Abstract
Secretory clusterin (sCLU) is a stress-induced, pro-survival glycoprotein elevated in early-stage cancers, in particular in APC/Min-defective colon cancers. sCLU is upregulated after exposure to various cytotoxic agents, including ionizing radiation (IR), leading to a survival advantage. We found that stimulation of insulin-like growth factor-1 (IGF-1)R–mitogen-activated protein kinase signaling was required for sCLU induction after IR exposure. Here, we show that activation of Ataxia telangiectasia-mutated kinase (ATM) by endogenous or exogenous forms of DNA damage was required to relieve basal repression of IGF-1 transcription by the p53/NF-YA complex, leading to sCLU expression. In contrast, ATM-deficient cells were unable to induce IGF-1 and sCLU after DNA damage. Although p53 levels were stabilized and elevated after DNA damage, dissociation of NF-YA and thereby p53 from the IGF-1 promoter resulted in IGF-1 induction, indicating that NF-YA was rate limiting. Cells with elevated endogenous DNA damage (deficient in H2AX, MDC1, NBS1, mTR or hMLH1) or cells exposed to DNA-damaging agents had elevated IGF-1 expression, resulting in activation of IGF-1R signaling and sCLU induction. In contrast, ATM-deficient cells were unable to induce sCLU after DNA damage. Our results integrate DNA damage from genetic instability and in response to IR or chemotherapeutic agents, to ATM activation and abrogation of p53/NF-YA-mediated IGF-1 transcriptional repression, leading to IGF-1–sCLU expression. Elucidation of this pathway is important to uncover new mechanisms for cancer progression and reveal new targets for drug development to overcome resistance to therapy.
Keywords: secretory clusterin, ATM, IGF-1, p53, ionizing radiation, genetic instability
Introduction
Before neoplastic transformation, initiated normal cells undergo a re-programming of inherent gene expression, permitting subsequent cancer promotion and progression. These changes typically result in upregulation of proteins that provide pro-survival signals and down-regulation of cell-death initiation events. One pro-survival pathway commonly and constitutively upregulated in human tumors is the insulin-like growth factor-1 (IGF-1)/IGF-1 receptor (IGF-1R) tyrosine kinase signaling cascade (Ryan and Goss, 2008). We found that exposure of cells to IGF-1 induced secretory clusterin (sCLU) expression, and that IGF-1R signaling was essential for sCLU upregulation after ionizing radiation (IR) exposure. In addition to IR, sCLU is induced after exposure to chemotherapeutic agents and ultraviolet (UV) radiation (Boothman et al., 1993; Miyake et al., 2000); however, the role of IGF-1/IGF-1R signaling in sCLU induction after these agents was not investigated. Although we noted that wild-type p53 repressed basal sCLU expression and partially inhibited sCLU upregulation after IR exposure (Criswell et al., 2003), a link between IGF-1R signaling and p53 suppression was not elucidated. The initiating signals from DNA damage to upregulation of IGF-1/IGF-1R signaling remain undefined.
sCLU is a heavily glycosylated, 80 kDa secreted protein that functions in both an extra- and intracellular manner to promote cell survival. sCLU is synthesized as a 60-kDa precursor protein (psCLU) that is targeted to the ER. Within the ER, psCLU is cleaved approximately into half, linked together by five disulfide bonds, and is heavily glycosylated to form mature sCLU (80 kDa). sCLU production appears as two major protein forms by western blotting. The band at ~60 kDa corresponds to psCLU and represents the protein before maturation. The ~40-kDa smear corresponds to one half of the mature sCLU at varying states of glycosylation (Jones and Jomary, 2002). Expression changes in psCLU and sCLU are always concordant, and both forms consistently match CLU promoter activity (Criswell et al., 2003), allowing any one of these proteins to be monitored for changes in sCLU expression.
Outside the cell, sCLU acts as a molecular chaperone, binding to stress-induced unfolded proteins, lipids and amyloid, among other molecules, and works to clear cell debris after tissue trauma (de Silva et al., 1990; Boggs et al., 1996; Wilson and Easterbrook-Smith, 2000). Within the cell, sCLU can bind Bax and prevent its translocation to the mitochondria, thereby blocking apoptosis (Zhang et al., 2005). Owing to these pro-survival functions, sCLU overexpression is responsible for increased resistance of cancer cells to various stresses, including doxorubicin, cisplatin and taxol (Miyake et al., 2000). Depletion of sCLU by antisense or small interfering RNA causes hypersensitization of cancer cells to paclitaxel and IR (Criswell et al., 2005; So et al., 2005). Aside from exogenous stress, sCLU is constitutively elevated in many early-stage cancers, during replicative senescence, and in Alzheimer’s disease (Chen et al., 2003). These observations strongly suggest that elevated sCLU levels in cancer, as well as induction of sCLU after cytotoxic agent exposures, may result in the consequent resistance of cancer cells to therapy, including cancer stem cells. Therefore, antisense constructs specific for sCLU (that is, OGX-011) were developed and are now in phase II/III clinical trials in combination with standard chemotherapies for prostate cancer (Chi et al., 2010). Thus, understanding sCLU regulation after DNA damage and in genetically unstable cells should lead to a better understanding of resistance to therapy and open avenues for improving cancer therapy.
Here, we show that p53 and NF-YA are necessary to suppress basal IGF-1 transcription by binding together at a single NF-Y consensus site within the IGF-1 promoter. Although p53 levels were stabilized and elevated after IR exposure, loss of p53/NF-YA binding from the IGF-1 promoter after IR exposure allows for IGF-1 induction to, in turn, activate IGF-1R signaling and sCLU induction. Interestingly, we noted that genetically unstable cells have heightened basal IGF-1 and sCLU levels and concomitant elevated Ataxia telangiectasia-mutated kinase (ATM) signaling, not unlike cells undergoing a DNA damage response. We found that ATM was the DNA damage sensor kinase responsible for IGF-1–sCLU upregulation after endogenous or exogenous DNA damage. Collectively, our findings show that the ATM–IGF-1–sCLU expression axis is an important pro-survival pathway activated in response DNA damage, which has important implications in carcinogenesis, tumor progression and resistance in cancer therapies.
Results
p53 negatively regulates basal IGF-1 expression
p53 has an important and essential role in responding to cell stress by either activating or repressing gene expression, controlling checkpoint responses, and inducing apoptosis. Data from our laboratory suggested that p53 suppressed basal sCLU expression; however, sCLU was moderately induced after IR exposure, even though p53 was stabilized (Criswell et al., 2003). The mechanism of p53-mediated repression of basal sCLU expression was not explored. As prior data suggested that IGF-1 signaling may regulate sCLU expression (Criswell et al., 2005) and that p53 was regulated, through Mdm-2 expression, by IGF-1 (Mayo and Donner, 2001), we examined the ability of p53 to regulate IGF-1 transcription. To test this, the IGF-1 promoter fused to luciferase reporter (IGF-1–LUC) (Mittanck et al., 1997) was transfected into a series of genetically matched cell lines altered in functional p53 (Figure 1a). p53-null (PC3 and HCT116 p53−/−) and p53 knockdown (RKO7 shp53, HCT116:3-6 shp53, HBEC 3kt shp53) cells had higher basal IGF-1 promoter activity compared with matched WT p53-expressing cells (white bars; PC3 (stably expressing WT p53), RKO7 SCR, HCT116 SCR, HCT116:3-6 SCR and HBEC 3kt). Immortalized human bronchial epithelial (HBEC 3kt, WT p53) cells expressing mutant R273H p53 (mp53) also had elevated basal IGF-1 promoter activity. Similarly, IGF-1 mRNA and protein expression was higher in HCT116:3-6 shp53 compared with SCR cells (Figures 1b and c). These data suggested that p53 transcriptionally repressed basal IGF-1 in a dose-dependent manner, as complete loss of functional p53 allowed for greater IGF-1 expression than in cells where p53 was knocked down.
Figure 1.
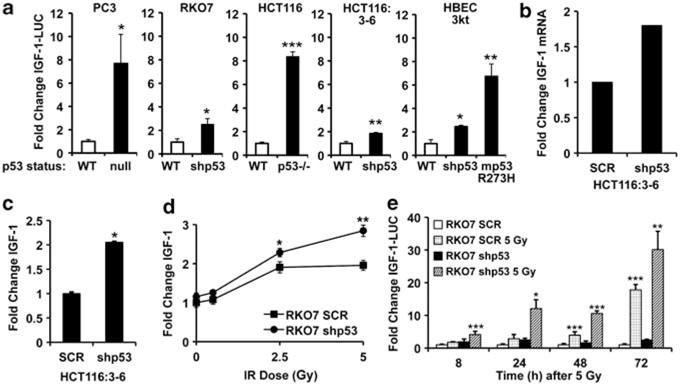
p53 suppresses IGF-1 expression. (a) PC3, PC-3 + p53; RKO7 SCR, RKO7 shp53; HCT116, HCT16 p53−/− ; HCT116:3-6, HCT116:3-6 shp53; and immortalized HBECs (WT, shp53, and mutant p53 (mp53)) were transiently transfected with IGF-1–LUC and RSV-β-Gal and monitored for luciferase and β-gal activities 72 h later. *WT vs null, shp53, p53−/−, or mp53. (b) HCT116:3-6 and HCT16:3-6 shp53 cells were analyzed for IGF-1 and actin mRNA using RT-PCR. Data were graphed as the relative density of stained bands from semi-quantitative RT-PCR. (c) Media were collected from HCT116:3-6 and HCT116:3-6 shp53 cells and analyzed by ELISA. (d) RKO7 SCR and shp53 cells were irradiated, media collected 72 h later, and IGF-1 measured by ELISA. Data were normalized to UT RKO7 SCR cells. */**, RKO7 SCR vs shp53. (e) RKO7 SCR and shp53 cells were transiently transfected with IGF-1–LUC and RSV-β-Gal, exposed to 5 Gy, and luciferase and β-Gal monitored. */**/***, mock vs 5 Gy. For data were graphed as means±s.d. *P≤0.05, **P≤0.01, ***P≤0.005.
As IGF-1R signaling is activated by IR exposure (Criswell et al., 2005), we examined whether IGF-1 was elevated after IR exposure, and if its expression was dependent on p53 status. In RKO7 SCR and shp53 cells, IGF-1 protein and promoter activity was induced in both cell lines after IR exposure. However, IGF-1 was induced to a significantly greater extent when p53 was knocked down (Figures 1d and e). These data strongly suggested that p53 transcriptionally repressed basal IGF-1 expression, while only partially blocking IR-induced IGF-1. The data also suggested that the p53 repression mechanism of IGF-1 (and sCLU) was fundamentally different than other p53 targets, such as Chk2, as Chk2 is further suppressed after IR exposure owing to stabilization of p53 (Matsui et al., 2004).
p53 and NF-Y-mediated transcriptional regulation of IGF-1
As IGF-1 was induced in WT p53 cells after IR exposure even though p53 was stabilized, we investigated the mechanism of p53 suppression, and whether this changed after IR. Interrogation of the human IGF-1 promoter for potential regulatory elements repressed by p53 led to the identification of a single NF-Y consensus site found at −438 bp in the IGF-1 promoter (Figure 2a). NF-Y sites are bound by NF-Y proteins, which are trimeric transcription factor complexes comprised of NF-YA, -YB and -YC proteins. NF-Y can interact with p53 to transcriptionally repress specific genes, including Cdc2 and Chk2 (Yun et al., 1999; Matsui et al., 2004). To determine whether the NF-Y consensus site in the IGF-1 promoter was required for transcriptional repression by p53, we mutated the site (Figures 2a and b). Dramatically increased activity in HCT116 cells was observed with the NF-Y mutant IGF-1 promoter (NF-Y MUT), similar to the activity of the WT IGF-1 promoter in HCT116 p53−/− cells (Figure 2b). Thus, p53 suppressed basal IGF-1 promoter activity through an NF-Y binding site in HCT116 cells.
Figure 2.
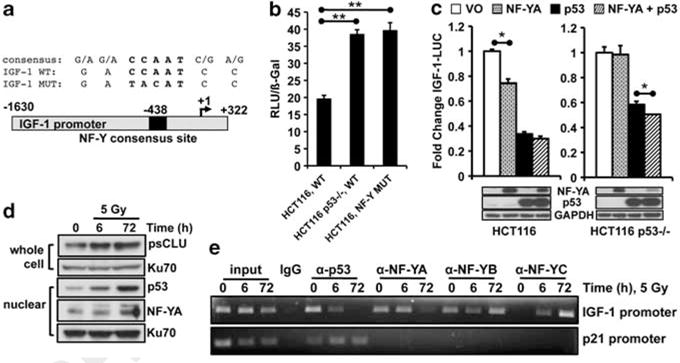
NF-YA/p53-mediated suppression of IGF-1. (a) Top, consensus sequence of NF-Y promoter binding site in the IGF-1 promoter. Bottom, schematic of IGF-1 promoter. (b) HCT116 or HCT116 p53−/− cells were transfected with IGF-1–LUC (WT) or IGF-1–LUC with a mutated NF-Y site (NF-Y MUT), and RSV-β-Gal. Cells were harvested 48 h later for luciferase measurements. RLU, relative light units. (c) HCT116 or HCT116 p53−/− cells were transfected with CMV-NF-YA, CMV-Flag-p53 or both, and IGF-1–LUC and RSV-β-gal. Cells were harvested at 24 h. Bottom, immunoblot of samples correspond to each bar graph. (d) HCT116 cells were irradiated and harvested by nuclear or whole-cell extraction for immunoblotting. (e) HCT116 cells were exposed to 5 Gy and harvested for ChIP to monitor binding to the IGF-1 or p21 promoters. For (b, c), data were graphed as means±s.d. *P≤0.05.
To examine whether NF-Y alone could repress the IGF-1 promoter in the absence of p53, NF-YA was overexpressed in HCT116 cells. NF-YA overexpression in HCT116 cells resulted in modest repression of the IGF-1 promoter; however, NF-YA was not able to suppress in the absence of p53 (HCT116 p53−/−, Figure 2c), consistent with studies showing that NF-YA mediates the interaction between p53 and DNA (Imbriano et al., 2005). Finally, chromatin immunoprecipitation analyses were performed to examine the binding of p53 and NF-YA, -YB or -YC to the IGF-1 promoter. After IR exposure, p53 protein levels were stabilized (Figure 2d); however, p53 and NF-YA binding to the IGF-1 promoter decreased. NF-YB and NF-YC binding increased 72 h after IR exposure (Figure 2e). Intriguingly, NF-YB binding to the IGF-1 promoter decreased 6 h after IR exposure; however, as IGF-1 and sCLU are most robustly induced 48–72 h after IR exposure, decreased binding at 6 h may not affect IGF-1 expression. As a positive control, increased p53 binding to the p21 promoter was noted after IR exposure (Figure 2e). These data suggest that IGF-1 induction after IR exposure in WT p53 cells was due to loss of p53 and NF-YA occupancy on the IGF-1 promoter, and thereby loss of repression. These data strongly suggest that NF-YA is rate limiting and p53 alone is not capable of repressing IGF-1.
p53 suppression of the IGF-1–sCLU pathway
To show that IGF-1 signaling was upstream of sCLU expression, cells were treated with AG1024, an IGF-1R tyrosine kinase inhibitor, before IR exposure. AG1024 blocked IR-induced CLU promoter activity in RKO7 shp53 cells, while having no effect on IGF-1 promoter induction (Figure 3a). The overall lower induction of IGF-1 and CLU promoter activity observed in the WT p53 RKO7 cells was probably due to the low serum conditions required for AG1024 efficacy. To determine if both IGF-1 and CLU expression were similarly regulated by p53, HCT116 and HCT116 p53−/− cells were transiently transfected with WT flag-tagged p53 cDNA (WT p53) and IGF-1– or CLU–LUC reporters before IR exposure. Basal IGF-1 and CLU promoter activities were significantly higher in HCT116 p53−/− cells compared with WT p53 cells (Figure 3b), consistent with previous results. Overexpression of non-lethal p53 levels partially blocked IR-induced IGF-1 and CLU promoter activities in both cell lines (Figure 3b). CLU and IGF-1 promoter activity and sCLU protein induction after IR exposure were also partially blocked by p53 in RKO7 cells (Supplementary Figures S1A–C). As both IGF-1 and CLU were similarly regulated by p53, an inter-dependent relationship between the expression patterns of these two genes was revealed, showing that upstream IGF-1 expression directly controlled sCLU synthesis.
Figure 3.
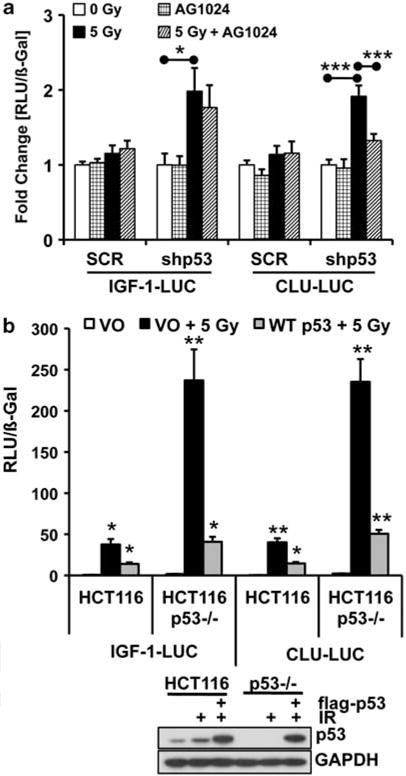
IGF-1 signaling is upstream of sCLU expression. (a) RKO7 SCR or shp53 cells were transiently transfected with IGF-1–LUC or CLU-LUC and RSV-β-gal. The next day, cells were pre-treated with AG1024 for 1 h before IR (5 Gy) and harvested 48 h later. Data were normalized to 0 Gy. (b) HCT116 and HCT116 p53−/− cells were co-transfected with vector or flag-WT p53 expression plasmid (5 ng), IGF-1–LUC or CLU-LUC, and RSV-β-Gal. Cells were either mock- or IR-treated and luciferase activities measured 48 h later. Samples were normalized to mock-treated, vector only transfected (VO). RLU, relative light units.
Induction of sCLU by DNA damage
As IGF-1R signaling was required for sCLU induction after IR exposure, we examined whether this pathway was activated after other forms of DNA damage. In RKO7 cells, induction of both the intracellular, ~60-kDa precursor sCLU (psCLU) and the ~80-kDa mature sCLU, appearing as an ~40-kDa smear, were noted after exposure to IR, H2O2 or topoisomerase I or IIa poisons, topotecan (TPT) and VP16, respectively (Figures 4a and c). Induction of sCLU was inhibited by AG1024 pre-treatment (Figure 4a). IGF-1 expression and phosphorylation of ERK were observed after DNA damage (Figure 4b and Supplementary Figures S2A–C), consistent with IGF-1R–mitogen-activated protein kinase (MAPK) activation after IR exposure (Criswell et al., 2005). These data suggested a common DNA damage-induced pathway of IGF-1 induction that activates IGF-1R–MAPK signaling, leading to sCLU expression.
Figure 4.
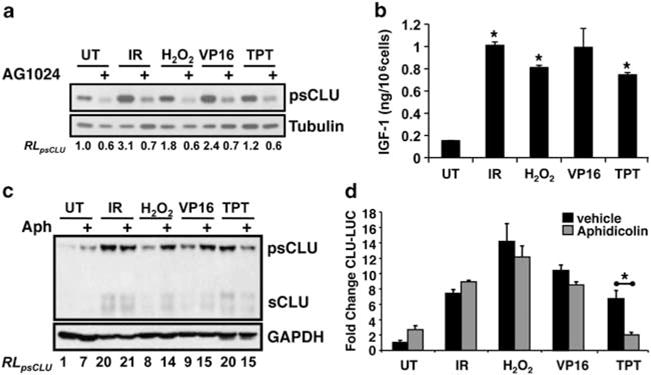
IGF-1–sCLU is induced after exogenous DNA damage. (a) Serum-starved MCF-7 cells were pre-treated with 4 μM AG1024 or DMSO and then treated with IR (5 Gy), H2O2 (50 μM), VP16 (10 μM) or TPT (2.2 μM) for 5 h or DMSO (UT). Whole-cell extracts were collected 72 h later for immunoblotting. (b) MCF-7 cells were treated as in (a), media collected at 48 h, and IGF-1 levels assessed by ELISA. *Treated vs untreated. (c) MCF-7 cells were pre-treated with Aph (100 ng/ml or DMSO for 2 h before the agents mentioned in (a), and extracts harvested at 48 h for western blotting. (d) MCF-7 cells were transiently transfected with CLU-LUC and RSV-β-gal and treated as in (c). For (b, d), data were graphed as means±s.d. *P≤0.05. RL, relative levels.
To determine whether DNA double-strand breaks (DSBs) were necessary for sCLU induction, cells were exposed to aphidicolin (Aph), a DNA polymerase α inhibitor, before treatment with various DNA-damaging agents. Topotecan, a topoisomerase-I poison, causes DNA–protein crosslinks that are converted to DSBs during DNA replication, which are prevented by Aph (D’Arpa et al., 1990). Aph abrogated TPT-induced sCLU protein expression and CLU promoter activity in RKO7 cells (Figures 4c and d). In contrast, Aph did not affect sCLU induction for the agents that directly caused DSBs. These data suggested that DSBs were minimally sufficient to induce sCLU.
The IGF-1–sCLU expression axis is upregulated in genetically unstable cells
Noting that DSBs were minimally required for IGF-1– sCLU expression (Figures 4c and d) and that genetically unstable cells commonly exhibit constitutive DSBs, we examined whether genetic instability, in general, led to an increase in sCLU expression. Various genetically unstable cells were examined for IGF-1 and sCLU expression compared with their stable, genetically matched cells (Figures 5a and b). Cells deficient in H2AX (H2AX−/−) have defective localization of BRCA1 and 53BP1, deficiencies in homologous recombination and persistent DSBs (Celeste et al., 2002). Elevated basal expression of sCLU was noted in H2AX−/− MEFs compared with their genetically matched and stable, WT counterparts. Similarly, sCLU expression was constitutively elevated in MDC1−/− MEFs, which have defects in ATM recruitment (but not activation), leading to increased DSBs and genetic instability (Lou et al., 2006). sCLU expression was also elevated in fifth generation MEFs defective for the RNA component of mouse telomerase, mTR (Figure 5a); mTR−/− MEFs are genetically unstable owing to a failure to extend telomere ends, revealing DSBs that lead to chromosomal rearrangements and end-to-end fusions (Hao and Greider, 2004). Genetically unstable NBS1−/− MEFs (Zhu et al., 2001) and mismatch repair-deficient (MMR, hMLH1-deficient) HCT116 cells (Wagner et al., 2008) also exhibited elevated basal sCLU expression compared with their matched, genetically stable WT counterparts (Figure 5b).
Figure 5.
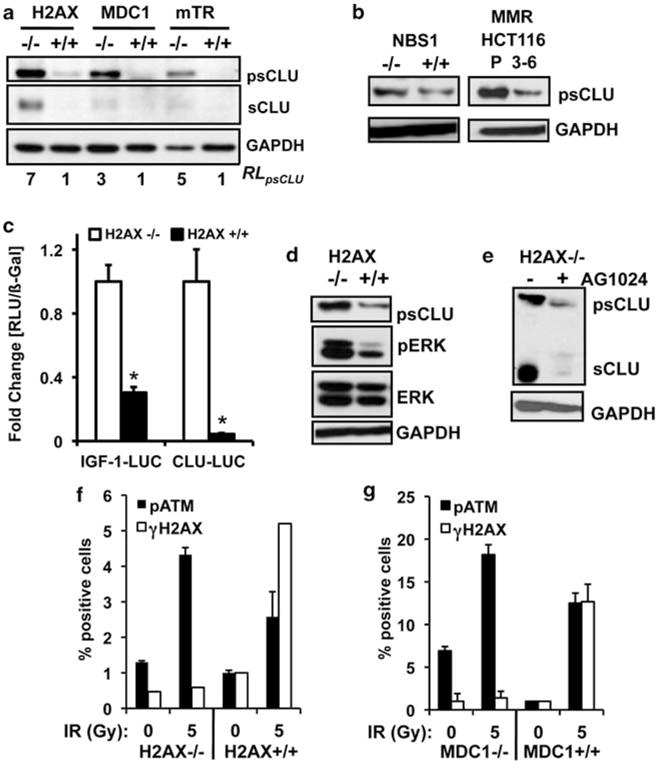
IGF-1–sCLU is upregulated in genetically unstable cells. (a) H2AX, MDC1, or mTR WT (+/+) or deficient (−/−) MEFs were harvested for immunoblotting. (b) NBS1+/+ and NBS1−/− MEFs and HCT116 parental (P) or chromosome 3 expressing 3–6 human colon cancer cells were analyzed for basal sCLU expression by immunoblotting. (c) H2AX−/− or H2AX+/+ MEFs were transiently transfected with CLU-LUC or IGF-1–LUC and RSV-β-gal and analyzed for luciferase activity and β-gal expression. (d) H2AX−/− and H2AX+/+ MEFs were analyzed for phospho–Erk-1/2 (pErk) levels by immunoblotting. (e) H2AX−/− and H2AX+/+ MEFs were treated with AG1024 (2 μM) and collected 48 h later. (f, g) H2AX (f) or MDC1 (g) WT (+/+) or deficient (−/−) MEFs were analyzed for pATMS1981 and γ-H2AX levels 1 h after 5 Gy by flow cytometry. For (c, f, g), data were graphed as means±s.d. *P≤0.05. RL, relative levels; RLU, relative light units.
We then examined whether IGF-1 signaling was elevated in cells with endogenous genetic instability. Elevated IGF-1 and CLU promoter activity was noted in H2AX−/− MEFs, compared with WT control cells (Figure 5c). Higher levels of intracellular IGF-1 in H2AX−/− MEFs were noted (Supplementary Figure S2D), with constitutively elevated basal phosphorylation of IGF-1R, AKT1 and ERK (Figure 5d and Supplementary Figure S2E). Exposure of both MEFs to exogenous IGF-1 induced phosphorylation of IGF-1R and its downstream target, AKT1 (Supplementary Figure S2E), confirming that IGF-1R signaling was intact in WT MEFs. Finally, AG1024 exposure reduced basal sCLU expression in H2AX−/−, MDC1−/− and mTR−/− MEFs (Figure 5e, and Supplementary Figures S2F and G), suggesting that IGF-1/IGF-1R signaling was constitutively active in unstable cells, leading to sCLU expression. These data strongly suggested that the same IGF-1/IGF-1R/MAPK/ERK signaling pathway that was activated after exogenous DNA damage was responsible for stimulating basal sCLU expression in genetically unstable cells.
sCLU induction following DNA damage is mediated by ATM
As IGF-1–sCLU expression was elevated in response to DNA damage, we wanted to delineate the DNA damage sensor responsible for upregulating this expression axis. As ATM and ATR signaling are activated in response to the DNA-damaging agents used, we suspected that these two PIKKs might be involved in IGF-1–sCLU expression. The genetically unstable MEFs were examined for upregulation of ATM signaling by analyzing phosphorylation of H2AX (γ-H2AX) and auto-activation of ATM (phosphorylation of S1981). Neither H2AX−/− nor MDC1−/− MEFs showed γ-H2AX staining due to their known abrogation of this response (Lou et al., 2006), but had elevated basal pATMS1981 levels (Figures 5f and g). WT MEFs were responsive to IR exposure, showing elevated pATMS1981 and γ-H2AX staining (Figures 5f and g). Additionally, both pATMS1981 and γ-H2AX were elevated in RKO7 cells exposed to TPT, which were abrogated by Aph pre-treatment (Supplementary Figure S3A). ATR signaling, as determined by Chk1 phosphorylation on serines 317 and 345, was elevated in H2AX-, MDC1- and mTR-deficient cells (Supplementary Figure S3B). These data suggested that both ATM and ATR signaling were elevated under the same conditions in which IGF-1–sCLU expression was upregulated.
The requirement for ATM or ATR activation in IGF-1/sCLU expression was examined by pre-treating cells with the dual ATM and ATR kinase inhibitor (see Bentle et al., 2006). ATR kinase inhibitor blocked pATMS1981 and γ-H2AX staining in irradiated MEF and human cells (Supplementary Figures S4A and B), and lowered basal sCLU protein expression in genetically unstable cells (Figure 6a). ATR kinase inhibitor also prevented sCLU induction in MCF-7 cells following IR or TPT exposure (Figure 6b). Thus, either ATM or ATR were possible DNA damage sensor kinases directing IGF-1–sCLU induction.
Figure 6.
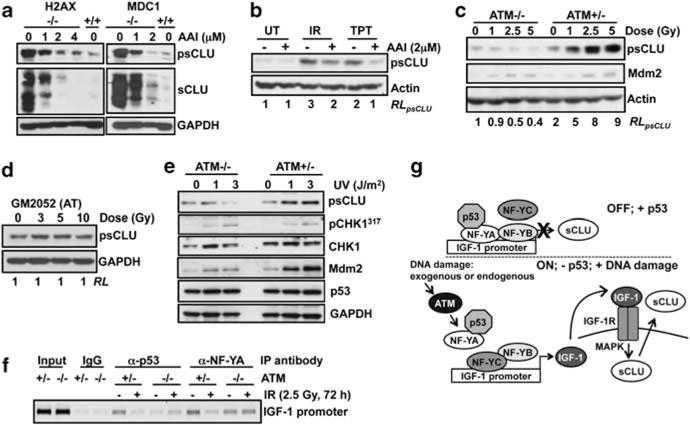
sCLU induction after DNA damage is mediated by ATM. (a) H2AX−/− and MDC1−/− cells were exposed to ATR kinase inhibitor (1–4 μM, 48 h) and harvested for immunoblotting. (b) MCF-7 cells were pretreated with ATR kinase inhibitor (2 μM, 1 h) before IR or TPT treatments, and then harvested at 72 h for western blotting. (c) Immortalized AT (ATM−/−) and AT reconstituted for ATM (ATM+/−) cells were exposed to 0–5 Gy and analyzed 48 h later by immunoblotting. (d) Primary human AT-deficient fibroblasts, GM2052, were treated with 0–10 Gy and analyzed 72 h later by immunoblotting. (e) ATM−/− and ATM+/− cells were exposed to UV (1 or 3 J/m2) or mock-treated, and harvested at 48 h for immunoblotting. (f) ATM−/− and ATM+/− were exposed to 2.5 Gy and harvested 72 h later for ChIP. p53 and NF-YA were immunoprecipitated, and the IGF-1 promoter was amplified using specific primers. (g) Model of ATM-IGF-1–MAPK–sCLU signaling pathway in the absence or presence of p53 and DNA damage. RL, relative levels.
To delineate between ATM and ATR, sCLU induction was examined in genetically defined, SV-40 immortalized AT fibroblasts (ATM−/−) compared with genetically matched, ATM-reconstituted AT fibroblasts (ATM+/−). Although ATM−/− cells are genetically unstable (Kojis et al., 1991) and sensitive to IR (Ziv et al., 1997), sCLU basal expression was equal to that of ATM-corrected (ATM+/−) fibroblasts (Figures 6c and e, and Supplementary Figure S5A), owing to the presence of IGF-1 in the culture medium. Importantly, ATM−/− fibroblasts did not induce sCLU after IR exposure, unlike ATM+/− fibroblasts (Figure 6c). Similarly, sCLU expression was not induced in GM2052 primary AT fibroblasts after IR exposure (Figure 6d). As an important control, both ATM−/− and ATM+/− fibroblasts were able to induce sCLU and showed phosphorylation of IGF-1R and Akt1 by exogenous IGF-1 (Supplementary Figures S5A and B), indicating that both cell lines were proficient in IGF-1–sCLU expression, but not in signaling from DNA damage to sCLU induction.
The role of ATR signaling in IGF-1–sCLU expression was examined by exposing ATM−/− and ATM+/− fibroblasts to UV (1 or 3 J/m2), which predominately activates ATR. Similar to IR, sCLU induction was observed only in ATM+/− cells after UV treatment (Figure 6e), probably due to ATR-dependent activation of ATM after UV exposure (Stiff et al., 2006). As a control, p53-mediated DNA damage-inducible expression of the human homolog of Mdm2 was noted in both ATM−/− and ATM+/− cells after UV exposure (Figure 6e) (Perry et al., 1993). Also, Chk1S317 phosphorylation in UV-irradiated ATM−/− and ATM+/− cells was noted (Figure 6e and Supplementary Figure S5C), confirming ATR activation (Zhao and Piwnica-Worms, 2001). These data strongly suggested that ATM, and not ATR, was necessary for sCLU induction after IR exposure.
As p53 and NF-YA binding to the IGF-1 promoter were lost after IR exposure in HCT116 cells, p53/NF-YA binding was examined in ATM+/− vs ATM−/− cells before and after IR. As in HCT116 cells (Figure 2e), both p53 and NF-YA were lost from the IGF-1 promoter after IR exposure in ATM+/−, although no change was detected in ATM−/− cells (Figure 6f). These data strongly suggested that ATM signaling was important for loss of NF-YA and p53 from the IGF-1 promoter after IR exposure, and that alterations are required for IGF-1–sCLU upregulation in response to DNA damage.
As ATM and ATM-activated proteins (for example, activated Chk2) phosphorylate p53 at serines 15 and 20 after IR exposure, we examined whether these post-translational modifications of p53 altered its ability to repress IGF-1 or sCLU. HCT116 p53−/− cells were transiently transfected with WT p53, or p53 constructs containing serine 15 or 20 mutated to non-phosphorylated alanine mutants (S15A, S20A), or phospho-mimic aspartate (S15D) or glutamate mutants (S15E, S20E). All serine 15 and 20 mutants repressed IGF-1 and CLU promoter activity similar to WT p53 (Supplementary Figures S5D and E), even though these are obvious direct and indirect targets of ATM kinase activity after DNA damage. As a positive control, the p21 promoter (el-Deiry et al., 1993) was induced by WT and to a lesser extent by S15A, S15E and S20A p53 mutants (Supplementary Figure S5F) (Dumaz and Meek, 1999). Mutation of S20 to glutamate enhanced p53-mediated activation of the p21 promoter over WT. Additionally, mutation of p53 at S20 blocked the ability of the DO1 p53 antibody to detect p53 (Supplementary Figure S5F, right). Thus, our data strongly suggested that activation of ATM signaling, and not ATR, was sufficient to induce IGF-1–sCLU expression, a common damage-inducible pro-survival expression axis initiated by genotoxic stress or genetic instability.
Discussion
Genetic instability is a hallmark of cancer initiation, triggering changes that induce a switch from normal to uncontrolled growth. Engagement of survival pathways within these aberrant cells is important for cancer promotion and progression. Our results integrate a novel signaling pathway, initiating from ATM, that activates the pro-survival IGF-1–sCLU axis due to endogenous or exogenous DNA damage (Figure 6g). All cytotoxic agents that induced DSBs and activated ATM kinase led to IGF-1 induction and subsequent activation of IGF-1R, leading to sCLU expression. In normal cells, transient upregulation of IGF-1–sCLU expression after DNA damage is most likely beneficial, blocking apoptosis, providing a mitogenic stimulus and mobilizing glucose utilization to maintain cellular homeostasis until DNA repair is concluded. When this pathway is uncontrolled, however, like in genetically unstable cells or by loss of p53 function, IGF-1–sCLU upregulation may promote resistance of tumors to IR or chemotherapies and allow initiated cells to progress.
Several studies have suggested links between ATM, p53 and IGF-1 signaling, but this is the first report to show that p53, in conjunction with NF-YA, directly suppresses the IGF-1 promoter (Figures 6f and g). Other studies suggested that p53 suppressed IGF-1; however, no mechanistic data were provided (Bocchetta et al., 2008; Sulkowski et al., 2009). Additionally, ATM upregulates IGF-1R expression, and loss of IGF-1R, in turn, blocks ATM activation after IR exposure (Macaulay et al., 2001; Shahrabani-Gargir et al., 2004). These observations appear to explain the biphasic activation of IGF-1R–MAPK signaling observed in MCF-7 cells after IR exposure (Criswell et al., 2005). Initial IGF-1 signaling by ATM activation could trigger a more robust positive feedback loop, wherein IR-induced IGF-1 signaling activates AKT1. AKT1 stimulation then leads to p53 degradation via activation of Mdm2 (Leri et al., 1999), resulting in robust but delayed synthesis of IGF-1–sCLU. This may also explain the slower kinetics of IGF-1 induction in RKO7 SCR cells, compared with RKO7 shp53 cells, because the RKO7 shp53 cells already have lowered p53 (Figure 1e).
NF-Y A–C proteins bind p53 and suppress transcription of target genes involved in cell cycle regulation, such as Cdc2 and Chk2 (Yun et al., 1999; Matsui et al., 2004). These genes are suppressed after DNA damage by p53 stabilization (Matsui et al., 2004). In contrast, IGF-1–sCLU expression was induced after low- (⩾2 cGy) and high-dose IR exposures, even though p53 was stabilized (Klokov et al., 2004). This was concomitant with the simultaneous loss of NF-YA and p53 binding to the IGF-1 promoter. NF-YB and NF-YC binding increased after IR exposure (Figure 2e), consistent with their abilities to bind TFIID independently of NF-YA and promote transcription (Bellorini et al., 1997). Even though the mechanism of IGF-1 suppression by p53 and NF-YA is different from other known targets, it is not incongruous with the known functions of the NF-Y subunits.
As we uncovered a novel mechanism of p53/NF-YA-mediated suppression of IGF-1 transcription, we propose several possibilities for this loss of suppression after IR exposure, including post-translational modification of NF-YA and/or p53. Cdk2 can phosphorylate NF-YA, and mutation of these sites blocked its ability to bind DNA (Yun et al., 2003). We propose a model whereby Cdk2 promotes NF-YA phosphorylation in the basal state, enhancing p53/NF-YA binding to DNA and repressing IGF-1 promoter transactivation. After IR exposure, ATM-mediated p53 stabilization promotes p21 expression. p21, in turn, suppresses Cdk2, blocking NF-YA phosphorylation and releasing NF-YA from the IGF-1 promoter (Figures 2e and 6f). Additionally, acetylation, phosphorylation or other post-translational modifications of p53 may have an impact on its ability to bind NF-YA and repress IGF-1 transcription. Studies to elucidate the roles of NF-YA and p53 post-translational modifications are ongoing. Collectively, our data indicate that ATM signaling results in NF-YA modification to prevent or reverse its binding to the IGF-1 promoter, suggesting that NF-YA is rate-limiting and cells retain the ability to induce IGF-1 expression and downstream sCLU production even after IR exposure, even though p53 is stabilized.
p53 null cells, or cells with p53 knockdown, induced IGF-1–sCLU to a much greater extent after IR exposure compared with WT p53 cells. These data suggest that a positive transcription factor, in addition to loss of p53/NF-YA from the IGF-1 promoter, is required for IGF-1 induction. There are several candidate factors, including NF-YB/-YC itself, AIB1 or TGFb1 signaling, that may regulate IGF-1 expression. NF-Y is a known IR-activated transcription factor (Peng et al., 2007), and NF-YB/-YC complexes may act as positive transcription factors for IGF-1 induction after IR exposure, consistent with increased NF-YB/-YC binding to the IGF-1 promoter (Figure 2e). On the other hand, levels of the steroid receptor coactivator, AIB1, are directly correlated with IGF-1 expression (Wang et al., 2000). AIB1 binds to the IGF-1 promoter with AP-1 (Yan et al., 2006), suggesting that changes in these complexes may regulate IGF-1 expression after IR exposure. Finally, as TGFb1 induces sCLU (Jin and Howe, 1997) and TGFb1 signaling can be activated after IR exposure (Andarawewa et al., 2007), TGFb1 signaling may be involved in IR-induced IGF-1. Current studies are ongoing in our lab to elucidate the positive factors in induction of IGF-1 after DNA damaging agents.
In conclusion, we elucidated the complete signaling pathway from DNA damage to sCLU expression. By determining the signaling pathways involved in IGF-1–sCLU upregulation, important targets for anti-tumor therapy emerge, as well as the possibility of using IGF-1 and sCLU as markers for genetic instability and cancer progression.
Materials and methods
Cell treatments
AG1024 IGF-1 was treated with etoposide, topotecan, Aph, N-acetyl cysteine, and ATR kinase inhibitor (CGK733). Cells were plated overnight, treated with TPT, H2O2 or VP16 (Sigma, St Louis, MO, USA) for 5 h at the indicated doses, and medium replaced. Pre-treatment with Aph (Sigma) was done for 2 h before treatment. For IGF-1 (R&D Systems, Minneapolis, MN, USA) and AG1024 treatments (EMD Chemicals, Gibbstown, NJ, USA), cells were serum-starved (0.5% FBS) overnight, and exposed to IGF-1 or AG1024 at the indicated doses. A JL Shepherd 137Cs Mark I-68 irradiator (3.87 Gy/min) was used for IR exposures. Mock and DMSO-treated cells were used as controls (UT).
Plasmids and site-directed mutagenesis
The IGF-1 promoter fused to luciferase was from Dr P Rotwein (Mittanck et al., 1997). The CLU promoter fused to luciferase was previously described (Criswell et al., 2005). Flag-tagged CMV-p53 cDNA was created by subcloning p53 cDNA into the pcDNA3.1-N-term-Flag construct. NF-YA cDNA was provided by Dr R Mantovani (Mantovani et al., 1994). Site-directed mutagenesis of p53 cDNAs and the IGF-1 promoter was performed using PCR-based mutagenesis, detailed in the Supplementary Material.
Luciferase reporter assays
Cells were transiently transfected with various promoter luciferase constructs as indicated, and with RSV-β-gal as a transfection control, using Fugene 6 (Roche). Treatments, where indicated, were performed 24 h after transfection. Luciferase activities were analyzed using Luciferase Assay Reagent (Promega, Madison, WI, USA). β-Galactosidase activity was determined using Galacto-Star reagent (Life Technologies, Carlsbad, CA, USA).
Enzyme-linked immunosorbent assays
IGF-1 was detected using capture and detection antibodies MAB291 and BAF291 (R&D Systems). Samples were normalized by cell number.
Flow cytometry
Cells were fixed with 1% formaldehyde, permeabilized with 100% ethanol, stained with pATMS1981 (Rockland, Gilbertsville, PA, USA), γ-H2AX (Millipore, Billerica, MA, USA), and IGF-1 (Santa Cruz Biotechnology, Santa Cruz, CA, USA) antibodies and FITC-tagged secondary antibodies (Life Technologies), and counterstained with propidium iodide. Data were graphed as percent cells staining positive ±s.d.
Chromatin immunoprecipitation
Cells were fixed in 1% formaldehyde, sonicated to shear chromatin, and incubated overnight with antibodies against p53, NF-YA, NF-YB and NF-YC (Santa Cruz). Protein A/G beads were added the next day, then washed extensively. Chromatin was eluted from beads, cross-links reversed and DNA purified. Primers are listed in Supplementary Material.
Western blotting
Western blotting was described previously (Criswell et al., 2005). GAPDH, β-actin or α-tubulin was probed as loading controls. Relative expression was calculated from X-ray films using Image J, comparing the relative density of experimental conditions with a loading control. Control values were set to 1.
Statistics
Statistics were calculated using paired Student’s t-tests (n⩾ 3), and each experiment was performed at least three times.
Supplementary Material
Acknowledgments
This work was supported by DOE grant (#DE-FG02-06ER64186-17) to DAB, a DOD BCRP pre-doctoral fellowship (W81XWH-06-0748) to EMG and a DOD PCRP postdoctoral fellowship (X8IXWH-09-1-0168) to XL. We are grateful to the Robert B and Virginia Payne Endowment for support of this work. We also thank the support received from the SAIR NIH U24 grant CA126608 and the Imaging Shared Resource of the Simmons Cancer Center. This is CSCN #024 and used the Flow Cytometry and Biostatistics cores of the Simmons Comprehensive Cancer Center.
Footnotes
Conflict of interest
The authors declare no conflict of interest.
References
- Andarawewa KL, Erickson AC, Chou WS, Costes SV, Gascard P, Mott JD, et al. Ionizing radiation predisposes nonmalignant human mammary epithelial cells to undergo transforming growth factor beta induced epithelial to mesenchymal transition. Cancer Res. 2007;67:8662–8670. doi: 10.1158/0008-5472.CAN-07-1294. [DOI] [PubMed] [Google Scholar]
- Bellorini M, Lee DK, Dantonel JC, Zemzoumi K, Roeder RG, Tora L, et al. CCAAT binding NF-Y-TBP interactions: NF-YB and NF-YC require short domains adjacent to their histone fold motifs for association with TBP basic residues. Nucleic Acids Res. 1997;25:2174–2181. doi: 10.1093/nar/25.11.2174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentle MS, Reinicke KE, Bey EA, Spitz DR, Boothman DA. Calcium-dependent modulation of poly(ADP-ribose) polymerase-1 alters cellular metabolism and DNA repair. J Biol Chem. 2006;281:33684–33696. doi: 10.1074/jbc.M603678200. [DOI] [PubMed] [Google Scholar]
- Bocchetta M, Eliasz S, De Marco MA, Rudzinski J, Zhang L, Carbone M. The SV40 large T antigen-p53 complexes bind and activate the insulin-like growth factor-I promoter stimulating cell growth. Cancer Res. 2008;68:1022–1029. doi: 10.1158/0008-5472.CAN-07-5203. [DOI] [PubMed] [Google Scholar]
- Boggs LN, Fuson KS, Baez M, Churgay L, McClure D, Becker G, et al. Clusterin (Apo J) protects against in vitro amyloid-beta (1–40) neurotoxicity. J Neurochem. 1996;67:1324–1327. doi: 10.1046/j.1471-4159.1996.67031324.x. [DOI] [PubMed] [Google Scholar]
- Boothman DA, Meyers M, Fukunaga N, Lee SW. Isolation of x-ray-inducible transcripts from radioresistant human melanoma cells. Proc Natl Acad Sci USA. 1993;90:7200–7204. doi: 10.1073/pnas.90.15.7200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celeste A, Petersen S, Romanienko PJ, Fernandez-Capetillo O, Chen HT, Sedelnikova OA, et al. Genomic instability in mice lacking histone H2AX. Science. 2002;296:922–927. doi: 10.1126/science.1069398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Halberg RB, Ehrhardt WM, Torrealba J, Dove WF. Clusterin as a biomarker in murine and human intestinal neoplasia. Proc Natl Acad Sci USA. 2003;100:9530–9535. doi: 10.1073/pnas.1233633100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi KN, Hotte SJ, Yu EY, Tu D, Eigl BJ, Tannock I, et al. Randomized phase II study of Docetaxel and Prednisone with or without OGX-011 in patients with metastatic castration-resistant prostate cancer. J Clin Oncol. 2010;28:4247–4254. doi: 10.1200/JCO.2009.26.8771. [DOI] [PubMed] [Google Scholar]
- Criswell T, Beman M, Araki S, Leskov K, Cataldo E, Mayo LD, et al. Delayed activation of insulin-like growth factor-1 receptor/Src/MAPK/Egr-1 signaling regulates clusterin expression, a pro-survival factor. J Biol Chem. 2005;280:14212–14221. doi: 10.1074/jbc.M412569200. [DOI] [PubMed] [Google Scholar]
- Criswell T, Klokov D, Beman M, Lavik JP, Boothman DA. Repression of IR-inducible clusterin expression by the p53 tumor suppressor protein. Cancer Biol Ther. 2003;2:372–380. doi: 10.4161/cbt.2.4.430. [DOI] [PubMed] [Google Scholar]
- D’Arpa P, Beardmore C, Liu LF. Involvement of nucleic acid synthesis in cell killing mechanisms of topoisomerase poisons. Cancer Res. 1990;50:6919–6924. [PubMed] [Google Scholar]
- de Silva HV, Harmony JA, Stuart WD, Gil CM, Robbins J. Apolipoprotein J: structure and tissue distribution. Biochemistry. 1990;29:5380–5389. doi: 10.1021/bi00474a025. [DOI] [PubMed] [Google Scholar]
- Dumaz N, Meek DW. Serine15 phosphorylation stimulates p53 transactivation but does not directly influence interaction with HDM2. EMBO J. 1999;18:7002–7010. doi: 10.1093/emboj/18.24.7002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- el-Deiry WS, Tokino T, Velculescu VE, Levy DB, Parsons R, Trent JM, et al. WAF1, a potential mediator of p53 tumor suppression. Cell. 1993;75:817–825. doi: 10.1016/0092-8674(93)90500-p. [DOI] [PubMed] [Google Scholar]
- Hao LY, Greider CW. Genomic instability in both wild-type and telomerase null MEFs. Chromosoma. 2004;113:62–68. doi: 10.1007/s00412-004-0291-7. [DOI] [PubMed] [Google Scholar]
- Imbriano C, Gurtner A, Cocchiarella F, Di Agostino S, Basile V, Gostissa M, et al. Direct p53 transcriptional repression: in vivo analysis of CCAAT-containing G2/M promoters. Mol Cell Biol. 2005;25:3737–3751. doi: 10.1128/MCB.25.9.3737-3751.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin G, Howe PH. Regulation of clusterin gene expression by transforming growth factor beta. J Biol Chem. 1997;272:26620–26626. doi: 10.1074/jbc.272.42.26620. [DOI] [PubMed] [Google Scholar]
- Jones SE, Jomary C. Clusterin. Int J Biochem Cell Biol. 2002;34:427–431. doi: 10.1016/s1357-2725(01)00155-8. [DOI] [PubMed] [Google Scholar]
- Klokov D, Criswell T, Leskov KS, Araki S, Mayo L, Boothman DA. IR-inducible clusterin gene expression: a protein with potential roles in ionizing radiation-induced adaptive responses, genomic instability, and bystander effects. Mutat Res. 2004;568:97–110. doi: 10.1016/j.mrfmmm.2004.06.049. [DOI] [PubMed] [Google Scholar]
- Kojis TL, Gatti RA, Sparkes RS. The cytogenetics of ataxia telangiectasia. Cancer Genet Cytogenet. 1991;56:143–156. doi: 10.1016/0165-4608(91)90164-p. [DOI] [PubMed] [Google Scholar]
- Leri A, Liu Y, Claudio PP, Kajstura J, Wang X, Wang S, et al. Insulin-like growth factor-1 induces Mdm2 and down-regulates p53, attenuating the myocyte renin-angiotensin system and stretch-mediated apoptosis. Am J Pathol. 1999;154:567–580. doi: 10.1016/S0002-9440(10)65302-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lou Z, Minter-Dykhouse K, Franco S, Gostissa M, Rivera MA, Celeste A, et al. MDC1 maintains genomic stability by participating in the amplification of ATM-dependent DNA damage signals. Mol Cell. 2006;21:187–200. doi: 10.1016/j.molcel.2005.11.025. [DOI] [PubMed] [Google Scholar]
- Macaulay VM, Salisbury AJ, Bohula EA, Playford MP, Smorodinsky NI, Shiloh Y. Downregulation of the type 1 insulin-like growth factor receptor in mouse melanoma cells is associated with enhanced radiosensitivity and impaired activation of Atm kinase. Oncogene. 2001;20:4029–4040. doi: 10.1038/sj.onc.1204565. [DOI] [PubMed] [Google Scholar]
- Mantovani R, Li XY, Pessara U, Hooft van Huisjduijnen R, Benoist C, Mathis D. Dominant negative analogs of NF-YA. J Biol Chem. 1994;269:20340–20346. [PubMed] [Google Scholar]
- Matsui T, Katsuno Y, Inoue T, Fujita F, Joh T, Niida H, et al. Negative regulation of Chk2 expression by p53 is dependent on the CCAAT-binding transcription factor NF-Y. J Biol Chem. 2004;279:25093–25100. doi: 10.1074/jbc.M403232200. [DOI] [PubMed] [Google Scholar]
- Mayo LD, Donner DB. A phosphatidylinositol 3-kinase/Akt pathway promotes translocation of Mdm2 from the cytoplasm to the nucleus. Proc Natl Acad Sci USA. 2001;98:11598–11603. doi: 10.1073/pnas.181181198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittanck DW, Kim SW, Rotwein P. Essential promoter elements are located within the 5′ untranslated region of human insulin-like growth factor-I exon I. Mol Cell Endocrinol. 1997;126:153–163. doi: 10.1016/s0303-7207(96)03979-2. [DOI] [PubMed] [Google Scholar]
- Miyake H, Nelson C, Rennie PS, Gleave ME. Acquisition of chemoresistant phenotype by overexpression of the antiapoptotic gene testosterone-repressed prostate message-2 in prostate cancer xenograft models. Cancer Res. 2000;60:2547–2554. [PubMed] [Google Scholar]
- Peng Y, Stewart D, Li W, Hawkins M, Kulak S, Ballermann B, et al. Irradiation modulates association of NF-Y with histone-modifying cofactors PCAF and HDAC. Oncogene. 2007;26:7576–7583. doi: 10.1038/sj.onc.1210565. [DOI] [PubMed] [Google Scholar]
- Perry ME, Piette J, Zawadzki JA, Harvey D, Levine AJ. The mdm-2 gene is induced in response to UV light in a p53-dependent manner. Proc Natl Acad Sci USA. 1993;90:11623–11627. doi: 10.1073/pnas.90.24.11623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan PD, Goss PE. The emerging role of the insulin-like growth factor pathway as a therapeutic target in cancer. Oncologist. 2008;13:16–24. doi: 10.1634/theoncologist.2007-0199. [DOI] [PubMed] [Google Scholar]
- Shahrabani-Gargir L, Pandita TK, Werner H. Ataxia-telangiectasia mutated gene controls insulin-like growth factor I receptor gene expression in a deoxyribonucleic acid damage response pathway via mechanisms involving zinc-finger transcription factors Sp1 and WT1. Endocrinology. 2004;145:5679–5687. doi: 10.1210/en.2004-0613. [DOI] [PubMed] [Google Scholar]
- So A, Sinnemann S, Huntsman D, Fazli L, Gleave M. Knockdown of the cytoprotective chaperone, clusterin, chemosensitizes human breast cancer cells both in vitro and in vivo. Mol Cancer Ther. 2005;4:1837–1849. doi: 10.1158/1535-7163.MCT-05-0178. [DOI] [PubMed] [Google Scholar]
- Stiff T, Walker SA, Cerosaletti K, Goodarzi AA, Petermann E, Concannon P, et al. ATR-dependent phosphorylation and activation of ATM in response to UV treatment or replication fork stalling. EMBO J. 2006;25:5775–5782. doi: 10.1038/sj.emboj.7601446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulkowski S, Wincewicz A, Zalewski B, Famulski W, Lotowska JM, Koda M, et al. Hypoxia related growth factors and p53 in preoperative sera from patients with colorectal cancer–evaluation of the prognostic significance of these agents. Clin Chem Lab Med. 2009;47:1439–1445. doi: 10.1515/CCLM.2009.305. [DOI] [PubMed] [Google Scholar]
- Wagner MW, Li LS, Morales JC, Galindo CL, Garner HR, Bornmann WG, et al. Role of c-Abl kinase in DNA mismatch repair-dependent G2 cell cycle checkpoint arrest responses. J Biol Chem. 2008;283:21382–21393. doi: 10.1074/jbc.M709953200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Rose DW, Hermanson O, Liu F, Herman T, Wu W, et al. Regulation of somatic growth by the p160 coactivator p/CIP. Proc Natl Acad Sci USA. 2000;97:13549–13554. doi: 10.1073/pnas.260463097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson MR, Easterbrook-Smith SB. Clusterin is a secreted mammalian chaperone. Trends Biochem Sci. 2000;25:95–98. doi: 10.1016/s0968-0004(99)01534-0. [DOI] [PubMed] [Google Scholar]
- Yan J, Yu CT, Ozen M, Ittmann M, Tsai SY, Tsai MJ. Steroid receptor coactivator-3 and activator protein-1 coordinately regulate the transcription of components of the insulin-like growth factor/AKT signaling pathway. Cancer Res. 2006;66:11039–11046. doi: 10.1158/0008-5472.CAN-06-2442. [DOI] [PubMed] [Google Scholar]
- Yun J, Chae HD, Choy HE, Chung J, Yoo HS, Han MH, et al. p53 negatively regulates cdc2 transcription via the CCAAT-binding NF-Y transcription factor. J Biol Chem. 1999;274:29677–29682. doi: 10.1074/jbc.274.42.29677. [DOI] [PubMed] [Google Scholar]
- Yun J, Chae HD, Choi TS, Kim EH, Bang YJ, Chung J, et al. Cdk2-dependent phosphorylation of the NF-Y transcription factor and its involvement in the p53-p21 signaling pathway. J Biol Chem. 2003;278:36966–36972. doi: 10.1074/jbc.M305178200. [DOI] [PubMed] [Google Scholar]
- Zhang H, Kim JK, Edwards CA, Xu Z, Taichman R, Wang CY. Clusterin inhibits apoptosis by interacting with activated Bax. Nat Cell Biol. 2005;7:909–915. doi: 10.1038/ncb1291. [DOI] [PubMed] [Google Scholar]
- Zhao H, Piwnica-Worms H. ATR-mediated checkpoint pathways regulate phosphorylation and activation of human Chk1. Mol Cell Biol. 2001;21:4129–4139. doi: 10.1128/MCB.21.13.4129-4139.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Petersen S, Tessarollo L, Nussenzweig A. Targeted disruption of the Nijmegen breakage syndrome gene NBS1 leads to early embryonic lethality in mice. Curr Biol. 2001;11:105–109. doi: 10.1016/s0960-9822(01)00019-7. [DOI] [PubMed] [Google Scholar]
- Ziv Y, Bar-Shira A, Pecker I, Russell P, Jorgensen TJ, Tsarfati I, et al. Recombinant ATM protein complements the cellular A-T phenotype. Oncogene. 1997;15:159–167. doi: 10.1038/sj.onc.1201319. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


