Abstract
Coxiella burnetii replicates within permissive host cells by employing a Dot/Icm type IV secretion system (T4SS) to translocate effector proteins that direct the formation of a parasitophorous vacuole. C57BL/6 mouse macrophages restrict the intracellular replication of the C. burnetii Nine Mile phase II (NMII) strain. However, eliminating Toll-like receptor 2 (TLR2) permits bacterial replication, indicating that the restriction of bacterial replication is immune mediated. Here, we examined whether additional innate immune pathways are employed by C57BL/6 macrophages to sense and restrict NMII replication. In addition to the known role of TLR2 in detecting and restricting NMII infection, we found that TLR4 also contributes to cytokine responses but is not required to restrict bacterial replication. Furthermore, the TLR signaling adaptors MyD88 and Trif are required for cytokine responses and restricting bacterial replication. The C. burnetii NMII T4SS translocates bacterial products into C57BL/6 macrophages. However, there was little evidence of cytosolic immune sensing of NMII, as there was a lack of inflammasome activation, T4SS-dependent cytokine responses, and robust type I interferon (IFN) production, and these pathways were not required to restrict bacterial replication. Instead, endogenous tumor necrosis factor (TNF) produced upon TLR sensing of C. burnetii NMII was required to control bacterial replication. Therefore, our findings indicate a primary role for TNF produced upon immune detection of C. burnetii NMII by TLRs, rather than cytosolic PRRs, in enabling C57BL/6 macrophages to restrict bacterial replication.
INTRODUCTION
To initiate innate immune defense against bacterial pathogens, infected host cells utilize pattern recognition receptors (PRRs) to detect pathogen-associated molecular patterns (PAMPs) (1–3). Toll-like receptors (TLRs) located at the cell surface and within endosomes detect extracellular PAMPs such as bacterial lipoproteins and lipopolysaccharide (LPS) (4). Downstream of TLRs, the adaptor proteins MyD88 and Trif activate several signaling pathways, including NF-κB, mitogen-activated protein kinases (MAPKs), and interferon (IFN) regulatory factor 3 (IRF3), which direct the expression of proinflammatory cytokines and other antimicrobial effectors (4). For intracellular bacterial pathogens, cytosolic PRRs, such as those of the nucleotide binding domain/leucine-rich repeat (NLR) and RIG-I-like receptor (RLR) families, often are critical for host defense as they respond to PAMPs introduced into the host cell cytosol by bacterial pore-forming toxins or specialized secretion systems (5–8). In addition, cytosolic sensing can lead to the assembly of a multiprotein complex termed the inflammasome, which activates the host proteases caspase-1 and caspase-11, resulting in the release of IL-1 family cytokines and a form of cell death known as pyroptosis (9–16). These innate immune pathways collaborate to restrict intracellular bacterial infection through both cell-intrinsic and -extrinsic mechanisms (17–22). Since intracellular pathogens have evolved to manipulate or evade a specific set of host defense pathways to facilitate their intracellular lifestyle, a particular subset of innate immune sensors would be expected to be efficacious in sensing and restricting a given pathogen. However, much remains to be known about the particular innate immune pathways employed by host cells to sense and control different intracellular bacterial pathogens.
Coxiella burnetii is a facultative intracellular, Gram-negative bacterium responsible for the zoonotic disease Q (query) fever, an acute flu-like illness that can progress to a severe, chronic disease that often manifests as severe endocarditis (23). Once C. burnetii enters the host and is taken up by macrophages or other host cells, the C. burnetii-containing vacuole is directed down the canonical endosomal pathway while also engaging the autophagy and early secretory pathways to form a lysosome-derived vacuole (24–30). Within this compartment, C. burnetii is able to resist the bactericidal activities of the lysosome and replicate for several days (24, 31, 32). To establish this unique niche, C. burnetii utilizes a Dot/Icm type IVB secretion system homologous to that of its evolutionary relative, Legionella pneumophila, and translocates over 100 effector proteins into the host cytosol (33–40). These effectors are predicted to target and manipulate host cell factors, with several effectors having known roles in intracellular membrane trafficking and the inhibition of host cell death (27, 41–45).
C. burnetii exists as two phase variants. Virulent phase I C. burnetii synthesizes LPS with a highly branched O-chain, which shields the bacteria from complement-mediated killing in serum (46, 47). In contrast, phase II C. burnetii, generated by laboratory passage, produces a truncated O-antigen polysaccharide and is avirulent in immunocompetent mice and guinea pigs (48, 49). A well-defined phase II variant of the C. burnetii Nine Mile reference strain (NMII; RSA493 clone 4) contains an ∼26-kb chromosomal deletion that eliminates several LPS biosynthesis genes (48, 50, 51). The NMII strain has served as a useful model for elucidating the molecular mechanisms underlying how C. burnetii interacts with and replicates within host cells. This is in part because NMII and the isogenic phase I strain (NMI) replicate in an indistinguishable vacuole in human macrophage lines and also replicate similarly in mouse macrophage cell lines and primary human macrophages in vitro (52–54). There is no difference in the ability of NMI and NMII to stimulate the production of the cytokines tumor necrosis factor (TNF) and interleukin-6 (IL-6) from human macrophages, but NMII elicits increased immune responses in other cell types, as NMII elicits IL-1β secretion from human alveolar macrophages, increased p38 MAPK activation in mouse macrophages and human dendritic cells, and increased human dendritic cell maturation and cytokine production (54, 55). These findings suggest that NMI and NMII elicit overlapping as well as distinct immune responses from human and mouse macrophages.
Primary mouse macrophages derived from C57BL/6 (B6) mice, in contrast to macrophages from other inbred mouse strains, are not permissive for intracellular C. burnetii NMII replication (56, 57). How B6 mouse macrophages detect and restrict NMII remains poorly understood. TLR2 was found to have a major role in detecting and controlling NMII, as eliminating TLR2 expression significantly reduces cytokine responses against NMII and renders B6 macrophages more permissive for NMII replication (58). This initial finding indicates that innate immune-driven mechanisms underlie the ability of B6 macrophages to restrict NMII replication and suggests that B6 macrophages can serve as a useful model for uncovering cell-intrinsic immune mechanisms that control C. burnetii NMII infection.
Whereas TLR2 is required, TLR4 is thought to be dispensable for immune responses against C. burnetii, as previous studies have shown that C. burnetii LPS is not stimulatory for TLR4 and behaves as a TLR4 antagonist in human peripheral blood mononuclear cells (PBMCs) (55, 58). However, whether there is a role for other TLRs in detecting and controlling C. burnetii NMII infection in the absence of TLR2 signaling is unknown. Similarly, whether the TLR signaling adaptors MyD88 and Trif mediate sensing and control of C. burnetii NMII also is unknown. Furthermore, C. burnetii NMII can translocate type 4 secretion system (T4SS) effectors into C57BL/6 macrophages (59), indicating that the block in bacterial replication is due to intracellular PRR sensing of T4SS-translocated products. Indeed, the cytosolic sensor NOD2 contributes to cytokine responses against C. burnetii phase I organisms, demonstrating the potential for C. burnetii to be detected by cytosolic surveillance pathways (60, 61). Whether cytosolic immune pathways detect and restrict C. burnetii NMII infection in B6 macrophages is not known.
In contrast, it is relatively well understood how C57BL/6 macrophages restrict the intracellular replication of the evolutionarily related pathogen Legionella pneumophila. L. pneumophila has evolved to infect protozoa primarily (62). Thus, it is thought that unlike C. burnetii, L. pneumophila has not evolved evasion strategies for purposely subverting mammalian innate immunity, so it has been a useful model for uncovering innate immune pathways involved in sensing and restricting intracellular pathogens (63). Whereas A/J macrophages are permissive for L. pneumophila replication, C57BL/6 macrophages restrict L. pneumophila replication (64). Studying how C57BL/6 macrophages sense and control L. pneumophila infection has allowed for the identification of multiple TLR- and cytosolic PRR-mediated responses involved in this process, such as the NAIP5 inflammasome and the proinflammatory cytokines TNF and type I IFNs (20, 65–70). Importantly, the activation of cytosolic PRRs involves the sensing of bacterial products, such as flagellin, that are translocated into the host cell cytosol by a functional L. pneumophila Dot/Icm T4SS (71, 72).
Here, we set out to identify whether additional TLRs or cytosolic innate immune pathways are employed by B6 macrophages to detect and restrict C. burnetii NMII. In addition to the established role for TLR2 in mediating cytokine responses and control of NMII replication, we find that TLR2 and TLR4 work in concert to mediate cytokine responses to NMII, but TLR4 does not significantly contribute to the control of NMII replication. Furthermore, we find that the TLR signaling adaptors MyD88 and Trif both are required for cytokine responses and to restrict bacterial replication. Although the C. burnetii NMII T4SS translocates bacterial products into B6 macrophages, we did not observe the T4SS-dependent production of cytokines, type I IFNs, or inflammasome activation, and these immune factors were not required to restrict bacterial replication. Instead, we find that TNF produced by infected macrophages upon TLR-mediated detection of NMII is essential for allowing macrophages to restrict bacterial replication, and that decreased TNF production accounts for the increased susceptibility of TLR2-deficient macrophages to NMII infection. These findings suggest that unlike the case for L. pneumophila, TLR sensing and subsequent TNF production, rather than cytosolic PRR-mediated responses, is a major mechanism used by B6 mouse macrophages to restrict C. burnetii NMII infection.
MATERIALS AND METHODS
Ethics statement.
All experiments performed in this study involving mice, used solely as a tissue source, were done in accordance with the Animal Welfare Act (AWA) and the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health (73). The Institutional Animal Care and Use Committee of the University of Pennsylvania approved all procedures (protocols 803465, 803459, 804714, and 804928).
Bacterial strains.
Coxiella burnetii Nine Mile phase II (plaque-purified clone 4; RSA 439) (50) and Legionella pneumophila serogroup 1-derived strains were used in all experiments (74). For C. burnetii infections, acidified citrate cysteine medium (ACCM-2) (75) was inoculated with wild-type (WT) C. burnetii, WT C. burnetii expressing mCherry (76), an icmL::Tn strain (33) that contains a transposon insertion in the icmL gene rendering the T4SS nonfunctional, and WT and icmL::Tn strains harboring plasmids carrying blaM alone or blaM fused to the gene encoding the known T4SS substrate CBU_0077 under the control of the CBU_1169 promoter (33). All NMII strains were grown at 37°C in 5% CO2 and 2.5% O2 for 6 days to late log phase (∼1.0 × 109 bacteria/ml). One day after inoculation, kanamycin (275 μg/ml) was added to icmL::Tn cultures and chloramphenicol (3 μg/ml) was added to strains harboring plasmids encoding BlaM or BlaM-0077 fusion proteins (30). To quantify C. burnetii genome equivalents (GEs) for infection of macrophages, C. burnetii genomic DNA was isolated with the illustra bacterial genomic prep mini spin kit (GE Healthcare), and genomic equivalents were measured by quantitative PCR (qPCR) of the C. burnetii dotA gene using SYBR green, the CFX96 qPCR machine (Bio-Rad Laboratories), and the following primers: dotA 5′ (GCGCAATACGCTCAATCACA) and dotA 3′ (CCATGGCCCCAATTCTCT). The L. pneumophila ΔdotA mutant (74), ΔflaA mutant (72), and ΔdotA and ΔflaA strains harboring pSS128, encoding a BlaM-RalF fusion protein (77) on the Lp02 (thyA) background, which is a thymidine auxotroph derived from strain Lp01, were cultured as a heavy patch on charcoal yeast extract agar containing thymidine for 48 h at 37°C. Lp02 strains harboring the BlaM reporter proteins were grown in the presence of chloramphenicol (6.75 μg/ml).
Mammalian cell culture.
Bone marrow was isolated from the femurs, tibia, and humeri of C57BL/6 (Jackson), Tlr2−/− (78), Tlr4−/− (79), Tlr2−/− Tlr4−/−, Myd88−/− (80), Trif−/− (81), Myd88−/− Trif−/−, Tnf−/− (82), Casp1−/− Casp11−/− (83), and Ifnar−/− (84) mice. Bone marrow cells were differentiated into macrophages for 7 to 8 days in RPMI containing 30% L929 cell supernatant and 20% fetal bovine serum (FBS) at 37°C in a humidified incubator. Macrophages were lifted with ice-cold phosphate-buffered saline (PBS) plus 2 mM EDTA and replated 1 day prior to infection in RPMI containing 15% L929 cell supernatant and 10% FBS. Macrophages were plated into 48- or 24-well plates at 1.5 × 105 or 2.0 × 105 cells per well, respectively. Macrophages were infected at various multiplicities of infection (MOIs) of C. burnetii NMII and L. pneumophila as indicated. C. burnetii NMII and L. pneumophila bacteria were washed once with PBS prior to infection of macrophages. After infection, cells were spun at 1,200 rpm for 5 min prior to incubation at 37°C. At designated time points, macrophage supernatants, whole-cell lysates, or mRNA was collected. For experiments involving the treatment of B6 BMDMs with purified C. burnetii NMII LPS, C. burnetii NMII LPS was isolated as previously described (58). B6 BMDMs were treated with C. burnetii LPS at 100 ng/ml or 1,000 ng/ml or Escherichia coli LPS at 1 or 10 ng/ml for 16 h. For experiments examining LPS antagonism, B6 BMDMs were pretreated with C. burnetii LPS at 100 ng/ml for 1 h prior to the addition of E. coli LPS at 10 ng/ml for 16 h. Supernatants were harvested and TNF was measured by enzyme-linked immunosorbent assay (ELISA).
Immunoblot analysis.
Cell lysates were collected with 1× SDS-PAGE sample buffer or supernatants were mixed 1:1 with 2× SDS-PAGE buffer containing protease and phosphatase inhibitors. Samples were boiled for 5 min, separated by SDS-PAGE, and transferred to Immobilon P membranes (Millipore). Primary antibodies specific for phospho-p38 MAPK (Cell Signaling Technology), total p38 MAPK (Cell Signaling Technology), caspase-1 p10 (Santa Cruz Biotechnology), IL-1β (R&D Systems), and β-actin (Sigma) were used. Detection was performed with horseradish peroxidase (HRP)-conjugated anti-rabbit antibodies (Cell Signaling Technology), anti-mouse antibodies (Cell Signaling Technology), or anti-rat antibodies (Jackson ImmunoResearch).
ELISA.
Harvested supernatants from infected macrophages were assayed for cytokine levels using capture and detection antibodies specific for TNF, IL-6, IL-1α, and IL-1β (BD Biosciences) according to the manufacturer's guidelines.
Cytotoxicity assay.
Cells were infected with WT C. burnetii NMII for 24 h or treated with 0.5 μg/ml LPS for 4 h and 2.5 mM ATP for 1 h. Supernatants were harvested and used to measure percent cytotoxicity via lactate dehydrogenase (LDH) release using the LDH cytotoxicity assay kit (Clontech).
Measuring intracellular Coxiella burnetii replication.
Measuring intracellular replication of C. burnetii in C57BL/6, Tlr2−/−, Tlr4−/−, Tlr2−/− Tlr4−/−, Myd88−/−, Trif−/−, Myd88−/− Trif−/−, Tnf−/−, or Ifnar−/− BMDMs was performed as follows. Macrophages were infected with C. burnetii Nine Mile phase II expressing mCherry at an MOI of 100 or 500 in 24-well plates. Twenty-four hours postinfection, cells were washed 3 times with PBS to remove extracellular bacteria and fresh medium was added to the cells every 2 days. At designated time points postinfection, the extracellular medium was collected and the adherent macrophages were lysed with 1 ml sterile distilled water. The lysed samples then were combined with the extracellular medium from the same well. Bacterial genomic DNA was purified and C. burnetii GEs were measured by qPCR using primers specific for the C. burnetii dotA gene. The fold change in bacterial GEs was calculated as the ratio of the number of GEs measured on a given day to the number of GEs measured on day 1. For experiments examining the role of TNF in restricting C. burnetii growth, 10 ng/ml recombinant mouse TNF (BioLegend) or supernatants from B6 macrophages infected with C. burnetii for 24 h were added to C. burnetii-infected macrophages on day 1 and day 5 postinfection.
Enumeration of Coxiella burnetii-containing vacuoles by microscopy.
A total of 2.0 × 105 cells were plated onto a glass coverslip per well in triplicate in 24-well plates for each condition. Seven days postinfection, cells were washed with PBS, stained with 4′,6-diamidino-2-phenylindole (DAPI), and fixed with 4% paraformaldehyde. Images were taken with a Nikon Eclipse 2000E-U epifluorescence microscope, and images were acquired with NIS Elements B4 4.10.01 software. Large vacuoles (>5 μM) containing mCherry-expressing C. burnetii were enumerated as a percentage of total DAPI-positive cells. For each coverslip, greater than 300 cells were counted.
Quantitative RT-PCR.
RNA was isolated from infected macrophages using the RNeasy minikit and DNase treated using the RNase-free DNase set (Qiagen) to remove contaminating genomic DNA. The isolated RNA then was reverse transcribed into cDNA using Superscript II reverse transcriptase (Invitrogen). Relative mRNA abundance was measured by qPCR using SYBR green and the CFX96 qPCR machine (Bio-Rad Laboratories). The following primer pairs were used: Ifnb 5′ (GCACTGGGTGGAATGAGACTATTG) and Ifnb 3′ (TTCTGAGGCATCAACTGACAGGTC), Ifna4 5′ (CCCACAGCCCAGAGAGTGACC) and Ifna4 3′ (GGCCCTCTTGTTCCCGAGGT), Nos2 5′ (ACATCGACCCGTCCAGTAT) and Nos2 3′ (CAGAGGGGTAGGCTTGTCT), and Hprt 5′ (GTTGGATACAGGCCAGACT) and Hprt 3′ (GAGGGTAGGCTGGCCTAT). To calculate relative fold induction using the ΔΔCT method (85), the cycle threshold (CT) of a given gene was normalized to the hypoxanthine phosphoribosyltransferase CT and compared to the normalized CT in uninfected cells.
Viral proliferation bioassay.
The viral proliferation bioassay was performed as previously described (86), with some modifications, as follows. Briefly, C57BL/6 macrophages were mock infected, treated with poly(I·C) (Invivogen), or infected with the L. pneumophila ΔflaA strain at an MOI of 5 or with WT C. burnetii NMII at an MOI of 50 for 24 h. Supernatants were collected and UV inactivated by exposure to 600 mJ · cm−2 UVA light in a Stratalinker 1800 (Stratagene). L2 mouse fibroblasts were treated with UV-inactivated supernatants for 24 h and then infected with Newcastle disease virus expressing green fluorescent protein (NDV-GFP) at an MOI of 1 PFU/cell. At 24 h postinfection, cells were fixed, stained with DAPI, and examined under an Eclipse TE2000-U epifluorescence microscope (Nikon Instruments, Inc.). Images were acquired using NIS Elements B4 4.10.01 software (Nikon Instruments, Inc.). Mean GFP fluorescence intensity was determined using ImageJ, with background fluorescence determined from mock virus-infected samples. For each sample, the means from three random fields of equal area were quantified. Images and quantification are representative of two independent experiments.
Measurement of nitric oxide.
Macrophages were infected at the indicated MOIs in 96-well plates at 1.5 × 105 cells per well in 150 μl media. At the indicated time points, nitrite (NO2−) levels were measured with the Griess reagent system according to the manufacturer's guidelines (Promega).
T4SS effector translocation assay.
T4SS-mediated translocation of BlaM-CBU_0077 fusion proteins into B6 BMDMs was assayed as follows. Briefly, 5.0 × 105 BMDMs were seeded into 12-well non-tissue culture-coated dishes 1 day prior to infection. Cells were infected with WT or icmL NMII strains expressing BlaM-CBU_0077 at the indicated MOIs. At 16 h postinfection, cells were loaded with the fluorescent substrate CCF4-AM for 90 min at room temperature using the LiveBLAzer fluorescent resonance energy transfer (FRET) B/G loading kit with 15 mM probenecid (Thermo Fisher Scientific). Cells then were analyzed using a BD LSRII flow cytometer (BD Biosciences) and FlowJo software.
Statistical analysis.
The plotting of data and statistical analysis were performed using GraphPad Prism software. Statistical significance was determined using unpaired, two-tailed Student's t test or one-way analysis of variance (ANOVA) with Tukey's posttest. Differences were considered statistically significant if the P value was <0.05.
RESULTS
Both TLR2 and TLR4 mediate cytokine responses to Coxiella burnetii Nine Mile phase II in C57BL/6 macrophages.
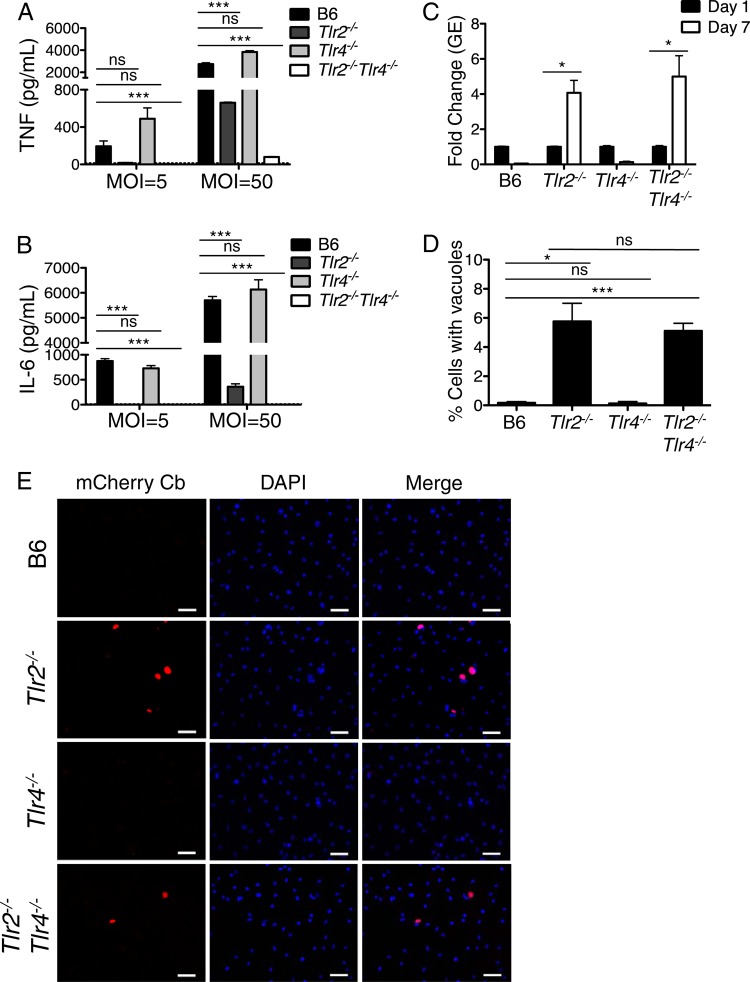
C57BL/6 (B6) bone marrow-derived macrophages (BMDMs) do not permit the robust intracellular replication of C. burnetii NMII (56). Restriction of NMII replication is mediated in part by TLR2 signaling (58), and TLR2 is required for the production of proinflammatory cytokines by human PBMCs in response to C. burnetii phase I infection and mouse macrophages in response to both C. burnetii phase I and phase II (58, 61). However, the roles of other TLRs or the signaling adaptor MyD88 or Trif in enabling B6 macrophages to restrict NMII replication or mount an effective cytokine response are largely unknown. C. burnetii lipid A is thought to be poorly stimulatory for TLR4 and is capable of behaving as a TLR4 antagonist in human PBMCs (58). Abolishing TLR4 alone does not impact cytokine responses to phase I or II infection in vitro, and the absence of TLR4 does not affect NMII replication within mouse macrophages (58, 61). Given that TLR2 responds so robustly to NMII, we wondered whether a role for TLR4 could be revealed in the absence of TLR2. Thus, we infected B6, Tlr2−/−, Tlr4−/−, and Tlr2−/− Tlr4−/− BMDMs with C. burnetii NMII and measured secreted levels of TNF and IL-6 at 24 h postinfection. We observed a significant reduction in TNF and IL-6 production in infected Tlr2−/− BMDMs and no difference in cytokine production in infected Tlr4−/− BMDMs, consistent with previously published studies (Fig. 1A and B) (58). However, BMDMs lacking TLR2 still produced substantial amounts of TNF and IL-6 following NMII infection at an MOI of 50. In contrast, BMDMs lacking both TLR2 and TLR4 did not produce detectable amounts of TNF and IL-6 following NMII infection at an MOI of 50, indicating a role for TLR4 in sensing NMII infection that is unmasked in the absence of TLR2.
FIG 1.
TLR2 and TLR4 mediate immune responses to Coxiella burnetii Nine Mile II in C57BL/6 macrophages. (A and B) C57BL/6, Tlr2−/−, Tlr4−/−, and Tlr2−/− Tlr4−/− bone marrow-derived macrophages (BMDMs) were infected with WT C. burnetii NMII at an MOI of 5 or 50 for 24 h. Levels of TNF and IL-6 in the supernatants were measured by ELISA. Graphs show the means ± standard errors of the means (SEM) from triplicate wells. Results are representative of three independent experiments. (C) C57BL/6, Tlr2−/−, Tlr4−/−, and Tlr2−/− Tlr4−/− BMDMs were infected with WT C. burnetii NMII at an MOI of 100. At days 1 and 7 postinfection, bacterial uptake and replication were measured as genomic equivalents (GEs) by qPCR. Graphs show the fold change in GEs on day 7 relative to GEs on day 1 ± SEM from triplicate wells. (D) Seven days postinfection, BMDMs of the indicated genotypes infected with mCherry-expressing WT C. burnetii NMII at an MOI of 100 were fixed, stained with DAPI, and examined by fluorescence microscopy. The number of large mCherry-expressing NMII-containing vacuoles greater than 5 μm in size was determined and calculated as a percentage of the total cell number. Graphs show the mean percentage of cells containing large NMII vacuoles ± SEM from triplicate coverslips. At least 300 cells were counted per coverslip. Results are representative of two independent experiments. (E) Representative fluorescence micrographs of mouse BMDMs of the indicated genotypes infected with mCherry-expressing C. burnetii (Cb) NMII at an MOI of 100 and fixed and stained with DAPI on day 7 postinfection. Images were taken at ×40 magnification. Scale bars represent 25 μm. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, no significance.
To test if TLR4 collaborates with TLR2 to restrict C. burnetii NMII replication, we infected B6, Tlr2−/−, Tlr4−/−, and Tlr2−/− Tlr4−/− BMDMs with C. burnetii expressing mCherry. Similar levels of bacterial uptake were observed for the different genotypes of mouse macrophages at day 1 postinfection (see Fig. S1A in the supplemental material). At day 7 postinfection, we measured intracellular replication (Fig. 1C) and the percentage of cells containing large NMII-containing vacuoles greater than 5 μm in diameter (Fig. 1D and E). In agreement with previous findings (58), B6 and Tlr4−/− BMDMs limited bacterial replication, whereas TLR2-deficient BMDMs were more permissive for intracellular C. burnetii replication (Fig. 1C to E). Tlr2−/− Tlr4−/− BMDMs did not exhibit increased C. burnetii replication or formation of large vacuoles compared to Tlr2−/− BMDMs (Fig. 1C to E), suggesting that although the concomitant deletion of TLR2 and TLR4 significantly decreases cytokine responses to NMII, it does not enhance NMII replication within mouse macrophages.
As our data suggested that TLR4 contributes to cytokine responses against C. burnetii NMII, we next asked whether this response could be attributed to TLR4 detecting NMII LPS. Previous findings with human PBMCs showed that purified NMII LPS is poorly stimulatory and can antagonize responses to E. coli LPS (58). Whether purified NMII LPS behaves similarly with C57BL/6 macrophages is unknown. Treatment of B6 macrophages with NMII LPS at 100 ng/ml or 1,000 ng/ml did not induce detectable TNF production, whereas treatment with Escherichia coli LPS at doses as low as 1 and 10 ng/ml elicited robust TNF production (see Fig. S2 in the supplemental material). Pretreatment of B6 BMDMs with 100 ng/ml NMII LPS antagonized 10 ng/ml E. coli LPS, as measured by significantly decreased TNF production (see Fig. S2), in agreement with previous findings in human PBMCs (58). These data show that purified NMII LPS alone is insufficient to stimulate cytokine production in C57BL/6 macrophages, indicating that TLR4-dependent responses to C. burnetii NMII preferentially occur during infection with intact NMII bacteria.
The TLR signaling adaptors MyD88 and Trif are required for cytokine responses and control of Coxiella burnetii Nine Mile phase II replication in C57BL/6 macrophages.
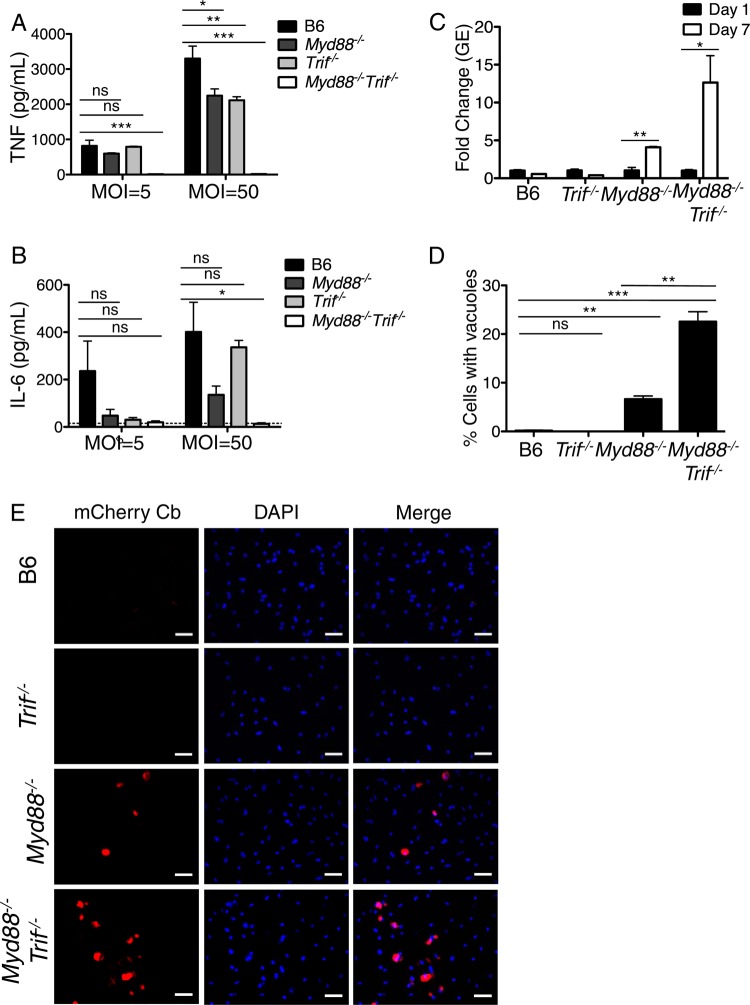
We next examined the contributions of the TLR signaling adaptors MyD88 and Trif to cytokine responses against C. burnetii NMII. Myd88−/− BMDMs produced significantly lower, but still substantial, levels of TNF and IL-6 compared to WT BMDMs following C. burnetii NMII infection at an MOI of 50 (Fig. 2A and B). NMII-infected Trif−/− BMDMs similarly produced significantly lower, but still substantial, levels of TNF and IL-6 compared to those of WT BMDMs. In contrast, infected Myd88−/− Trif−/− BMDMs did not produce detectable levels of TNF or IL-6, suggesting that both MyD88 and Trif signaling are required for cytokine production during C. burnetii infection. These data are consistent with the known role for TLR2 in sensing NMII, as TLR2 signaling requires MyD88, and also implicate a role for TLR4 and/or another Trif-dependent TLR in mediating cytokine responses against C. burnetii NMII.
FIG 2.
Myd88 and Trif are required for cytokine production and the restriction of intracellular Coxiella burnetii Nine Mile II replication in C57BL/6 macrophages. (A and B) C57BL/6, Trif−/−, Myd88−/−, and Myd88−/− Trif−/− BMDMs were infected with WT C. burnetii NMII at MOIs of 5 and 50 for 24 h. Levels of TNF and IL-6 in the supernatants were measured by ELISA. Graphs show the means ± SEM from triplicate wells. Results are representative of two independent experiments. (C) C57BL/6, Trif−/−, Myd88−/−, and Myd88−/− Trif−/− BMDMs were infected with WT C. burnetii NMII at an MOI of 100. At days 1 and 7 postinfection, bacterial uptake and replication were measured as genomic equivalents (GEs) by qPCR. Graphs show the fold change in GEs relative to the GEs measured on day 1 ± SEM from triplicate wells. (D) Seven days postinfection, BMDMs of the indicated genotypes infected with mCherry-expressing WT C. burnetii NMII at an MOI of 100 were fixed, stained with DAPI, and examined by fluorescence microscopy. The number of large NMII-containing vacuoles was determined and calculated as a percentage of the total cell number on day 7 postinfection. Graphs show the mean percentage of cells containing C. burnetii vacuoles ± SEM from triplicate coverslips. At least 300 cells were counted per coverslip. Results are representative of two independent experiments. (E) Representative fluorescence micrographs of BMDMs of the indicated genotypes infected with mCherry-expressing C. burnetii (Cb) NMII at an MOI of 100 and fixed and stained with DAPI on day 7 postinfection. Images were taken at ×40 magnification. Scale bars represent 25 μm. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, no significance.
To test if MyD88 and Trif are required to restrict C. burnetii NMII replication, we infected B6, Myd88−/−, Trif−/−, and Myd88−/− Trif−/− BMDMs with C. burnetii NMII expressing mCherry. Similar levels of bacterial uptake were observed for the different genotypes of BMDMs at day 1 postinfection (see Fig. S1B in the supplemental material). When we examined intracellular C. burnetii NMII replication at day 7 postinfection, we found that Myd88−/− BMDMs were significantly more permissive than B6 BMDMs, whereas Trif−/− BMDMs were not (Fig. 2C to E). BMDMs deficient in both Myd88 and Trif were more permissive than MyD88-deficient BMDMs, as measured by increases in both GEs and the number of large vacuoles (Fig. 2C to E). These data indicate that MyD88 and Trif collaborate to mediate cytokine production and restrict intracellular C. burnetii NMII replication in B6 macrophages.
Coxiella burnetii Nine Mile II T4SS activity does not enhance cytokine production in C57BL/6 macrophages.
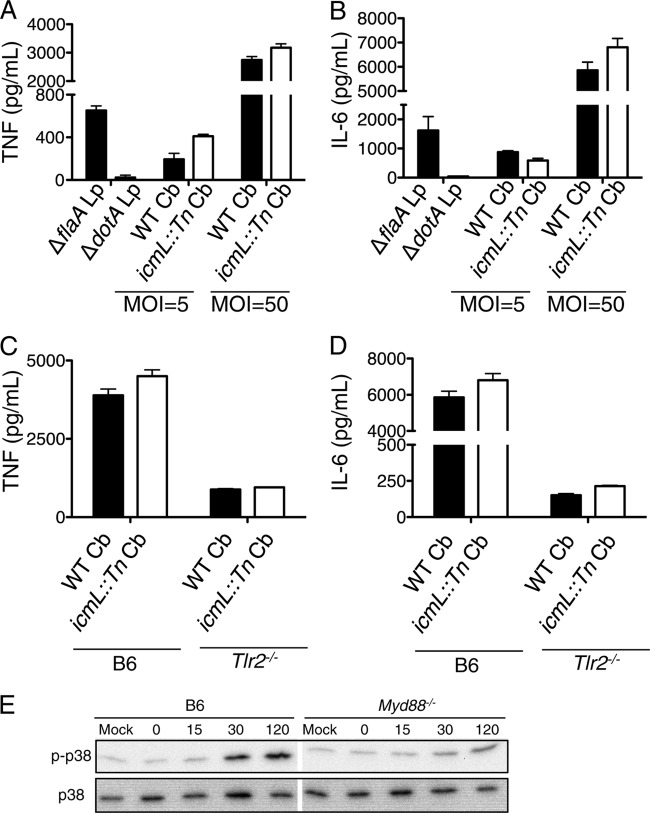
Cytosolic sensing of bacterial products translocated into the host cell cytosol by specialized secretion systems can enhance cytokine responses, as has been observed for macrophages infected with L. pneumophila expressing a functional T4SS compared to those infected with T4SS mutants (87–89). C. burnetii NMII translocates T4SS effectors into B6 macrophages (59), but whether cytosolic sensing of C. burnetii T4SS activity leads to increased cytokine production or aid in restricting bacterial replication is unknown. To test whether cytosolic sensing of C. burnetii NMII occurs in B6 macrophages, we first determined whether we could observe T4SS-dependent effector translocation into B6 macrophages under our experimental conditions. We utilized a FRET-based reporter system in which C. burnetii NMII expresses TEM-1 β-lactamase (BlaM) translationally fused to the T4SS effector CBU_0077 (33). As a positive control for T4SS-mediated translocation, we used the L. pneumophila ΔflaA mutant expressing BlaM translationally fused to the T4SS effector RalF and lacking flagellin, thereby bypassing NAIP5 inflammasome activation and cell death in B6 BMDMs (71, 72, 77). Furthermore, as C. burnetii NMII does not replicate within macrophages by 24 h postinfection whereas L. pneumophila normally replicates to large numbers by 24 h postinfection, we utilized L. pneumophila thymidine auxotroph (thyA) strains for comparison in order to avoid the confounding effects of bacterial replication. Following infection, cells were loaded with the fluorescent reagent CCF4-AM, and flow cytometry was performed to quantify the percentage of B6 BMDMs receiving T4SS-translocated effectors, as measured by increased coumarin (blue) fluorescence following BlaM-mediated cleavage of CCF4-AM. As expected, the L. pneumophila ΔflaA mutant robustly translocated BlaM-RalF into B6 BMDMs, whereas the L. pneumophila ΔdotA mutant lacking a functional Dot/Icm T4SS did not (see Fig. S3 in the supplemental material). In agreement with previous findings (59), we observed substantial CBU_0077 translocation into B6 BMDMs at 24 h postinfection, as approximately 11% or 23% of cells were positive for translocation at an MOI of 50 or 100, respectively (see Fig. S3). In contrast, the icmL::Tn mutant containing an inactivating transposon insertion in the icmL gene was unable to translocate CBU_0077 into macrophages, in agreement with previous findings that CBU_0077 translocation is T4SS dependent (see Fig. S3) (30, 53).
We next asked whether there is cytosolic sensing of C. burnetii NMII T4SS activity and enhanced cytokine production. As previously demonstrated (87), B6 BMDMs infected with the L. pneumophila ΔflaA thymidine auxotroph strain produced increased amounts of TNF and IL-6 compared to those of BMDMs infected with the L. pneumophila ΔdotA mutant (Fig. 3A and B). In contrast, we did not observe enhanced TNF or IL-6 production in B6 BMDMs infected with WT NMII compared to that of icmL::Tn mutant-infected BMDMs (Fig. 3A and B). To determine whether the failure to observe T4SS-enhanced cytokine production was due to the restriction of WT C. burnetii NMII replication by B6 BMDMs, we next examined TNF and IL-6 production in replication-permissive Tlr2−/− macrophages infected with WT or icmL::Tn NMII. Although substantial amounts of TNF and IL-6 were produced by infected Tlr2−/− BMDMs, we still observed no difference in cytokine production between Tlr2−/− BMDMs infected with the WT and those infected with the icmL::Tn NMII strain (Fig. 3C and D). These findings suggest cytosolic sensing of NMII T4SS activity does not enhance proinflammatory cytokine production in C57BL/6 macrophages.
FIG 3.
C. burnetii Nine Mile II T4SS activity does not enhance cytokine production or early p38 MAPK phosphorylation in C57BL/6 macrophages. (A and B) C57BL/6 BMDMs were infected with the L. pneumophila (Lp) ΔflaA or ΔdotA mutant at an MOI of 5 or with WT C. burnetii (Cb) or the icmL::Tn mutant NMII at an MOI of 5 or 50 for 24 h. Levels of TNF (A) and IL-6 (B) in the supernatants were measured by ELISA. (C and D) C57BL/6 and TLR2−/− BMDMs were infected with WT C. burnetii (Cb) or the icmL::Tn NMII mutant at an MOI of 50 for 24 h. Levels of TNF (C) and IL-6 (D) in the supernatants were measured by ELISA. Graphs show the means ± SEM from triplicate wells. Results are representative of three (A and B) or two (C and D) independent experiments. (E) Immunoblot analysis of phosphorylated and total p38 MAPK in cell lysates from C57BL/6 and Myd88−/− BMDMs that were mock infected or infected with WT C. burnetii NMII at an MOI of 50 for 0, 15, 30, or 120 min. Results are representative of two independent experiments.
Early p38 MAPK activation in C57BL/6 macrophages during Coxiella burnetii Nine Mile II infection is TLR dependent.
Upon the detection of PAMPs by TLRs and cytosolic PRRs, signaling events that involve phosphorylation and activation of host MAPKs contribute to the generation of a proinflammatory immune response. L. pneumophila induces rapid p38 MAPK activation within 20 min postinfection that involves both TLR signaling as well as cytosolic sensing of T4SS-translocated bacterial effectors (89, 90). p38 MAPK activation contributes to cytokine production in cells infected with C. burnetii phase I or phase II (61, 91–93), but whether p38 MAPK activation involves TLR or cytosolic sensing of C. burnetii is unknown. To test if p38 MAPK phosphorylation shortly after C. burnetii NMII infection requires TLR signaling, we infected B6 and Myd88−/− BMDMs with NMII and examined p38 MAPK phosphorylation up to 2 h postinfection (Fig. 3E). We observed robust p38 MAPK phosphorylation in infected B6 macrophages. In contrast, p38 MAPK phosphorylation was substantially attenuated in infected MyD88-deficient BMDMs, suggesting that early p38 MAPK activation in response to C. burnetii NMII is largely TLR dependent and is not robustly triggered by cytosolic immunosurveillance pathways.
Inflammasomes are not activated by Coxiella burnetii Nine Mile II and are not required to restrict bacterial replication in C57BL/6 macrophages.
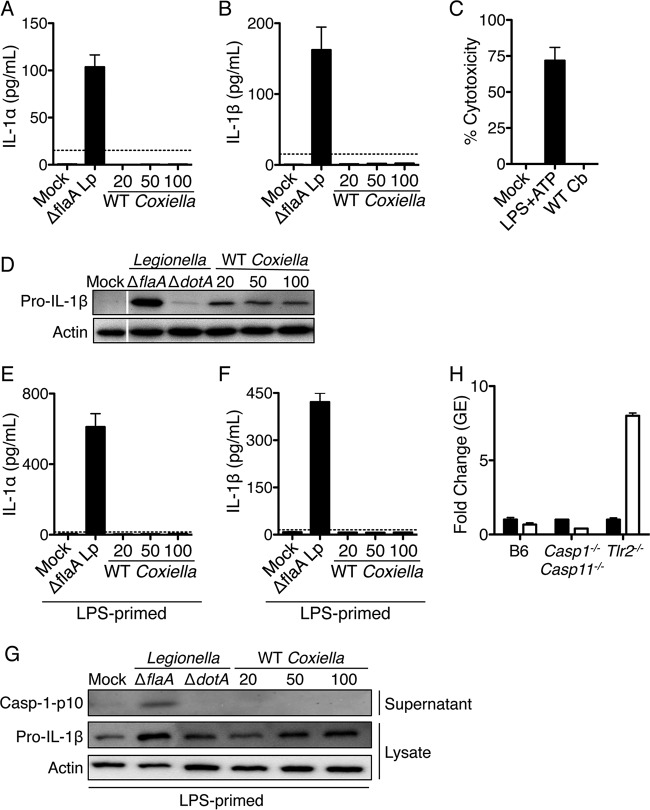
Bacterial ligands introduced into the host cell cytoplasm by virulence-associated specialized secretion systems can be sensed by cytosolic PRRs. A subset of cytosolic PRRs trigger inflammasome-dependent activation of caspase-1 and caspase-11, resulting in pyroptotic cell death and release of IL-1 family cytokines, including IL-1α and IL-1β. Infection with T4SS-expressing L. pneumophila activates multiple inflammasomes, including a flagellin-dependent NAIP5 inflammasome, an NLRP3 inflammasome that responds to an unidentified T4SS-dependent signal, and a noncanonical caspase-11 inflammasome that detects hexa-acylated or penta-acylated lipid A (20, 71, 72, 94–101). Unlike L. pneumophila, C. burnetii does not produce flagellin and would not be expected to activate the NAIP5 inflammasome (102). Furthermore, C. burnetii expresses tetra-acylated lipid A, which is thought to be nonstimulatory for caspase-11 and would not be expected to activate caspase-11 (58). Whether C. burnetii NMII activates NLRP3 or other inflammasome sensors in C57BL/6 macrophages and whether inflammasomes enable C57BL/6 macrophages to restrict NMII replication is unclear. Thus, we infected B6 BMDMs with NMII and measured the secretion of the inflammasome-dependent cytokines IL-1α and IL-1β and cell death (Fig. 4A to C). As a positive control for inflammasome activation, we infected B6 BMDMs in parallel with the L. pneumophila ΔflaA thyA mutant. This flagellin-deficient strain cannot activate NAIP5 but still activates the NLRP3 and caspase-11 inflammasomes (94, 95). As expected, infection with the L. pneumophila ΔflaA mutant at an MOI of 5 induced the robust release of both IL-1α and IL-1β. In contrast, we were unable to detect IL-1α and IL-1β in the supernatant of C. burnetii NMII-infected cells even at a high MOI of 100 at 24 h postinfection (Fig. 4A and B), despite the ability of NMII to translocate T4SS effectors into ∼24% of B6 macrophages under these conditions (see Fig. S3 in the supplemental material). Furthermore, we did not detect cell death in NMII-infected B6 BMDMs, whereas treatment of B6 BMDMs with the NLRP3 inflammasome stimulus of LPS plus ATP induced robust cell death (Fig. 4C). These findings suggest that C. burnetii NMII does not induce inflammasome activation in B6 macrophages, in agreement with a recent study (45).
FIG 4.
Coxiella burnetii Nine Mile II infection does not induce inflammasome activation in C57BL/6 macrophages. (A and B) Unprimed C57BL/6 BMDMs were mock infected, infected with the L. pneumophila ΔflaA mutant at an MOI of 5, or infected with WT C. burnetii NMII at an MOI of 20, 50, or 100 for 24 h. Levels of IL-1α (A) and IL-1β (B) were measured in the supernatants by ELISA. Graphs show the means ± SEM from triplicate wells. Results are representative of three independent experiments. (C) C57BL/6 BMDMs were treated with 0.5 μg/ml LPS for 4 h followed by 2.5 mM ATP for 1 h or infected with WT C. burnetii NMII at an MOI of 50 for 24 h. Percent cytotoxicity was measured via LDH release. The graph shows the means ± SEM from triplicate wells. (D) Immunoblot analysis of pro-IL-1β and actin in cell lysates from C57BL/6 BMDMs that were mock infected or infected with the L. pneumophila ΔflaA or ΔdotA mutant at an MOI of 5 or WT C. burnetii NMII at an MOI of 20, 50, or 100 for 24 h. Results are representative of two independent experiments. (E, F, and G) C57BL/6 BMDMs first were primed with 0.5 μg/ml LPS for 4 h and then either mock infected, infected with the L. pneumophila ΔflaA mutant at an MOI of 5, or infected with WT C. burnetii at MOIs of 20, 50, and 100 for 24 h. Levels of IL-1α (E) and IL-1β (F) were measured in the supernatants by ELISA. Graphs show the means ± SEM from triplicate wells. Results are representative of two independent experiments. (G) Immunoblot analysis of processed caspase-1 (casp-1 p10) in the supernatant and pro-IL-1β and actin in the cell lysate. Results are representative of two independent experiments. (H) C57BL/6, Tlr2−/−, and Casp1−/− Casp11−/− BMDMs were infected with WT NMII C. burnetii at an MOI of 100. At days 1 (black bars) and 7 (white bars) postinfection, bacterial uptake and replication were measured as genomic equivalents (GEs) by qPCR. Graphs show the fold change in GEs on day 7 relative to GEs on day 1 ± SEM from triplicate wells.
Inflammasome activation is a two-step process that requires an initial priming signal, often TLR dependent, to upregulate the expression of pro-IL-1β and other inflammasome components, followed by a second activation signal triggered upon the cytosolic detection of bacterial PAMPs (16). C. burnetii NMII is capable of priming macrophages, as pro-IL-1β is upregulated in B6 BMDMs infected with NMII at MOIs ranging from 20 to 100 (Fig. 4D). However, the levels of pro-IL-1β induced by NMII even at an MOI of 100 were substantially lower than those in BMDMs infected with L. pneumophila at an MOI of 5, indicating that C. burnetii NMII does not prime BMDMs as robustly as L. pneumophila. Thus, it is possible that the level of priming induced by NMII infection is insufficient for inflammasome activation. To test this possibility, we preprimed B6 BMDMs with E. coli LPS for 4 h to induce the robust expression of pro-IL-1β and other inflammasome components. We then infected LPS-primed cells with the L. pneumophila ΔflaA mutant or WT C. burnetii NMII and measured secreted IL-1α and IL-1β 24 h later. Mock-infected, LPS-primed BMDMs did not release IL-1α or IL-1β into the supernatant, as LPS priming alone is insufficient for inflammasome activation, whereas L. pneumophila ΔflaA mutant-infected primed BMDMs released substantial amounts of IL-1α and IL-1β (Fig. 4E and F). In contrast, we were unable to detect secreted IL-1α or IL-1β in the supernatants of LPS-primed cells infected with C. burnetii NMII at MOIs ranging from 20 to 100 for 24 h (Fig. 4E and F), suggesting that even following LPS priming, NMII infection fails to induce inflammasome-dependent cytokine secretion. We next examined whether the failure to detect inflammasome-dependent cytokine secretion in LPS-primed, infected macrophages was due to an upstream defect in caspase-1 activation. To test this, we assayed for caspase-1 processing into a mature 10-kDa-sized fragment (p10) and its release into the supernatant of infected cells (Fig. 4G). Mock infection of LPS-primed BMDMs did not induce caspase-1 processing, whereas L. pneumophila ΔflaA mutant infection led to caspase-1 processing. In contrast, caspase-1 processing was undetectable following C. burnetii NMII infection at all MOIs tested. To determine whether we could observe inflammasome activation in mouse macrophages permissive for growth, we further measured IL-1α and IL-1β release in primed and unprimed Tlr2−/− macrophages infected with NMII at an MOI of 100 and again saw no IL-1α or IL-1β secretion, whereas the canonical inflammasome stimulus of LPS plus ATP elicited both IL-1α and IL-1β secretion (see Fig. S4A and B in the supplemental material), indicating that NMII infection of permissive Tlr2−/− macrophages does not activate the inflammasome. Moreover, despite similar levels of internalized bacteria at 24 h postinfection (see Fig. S4C), we did not detect increased replication of NMII in Casp1−/− Casp11−/− BMDMs by 7 days postinfection, whereas NMII was able to replicate within Tlr2−/− BMDMs (Fig. 4H). Collectively, these data indicate that although C. burnetii NMII infection upregulates the expression of inflammasome-related proteins, such as pro-IL-1β, C. burnetii T4SS activity does not trigger inflammasome activation in restrictive WT macrophages or permissive Tlr2−/− macrophages of the B6 background, and inflammasomes are not required to restrict intracellular NMII replication.
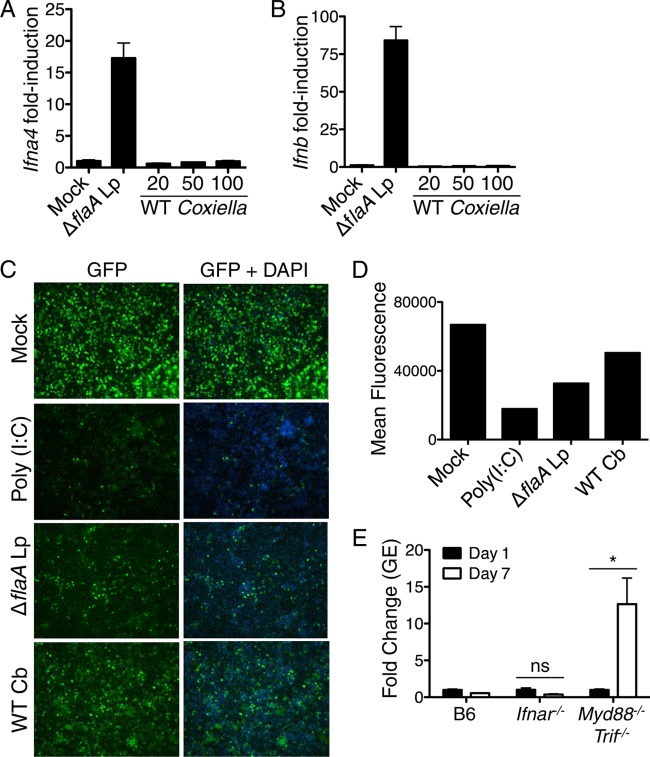
Type I interferons are not robustly induced by Coxiella burnetii Nine Mile II and are not required to restrict bacterial replication within C57BL/6 macrophages.
Many intracellular bacterial pathogens stimulate the production of type I IFN by host cells (103). The induction of type I IFNs is regulated in part by extracellular sensors, such as TLR4, as well as cytosolic sensors, such as the RNA sensors RIG-I and MDA-5, the DNA sensor cGAS, and the cyclic dinucleotide sensor STING (103–105). Type I IFNs are elicited during L. pneumophila infection and aid in restricting intracellular bacterial replication (67–70, 106–108). Whether type I IFNs are elicited by C. burnetii NMII and restrict NMII replication within B6 macrophages is unknown. To test if C. burnetii NMII activates a type I IFN response, we infected B6 BMDMs with C. burnetii NMII and measured levels of Ifna4 or Ifnb mRNAs by quantitative RT-PCR (Fig. 5A and B). We were unable to detect transcriptional induction of Ifna4 or Ifnb 16 h postinfection, suggesting that NMII does not robustly induce expression of type I IFNs. In contrast, we observed robust upregulation of the Ifna4 or Ifnb mRNAs in response to L. pneumophila ΔflaA thyA mutant infection (Fig. 5A and B). However, it is possible that undetectable but biologically active amounts of type I IFNs are produced during NMII infection. Alternatively, NMII may induce the expression of one of the other nine known type I IFNs. Therefore, we used a bioassay that detects type I IFN antiviral activity by measuring the ability of supernatants from NMII-infected BMDMs to inhibit replication of GFP-expressing Newcastle disease virus (NDV) in mouse fibroblasts (86). As expected, supernatants from BMDMs treated with the TLR3 ligand poly(I·C) or L. pneumophila ΔflaA mutant, which both robustly induce type I IFN, led to a marked decrease in viral replication as measured by decreased GFP intensity compared to that of mock-treated cells (Fig. 5C and D). Supernatants from C. burnetii NMII-infected BMDMs also led to decreased viral replication, although to a lesser degree, suggesting that low, biologically active levels of type I IFN are produced during NMII infection. We next tested if type I IFN signaling restricts C. burnetii NMII by measuring bacterial replication in B6 BMDMs or Ifnar−/− BMDMs lacking the type I IFN receptor (IFNAR) (84). NMII replication did not increase in IFNAR-deficient BMDMs by day 7 postinfection, whereas NMII replication significantly increased in Myd88−/− Trif−/− BMDMs (Fig. 5E). These data demonstrate that C. burnetii NMII does not induce robust production of type I IFNs, and type I IFNs are not required to restrict intracellular NMII replication in B6 macrophages.
FIG 5.
Type I interferons are not robustly induced and do not restrict Coxiella burnetii Nine Mile II replication. (A and B) C57BL/6 BMDMs were mock infected, infected with the L. pneumophila ΔflaA mutant at an MOI of 10, or infected with WT C. burnetii NMII at an MOI of 10, 50, or 100 for 16 h. Fold induction of Ifna4 and Ifnb mRNAs was measured via qRT-PCR. Graphs show the means ± SEM from triplicate wells. Results are representative of two independent experiments. (C) Supernatants from C57BL/6 BMDMs that were mock infected, infected with the L. pneumophila ΔflaA mutant at an MOI of 10, or infected with WT C. burnetii NMII at an MOI of 50 for 16 h were incubated with L2 mouse fibroblasts for 24 h. L2 cells then were infected with NDV-GFP at an MOI of 1. As a positive control, L2 cells were treated with supernatants from C57BL/6 BMDMs stimulated with poly(I·C) for 16 h. At 24 h postinfection, cells were fixed, stained with DAPI, and examined for levels of NDV-GFP replication by fluorescence microscopy. Shown are representative fluorescence micrographs. (D) Graphs of mean GFP fluorescence from each sample quantified with ImageJ software. Results are representative of two independent experiments. (E) C57BL/6, Ifnar−/−, or Myd88−/− Trif−/− BMDMs were infected with WT C. burnetii NMII at an MOI of 100. C. burnetii GEs were measured by qPCR on days 1 and 7 postinfection. Graphs show the fold change in GEs relative to those at day 1 ± SEM from triplicate wells. Results are representative of two independent experiments. *, P < 0.05; ns, no significance.
TLR2-mediated production of TNF is required to restrict Coxiella burnetii Nine Mile II replication within macrophages.
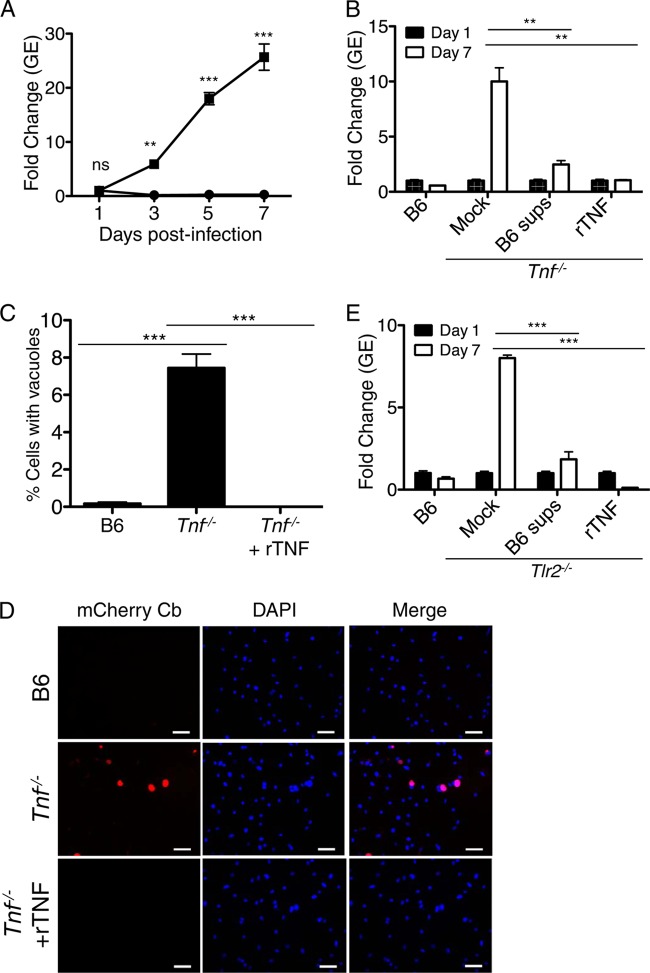
The addition of exogenous TNF along with IFN-γ to human monocytes or a mouse fibroblast cell line restricts intracellular replication of C. burnetii phase I and phase II, respectively (109, 110). Furthermore, antibody-mediated neutralization of TNF increases C. burnetii phase I replication in human monocytes (110). Whether endogenous TNF produced upon TLR-mediated detection of NMII accounts for the ability of B6 BMDMs to restrict NMII replication is unknown. To test this, we infected Tnf−/− BMDMs with NMII and measured intracellular replication and vacuole formation at day 7 postinfection. Although the levels of bacterial uptake by B6 and Tnf−/− BMDMs were similar at day one postinfection (see Fig. S5 in the supplemental material), we observed robust bacterial replication within Tnf−/− BMDMs, with a greater than 25-fold increase in GEs and a significant increase in large NMII-containing vacuoles by day seven postinfection (Fig. 6A to D), suggesting that TNF is required for B6 BMDMs to restrict NMII replication. To determine whether restriction was indeed mediated by TNF, we added 10 ng/ml recombinant TNF (rTNF) to NMII-infected Tnf−/− BMDMs at days 1 and 5 postinfection. We found that the addition of rTNF restored the ability of Tnf−/− BMDMs to restrict NMII replication, as measured by a decrease in both GEs and the percentage of large NMII-containing vacuoles at day 7 postinfection (Fig. 6B to E), suggesting that the deficiency in TNF production accounts for the increased susceptibility of Tnf−/− BMDMs. Furthermore, the addition of supernatants from B6 BMDMs infected with NMII for 24 h restored the ability of Tnf−/− BMDMs to restrict intracellular NMII replication, further supporting that intracellular restriction of NMII replication by B6 BMDMs is mediated by TNF secreted in response to infection. In agreement with the known role of TNF to enhance the expression of nitric oxide synthase (Nos2) (109), we found that compared to NMII-infected B6 BMDMs, infected Tnf−/− BMDMs induced significantly less Nos2 mRNA, as determined by qRT-PCR (see Fig. S6A). Furthermore, infected Tnf−/− BMDMs produced smaller amounts of nitric oxide, as measured by decreased nitrite levels in the supernatants of infected cells (see Fig. S6B). As nitric oxide is known to limit the replication of both C. burnetii phase I and phase II bacteria within macrophages and fibroblasts (109, 111, 112), the decreased inducible nitric oxide synthase (iNOS) expression and nitric oxide production observed in Tnf−/− macrophages may contribute to the inability of these cells to restrict NMII replication.
FIG 6.
Endogenously produced TNF restricts Coxiella burnetii Nine Mile II replication in C57BL/6 macrophages. (A) C57BL/6 and Tnf−/− BMDMs were infected with WT C. burnetii NMII expressing mCherry at an MOI of 100. C. burnetii GEs were determined by qPCR on days 1, 3, 5, and 7 postinfection. Graphs show the fold change in GEs relative to those at day 1 ± SEM from triplicate wells. Results are representative of three independent experiments. (B) Ten ng/ml rTNF or supernatants from C57BL/6 BMDMs infected with WT C. burnetii NMII for 24 h were added to WT C. burnetii-infected Tnf−/− BMDMs on days 1 and 5 postinfection. C. burnetii GEs were measured by qPCR, and the fold increase in GEs on day 7 postinfection was determined relative to the GEs on day 1. Graphs show the fold change in GEs relative to those on day 1 ± SEM from triplicate wells. Results are representative of two independent experiments. (C) B6 or Tnf−/− BMDMs were infected with mCherry-expressing WT C. burnetii or 10 ng/ml rTNF was added to mCherry-expressing WT C. burnetii-infected Tnf−/− BMDMs on days 1 and 5 postinfection. On day 7 postinfection, cells were fixed, stained with DAPI, and imaged by fluorescence microscopy. The number of mCherry-expressing C. burnetii-containing vacuoles was determined and calculated as a percentage of the total cell number. Graphs show mean percentages of cells containing C. burnetii vacuoles ± SEM from triplicate coverslips. At least 300 cells were counted per coverslip. (D) Representative images (40×) of mCherry-expressing C. burnetii-containing vacuoles in infected BMDMs treated as described for panel C. Scale bars represent 25 μM. Results are representative of two independent experiments. (E) Ten ng/ml rTNF or supernatants from B6 BMDMs infected with WT C. burnetii NMII for 24 h was added to WT C. burnetii-infected Tlr2−/− BMDMs on days 1 and 5 postinfection. C. burnetii GEs were measured by qPCR, and the fold increase in GEs on day 7 postinfection was determined relative to the GEs on day 1. Graphs show the fold change in GEs relative to those on day 1 ± SEM from triplicate wells. Results are representative of two independent experiments. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, no significance.
We observed that the percentage of Tnf−/− BMDMs harboring large NMII-containing vacuoles at day 7 postinfection with MOIs of 100 and 500 was similar to the percentage of Tlr2−/− BMDMs containing large vacuoles (see Fig. S7 in the supplemental material). This finding, along with the decreased TNF production observed in Tlr2−/− BMDMs, led us to hypothesize that the inability of Tlr2−/− BMDMs to control intracellular NMII replication was due to insufficient TNF production. To test this hypothesis, we examined whether the ability of Tlr2−/− BMDMs to control NMII could be restored by exogenous treatment with rTNF or supernatants from NMII-infected B6 BMDMs. In contrast to mock-treated Tlr2−/− BMDMs, Tlr2−/− macrophages treated with rTNF or supernatants from NMII-infected B6 BMDMs restricted bacterial replication at levels comparable to those of B6 BMDMs (Fig. 6E). These data indicate that the inability of Tlr2−/− BMDMs to control NMII replication is due primarily to defective TNF production, as the addition of exogenous TNF restored the ability of these cells to limit NMII replication. Taken together, our data suggest that TNF produced upon TLR sensing of NMII infection critically enables B6 macrophages to control intracellular replication of NMII.
DISCUSSION
B6 macrophages are not permissive for intracellular replication of C. burnetii NMII (56). Past studies identified a critical role for TLR2 in enabling B6 macrophages to detect and limit NMII replication (58), suggesting that innate immunity aids in controlling NMII infection in this cell type. The roles of other TLRs and the signaling adaptors MyD88 and Trif in detecting and restricting C. burnetii NMII B6 macrophages remained poorly understood. Furthermore, whether cytosolic immunosurveillance pathways detect bacterial products translocated by the C. burnetii T4SS and limit C. burnetii growth in B6 macrophages was unclear. In our study, we aimed to identify additional innate immune pathways used by B6 macrophages to detect and restrict C. burnetii NMII. Here, we find that TLR2 and TLR4 signal in concert to mediate proinflammatory cytokine production in response to C. burnetii NMII, although TLR4 was dispensable for restricting NMII replication. The TLR signaling adaptors MyD88 and Trif were required for this cytokine response and also contributed to the restriction of NMII replication. Despite published findings and data presented here showing that C. burnetii NMII can translocate T4SS effectors into the cytosol of B6 macrophages (59), we find that NMII does not induce inflammasome activation, T4SS-dependent enhancement of proinflammatory cytokine production, or robust type I IFN production, and inflammasomes or type I IFNs are not required to limit C. burnetii replication within B6 macrophages. These data indicate that NMII infection does not robustly trigger cytosolic PRRs in B6 macrophages. Instead, we found that endogenous TNF produced as a consequence of TLR detection is required by B6 macrophages to restrict intracellular replication of NMII.
We first set out to examine the contributions of TLRs and their signaling adaptors in mediating proinflammatory cytokine production and bacterial restriction in B6 BMDMs infected with C. burnetii NMII. In agreement with previous studies, we found TNF and IL-6 production to be mediated largely by TLR2. Interestingly, we also uncovered a role for TLR4 in the production of these cytokines. Deletion of TLR4 alone did not impact cytokine production, consistent with previous findings (58), whereas concomitant ablation of TLR2 and TLR4 significantly decreased cytokine production compared to that of TLR2 deficiency alone but did not result in increased NMII replication. Consistent with a role for both TLR2 and TLR4 in immune sensing of C. burnetii NMII, we found that the adaptor proteins MyD88, which signals downstream of both TLR2 and TLR4, and Trif, which signals downstream of TLR4, both are required for cytokine production and restriction of NMII replication in macrophages. A role for TLR4 was revealed only in the absence of TLR2, which suggests that TLR4 is not essential for macrophage responses to NMII, consistent with previous findings and data shown here demonstrating that C. burnetii LPS alone is not stimulatory (55, 58). However, it is possible that in the context of infection with whole bacteria, TLR4 somehow responds to NMII, either because lipid A variants that are more stimulatory are expressed during the intracellular life cycle or because TLR4 senses another bacterial or host-derived PAMP. Nevertheless, our data point to a more dominant role for TLR2 in sensing NMII. Furthermore, we found a dominant role for TLR signaling in regulating p38 MAPK activation during C. burnetii NMII infection. These results show that macrophage responses to NMII infection are highly dependent on TLR sensing. As virulent phase I C. burnetii, in contrast to avirulent phase II C. burnetii, does not stimulate p38 MAPK signaling (55, 113), and as it inhibits p38 MAPK signaling in a TLR4-dependent manner (92), further studies are necessary to test if TLR2 and TLR4 act in concert to respond to phase I C. burnetii as well.
To mount an immune response tailored against intracellular pathogens, the immune system often relies on cytosolic PRRs that respond to bacterial products introduced into the host cytosol by intracellular bacteria. Studies on the evolutionarily related pathogen L. pneumophila have shown that its Dot/Icm T4SS is required for bacterial survival within macrophages but also inadvertently exposes L. pneumophila to an array of cytosolic immunosurveillance pathways. Infection with L. pneumophila expressing a functional T4SS significantly increases the production of TNF, IL-6, and IL-12 and robustly activates host MAPK signaling, type I IFN production, and several inflammasome pathways compared to levels for avirulent mutants lacking a functional T4SS (114). Given that C. burnetii NMII uses a Dot/Icm T4SS that is functionally similar to that of L. pneumophila to translocate effector proteins into B6 BMDMs (59, 115), we next asked whether B6 BMDMs utilize cytosolic immunosurveillance pathways to detect and restrict NMII infection. In contrast to what was observed for L. pneumophila, we did not observe a clear requirement for the C. burnetii NMII T4SS in enhancing cytokine responses. Instead, we observed a slight decrease in TNF and IL-6 production in response to T4SS-expressing NMII compared to that of a T4SS-deficient strain. While the difference was not robust, it suggests the intriguing possibility that NMII T4SS effectors target immune signaling pathways to dampen proinflammatory cytokine production.
The inflammasome is another cytosolic immune response that is robustly activated and plays a critical role in controlling a variety of pathogens, including L. pneumophila (116). Unlike L. pneumophila, C. burnetii does not possess flagellin (117), a potent activator of the NAIP5 inflammasome (71, 72). It was unknown, however, if C. burnetii, like L. pneumophila, possesses other stimulators of inflammasome activation. We found that while C. burnetii NMII induces pro-IL-1β production, it does not induce inflammasome activation in WT B6 macrophages, in agreement with a recently published study (45). Furthermore, NMII did not induce inflammasome activation in Tlr2−/− macrophages that support NMII replication, suggesting that the inability of NMII to trigger inflammasome activation in B6 macrophages is not due to an inability to replicate in this cell type. One possibility for the lack of inflammasome activation is that C. burnetii may not express PAMPs that are recognized by inflammasome-associated PRRs. For example, caspase-11 recognizes hexa- or penta-acylated LPS (100, 101), whereas C. burnetii generates tetra-acylated LPS (58), although a recent study indicated that purified C. burnetii LPS transfected into macrophages could activate caspase-11 (45). Alternatively, C. burnetii may employ T4SS effectors to actively inhibit inflammasome activation. It was recently shown that the C. burnetii T4SS effector protein, IcaA, can inhibit caspase-11 activation in mouse macrophages when expressed in L. pneumophila (45). Interestingly, an IcaA-deficient C. burnetii NMII strain is unable to inhibit noncanonical inflammasome activation induced by L. pneumophila coinfection, but there still is no evidence of inflammasome activation in mouse macrophages infected with IcaA-deficient C. burnetii alone, suggesting that there are additional mechanisms employed by C. burnetii to evade inflammasome activation (45). It has been shown, however, that primary human alveolar macrophages produce mature IL-1β in response to NMII but not phase I C. burnetii, suggesting that human and mouse inflammasome sensors or various cell types differ in their responses to NMII infection (54). The finding that NMII does not induce inflammasome activation in mouse cells may provide some insight into the observation that neutrophil recruitment is delayed in mice infected with phase I or phase II C. burnetii until 1 week postinfection (118), as inflammasome-derived IL-1 is a critical regulator of neutrophil recruitment during other bacterial infections, such as pulmonary L. pneumophila infection (68, 90–92). Further studies are warranted to determine the additional mechanisms by which C. burnetii evades inflammasome activation in mouse macrophages.
Many intracellular bacteria trigger cytosolic nucleic acid sensors that lead to type I IFN production, and this response can limit bacterial replication, as is the case for L. pneumophila (103). It was unknown, however, if type I IFNs are induced or play a role in controlling NMII infection in B6 macrophages. Our studies indicate that although NMII infection induces very low but biologically active levels of type I IFNs, BMDMs lacking the type I IFN receptor did not exhibit increased NMII replication. These data suggest that although small amounts of type I IFNs are produced during infection, they do not participate in restricting NMII replication in B6 macrophages.
As our studies suggested a more prominent role for TLRs than cytosolic immune sensing in detecting and restricting C. burnetii NMII, we next examined the mechanistic basis underlying TLR-mediated control of bacterial replication. Given that our data and other published studies indicate a critical role for TLR signaling in mediating the production of proinflammatory cytokines during NMII infection, we wondered whether these cytokines participated in restricting NMII replication. We focused on TNF, as TNF is known to restrict the intracellular replication of several vacuolar pathogens, including L. pneumophila (67), Mycobacterium species (119), and Salmonella enterica serovar Typhimurium (120), within macrophages, and a previous study showed that antibody-mediated neutralization of TNF led to increased C. burnetii phase I replication in human monocytes (110). We found that Tnf−/− BMDMs are highly permissive for intracellular replication of NMII. Furthermore, the exogenous addition of recombinant TNF or supernatants from NMII-infected B6 BMDMs restored the ability of both TNF-deficient and TLR2-deficient BMDMs to contain NMII replication. These data suggest that Tlr2−/− BMDMs are unable to control NMII replication due to defective TNF production. Interestingly, BMDMs lacking only Trif still are able to limit intracellular NMII replication despite having a defect in TNF production, whereas MyD88-deficient BMDMs have a defect in both TNF production and control of NMII replication, similar to the phenotype observed in Tlr2−/− BMDMs. We were able to uncover a role for Trif in limiting NMII replication only when we deleted both Trif and MyD88, as this led to increased bacterial replication compared to deletion of MyD88 alone. These findings suggest that in addition to TNF production, TLR2-dependent MyD88 signaling regulates other antibacterial mechanisms that collaborate with TNF signaling to restrict NMII replication within B6 macrophages.
Other studies also have implicated a role for TNF in inhibiting C. burnetii replication within host cells, but the majority of these studies examined TNF in the context of the exogenous addition of TNF and IFN-γ simultaneously, as these cytokines are known to synergize for potent activation of macrophages (109, 110, 121). During in vitro and in vivo infection, macrophages do not produce IFN-γ. Instead, IFN-γ is produced by other cell types, such as natural killer (NK) cells and T cells, during in vivo infection (122). Thus, our studies suggest a key role for TNF endogenously produced downstream of TLR detection in inhibiting NMII replication. Our data show that TNF-deficient macrophages have a defect in iNOS expression and nitric oxide production following C. burnetii infection. As nitric oxide contributes to the restriction of C. burnetii replication in mouse L929 fibroblasts and B6 macrophages (109, 111), nitric oxide is one possible mechanism by which endogenously produced TNF controls NMII infection. A previous study found that C. burnetii infection of THP-1 cells induces TNF-dependent, caspase-independent cell death (123), suggesting another mechanism by which TNF restricts C. burnetii replication. We were unable to observe cell death in NMII-infected B6 macrophages using bulk cytotoxicity assays, suggesting that cell death is not a major mechanism by which TNF restricts NMII replication within B6 macrophages. Sensitive single-cell-based assays would provide a more definitive means of assessing whether TNF induces similar caspase-independent cell death to restrict NMII replication in B6 macrophages. Further studies are necessary to determine the exact mechanisms by which TNF restricts NMII infection in B6 macrophages and whether TNF similarly restricts NMII replication in mouse macrophages from other backgrounds or human macrophages or other cell types. Furthermore, it would be of interest to determine whether the intracellular replication of NMI or other phase I pathotypes of C. burnetii is similarly restricted by TNF.
Virulent phase I C. burnetii is highly infectious, and one bacterium is sufficient to cause disease in immunocompetent individuals (124). Phase II C. burnetii is unable to cause disease, presumably because it is more immunostimulatory. Despite the more immunostimulatory nature of phase II C. burnetii, we were unable to observe the activation of the cytosolic immunosurveillance pathways surveyed here. This suggests that the limited activation of cytosolic immune pathways is a feature also shared by phase I C. burnetii and contributes to the virulence of phase I organisms. In conclusion, our findings demonstrate the importance of TLR signaling in enabling B6 macrophages to restrict C. burnetii NMII, and we identify TNF as a key mechanism by which TLR signaling restricts intracellular replication in this nonpermissive cell type. Our data also indicate that C. burnetii NMII T4SS activity does not robustly trigger several cytosolic immunosurveillance pathways that we examined, and these pathways do not play a major role in the control of intracellular NMII replication in B6 macrophages, suggesting that C. burnetii evades the activation of cytosolic immune sensors. Our studies provide further insight into the innate immune responses that underlie the interactions between mouse macrophages and C. burnetii and enable macrophages to sense and restrict infection.
Supplementary Material
ACKNOWLEDGMENTS
We thank Susan Ross for providing Tlr2−/−, Tlr4−/−, Tlr2−/− Tlr4−/−, and Myd88−/− mice, Jeffrey Weiser for providing Tlr2−/− mice, and Igor Brodsky for providing Tlr2−/− Tlr4−/− and Myd88−/− Trif−/− mouse bone marrow. We thank Karen Fowler for technical assistance with the C. burnetii NMII BlaM-CBU_0077 strains, Alan Copenhaver and Martin Naradikian for technical advice on flow cytometry and effector translocation assays, and members of the Shin, Brodsky, and Roy laboratories for helpful discussions and advice.
Funding Statement
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/IAI.01536-15.
REFERENCES
- 1.Janeway CA Jr, Medzhitov R. 2002. Innate immune recognition. Annu Rev Immunol 20:197–216. doi: 10.1146/annurev.immunol.20.083001.084359. [DOI] [PubMed] [Google Scholar]
- 2.Janeway CA., Jr 1989. Approaching the asymptote? Evolution and revolution in immunology. Cold Spring Harbor Symp Quant Biol 54(Part 1):1–13. doi: 10.1101/SQB.1989.054.01.003. [DOI] [PubMed] [Google Scholar]
- 3.Medzhitov R. 2007. Recognition of microorganisms and activation of the immune response. Nature 449:819–826. doi: 10.1038/nature06246. [DOI] [PubMed] [Google Scholar]
- 4.Akira S, Uematsu S, Takeuchi O. 2006. Pathogen recognition and innate immunity. Cell 124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 5.Ting JP, Kastner DL, Hoffman HM. 2006. CATERPILLERs, pyrin and hereditary immunological disorders. Nat Rev Immunol 6:183–195. doi: 10.1038/nri1788. [DOI] [PubMed] [Google Scholar]
- 6.Fritz JH, Ferrero RL, Philpott DJ, Girardin SE. 2006. Nod-like proteins in immunity, inflammation and disease. Nat Immunol 7:1250–1257. doi: 10.1038/ni1412. [DOI] [PubMed] [Google Scholar]
- 7.Inohara C, McDonald C, Nunez G. 2005. NOD-LRR proteins: role in host-microbial interactions and inflammatory disease. Annu Rev Biochem 74:355–383. doi: 10.1146/annurev.biochem.74.082803.133347. [DOI] [PubMed] [Google Scholar]
- 8.Vance RE, Isberg RR, Portnoy DA. 2009. Patterns of pathogenesis: discrimination of pathogenic and nonpathogenic microbes by the innate immune system. Cell Host Microbe 6:10–21. doi: 10.1016/j.chom.2009.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Martinon F, Burns K, Tschopp J. 2002. The inflammasome: a molecular platform triggering activation of inflammatory caspases and processing of proIL-beta. Mol Cell 10:417–426. doi: 10.1016/S1097-2765(02)00599-3. [DOI] [PubMed] [Google Scholar]
- 10.Kayagaki N, Warming S, Lamkanfi M, Vande Walle L, Louie S, Dong J, Newton K, Qu Y, Liu J, Heldens S, Zhang J, Lee WP, Roose-Girma M, Dixit VM. 2011. Non-canonical inflammasome activation targets caspase-11. Nature 479:117–121. doi: 10.1038/nature10558. [DOI] [PubMed] [Google Scholar]
- 11.Rathinam VA, Vanaja SK, Waggoner L, Sokolovska A, Becker C, Stuart LM, Leong JM, Fitzgerald KA. 2012. TRIF licenses caspase-11-dependent NLRP3 inflammasome activation by gram-negative bacteria. Cell 150:606–619. doi: 10.1016/j.cell.2012.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Broz P, Ruby T, Belhocine K, Bouley DM, Kayagaki N, Dixit VM, Monack DM. 2012. Caspase-11 increases susceptibility to Salmonella infection in the absence of caspase-1. Nature 490:288–291. doi: 10.1038/nature11419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gurung P, Malireddi RK, Anand PK, Demon D, Vande Walle L, Liu Z, Vogel P, Lamkanfi M, Kanneganti TD. 2012. Toll or interleukin-1 receptor (TIR) domain-containing adaptor inducing interferon-beta (TRIF)-mediated caspase-11 protease production integrates Toll-like receptor 4 (TLR4) protein- and Nlrp3 inflammasome-mediated host defense against enteropathogens. J Biol Chem 287:34474–34483. doi: 10.1074/jbc.M112.401406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rathinam VA, Vanaja SK, Fitzgerald KA. 2012. Regulation of inflammasome signaling. Nat Immunol 13:333–332. doi: 10.1038/ni.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Broz P, Monack DM. 2013. Noncanonical inflammasomes: caspase-11 activation and effector mechanisms. PLoS Pathog 9:e1003144. doi: 10.1371/journal.ppat.1003144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lamkanfi M, Dixit VM. 2014. Mechanisms and functions of inflammasomes. Cell 157:1013–1022. doi: 10.1016/j.cell.2014.04.007. [DOI] [PubMed] [Google Scholar]
- 17.Fontana MF, Vance RE. 2011. Two signal models in innate immunity. Immunol Rev 243:26–39. doi: 10.1111/j.1600-065X.2011.01037.x. [DOI] [PubMed] [Google Scholar]
- 18.Abdullah Z, Knolle PA. 2014. Scaling of immune responses against intracellular bacterial infection. EMBO J 33:2283–2294. doi: 10.15252/embj.201489055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miao EA, Leaf IA, Treuting PM, Mao DP, Dors M, Sarkar A, Warren SE, Wewers MD, Aderem A. 2010. Caspase-1-induced pyroptosis is an innate immune effector mechanism against intracellular bacteria. Nat Immunol 11:1136–1142. doi: 10.1038/ni.1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zamboni DS, Kobayashi KS, Kohlsdorf T, Ogura Y, Long EM, Vance RE, Kuida K, Mariathasan S, Dixit VM, Flavell RA, Dietrich WF, Roy CR. 2006. The Birc1e cytosolic pattern-recognition receptor contributes to the detection and control of Legionella pneumophila infection. Nat Immunol 7:318–325. doi: 10.1038/ni1305. [DOI] [PubMed] [Google Scholar]
- 21.Amer A, Franchi L, Kanneganti TD, Body-Malapel M, Ozoren N, Brady G, Meshinchi S, Jagirdar R, Gewirtz A, Akira S, Nunez G. 2006. Regulation of Legionella phagosome maturation and infection through flagellin and host Ipaf. J Biol Chem 281:35217–35223. doi: 10.1074/jbc.M604933200. [DOI] [PubMed] [Google Scholar]
- 22.Akhter A, Caution K, Abu Khweek A, Tazi M, Abdulrahman BA, Abdelaziz DH, Voss OH, Doseff AI, Hassan H, Azad AK, Schlesinger LS, Wewers MD, Gavrilin MA, Amer AO. 2012. Caspase-11 promotes the fusion of phagosomes harboring pathogenic bacteria with lysosomes by modulating actin polymerization. Immunity 37:35–47. doi: 10.1016/j.immuni.2012.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Raoult D, Marrie T, Mege J. 2005. Natural history and pathophysiology of Q fever. Lancet Infect Dis 5:219–226. doi: 10.1016/S1473-3099(05)70052-9. [DOI] [PubMed] [Google Scholar]
- 24.Heinzen RA, Scidmore MA, Rockey DD, Hackstadt T. 1996. Differential interaction with endocytic and exocytic pathways distinguish parasitophorous vacuoles of Coxiella burnetii and Chlamydia trachomatis. Infect Immun 64:796–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Beron W, Gutierrez MG, Rabinovitch M, Colombo MI. 2002. Coxiella burnetii localizes in a Rab7-labeled compartment with autophagic characteristics. Infect Immun 70:5816–5821. doi: 10.1128/IAI.70.10.5816-5821.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Winchell CG, Graham JG, Kurten RC, Voth DE. 2014. Coxiella burnetii type IV secretion-dependent recruitment of macrophage autophagosomes. Infect Immun 82:2229–2238. doi: 10.1128/IAI.01236-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Newton HJ, Kohler LJ, McDonough JA, Temoche-Diaz M, Crabill E, Hartland EL, Roy CR. 2014. A screen of Coxiella burnetii mutants reveals important roles for Dot/Icm effectors and host autophagy in vacuole biogenesis. PLoS Pathog 10:e1004286. doi: 10.1371/journal.ppat.1004286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Romano PS, Gutierrez MG, Beron W, Rabinovitch M, Colombo MI. 2007. The autophagic pathway is actively modulated by phase II Coxiella burnetii to efficiently replicate in the host cell. Cell Microbiol 9:891–909. doi: 10.1111/j.1462-5822.2006.00838.x. [DOI] [PubMed] [Google Scholar]
- 29.Gutierrez MG, Vazquez CL, Munafo DB, Zoppino FC, Beron W, Rabinovitch M, Colombo MI. 2005. Autophagy induction favours the generation and maturation of the Coxiella-replicative vacuoles. Cell Microbiol 7:981–993. doi: 10.1111/j.1462-5822.2005.00527.x. [DOI] [PubMed] [Google Scholar]
- 30.Campoy EM, Zoppino FC, Colombo MI. 2011. The early secretory pathway contributes to the growth of the Coxiella-replicative niche. Infect Immun 79:402–413. doi: 10.1128/IAI.00688-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Coleman SA, Fischer ER, Howe D, Mead DJ, Heinzen RA. 2004. Temporal analysis of Coxiella burnetii morphological differentiation. J Bacteriol 186:7344–7352. doi: 10.1128/JB.186.21.7344-7352.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Voth DE, Heinzen RA. 2007. Lounging in a lysosome: the intracellular lifestyle of Coxiella burnetii. Cell Microbiol 9:829–840. doi: 10.1111/j.1462-5822.2007.00901.x. [DOI] [PubMed] [Google Scholar]
- 33.Carey KL, Newton HJ, Luhrmann A, Roy CR. 2011. The Coxiella burnetii Dot/Icm system delivers a unique repertoire of type IV effectors into host cells and is required for intracellular replication. PLoS Pathog 7:e1002056. doi: 10.1371/journal.ppat.1002056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pan X, Luhrmann A, Satoh A, Laskowski-Arce MA, Roy CR. 2008. Ankyrin repeat proteins comprise a diverse family of bacterial type IV effectors. Science 320:1651–1654. doi: 10.1126/science.1158160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen C, Banga S, Mertens K, Weber MM, Gorbaslieva I, Tan Y, Luo ZQ, Samuel JE. 2010. Large-scale identification and translocation of type IV secretion substrates by Coxiella burnetii. Proc Natl Acad Sci U S A 107:21755–21760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Graham JG, Winchell CG, Sharma UM, Voth DE. 2015. Identification of ElpA, a Coxiella burnetii pathotype-specific Dot/Icm type IV secretion system substrate. Infect Immun 83:1190–1198. doi: 10.1128/IAI.02855-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Larson CL, Beare PA, Voth DE, Howe D, Cockrell DC, Bastidas RJ, Valdivia RH, Heinzen RA. 2015. Coxiella burnetii effector proteins that localize to the parasitophorous vacuole membrane promote intracellular replication. Infect Immun 83:661–670. doi: 10.1128/IAI.02763-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Beare PA, Gilk SD, Larson CL, Hill J, Stead CM, Omsland A, Cockrell DC, Howe D, Voth DE, Heinzen RA. 2011. Dot/Icm type IVB secretion system requirements for Coxiella burnetii growth in human macrophages. mBio 2:e00175-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Voth DE, Beare PA, Howe D, Sharma UM, Samoilis G, Cockrell DC, Omsland A, Heinzen RA. 2011. The Coxiella burnetii cryptic plasmid is enriched in genes encoding type IV secretion system substrates. J Bacteriol 193:1493–1503. doi: 10.1128/JB.01359-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Voth DE, Howe D, Beare PA, Vogel JP, Unsworth N, Samuel JE, Heinzen RA. 2009. The Coxiella burnetii ankyrin repeat domain-containing protein family is heterogeneous, with C-terminal truncations that influence Dot/Icm-mediated secretion. J Bacteriol 191:4232–4242. doi: 10.1128/JB.01656-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Luhrmann A, Nogueira CV, Carey KL, Roy CR. 2010. Inhibition of pathogen-induced apoptosis by a Coxiella burnetii type IV effector protein. Proc Natl Acad Sci U S A 107:18997–19001. doi: 10.1073/pnas.1004380107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Klingenbeck L, Eckart RA, Berens C, Luhrmann A. 2013. The Coxiella burnetii type IV secretion system substrate CaeB inhibits intrinsic apoptosis at the mitochondrial level. Cell Microbiol 15:675–687. doi: 10.1111/cmi.12066. [DOI] [PubMed] [Google Scholar]
- 43.Larson CL, Beare PA, Howe D, Heinzen RA. 2013. Coxiella burnetii effector protein subverts clathrin-mediated vesicular trafficking for pathogen vacuole biogenesis. Proc Natl Acad Sci U S A 110:E4770–E4779. doi: 10.1073/pnas.1309195110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Eckart RA, Bisle S, Schulze-Luehrmann J, Wittmann I, Jantsch J, Schmid B, Berens C, Luhrmann A. 2014. Antiapoptotic activity of Coxiella burnetii effector protein AnkG is controlled by p32-dependent trafficking. Infect Immun 82:2763–2771. doi: 10.1128/IAI.01204-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cunha LD, Ribeiro JM, Fernandes TD, Massis LM, Khoo CA, Moffatt JH, Newton HJ, Roy CR, Zamboni DS. 2015. Inhibition of inflammasome activation by Coxiella burnetii type IV secretion system effector IcaA. Nat Commun 6:10205. doi: 10.1038/ncomms10205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hackstadt T. 1988. Steric hindrance of antibody binding to surface proteins of Coxiella burnetti by phase I lipopolysaccharide. Infect Immun 56:802–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vishwanath S, Hackstadt T. 1988. Lipopolysaccharide phase variation determines the complement-mediated serum susceptibility of Coxiella burnetii. Infect Immun 56:40–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Moos A, Hackstadt T. 1987. Comparative virulence of intra- and interstrain lipopolysaccharide variants of Coxiella burnetii in the guinea pig model. Infect Immun 55:1144–1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ormsbee R, Peacock M, Gerloff R, Tallent G, Wike D. 1978. Limits of rickettsial infectivity. Infect Immun 19:239–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Amano K, Williams JC. 1984. Chemical and immunological characterization of lipopolysaccharides from phase I and phase II Coxiella burnetii. J Bacteriol 160:994–1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hoover TA, Culp DW, Vodkin MH, Williams JC, Thompson HA. 2002. Chromosomal DNA deletions explain phenotypic characteristics of two antigenic variants, phase II and RSA 514 (crazy), of the Coxiella burnetii Nine Mile strain. Infect Immun 70:6726–6733. doi: 10.1128/IAI.70.12.6726-2733.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Howe D, Shannon JG, Winfree S, Dorward DW, Heinzen RA. 2010. Coxiella burnetii phase I and II variants replicate with similar kinetics in degradative phagolysosome-like compartments of human macrophages. Infect Immun 78:3465–3474. doi: 10.1128/IAI.00406-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Baca OG, Akporiaye ET, Aragon AS, Martinez IL, Robles MV, Warner NL. 1981. Fate of phase I and phase II Coxiella burnetii in several macrophage-like tumor cell lines. Infect Immun 33:258–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Graham JG, MacDonald LJ, Hussain SK, Sharma UM, Kurten RC, Voth DE. 2013. Virulent Coxiella burnetii pathotypes productively infect primary human alveolar macrophages. Cell Microbiol 15:1012–1025. doi: 10.1111/cmi.12096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shannon JG, Howe D, Heinzen RA. 2005. Virulent Coxiella burnetii does not activate human dendritic cells: role of lipopolysaccharide as a shielding molecule. Proc Natl Acad Sci U S A 102:8722–8727. doi: 10.1073/pnas.0501863102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zamboni DS. 2004. Genetic control of natural resistance of mouse macrophages to Coxiella burnetii infection in vitro: macrophages from restrictive strains control parasitophorous vacuole maturation. Infect Immun 72:2395–2399. doi: 10.1128/IAI.72.4.2395-2399.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zamboni DS, Mortara RA, Freymuller E, Rabinovitch M. 2002. Mouse resident peritoneal macrophages partially control in vitro infection with Coxiella burnetii phase II. Microbes Infect 4:591–598. doi: 10.1016/S1286-4579(02)01577-0. [DOI] [PubMed] [Google Scholar]
- 58.Zamboni DS, Campos MA, Torrecilhas AC, Kiss K, Samuel JE, Golenbock DT, Lauw FN, Roy CR, Almeida IC, Gazzinelli RT. 2004. Stimulation of toll-like receptor 2 by Coxiella burnetii is required for macrophage production of proinflammatory cytokines and resistance to infection. J Biol Chem 279:54405–54415. doi: 10.1074/jbc.M410340200. [DOI] [PubMed] [Google Scholar]
- 59.Newton HJ, McDonough JA, Roy CR. 2013. Effector protein translocation by the Coxiella burnetii Dot/Icm type IV secretion system requires endocytic maturation of the pathogen-occupied vacuole. PLoS One 8:e54566. doi: 10.1371/journal.pone.0054566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Benoit M, Bechah Y, Capo C, Murray PJ, Mege JL, Desnues B. 2009. Role of the cytoplasmic pattern recognition receptor Nod2 in Coxiella burnetii infection. Clin Microbiol Infect 15(Suppl 2):S154–S155. [DOI] [PubMed] [Google Scholar]
- 61.Ammerdorffer A, Schoffelen T, Gresnigt MS, Oosting M, den Brok MH, Abdollahi-Roodsaz S, Kanneganti TD, de Jong DJ, van Deuren M, Roest HJ, Rebel JM, Netea MG, Joosten LA, Sprong T. 2015. Recognition of Coxiella burnetii by toll-like receptors and nucleotide-binding oligomerization domain-like receptors. J Infect Dis 211:978–987. doi: 10.1093/infdis/jiu526. [DOI] [PubMed] [Google Scholar]
- 62.Fields BS. 1996. The molecular ecology of legionellae. Trends Microbiol 4:286–290. doi: 10.1016/0966-842X(96)10041-X. [DOI] [PubMed] [Google Scholar]
- 63.Vance RE. 2010. Immunology taught by bacteria. J Clin Immunol 30:507–511. doi: 10.1007/s10875-010-9389-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yoshida S, Goto Y, Mizuguchi Y, Nomoto K, Skamene E. 1991. Genetic control of natural resistance in mouse macrophages regulating intracellular Legionella pneumophila multiplication in vitro. Infect Immun 59:428–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Diez E, Lee SH, Gauthier S, Yaraghi Z, Tremblay M, Vidal S, Gros P. 2003. Birc1e is the gene within the Lgn1 locus associated with resistance to Legionella pneumophila. Nat Genet 33:55–60. [DOI] [PubMed] [Google Scholar]
- 66.Wright EK, Goodart SA, Growney JD, Hadinoto V, Endrizzi MG, Long EM, Sadigh K, Abney AL, Bernstein-Hanley I, Dietrich WF. 2003. Naip5 affects host susceptibility to the intracellular pathogen Legionella pneumophila. Curr Biol 13:27–36. doi: 10.1016/S0960-9822(02)01359-3. [DOI] [PubMed] [Google Scholar]
- 67.Coers J, Vance RE, Fontana MF, Dietrich WF. 2007. Restriction of Legionella pneumophila growth in macrophages requires the concerted action of cytokine and Naip5/Ipaf signalling pathways. Cell Microbiol 9:2344–2357. doi: 10.1111/j.1462-5822.2007.00963.x. [DOI] [PubMed] [Google Scholar]
- 68.Monroe KM, McWhirter SM, Vance RE. 2009. Identification of host cytosolic sensors and bacterial factors regulating the type I interferon response to Legionella pneumophila. PLoS Pathog 5:e1000665. doi: 10.1371/journal.ppat.1000665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lippmann J, Muller HC, Naujoks J, Tabeling C, Shin S, Witzenrath M, Hellwig K, Kirschning CJ, Taylor GA, Barchet W, Bauer S, Suttorp N, Roy CR, Opitz B. 2011. Dissection of a type I interferon pathway in controlling bacterial intracellular infection in mice. Cell Microbiol 13:1668–1682. doi: 10.1111/j.1462-5822.2011.01646.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Plumlee CR, Lee C, Beg AA, Decker T, Shuman HA, Schindler C. 2009. Interferons direct an effective innate response to Legionella pneumophila infection. J Biol Chem 284:30058–30066. doi: 10.1074/jbc.M109.018283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Molofsky AB, Byrne BG, Whitfield NN, Madigan CA, Fuse ET, Tateda K, Swanson MS. 2006. Cytosolic recognition of flagellin by mouse macrophages restricts Legionella pneumophila infection. J Exp Med 203:1093–1104. doi: 10.1084/jem.20051659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ren T, Zamboni DS, Roy CR, Dietrich WF, Vance RE. 2006. Flagellin-deficient Legionella mutants evade caspase-1- and Naip5-mediated macrophage immunity. PLoS Pathog 2:e18. doi: 10.1371/journal.ppat.0020018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.National Research Council. 2011. Guide for the care and use of laboratory animals, 8th ed National Academies Press, Washington, DC. [Google Scholar]
- 74.Berger KH, Isberg RR. 1993. Two distinct defects in intracellular growth complemented by a single genetic locus in Legionella pneumophila. Mol Microbiol 7:7–19. doi: 10.1111/j.1365-2958.1993.tb01092.x. [DOI] [PubMed] [Google Scholar]
- 75.Omsland A, Beare PA, Hill J, Cockrell DC, Howe D, Hansen B, Samuel JE, Heinzen RA. 2011. Isolation from animal tissue and genetic transformation of Coxiella burnetii are facilitated by an improved axenic growth medium. Appl Environ Microbiol 77:3720–3725. doi: 10.1128/AEM.02826-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Beare PA, Howe D, Cockrell DC, Omsland A, Hansen B, Heinzen RA. 2009. Characterization of a Coxiella burnetii ftsZ mutant generated by Himar1 transposon mutagenesis. J Bacteriol 191:1369–1381. doi: 10.1128/JB.01580-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Copenhaver AM, Casson CN, Nguyen HT, Fung TC, Duda MM, Roy CR, Shin S. 2014. Alveolar macrophages and neutrophils are the primary reservoirs for legionella pneumophila and mediate cytosolic surveillance of type IV secretion. Infect Immun 82:4325–4336. doi: 10.1128/IAI.01891-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Takeuchi O, Hoshino K, Kawai T, Sanjo H, Takada H, Ogawa T, Takeda K, Akira S. 1999. Differential roles of TLR2 and TLR4 in recognition of gram-negative and gram-positive bacterial cell wall components. Immunity 11:443–451. doi: 10.1016/S1074-7613(00)80119-3. [DOI] [PubMed] [Google Scholar]
- 79.Hoshino K, Takeuchi O, Kawai T, Sanjo H, Ogawa T, Takeda Y, Takeda K, Akira S. 1999. Cutting edge: Toll-like receptor 4 (TLR4)-deficient mice are hyporesponsive to lipopolysaccharide: evidence for TLR4 as the Lps gene product. J Immunol 162:3749–3752. [PubMed] [Google Scholar]
- 80.Adachi O, Kawai T, Takeda K, Matsumoto M, Tsutsui H, Sakagami M, Nakanishi K, Akira S. 1998. Targeted disruption of the MyD88 gene results in loss of IL-1- and IL-18-mediated function. Immunity 9:143–150. doi: 10.1016/S1074-7613(00)80596-8. [DOI] [PubMed] [Google Scholar]
- 81.Yamamoto M, Sato S, Hemmi H, Hoshino K, Kaisho T, Sanjo H, Takeuchi O, Sugiyama M, Okabe M, Takeda K, Akira S. 2003. Role of adaptor TRIF in the MyD88-independent toll-like receptor signaling pathway. Science 301:640–643. doi: 10.1126/science.1087262. [DOI] [PubMed] [Google Scholar]
- 82.Pasparakis M, Alexopoulou L, Episkopou V, Kollias G. 1996. Immune and inflammatory responses in TNF alpha-deficient mice: a critical requirement for TNF alpha in the formation of primary B cell follicles, follicular dendritic cell networks and germinal centers, and in the maturation of the humoral immune response. J Exp Med 184:1397–1411. doi: 10.1084/jem.184.4.1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kuida K, Lippke JA, Ku G, Harding MW, Livingston DJ, Su MS, Flavell RA. 1995. Altered cytokine export and apoptosis in mice deficient in interleukin-1 beta converting enzyme. Science 267:2000–2003. doi: 10.1126/science.7535475. [DOI] [PubMed] [Google Scholar]
- 84.Muller U, Steinhoff U, Reis LF, Hemmi S, Pavlovic J, Zinkernagel RM, Aguet M. 1994. Functional role of type I and type II interferons in antiviral defense. Science 264:1918–1921. doi: 10.1126/science.8009221. [DOI] [PubMed] [Google Scholar]
- 85.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−ΔΔCT) method. Methods 25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 86.Park MS, Shaw ML, Munoz-Jordan J, Cros JF, Nakaya T, Bouvier N, Palese P, Garcia-Sastre A, Basler CF. 2003. Newcastle disease virus (NDV)-based assay demonstrates interferon-antagonist activity for the NDV V protein and the Nipah virus V, W, and C proteins. J Virol 77:1501–1511. doi: 10.1128/JVI.77.2.1501-1511.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Shin S, Case CL, Archer KA, Nogueira CV, Kobayashi KS, Flavell RA, Roy CR, Zamboni DS. 2008. Type IV secretion-dependent activation of host MAP kinases induces an increased proinflammatory cytokine response to Legionella pneumophila. PLoS Pathog 4:e1000220. doi: 10.1371/journal.ppat.1000220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Fontana MF, Banga S, Barry KC, Shen X, Tan Y, Luo ZQ, Vance RE. 2011. Secreted bacterial effectors that inhibit host protein synthesis are critical for induction of the innate immune response to virulent Legionella pneumophila. PLoS Pathog 7:e1001289. doi: 10.1371/journal.ppat.1001289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Fontana MF, Shin S, Vance RE. 2012. Activation of host mitogen-activated protein kinases by secreted Legionella pneumophila effectors that inhibit host protein translation. Infect Immun 80:3570–3575. doi: 10.1128/IAI.00557-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Shin S, Roy CR. 2008. Host cell processes that influence the intracellular survival of Legionella pneumophila. Cell Microbiol 10:1209–1220. [DOI] [PubMed] [Google Scholar]
- 91.Voth DE, Heinzen RA. 2009. Sustained activation of Akt and Erk1/2 is required for Coxiella burnetii antiapoptotic activity. Infect Immun 77:205–213. doi: 10.1128/IAI.01124-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Barry AO, Boucherit N, Mottola G, Vadovic P, Trouplin V, Soubeyran P, Capo C, Bonatti S, Nebreda A, Toman R, Lemichez E, Mege JL, Ghigo E. 2012. Impaired stimulation of p38alpha-MAPK/Vps41-HOPS by LPS from pathogenic Coxiella burnetii prevents trafficking to microbicidal phagolysosomes. Cell Host Microbe 12:751–763. doi: 10.1016/j.chom.2012.10.015. [DOI] [PubMed] [Google Scholar]
- 93.Boucherit N, Barry AO, Mottola G, Trouplin V, Capo C, Mege JL, Ghigo E. 2012. Effects of Coxiella burnetii on MAPKinases phosphorylation. FEMS Immunol Med Microbiol 64:101–103. doi: 10.1111/j.1574-695X.2011.00852.x. [DOI] [PubMed] [Google Scholar]
- 94.Casson CN, Copenhaver AM, Zwack EE, Nguyen HT, Strowig T, Javdan B, Bradley WP, Fung TC, Flavell RA, Brodsky IE, Shin S. 2013. Caspase-11 activation in response to bacterial secretion systems that access the host cytosol. PLoS Pathog 9:e1003400. doi: 10.1371/journal.ppat.1003400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Case CL, Kohler LJ, Lima JB, Strowig T, de Zoete MR, Flavell RA, Zamboni DS, Roy CR. 2013. Caspase-11 stimulates rapid flagellin-independent pyroptosis in response to Legionella pneumophila. Proc Natl Acad Sci U S A 110:1851–1856. doi: 10.1073/pnas.1211521110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Case CL, Shin S, Roy CR. 2009. Asc and Ipaf inflammasomes direct distinct pathways for caspase-1 activation in response to Legionella pneumophila. Infect Immun 77:1981–1991. doi: 10.1128/IAI.01382-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Lightfield KL, Persson J, Brubaker SW, Witte CE, von Moltke J, Dunipace EA, Henry T, Sun YH, Cado D, Dietrich WF, Monack DM, Tsolis RM, Vance RE. 2008. Critical function for Naip5 in inflammasome activation by a conserved carboxy-terminal domain of flagellin. Nat Immunol 9:1171–1178. doi: 10.1038/ni.1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Cerqueira DM, Pereira MS, Silva AL, Cunha LD, Zamboni DS. 2015. Caspase-1 but not caspase-11 is required for NLRC4-mediated pyroptosis and restriction of infection by flagellated legionella species in mouse macrophages and in vivo. J Immunol 195:2303–2311. doi: 10.4049/jimmunol.1501223. [DOI] [PubMed] [Google Scholar]
- 99.Hagar JA, Powell DA, Aachoui Y, Ernst RK, Miao EA. 2013. Cytoplasmic LPS activates caspase-11: implications in TLR4-independent endotoxic shock. Science 341:1250–1253. doi: 10.1126/science.1240988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kayagaki N, Wong MT, Stowe IB, Ramani SR, Gonzalez LC, Akashi-Takamura S, Miyake K, Zhang J, Lee WP, Muszynski A, Forsberg LS, Carlson RW, Dixit VM. 2013. Noncanonical inflammasome activation by intracellular LPS independent of TLR4. Science 341:1246–1249. doi: 10.1126/science.1240248. [DOI] [PubMed] [Google Scholar]
- 101.Shi J, Zhao Y, Wang Y, Gao W, Ding J, Li P, Hu L, Shao F. 2014. Inflammatory caspases are innate immune receptors for intracellular LPS. Nature 514:187–192. [DOI] [PubMed] [Google Scholar]
- 102.Seshadri R, Paulsen IT, Eisen JA, Read TD, Nelson KE, Nelson WC, Ward NL, Tettelin H, Davidsen TM, Beanan MJ, Deboy RT, Daugherty SC, Brinkac LM, Madupu R, Dodson RJ, Khouri HM, Lee KH, Carty HA, Scanlan D, Heinzen RA, Thompson HA, Samuel JE, Fraser CM, Heidelberg JF. 2003. Complete genome sequence of the Q-fever pathogen Coxiella burnetii. Proc Natl Acad Sci U S A 100:5455–5460. doi: 10.1073/pnas.0931379100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Monroe KM, McWhirter SM, Vance RE. 2010. Induction of type I interferons by bacteria. Cell Microbiol 12:881–890. doi: 10.1111/j.1462-5822.2010.01478.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Burdette DL, Monroe KM, Sotelo-Troha K, Iwig JS, Eckert B, Hyodo M, Hayakawa Y, Vance RE. 2011. STING is a direct innate immune sensor of cyclic di-GMP. Nature 478:515–518. doi: 10.1038/nature10429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Cai X, Chiu YH, Chen ZJ. 2014. The cGAS-cGAMP-STING pathway of cytosolic DNA sensing and signaling. Mol Cell 54:289–296. doi: 10.1016/j.molcel.2014.03.040. [DOI] [PubMed] [Google Scholar]
- 106.Schiavoni G, Mauri C, Carlei D, Belardelli F, Pastoris MC, Proietti E. 2004. Type I IFN protects permissive macrophages from Legionella pneumophila infection through an IFN-gamma-independent pathway. J Immunol 173:1266–1275. doi: 10.4049/jimmunol.173.2.1266. [DOI] [PubMed] [Google Scholar]
- 107.Opitz B, Vinzing M, van Laak V, Schmeck B, Heine G, Gunther S, Preissner R, Slevogt H, N′Guessan PD, Eitel J, Goldmann T, Flieger A, Suttorp N, Hippenstiel S. 2006. Legionella pneumophila induces IFNbeta in lung epithelial cells via IPS-1 and IRF3, which also control bacterial replication. J Biol Chem 281:36173–36179. doi: 10.1074/jbc.M604638200. [DOI] [PubMed] [Google Scholar]
- 108.Stetson DB, Medzhitov R. 2006. Recognition of cytosolic DNA activates an IRF3-dependent innate immune response. Immunity 24:93–103. doi: 10.1016/j.immuni.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 109.Howe D, Barrows LF, Lindstrom NM, Heinzen RA. 2002. Nitric oxide inhibits Coxiella burnetii replication and parasitophorous vacuole maturation. Infect Immun 70:5140–5147. doi: 10.1128/IAI.70.9.5140-5147.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Ghigo E, Capo C, Raoult D, Mege JL. 2001. Interleukin-10 stimulates Coxiella burnetii replication in human monocytes through tumor necrosis factor down-modulation: role in microbicidal defect of Q fever. Infect Immun 69:2345–2352. doi: 10.1128/IAI.69.4.2345-2352.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Zamboni DS, Rabinovitch M. 2003. Nitric oxide partially controls Coxiella burnetii phase II infection in mouse primary macrophages. Infect Immun 71:1225–1233. doi: 10.1128/IAI.71.3.1225-1233.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Brennan RE, Russell K, Zhang G, Samuel JE. 2004. Both inducible nitric oxide synthase and NADPH oxidase contribute to the control of virulent phase I Coxiella burnetii infections. Infect Immun 72:6666–6675. doi: 10.1128/IAI.72.11.6666-6675.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Shannon JG, Howe D, Heinzen RA. 2005. Lack of dendritic cell maturation following infection by Coxiella burnetii synthesizing different lipopolysaccharide chemotypes. Ann N Y Acad Sci 1063:154–160. doi: 10.1196/annals.1355.024. [DOI] [PubMed] [Google Scholar]
- 114.Shin S. 2012. Innate immunity to intracellular pathogens: lessons learned from Legionella pneumophila. Adv Appl Microbiol 79:43–71. doi: 10.1016/B978-0-12-394318-7.00003-6. [DOI] [PubMed] [Google Scholar]
- 115.Zamboni DS, McGrath S, Rabinovitch M, Roy CR. 2003. Coxiella burnetii express type IV secretion system proteins that function similarly to components of the Legionella pneumophila Dot/Icm system. Mol Microbiol 49:965–976. doi: 10.1046/j.1365-2958.2003.03626.x. [DOI] [PubMed] [Google Scholar]
- 116.Casson CN, Shin S. 2013. Inflammasome-mediated cell death in response to bacterial pathogens that access the host cell cytosol: lessons from legionella pneumophila. Front Cell Infect Microbiol 3:111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Seshadri R, Samuel J. 2005. Genome analysis of Coxiella burnetii species: insights into pathogenesis and evolution and implications for biodefense. Ann N Y Acad Sci 1063:442–450. doi: 10.1196/annals.1355.063. [DOI] [PubMed] [Google Scholar]
- 118.Elliott A, Peng Y, Zhang G. 2013. Coxiella burnetii interaction with neutrophils and macrophages in vitro and in SCID mice following aerosol infection. Infect Immun 81:4604–4614. doi: 10.1128/IAI.00973-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Bekker LG, Freeman S, Murray PJ, Ryffel B, Kaplan G. 2001. TNF-alpha controls intracellular mycobacterial growth by both inducible nitric oxide synthase-dependent and inducible nitric oxide synthase-independent pathways. J Immunol 166:6728–6734. doi: 10.4049/jimmunol.166.11.6728. [DOI] [PubMed] [Google Scholar]
- 120.Ables GP, Takamatsu D, Noma H, El-Shazly S, Jin HK, Taniguchi T, Sekikawa K, Watanabe T. 2001. The roles of Nramp1 and Tnfa genes in nitric oxide production and their effect on the growth of Salmonella typhimurium in macrophages from Nramp1 congenic and tumor necrosis factor-alpha-/- mice. J Interferon Cytokine Res 21:53–62. doi: 10.1089/107999001459169. [DOI] [PubMed] [Google Scholar]
- 121.Dellacasagrande J, Capo C, Raoult D, Mege JL. 1999. IFN-gamma-mediated control of Coxiella burnetii survival in monocytes: the role of cell apoptosis and TNF. J Immunol 162:2259–2265. [PubMed] [Google Scholar]
- 122.Schroder K, Hertzog PJ, Ravasi T, Hume DA. 2004. Interferon-gamma: an overview of signals, mechanisms and functions. J Leukoc Biol 75:163–189. [DOI] [PubMed] [Google Scholar]
- 123.Zhang Y, Zhang G, Hendrix LR, Tesh VL, Samuel JE. 2012. Coxiella burnetii induces apoptosis during early stage infection via a caspase-independent pathway in human monocytic THP-1 cells. PLoS One 7:e30841. doi: 10.1371/journal.pone.0030841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Brooke RJ, Kretzschmar ME, Mutters NT, Teunis PF. 2013. Human dose response relation for airborne exposure to Coxiella burnetii. BMC Infect Dis 13:488. doi: 10.1186/1471-2334-13-488. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.