Abstract
Enoyl-acyl carrier protein reductase (ENR), a critical enzyme in type II fatty acid biosynthesis, is a promising target for drug discovery against hepatocyte-stage Plasmodium falciparum. In order to identify PfENR-specific inhibitors, we docked 70 FDA-approved, bioactive, and/or natural product small molecules known to inhibit the growth of whole-cell blood-stage P. falciparum into several PfENR crystallographic structures. Subsequent in vitro activity assays identified a noncompetitive low-micromolar PfENR inhibitor, celastrol, from this set of compounds.
Keywords: malaria, Plasmodium falciparum, enoyl-acyl carrier protein reductase, in silico, celastrol
Graphical Abstract
1. Introduction
Malaria, caused by eukaryotic protists of the genus Plasmodium, is a devastating disease that infects a quarter of a billion people annually, costing nearly a million lives.1 Resistance to most of the commonly used antimalarials like chloroquine and sulfadoxine/pyrimethamine is widespread, the result of point mutations in dihydrofolate reductase and dihydropteroate synthase, respectively.2 Artemisinin combination therapies, which inhibit multiple stages of the malarial life cycle,2 are currently the first line of defense.3 Given the recent emergence of artemisinin-resistant P. falciparum on the Cambodia-Thailand border, new drugs for combating multidrug resistant malaria are urgently needed.4,5
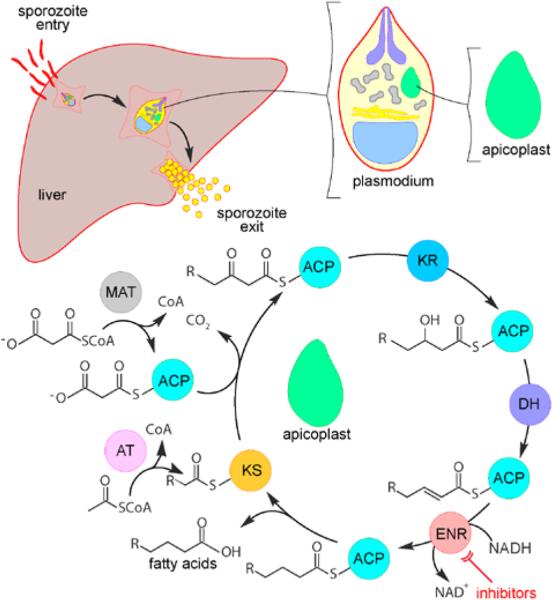
The Plasmodium life cycle consists of three distinctive stages: the mosquito, liver, and blood stage. While the continued development of novel therapeutics effective against the infectious and symptomatic blood stage is critical, targeting the preceding liver stage may be required for effective prophylaxis.6–9 Notably, all malarial parasites that infect humans (Plasmodium falciparum, Plasmodium vivax, Plasmodium malariae, and Plasmodium ovale) undergo extensive replication in the liver stage,7 and P. malariae and P. ovale also have a non-dividing dormant hepatic stage responsible for relapse. Although some existing antimalarial drugs (e.g., primaquine, anti-folates, and atovaquone) are effective against both the blood- and liver-stage parasites,7,8 the continual threat of growing resistance requires new, broadly effective medications.
A search of the TDR Targets Database,10 an online resource that prioritizes neglected-disease drug targets, suggests that inhibitors of P. falciparum fatty-acid biosynthesis might serve as effective anti-malarial therapeutics. Given that the study of fatty-acid biosynthesis is one of our lab's primary emphases, we found this drug-discovery approach particularly interesting. Fatty acids (FA) are critical for Plasmodium survival during both the liver and blood stages.11 In the blood stage, merozoites infect red-blood cells, initiating a replication process that requires exogenous (host-derived) FA to support membrane biogenesis.12 It was previously thought that both the liver and blood stages of the malarial parasite relied on the uptake of FA from the host.13,14 However, this paradigm was challenged when FA enzymes (FabH, FabZ, FabF) capable of de novo biosynthesis were found targeted to the apicoplast, a vestigial nonphotosynthetic plastid.15,16 The activity of these enzymes was confirmed using radio-labeled acetate and malonyl-CoA, which were successfully incorporated into synthesized FA in vivo.17 Subsequent studies revealed that P. falciparum FA synthesis is required only in the liver-stage of the parasitic life cycle.18,19
Given that bacterial FA synthesis (FAS) is the target of several antibiotics (e.g. isoniazid,20,21 diazaborines,22 triclosan23 and thiolactomycin24–27), several studies have sought to similarly inhibit P. falciparum FAS.19,28–30 Though the efficacy of some FAS inhibitors against the blood stage is poorly understood,19 inhibition of the late liver stage, which requires FAS, is thought to be promising for prophylactics.8,11,12
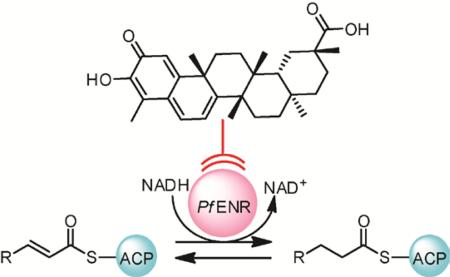
Unlike most eukaryotes (including humans), Plasmodium uses a segmented type II FAS typical of bacteria (Fig. 1) rather than a type I FAS megasynthase.31 Consequently, malarial type II FAS enzymes are structurally different from the functionally equivalent human type I FAS, making P. falciparum FAS a promising target for drug development. The rate-limiting enzyme in malarial FAS, enoyl-acyl carrier protein reductase (ENR),32 is responsible for the reduction of trans-2-acyl-ACP to acyl-ACP (Fig. 1). The prodrug isoniazid, whose active metabolite isonicotinyl-NAD inhibits ENR, is FDA-approved for the treatment of Mycobacterium tuberculosis infection, further attesting to the viability of ENR as a drug target. ENR inhibitors may prove effective against other pathogens as well.20 As ENR inhibitors have been shown to impede the growth of P. falciparum, several antimalarial drug-discovery projects have focused on ENR as a potential target.19,28–30
Figure 1.
Illustration of fatty-acid biosynthesis in the P. falciparum apicoplast and a proposed strategy to target ENR with inhibitors. Abbreviations are as follows: acyl carrier protein (ACP), malonyl transferase (MAT), β-ketoacyl acyl carrier protein reductase (KR), 3-hydroxyacyl ACP dehydrase (DH), enoyl-acyl carrier protein reductase (ENR), β–ketoacyl acyl carrier protein synthetase (KS), and coenzyme A (CoA).
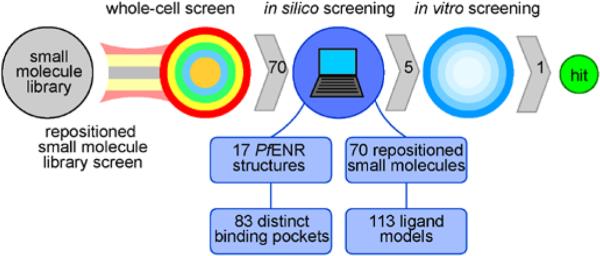
In 2006, Weisman et al. identified 70 bioactive molecules, natural products, and approved drugs from the MicroSource Spectrum33 and Killer Collections33 that retard the growth of whole-cell blood-stage P. falciparum in cell culture by 70% or more at 1 μM.33 Compounds from these specific collections are useful as potential leads because of available pharmacokinetic and toxicological reports, as well as demonstrated P. falciparum whole-cell inhibition. In order to better elucidate the mechanism of these compounds and assess their potential efficacy against the liver-stage parasite, we used complementary computational and experimental methods to test for P. falciparum ENR inhibition (Fig. 2). We demonstrate that celastrol33 (compound 1) inhibits PfENR with an IC50 in the low-micromolar range. We also characterize celastrol inhibition behavior in hopes of facilitating future drug-discovery efforts.
Figure 2.
Overview of the methodology used to identify celastrol.
2. Results and Discussion
By coupling in silico and in vitro techniques, we determined that the pentacyclic triterpenoid celastrol, a compound known to inhibit the growth of whole-cell blood-stage P. falciparum,33 also inhibits the enzymatic activity of ENR, a critical enzyme in malarial FAS that is essential during the liver stage of the parasitic life cycle.6
2.1. Virtual screening of 70 small-molecule inhibitors
To verify that our AutoDock-Vina-based docking protocol34 was well suited to the ENR system, we first considered the three wild-type, triclosan (TCL)-bound P. falciparum ENR structures deposited in the PDB (1NHG,31 1UH5,35 and 2O2Y36), which collectively contained eight co-crystallized TCL inhibitors. As optimal TCL binding is dependent on NAD+ (see later discussion), the cofactor was retained, and each TCL molecule was redocked into its respective pocket. All eight dockings successfully recaptured the crystallographic pose, with heavy-atom RMSD values ranging from 0.42 to 0.53 Å and predicted free energies of binding ranging from −9.0 to −9.8 kcal/mol.
As a second step, we identified an additional 14 ligand-bound PfENR structures with varied active-site geometries from the Protein Data Bank.37 As many of these structures included multiple chains and multiple bound ligands (e.g., triclosan and analogs, NAD+, and NADH), 83 distinct pockets were ultimately considered. Both the co-crystallized ligands and cofactors were removed in preparation for docking.
To assess the performance of our docking protocol, we next performed a benchmark virtual screen that included 36 potent PfENR inhibitors, as well as 1594 decoy molecules presumed to be non-binders. Each of these compounds were docked into the 83 ENR pockets, and the single best score, independent of the receptor, was used for ranking. The area under the receiver operator characteristic (ROC) curve (data not shown)38 associated with this screen suggested that our protocol could in fact discriminate between ligands and decoys. If a known inhibitor and decoy were picked at random, our protocol would have ranked the inhibitor better than the decoy 69% percent of the time, far better than the 50% expected of random sorting. There is, of course, no perfect virtual-screening methodology, and false negatives can never be entirely eliminated, but our methodology does in fact enrich the pool of top-ranked compounds with true ENR inhibitors.
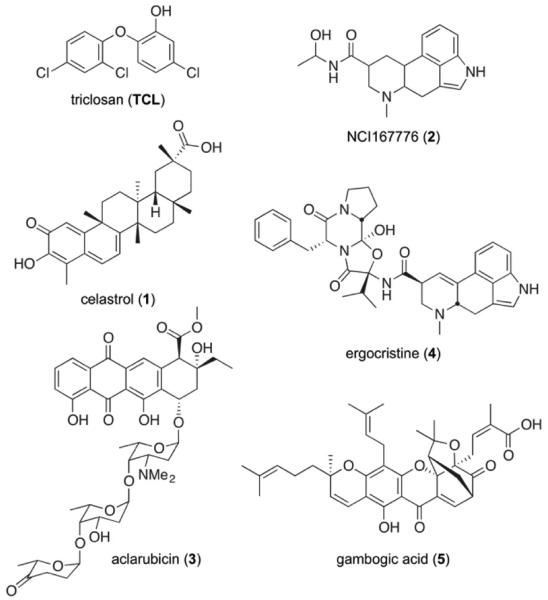
Having verified that our protocol can 1) reproduce the crystallographic pose of TCL and 2) adequately separate ENR inhibitors from decoys, we next applied the same protocol to one hundred and thirteen models of 70 experimentally validated whole-cell P. falciparum inhibitors.33 The ranked compounds are listed in Table S1. The top ~5%, which all had docking scores better than −12 kcal/mol, were tested experimentally for PfENR inhibition. These small molecules include: celastrol, aclarubicin/aclacinomycin Y, dihydroergotamine, and gambogic acid (Fig. 3). Compounds 1-4 were available through the NCI: celastrol (NSC70931, 1); aclacinomycin A (aclarubicin), the biochemical precursor of aclacinomycin Y; dihydroergotoxine (NSC409663, 3); and gambogic acid (4).
Figure 3.
The five compounds that were subjected to in vitro validation. Compounds 1-4, from the MicroSource Spectrum32 and Killer Collections32 libraries, were identified by in silico screening.
2.2. Kinetic evaluation of PfENR
Prior to experimentally validating the putative inhibitors identified in our in silico screen, we first confirmed that the kinetic behavior of PfENR expressed in our lab was consistent with that published in the literature.39,40 PfENR activity was tested by individually varying the concentration of crotonyl-CoA and NADH without NAD+ preincubation (Table S1). The Km and kcat values for crotonyl-CoA were determined to be 0.17 ± 0.06 mM and 1.0 ± 0.2 sec−1, respectively, by varying the concentration of crotonyl-CoA from 20 to 300 μM while holding the concentration of NADH constant (Fig. S2a). Similarly, the NADH Km and kcat values were found to be 0.24 ± 0.04 mM and 3.8 ± 1.0 sec−1, respectively, by varying the concentration of NADH from 80 to 500 μM while keeping the concentration of crotonyl-CoA constant (Fig. S2b). These Km values are both comparable to those reported in the literature.39 The kcat/Km ratio for NADH and crotonyl-CoA was 2.6 ± 0.7 sec−1 mM−1 and 6.0 ± 1.1 sec−1 mM−1, respectively (Table S2).
Various volumes of DMSO were tested in order to determine if DMSO had an effect on PfENR activity. DMSO affected ENR activity only when it constituted greater than 8% of the total reaction mixture volume (data not shown).
2.3. Confirmation of ENR inhibition via enzymatic assays
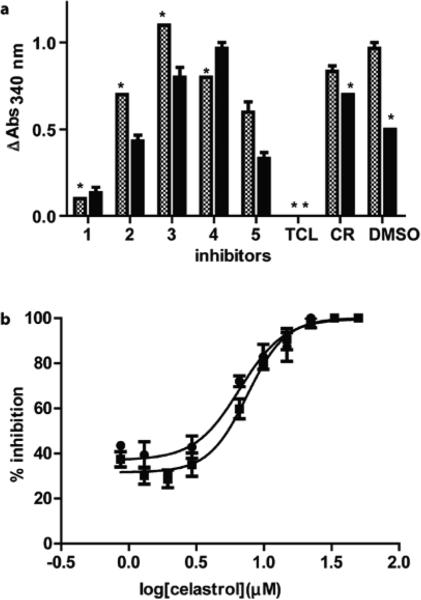
Having verified the activity of PfENR expressed in our lab, we next validated our continuous ENR-inhibition assay (Fig. 4a). The assay measures the change in NADH absorbance at 340 nm over time in the presence of potential inhibitors. In the case of complete inhibition, ENR catalysis is entirely halted, and NADH is not consumed. As a positive control, we tested triclosan (TCL), a known picomolar PfENR inhibitor,41 and observed no change in NADH consumption.39 As a negative control, we tested cerulenin (CR), a commercially available inhibitor of β-ketoacyl-acyl-carrier protein synthase,42 and observed no PfENR inhibition, similar to the results obtained in the presence of DMSO alone (Fig. 4a).
Figure 4.
a) In vitro assay results using 25 μM of each candidate inhibitor, with (checkered box) and without (solid box) NAD+ preincubation. Triclosan (TCL) and cerulenin (CR) served as the positive and negative controls, respectively. b) The average IC50 values of compound 1 were calculated to be 5.9 and 4.7 μM with (●) and without (■) NAD+ preincubation, respectively. Data was collected in triplicate and the individual values were within 10% error. (*) Indicates minimal change error bars.
2.4. Experimental validation of the top virtual-screen hits
This same real-time assay was next used to evaluate compounds 1-4 from the in silico screen. Compounds 3 and 4 showed no ENR inhibition. Paradoxically, compound 2 showed an increase in absorbance relative to the DMSO and cerulenin controls, suggesting PfENR agonism. However, upon further inspection, compound 2 also showed a higher absorbance relative to the NADH negative control. As this compound is insoluble above 25 μM, we hypothesize that it precipitates under the reaction conditions used, resulting in a higher absorbance at 340 nm.
Our enzymatic assay demonstrated that celastrol (compound 1), a triterpene quinone methide isolated from the roots of the celastraceae Tripterygium wilfordi, inhibits PfENR with average IC50 values of 5.9 and 4.7 μM with and without NAD+ preincubation, respectively (Fig. 4b). When compound 1 was tested in the presence of 0.01% Triton-X to rule out non-specific inhibition by aggregation, a similar IC50 value was obtained (3.4 μM, Figure S8).
2.5. Celastrol inhibition kinetics
In order to understand the kinetic behavior of celastrol in context, it is important to first understand the binding kinetics of TCL, a well characterized and prototypical PfENR inhibitor. As a slow-onset, tight-binding inhibitor,40,43 TCL is desirable because of its sustained efficacy.44 TCL potency depends on NAD+ binding, and the concentration of NAD+, a product of ENR catalysis, increases only as the reaction proceeds.40,45 This NAD+ dependence appears to be the result of two distinct factors. First, the formation of a ternary ENR-NAD+-TCL complex stabilizes a normally flexible substrate-binding loop and is postulated to be the slowest step in the formation of the enzyme-inhibitor complex.45–49 Second, once bound, TCL is stabilized via π-stacking interactions between the TCL phenolic ring and the NAD+ nicotinamide ring.40 Indeed, kinetic studies have demonstrated that the IC50 value of TCL improves with NAD+ preincubation (i.e., TCL is an uncompetitive inhibitor with respect to varying NAD+ concentrations).45,47,48
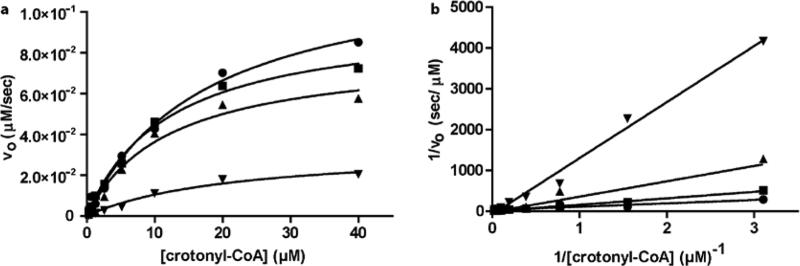
In order to determine whether or not celastrol and TCL binding kinetics are similar, we performed extensive kinetic studies, both with and without NAD+ preincubation. First, the mechanism of celastrol inhibition was studied by observing the maximum reaction velocity (Vmax) at varied crotonyl-CoA concentrations with fixed NADH concentration (Fig. 5a). Plotting the reaction velocity as a function of crotonyl-CoA concentration (Fig. 5a) revealed that the Vmax decreased in the presence of increasing concentrations of 1, consistent with noncompetitive inhibition. The reciprocal plots for velocity and varying crotonyl-CoA concentration at fixed concentrations of NADH similarly suggested line behaviors consistent with noncompetitive inhibition (Fig. 5b). Additionally, Km, app was independent of celastrol concentration in the presence of NAD+, as no change in Km, app was observed with varying celastrol concentrations (Fig. S7a), also consistent with noncompetitive inhibition. The Ki (5.3 μM) for celastrol was determined by the slope of the line when the Vmax, app was plotted as a function of celastrol concentration (Fig. S7b). These studies suggest that celastrol binds both with and without bound crotonyl-CoA.
Figure 5.
a) Progress curve analysis of PfENR inhibition at various concentrations of compound 1 in the presence of varying concentrations of crotonyl-CoA (3.22 × 10−4 – 40 μM) and 50 μM NAD+ preincubation. b) Double-reciprocal plot of varying celastrol concentrations (● 0 μM, ■ 9.7 × 10−2 μM, ▲7.8 × 10−2 μM, and ▼ 12.5 μM) in order to access inhibition behavior. The average values were plotted and all experiments were collected in triplicate.
It is reported that the quinone methide moiety of celastrol can form Michael adducts with nucleophilic residues such as cysteine.50,51 To rule out the possibility of non-specific PfENR inhibition via this mechanism, PfENR was incubated with celastrol at a concentration 5 times the observed IC50 value. Following incubation, the protein was rapidly diluted 100-fold, and the reaction was initiated with crotonyl-CoA and NADH. The activity of compound 1 was identical to the DMSO control (Figure S9), suggesting that celastrol does not covalently bind to PfENR.
As an additional test to rule out covalent binding to Cys100 and Cys249, we incubated PfENR with iodoacetamide to cap all potentially reactive cysteine residues. After confirming that this treatment did not alter PfENR activity (Figure S10), celastrol inhibition of iodoacetamide-treated PfENR at high concentrations was tested and found to be comparable to that of the untreated protein (Figure S10).
2.6. Celastrol may bind in the combined substrate-cofactor pocket
Our docking study suggests that celastrol may occupy both the NAD+ and substrate-binding pocket, without requiring bound cofactor or substrate. This proposed mechanism of inhibition by combined substrate-cofactor displacement is not without precedent. Recently, pyridomycin, which inhibits the type II ENR (InhA) from Mycobacterium tuberculosis, was discovered to similarly bind in the combined substrate-cofactor pocket,52 a mode of ENR inhibition that has not been previously described.52
Binding to the combined substrate-cofactor pocket may seem incompatible with the noncompetitive inhibition we observed. However, some enzymes, including bisubstrate/product enzymes like ENR, exhibit noncompetitive inhibition with respect to both substrates even though the relevant inhibitors bind in the primary pocket.53 It may be that at high celastrol concentrations this bi-substrate binding mode alters the conformation of the binding pocket, essentially sequestering some ENR molecules by placing them in a state that prevents NADH and crotonyl-CoA binding. This may effectively reduce the concentration of the catalytically active enzyme, thereby reducing Vmax without affecting Km, consistent with noncompetitive inhibition. Further crystallographic and analytical gel filtration studies are required to confirm and elucidate the proposed celastrol-induced changes in pocket geometry.
To better understand the predicted celastrol binding mode, we used the BINANA algorithm54 to analyze the best-scoring docked celastrol pose. Implemented as a stand-alone Python script, BINANA analyzes the geometries of the receptor and docked ligand in order to predict key electrostatic, hydrogen-bond, hydrophobic, and π-π intermolecular interactions. Together with visual inspection, this analysis suggested that celastrol binding is primarily governed by high shape complementarity and nonspecific hydrophobic interactions. However, a few specific ligand-receptor interactions may contribute to overall molecular recognition. For example, a charge-charge interaction may form between the celastrol carboxylate group and the Lys285 side chain, and a T-shaped π-stacking interaction may form between the celastrol aromatic ring and Tyr277. While less certain, the Tyr267 side chain may also participate in a parallel-displaced π-stacking interaction (Fig. S11).
While the possibility of celastrol binding by cofactor displacement is intriguing, allosteric inhibition must also be considered given the observed noncompetitive inhibition. To explore this possibility, we used computer docking to computationally flood the entire surface of a 2OL4 monomer with celastrol molecules, without the NAD+ cofactor present. Of the 1,892 celastrol poses generated, only three unique poses had docking scores better than −10 kcal/mol, and all three of these placed celastrol within the combined substrate-cofactor pocket. While intriguing, these computational results cannot entirely rule out allosteric binding.
Finally, the possibility that 1 binds exclusively in the substrate pocket without occupying the cofactor site, much like TCL, must also be considered. Visual inspection of PfENR crystal structures with co-crystallized TCL molecules (e.g. PDB ID: 3AM3)55 suggests that TCL, with a volume of 817 Å3 (LigPrep version 2.5),56 occupies the entirety of the substrate-binding pocket. In contrast, 1 has a calculated volume of 1312 Å3, and the combined volume of TCL and NAD+ is 2508 Å3. The volume of docked celastrol is thus closer to that of ENR-bound pyridomycin (1508 Å3), which is known to span the combined substrate-cofactor pocket,52 than TCL. While the suggestion that 1 binds in the combined pocket is interesting, we note that this volumetric analysis does not preclude the possibility of large-scale ENR conformational changes that could substantially alter pocket volumes. Regardless, coupling our in silico and in vitro results suggests an inhibition behavior that is difficult to describe with canonical inhibitor characterization.
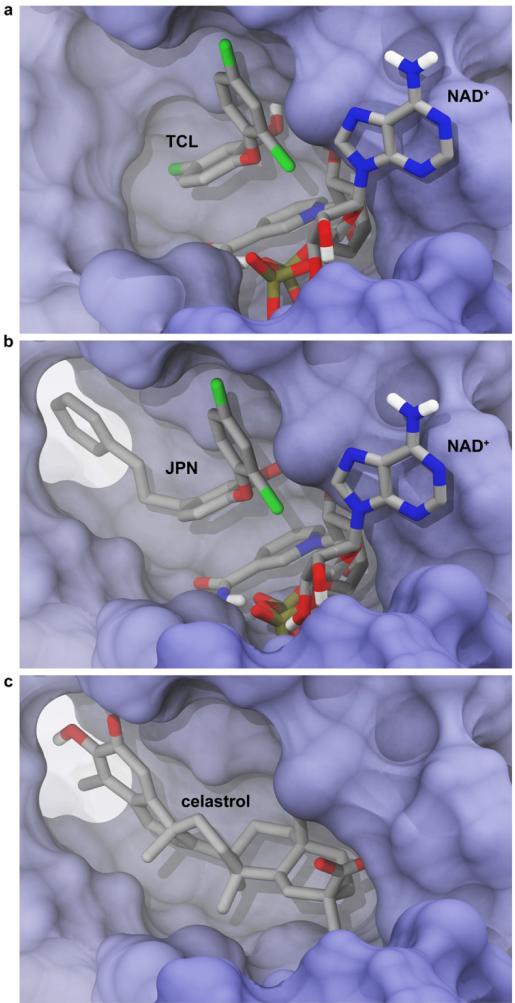
2.7. Computer docking: accounting for receptor flexibility
The current study highlights the importance of accounting for receptor flexibility when performing computer-docking studies. Most ENR crystal structures with co-crystallized TCL and NAD+ ligands have closed pocket conformations that are not compatible with celastrol binding (e.g. PDB: 3AM3) (Fig. 6a).55 However, 2OL4 was co-crystallized with a TCL analogue, 2-(2,4-dichlorophenoxy)-5-(3-phenylpropyl)phenol (JPN), that displaces the F368 side chain, opening up a back pocket that may be critical for celastrol binding (Fig. 6b). Celastrol was identified as an ENR inhibitor only because of the unique geometry of the 2OL4 pocket (Fig. 6c).30 Had we not considered multiple structures with varied geometries, it is unlikely that celastrol would have been identified.
Figure 6.
The ENR binding pocket is flexible and can accommodate various ligands. Some protein residues were removed in order to facilitate visualization. a) The crystallographic pose of triclosan and the NAD+ cofactor (PDB ID: 3AM3).51 b) The crystallographic pose of a triclosan analogue, 2-(2,4-dichlorophenoxy)-5-(3-phenylpropyl)phenol (JPN) (PDB ID: 2OL4).29 Note that the aromatic moiety of this analogue alters the binding-pocket geometry by displacing the F368 side chain relative to its position when TCL is bound. c) The best-predicted celastrol docking pose, obtained when the molecule was docked into the 2OL4 structure.
Traditionally, computer docking has ignored flexibility, despite the central role that macromolecular (e.g., protein) dynamics plays in modern theories of small-molecule binding. Protein binding pockets adopt many different conformations as they move dynamically in solution. Only some of these possible conformations position key interacting moieties such that they can form ideal van der Waals, hydrogen-bond, electrostatic, and/or other interactions with a given ligand. While docking into a single crystal structure may lead to the identification of ligands that are amenable to that particular conformation, other ligands that bind alternate conformations may be overlooked.57
3. Conclusion
As FAS is reportedly only required for parasite survival during the liver stage,19 celastrol efficacy in the blood stage suggests additional target(s). As further evidence of this possibility, we note that the IC50 values of celastrol against PfENR (~5 μM) are higher than the whole-cell growth inhibition measured by Weisman et. al (70% or more at 1 μM).32 However, Figueiredo and co-workers reported whole-cell IC50 values higher than our PfENR values (400 and 560 μM against chloroquine-resistant P. falciparum K1 and chloroquine-sensitive P. falciparum NF, respectively58), so the possibility that ENR is the primary celastrol target cannot be entirely ruled out based on published whole-cell potencies alone.
Celastrol is one of several reported FAS inhibitors that are paradoxically effective against the blood-stage parasite.11,17,59–64 Given the substantial structural diversity of these small-molecule inhibitors, as well as the diverse FAS proteins to which they bind, the idea that they all act on a common secondary blood-stage target seems unlikely.65 And yet genetic studies have demonstrated that parasitic FAS is only required in the liver stage,18,19 suggesting FAS-exclusive inhibitors should not be effective against blood-stage parasites. Given this conflicting evidence, it is unclear whether or not pharmacological FAS inhibition is useful against the blood stage, though its utility in treating liver-stage malaria is widely supported. Further study is required to determine whether or not celastrol has any significant additional blood-stage targets.
While the possibility of binding to multiple malarial targets is promising, binding to human targets should be avoided in order to minimize side effects. We performed a UniProt66 BLAST search67 of PfENR (UniProt ID Q9BJJ9) to identify human homologs. This search revealed that mitochondrial 2,4-dienoyl-CoA reductase (DECR1, UniProt ID B7Z6B8) is the most homologous human protein. The PfENR residues Lys285, Tyr277, and Tyr267, which are predicted to form specific interactions with celastrol, are equivalent to the DECR1 residues Lys182, Phe175, and Thr165, respectively. Differences at positions 277/175 and 267/165 could potentially be exploited to improve specificity. For example, a hydrogen-bond acceptor could be added to celastrol to exploit the hydroxyl group present on PfENR Tyr277 but absent on DECR1 Phe175. Similarly, changes to celastrol that enhance the potential π-π interaction with PfENR Tyr267 could also improve specificity, given that DECR1 Thr165 is not aromatic. Specificity should be an important concern throughout the optimization process, but we do note that celastrol has been used in traditional Chinese medicine for centuries,68,69 suggesting it is not particularly toxic.
Based on a thorough search of the ENR inhibitors deposited in the BindingDB database,70 celastrol appears to be structurally distinct from other reported ENR ligands. Additionally, given its potential effectiveness against multiple stages of the malarial life cycle, we are hopeful that celastrol and its derivatives may be more broadly effective than most current antimalarials, especially given the potential for prophylactic use.
4. Materials and Methods
3.1. In silico screening
We first used computer docking to identify predicted ENR inhibitors. To validate our AutoDock-Vina-based docking protocol34, we obtained three structures of P. falciparum ENR from the Protein Data Bank37 that included co-crystallized triclosan (TCL) molecules. Structures 1NHG, 1UH5, and 2O2Y, with resolutions ranging from 2.20 to 2.43 Å, included two, two, and four chains, respectively, providing a total of eight TCL-bound ENR pockets for validation.31,35,36 Hydrogen atoms were added to the ENR structures using Schrodinger's Protein Preparation Wizard to ensure that the hydrogen-bond network was optimized for TCL binding. The receptors and ligands were subsequently processed with AutoDockTools71 to compute Gasteiger partial charges for each atom,72 to assign AutoDock atom types, and to merge non-polar hydrogen atoms with their parent heteroatoms. Ligand torsions were assigned using AutoDock's AutoTors utility to permit full ligand flexibility during docking. In all cases, the docking box was centered on the corresponding binding pocket. The dimensions of each docking box were chosen so that it extended 10 Å beyond the co-crystallized TCL molecule in all three dimensions. Vina's exhaustiveness parameter was set to 8.
Following this initial pose-prediction validation step, we next prepared additional ENR receptor structures. Rather than relying on a single crystal structure, we considered multiple binding pockets to better account for receptor flexibility. We identified 17 structures of P. falciparum ENR from the Protein Data Bank37 that included co-crystallized ligands and/or cofactors, with resolutions ranging from 2.1 to 3.0 Å (2.5 Å average): 1NHG, 1NHW, 1NNU, 1UH5, 1V35, 1VRW, 1ZSN, 1ZW1, 1ZXB, 1ZXL, 2FOI, 2NQ8, 2O2Y, 2OL4, 2OOS, 2OP0, and 2OP1.30,31,35,36,73 Among these 17 crystal structures there were a total of 48 individual ENR monomers (i.e., eleven crystal structures included two chains, and six structures included four chains). Thirty-eight of the 48 individual chains had ligands bound in both the substrate and NAD+ pockets, and nine had ligands bound in the NAD+ pocket alone. Curiously, unlike chain B from the 2OP0 structure, 2OP0:A contained no bound ligands. For this chain alone we therefore considered the entire protein surface to be a single “pocket” for the purposes of docking. In total, 83 distinct ENR pockets were ultimately selected. All receptors were processed with AutoDockTools71 as described above.
We next performed a benchmark screen to verify that our computational protocol was able to adequately separate known ENR inhibitors from decoy molecules. The BindingDB70,74 was used to identify 36 PfENR inhibitors from the literature with IC50 values less than 500 nM.30,73,75,76 As decoys (i.e., presumed non-binders), we selected 1594 compounds from the National Cancer Institute's Diversity Set IV. The LigPrep computer program (LigPrep version 2.5)56 was used to generate 3D models of these molecules for docking. Within LigPrep, one low-energy ring conformation was generated per compound, protonation was assigned at pH 7.0 using Schrödinger's Epik module, the OPLS 2005 force field77 was used for geometry optimization, and chiralities were determined from the structures themselves.
These compounds were docked into the 83 distinct ENR binding pockets using AutoDock Vina.34 The docking boxes were generally centered on the geometric center of the corresponding crystallographic ligand, though all ligands (including cofactors) were ultimately removed prior to the actual docking. The dimensions of each docking box were chosen so that the box extended 10 Å beyond the crystallographic ligand in all three dimensions. 2OP0:A was the one exception; the docking box associated with this receptor encompassed the entire monomer, as described above. Docking boxes centered on the ENR substrate-binding pocket were generally large enough to encompass the NAD+ pocket, and boxes centered on the NAD+ pocket were large enough to encompass the substrate-binding pocket. The same Vina parameters described previously were used for docking. Receiver operator characteristic (ROC) curves were generated by considering the single pose with the best docking score, independent of receptor conformation.
Having verified that our docking protocol 1) recaptures the TCL crystallographic pose and 2) adequately separates known ENR inhibitors from decoys, we next applied the same protocol to 70 FDA-approved, bioactive, and/or natural-product small molecules previously found to inhibit the growth of whole-cell blood-stage P. falciparum by 70% or more at 1 μM.33 The same LigPrep parameters were again used to prepare these molecules for docking, except varied conformational and protonation states were considered. Specifically, multiple protonation states were assigned for pH values ranging from 5.0 to 9.0, rather than just pH 7.0, and varied ring conformational states were also calculated.
Ultimately, 113 three-dimensional structures with varied conformational and protonation states were generated from these 70 molecules. Two or more protonation states were possible for 13 compounds, two plausible ring conformations were identified for one compound, and multiple combinations of ring-conformation/protonation states were possible for three compounds. The remaining 53 small molecules had no alternate protonation or conformational states. The resulting 113 small-molecule models were further processed with AutoDockTools71 to compute Gasteiger partial charges for each atom,72 to assign AutoDock atom types, and to merge non-polar hydrogen atoms with their parent heteroatoms. Ligand torsions were again assigned using AutoDock's AutoTors utility.
3.2. Computational flooding to identify alternative binding pockets
In order to computationally identify potential allosteric sites, a three-dimensional grid of points spaced 10 Å apart was positioned over chain A of the 2OL4 ENR model, with and without the co-crystallized NAD+, as appropriate. Celastrol was then docked into cubic boxes with sides of length 30.0 Å, centered on each of these points. Each Vina docking produced multiple candidate poses. Any pose obtained, regardless of the docking box used, was considered for subsequent analysis.
3.3. PfENR expression and purification
N-terminus 6×His-tag PfENR encoded in a pET28a plasmid in E. coli BL21 cells was expressed and purified for the in vitro studies, as described previously.31 Cells were grown for 12 hours at 37 °C in 1L TB medium containing 100 mg/L kanamycin sulfate to an OD of 1.0 without the addition of IPTG. The cells were harvested by centrifugation at 1,000 g for 30 minutes, resuspended in lysis buffer (20 mM Tris/HCl buffer (pH 7.4) with 150 mM NaCl), and supplemented with 0.1 mg/mL lysozyme (Worthington Biochemical Corp), 5 μg/mL DNAse I (Sigma), and 5 μg/mL RNAse (Worthington Biochemical Corp.). The cells were lysed by French pressure cell press between 500-1,000 psi. The lysate was subsequently centrifuged at 12,000 g for 45 minutes, and the supernatant was bound with Ni-NTA resin (Qiagen). The column was sequentially washed with 20 mM Tris/HCl buffer (pH 7.4) with 150 mM NaCl. The protein was eluted using a step gradient from 60 to 500 mM buffered imidazole, and fractions were tested for purity by SDS-PAGE (Fig. S1). A size exclusion column (GE HiPrep Sephacryl 16/60 S-200) was used to elute fractions in 20 mM Tris/HCl buffer (pH 7.4) with 150 mM NaCl. PfENR was concentrated with a 10-kDa Amicon spin filter (Milipore) to 2.4 mg/mL. The concentrated sample was subsequently stored in 40% glycerol, 20 mM Tris/HCl buffer (pH 7.4) with 150 mM NaCl at −80°C after flash freezing in liquid nitrogen.
3.4. PfENR continuous assay
The purified ENR was assayed at 27 °C by monitoring the consumption of NADH at an absorbance of 340 nm78 (Perkin Elmer HTS 7000 Plus Bio Assay Reader), using crotonyl coenzyme A trilithium salt (crotonyl-CoA) as the substrate (Sigma) (Fig. S2). The 100 μL reaction volume contained a final concentration of 0.25 μM ENR, 20 mM Tris/HCl buffer (pH 7.4) with 150 mM NaCl, 100 μM crotonyl-CoA, and 100 μM NADH (Sigma). The enzyme was preincubated in 20 mM Tris/HCl buffer (pH 7.4), 150 mM NaCl, and crotonyl-CoA. The reaction was initiated with NADH. The Km for crotonyl-CoA was determined by varying the concentration of crotonyl-CoA (10-300 μM) while keeping the NADH concentration constant at 100 μM. The Km for NADH was determined by titrating NADH (20-500 μM) and keeping the crotonyl-CoA concentration constant at 100 μM.
3.5. PfENR triclosan validation
PfENR inhibition was observed by calculating the consumption of NADH at 340 nm in the presence of varied concentrations of triclosan (Sigma) (Fig. S3). The 100 μL reaction volume contained a final concentration of 50 μM NAD+, 20 mM Tris/HCl buffer (pH 7.4), 150 mM NaCl, 200 μM crotonyl-CoA, 100 μM NADH, and 0.05 μM ENR. ENR was preincubated at 25 °C for 45 minutes with a final concentration of 50 μM NAD+, 20 mM Tris/HCl buffer (pH 7.4), 150 mM NaCl, and varying concentrations of triclosan (300 nM to 0.3 nM). TCL took up 5% of the total reaction volume after dissolving in DMSO. Following this preincubation period, the reaction was initiated with a final concentration of 200 μM crotonyl-CoA and 100 μM NADH.
3.6. Validation of the in silico compounds by continuous assay
Four potential inhibitors identified in silico were subsequently tested experimentally. The final inhibitor concentration was 25 μM in a total reaction volume of 100 μL. Compounds 1-4 (NCI Development Therapeutics Program and Sigma) were tested for PfENR inhibition by observing the consumption of NADH. The total 100 μL reaction volume contained 20 mM Tris/HCl buffer (pH 7.4), 150 mM NaCl, 200 μM crotonyl-CoA, 100 μM NADH, and 0.05 μM ENR. ENR was preincubated with NAD+, 100 mM Tris/HCl buffer (pH 7.4) with 150 mM NaCl, and inhibitor at 25 °C for 45 minutes. Following this preincubation period, the reaction was initiated using 200 μM crotonyl-CoA and 100 μM NADH. To determine if celastrol inhibition was NAD+ dependent, PfENR inhibition was also observed without NAD+ preincubation.
3.7. PfENR inhibition assay and kinetic studies with compound 1
To further characterize the kinetics of compound 1 inhibition, the consumption of NADH in varying concentrations of 1 was observed. The final 100 μL reaction volume contained 50 μM NAD+, 20 mM Tris/HCl buffer (pH 7.4), 150 mM NaCl, 200 μM crotonyl-CoA, 100 μM NADH, and 0.05 μM ENR. ENR was preincubated at 25 °C for 45 minutes with a final concentration of 50 μM NAD+, 20 mM Tris/HCl buffer (pH 7.4), 150 mM NaCl, and varying concentrations (0.9-50 μM) of 1. Following this preincubation period, the reaction was initiated with a final concentration of 200 μM crotonyl-CoA and 100 μM NADH.
The inhibition mechanism of 1 was studied by varying the concentrations of the crotonyl-CoA substrate under Michaelis-Menten conditions with and without NAD+ preincubation (50 μM) in 20 mM Tris/HCl buffer (pH 7.4), 150 mM NaCl, and 0.05 μM ENR. In order to determine the inhibition mechanism with respect to crotonyl-CoA, PfENR was incubated in a total reaction volume of 100 μL with and without NAD+ (50 μM) with a constant saturating concentration of NADH (100 μM) and varying concentrations of 1 dissolved in DMSO. The reaction was subsequently initiated with varying crotonyl-CoA concentrations (3-300 μM). Kinetic data were calculated using GraphPad Prism and fitted to the Michaelis-Menten equation, Y = Vmax*X/(Km + X), where Km is the Michaelis-Menten constant, X is the substrate concentration (e.g., [crotonyl-CoA] or [NADH]), Vmax is the maximum enzyme velocity, and Y is the velocity of the enzyme (SI Table S2). The behavior of celastrol with respect to varying crotonyl-CoA was plotted using Michaelis-Menten and Lineweaver-Burk plots to visualize any changes in Vmax,app and Km. The Ki was calculated by plotting the Vmax,app as a function of varying celastrol concentration. The data was collected in triplicate. To rule out non-specific inhibition by aggregation, a similar experiment was conducted using 0.01% Triton-X during PfENR preincubation.
3.8 Testing for the reversibility of celastrol binding
PfENR was incubated at a final concentration of 3 μM with 150 mM NaCl, 0.01% Triton-X, 100 mM NAD+, and 5-fold the IC50 concentration of celastrol (15 μM) in 20 mM Tris/HCl buffer (pH 7.4), for 45 minutes at 25 °C. Following incubation, the enzyme was diluted 100-fold with 200 μM crotonyl-CoA and 100 μM NADH to initiate the reaction.
In a separate experiment, the possibility of covalent celastrol-cysteine adduct formation was excluded. The enzyme was incubated with 1 mM iodoacetamide (Sigma) in 100 mM ammonium bicarbonate for 60 minutes at 25 °C in the dark. A desalting column (GE Healthcare Illustra Nap-5) was used to separate excess iodoacetamide from the protein. The activity of iodoacetmide-treated PfENR was tested with and without DMSO (5% total volume), in the presence of celastrol at a final concentration of 50 μM. PfENR was preincubated with celastrol at 25 °C for 45 minutes with a final concentration of 50 μM NAD+, 20 mM Tris/HCl buffer (pH 7.4), 150 mM NaCl, and 0.01% Triton-X. Following this preincubation period, the reaction was initiated with a final concentration of 200 μM crotonyl-CoA and 100 μM NADH.
3.9. Compound purity
All NCI compounds, except for compound 1, had confirmed identities and showed greater than 97% purity by spectral analysis (data not shown). LC-MS showed that the initial NCI celastrol sample was only 77% pure. To rule out the possibility that uncharacterized impurities were responsible for ENR inhibition, we obtained a second sample of compound 1 from Sigma-Aldrich. Spectral analysis confirmed the identity of celastrol and LC-MS showed greater than 97% purity (Fig. S4-6) The normalized Sigma-Aldrich and NCI celastrol data showed comparable PfENR inhibition (data not reported).
Supplementary Material
Acknowledgments
This project was supported by the San Diego Match Fellowship and the University of California San Diego Interfaces Training Grant. Funding was also provided by NIH R21A090213, NIH R01GM094924, and NIH R01GM095970 to MDB, and NIH GM31749, NSF MCB-1020765, and MCA93S013 to JAM. Support from the Howard Hughes Medical Institute, the NSF Supercomputer Centers, the San Diego Supercomputer Center, the W.M. Keck Foundation, the National Biomedical Computational Resource, and the Center for Theoretical Biological Physics is gratefully acknowledged.
We would like to acknowledge the UCSD Chemistry & Biochemistry Mass Spectrometry Facility; Prof. P. Dorrestein and Dr. J. Yang for providing the P. falciparum ENR plasmid; the National Cancer Institute (NCI) chemical repository for providing some of the inhibitors for the ENR-inhibition screening assay; Dr. J. La Clair and C. Vickery for figure design; Dr. J. Beld for manuscript design and experimental guidance; Dr. N. Kosa for enzymatic assay and protein expression consulting; and Dr. H. Rivera and J. Hammons for their assistance determining the purity of the inhibitors.
Footnotes
Supplementary Material
Supplementary data associated with this article can be found in the online version.
References and notes
- 1.WHO World Malaria Report. World Health Organization; Geneva Switzerland: 2010. < http://www.who.int/malaria/publications/atoz/9789241564106/en/>. [Google Scholar]
- 2.Klonis N, Crespo-Ortiz MP, Bottova I, Abu-Bakar N, Kenny S, Rosenthal PJ, Tilley L. Proc. Natl. Acad. Sci. 2011 doi: 10.1073/pnas.1104063108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dondorp AM, Yeung S, White L, Nguon C, Day NPJ, Socheat D, von Seidlein L. Nat. Rev. Microbiol. 2010;8:272–280. doi: 10.1038/nrmicro2331. [DOI] [PubMed] [Google Scholar]
- 4.Dondorp AM, Nosten F, Yi P, Das D, Phyo AP, Tarning J, Lwin KM, Ariey F, Hanpithakpong W, Lee SJ, Ringwald P, Silamut K, Imwong M, Chotivanich K, Lim P, Herdman T, An SS, Yeung S, Singhasivanon P, Day NPJ, Lindegardh N, Socheat D, White NJ. N. Engl. J. Med. 2009;361:455–467. doi: 10.1056/NEJMoa0808859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Afonso A, Hunt P, Cheesman S, Alves AC, Cunha CV, do Rosário V, Cravo P. Antimicrob. Agents Chemother. 2006;50:480–489. doi: 10.1128/AAC.50.2.480-489.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tarun AS, Vaughan AM, Kappe SH. I. Trends Parasitol. 2009;25:545–550. doi: 10.1016/j.pt.2009.09.002. [DOI] [PubMed] [Google Scholar]
- 7.Mazier D, Rénia L, Snounou G. Nat. Rev. Drug Discov. 2009;8:854–864. doi: 10.1038/nrd2960. [DOI] [PubMed] [Google Scholar]
- 8.Derbyshire ER, Mota MM, Clardy J. PLoS Pathog. 2011;7:e1002178. doi: 10.1371/journal.ppat.1002178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Flannery EL, Fidock DA, Winzeler EA. J. Med. Chem. 2013;56:7761–7771. doi: 10.1021/jm400325j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Agüero F, Al-Lazikani B, Aslett M, Berriman M, Buckner FS, Campbell RK, Carmona S, Carruthers IM, Chan AE, Chen F. Nat. Rev. Drug Discov. 2008;7:900–907. doi: 10.1038/nrd2684. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schrader FC, Glinca S, Sattler JM, Dahse H-M, Afanador GA, Prigge ST, Lanzer M, Mueller A-K, Klebe G, Schlitzer M. ChemMedChem. 2013;8:442–461. doi: 10.1002/cmdc.201200407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Qidwai T, Khan F. Chem. Biol. Drug Des. 2012;80:155–172. doi: 10.1111/j.1747-0285.2012.01389.x. [DOI] [PubMed] [Google Scholar]
- 13.Vial HJ, Thuet MJ, PhilIppot JR. J. Eukaryot. Microbiol. 1982;29:258–263. doi: 10.1111/j.1550-7408.1982.tb04023.x. [DOI] [PubMed] [Google Scholar]
- 14.Elabbadi N, Ancelin ML, Vial HJ. Antimicrob. Agents Chemother. 1992;36:50–55. doi: 10.1128/aac.36.1.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Waller RF, Keeling PJ, Donald RG, Striepen B, Handman E, Lang-Unnasch N, Cowman AF, Besra GS, Roos DS, McFadden GI. Proc. Natl. Acad. Sci. 1998;95:12352–12357. doi: 10.1073/pnas.95.21.12352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ralph SA, Dooren G. G. van, Waller RF, Crawford MJ, Fraunholz MJ, Foth BJ, Tonkin CJ, Roos DS, McFadden GI. Nat. Rev. Microbiol. 2004;2:203–216. doi: 10.1038/nrmicro843. [DOI] [PubMed] [Google Scholar]
- 17.Surolia N, Surolia A. Nat Med. 2001;7:167–173. doi: 10.1038/84612. [DOI] [PubMed] [Google Scholar]
- 18.Vaughan AM, O'Neill MT, Tarun AS, Camargo N, Phuong TM, Aly ASI, Cowman AF, Kappe SHI. Cell. Microbiol. 2008;11:506–520. doi: 10.1111/j.1462-5822.2008.01270.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yu M, Kumar TRS, Nkrumah LJ, Coppi A, Retzlaff S, Li CD, Kelly BJ, Moura PA, Lakshmanan V, Freundlich JS, Valderramos J-C, Vilcheze C, Siedner M, Tsai JH-C, Falkard B, Sidhu A. bir S., Purcell LA, Gratraud P, Kremer L, Waters AP, Schiehser G, Jacobus DP, Janse CJ, Ager A, Jacobs WR, Jr., Sacchettini JC, Heussler V, Sinnis P, Fidock DA. Cell Host Microbe. 2008;4:567–578. doi: 10.1016/j.chom.2008.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Baldock C, Rafferty JB, Sedelnikova SE, Baker PJ, Stuitje AR, Slabas AR, Hawkes TR, Rice DW. Science. 1996;274:2107–2110. doi: 10.1126/science.274.5295.2107. [DOI] [PubMed] [Google Scholar]
- 21.Parikh SL, Xiao G, Tonge PJ. Biochem. 2000;39:7645–7650. doi: 10.1021/bi0008940. [DOI] [PubMed] [Google Scholar]
- 22.Roujeinikova A, Sedelnikova S, Boer G.-J. de, Stuitje AR, Slabas AR, Rafferty JB, Rice DW. J. Biol. Chem. 1999;274:30811–30817. doi: 10.1074/jbc.274.43.30811. [DOI] [PubMed] [Google Scholar]
- 23.Heath RJ, Yu Y-T, Shapiro MA, Olson E, Rock CO. J. Biol. Chem. 1998;273:30316–30320. doi: 10.1074/jbc.273.46.30316. [DOI] [PubMed] [Google Scholar]
- 24.Hayashi T, Yamamoto O, Sasaki H, Kawaguchi A, Okazaki H. Biochem. Biophys. Res. Commun. 1983;115:1108–1113. doi: 10.1016/s0006-291x(83)80050-3. [DOI] [PubMed] [Google Scholar]
- 25.Jackowski S, Murphy CM, Cronan JE, Rock CO. J. Biol. Chem. 1989;264:7624–7629. [PubMed] [Google Scholar]
- 26.Nishida I, Kawaguchi A, Yamada M. J. Biochem. (Tokyo) 1986;99:1447–1454. doi: 10.1093/oxfordjournals.jbchem.a135614. [DOI] [PubMed] [Google Scholar]
- 27.Kuo MR, Morbidoni HR, Alland D, Sneddon SF, Gourlie BB, Staveski MM, Leonard M, Gregory JS, Janjigian AD, Yee C, Musser JM, Kreiswirth B, Iwamoto H, Perozzo R, Jacobs WR, Sacchettini JC, Fidock DA. J. Biol. Chem. 2003;278:20851–20859. doi: 10.1074/jbc.M211968200. [DOI] [PubMed] [Google Scholar]
- 28.McLeod R, Muench SP, Rafferty JB, Kyle DE, Mui EJ, Kirisits MJ, Mack DG, Roberts CW, Samuel BU, Lyons RE, Dorris M, Milhous WK, Rice DW. Int. J. Parasitol. 2001;31:109–113. doi: 10.1016/s0020-7519(01)00111-4. [DOI] [PubMed] [Google Scholar]
- 29.Kumar G, Parasuraman P, Sharma SK, Banerjee T, Karmodiya K, Surolia N, Surolia AJ. Med. Chem. 2007;50:2665–2675. doi: 10.1021/jm061257w. [DOI] [PubMed] [Google Scholar]
- 30.Freundlich JS, Wang F, Tsai H-C, Kuo M, Shieh H-M, Anderson JW, Nkrumah LJ, Valderramos J-C, Yu M, Kumar TRS, Valderramos SG, Jacobs WR, Schiehser GA, Jacobus DP, Fidock DA, Sacchettini JC. J. Biol. Chem. 2007;282:25436–25444. doi: 10.1074/jbc.M701813200. [DOI] [PubMed] [Google Scholar]
- 31.Perozzo R, Kuo M, Sidhu A. bir S., Valiyaveettil JT, Bittman R, Jacobs WR, Fidock DA, Sacchettini JC. J. Biol. Chem. 2002;277:13106–13114. doi: 10.1074/jbc.M112000200. [DOI] [PubMed] [Google Scholar]
- 32.Heath RJ, Rock CO. J. Biol. Chem. 1995;270:26538–26542. doi: 10.1074/jbc.270.44.26538. [DOI] [PubMed] [Google Scholar]
- 33.Weisman JL, Liou AP, Shelat AA, Cohen FE, Kiplin Guy R, DeRisi JL. Chem. Biol. Drug Des. 2006;67:409–416. doi: 10.1111/j.1747-0285.2006.00391.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Trott O, Olson AJ. J. Comput. Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pidugu LS, Kapoor M, Surolia N, Surolia A, Suguna K. J. Mol. Biol. 2004;343:147–155. doi: 10.1016/j.jmb.2004.08.033. [DOI] [PubMed] [Google Scholar]
- 36.Muench SP, Prigge ST, McLeod R, Rafferty JB, Kirisits MJ, Roberts CW, Mui EJ, Rice DW. Acta Crystallogr. D Biol. Crystallogr. 2007;63:328–338. doi: 10.1107/S0907444906053625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Triballeau N, Acher F, Brabet I, Pin J-P, Bertrand H-OJ. Med. Chem. 2005;48:2534–2547. doi: 10.1021/jm049092j. [DOI] [PubMed] [Google Scholar]
- 39.Kapoor M, Jamal Dar M, Surolia A, Surolia N. Biochem. Biophys. Res. Commun. 2001;289:832–837. doi: 10.1006/bbrc.2001.6061. [DOI] [PubMed] [Google Scholar]
- 40.Kapoor M, Reddy CC, Krishnasastry MV, Surolia N, Surolia A. Biochem. J. 2004;381:719–724. doi: 10.1042/BJ20031821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kapoor M, Gopalakrishnapai J, Surolia N, Surolia A. Biochem. J. 2004;381:735–741. doi: 10.1042/BJ20040302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Choi K-H, Kremer L, Besra GS, Rock CO. J. Biol. Chem. 2000;275:28201–28207. doi: 10.1074/jbc.M003241200. [DOI] [PubMed] [Google Scholar]
- 43.Swinney DC. Nat. Rev. Drug Discov. 2004;3:801–808. doi: 10.1038/nrd1500. [DOI] [PubMed] [Google Scholar]
- 44.Chang A, Schiebel J, Yu W, Bommineni GR, Pan P, Baxter MV, Khanna A, Sotriffer CA, Kisker CF, Tonge PJ. Biochem. 2013 doi: 10.1021/bi400413c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tonge P, Kisker C, Slayden R. Curr. Top. Med. Chem. 2007;7:489–498. doi: 10.2174/156802607780059781. [DOI] [PubMed] [Google Scholar]
- 46.Lu H, England K, am Ende C, Truglio JJ, Luckner S, Reddy BG, Marlenee NL, Knudson SE, Knudson DL, Bowen RA, Kisker C, Slayden RA, Tonge PJ. ACS Chem Biol. 2009;4:221–231. doi: 10.1021/cb800306y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sullivan TJ, Truglio JJ, Boyne ME, Novichenok P, Zhang X, Stratton CF, Li H-J, Kaur T, Amin A, Johnson F, Slayden RA, Kisker C, Tonge PJ. ACS Chem. Biol. 2006;1:43–53. doi: 10.1021/cb0500042. [DOI] [PubMed] [Google Scholar]
- 48.Xu H, Sullivan TJ, Sekiguchi J, Kirikae T, Ojima I, Stratton CF, Mao W, Rock FL, Alley MRK, Johnson F, Walker SG, Tonge PJ. Biochem. 2008;47:4228–4236. doi: 10.1021/bi800023a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lindert S, McCammon JA. Protein Sci. 2012;21:1734–1745. doi: 10.1002/pro.2155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sreeramulu S, Gande SL, Göbel M, Schwalbe H. Angew. Chem. Int. Ed. 2009;48:5853–5855. doi: 10.1002/anie.200900929. [DOI] [PubMed] [Google Scholar]
- 51.Salminen A, Lehtonen M, Paimela T, Kaarniranta K. Biochem. Biophys. Res. Commun. 2010;394:439–442. doi: 10.1016/j.bbrc.2010.03.050. [DOI] [PubMed] [Google Scholar]
- 52.Hartkoorn RC, Pojer F, Read JA, Gingell H, Neres J, Horlacher OP, Altmann K-H, Cole ST. Nat. Chem. Biol. 2014;10:96–98. doi: 10.1038/nchembio.1405. [DOI] [PubMed] [Google Scholar]
- 53.Blat Y. Chem. Biol. Drug Des. 2010;75:535–540. doi: 10.1111/j.1747-0285.2010.00972.x. [DOI] [PubMed] [Google Scholar]
- 54.Durrant JD, McCammon JA. J. Mol. Graph. Model. 2011;29:888–893. doi: 10.1016/j.jmgm.2011.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Maity K, Banerjee T, Prabakaran N, Surolia N, Surolia A, Suguna K. IUBMB Life. 2011;63:30–41. doi: 10.1002/iub.412. [DOI] [PubMed] [Google Scholar]
- 56.Suite 2012: LigPrep, version 2.5. Schrödinger, LLC; New York, NY: 2012. [Google Scholar]
- 57.Amaro RE, Baron R, McCammon JA. J. Comput. Aided Mol. Des. 2008;22:693–705. doi: 10.1007/s10822-007-9159-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Figueiredo JN, Räz B, Séquin U. J. Nat. Prod. 1998;61:718–723. doi: 10.1021/np9704157. [DOI] [PubMed] [Google Scholar]
- 59.Waller RF, Ralph SA, Reed MB, Su V, Douglas JD, Minnikin DE, Cowman AF, Besra GS, McFadden GI. Antimicrob. Agents Chemother. 2003;47:297. doi: 10.1128/AAC.47.1.297-301.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Prigge ST, He X, Gerena L, Waters NC, Reynolds KA. Biochem. 2003;42:1160–1169. doi: 10.1021/bi026847k. [DOI] [PubMed] [Google Scholar]
- 61.Sharma SK, Kapoor M, Ramya TNC, Kumar S, Kumar G, Modak R, Sharma S, Surolia N, Surolia A. J. Biol. Chem. 2003;278:45661–45671. doi: 10.1074/jbc.M304283200. [DOI] [PubMed] [Google Scholar]
- 62.Tasdemir D, Lack G, Brun R, Rüedi P, Scapozza L, Perozzo R. J. Med. Chem. 2006;49:3345–3353. doi: 10.1021/jm0600545. [DOI] [PubMed] [Google Scholar]
- 63.Surolia A, Ramya TNC, Ramya V, Surolia N. Biochem. J. 2004;383:401. doi: 10.1042/BJ20041051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tasdemir D, Topaloglu B, Perozzo R, Brun R, O'Neill R, Carballeira NM, Zhang X, Tonge PJ, Linden A, Rüedi P. Bioorg. Med. Chem. 2007;15:6834–6845. doi: 10.1016/j.bmc.2007.07.032. [DOI] [PubMed] [Google Scholar]
- 65.Spalding MD, Prigge ST. Cell Host Microbe. 2008;4:509–511. doi: 10.1016/j.chom.2008.11.006. [DOI] [PubMed] [Google Scholar]
- 66.Bairoch A, Apweiler R, Wu CH, Barker WC, Boeckmann B, Ferro S, Gasteiger E, Huang H, Lopez R, Magrane M, Martin MJ, Natale DA, O'Donovan C, Redaschi N, Yeh L-SL. Nucleic Acids Res. 2005;33:D154–D159. doi: 10.1093/nar/gki070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 68.Yang H, Chen D, Cui QC, Yuan X, Dou QP. Cancer Res. 2006;66:4758–4765. doi: 10.1158/0008-5472.CAN-05-4529. [DOI] [PubMed] [Google Scholar]
- 69.Kutney JP, Hewitt GM, Lee G, Piotrowska K, Roberts M, Rettig SJ. Can. J. Chem. 1992;70:1455–1480. [Google Scholar]
- 70.Liu T, Lin Y, Wen X, Jorissen RN, Gilson MK. Nucleic Acids Res. 2007;35:D198–D201. doi: 10.1093/nar/gkl999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, Olson AJ. J. Comput. Chem. 2009;30:2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gasteiger J, Marsili M. Tetrahedron. 1980;36:3219–3228. [Google Scholar]
- 73.Freundlich JS, Anderson JW, Sarantakis D, Shieh H-M, Yu M, Valderramos J-C, Lucumi E, Kuo M, Jacobs WR, Jr., Fidock DA, Schiehser GA, Jacobus DP, Sacchettini JC. Bioorg. Med. Chem. Lett. 2005;15:5247–5252. doi: 10.1016/j.bmcl.2005.08.044. [DOI] [PubMed] [Google Scholar]
- 74.Chen X, Lin Y, Liu M, Gilson MK. Bioinformatics. 2002;18:130–139. doi: 10.1093/bioinformatics/18.1.130. [DOI] [PubMed] [Google Scholar]
- 75.Freundlich JS, Yu M, Lucumi E, Kuo M, Tsai H-C, Valderramos J-C, Karagyozov L, Jacobs WR, Jr., Schiehser GA, Fidock DA, Jacobus DP, Sacchettini JC. Bioorg. Med. Chem. Lett. 2006;16:2163–2169. doi: 10.1016/j.bmcl.2006.01.051. [DOI] [PubMed] [Google Scholar]
- 76.Frecer V, Megnassan E, Miertus S. Eur. J. Med. Chem. 2009;44:3009–3019. doi: 10.1016/j.ejmech.2008.12.028. [DOI] [PubMed] [Google Scholar]
- 77.Banks JL, Beard HS, Cao Y, Cho AE, Damm W, Farid R, Felts AK, Halgren TA, Mainz DT, Maple JR, Murphy R, Philipp DM, Repasky MP, Zhang LY, Berne BJ, Friesner RA, Gallicchio E, Levy RM. J. Comput. Chem. 2005;26:1752–1780. doi: 10.1002/jcc.20292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bergmeyer HU, Gawehn K. Methods of enzymatic analysis. Verlag Chemie; 1974. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.