med 338:1861–1868. doi: 10.1056/NEJM199806253382601. [DOI] [PubMed] [Google Scholar]
- 11.Krone CL, Wyllie AL, van Beek J, Rots NY, Oja AE, Chu MLJN, Bruin JP, Bogaert D, Sanders EAM, Trzciński K. 2015. Carriage of Streptococcus pneumoniae in aged adults with influenza-like-illness. PLoS One 10:e0119875. doi: 10.1371/journal.pone.0119875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Perez-Trallero E, Marimon JM, Larruskain J, Alonso M, Ercibengoa M. 2011. Antimicrobial susceptibilities and serotypes of Streptococcus pneumoniae isolates from elderly patients with pneumonia and acute exacerbation of chronic obstructive pulmonary disease. Antimicrob Agents Chemother 55:2729–2734. doi: 10.1128/AAC.01546-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stupka JE, Mortensen EM, Anzueto A, Restrepo MI. 2009. Community-acquired pneumonia in elderly patients. Aging Health 5:763–774. doi: 10.2217/ahe.09.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Janssens J, Krause K. 2004. Pneumonia in the very old. Lancet Infect Dis 4:112–124. doi: 10.1016/S1473-3099(04)00931-4. [DOI] [PubMed] [Google Scholar]
- 15.Wroe PC, Finkelstein JA, Ray GT, Linder JA, Johnson KM, Rifas-Shiman S, Moore MR, Huang SS. 2012. Aging population and future burden of pneumococcal pneumonia in the United States. J Infect Dis 205:1589–1592. doi: 10.1093/infdis/jis240. [DOI] [PubMed] [Google Scholar]
- 16.Bosch AATM, Biesbroek G, Trzcinski K, Sanders EAM, Bogaert D. 2013. Viral and bacterial interactions in the upper respiratory tract. PLoS Pathog 9:e1003057. doi: 10.1371/journal.ppat.1003057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Regev-Yochay G, Dagan R, Raz M, Carmeli Y, Shainberg B, Derazne E, Rahav G, Rubinstein E. 2004. Association between carriage of Streptococcus pneumoniae and Staphylococcus aureus in children. JAMA 292:716–720. doi: 10.1001/jama.292.6.716. [DOI] [PubMed] [Google Scholar]
- 18.Margolis E, Yates A, Levin BR. 2010. The ecology of nasal colonization of Streptococcus pneumoniae, Haemophilus influenzae and Staphylococcus aureus: the role of competition and interactions with host's immune response. BMC Microbiol 10:59. doi: 10.1186/1471-2180-10-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shiri T, Nunes MC, Adrian PV, Van Niekerk N, Klugman KP, Madhi SA. 2013. Interrelationship of Streptococcus pneumoniae, Haemophilus influenzae and Staphylococcus aureus colonization within and between pneumococcal-vaccine naïve mother-child dyads. BMC Infect Dis 13:483. doi: 10.1186/1471-2334-13-483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Regev-Yochay G, Trzcinski K, Thompson CM, Malley R, Lipsitch M. 2006. Interference between Streptococcus pneumoniae and Staphylococcus aureus: in vitro hydrogen peroxide-mediated killing by Streptococcus pneumoniae. J Bacteriol 188:4996–5001. doi: 10.1128/JB.00317-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Quintero B, Araque M, van der Gaast-de Jongh C, Escalona F, Correa M, Morillo-Puente S, Vielma S, Hermans PWM. 2011. Epidemiology of Streptococcus pneumoniae and Staphylococcus aureus colonization in healthy Venezuelan children. Eur J Clin Microbiol Infect Dis 30:7–19. doi: 10.1007/s10096-010-1044-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Murphy TF. 2006. Otitis media, bacterial colonization, and the smoking parent. Clin Infect Dis 42:904–906. doi: 10.1086/500942. [DOI] [PubMed] [Google Scholar]
- 23.Chien YW, Vidal JE, Grijalva CG, Bozio C, Edwards KM, Williams JV, Griffin MR, Verastegui H, Hartinger SM, Gil AI, Lanata CF, Klugman KP. 2012. Density interactions between Streptococcus pneumoniae, Haemophilus influenzae and Staphylococcus aureus in the nasopharynx of young Peruvian children. Pediatr Infect Dis J 32:72–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tikhomirova A, Kidd SP. 2013. Haemophilus influenzae and Streptococcus pneumoniae: living together in a biofilm. Pathog Dis 69:114–126. doi: 10.1111/2049-632X.12073. [DOI] [PubMed] [Google Scholar]
- 25.Puchta A, Verschoor CP, Thurn T, Bowdish DME. 2014. Characterization of inflammatory responses during intranasal colonization with Streptococcus pneumoniae. J Vis Exp 2014:e50490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Puchta A, Naidoo A, Verschoor CP, Loukov D, Thevaranjan N, Mandur TS, Nguyen PS, Jordana M, Loeb M, Xing Z, Kobzik L, Larche MJ, Bowdish DM. 2015. TNF drives monocyte dysfunction with age and results in impaired anti-pneumococcal immunity. PLoS Pathog 12:e1005368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Siegel SJ, Tamashiro E, Weiser JN. 2015. Clearance of pneumococcal colonization in infants is delayed through altered macrophage trafficking. PLoS Pathog 11:e1005004. doi: 10.1371/journal.ppat.1005004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dorrington MG, Roche AM, Chauvin SE, Tu Z, Mossman KL, Weiser JN, Bowdish DME. 2013. MARCO is required for TLR2- and Nod2-mediated responses to Streptococcus pneumoniae and clearance of pneumococcal colonization in the murine nasopharynx. J Immunol 190:250–258. doi: 10.4049/jimmunol.1202113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stearns JC, Davidson CJ, McKeon S, Whelan FJ, Fontes ME, Schryvers AB, Bowdish DME, Kellner JD, Surette MG. 2015. Culture and molecular-based profiles show shifts in bacterial communities of the upper respiratory tract that occur with age. ISME J 9:1246–1259. doi: 10.1038/ismej.2014.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bartram AK, Lynch MDJ, Stearns JC, Moreno-Hagelsieb G, Neufeld JD. 2011. Generation of multimillion-sequence 16S rRNA gene libraries from complex microbial communities by assembling paired-end Illumina reads. Appl Environ Microbiol 77:3846–3852. doi: 10.1128/AEM.02772-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Martin M. 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J 17:10. doi: 10.14806/ej.17.1.200. [DOI] [Google Scholar]
- 32.Masella AP, Bartram AK, Truszkowski JM, Brown DG, Neufeld JD. 2012. PANDAseq: paired-end assembler for Illumina sequences. BMC Bioinformatics 13:31. doi: 10.1186/1471-2105-13-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ye Y. 2010. Identification and quantification of abundant species from pyrosequences of 16S rRNA by consensus alignment. Proc IEEE Int Conf Bioinformatics Biomed 2010:153–157. doi: 10.1109/BIBM.2010.5706555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Carvalho MDGS, Tondella ML, McCaustland K, Weidlich L, McGee L, Mayer LW, Steigerwalt A, Whaley M, Facklam RR, Fields B, Carlone G, Ades EW, Dagan R, Sampson JS. 2007. Evaluation and improvement of real-time PCR assays targeting lytA, ply, and psaA genes for detection of pneumococcal DNA. J Clin Microbiol 45:2460–2466. doi: 10.1128/JCM.02498-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sauder LA, Engel K, Stearns JC, Masella AP, Pawliszyn R, Neufeld JD. 2011. Aquarium nitrification revisited: Thaumarchaeota are the dominant ammonia oxidizers in freshwater aquarium biofilters. PLoS One 6:e23281. doi: 10.1371/journal.pone.0023281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McMurdie PJ, Holmes S. 2014. Waste not, want not: why rarefying microbiome data is inadmissible. PLoS Comput Biol 10:e1003531. doi: 10.1371/journal.pcbi.1003531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Park H, Shin JW, Park S-G, Kim W. 2014. Microbial communities in the upper respiratory tract of patients with asthma and chronic obstructive pulmonary disease. PLoS One 9:e109710. doi: 10.1371/journal.pone.0109710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bogaert D, De Groot R, Hermans PWM. 2004. Streptococcus pneumoniae colonisation: the key to pneumococcal disease. Lancet Infect Dis 4:144–154. doi: 10.1016/S1473-3099(04)00938-7. [DOI] [PubMed] [Google Scholar]
- 40.Ridda I, Macintyre CR, Lindley R, McIntyre PB, Brown M, Oftadeh S, Sullivan J, Gilbert GL. 2010. Lack of pneumococcal carriage in the hospitalised elderly. Vaccine 28:3902–3904. doi: 10.1016/j.vaccine.2010.03.073. [DOI] [PubMed] [Google Scholar]
- 41.Ahmed AE, Nicholson KG, Nguyen-Van-Tam JS. 1995. Reduction in mortality associated with influenza vaccine during 1989-90 epidemic. Lancet 346:591–595. doi: 10.1016/S0140-6736(95)91434-X. [DOI] [PubMed] [Google Scholar]
- 42.File TM. 2003. Community-acquired pneumonia. Lancet 362:1991–2001. doi: 10.1016/S0140-6736(03)15021-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Polverino E, Dambrava P, Cillóniz C, Balasso V, Marcos MA, Esquinas C, Mensa J, Ewig S, Torres A. 2010. Nursing home-acquired pneumonia: a 10 year single-centre experience. Thorax 65:354–359. doi: 10.1136/thx.2009.124776. [DOI] [PubMed] [Google Scholar]
- 44.Rudnick W, Liu Z, Shigayeva A, Low DE, Green K, Plevneshi A, Devlin R, Downey J, Katz K, Kitai I, Krajden S, Ostrowska K, Richardson D, Richardson S, Sarabia A, Silverman M, Simor AE, Tyrrell G, McGeer A. 2013. Pneumococcal vaccination programs and the burden of invasive pneumococcal disease in Ontario, Canada, 1995-2011. Vaccine 31:5863–5871. doi: 10.1016/j.vaccine.2013.09.049. [DOI] [PubMed] [Google Scholar]
- 45.Leventer-Roberts M, Feldman BS, Brufman I, Cohen-Stavi CJ, Hoshen M, Balicer RD. 2015. Effectiveness of 23-valent pneumococcal polysaccharide vaccine against invasive disease and hospital-treated pneumonia among people aged ≥65 years: a retrospective case-control study. Clin Infect Dis 60:1472–1480. [DOI] [PubMed] [Google Scholar]
- 46.Bogaert D, Keijser B, Huse S, Rossen J, Veenhoven R, van Gils E, Bruin J, Montijn R, Bonten M, Sanders E. 2011. Variability and diversity of nasopharyngeal microbiota in children: a metagenomic analysis. PLoS One 6:e17035. doi: 10.1371/journal.pone.0017035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lijek RS, Luque SL, Liu Q, Parker D, Bae T, Weiser JN. 2012. Protection from the acquisition of Staphylococcus aureus nasal carriage by cross-reactive antibody to a pneumococcal dehydrogenase. Proc Natl Acad Sci U S A 109:13823–13828. doi: 10.1073/pnas.1208075109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lijek RS, Weiser JN. 2012. Co-infection subverts mucosal immunity in the upper respiratory tract. Curr Opin Immunol 24:417–423. doi: 10.1016/j.coi.2012.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lysenko ES, Lijek RS, Brown SP, Weiser JN. 2010. Within-host competition drives selection for the capsule virulence determinant of streptococcus pneumoniae. Curr Biol 20:1222–1226. doi: 10.1016/j.cub.2010.05.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Marks LR, Davidson BA, Knight PR, Hakansson AP. 2013. Interkingdom signaling induces Streptococcus pneumoniae biofilm dispersion and transition from asymptomatic colonization to disease. mBio 4:1–13. doi: 10.3391/mbi.2013.4.1.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lysenko ES, Ratner AJ, Nelson AL, Weiser JN. 2005. The role of innate immune responses in the outcome of interspecies competition for colonization of mucosal surfaces. PLoS Pathog 1:0003–0011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hathaway LJ, Bättig P, Reber S, Rotzetter JU, Aebi S, Hauser C, Heller M, Kadioglu A, Mühlemann K. 2014. Streptococcus pneumoniae detects and responds to foreign bacterial peptide fragments in its environment. Open Biol 4:130224. doi: 10.1098/rsob.130224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cremers AJ, Zomer AL, Gritzfeld JF, Ferwerda G, van Hijum SA, Ferreira DM, Shak JR, Klugman KP, Boekhorst J, Timmerman HM, de Jonge MI, Gordon SB, Hermans PW. 2014. The adult nasopharyngeal microbiome as a determinant of pneumococcal acquisition. Microbiome 2:44. doi: 10.1186/2049-2618-2-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gao Z, Kang Y, Yu J, Ren L. 2014. Human pharyngeal microbiome may play a protective role in respiratory tract infections. Genomics Proteomics Bioinformatics 12:144–150. doi: 10.1016/j.gpb.2014.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lemon KP, Klepac-Ceraj V, Schiffer HK, Brodie EL, Lynch SV, Kolter R. 2010. Comparative analyses of the bacterial microbiota of the human nostril and oropharynx. mBio 1:e00129-10. doi: 10.1128/mBio.00129-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pei Z, Bini EJ, Yang L, Zhou M, Francois F, Blaser MJ. 2004. Bacterial biota in the human distal esophagus. Proc Natl Acad Sci U S A 101:4250–4255. doi: 10.1073/pnas.0306398101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Spor A, Koren O, Ley R. 2011. Unravelling the effects of the environment and host genotype on the gut microbiome. Nat Rev Microbiol 9:279–290. doi: 10.1038/nrmicro2540. [DOI] [PubMed] [Google Scholar]
- 58.Hibbing ME, Fuqua C, Parsek MR, Peterson SB. 2010. Bacterial competition: surviving and thriving in the microbial jungle. Nat Rev Microbiol 8:15–25. doi: 10.1038/nrmicro2259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.O'Sullivan O, Coakley M, Lakshminarayanan B, Conde S, Claesson MJ, Cusack S, Fitzgerald AP, O'Toole PW, Stanton C, Ross RP. 2013. Alterations in intestinal microbiota of elderly Irish subjects post-antibiotic therapy. J Antimicrob Chemother 68:214–221. doi: 10.1093/jac/dks348. [DOI] [PubMed] [Google Scholar]
- 60.Claesson MJ, Jeffery IB, Conde S, Power SE, O'Connor EM, Cusack S, Harris HMB, Coakley M, Lakshminarayanan B, O'Sullivan O, Fitzgerald GF, Deane J, O'Connor M, Harnedy N, O'Connor K, O'Mahony D, van Sinderen D, Wallace M, Brennan L, Stanton C, Marchesi JR, Fitzgerald AP, Shanahan F, Hill C, Ross RP, O'Toole PW. 2012. Gut microbiota composition correlates with diet and health in the elderly. Nature 488:178–184. doi: 10.1038/nature11319. [DOI] [PubMed] [Google Scholar]
- 61.Claesson MJ, Cusack S, O'Sullivan O, Greene-Diniz R, de Weerd H, Flannery E, Marchesi JR, Falush D, Dinan T, Fitzgerald G, Stanton C, van Sinderen D, O'Connor M, Harnedy N, O'Connor K, Henry C, O'Mahony D, Fitzgerald AP, Shanahan F, Twomey C, Hill C, Ross RP, O'Toole PW. 2011. Composition, variability, and temporal stability of the intestinal microbiota of the elderly. Proc Natl Acad Sci U S A 108(Suppl):4586–4591. doi: 10.1073/pnas.1000097107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Krone CL, Biesbroek G, Trzciński K, Sanders EAM, Bogaert D. 2014. Respiratory microbiota dynamics following Streptococcus pneumoniae acquisition in young and elderly mice. Infect Immun 82:1725–1731. doi: 10.1128/IAI.01290-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lexau CA, Lynfield R, Danila R, Pilishvili T, Facklam R, Farley MM, Harrison LH, Schaffner W, Reingold A, Bennett NM, Hadler J, Cieslak PR, Whitney CG. 2005. Changing epidemiology of invasive pneumococcal disease among older adults in the era of pediatric pneumococcal conjugate vaccine. JAMA 294:2043–2051. doi: 10.1001/jama.294.16.2043. [DOI] [PubMed] [Google Scholar]
- 64.El-Solh AA, Sikka P, Ramadan F, Davies J. 2001. Etiology of severe pneumonia in the very elderly. Am J Respir Crit Care Med 163:645–651. doi: 10.1164/ajrccm.163.3.2005075. [DOI] [PubMed] [Google Scholar]
- 65.Palmer LB, Albulak K, Fields S, Filkin AM, Simon S, Smaldone GC. 2001. Oral clearance and pathogenic oropharyngeal colonization in the elderly. Am J Respir Crit Care Med 164:464–468. doi: 10.1164/ajrccm.164.3.2008149. [DOI] [PubMed] [Google Scholar]
- 66.Hinojosa E, Boyd AR, Orihuela CJ. 2009. Age-associated inflammation and Toll-like receptor dysfunction prime the lungs for pneumococcal pneumonia. J Infect Dis 200:546–554. doi: 10.1086/600870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cremers AJH, Lut J, Hermans PWM, Meis JF, de Jonge MI, Ferwerda G. 2014. Avidity of antibodies against infecting pneumococcal serotypes increases with age and severity of disease. Clin Vaccine Immunol 21:904–907. doi: 10.1128/CVI.00147-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nicoletti C, Borghesi-Nicoletti C, Yang XH, Schulze DH, Cerny J. 1991. Repertoire diversity of antibody response to bacterial antigens in aged mice. II. Phosphorylcholine-antibody in young and aged mice differ in both VH/VL gene repertoire and in specificity. J Immunol 147:2750–2755. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.