Abstract
Diabetes and its complications are hyperglycemic toxicity diseases. Many metabolic pathways in this array of diseases become aberrant, which is accompanied with a variety of posttranslational protein modifications that in turn reflect diabetic glucotoxicity. In this review, we summarize some of the most widely studied protein modifications in diabetes and its complications. These modifications include glycation, carbonylation, nitration, cysteine S-nitrosylation, acetylation, sumoylation, ADP-ribosylation, O-GlcNAcylation, and succination. All these posttranslational modifications can be significantly attributed to oxidative stress and/or carbon stress induced by diabetic redox imbalance that is driven by activation of pathways, such as the polyol pathway and the ADP-ribosylation pathway. Exploring the nature of these modifications should facilitate our understanding of the pathological mechanisms of diabetes and its associated complications.
Keywords: diabetes, hyperglycemia, redox imbalance, glucotoxicity, NAD+, posttranslational modifications, oxidative stress, carbon stress
Introduction
Glucose is a fundamental molecule for life, and its combustion is exploited in all ways to sustain life. While glucose is essential for cellular survival, too much of it is detrimental.1–3 This is the case in diabetes that either originates from or manifests the dysregulation of glucose metabolism.4 In type 1 diabetes, pancreatic β-cells are destroyed by autoimmune response, and hence no insulin would be available for stimulating glucose metabolism, leading to diabetic hyperglycemia.4–6 In type 2 diabetes, insulin resistance usually precedes β-cell dysfunction via a failure of compensation mechanism.7–9 Initially, insulin resistance would aggravate more insulin secretion by increasing β-cell mass.1,8,10–12 However, such an increase has a limit and will eventually fail to meet the needs for more insulin secretion.9,13,14 Under this circumstance, β-cells die, insulin levels decrease, and frank type 2 diabetes mellitus develops and progresses.15–18 Regardless of the types of diabetes, it is the persistent level of hyperglycemia that causes all the metabolic problems manifested by diabetic complications, such as blindness, peripheral neuropathy, and chronic kidney disease.6,19,20 Indeed, all the metabolic problems can be attributed to hyperglycemic glucotoxicity.1,2,21–25
Therefore, how glucotoxicity is attained in diabetes? Protein modifications induced directly or indirectly by hyperglycemia manifest glucotoxicity. In this review, we attempt to summarize a variety of protein modifications in diabetes. We believe that many of these protein modification processes could serve as therapeutic targets or have therapeutic values. We focus on diabetic protein modifications, including glycation, carbonylation, nitration, nitrosylation, acetylation, ADP-ribosylation, and succination. But before expanding on these modifications, we would like to briefly overview the dysregulated glucose metabolic pathways in diabetes.
Glucose Metabolism and Redox Imbalance in Diabetes
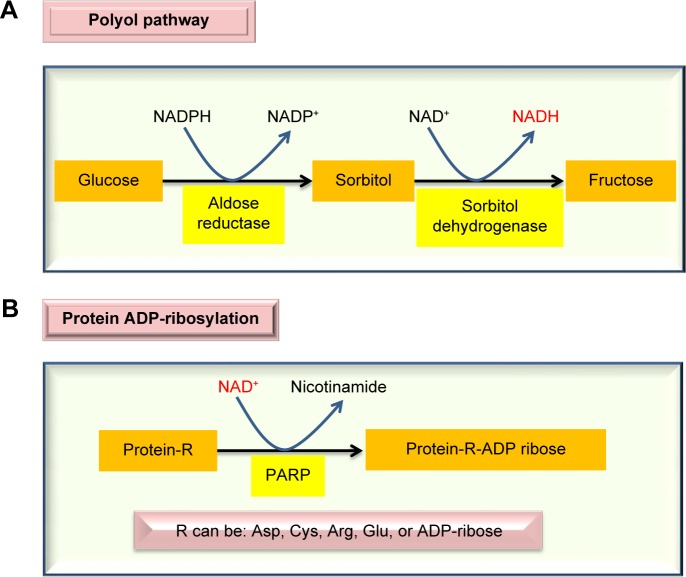
When blood glucose level is persistently high, the body will attempt to mobilize all the possible pathways involved in glucose clearance. One such significant pathway is the polyol pathway.26–29 This pathway is usually dormant in nondiabetic state but can be activated to metabolize up to 30% of the glucose pool in diabetes.30 The pathway involves two reactions, catalyzed by aldose reductase and sorbitol dehydrogenase, respectively. As shown in Figure 1A, the pathway makes excess NADH by consuming NADPH, hence breaking the redox balance between NADH and NAD+. As the aldose reductase reaction is rate limiting, inhibition of aldose reductase has been shown to prevent the occurrence of diabetes and diabetic complications.31–34 Additionally, glucose is converted into fructose, a sugar molecule whose metabolism bypasses glucokinase and phosphorfructokinase-1 in the glycolytic pathway and thus is less regulated,35–37 thereby inducing metabolic stress.35 Excess NADH can overload the mitochondrial electron transport chain and drive overproduction of reactive oxygen species (ROS), which can attack proteins and induce protein modifications.35,38 Additionally, consumption of NADPH by the polyol pathway can impair the function of glutathione reductase that uses NADPH to regenerate the reduced form of glutathione (GSH) from the oxidized form of glutathione (GSSG),39 thus further aggravating cellular redox imbalance.40
Figure 1.
Major enzymatic pathways activated by diabetic hyperglycemia that can impair cellular redox imbalance between NADH and NAD+. The polyol pathway (A) produces NADH, while the ADP-ribosylation pathway (B) can potentially deplete NAD+, accentuating the redox imbalance status between NADH and NAD+.
Also in diabetes, chronic production of ROS can cause DNA damage.41–44 This damage will activate poly-ADP-ribose polymerase that is evolved to repair the damaged DNA molecules.45–47 As poly-ADP-ribose polymerase uses NAD+ as its substrate (Fig. 1B) and is often overactivated,48 its activation usually can deplete NAD+ and leads to the further accentuation of redox imbalance, thereby, causing cell death.49–52 It should be pointed out that while activation of both the polyol pathway and the ADP-ribosylation pathway by diabetic hyperglycemia initially appears to be defensive and adaptive, the eventual consequences are lethal. Therefore, diabetes and its complications could be considered as a failure of compensation diseases.53–55
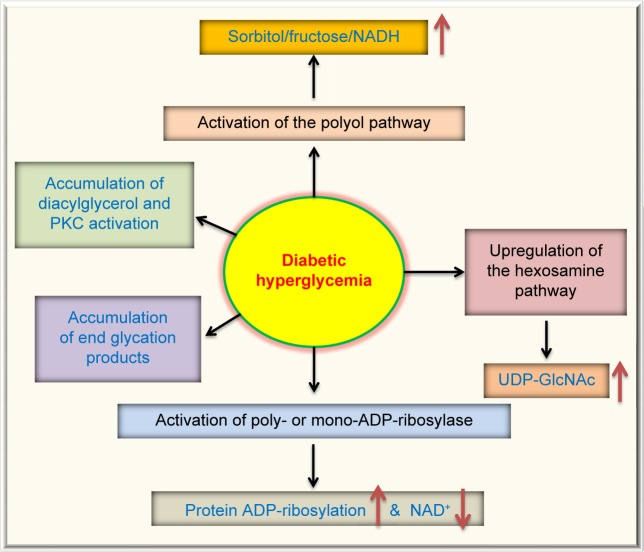
Moreover, diabetic hyperglycemia can also activate other metabolic or signaling pathways. These are summarized in Figure 2, which, in addition to the polyol pathway27,56 and the ADP-ribosylation pathway mentioned earlier, also include the glycation pathway,57,58 the hexosamine pathway,59,60 and the PKC activation pathway.61,62 All these aberrant pathways have been shown to eventually elevate cellular ROS levels,63,64 hence further aggravating cellular redox imbalance and oxidative stress.38 This redox imbalance is probably the driving force for diabetic ROS production and oxidative stress, which are involved in a variety of protein posttranslational modifications.63
Figure 2.
Pathways that are activated or upregulated by diabetic hyperglycemia. In addition to the two pathways shown in Figure 1, diabetic hyperglycemia can also cause activation of the protein kinase C pathway, accumulation of advanced glycation end-products, and upregulation of the hexosamine pathway that fuels the substrate for protein GlcNAcylation. All these pathways have been suggested to be involved in reactive oxygen species production and oxidative stress in the pathogenesis of diabetes and its complications.64,192,193
Protein Modifications in Diabetes
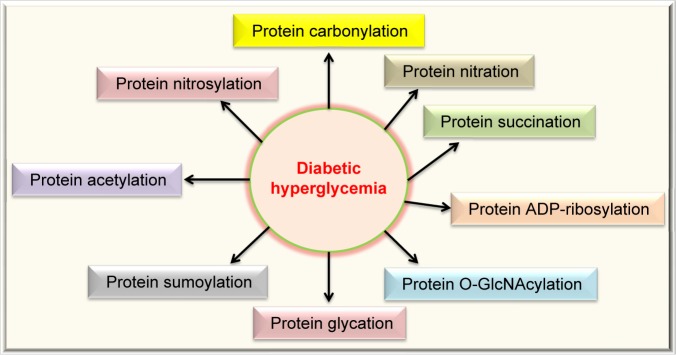
Protein modifications are strategies routinely used by cells to expand their function65–67 but can also reflect the status quo of struggled cellular functions under stressed conditions.68–71 Figure 3 summarizes the types of posttranslational protein modifications in diabetes that are covered in this review. The modifications can be classified into two categories: irreversible and reversible. Irreversible protein modifications include carbonylation, nitration, and glycation, and reversible protein modifications include nitrosylation, acetylation, sumoylation, O-GlcNAcylation, ADP-ribosylation, and succination.
Figure 3.
Protein modifications in diabetes reviewed in this article. These modifications include carbonylation, nitration, succination, ADP-ribosylation, O-GlcNAcylation, glycation (AGEs), sumoylation, acetylation, and nitrosylation. All these protein modifications can manifest glucotoxicity in diabetes.
Advanced glycation end products
Glucose, in its reduced form, can directly react with proteins.72,73 The reaction usually takes place between the glucose’s aldehyde group and the side chain of lysine residues as well as the N-terminal amino groups for given proteins.72,73 The initial species is a Schiff base that can rearrange to form an Amadori intermediate. This intermediate can further rearrange to form varying forms of advanced glycation end products (AGEs).72,73
Glucose can also undergo autoxidation to form ketoaldehyde and hydrogen peroxide in the presence of transition metals.58,74,75 The resulting ketoaldehyde can further react with the amino groups in proteins. This is followed by the formation of ketoimine via Schiff’s base. The ketoimine is then involved in the formation of protein-linked AGEs.58,75,76 It should be noted that fructose can also induce protein glycation.77
Protein can also be modified by methylglyoxal (MGO), a reactive product in the glycolytic pathway.78–81 MGO is a carbonyl-containing compound and mainly reacts with lysine, arginine, and cysteine residues.79,80,82 The eventual protein adducts are a variety of AGEs that could be structurally distinct.83,84 It has also been reported that MGO can have profound detrimental effects in diabetes.79,85 For example, MGO can impair mitochondrial function in diabetes via modifications of a variety of mitochondrial proteins.83
Protein carbonylation
Protein carbonylation is an irreversible process.39,86 Protein carbonyls can be formed directly by ROS attack or indirectly by conjugating to lipid peroxidation byproducts, such as hydroxynonenal.20,87,88 Protein carbonylation can be formed on a variety of amino acid residues, including histidine, cysteine, lysine, arginine, proline, and threonine.87 Protein carbonyls not only have been used as a biomarker for protein oxidation in aging and disease89 but have also been shown to impair protein structure and function.90,91 In diabetes, it has been shown that protein carbonylation is increased in red blood cell membranes in diabetic retinopathy.92 It has also been reported that more plasma proteins show elevated protein carbonyl content in type 2 diabetes.93 In our own studies, we have shown that mitochondrial complex I isolated from diabetic kidneys exhibited selective protein carbonylation via the conjugation with lipid peroxidation product hydroxynonenal that contains a carbonyl group.20 As protein carbonyls are toxic protein adducts impairing protein function and carbonylation can occur to proteins involved in insulin signaling,94 insulin signaling pathways can be disrupted.94 Indeed, protein carbonylation has been suggested to be implicated in insulin resistance,94–96 which is an early event in the development of type 2 diabetes.97–99
Protein nitration
Protein nitration is also an irreversible protein modification. It occurs on protein tyrosine residues due to attack by peroxynitrite.100,101 As peroxynitrite is formed by reaction between superoxide and nitric oxide,102,103 this modification is related to both ROS and reactive nitrogen species. Glucose is known to be implicated in the formation of nitrotyrosine.104,105 Elegant studies by Koeck et al104,105 have demonstrated that glucose can mediate tyrosine nitration in both adipocytes and β-cells, suggesting a role of glucose-modulated nitration in obesity, insulin resistance, and β-cell dysfunction. Importantly, in both adipocytes and β-cells, specific proteins that underwent nitration have been identified; many of them are involved in glucose metabolism and bioenergetics.104,105
O-GlcNAcylation
This posttranslational modification is a reversible modification occurring on serine or threonine residues.106 The substrate for this modification is uridine diphospho-N-acetylglucosamine, the end product of the hexosamine pathway.59,60,107,108 As glucose level becomes higher in diabetes, more glucose is fluxed into the hexosamine pathway, resulting in elevated levels of uridine diphospho-N-acetylglucosamine that can attach to proteins.107,109 Protein O-GlcNAcylation has been found to be involved in numerous biological processes, such as transcription, redox signaling, apoptosis, autophagy, and protein degradation.110–112 Many proteins involved in insulin signaling can undergo this modification. Moreover, O-GlcNAcylation can worsen glucotoxicity in the liver. For example, O-GlcNAcylation of FoxO1 in hepatocytes can increase its transcriptional activity that then upregulates the expression of glucose 6-phosphotase, leading to hyperglycemia by increasing hepatic glucose production.113 Therefore, protein O-GlcNAcylation has been regarded as a major factor in the development of insulin resistance and diabetes and diabetic complications.109,114,115
Protein S-nitrosylation
This modification occurs on cysteine residues and is also a reversible modification.71 As cysteine oxidation status can reflect cellular redox status, this modification is tightly linked to oxidative stress and glutathione content.65 As the modification is reversible, it can regulate protein function either beneficially or detrimentally.65,116 In fact, many studies are now being conducted to explore the beneficial role of this modification in aging and disease.117–121 Nonetheless, S-nitrosylation can play a deleterious role in diabetes.122 For example, it has been reported that in the early phase of diabetes, the level of protein S-nitrosylation is increased that might lead to mitochondrial dysfunction.123 It has also been reported that S-nitrosylation is involved in insulin resistance via the modification and inactivation of protein kinase B.124 It should be mentioned that, similar to this modification, other types of cysteine modifications, such as S-glutathionylation, have also been shown to be involved in the pathogenesis of diabetes and its complications.125,126 For example, hemoglobin shows increased levels of glutathionylation in type 2 diabetes.127
Protein acetylation
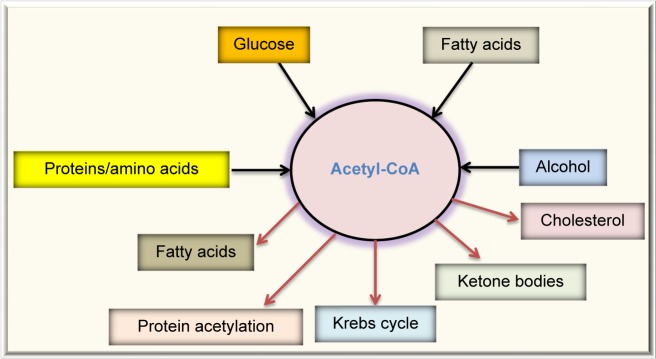
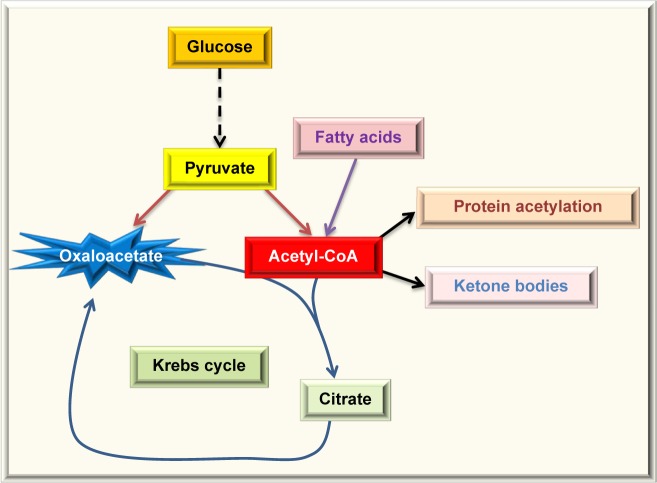
Protein acetylation is the attachment of an acetyl group onto a lysine side chain in a target protein, and the acetyl group usually comes from acetyl-CoA,128,129 which is a central molecule in metabolism. As shown in Figure 4, acetyl-CoA can be derived from combustion of glucose, fatty acids, alcohol, and amino acids. Under normal condition, acetyl-CoA is channeled into the Krebs cycle for ATP production and is also used for the synthesis of cholesterol and fatty acids. Excess acetyl-CoA usually leads to ketone body production130–132 and nonenzymatic protein acetylation.133 This modification often occurs on lysine residues134,135 and has been referred to as carbon stress.108,136–138 Except histone acetylation and enzyme-catalyzed acetylation that are well-regulated processes,139 protein acetylation occurring in cytosol and mitochondria has been widely considered as a pure chemical, nonenzymatic reaction,128,129,133,140,141 although the removal of the lysine-conjugated acetyl groups requires deacetylating enzymes, such as sirtuins.142–144 When the glucose level is high, so is acetyl-CoA that is used as the substrate of acetylation. Hence, proteins can be highly acetylated under hyperglycemic or overnutritional conditions.145,146 When cells switch to use fatty acids as their major energy source, such as under the condition of insulin resistance, whereby glucose cannot enter the cells,147 the levels of acetyl-CoA can increase dramatically (Fig. 5) and protein acetylation can concomitantly increase.146 Thus, it has been reported that increased fatty acid oxidation leads to elevation in protein acetylation in the diabetic heart.148 Additionally, over consumption of alcohol that fuels the production of acetyl-CoA can also elevate protein acetylation.133,149 It should be noted that removal of the acetyl group by enzymes, such as sirtuins, requires the presence of NAD+, which is used as the substrate for deacetylases.150,151 Therefore, a lower level of NAD+ would inhibit protein deacetylation and increase protein acetylation.152 Hence, protein acetylation is a modification that is highly governed by the availability of fuels and NAD+, the latter being tightly linked to cellular redox balance.153–155 In this regard, it is no surprising that aldose reductase can increase protein acetylation via diminishing the NAD+ levels.156
Figure 4.
Fates of acetyl-CoA, a central molecule in fuel metabolism. Acetyl-CoA can be derived by combustion of glucose, fatty acids, proteins or amino acids, and alcohol. In normal condition, acetyl-CoA is mainly channeled into the Krebs cycle for energy production. In overnutrition state, acetyl-CoA can be used to store excess energy by forming fatty acids. Acetyl-CoA is also the source for cholesterol synthesis. In starved state, acetyl-CoA is converted into ketone bodies. Acetyl-CoA is also the substrate used for protein acetylation.
Figure 5.
Excess acetyl-CoA drives nonenzymatic protein acetylation. For noninsulin-dependent cells, diabetic hyperglycemia can overload them with glucose, causing the oversupply of acetyl-CoA. For insulin-dependent tissues in diabetes, the cell cannot get enough glucose and will have to use fatty acids as the source of energy. Because oxaloacetate cannot be continuously formed due to lack of glucose, the level of acetyl-CoA could be extremely high, leading to ketone body production and protein acetylation.
Protein succination
This modification, along with protein acetylation, has also been categorized under carbon stress.138 Protein succination is due to a conjugation reaction between fumerate and proteins and often occurs on protein cysteine residues.157,158 Any fuel source that would elevate the level of fumerate, an intermediate in the Krebs cycle, would theoretically facilitate protein succination.158–161 Similar to S-nitrosylation, protein succination has been shown to increase in diabetes and its complications.161,162 Protein succination can also impair protein function and cellular redox signaling.38,65,71,163–167 Indeed, it has been reported that protein succination is a manifestation of glucotoxicity in both the glycolytic pathway and the mitochondrial bioenergetics pathway.158,161
Protein sumoylation
This posttranslational modification refers to the attachment of a small protein, called small ubiquitin-like modifier (SUMO) protein.168,169 SUMOs are covalently attached onto target proteins and can also be detached.170,171 Hence, sumoylation is also a reversible process. Protein sumoylation is known to be involved in protein translocation, protein stabilization, inflammation, redox imbalance, and oxidative stress.172 In diabetes, SUMO-4 has been implicated in the development of diabetes.173 The target proteins of SUMO-4 include IKBα, STAT, AP-1, and heat shock transcription factors.174 Moreover, SUMO-4 seems to restrict its action in pancreas and immune systems as well as in kidneys.175,176 With respect to regulation of blood glucose levels, sumoylation is known to occur in Glut4, thereby facilitating its translocation onto cell membranes.177 Sumoylation is also known to occur in protein-tyrosine phosphatase 1B (PTP1B), whereby PTP1B function is inhibited.178 As PTP1B is involved in a negative regulation of insulin receptor, PTP1B sumoylation is considered to positively regulate insulin signaling.169 While some of these studies indicate the beneficial role of protein sumoylation, the modification has also been shown to be involved in diabetic pathogenesis.176,179,180 For example, high glucose has been shown to induce sumoylation of Smad4 in mesangial cells, a process likely involved in renal fibrosis in diabetic kidney.181 Additionally, protein sumoylation has been linked to increased endothelial inflammation, a process known to occur in diabetes and its complications.182
Protein ADP-ribosylation
This posttranslational modification occurring in several amino acid residues, such as cysteine, arginine, and asparagine, is the transfer of the ADP-ribose moiety of the NAD+ molecule onto a target protein,183,184 and either mono-ADP-ribosylation or poly-ADP-ribosylation can occur.185 Because NAD+ is used as a substrate for ADP-ribosylases, the process is also highly dependent on NAD+ availability, and activation of ADP-ribosylases can actually deplete NAD+.41–44 This is indeed the case in diabetes as over-activation of poly-ADP-ribosylases and NAD+ depletion have been observed.41,186,187 Accordingly, inhibition of poly-ADP-ribosylases has been demonstrated to prevent the development of diabetes and its complications.188–190 As ROS-induced DNA damage can activate poly-ADP-ribosylases, ADP-ribosylation is thought to be deeply involved in oxidative stress and glucotoxicity.191
Conclusion
In this review, we have summarized the evidence that posttranslational protein modifications can manifest glucotoxicity in diabetes. We have discussed the types of protein modifications that have been, and are still being, intensively investigated in the field of diabetes research. These modifications, including carbonylation, nitration, glycation, O-GlcNAcylation, nitrosylation, succination, acetylation, and ADP-ribosylation, can affect or modulate the function of the modified proteins, with consequences that are more often detrimental than beneficial. Importantly, the driving force behind all these modifications is dysregulation of glucose metabolism in diabetes that results in persistent hyperglycemia. Further studies on these protein modifications in diabetes will continue to help our understanding of the pathogenic mechanisms of diabetes and its complications.
Footnotes
ACADEMIC EDITOR: Gabor Mocz, Editor in Chief
PEER REVIEW: Three peer reviewers contributed to the peer review report. Reviewers’ reports totaled 1,051 words, excluding any confidential comments to the academic editor.
FUNDING: LJY was supported in part by the National Institute of Neurological Disorders and Stroke (Grant number: R01NS079792). The authors confirm that the funder had no influence over the study design, content of the article, or selection of this journal.
COMPETING INTERESTS: Authors disclose no potential conflicts of interest.
Paper subject to independent expert single-blind peer review. All editorial decisions made by independent academic editor. Upon submission manuscript was subject to anti-plagiarism scanning. Prior to publication all authors have given signed confirmation of agreement to article publication and compliance with all applicable ethical and legal requirements, including the accuracy of author and contributor information, disclosure of competing interests and funding sources, compliance with ethical requirements relating to human and animal study participants, and compliance with any copyright requirements of third parties. This journal is a member of the Committee on Publication Ethics (COPE).
Provenance: the authors were invited to submit this paper.
Author Contributions
Conceived the idea and wrote the first draft of the article: L-JY. Contributed to the preparation and design of the article and reviewed and approved the final form of the article: HZ, JW, ZJ, L-JY. All authors reviewed and approved of the final manuscript.
REFERENCES
- 1.Kaiser N, Leibowitz G, Nesher R. Glucotoxicity and beta-cell failure in type 2 diabetes mellitus. J Pediatr Endocrinol Metab. 2003;16(1):5–22. doi: 10.1515/jpem.2003.16.1.5. [DOI] [PubMed] [Google Scholar]
- 2.Del Prato S. Role of glucotoxicity and lipotoxicity in the pathophysiology of Type 2 diabetes mellitus and emerging treatment strategies. Diabet Med. 2009;26(12):1185–1192. doi: 10.1111/j.1464-5491.2009.02847.x. [DOI] [PubMed] [Google Scholar]
- 3.Brunner Y, Schvartz D, Priego-Capote F, Coute Y, Sanchez JC. Glucotoxicity and pancreatic proteomics. J Proteomics. 2009;71(6):576–591. doi: 10.1016/j.jprot.2008.10.002. [DOI] [PubMed] [Google Scholar]
- 4.Tuch B, Dunlop M, Proietto J. Diabetes Research: A Guide for Postgraduates. Harwood Academic Publishers; Amsterdam, The Netherland: 2000. [Google Scholar]
- 5.Funk SD, Yurdagul A, Jr, Orr AW. Hyperglycemia and endothelial dysfunction in atherosclerosis: lessons from type 1 diabetes. Int J Vasc Med. 2012;2012:569654. doi: 10.1155/2012/569654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eiselein L, Schwartz HJ, Rutledge JC. The challenge of type 1 diabetes mellitus. ILAR J. 2004;45(3):231–236. doi: 10.1093/ilar.45.3.231. [DOI] [PubMed] [Google Scholar]
- 7.Leibowitz G, Kaiser N, Cerasi E. Beta-cell failure in type 2 diabetes. J Diabetes Investig. 2011;2(2):82–91. doi: 10.1111/j.2040-1124.2010.00094.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Butler AE, Janson J, Bonner-Weir S, Ritzel R, Rizza RA, Butler PC. Beta-cell deficit and increased beta-cell apoptosis in humans with type 2 diabetes. Diabetes. 2003;52(1):102–110. doi: 10.2337/diabetes.52.1.102. [DOI] [PubMed] [Google Scholar]
- 9.Chang-Chen KJ, Mullur R, Bernal-Mizrachi E. Beta-cell failure as a complication of diabetes. Rev Endocr Metab Disord. 2008;9(4):329–343. doi: 10.1007/s11154-008-9101-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Larsen MO. Beta-cell function and mass in type 2 diabetes. Dan Med Bull. 2009;56(3):153–164. [PubMed] [Google Scholar]
- 11.Abdul-Ghani MA, DeFronzo RA. Oxidative stress in type 2 diabetes. Totowa, New Jersey, USA. In: Miwa S, Beckman KB, Muller FL, editors. Oxidative Stress in Aging. Humana Press; 2008. pp. 191–212. [Google Scholar]
- 12.DeFronzo RA. Insulin resistance: a multifaceted syndrome responsible for NIDDM, obesity, hypertension, dyslipidaemia and atherosclerosis. Neth J Med. 1997;50(5):191–197. doi: 10.1016/s0300-2977(97)00012-0. [DOI] [PubMed] [Google Scholar]
- 13.Maedler K, Donath MY. Beta-cells in type 2 diabetes: a loss of function and mass. Horm Res. 2004;62(suppl 3):67–73. doi: 10.1159/000080503. [DOI] [PubMed] [Google Scholar]
- 14.Muoio DM, Newgard CB. Mechanisms of disease: molecular and metabolic mechanisms of insulin resistance and beta-cell failure in type 2 diabetes. Nat Rev Mol Cell Biol. 2008;9(3):193–205. doi: 10.1038/nrm2327. [DOI] [PubMed] [Google Scholar]
- 15.Lee SA, Lee WJ, Kim EH, et al. Progression to insulin deficiency in Korean patients with type 2 diabetes mellitus positive for anti-GAD antibody. Diabet Med. 2011;28(3):319–324. doi: 10.1111/j.1464-5491.2010.03186.x. [DOI] [PubMed] [Google Scholar]
- 16.Gallwitz B, Kazda C, Kraus P, Nicolay C, Schernthaner G. Contribution of insulin deficiency and insulin resistance to the development of type 2 diabetes: nature of early stage diabetes. Acta Diabetol. 2013;50(1):39–45. doi: 10.1007/s00592-011-0319-4. [DOI] [PubMed] [Google Scholar]
- 17.Cnop M, Welsh N, Jonas JC, Jorns A, Lenzen S, Eizirik DL. Mechanisms of pancreatic beta-cell death in type 1 and type 2 diabetes: many differences, few similarities. Diabetes. 2005;54(suppl 2):S97–S107. doi: 10.2337/diabetes.54.suppl_2.s97. [DOI] [PubMed] [Google Scholar]
- 18.Szendroedi J, Phielix E, Roden M. The role of mitochondria in insulin resistance and type 2 diabetes mellitus. Nat Rev Endocrinol. 2012;8(2):92–103. doi: 10.1038/nrendo.2011.138. [DOI] [PubMed] [Google Scholar]
- 19.Gholap NN, Davies MJ, Mostafa SA, Khunti K. Diagnosing type 2 diabetes and identifying high-risk individuals using the new glycated haemoglobin (HbA1c) criteria. Br J Gen Pract. 2013;63(607):e165–e167. doi: 10.3399/bjgp13X663244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu J, Luo X, Yan LJ. Two dimensional blue native/SDS-PAGE to identify mitochondrial complex I subunits modified by 4-hydroxynonenal (HNE) Front Physiol. 2015;6:98. doi: 10.3389/fphys.2015.00098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Korsgren O, Jansson L, Sandler S, Andersson A. Hyperglycemia-induced B cell toxicity. The fate of pancreatic islets transplanted into diabetic mice is dependent on their genetic background. J Clin Invest. 1990;86(6):2161–2168. doi: 10.1172/JCI114955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Poitout V, Robertson RP. Minireview: secondary beta-cell failure in type 2 diabetes—a convergence of glucotoxicity and lipotoxicity. Endocrinology. 2002;143(2):339–342. doi: 10.1210/endo.143.2.8623. [DOI] [PubMed] [Google Scholar]
- 23.Roseman HM. Progression from obesity to type 2 diabetes: lipotoxicity, glucotoxicity, and implications for management. J Manag Care Pharm. 2005;11(6 suppl B):S3–S11. [PubMed] [Google Scholar]
- 24.Weir GC, Marselli L, Marchetti P, Katsuta H, Jung MH, Bonner-Weir S. Towards better understanding of the contributions of overwork and glucotoxicity to the beta-cell inadequacy of type 2 diabetes. Diabetes Obes Metab. 2009;11(suppl 4):82–90. doi: 10.1111/j.1463-1326.2009.01113.x. [DOI] [PubMed] [Google Scholar]
- 25.Wu J, Yan LJ. Streptozotocin-induced type 1 diabetes in rodents as a model for studying mitochondrial mechanisms of diabetic beta cell glucotoxicity. Diabetes Metab Syndr Obes. 2015;8:181–188. doi: 10.2147/DMSO.S82272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chung SS, Chung SK. Aldose reductase in diabetic microvascular complications. Curr Drug Targets. 2005;6(4):475–486. doi: 10.2174/1389450054021891. [DOI] [PubMed] [Google Scholar]
- 27.Dunlop M. Aldose reductase and the role of the polyol pathway in diabetic nephropathy. Kidney Int Suppl. 2000;77:S3–S12. doi: 10.1046/j.1523-1755.2000.07702.x. [DOI] [PubMed] [Google Scholar]
- 28.Hodgkinson AD, Sondergaard KL, Yang B, Cross DF, Millward BA, Demaine AG. Aldose reductase expression is induced by hyperglycemia in diabetic nephropathy. Kidney Int. 2001;60(1):211–218. doi: 10.1046/j.1523-1755.2001.00788.x. [DOI] [PubMed] [Google Scholar]
- 29.Iwata K, Nishinaka T, Matsuno K, et al. The activity of aldose reductase is elevated in diabetic mouse heart. J Pharmacol Sci. 2007;103(4):408–416. doi: 10.1254/jphs.fp0070136. [DOI] [PubMed] [Google Scholar]
- 30.Fantus IG. The pathogenesis of the chronic complications of the diabetes mellitus. Endocrinol Rounds. 2002;2(4):1–8. [Google Scholar]
- 31.Yasunari K, Kohno M, Kano H, Minami M, Yoshikawa J. Aldose reductase inhibitor improves insulin-mediated glucose uptake and prevents migration of human coronary artery smooth muscle cells induced by high glucose. Hypertension. 2000;35(5):1092–1098. doi: 10.1161/01.hyp.35.5.1092. [DOI] [PubMed] [Google Scholar]
- 32.Yasunari K, Kohno M, Kano H, Yokokawa K, Horio T, Yoshikawa J. Aldose reductase inhibitor prevents hyperproliferation and hypertrophy of cultured rat vascular smooth muscle cells induced by high glucose. Arterioscler Thromb Vasc Biol. 1995;15(12):2207–2212. doi: 10.1161/01.atv.15.12.2207. [DOI] [PubMed] [Google Scholar]
- 33.Reddy AB, Ramana KV. Aldose reductase inhibition: emerging drug target for the treatment of cardiovascular complications. Recent Pat Cardiovasc Drug Discov. 2010;5(1):25–32. doi: 10.2174/157489010790192683. [DOI] [PubMed] [Google Scholar]
- 34.Lo AC, Cheung AK, Hung VK, et al. Deletion of aldose reductase leads to protection against cerebral ischemic injury. J Cereb Blood Flow Metab. 2007;27(8):1496–1509. doi: 10.1038/sj.jcbfm.9600452. [DOI] [PubMed] [Google Scholar]
- 35.Luo X, Li R, Yan LJ. Roles of pyruvate, NADH, and mitochondrial complex I in redox balance and imbalance in β cell function and dysfunction. J Diabetes Res. 2015;2015:512618. doi: 10.1155/2015/512618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yabe-Nishimura C. Aldose reductase in glucose toxicity: a potential target for the prevention of diabetic complications. Pharmacol Rev. 1998;50(1):21–33. [PubMed] [Google Scholar]
- 37.Tang WH, Martin KA, Hwa J. Aldose reductase, oxidative stress, and diabetic mellitus. Front Pharmacol. 2012;3:87. doi: 10.3389/fphar.2012.00087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yan LJ. Positive oxidative stress in aging and aging-related disease tolerance. Redox Biol. 2014;2C:165–169. doi: 10.1016/j.redox.2014.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yan LJ, Christians ES, Liu L, Xiao X, Sohal RS, Benjamin IJ. Mouse heat shock transcription factor 1 deficiency alters cardiac redox homeostasis and increases mitochondrial oxidative damage. EMBO J. 2002;21(19):5164–5172. doi: 10.1093/emboj/cdf528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.De Mattia G, Laurenti O, Bravi C, Ghiselli A, Iuliano L, Balsano F. Effect of aldose reductase inhibition on glutathione redox status in erythrocytes of diabetic patients. Metabolism. 1994;43(8):965–968. doi: 10.1016/0026-0495(94)90175-9. [DOI] [PubMed] [Google Scholar]
- 41.Dolle C, Rack JG, Ziegler M. NAD and ADP-ribose metabolism in mitochondria. FEBS J. 2013;280(15):3530–3541. doi: 10.1111/febs.12304. [DOI] [PubMed] [Google Scholar]
- 42.Ying W. NAD+/NADH and NADP+/NADPH in cellular functions and cell death: regulation and biological consequences. Antioxid Redox Signal. 2008;10(2):179–206. doi: 10.1089/ars.2007.1672. [DOI] [PubMed] [Google Scholar]
- 43.Szabados E, Fischer GM, Gallyas F, Jr, Kispal G, Sumegi B. Enhanced ADP-ribosylation and its diminution by lipoamide after ischemia-reperfusion in perfused rat heart. Free Radic Biol Med. 1999;27(9–10):1103–1113. doi: 10.1016/s0891-5849(99)00151-3. [DOI] [PubMed] [Google Scholar]
- 44.Szabo C. Roles of poly(ADP-ribose) polymerase activation in the pathogenesis of diabetes mellitus and its complications. Pharmacol Res. 2005;52(1):60–71. doi: 10.1016/j.phrs.2005.02.015. [DOI] [PubMed] [Google Scholar]
- 45.Pittelli M, Felici R, Pitozzi V, et al. Pharmacological effects of exogenous NAD on mitochondrial bioenergetics, DNA repair, and apoptosis. Mol Pharmacol. 2011;80(6):1136–1146. doi: 10.1124/mol.111.073916. [DOI] [PubMed] [Google Scholar]
- 46.Szabo C, Zanchi A, Komjati K, et al. Poly(ADP-ribose) polymerase is activated in subjects at risk of developing type 2 diabetes and is associated with impaired vascular reactivity. Circulation. 2002;106(21):2680–2686. doi: 10.1161/01.cir.0000038365.78031.9c. [DOI] [PubMed] [Google Scholar]
- 47.Pacher P, Liaudet L, Soriano FG, Mabley JG, Szabo E, Szabo C. The role of poly(ADP-ribose) polymerase activation in the development of myocardial and endothelial dysfunction in diabetes. Diabetes. 2002;51(2):514–521. doi: 10.2337/diabetes.51.2.514. [DOI] [PubMed] [Google Scholar]
- 48.Boesten DM, von Ungern-Sternberg SN, den Hartog GJ, Bast A. Protective pleiotropic effect of flavonoids on NAD(+) levels in endothelial cells exposed to high glucose. Oxid Med Cell Longev. 2015;2015:894597. doi: 10.1155/2015/894597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pacher P, Szabo C. Role of poly(ADP-ribose) polymerase-1 activation in the pathogenesis of diabetic complications: endothelial dysfunction, as a common underlying theme. Antioxid Redox Signal. 2005;7(11–12):1568–1580. doi: 10.1089/ars.2005.7.1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Obrosova IG, Drel VR, Pacher P, et al. Oxidative-nitrosative stress and poly(ADP-ribose) polymerase (PARP) activation in experimental diabetic neuropathy: the relation is revisited. Diabetes. 2005;54(12):3435–3441. doi: 10.2337/diabetes.54.12.3435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chiu J, Xu BY, Chen S, Feng B, Chakrabarti S. Oxidative stress-induced, poly(ADP-ribose) polymerase-dependent upregulation of ET-1 expression in chronic diabetic complications. Can J Physiol Pharmacol. 2008;86(6):365–372. doi: 10.1139/Y08-033. [DOI] [PubMed] [Google Scholar]
- 52.Puthanveetil P, Zhang D, Wang Y, et al. Diabetes triggers a PARP1 mediated death pathway in the heart through participation of FoxO1. J Mol Cell Cardiol. 2012;53(5):677–686. doi: 10.1016/j.yjmcc.2012.08.013. [DOI] [PubMed] [Google Scholar]
- 53.Patti ME, Corvera S. The role of mitochondria in the pathogenesis of type 2 diabetes. Endocr Rev. 2010;31(3):364–395. doi: 10.1210/er.2009-0027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Beaudry JL, Riddell MC. Effects of glucocorticoids and exercise on pancreatic beta-cell function and diabetes development. Diabetes Metab Res Rev. 2012;28(7):560–573. doi: 10.1002/dmrr.2310. [DOI] [PubMed] [Google Scholar]
- 55.Bergman RN. The two-edged sword. Endocrinol Nutr. 2009;56(suppl 4):5–7. doi: 10.1016/S1575-0922(09)73507-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chung SS, Ho EC, Lam KS, Chung SK. Contribution of polyol pathway to diabetes-induced oxidative stress. J Am Soc Nephrol. 2003;14(8 suppl 3):S233–S236. doi: 10.1097/01.asn.0000077408.15865.06. [DOI] [PubMed] [Google Scholar]
- 57.Vlassara H, Striker GE. Advanced glycation endproducts in diabetes and diabetic complications. Endocrinol Metab Clin North Am. 2013;42(4):697–719. doi: 10.1016/j.ecl.2013.07.005. [DOI] [PubMed] [Google Scholar]
- 58.Wolff SP, Jiang ZY, Hunt JV. Protein glycation and oxidative stress in diabetes mellitus and ageing. Free Radic Biol Med. 1991;10(5):339–352. doi: 10.1016/0891-5849(91)90040-a. [DOI] [PubMed] [Google Scholar]
- 59.Beyer AM, Weihrauch D. Hexosamine pathway activation and O-linked-N-acetylglucosamine: novel mediators of endothelial dysfunction in hyperglycemia and diabetes. Vascul Pharmacol. 2012;56(3–4):113–114. doi: 10.1016/j.vph.2012.01.005. [DOI] [PubMed] [Google Scholar]
- 60.Schleicher ED, Weigert C. Role of the hexosamine biosynthetic pathway in diabetic nephropathy. Kidney Int Suppl. 2000;77:S13–S18. doi: 10.1046/j.1523-1755.2000.07703.x. [DOI] [PubMed] [Google Scholar]
- 61.Xia L, Wang H, Munk S, et al. Reactive oxygen species, PKC-beta1, and PKC-zeta mediate high-glucose-induced vascular endothelial growth factor expression in mesangial cells. Am J Physiol Endocrinol Metab. 2007;293(5):E1280–E1288. doi: 10.1152/ajpendo.00223.2007. [DOI] [PubMed] [Google Scholar]
- 62.Xia L, Wang H, Munk S, et al. High glucose activates PKC-zeta and NADPH oxidase through autocrine TGF-beta1 signaling in mesangial cells. Am J Physiol Renal Physiol. 2008;295(6):F1705–F1714. doi: 10.1152/ajprenal.00043.2008. [DOI] [PubMed] [Google Scholar]
- 63.Yan LJ. Pathogenesis of chronic hyperglycemia: from reductive stress to oxidative stress. J Diabetes Res. 2014;2014:137919. doi: 10.1155/2014/137919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Robertson RP. Chronic oxidative stress as a central mechanism for glucose toxicity in pancreatic islet beta cells in diabetes. J Biol Chem. 2004;279(41):42351–42354. doi: 10.1074/jbc.R400019200. [DOI] [PubMed] [Google Scholar]
- 65.Yan LJ. Protein redox modification as a cellular defense mechanism against tissue ischemic injury. Oxid Med Cell Longev. 2014;2014:12. doi: 10.1155/2014/343154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Knorre DG, Kudryashova NV, Godovikova TS. Chemical and functional aspects of posttranslational modification of proteins. Acta Naturae. 2009;1(3):29–51. [PMC free article] [PubMed] [Google Scholar]
- 67.Walsh CT. Posttranslational Modification of Proteins: Expanding Nature’s Inventory. Eaglewood, CO: Robert and Company Publishers; 2006. [Google Scholar]
- 68.Berlett BS, Stadtman ER. Protein oxidation in aging, disease, and oxidative stress. J Biol Chem. 1997;272(33):20313–20316. doi: 10.1074/jbc.272.33.20313. [DOI] [PubMed] [Google Scholar]
- 69.Stadtman ER. Protein oxidation in aging and age-related diseases. Ann N Y Acad Sci. 2001;928:22–38. doi: 10.1111/j.1749-6632.2001.tb05632.x. [DOI] [PubMed] [Google Scholar]
- 70.Chakravarti B, Chakravarti DN. Oxidative modification of proteins: age-related changes. Gerontology. 2007;53(3):128–139. doi: 10.1159/000097865. [DOI] [PubMed] [Google Scholar]
- 71.Cai Z, Yan LJ. Protein oxidative modifications: beneficial roles in disease and health. J Biochem Pharmacol Res. 2013;1(1):15–26. [PMC free article] [PubMed] [Google Scholar]
- 72.Nass N, Bartling B, Navarrete Santos A, et al. Advanced glycation end products, diabetes and ageing. Z Gerontol Geriatr. 2007;40(5):349–356. doi: 10.1007/s00391-007-0484-9. [DOI] [PubMed] [Google Scholar]
- 73.Nass N, Kukat A, Seibel P, et al. Advanced glycation end product accumulation in rho(0) cells without a functional respiratory chain. Biol Chem. 2009;390(9):915–919. doi: 10.1515/BC.2009.083. [DOI] [PubMed] [Google Scholar]
- 74.Chetyrkin S, Mathis M, Pedchenko V, et al. Glucose autoxidation induces functional damage to proteins via modification of critical arginine residues. Biochemistry. 2011;50(27):6102–6112. doi: 10.1021/bi200757d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wolff SP, Dean RT. Glucose autoxidation and protein modification. The potential role of ‘autoxidative glycosylation’ in diabetes. Biochem J. 1987;245(1):243–250. doi: 10.1042/bj2450243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lyons TJ, Jenkins AJ. Glycation, oxidation, and lipoxidation in the development of the complications of diabetes: a carbonyl stress hypothesis. Diabetes Rev (Alex) 1997;5(4):365–391. [PMC free article] [PubMed] [Google Scholar]
- 77.Takagi Y, Kashiwagi A, Tanaka Y, Asahina T, Kikkawa R, Shigeta Y. Significance of fructose-induced protein oxidation and formation of advanced glycation end product. J Diabetes Complications. 1995;9(2):87–91. doi: 10.1016/1056-8727(94)00022-g. [DOI] [PubMed] [Google Scholar]
- 78.Dmitriev LF, Dugin SF. Aldehydes and disturbance of carbohydrate metabolism: some consequences and possible approaches to its normalization. Arch Physiol Biochem. 2007;113(2):87–95. doi: 10.1080/13813450701384783. [DOI] [PubMed] [Google Scholar]
- 79.Allaman I, Belanger M, Magistretti PJ. Methylglyoxal, the dark side of glycolysis. Front Neurosci. 2015;9:23. doi: 10.3389/fnins.2015.00023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Maessen DE, Stehouwer CD, Schalkwijk CG. The role of methylglyoxal and the glyoxalase system in diabetes and other age-related diseases. Clin Sci (Lond) 2015;128(12):839–861. doi: 10.1042/CS20140683. [DOI] [PubMed] [Google Scholar]
- 81.Queisser MA, Yao D, Geisler S, et al. Hyperglycemia impairs proteasome function by methylglyoxal. Diabetes. 2010;59(3):670–678. doi: 10.2337/db08-1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Thornalley PJ, Langborg A, Minhas HS. Formation of glyoxal, methylglyoxal and 3-deoxyglucosone in the glycation of proteins by glucose. Biochem J. 1999;344(pt 1):109–116. [PMC free article] [PubMed] [Google Scholar]
- 83.Rosca MG, Mustata TG, Kinter MT, et al. Glycation of mitochondrial proteins from diabetic rat kidney is associated with excess superoxide formation. Am J Physiol Renal Physiol. 2005;289(2):F420–F430. doi: 10.1152/ajprenal.00415.2004. [DOI] [PubMed] [Google Scholar]
- 84.Ozdemir AM, Hopfer U, Rosca MV, Fan XJ, Monnier VM, Weiss MF. Effects of advanced glycation end product modification on proximal tubule epithelial cell processing of albumin. Am J Nephrol. 2008;28(1):14–24. doi: 10.1159/000108757. [DOI] [PubMed] [Google Scholar]
- 85.Turk Z. Glycotoxines, carbonyl stress and relevance to diabetes and its complications. Physiol Res. 2010;59(2):147–156. doi: 10.33549/physiolres.931585. [DOI] [PubMed] [Google Scholar]
- 86.Yan LJ. Analysis of oxidative modification of proteins. Curr Protoc Protein Sci. 2009 doi: 10.1002/0471140864.ps1404s55. Chapter 14:Unit 14.4. [DOI] [PubMed] [Google Scholar]
- 87.Stadtman ER, Levine RL. Free radical-mediated oxidation of free amino acids and amino acid residues in proteins. Amino Acids. 2003;25(3–4):207–218. doi: 10.1007/s00726-003-0011-2. [DOI] [PubMed] [Google Scholar]
- 88.Yan LJ, Sohal RS. Mitochondrial adenine nucleotide translocase is modified oxidatively during aging. Proc Natl Acad Sci U S A. 1998;95:12896–12901. doi: 10.1073/pnas.95.22.12896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Yan LJ, Levine RL, Sohal RS. Oxidative damage during aging targets mitochondrial aconitase. Proc Natl Acad Sci U S A. 1997;94:11168–11172. doi: 10.1073/pnas.94.21.11168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Dalle-Donne I, Aldini G, Carini M, Colombo R, Rossi R, Milzani A. Protein carbonylation, cellular dysfunction, and disease progression. J Cell Mol Med. 2006;10(2):389–406. doi: 10.1111/j.1582-4934.2006.tb00407.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Fedorova M, Bollineni RC, Hoffmann R. Protein carbonylation as a major hallmark of oxidative damage: update of analytical strategies. Mass Spectrom Rev. 2014;33(2):79–97. doi: 10.1002/mas.21381. [DOI] [PubMed] [Google Scholar]
- 92.Margetis PI, Antonelou MH, Petropoulos IK, Margaritis LH, Papassideri IS. Increased protein carbonylation of red blood cell membrane in diabetic retinopathy. Exp Mol Pathol. 2009;87(1):76–82. doi: 10.1016/j.yexmp.2009.04.001. [DOI] [PubMed] [Google Scholar]
- 93.Bollineni RC, Fedorova M, Bluher M, Hoffmann R. Carbonylated plasma proteins as potential biomarkers of obesity induced type 2 diabetes mellitus. J Proteome Res. 2014;13(11):5081–5093. doi: 10.1021/pr500324y. [DOI] [PubMed] [Google Scholar]
- 94.Boden G, Homko C, Barrero CA, et al. Excessive caloric intake acutely causes oxidative stress, GLUT4 carbonylation, and insulin resistance in healthy men. Sci Transl Med. 2015;7(304):304re307. doi: 10.1126/scitranslmed.aac4765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ruskovska T, Bernlohr DA. Oxidative stress and protein carbonylation in adipose tissue—implications for insulin resistance and diabetes mellitus. J Proteomics. 2013;92:323–334. doi: 10.1016/j.jprot.2013.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Mendez L, Pazos M, Molinar-Toribio E, et al. Protein carbonylation associated to high-fat, high-sucrose diet and its metabolic effects. J Nutr Biochem. 2014;25(12):1243–1253. doi: 10.1016/j.jnutbio.2014.06.014. [DOI] [PubMed] [Google Scholar]
- 97.Taganis T. Reactive oxygen species and insulin resistance: the good, the bad and the ugly. Trends Pharmacol Sci. 2011;32(2):82–89. doi: 10.1016/j.tips.2010.11.006. [DOI] [PubMed] [Google Scholar]
- 98.Henriksen EJ, Diamond-Stanic MK, Marchionne EM. Oxidative stress and the etiology of insulin resistance and type 2 diabetes. Free Radic Biol Med. 2011;51(5):993–999. doi: 10.1016/j.freeradbiomed.2010.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Barnett AH. Type 2 Diabetes. 2nd ed. Oxford University Press; Oxford, United Kindom: 2012. [Google Scholar]
- 100.Radi R. Protein tyrosine nitration: biochemical mechanisms and structural basis of functional effects. Acc Chem Res. 2013;46(2):550–559. doi: 10.1021/ar300234c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Radi R, Cassina A, Hodara R, Quijano C, Castro L. Peroxynitrite reactions and formation in mitochondria. Free Radic Biol Med. 2002;33(11):1451–1464. doi: 10.1016/s0891-5849(02)01111-5. [DOI] [PubMed] [Google Scholar]
- 102.Ferrer-Sueta G, Radi R. Chemical biology of peroxynitrite: kinetics, diffusion, and radicals. ACS Chem Biol. 2009;4(3):161–177. doi: 10.1021/cb800279q. [DOI] [PubMed] [Google Scholar]
- 103.Goldstein S, Merenyi G. The chemistry of peroxynitrite: implications for biological activity. Methods Enzymol. 2008;436:49–61. doi: 10.1016/S0076-6879(08)36004-2. [DOI] [PubMed] [Google Scholar]
- 104.Koeck T, Corbett JA, Crabb JW, Stuehr DJ, Aulak KS. Glucose-modulated tyrosine nitration in beta cells: targets and consequences. Arch Biochem Biophys. 2009;484(2):221–231. doi: 10.1016/j.abb.2009.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Koeck T, Willard B, Crabb JW, Kinter M, Stuehr DJ, Aulak KS. Glucose-mediated tyrosine nitration in adipocytes: targets and consequences. Free Radic Biol Med. 2009;46(7):884–892. doi: 10.1016/j.freeradbiomed.2008.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Lima VV, Spitler K, Choi H, Webb RC, Tostes RC. O-GlcNAcylation and oxidation of proteins: is signalling in the cardiovascular system becoming sweeter? Clin Sci (Lond) 2012;123(8):473–486. doi: 10.1042/CS20110638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Yki-Jarvinen H, Vogt C, Iozzo P, et al. UDP-N-acetylglucosamine transferase and glutamine: fructose 6-phosphate amidotransferase activities in insulin-sensitive tissues. Diabetologia. 1997;40(1):76–81. doi: 10.1007/s001250050645. [DOI] [PubMed] [Google Scholar]
- 108.Luo X, Wu J, Jing S, Yan LJ. Hyperglycemic stress and carbon stress in diabetic glucotoxicity. Aging Dis. 2016;7(1):90–110. doi: 10.14336/AD.2015.0702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Ma J, Hart GW. Protein O-GlcNAcylation in diabetes and diabetic complications. Expert Rev Proteomics. 2013;10(4):365–380. doi: 10.1586/14789450.2013.820536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Zachara NE, Hart GW. Cell signaling, the essential role of O-GlcNAc! Biochim Biophys Acta. 2006;1761(5–6):599–617. doi: 10.1016/j.bbalip.2006.04.007. [DOI] [PubMed] [Google Scholar]
- 111.Hart GW. Three decades of research on O-GlcNAcylation—a major nutrient sensor that regulates signaling, transcription and cellular metabolism. Front Endocrinol (Lausanne) 2014;5:183. doi: 10.3389/fendo.2014.00183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Hardiville S, Hart GW. Nutrient regulation of signaling, transcription, and cell physiology by O-GlcNAcylation. Cell Metab. 2014;20(2):208–213. doi: 10.1016/j.cmet.2014.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Kuo M, Zilberfarb V, Gangneux N, Christeff N, Issad T. O-GlcNAc modification of FoxO1 increases its transcriptional activity: a role in the glucotoxicity phenomenon? Biochimie. 2008;90(5):679–685. doi: 10.1016/j.biochi.2008.03.005. [DOI] [PubMed] [Google Scholar]
- 114.Banerjee PS, Ma J, Hart GW. Diabetes-associated dysregulation of O-GlcNAcylation in rat cardiac mitochondria. Proc Natl Acad Sci U S A. 2015;112(19):6050–6055. doi: 10.1073/pnas.1424017112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Fardini Y, Masson E, Boudah O, et al. O-GlcNAcylation of FoxO1 in pancreatic beta cells promotes Akt inhibition through an IGFBP1-mediated autocrine mechanism. FASEB J. 2014;28(2):1010–1021. doi: 10.1096/fj.13-238378. [DOI] [PubMed] [Google Scholar]
- 116.Tong G, Aponte AM, Kohr MJ, Steenbergen C, Murphy E, Sun J. Postconditioning leads to an increase in protein S-nitrosylation. Am J Physiol Heart Circ Physiol. 2014;306(6):H825–H832. doi: 10.1152/ajpheart.00660.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Sun J, Murphy E. Protein S-nitrosylation and cardioprotection. Circ Res. 2010;106(2):285–296. doi: 10.1161/CIRCRESAHA.109.209452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Sun J, Morgan M, Shen RF, Steenbergen C, Murphy E. Preconditioning results in S-nitrosylation of proteins involved in regulation of mitochondrial energetics and calcium transport. Circ Res. 2007;101(11):1155–1163. doi: 10.1161/CIRCRESAHA.107.155879. [DOI] [PubMed] [Google Scholar]
- 119.Lima B, Forrester MT, Hess DT, Stamler JS. S-nitrosylation in cardiovascular signaling. Circ Res. 2010;106(4):633–646. doi: 10.1161/CIRCRESAHA.109.207381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Nakamura T, Lipton SA. Protein S-nitrosylation as a therapeutic target for neurodegenerative diseases. Trends Pharmacol Sci. 2016;37(1):73–84. doi: 10.1016/j.tips.2015.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Chouchani ET, Methner C, Nadtochiy SM, et al. Cardioprotection by S-nitrosation of a cysteine switch on mitochondrial complex I. Nat Med. 2013;19(6):753–759. doi: 10.1038/nm.3212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Kaneki M, Shimizu N, Yamada D, Chang K. Nitrosative stress and pathogenesis of insulin resistance. Antioxid Redox Signal. 2007;9(3):319–329. doi: 10.1089/ars.2006.1464. [DOI] [PubMed] [Google Scholar]
- 123.Noriega-Cisneros R, Cortes-Rojo C, Manzo-Avalos S, et al. Mitochondrial response to oxidative and nitrosative stress in early stages of diabetes. Mitochondrion. 2013;13(6):835–840. doi: 10.1016/j.mito.2013.05.012. [DOI] [PubMed] [Google Scholar]
- 124.Yasukawa T, Tokunaga E, Ota H, Sugita H, Martyn JA, Kaneki M. S-nitrosylation-dependent inactivation of Akt/protein kinase B in insulin resistance. J Biol Chem. 2005;280(9):7511–7518. doi: 10.1074/jbc.M411871200. [DOI] [PubMed] [Google Scholar]
- 125.Mieyal JJ, Gallogly MM, Qanungo S, Sabens EA, Shelton MD. Molecular mechanisms and clinical implications of reversible protein S-glutathionylation. Antioxid Redox Signal. 2008;10(11):1941–1988. doi: 10.1089/ars.2008.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Sanchez-Gomez FJ, Espinosa-Diez C, Dubey M, Dikshit M, Lamas S. S-glutathionylation: relevance in diabetes and potential role as a biomarker. Biol Chem. 2013;394(10):1263–1280. doi: 10.1515/hsz-2013-0150. [DOI] [PubMed] [Google Scholar]
- 127.Sampathkumar R, Balasubramanyam M, Sudarslal S, Rema M, Mohan V, Balaram P. Increased glutathionylated hemoglobin (HbSSG) in type 2 diabetes subjects with microangiopathy. Clin Biochem. 2005;38(10):892–899. doi: 10.1016/j.clinbiochem.2005.06.009. [DOI] [PubMed] [Google Scholar]
- 128.Wagner GR, Payne RM. Widespread and enzyme-independent Nε-acetylation and Nε-succinylation of proteins in the chemical conditions of the mitochondrial matrix. J Biol Chem. 2013;288(40):29036–29045. doi: 10.1074/jbc.M113.486753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Baeza J, Smallegan MJ, Denu JM. Site-specific reactivity of nonenzymatic lysine acetylation. ACS Chem Biol. 2015;10(1):122–128. doi: 10.1021/cb500848p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Prins ML. Cerebral metabolic adaptation and ketone metabolism after brain injury. J Cereb Blood Flow Metab. 2008;28(1):1–16. doi: 10.1038/sj.jcbfm.9600543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.White H, Venkatesh B. Clinical review: ketones and brain injury. Crit Care. 2011;15(2):219. doi: 10.1186/cc10020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Cotter DG, Schugar RC, Crawford PA. Ketone body metabolism and cardiovascular disease. Am J Physiol Heart Circ Physiol. 2013;304(8):H1060–H1076. doi: 10.1152/ajpheart.00646.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Fritz KS, Galligan JJ, Hirschey MD, Verdin E, Petersen DR. Mitochondrial acetylome analysis in a mouse model of alcohol-induced liver injury utilizing SIRT3 knockout mice. J Proteome Res. 2012;11(3):1633–1643. doi: 10.1021/pr2008384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Iyer A, Fairlie DP, Brown L. Lysine acetylation in obesity, diabetes and metabolic disease. Immunol Cell Biol. 2012;90(1):39–46. doi: 10.1038/icb.2011.99. [DOI] [PubMed] [Google Scholar]
- 135.Kosanam H, Thai K, Zhang Y, et al. Diabetes induces lysine acetylation of intermediary metabolism enzymes in the kidney. Diabetes. 2014;63(7):2432–2439. doi: 10.2337/db12-1770. [DOI] [PubMed] [Google Scholar]
- 136.Fernandes J, Weddle A, Kinter CS, et al. Lysine acetylation activates mitochondrial aconitase in the heart. Biochemistry. 2015;54(25):4008–4018. doi: 10.1021/acs.biochem.5b00375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Muoio DM. Metabolic inflexibility: when mitochondrial indecision leads to metabolic gridlock. Cell. 2014;159(6):1253–1262. doi: 10.1016/j.cell.2014.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Wagner GR, Hirschey MD. Nonenzymatic protein acylation as a carbon stress regulated by sirtuin deacylases. Mol Cell. 2014;54(1):5–16. doi: 10.1016/j.molcel.2014.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Pham TX, Lee J. Dietary regulation of histone acetylases and deacetylases for the prevention of metabolic diseases. Nutrients. 2012;4(12):1868–1886. doi: 10.3390/nu4121868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Paik WK, Pearson D, Lee HW, Kim S. Nonenzymatic acetylation of histones with acetyl-CoA. Biochim Biophys Acta. 1970;213(2):513–522. doi: 10.1016/0005-2787(70)90058-4. [DOI] [PubMed] [Google Scholar]
- 141.Ramponi G, Manao G, Camici G. Nonenzymatic acetylation of histones with acetyl phosphate and acetyl adenylate. Biochemistry. 1975;14(12):2681–2685. doi: 10.1021/bi00683a018. [DOI] [PubMed] [Google Scholar]
- 142.Wang Y. Molecular links between caloric restriction and Sir2/SIRT1 activation. Diabetes Metab J. 2014;38(5):321–329. doi: 10.4093/dmj.2014.38.5.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Ramis MR, Esteban S, Miralles A, Tan DX, Reiter RJ. Caloric restriction, resveratrol and melatonin: role of SIRT1 and implications for aging and related-diseases. Mech Ageing Dev. 2015;146–148C:28–41. doi: 10.1016/j.mad.2015.03.008. [DOI] [PubMed] [Google Scholar]
- 144.Kincaid B, Bossy-Wetzel E. Forever young: SIRT3 a shield against mitochondrial meltdown, aging, and neurodegeneration. Front Aging Neurosci. 2013;5:48. doi: 10.3389/fnagi.2013.00048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Verdin E, Ott M. 50 years of protein acetylation: from gene regulation to epigenetics, metabolism and beyond. Nat Rev Mol Cell Biol. 2015;16(4):258–264. doi: 10.1038/nrm3931. [DOI] [PubMed] [Google Scholar]
- 146.Seely L, Olefsky JM. Potential cellular and genetic mechanisms for insulin resistance in the common disorders of diabetes and obesity. In: Moller DE, editor. Insulin resistance. New York: Wiley; 1993. pp. 187–252. [Google Scholar]
- 147.Lieberman M, Marks AD. Marks’ Basic Medical Biochemistry: A Clinical Approach. 4th ed. Philadelphia: Wolters Kluwer/Lippincott Williams & Wilkins; 2013. [Google Scholar]
- 148.Vazquez EJ, Berthiaume JM, Kamath V, et al. Mitochondrial complex I defect and increased fatty acid oxidation enhance protein lysine acetylation in the diabetic heart. Cardiovasc Res. 2015;107(4):453–465. doi: 10.1093/cvr/cvv183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Fritz KS, Green MF, Petersen DR, Hirschey MD. Ethanol metabolism modifies hepatic protein acylation in mice. PLoS One. 2013;8(9):e75868. doi: 10.1371/journal.pone.0075868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Huynh FK, Hershberger KA, Hirschey MD. Targeting sirtuins for the treatment of diabetes. Diabetes Manag (Lond) 2013;3(3):245–257. doi: 10.2217/dmt.13.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Jing E, Emanuelli B, Hirschey MD, et al. Sirtuin-3 (Sirt3) regulates skeletal muscle metabolism and insulin signaling via altered mitochondrial oxidation and reactive oxygen species production. Proc Natl Acad Sci U S A. 2011;108(35):14608–14613. doi: 10.1073/pnas.1111308108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Morris BJ. Seven sirtuins for seven deadly diseases of aging. Free Radic Biol Med. 2013;56:133–171. doi: 10.1016/j.freeradbiomed.2012.10.525. [DOI] [PubMed] [Google Scholar]
- 153.Nikiforov A, Kulikova V, Ziegler M. The human NAD metabolome: functions, metabolism and compartmentalization. Crit Rev Biochem Mol Biol. 2015;50(4):284–297. doi: 10.3109/10409238.2015.1028612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Turkmen K, Karagoz A, Kucuk A. Sirtuins as novel players in the pathogenesis of diabetes mellitus. World J Diabetes. 2014;5(6):894–900. doi: 10.4239/wjd.v5.i6.894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Yang T, Sauve AA. NAD metabolism and sirtuins: metabolic regulation of protein deacetylation in stress and toxicity. AAPS J. 2006;8(4):E632–E643. doi: 10.1208/aapsj080472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Vedantham S, Thiagarajan D, Ananthakrishnan R, et al. Aldose reductase drives hyperacetylation of Egr-1 in hyperglycemia and consequent upregulation of proinflammatory and prothrombotic signals. Diabetes. 2014;63(2):761–774. doi: 10.2337/db13-0032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Nagai R, Brock JW, Blatnik M, et al. Succination of protein thiols during adipocyte maturation: a biomarker of mitochondrial stress. J Biol Chem. 2007;282(47):34219–34228. doi: 10.1074/jbc.M703551200. [DOI] [PubMed] [Google Scholar]
- 158.Frizzell N, Thomas SA, Carson JA, Baynes JW. Mitochondrial stress causes increased succination of proteins in adipocytes in response to glucotoxicity. Biochem J. 2012;445(2):247–254. doi: 10.1042/BJ20112142. [DOI] [PubMed] [Google Scholar]
- 159.Merkley ED, Metz TO, Smith RD, Baynes JW, Frizzell N. The succinated proteome. Mass Spectrom Rev. 2014;33(2):98–109. doi: 10.1002/mas.21382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Yang M, Ternette N, Su H, et al. The succinated proteome of FH-mutant tumours. Metabolites. 2014;4(3):640–654. doi: 10.3390/metabo4030640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Blatnik M, Thorpe SR, Baynes JW. Succination of proteins by fumarate: mechanism of inactivation of glyceraldehyde-3-phosphate dehydrogenase in diabetes. Ann N Y Acad Sci. 2008;1126:272–275. doi: 10.1196/annals.1433.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Frizzell N, Lima M, Baynes JW. Succination of proteins in diabetes. Free Radic Res. 2011;45(1):101–109. doi: 10.3109/10715762.2010.524643. [DOI] [PubMed] [Google Scholar]
- 163.Winterbourn CC, Hampton MB. Thiol chemistry and specificity in redox signaling. Free Radic Biol Med. 2008;45(5):549–561. doi: 10.1016/j.freeradbiomed.2008.05.004. [DOI] [PubMed] [Google Scholar]
- 164.Ying J, Clavreul N, Sethuraman M, Adachi T, Cohen RA. Thiol oxidation in signaling and response to stress: detection and quantification of physiological and pathophysiological thiol modifications. Free Radic Biol Med. 2007;43(8):1099–1108. doi: 10.1016/j.freeradbiomed.2007.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Forman HJ, Fukuto JM, Torres M. Redox signaling: thiol chemistry defines which reactive oxygen and nitrogen species can act as second messengers. Am J Physiol Cell Physiol. 2004;287(2):C246–C256. doi: 10.1152/ajpcell.00516.2003. [DOI] [PubMed] [Google Scholar]
- 166.Fomenko DE, Marino SM, Gladyshev VN. Functional diversity of cysteine residues in proteins and unique features of catalytic redox-active cysteines in thiol oxidoreductases. Mol Cells. 2008;26(3):228–235. [PMC free article] [PubMed] [Google Scholar]
- 167.Biswas S, Chida AS, Rahman I. Redox modifications of protein-thiols: emerging roles in cell signaling. Biochem Pharmacol. 2006;71(5):551–564. doi: 10.1016/j.bcp.2005.10.044. [DOI] [PubMed] [Google Scholar]
- 168.Feligioni M, Brambilla E, Camassa A, et al. Crosstalk between JNK and SUMO signaling pathways: deSUMOylation is protective against H2O2-induced cell injury. PLoS One. 2011;6(12):e28185. doi: 10.1371/journal.pone.0028185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Feligioni M, Nistico R. SUMO: a (oxidative) stressed protein. Neuromolecular Med. 2013;15(4):707–719. doi: 10.1007/s12017-013-8266-6. [DOI] [PubMed] [Google Scholar]
- 170.Heo KS, Chang E, Le NT, et al. De-SUMOylation enzyme of sentrin/SUMO-specific protease 2 regulates disturbed flow-induced SUMOylation of ERK5 and p53 that leads to endothelial dysfunction and atherosclerosis. Circ Res. 2013;112(6):911–923. doi: 10.1161/CIRCRESAHA.111.300179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Huang CJ, Wu D, Khan FA, Huo LJ. DeSUMOylation: an important therapeutic target and protein regulatory event. DNA Cell Biol. 2015;34(11):652–660. doi: 10.1089/dna.2015.2933. [DOI] [PubMed] [Google Scholar]
- 172.Zhao J. Sumoylation regulates diverse biological processes. Cell Mol Life Sci. 2007;64(23):3017–3033. doi: 10.1007/s00018-007-7137-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Guo D, Li M, Zhang Y, et al. A functional variant of SUMO4, a new I kappa B alpha modifier, is associated with type 1 diabetes. Nat Genet. 2004;36(8):837–841. doi: 10.1038/ng1391. [DOI] [PubMed] [Google Scholar]
- 174.Guo D, Han J, Adam BL, et al. Proteomic analysis of SUMO4 substrates in HEK293 cells under serum starvation-induced stress. Biochem Biophys Res Commun. 2005;337(4):1308–1318. doi: 10.1016/j.bbrc.2005.09.191. [DOI] [PubMed] [Google Scholar]
- 175.Li M, Guo D, Isales CM, et al. SUMO wrestling with type 1 diabetes. J Mol Med (Berl) 2005;83(7):504–513. doi: 10.1007/s00109-005-0645-5. [DOI] [PubMed] [Google Scholar]
- 176.Bohren KM, Nadkarni V, Song JH, Gabbay KH, Owerbach D. A M55V polymorphism in a novel SUMO gene (SUMO-4) differentially activates heat shock transcription factors and is associated with susceptibility to type I diabetes mellitus. J Biol Chem. 2004;279(26):27233–27238. doi: 10.1074/jbc.M402273200. [DOI] [PubMed] [Google Scholar]
- 177.Giorgino F, de Robertis O, Laviola L, et al. The sentrin-conjugating enzyme mUbc9 interacts with GLUT4 and GLUT1 glucose transporters and regulates transporter levels in skeletal muscle cells. Proc Natl Acad Sci U S A. 2000;97(3):1125–1130. doi: 10.1073/pnas.97.3.1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Dadke S, Cotteret S, Yip SC, et al. Regulation of protein tyrosine phosphatase 1B by sumoylation. Nat Cell Biol. 2007;9(1):80–85. doi: 10.1038/ncb1522. [DOI] [PubMed] [Google Scholar]
- 179.Wang CY, She JX. SUMO4 and its role in type 1 diabetes pathogenesis. Diabetes Metab Res Rev. 2008;24(2):93–102. doi: 10.1002/dmrr.797. [DOI] [PubMed] [Google Scholar]
- 180.Lin HY, Wang CL, Hsiao PJ, et al. SUMO4 M55V variant is associated with diabetic nephropathy in type 2 diabetes. Diabetes. 2007;56(4):1177–1180. doi: 10.2337/db06-1283. [DOI] [PubMed] [Google Scholar]
- 181.Zhou X, Gao C, Huang W, et al. High glucose induces sumoylation of Smad4 via SUMO2/3 in mesangial cells. Biomed Res Int. 2014;2014:782625. doi: 10.1155/2014/782625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Le NT, Corsetti JP, Dehoff-Sparks JL, Sparks CE, Fujiwara K, Abe J. Reactive oxygen species, SUMOylation, and endothelial inflammation. Int J Inflam. 2012;2012:678190. doi: 10.1155/2012/678190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Mueller-Dieckmann C, Kernstock S, Lisurek M, et al. The structure of human ADP-ribosylhydrolase 3 (ARH3) provides insights into the reversibility of protein ADP-ribosylation. Proc Natl Acad Sci U S A. 2006;103(41):15026–15031. doi: 10.1073/pnas.0606762103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Hassa PO, Haenni SS, Elser M, Hottiger MO. Nuclear ADP-ribosylation reactions in mammalian cells: where are we today and where are we going? Microbiol Mol Biol Rev. 2006;70(3):789–829. doi: 10.1128/MMBR.00040-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Butepage M, Eckei L, Verheugd P, Luscher B. Intracellular mono-ADP-ribosylation in signaling and disease. Cells. 2015;4(4):569–595. doi: 10.3390/cells4040569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Horvath EM, Magenheim R, Kugler E, et al. Nitrative stress and poly(ADP-ribose) polymerase activation in healthy and gestational diabetic pregnancies. Diabetologia. 2009;52(9):1935–1943. doi: 10.1007/s00125-009-1435-3. [DOI] [PubMed] [Google Scholar]
- 187.Du X, Matsumura T, Edelstein D, et al. Inhibition of GAPDH activity by poly(ADP-ribose) polymerase activates three major pathways of hyperglycemic damage in endothelial cells. J Clin Invest. 2003;112(7):1049–1057. doi: 10.1172/JCI18127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Masutani M, Suzuki H, Kamada N, et al. Poly(ADP-ribose) polymerase gene disruption conferred mice resistant to streptozotocin-induced diabetes. Proc Natl Acad Sci U S A. 1999;96(5):2301–2304. doi: 10.1073/pnas.96.5.2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Pieper AA, Brat DJ, Krug DK, et al. Poly(ADP-ribose) polymerase-deficient mice are protected from streptozotocin-induced diabetes. Proc Natl Acad Sci U S A. 1999;96(6):3059–3064. doi: 10.1073/pnas.96.6.3059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Virag L, Szabo C. The therapeutic potential of poly(ADP-ribose) polymerase inhibitors. Pharmacol Rev. 2002;54(3):375–429. doi: 10.1124/pr.54.3.375. [DOI] [PubMed] [Google Scholar]
- 191.Daniels CM, Ong SE, Leung AK. The promise of proteomics for the study of ADP-ribosylation. Mol Cell. 2015;58(6):911–924. doi: 10.1016/j.molcel.2015.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Brownlee M. Biochemistry and molecular cell biology of diabetic complications. Nature. 2001;414(6865):813–820. doi: 10.1038/414813a. [DOI] [PubMed] [Google Scholar]
- 193.Brownlee M. The pathobiology of diabetic complications: a unifying mechanism. Diabetes. 2005;54(6):1615–1625. doi: 10.2337/diabetes.54.6.1615. [DOI] [PubMed] [Google Scholar]