Abstract
Deficient hydroxylation of debrisoquine is an autosomal recessive trait that affects approximately 7% of the Caucasian population. These individuals (poor metabolizers) carry deficient CYP2D6 gene variants and have an impaired metabolism of severely commonly used drugs. The opposite phenomenon also exists, and certain individuals metabolize the drugs very rapidly, resulting in subtherapeutic plasma concentrations at normal doses. In the present study, we have investigated the molecular genetic basis for ultrarapid metabolism of debrisoquine. Restriction fragment length polymorphism analysis of the CYP2D locus in two families with very rapid metabolism of debrisoquine [metabolic ratio (MR) for debrisoquine = 0.01-0.1] revealed the variant CYP2D6 gene CYP2D6L. Eco RI RFLP and Xba I pulsed-field gel electrophoresis analyses showed that this gene had been amplified 12-fold in three members (father and his two children) of one of the families, and two copies were present among members of the other family. The CYP2D6L gene had an open reading frame and carried two mutations causing amino acid substitutions: one in exon 6, yielding an Arg-296-->Cys exchange and one in exon 9 causing Ser-486-->Thr. The MR of subjects carrying one copy of the CYP2D6L gene did not significantly differ from that of those with the wild-type gene, indicating that the structural alterations were not of importance of the catalytic properties of the gene product. Examination of the MR among subjects carrying wild-type CYP2D6, CYP2D6L, or deficient alleles revealed a relationship between the number of active genes and MR. The data show the principle of inherited amplification of an active gene. Furthermore, the finding of a specific haplotype with two or more active CYP2D6 genes allows genotyping for ultrarapid drug metabolizers. This genotyping could be of predictive value for individualized and more efficient drug therapy.
Full text
PDF

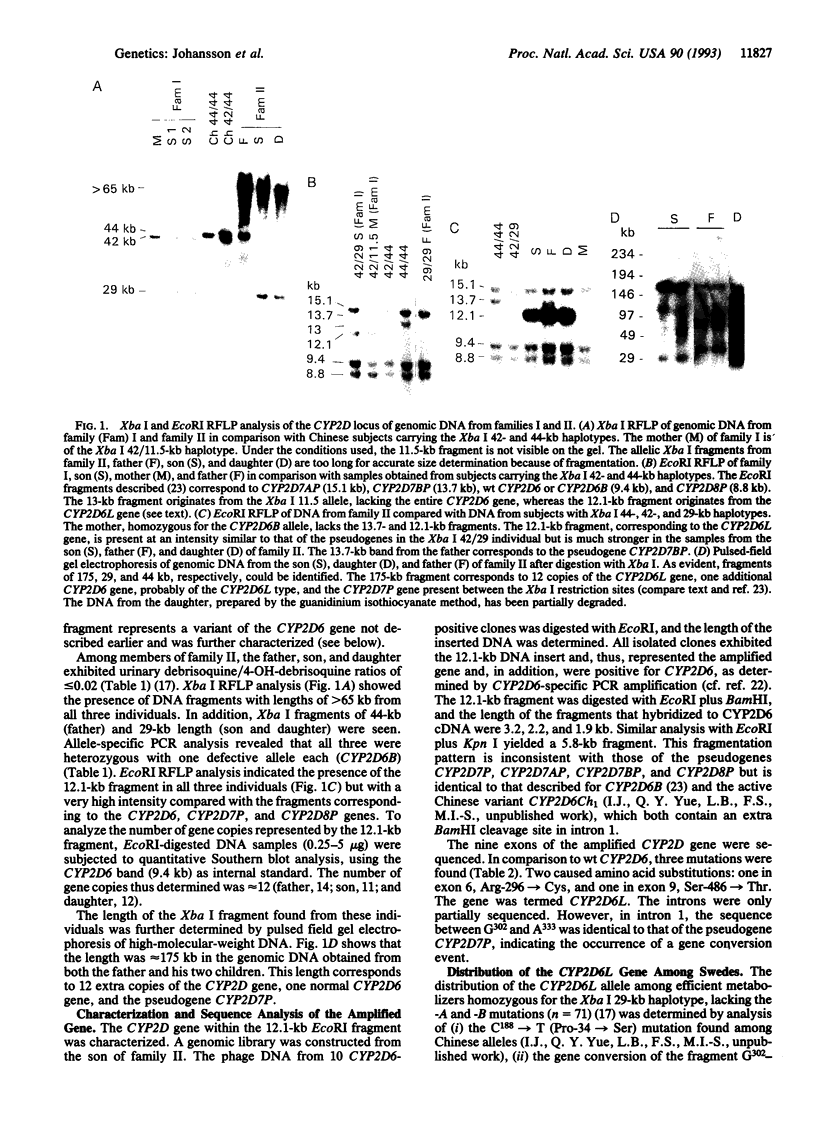
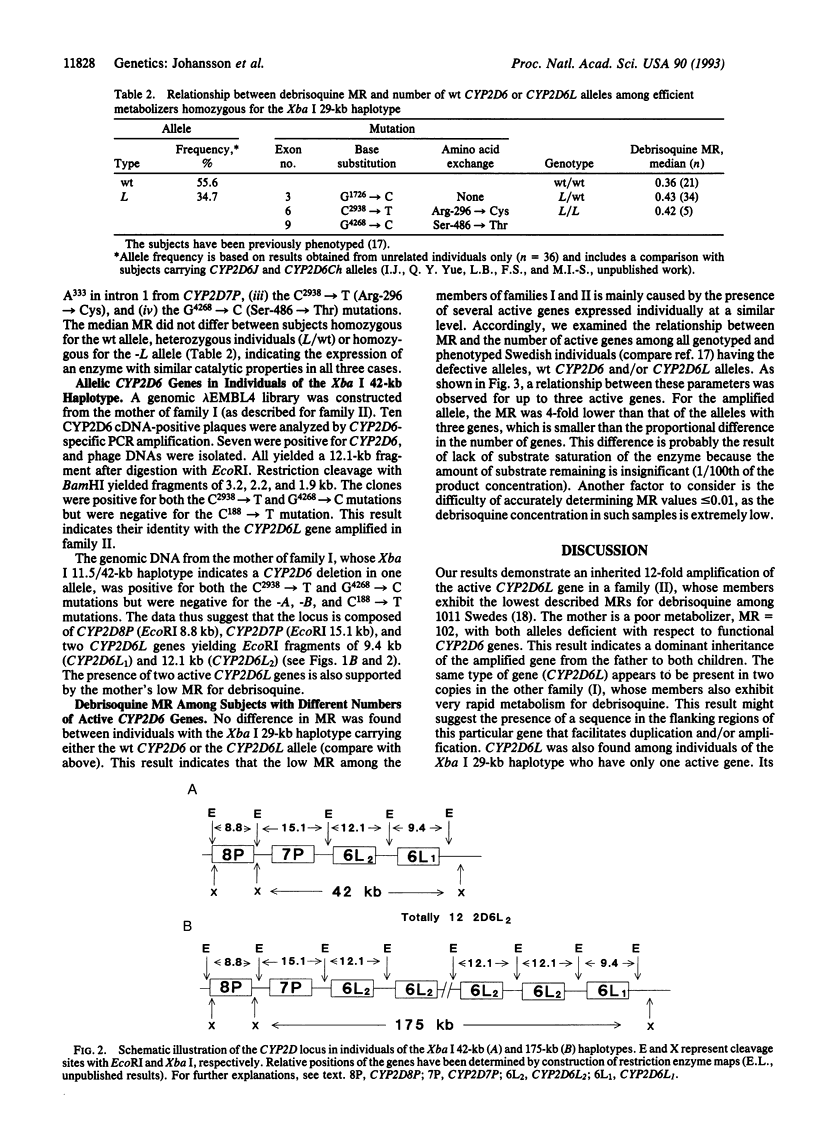

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alt F. W., Kellems R. E., Bertino J. R., Schimke R. T. Selective multiplication of dihydrofolate reductase genes in methotrexate-resistant variants of cultured murine cells. J Biol Chem. 1978 Mar 10;253(5):1357–1370. [PubMed] [Google Scholar]
- Bertilsson L., Aberg-Wistedt A., Gustafsson L. L., Nordin C. Extremely rapid hydroxylation of debrisoquine: a case report with implication for treatment with nortriptyline and other tricyclic antidepressants. Ther Drug Monit. 1985;7(4):478–480. [PubMed] [Google Scholar]
- Bertilsson L., Dahl M. L., Sjöqvist F., Aberg-Wistedt A., Humble M., Johansson I., Lundqvist E., Ingelman-Sundberg M. Molecular basis for rational megaprescribing in ultrarapid hydroxylators of debrisoquine. Lancet. 1993 Jan 2;341(8836):63–63. doi: 10.1016/0140-6736(93)92546-6. [DOI] [PubMed] [Google Scholar]
- Bertilsson L., Lou Y. Q., Du Y. L., Liu Y., Kuang T. Y., Liao X. M., Wang K. Y., Reviriego J., Iselius L., Sjöqvist F. Pronounced differences between native Chinese and Swedish populations in the polymorphic hydroxylations of debrisoquin and S-mephenytoin. Clin Pharmacol Ther. 1992 Apr;51(4):388–397. doi: 10.1038/clpt.1992.38. [DOI] [PubMed] [Google Scholar]
- Dahl M. L., Johansson I., Palmertz M. P., Ingelman-Sundberg M., Sjöqvist F. Analysis of the CYP2D6 gene in relation to debrisoquin and desipramine hydroxylation in a Swedish population. Clin Pharmacol Ther. 1992 Jan;51(1):12–17. doi: 10.1038/clpt.1992.2. [DOI] [PubMed] [Google Scholar]
- Gaedigk A., Blum M., Gaedigk R., Eichelbaum M., Meyer U. A. Deletion of the entire cytochrome P450 CYP2D6 gene as a cause of impaired drug metabolism in poor metabolizers of the debrisoquine/sparteine polymorphism. Am J Hum Genet. 1991 May;48(5):943–950. [PMC free article] [PubMed] [Google Scholar]
- Gonzalez F. J. The molecular biology of cytochrome P450s. Pharmacol Rev. 1988 Dec;40(4):243–288. [PubMed] [Google Scholar]
- Gonzalez F. J., Vilbois F., Hardwick J. P., McBride O. W., Nebert D. W., Gelboin H. V., Meyer U. A. Human debrisoquine 4-hydroxylase (P450IID1): cDNA and deduced amino acid sequence and assignment of the CYP2D locus to chromosome 22. Genomics. 1988 Feb;2(2):174–179. doi: 10.1016/0888-7543(88)90100-0. [DOI] [PubMed] [Google Scholar]
- Heim M. H., Meyer U. A. Evolution of a highly polymorphic human cytochrome P450 gene cluster: CYP2D6. Genomics. 1992 Sep;14(1):49–58. doi: 10.1016/s0888-7543(05)80282-4. [DOI] [PubMed] [Google Scholar]
- Heim M., Meyer U. A. Genotyping of poor metabolisers of debrisoquine by allele-specific PCR amplification. Lancet. 1990 Sep 1;336(8714):529–532. doi: 10.1016/0140-6736(90)92086-w. [DOI] [PubMed] [Google Scholar]
- Kagimoto M., Heim M., Kagimoto K., Zeugin T., Meyer U. A. Multiple mutations of the human cytochrome P450IID6 gene (CYP2D6) in poor metabolizers of debrisoquine. Study of the functional significance of individual mutations by expression of chimeric genes. J Biol Chem. 1990 Oct 5;265(28):17209–17214. [PubMed] [Google Scholar]
- Kalow W. Interethnic variation of drug metabolism. Trends Pharmacol Sci. 1991 Mar;12(3):102–107. doi: 10.1016/0165-6147(91)90516-u. [DOI] [PubMed] [Google Scholar]
- Kimura S., Umeno M., Skoda R. C., Meyer U. A., Gonzalez F. J. The human debrisoquine 4-hydroxylase (CYP2D) locus: sequence and identification of the polymorphic CYP2D6 gene, a related gene, and a pseudogene. Am J Hum Genet. 1989 Dec;45(6):889–904. [PMC free article] [PubMed] [Google Scholar]
- Nishimura S., Sekiya T. Human cancer and cellular oncogenes. Biochem J. 1987 Apr 15;243(2):313–327. doi: 10.1042/bj2430313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prody C. A., Dreyfus P., Zamir R., Zakut H., Soreq H. De novo amplification within a "silent" human cholinesterase gene in a family subjected to prolonged exposure to organophosphorous insecticides. Proc Natl Acad Sci U S A. 1989 Jan;86(2):690–694. doi: 10.1073/pnas.86.2.690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schimke R. T. Gene amplification in cultured animal cells. Cell. 1984 Jul;37(3):705–713. doi: 10.1016/0092-8674(84)90406-9. [DOI] [PubMed] [Google Scholar]
- Srivastava A., Norris J. S., Shmookler Reis R. J., Goldstein S. c-Ha-ras-1 proto-oncogene amplification and overexpression during the limited replicative life span of normal human fibroblasts. J Biol Chem. 1985 May 25;260(10):6404–6409. [PubMed] [Google Scholar]
- Stark G. R. DNA amplification in drug resistant cells and in tumours. Cancer Surv. 1986;5(1):1–23. [PubMed] [Google Scholar]
- Stark G. R., Wahl G. M. Gene amplification. Annu Rev Biochem. 1984;53:447–491. doi: 10.1146/annurev.bi.53.070184.002311. [DOI] [PubMed] [Google Scholar]
- Turner D. R., Morley A. A., Haliandros M., Kutlaca R., Sanderson B. J. In vivo somatic mutations in human lymphocytes frequently result from major gene alterations. Nature. 1985 May 23;315(6017):343–345. doi: 10.1038/315343a0. [DOI] [PubMed] [Google Scholar]
- Wahl G. M., Padgett R. A., Stark G. R. Gene amplification causes overproduction of the first three enzymes of UMP synthesis in N-(phosphonacetyl)-L-aspartate-resistant hamster cells. J Biol Chem. 1979 Sep 10;254(17):8679–8689. [PubMed] [Google Scholar]
- Wright J. A., Smith H. S., Watt F. M., Hancock M. C., Hudson D. L., Stark G. R. DNA amplification is rare in normal human cells. Proc Natl Acad Sci U S A. 1990 Mar;87(5):1791–1795. doi: 10.1073/pnas.87.5.1791. [DOI] [PMC free article] [PubMed] [Google Scholar]