Abstract
To determine the occurrence of carbapenem-resistant Acinetobacter baumannii in fish fished from the Mediterranean Sea near the Bejaia coast (Algeria), we studied 300 gills and gut samples that had been randomly and prospectively collected during 1 year. After screening on selective agar media, using PCR arrays and whole-genome sequencing, we identified for the first time two OXA-23-producing A. baumannii strains belonging to the widespread sequence type 2 (ST2)/international clone II and harboring aminoglycoside-modifying enzymes [aac(6′)-Ib and aac(3′)-I genes].
TEXT
Acinetobacter baumannii is an opportunistic aerobic nonfermentative Gram-negative rod found ubiquitously in the environment (1). This bacterium has emerged as an important cause of nosocomial infections, most notably ventilator-associated pneumonia and bacteremia associated with high mortality, urinary tract infections, and endocarditis (2). Moreover, it is highly capable of developing resistance to antimicrobial agents (1). Over the last 10 years, an increase in carbapenem-resistant A. baumannii strains has been observed worldwide; in particular, we noted a high prevalence in different countries in the south of Europe (3). The most common mechanism of carbapenem resistance in Acinetobacter species is the production of acquired carbapenem-hydrolyzing OXA-type class D β-lactamases (4). They are represented worldwide by six gene clusters: intrinsic chromosomal OXA-51-like, of which there are over 70 variants, and the acquired OXA-23-like, OXA-24/40-like, OXA-58-like, OXA-143-like, and OXA-235-like β-lactamases (5–7). Worldwide dissemination of OXA-23-producing A. baumannii is now well established and, notably, has been observed among Algerian hospitals in recent years (8–10). While A. baumannii is isolated from patients and hospital environmental sources during outbreaks, the reservoir outside the hospital is not well delineated. Several investigators have suspected that the survival of A. baumannii in the environment (in particular in water) could contribute to the transmission of the organism during outbreaks (3). Moreover, recent reports have also described the presence of carbapenem-resistant Acinetobacter spp. from animals. The OXA-23 carbapenemase has been found in Acinetobacter spp. from cattle, horses, and cats (11–13). NDM-1 has also been reported in Acinetobacter spp. from food animals (chicken and pig farms) in China (14, 15). However, knowledge about carbapenemase-producing Acinetobacter spp. of animal origin remains very limited, making it difficult to assess its impact on public health.
Between 1 March 2012 and 28 February 2013, we randomly and prospectively screened a total of 300 samples from different fish fished in the Mediterranean Sea (2 km from the Bejaia coast, Algeria). Sampling was carried out from Sardina pilchardus (n = 62), Engraulis encrasicolus (n = 38), Trachurus trachurus (n = 45), Sarpa salpa (n = 60), Pagellus acarne (n = 55), and Boops boops (n = 40). The gills and gut of each fish sample were collected by opening the gut using a sterile scalpel following washing of the gut surface with sterile saline. Samples were placed in 1 ml sterile 0.9% saline and then vortexed. Cultures were inoculated by streaking 100 μl of the suspensions onto MacConkey agar plates supplemented with 2 μg/ml of ceftazidime and incubated 24 h at 37°C under aerobic conditions. One colony per sample was retained for further investigation. Bacterial identification was carried out using matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) (Vitek MS; bioMérieux). Susceptibility testing was performed by the disk diffusion procedure (Bio-Rad) and E-tests (bioMérieux) according to the recommendations of the Antibiotic Committee of the French Society for Microbiology (http://www.sfm-microbiologie.org/page/page/showpage/page_id/90.html). Isolates were screened for carbapenemase production using the modified Hodge test (MHT) (16) and by the imipenem-EDTA method (17). Multiplex PCR detection and sequencing of genes that encode carbapenem-hydrolyzing class D β-lactamases were used (18). The presence of genes encoding the aminoglycoside-modifying enzymes (AMEs) was also detected using PCR (19). Finally, the genetic relationship was investigated by repetitive sequence-based PCR (rep-PCR) (using the DiversiLab system) and multilocus sequence typing (MLST) (20). Plasmid electroporation assays were performed with A. baumannii ATCC 19606. To identify the clonal lineage of the OXA-23-producing A. baumannii isolates, we used a previously described PCR (21). Genomic DNA of A. baumannii IM2 was sequenced using a NextSeq 500 Illumina platform by Helixio (St-Beauzire, France).
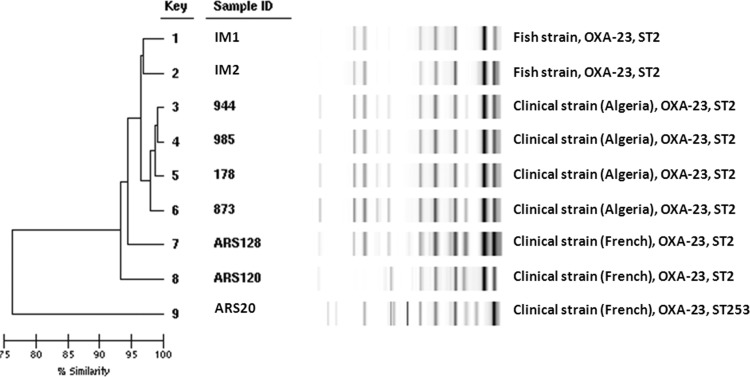
Of the 300 samples analyzed, A. baumannii isolates IM1 and IM2 (0.7%) were recovered from two fish (Pagellus acarne). The two isolates were resistant to almost all the antibiotics tested, including carbapenems. They remained susceptible only to amikacin and netilmicin (Table 1). The phenotypic assays showed that the two isolates were positive according to the MHT but the activity of β-lactamases was not inhibited by EDTA. PCR detection showed that the two A. baumannii strains harbored the naturally occurring blaOXA-51-like gene and the acquired OXA-carbapenemase blaOXA-23-like gene. Sequencing confirmed that the isolates harbored the β-lactamase OXA-23. Moreover, we detected the presence of aac(6′)-Ib and aac(3′)-I in the two isolates. These two isolates and different human clinical strains isolated in France (Languedoc-Roussillon, southern France) and Algeria (Annaba, eastern Algerian) shared the same genotype (Fig. 1). Assays to transfer plasmids by electroporation were unsuccessful. MLST assigned the isolates to sequence type 2 (ST2). Multiplex PCR to identify clonal lineages was positive for group 1, showing that the strains belonged to the widespread ST2/international clone II. Whole-genome sequencing of IM2 indicated that the transposon Tn2006 was the vehicle of the resistance gene blaOXA-23. The 5,292 bp of this transposon were found in the genomic DNA of IM2 (in position 2759078, after a hypothetical-protein-coding gene), confirming the chromosomal insertion of Tn2006 (GenBank accession no. KU168371).
TABLE 1.
Characteristics of carbapenem-resistant A. baumanii isolated from wild P. acarne fished from the Mediterranean Sea (2 km from Bejaia coast, Algeria)a
| Strain | Isolation date (mo/day/yr) | Organ | Resistance phenotype antibiotics (MIC, μg/ml)b |
|---|---|---|---|
| IM1 | 11/04/2012 | Gills | CTX (32), CAZ (32), FEP (32), IMP (>32), MEM (32), DOR (>32), AMK (2), GEN (16), KAN(>32), NET (1), OFX (>32), CIP (>32), SXT (8) |
| IM2 | 04/14/2012 | Gut | CTX (32), CAZ (32), FEP (32), IMP (>32), MEM (32), DOR (>32), AMK (2), GEN (16), KAN(>32), NET (1), OFX (>32), CIP (>32), SXT (8) |
Both strain were ST2, harbored Tn2006, contained the β-lactamases OXA-23 and OXA-51, and carried the associated resistance genes aac(6′)-Ib and aac(3′)-I.
CTX, cefotaxime; CAZ, ceftazidime; FEP, cefepime; IMP, imipenem; MEM, meropenem; DOR, doripenem; AMK, amikacin; GEN, gentamicin; KAN, kanamycin; NET, netilmycin; OFX, ofloxacin; CIP, ciprofloxacin; SXT, co-trimoxazole.
FIG 1.
Dendrogram of Rep-PCR and MLST of the two OXA-23-producing Acinetobacter baumannii strains isolated from wild fish in the Mediterranean Sea, 3 representative human clinical strains isolated from a French hospital, and 3 representative human clinical strains isolated from an Algerian hospital. For the purpose of predicting different clones, the top match feature at 95% similarity was used.
This study highlighted for the first time that wild fish in the Mediterranean Sea can be contaminated with carbapenem- and aminoglycoside-resistant A. baumannii isolates belonging to the worldwide clone ST2. The emergence and spread of several outbreak or sporadic A. baumannii strains producing OXA-23-like enzymes have been reported around the world, and these strains were assigned to international clonal lineage I or II (4). Over a long period, the blaOXA-58 carbapenemase gene has been predominant among carbapenem-resistant A. baumannii isolates in various Mediterranean countries. Since 2009, a replacement of blaOXA-58 with blaOXA-23 has been reported, and it became the most prevalent carbapenemase-encoding gene circulating in the Mediterranean region (5). The replacement of OXA-58 by OXA-23 might be explained by the selective advantage associated with the higher carbapenemase activity of OXA-23 (22). Recently, different reports showed the dissemination of multidrug-resistant pathogenic bacteria in food products and in food-producing animals (5, 23, 24). Comparison of human and animal carbapenem-resistant Acinetobacter isolates is important to enhance the knowledge of the potential routes of transfer of these bacteria and resistance genes in different ecosystems. Few studies have been published describing the dissemination of carbapenem-resistant Acinetobacter isolates in food animals and wild animals (11–15). All these points and the clonality between a panel of clinical strains showed the possible exchange place between the A. baumannii populations in infected humans and water. We suggest that these isolates were most likely derived from contamination of the fish from human sewage via river water and a growing amount of waste from land urban, industrial, and agricultural operations that has been discharged untreated into the sea near the coast in regions of the Mediterranean Sea.
In conclusion, our study highlighted the idea that OXA-23-producing A. baumannii may be isolated from wild animals in rare cases. These findings emphasize the ability of these isolates to spread in the environment. More studies should be performed in the future to track the evolution of carbapenem-resistant Acinetobacter isolates and their frequencies in different ecosystems.
Nucleotide sequence accession number.
Sequence information for A. baumannii strain IM2 transposon Tn2006 was deposited in GenBank under accession no. KU168371.
ACKNOWLEDGMENTS
INSERM, U1047, is supported by INSERM and Université de Montpellier.
We have no conflicts of interest to report.
Funding Statement
This research received no specific grant from any funding agency in the public, commercial, or not-for-profit sectors.
REFERENCES
- 1.Dent LL, Marshall DR, Pratap S, Hulette RB. 2010. Multidrug resistant Acinetobacter baumannii: a descriptive study in a city hospital. BMC Infect Dis 10:196. doi: 10.1186/1471-2334-10-196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Garcia-Quintanilla M, Pulido MR, López-Rojas R, Pachón J, McConnell MJ. 2013. Emerging therapies for multidrug resistant Acinetobacter baumannii. Trends Microbiol 21:1157–1163. doi: 10.1016/j.tim.2012.12.002. [DOI] [PubMed] [Google Scholar]
- 3.Kempf M, Rolain JM. 2012. Emergence of resistance to carbapenems in Acinetobacter baumannii in Europe: clinical impact and therapeutic options. Int J Antimicrob Agents 39:105–114. doi: 10.1016/j.ijantimicag.2011.10.004. [DOI] [PubMed] [Google Scholar]
- 4.Zarrilli R, Pournaras S, Giannouli M, Tsakris A. 2013. Global evolution of multidrug-resistant Acinetobacter baumannii clonal lineages. Int J Antimicrob Agents 41:11–19. doi: 10.1016/j.ijantimicag.2012.09.008. [DOI] [PubMed] [Google Scholar]
- 5.Djahmi N, Dunyach-Remy C, Pantel A, Dekhil M, Sotto A, Lavigne JP. 2014. Epidemiology of carbapenemase-producing Enterobacteriaceae and Acinetobacter baumannii in Mediterranean countries. Biomed Res Int 2014:305784. doi: 10.1155/2014/305784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Higgins PG, Poirel L, Lehmann M, Nordmann P, Seifert H. 2009. OXA-143, a novel carbapenem-hydrolyzing class D beta-lactamase in Acinetobacter baumannii. Antimicrob Agents Chemother 53:5035–5038. doi: 10.1128/AAC.00856-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Higgins PG, Pérez-Llarena FJ, Zander E, Fernández A, Bou G, Seifert H. 2013. OXA-235, a novel class D β-lactamase involved in resistance to carbapenems in Acinetobacter baumannii. Antimicrob Agents Chemother 57:2121–2126. doi: 10.1128/AAC.02413-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mugnier P, Poirel L, Naas T, Nordmann P. 2010. Worldwide dissemination of the blaOXA-23 carbapenemase gene of Acinetobacter baumannii. Emerg Infect Dis 16:35–40. doi: 10.3201/eid1601.090852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mesli E, Berrazeg M, Drissi M, Rolain J. 2013. Prevalence of carbapenemase-encoding genes including New Delhi metallo-β-lactamase in Acinetobacter species, Algeria. Int J Infect Dis 17:e739–e743. doi: 10.1016/j.ijid.2013.02.024. [DOI] [PubMed] [Google Scholar]
- 10.Bakour S, Kempf M, Touati A, Ait Ameur A, Haouchine D, Sahli F, Rolain JM. 2012. Carbapenemase-producing Acinetobacter baumannii in two university hospitals in Algeria. J Med Microbiol 61:1341–1343. doi: 10.1099/jmm.0.045807-0. [DOI] [PubMed] [Google Scholar]
- 11.Poirel L, Berçot B, Millemann Y, Bonnin RA, Pannaux G, Nordmann P. 2012. Carbapenemase-producing Acinetobacter spp. in cattle, France. Emerg Infect Dis 18:523–525. doi: 10.3201/eid1803.111330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Smet A, Boyen F, Pasmans F, Pasmans F, Butaye P, Martens A, Nemec A, Deschaght P, Vaneechoutte M, Haesebrouck F. 2012. OXA-23-producing Acinetobacter species from horses: a public health hazard? J Antimicrob Chemother 67:3009–3010. doi: 10.1093/jac/dks311. [DOI] [PubMed] [Google Scholar]
- 13.Pomba C, Endimiani A, Rossano A, Saial D, Couto N, Perreten V. 2014. First report of OXA-23-mediated carbapenem resistance in sequence type 2 multidrug-resistant Acinetobacter baumannii associated with urinary tract infection in a cat. Antimicrob Agents Chemother 58:1267–1268. doi: 10.1128/AAC.02527-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang Y, Wu C, Zhang Q, Qi J, Liu H, Wang Y, He T, Ma L, Lai J, Shen Z, Liu Y, Shen J. 2012. Identification of New Delhi metallo-β-lactamase 1 in Acinetobacter lwoffii of food animal origin. PLoS One 7:e37152. doi: 10.1371/journal.pone.0037152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang WJ, Lu Z, Schwarz S, Zhang RM, Wang XM, Si W, Yu S, Chen L, Liu S. 2013. Complete sequence of the bla(NDM-1)-carrying plasmid pNDM-AB from Acinetobacter baumannii of food animal origin. J Antimicrob Chemother 68:1681–1682. doi: 10.1093/jac/dkt066. [DOI] [PubMed] [Google Scholar]
- 16.Lee K, Kim CK, Yong D, Jeong SH, Yum JH, Seo YH, Docquier JD, Chong Y. 2010. Improved performance of the modified Hodge test with MacConkey agar for screening carbapenemase-producing Gram-negative bacilli. J Microbiol Methods 83:149–152. doi: 10.1016/j.mimet.2010.08.010. [DOI] [PubMed] [Google Scholar]
- 17.Lee K, Chong YÃ, Shin HB, Kim YA, Yong D, Yum JH. 2001. Modified Hodge and EDTA-disk synergy tests to screen metallo-β-lactamase-producing strains of Pseudomonas and Acinetobacter species. Clin Microbiol Infect 7:88–91. doi: 10.1046/j.1469-0691.2001.00204.x. [DOI] [PubMed] [Google Scholar]
- 18.Woodford N, Ellington MJ, Coelho JM, Turton JF, Ward ME, Brown S, Amyes SG, Livermore DM. 2006. Multiplex PCR for genes encoding prevalent OXA carbapenemases in Acinetobacter spp. Int J Antimicrob Agents 27:351–353. doi: 10.1016/j.ijantimicag.2006.01.004. [DOI] [PubMed] [Google Scholar]
- 19.Yin XL, Hou TW, Xu SB, Ma CQ, Yao ZY, Li W, Wei L. 2008. Detection of drug resistance-associated genes of multidrug-resistant Acinetobacter baumannii. Microb Drug Resist 14:145–150. doi: 10.1089/mdr.2008.0799. [DOI] [PubMed] [Google Scholar]
- 20.Diancourt L, Passet V, Nemec A, Dijkshoorn L, Brisse S. 2010. The population structure of Acinetobacter baumannii: expanding multiresistant clones from an ancestral susceptible genetic pool. PLoS One 5:e10034. doi: 10.1371/journal.pone.0010034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Turton JF, Gabriel SN, Valderrey C, Kaufmann ME, Pitt TL. 2007. Use of sequence-based typing and multiplex PCR to identify clonal lineages of outbreak strains of Acinetobacter baumannii. Clin Microbiol Infect 13:807–815. doi: 10.1111/j.1469-0691.2007.01759.x. [DOI] [PubMed] [Google Scholar]
- 22.Espinal P, Macià MD, Roca I, Gato E, Ruíz E, Fernández-Cuenca F, Oliver A, Rodríguez-Baño J, Bou G, Tomás M, Vila J. 2013. First report of an OXA-23 carbapenemase-producing Acinetobacter baumannii clinical isolate related to Tn2006 in Spain. Antimicrob Agents Chemother 57:589–591. doi: 10.1128/AAC.01157-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Woodford N, Wareham DW, Guerra B, Teale C. 2014. Carbapenemase-producing Enterobacteriaceae and non-Enterobacteriaceae from animals and the environment: an emerging public health risk of our own making? J Antimicrob Chemother 69:287–291. doi: 10.1093/jac/dkt392. [DOI] [PubMed] [Google Scholar]
- 24.Jouini A, Slama Ben K, Klibi N, Sallem RB, Estepa V, Vinué L, Sáenz Y, Ruiz-Larrea F, Boudabous A, Torres C. 2013. Lineages and virulence gene content among extended-spectrum β-lactamase-producing Escherichia coli strains of food origin in Tunisia. J Food Prot 76:323–327. doi: 10.4315/0362-028X.JFP-12-251. [DOI] [PubMed] [Google Scholar]