Abstract
In Enterobacter cloacae, the genetic lesions associated with derepression of the AmpC β-lactamase include diverse single nucleotide polymorphisms (SNPs) and/or indels in the ampD and ampR genes and SNPs in ampC, while diverse SNPs in the promoter region or SNPs/indels within the coding sequence of outer membrane proteins have been described to alter porin production leading to carbapenem resistance. We sought to define the underlying mechanisms conferring cephalosporin and carbapenem resistance in a collection of E. cloacae isolates with unusually high carbapenem resistance and no known carbapenemase and, in contrast to many previous studies, considered the SNPs we detected in relation to the multilocus sequence type (MLST)-based phylogeny of our collection. Whole-genome sequencing was applied on the most resistant isolates to seek novel carbapenemases, expression of ampC was measured by reverse transcriptase PCR, and porin translation was detected by SDS-PAGE. SNPs occurring in ampC, ampR, ompF, and ompC genes (and their promoter regions) were mostly phylogenetic variations, relating to the isolates' sequence types, whereas nonsynonymous SNPs in ampD were associated with derepression of AmpC and cephalosporin resistance. The additional loss of porins resulted in high-level carbapenem resistance, underlining the clinical importance of chromosomal mutations among carbapenem-resistant E. cloacae.
INTRODUCTION
Enterobacter spp. can cause a wide variety of nosocomial infections, including invasive and device-associated diseases (1). Strains resistant to multiple classes of antimicrobials have caused numerous outbreaks in hospitals and intensive care units worldwide (2, 3). The recent emergence of resistance to carbapenems threatens our last good therapeutic option for many infections, including those caused by Enterobacter spp., and constitutes a global public health concern (4).
Although carbapenem resistance due to the production of acquired carbapenemase genes is increasingly reported among members of Enterobacteriaceae, these mechanisms are still less commonly observed in Enterobacter spp. than in Klebsiella spp. or Escherichia coli (5, 6). An alternative carbapenem resistance strategy for these species is variously to combine extended-spectrum β-lactamases (ESBLs), increased efflux, porin alteration, and a strongly expressed (derepressed) endogenous AmpC enzyme (7–9). The genetic lesions associated with the derepression of the AmpC β-lactamase have been described to occur as diverse single nucleotide polymorphisms (SNPs) and/or indels in the N-acetylmuramyl-l-alanine amidase and in transcriptional regulator gene ampD and ampR, respectively (10–13), while outer membrane protein (porin) production is altered by diverse SNPs in the promoter region or by SNPs/indels within the coding sequence (8).
In Enterobacter cloacae, a wide range of carbapenem MICs have been attributed to these alternative, noncarbapenemase-mediated mechanisms, but hitherto-detected SNPs have not been related to the isolates' phylogeny. Furthermore, the exact interplay and contributions of AmpC and porin loss to high-level carbapenem resistance remain unclear. To understand these mechanisms better, we investigated the presence of novel carbapenemases among a collection of carbapenem-resistant E. cloacae isolates from the United Kingdom and Switzerland, detected lesions in ampC, ampR, ampD, ompF, and ompC and their respective promoter regions among the United Kingdom isolates, and considered SNPs that we detected in relation to the multilocus sequence type (MLST)-based phylogeny of our collection. Additionally, the relationship between levels of AmpC transcription, production of the porins, and carbapenem MICs was investigated.
MATERIALS AND METHODS
Bacterial isolates and antimicrobial susceptibility testing.
Thirty-three E. cloacae isolates were included, consisting of 25 ertapenem-resistant clinical isolates (MICs, ≥2 mg/liter) from 25 patients. Sixteen of these were submitted from 15 United Kingdom laboratories to Public Health England's Antimicrobial Resistance and Healthcare Associated Infections (AMRHAI) Reference Unit between October 2012 and October 2013, and nine were from different patients in Basel, Switzerland (CH isolates 4, 6 to 8, 10, 14, 15, 17, 18). Of the remaining eight isolates, seven (S isolates S1, S8 to S12, and wild-type strain E. cloacae NCTC 13405) that were susceptible to carbapenems and most cephalosporins (except cefoxitin) were used as comparators for outer membrane protein (OMP) extraction experiments and PCR controls, whereas NCTC 13406, the AmpC-derepressed mutant of NCTC 13405, was used as a control for AmpC activity.
MICs were determined by British Society for Antimicrobial Chemotherapy (BSAC) agar dilution methodology (14) or by Etest and were interpreted according to BSAC (15) and EUCAST guidelines. All isolates were tested for cefotaxime/cloxacillin synergy to confirm the role of AmpC in their resistance phenotype.
Strain typing.
Pulsed-field gel electrophoresis (PFGE) of XbaI-digested genomic DNA was performed on all isolates using the following conditions: 6 V cm−1, 12°C, 1.2% (wt/vol) agarose, and 30 h with ramping times of 5 to 35 s. Restriction patterns were analyzed, and a dendrogram was generated using BioNumerics v. 6.1 software (Applied Maths, Kortrijk, Belgium), as described previously (16), using a 1.3% tolerance in band position. MLST was undertaken as described by Miyoshi-Akiyama et al. (17), and sequence types (STs) were assigned using the PubMLST website (http://pubmlst.org/perl/bigsdb/bigsdb.pl?db=pubmlst_ecloacae_seqdef&page=sequenceQuery).
Detection of β-lactamases by PCR.
PCR from boiled lysates was used to detect genes encoding TEM, SHV, and CTX-M β-lactamases and plasmidic ampC genes of the ACC, CIT, DHA, ENT/EBC, FOX, and MOX groups (18–21). A carbapenemase screening test (Rapid CARB; Rosco Diagnostica, Taastrup, Denmark) was performed on isolates with meropenem MICs of >0.125 mg/liter. The genes blaKPC, blaNDM, blaVIM, blaIMP, blaIMI, blaOXA-48, blaGES, and blaFRI-1 were also sought via PCR (22–24).
Detection of sequence alterations.
The United Kingdom isolates plus the susceptible controls (NCTC strains and S strains) were subject to DNA sequencing of the ampC and ampR coding and intergenic regions as well as the ampD, ompC, and ompF open reading frames and their promoter regions using the primers listed in Table S1 in the supplemental material. DNA sequences of the amplicons were determined as described previously (25); they were compared with our reference strain NCTC 13405, with each other, and with known wild-type sequences (ampC, GenBank accession number X07274.1; ampD, GenBank accession number Z14003; and ampR, GenBank accession numbers X04730 and M27222.1) using BioNumerics v. 6.1 software (Applied Maths) and BioEdit (Ibis Biosciences, Carlsbad, CA, USA) to facilitate comparison with previous studies. The ampD sequences and concatenated ampC, ampR, and ompF (with promoter regions) from these isolates were aligned, clustered, and compared using the unweighted pair group method with arithmetic mean (UPGMA) to generate a phylogenetic tree (MEGA6; megasoftware.net) (26). The sequences of ompC were not included in the generation of the phylogenetic tree due to six isolates having large disruptions/deletions (see Results) within the coding sequence.
Whole-genome sequencing to seek novel carbapenemases.
The whole-genome sequences (WGS) of the two most resistant isolates (isolates 2 and 6) with meropenem MICs of ≥16 mg/liter were determined on a HiSeq Illumina instrument (San Diego, CA, USA) to over 30× coverage using the Nextera sample preparation method. Reads were assembled into contigs using VelvetOptimiser (http://bioinformatics.net.au/software.velvetoptimiser.shtml) with k-mer values from 47 to 71. Contigs were translated into six different frames and were checked by BLAST for any hit with known β-lactamase sequences. Predicted proteins from sequenced genomes were also scanned by HMMER v. 3 using hidden Markov models (HMMs) that were built with hmmbuild using major carbapenemase sequence alignments.
Analysis of ampC expression.
Samples (0.5 ml) of mid-log-phase cultures were taken and treated with RNAprotect reagent (Qiagen, Crawley, United Kingdom). RNA was extracted (Total RNA purification, 96-well kit; Norgen Biotek, Canada), and 35 μl of the eluate was treated with DNase I (New England BioLabs, Hitchin, United Kingdom). Reverse transcriptase PCR (RT-PCR) was performed using the TaqMan RNA-to-CT 1-step kit and an Applied Biosystems 7500 instrument (Life Technologies Ltd., Paisley, United Kingdom) as described previously (8). Transcript measurement was carried out in triplicate, and relative quantification of target genes using rspL as a reference (see Table S1 in the supplemental material) were calculated with the 2−ΔΔCT method as described previously (8, 27).
Examination of porin expression.
Outer membrane proteins were extracted according to the rapid procedure of Carlone et al. from cells grown in nutrient broth instead of LB (28, 29). Briefly, cells were lysed with a FastPrep homogenizer (MP Biosciences, Derby, United Kingdom) and glass beads (Sigma, Dorset, United Kingdom) for four cycles of 50 s at 4 m/s. After washing steps with 10 mM HEPES, the membrane pellets were suspended in 50 μl of 10 mM HEPES and stored at −70°C. This procedure routinely yields preparations containing 2 to 4 mg of protein per milliliter. Before loading the proteins on the gel, they were precipitated with acetone and resuspended in 40 μl of solubilization buffer. Five microliters of protein-buffer solution was separated on a 12% precast SDS-PAGE gel (Bio-Rad, Herts, United Kingdom).
RT-PCR was performed for each isolate to determine levels of ompF and ompC transcription as described above.
Nucleotide sequence accession numbers.
The nucleotide sequence data in this study will appear in the EMBL/GenBank/DDBJ nucleotide sequence databases under accession numbers KT780418 for ampC, KT780419 for ampR, KT780420 for ampD, KT780421 for ompF, and KT780422 for ompC from E. cloacae NCTC 13405.
RESULTS
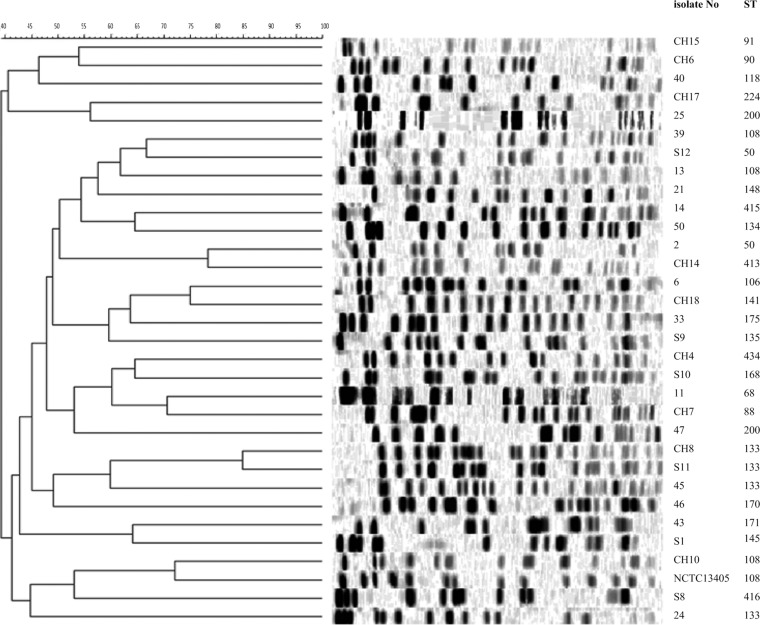
The 33 E. cloacae isolates represented 21 known and three novel STs (ST415, ST416, and ST434) and were genetically diverse by PFGE (Fig. 1).
FIG 1.
PFGE patterns of E. cloacae isolates and the corresponding STs.
Of the 25 ertapenem-resistant isolates (MIC range of 2 to >16 mg/liter and median of 16 mg/liter), 15 and 9 were also nonsusceptible to meropenem and imipenem, respectively (meropenem MIC range of <0.5 to ≥32 mg/liter and median of 8 mg/liter; imipenem MIC range of 0.5 to ≥32 mg/liter and median of 2 mg/liter); they were all resistant to third-generation cephalosporins. Nonsusceptibility to the fourth-generation cephalosporin cefepime was observed in 20 isolates (Tables 1 and 2). Despite their carbapenem resistance, 24 isolates were negative in the Rapid CARB test for carbapenemase activity, and the test for one isolate was uninterpretable. None of these 25 isolates produced a carbapenemase, and none had any of the ESBLs or plasmid-borne AmpC-type β-lactamases sought. WGS of the two isolates that were most resistant to meropenem (isolates 2 and 6) did not detect an acquired carbapenemase.
TABLE 1.
Carbapenem distribution among E. cloacae isolates with ertapenem MICs of ≥2 mg/litera
| Carbapenem | No. of isolates according to MIC (mg/liter) |
|||||||
|---|---|---|---|---|---|---|---|---|
| ≤0.25 | 0.5 | 1 | 2 | 4 | 8 | 16 | ≥32 | |
| Ertapenem | 6 | 4 | 4 | 11 | ||||
| Imipenem | 2 | 8 | 6 | 3 | 3 | 2 | 1 | |
| Meropenem | 2 | 3 | 1 | 4 | 9 | 1 | 1 | |
There are a total of 25 isolates.
TABLE 2.
Cephalosporin MIC distribution among E. cloacae isolates with ertapenem MICs of ≥2 mg/litera
| Cephalosporin | No. of isolates according to MIC (mg/liter) |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| ≤1 | 2 | 4 | 8 | 16 | 32 | 64 | 128 | 256 | ≥512 | |
| Cefotaxime | 3 | 3 | 3 | 6 | 10 | |||||
| Ceftazidime | 2 | 3 | 9 | 7 | 4 | |||||
| Cefepime | 2 | 3 | 1 | 5 | 3 | 3 | 7 | 1 | ||
There are a total of 25 isolates.
Sequence alterations.
Compared with our wild-type strain NCTC 13405, multiple sequence variants of the ampC, ampR, ompF, and ompC genes were detected in the 24 isolates tested (which consisted of eight carbapenem-susceptible comparators and the 16 carbapenem-resistant isolates from the United Kingdom), but most SNPs correlated with the STs of the isolates rather than with carbapenem resistance (see Fig. S1 in the supplemental material) and, when concatenated, revealed a tree with a topology similar to that derived from concatenated sequences of the seven MLST targets (see Fig. S2 in the supplemental material).
(i) ampC.
Compared with our reference sequence of NCTC 13405 and compared with E. cloacae P99 (X07274.1), 6 to 11 SNPs and 3 to 21 SNPs, respectively, leading to amino acid changes were identified in ampC sequences from the 24 isolates tested. Nevertheless, none were located in the SISK serine active site, in the YAN, KTG, and DAQA conserved motifs (30), or the β-lactam binding sites (Gln120, Ser289, Ser343, Asn346, Arg349), but all were detected in cephalosporin- and carbapenem-susceptible isolates alike. Thr14Leu, Lys41Ile, Pro68Ser, Trp221Arg, and Ile154Val were exclusive to two carbapenem-resistant isolates, but both belonged to the same ST (ST200).
(ii) ampR.
The only amino acid changes that were exclusive to carbapenem-resistant strains and not related to the isolates' STs were Val54Ile and Glu273Lys, each of which was observed in only 2/16 resistant isolates (isolates 6 and 46 and isolates 11 and 21, respectively).
(iii) ampD.
Amino acid changes in AmpD were detected across the phylogenetic tree. Among the mutations identified across the sequences of ampD in all 24 isolates (including control isolates), 14 substitutions (Ala9Thr, His18Pro, Pro40Leu, Trp42Arg, Ile48Ser, Arg78Ser, Val84Leu, Gln86Leu, Tyr87Ser, Met101Lys, Gly114Ala, Tyr130Asp, Leu133Gln, and Asp149Gly) were uniquely identified in the cephalosporin-resistant isolates, while isolate 2 had AmpD truncated by mutation leading to a premature stop codon at position 84. All of these amino acid substitutions were also found when our isolates were compared with wild-type ampD Z14003. Of these amino acid changes, only alterations at positions 40, 48, 86, 87, and 114 were considered significant in terms of being detected in isolates belonging to STs where the susceptible control isolates of the same ST did not harbor the same mutation. Met101Lys was identified more than once in two different STs, but we did not have susceptible control isolates of these STs with which to compare them. All other amino acid changes were detected in isolates belonging to different STs.
(iv) ompF and ompC.
Sequence variation was observed in the OMP genes ompF and ompC compared with NCTC 13405. Amino acid changes in the long “extracellular” loops (L1 to L5) of OmpF were found in carbapenem-susceptible and in carbapenem-resistant isolates alike, indicating that the changes were not associated with carbapenem resistance. Detection of these hypervariable domains in the cell-surface-exposed loops is in line with previously published data (31). Likewise, there was no association with carbapenem resistance in the small numbers of changes in the transmembrane domains, of which β1 to β13 and β15 and β16 were highly conserved (see Fig. S1 in the supplemental material). A premature stop codon was seen in two carbapenem-resistant isolates (2 and 21, Leu242X and Val301X, respectively), and in the carbapenem-resistant isolate 11, the first 111 bp were deleted. In OmpC, loop 3, which folds into the porin channel, was wild type in all isolates, while amino acid changes occurred in loops L4 to L8. An Arg191Cys change in isolate 46 was the only amino acid change that was not observed in any of the carbapenem-susceptible isolates (see Fig. S1). Gene disruptions due to indels were observed in six isolates; in isolates 39 and 2, the beginning of the gene and the promoter region were disrupted by IS3 and IS1, respectively. In two isolates (11 and 14), the beginning of the gene was deleted from 7 up to 54 nucleotides and was replaced by a transposon; in isolate 6, the beginning of the gene (485 nucleotides) was deleted and was fused to the adjacent two-component system. The ompC gene of isolate 33 was not amplifiable, and we were only able to extract the end of the gene with whole-genome sequencing (first 1,218 nucleotides missing). A summary of the above-mentioned mutations in the genes sequenced, which were associated most strongly with a cephalosporin- and/or carbapenem-resistant phenotype, is listed in Table 3.
TABLE 3.
Summary of mutations considered significant for being associated with AmpC derepression and/or carbapenem resistance
| Isolate no. | MIC (mg/liter) of antibiotic testeda |
AmpD amino acid | OmpF |
OmpC nucleotide | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| CLAX | CTX | FEP | ETP | IPM | MEM | Amino acid | Nucleotide | |||
| 2 | 16 | >256 | 64 | >16 | 16 | 16 | Val84X | Leu242X | IS1 (−15)b | |
| 6 | 8 | >256 | 64 | >16 | 32 | 32 | Δ0–485c | |||
| 11 | 16 | >256 | 64 | >16 | 8 | 8 | Δ1–111 | Δ1–7 | ||
| 13 | 4 | 256 | 64 | >16 | 8 | 8 | Pro40Leu | |||
| 14 | 16 | >256 | 16 | >16 | 8 | 8 | Δ1–54 | |||
| 21 | 32 | 64 | 64 | 16 | 4 | 8 | Val301X | |||
| 24 | 4 | >256 | 32 | 16 | 1 | 2 | Tyr87Ser | |||
| 25 | 64 | 256 | 8 | >16 | 2 | 8 | Ile48Ser | |||
| 33 | 8 | >256 | 8 | >16 | 8 | 8 | Δ1–1218 | |||
| 39 | >256 | >256 | 64 | 2 | 1 | 0.25 | Gln86Leu | |||
| 45 | 32 | 256 | 32 | >16 | 2 | 4 | Ile48Ser | |||
| 47 | >256 | >256 | 16 | >16 | 1 | 2 | Gly114Ala | |||
| NCTC 13406d | 4 | 256 | 2 | 1 | 0.5 | 0.125 | Gln86Leu | |||
CLAX, cloxacillin; CTX, cefotaxime; FEP, cefepime; ETP, ertapenem; IPM, imipenem; MEM, meropenem.
−, Position of insertions downstream of the A of the start codon.
Nucleotide 485 is fused to the adjacent two-component system.
NCTC13406, AmpC β-lactamase derepressed (constitutive hyper-producing) mutant of NCTC 13405.
Gene expression. (i) ampC expression in correlation to ampR and ampD sequences.
The ampC expression levels were measured in the wild-type, AmpC-inducible strain NCTC 13405 and its AmpC hyper-producing mutant NCTC 13406 as well as in three clinical isolates that were resistant to all carbapenems (isolate 2 belonging to ST50, isolate 45 belonging to ST133, and isolate 6). Two clinical carbapenem-susceptible isolates (S12 belonging to ST50 and S11 belonging to ST133) were chosen as comparators for resistant isolates 2 and 45. The ampR and ampD sequences of all isolates were related to the ampC expression level in order to find contributing SNPs that might lead to changes in ampC expression.
A single amino acid substitution (Gln86Leu) in ampD distinguished NCTC 13405 from NCTC 13406. There were no differences found between these isolates in any other genes sequenced, and an approximate 40-fold difference in ampC transcription was observed between this pair.
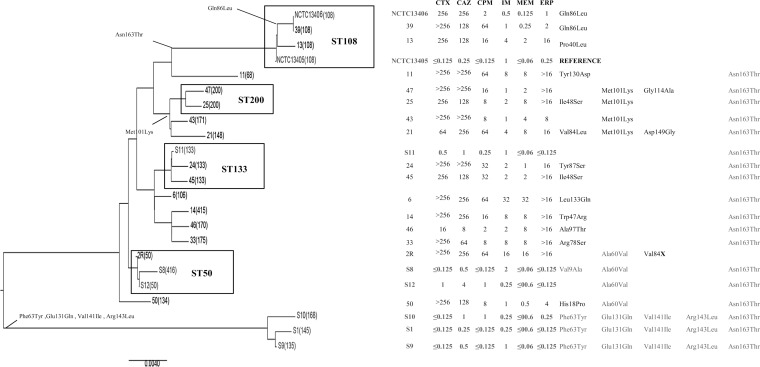
The susceptible and resistant ST50 isolates 2 and S12 had identical ampC and ampR sequences but differed in AmpD due to an early stop codon (Val84X) in isolate 2. The carbapenem-susceptible and carbapenem-resistant ST133 isolates (S11 and 45) also differed only in AmpD (Ile48Ser in isolate 45), and the AmpD sequence of isolate 6 had a Leu133Gln change (Table 4 and Fig. 2; see also Fig. S1 in the supplemental material).
TABLE 4.
Lesions associated with AmpC derepression and cephalosporin and carbapenem resistance in the isolates tested in detail, and ampC gene transcription compared to NCTC 13405 and ompF-ompC transcription compared to NCTC 13406
| Isolate no. | ST | MICs (mg/liter) of antibiotics tested |
Genes affected, changes compared to NCTC 13405 |
Gene transcription levelsa |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
ampC |
ampD |
ompF |
ompC nucleotide | ampC | ompF | ompC | |||||||||||
| CLAX | CTX | FEP | ETP | IPM | MEM | Nucleotide | Amino acid | Nucleotide | Amino acid | Nucleotide | Amino acid | ||||||
| 2 | 50 | 16 | >256 | 64 | >16 | 16 | 16 | 203_214dupb | Val84X | 695delA | Leu242X | IS1 (−15)c | 21×↑ | 2.5×↓ | 160×↓ | ||
| 6 | 106 | 8 | >256 | 64 | >16 | 32 | 32 | T399A | Leu133Gln | Δ0–485d | 29×↑ | 2.5×↓ | N/A | ||||
| 45 | 133 | 32 | 256 | 32 | >16 | 2 | 4 | T143G | Ile48Ser | 19×↑ | = | 3×↓ | |||||
| NCTC | 108 | <0.125 | <0.25 | <0.25 | 0.25 | 1 | ≤0.06 | N/A | N/A | ||||||||
| 13405 | |||||||||||||||||
| NCTC | 108 | 4 | 256 | 2 | 1 | 0.5 | 0.125 | A256T | Gln86Leu | 40×↑ | |||||||
| 13406 | |||||||||||||||||
↑, Elevated; ↓, reduced; =, equivalent; N/A, not available/not performed.
GCGCATCTGCGC.
−, The position of insertions downstream of the A of the start codon.
Nucleotide 485 is fused to the adjacent two component system.
FIG 2.
UPGMA tree of ampD sequences with cephalosporin MICs and amino acid changes. Multilocus sequence types are indicated in parentheses. CTX, cefotaxime; CAZ, ceftazidime; FEP, cefepime; IPM, imipenem; MEM, meropenem; ERP, ertapenem. MICs are displayed in milligrams per liter. NCTC 13405 is the reference wild-type E. cloacae strain, and NCTC 13406 is the AmpC β-lactamase-derepressed (constitutive hyper-producing) mutant of NCTC 13405. S isolates and NCTC 13405 are cephalosporin and carbapenem sensitive. NCTC 13406 is carbapenem sensitive. Bold squares indicate that isolates belong to the same ST. Left columns show amino acid changes. Gray fonts indicate amino acid changes that also occur in cephalosporin/carbapenem-susceptible isolates.
Isolates 2 and 45 had approximately 27- and 10-fold higher levels of ampC transcription than isolates S12 and S11, respectively. Comparison of ampC transcription for the three carbapenem-resistant isolates with the AmpC-inducible NCTC 13405 indicated that transcription levels in these isolates varied up to 29-fold.
(ii) Porin expression and DNA sequences of ompC and ompF.
OMP analysis of the same five isolates in which we studied ampC transcription showed that the susceptible isolates S11, S12, and NCTC 13406 produced more OMP bands than the resistant isolates 2 and 6 (see Fig. S3 in the supplemental material), which had disruption lesions in the sequences of ompF and/or ompC (Table 4; see also Fig. S1 in the supplemental material). Porin gene sequences for isolate 45 were identical to those from carbapenem-susceptible isolate S11. In concordance with the sequence information, all isolates retained the expression of OmpA. Transcript levels for ompF were up to 2.5-fold lower in clinical isolates 2 and 6 compared with those of NCTC 13406 and were nearly equivalent in isolate 45 compared with those of NCTC 13406. Transcription of ompC was noticeably lower in isolate 2, with levels 160-fold lower than those of NCTC 13406 and 3-fold lower in isolate 45. Measurement of ompC transcription in isolate 6 was not possible, as the primers used would not bind and extend due to a 485-bp deletion in the beginning of the gene.
DISCUSSION
Among a collection of E. cloacae clinical isolates with unusually high carbapenem MICs but no known carbapenemases, we found considerable genetic diversity indicated by PFGE, which was greater than that indicated by MLST, contrasting with previous findings (32). This is likely due to the higher discriminatory power of PFGE (33).
We sought to define the underlying mechanisms conferring cephalosporin and carbapenem resistance in this collection through a sequencing approach and, in contrast to many previous studies, considered the SNPs that we detected in relation to the MLST-based phylogeny of our collection to ascertain which were associated most strongly with a carbapenem-resistant phenotype and whether these related simply to the phylogeny of the isolates.
Previous data for low-level resistance and decreased susceptibility to carbapenems (meropenem and imipenem median MICs of 2 mg/liter and 4 mg/liter, respectively) (8, 9) have indicated that reduced outer membrane permeability combined with a strongly expressed AmpC is mechanistically important. Our data demonstrate that these features also occur in isolates with meropenem MICs of up to 32 mg/liter.
An AmpC overexpressing phenotype was a feature of the carbapenem-resistant strains that we analyzed; clearly though, as shown by NCTC 13406—whose ampC expression level was essentially equivalent to that of the carbapenem-resistant isolates—this is an association that is also held by carbapenem-susceptible, cephalosporin-resistant isolates. Loss of porins seemed to be the main contributor to carbapenem resistance and related to the resistance level in our isolates; the underlying mechanism of porin loss, however, could not be clearly elucidated. The transcriptional activity of ompC was significantly affected in the highly carbapenem-resistant isolate 2, whose promoter region was disrupted by an insertion sequence, and its ompF gene was disrupted by an early stop codon. In isolate 6, the beginning of ompC was disrupted; however, there was no early termination of the peptide that would explain the loss of OmpF, which was clearly observed. The presence of an intact ompF gene in this isolate leads to the assumption that the insertion of the porin into the outer membrane might be disrupted by unidentified mechanisms. The reduced expression of the porins in isolate 45 could not be explained by transcriptional regulation or sequence variability.
SNPs in the promoter regions of porin genes have been associated with porin loss (8) leading to carbapenem resistance, but the results of relating the observed SNPs to the MLST-based phylogeny revealed for the first time that most variations were phylogenetically related, occurring across carbapenem-susceptible and carbapenem-resistant isolates, and thus were not major contributors to resistance. Likewise, most SNPs detected in ampC, ampR, and their promoter regions and ompF and ompC were phylogenetic variations and had no correlation to the phenotypic resistance of the isolates.
The role of ampD mutations in ampC regulation has recently been confirmed in the study of Guérin et al. (34), in which ΔampD mutant E. cloacae showed much higher ampC expression levels than those of the wild-type strain. The association of AmpC overproduction in our carbapenem-resistant E. cloacae with nonsynonymous SNPs in ampD agreed with previous data (35). The truncation of AmpD in isolate 2 removed the carboxy-terminal region, which is required (as well as Asp164, His34, and His154) (36) for repression of ampC (13), and correlated with the stable derepression of AmpC in that isolate. In isolate 45, the potential significance of the Ile48Ser substitution in AmpC overproduction was supported by the observation of Ile48 in a carbapenem-susceptible comparator isolate of the same ST (isolate S11, ST133).
Amino acid substitutions potentially leading to AmpC overproduction were also observed in cephalosporin-resistant but carbapenem-susceptible isolates; NCTC 13405 and its mutant only differed in AmpD (Gln86Leu) where the mutant strain had a 40-fold increase in AmpC activity. Other amino acid substitutions that can be clearly correlated with cephalosporin and/or carbapenem resistance were Pro40Leu, Tyr87Ser, Val84X, and Gly114Ala. However, other substitutions in AmpD (Ala60Val and Phe63Tyr) were observed in carbapenem- and ceftazidime-susceptible strains; this contradicts previous reports of their involvement in ampC overexpression (12).
In AmpR, we found Glu273Lys, which was only detected in two highly cephalosporin-resistant strains. The observation of Val54Ile, described in highly cephalosporin-resistant isolates, was made in two isolates, of which one had only moderate level cephalosporin resistance. The role of these amino acid changes in AmpR for phenotypic resistance is unclear, however, as the activity of AmpR is dependent on the interaction with coregulators rather than on the expression of ampR (34).
In conclusion, this work has, for the first time, related SNPs observed in carbapenem-resistant and carbapenem-susceptible clinical isolates of E. cloacae to their MLST-based phylogeny and revealed hitherto unknown mutations in AmpD, which are responsible for cephalosporin and/or carbapenem resistance. SNPs detected in ampD were associated with constitutive AmpC overexpression, whereas most genetic differences in the other genes were phylogenetic variations and did not support any association with cephalosporin and/or carbapenem resistance. Although amino acid changes in AmpD correlated with AmpC overexpression and cephalosporin resistance, high levels of carbapenem resistance were associated most closely with porin loss occurring mainly due to insertion sequences and deletions disrupting their coding sequences and/or promoter regions. These data are observational and, along with explorations of the role of efflux, require further empirical molecular genetic proof via, e.g., complementation experiments, but nevertheless do indicate the ongoing importance of mutational carbapenem resistance in this species.
Supplementary Material
ACKNOWLEDGMENTS
We acknowledge Laurent Poirel and Patrice Nordmann for sharing the prepublication sequences of blaFRI-1. We also thank Jayesh Shah, Claire Perry, Marina Warner, Ayisha Chaudry, Rachael Adkin, Jacqueline Findlay, Tabassum Noorie, Rachel Pike, and Shazad Mushtaq for technical advice and assistance.
This study was supported by an award to B.B.F. from the Margarete und Walter Lichtenstein Stiftung, University of Basel, and Freiwillige Akademische Gesellschaft Basel, Switzerland.
N.W. has no personal interests to declare. However, P.H.E.'s AMRHAI Reference Unit has received financial support for conference attendance, lectures, research projects, or contracted evaluations from numerous sources, including Achaogen Inc., Allecra Antiinfectives GmbH, Amplex, AstraZeneca UK Ltd., Becton Dickinson Diagnostics, The British Society for Antimicrobial Chemotherapy (BSAC), Cepheid, Check-Points B.V., Cubist Pharmaceuticals, Department of Health, Enigma Diagnostics Ltd., Food Standards Agency, GlaxoSmithKline Services Ltd., Henry Stewart Talks, IHMA Ltd., Merck Sharpe & Dohme Corp., Meiji Seika Kiasya Ltd., Momentum Bioscience Ltd., Nordic Pharma Ltd., Norgine Pharmaceuticals, Rempex Pharmaceuticals Ltd., Rokitan Ltd., Smith & Nephew UK Ltd., Trius Therapeutics, VenatoRx, and Wockhardt Ltd. None of these poses a conflict of interest with this work.
The remaining authors declare no conflicts of interest.
Funding Statement
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.02835-15.
REFERENCES
- 1.Russo TA, Johnson JR. 2008. Diseases caused by Gram-negative enteric bacilli. In Longo DL, Fauci AS, Kasper DL, Hauser SL, Jameson JL, Loscalzo J (ed), Harrison's principles of internal medicine. McGraw-Hill, New York, NY. [Google Scholar]
- 2.Davin-Regli A, Monnet D, Saux P, Bosi C, Charrel R, Barthelemy A, Bollet C. 1996. Molecular epidemiology of Enterobacter aerogenes acquisition: one-year prospective study in two intensive care units. J Clin Microbiol 34:1474–1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kartali G, Tzelepi E, Pournaras S, Kontopoulou C, Kontos F, Sofianou D, Maniatis AN, Tsakris A. 2002. Outbreak of infections caused by Enterobacter cloacae producing the integron-associated beta-lactamase IBC-1 in a neonatal intensive care unit of a Greek hospital. Antimicrob Agents Chemother 46:1577–1580. doi: 10.1128/AAC.46.5.1577-1580.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lodato EM, Kaplan W. 2013. Priority medicines for Europe and the world. A public health approach to innovation. Background paper 6.1: antimicrobial resistance. World Health Organization, Geneva, Switzerland. [Google Scholar]
- 5.Pollett S, Miller S, Hindler J, Uslan D, Carvalho M, Humphries RM. 2014. Phenotypic and molecular characteristics of carbapenem-resistant Enterobacteriaceae in a health care system in Los Angeles, California, from 2011 to 2013. J Clin Microbiol 52:4003–4009. doi: 10.1128/JCM.01397-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pantel A, Boutet-Dubois A, Jean-Pierre H, Marchandin H, Sotto A, Lavigne JP, CARB-LR group . 2014. French regional surveillance program of carbapenemase-producing Gram-negative bacilli: results from a 2-year period. Eur J Clin Microbiol Infect Dis 33:2285–2292. doi: 10.1007/s10096-014-2189-5. [DOI] [PubMed] [Google Scholar]
- 7.Szabo D, Silveira F, Hujer AM, Bonomo RA, Hujer KM, Marsh JW, Bethel CR, Doi Y, Deeley K, Paterson DL. 2006. Outer membrane protein changes and efflux pump expression together may confer resistance to ertapenem in Enterobacter cloacae. Antimicrob Agents Chemother 50:2833–2835. doi: 10.1128/AAC.01591-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Doumith M, Ellington MJ, Livermore DM, Woodford N. 2009. Molecular mechanisms disrupting porin expression in ertapenem-resistant Klebsiella and Enterobacter spp. clinical isolates from the UK. J Antimicrob Chemother 63:659–667. doi: 10.1093/jac/dkp029. [DOI] [PubMed] [Google Scholar]
- 9.Yang FC, Yan JJ, Hung KH, Wu JJ. 2012. Characterization of ertapenem-resistant Enterobacter cloacae in a Taiwanese university hospital. J Clin Microbiol 50:223–226. doi: 10.1128/JCM.01263-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barnaud G, Labia R, Raskine L, Sanson-Le Pors MJ, Philippon A, Arlet G. 2001. Extension of resistance to cefepime and cefpirome associated to a six amino acid deletion in the H-10 helix of the cephalosporinase of an Enterobacter cloacae clinical isolate. FEMS Microbiol Lett 195:185–190. doi: 10.1111/j.1574-6968.2001.tb10519.x. [DOI] [PubMed] [Google Scholar]
- 11.Kuga A, Okamoto R, Inoue M. 2000. ampR gene mutations that greatly increase class C beta-lactamase activity in Enterobacter cloacae. Antimicrob Agents Chemother 44:561–567. doi: 10.1128/AAC.44.3.561-567.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kaneko K, Okamoto R, Nakano R, Kawakami S, Inoue M. 2005. Gene mutations responsible for overexpression of AmpC beta-lactamase in some clinical isolates of Enterobacter cloacae. J Clin Microbiol 43:2955–2958. doi: 10.1128/JCM.43.6.2955-2958.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kopp U, Wiedemann B, Lindquist S, Normark S. 1993. Sequences of wild-type and mutant ampD genes of Citrobacter freundii and Enterobacter cloacae. Antimicrob Agents Chemother 37:224–228. doi: 10.1128/AAC.37.2.224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Andrews JM. 2001. Determination of minimum inhibitory concentrations. J Antimicrob Chemother 48(Suppl):S5–S16. [DOI] [PubMed] [Google Scholar]
- 15.British Society for Antimicrobial Chemotherapy. 2013. BSAC methods for antimicrobial susceptibility testing. British Society for Antimicrobial Chemotherapy, Birmingham, UK: http://bsac.org.uk/wp-content/uploads/2012/02/Version-12-Apr-2013_final.pdf. [Google Scholar]
- 16.Turton JF, Kaufmann ME, Warner M, Coelho J, Dijkshoorn L, van der Reijden T, Pitt TL. 2004. A prevalent, multiresistant clone of Acinetobacter baumannii in Southeast England. J Hosp Infect 58:170–179. doi: 10.1016/j.jhin.2004.05.011. [DOI] [PubMed] [Google Scholar]
- 17.Miyoshi-Akiyama T, Hayakawa K, Ohmagari N, Shimojima M, Kirikae T. 2013. Multilocus sequence typing (MLST) for characterization of Enterobacter cloacae. PLoS One 8:e66358. doi: 10.1371/journal.pone.0066358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Walker RA, Lindsay E, Woodward MJ, Ward LR, Threlfall EJ. 2001. Variation in clonality and antibiotic-resistance genes among multiresistant Salmonella enterica serotype typhimurium phage-type U302 (MR U302) from humans, animals, and foods. Microb Drug Resist 7:13–21. doi: 10.1089/107662901750152701. [DOI] [PubMed] [Google Scholar]
- 19.Woodford N, Fagan EJ, Ellington MJ. 2006. Multiplex PCR for rapid detection of genes encoding CTX-M extended-spectrum (beta)-lactamases. J Antimicrob Chemother 57:154–155. [DOI] [PubMed] [Google Scholar]
- 20.Perez-Perez FJ, Hanson ND. 2002. Detection of plasmid-mediated AmpC beta-lactamase genes in clinical isolates by using multiplex PCR. J Clin Microbiol 40:2153–2162. doi: 10.1128/JCM.40.6.2153-2162.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hopkins KL, Deheer-Graham A, Threlfall EJ, Batchelor MJ, Liebana E. 2006. Novel plasmid-mediated CTX-M-8 subgroup extended-spectrum beta-lactamase (CTX-M-40) isolated in the UK. Int J Antimicrob Agents 27:572–575. doi: 10.1016/j.ijantimicag.2006.03.003. [DOI] [PubMed] [Google Scholar]
- 22.Liao IC, Chen HM, Wu JJ, Tsai PF, Wang LR, Yan JJ. 2011. Metallo-beta-lactamase-producing Enterobacteriaceae isolates at a Taiwanese hospital: lack of distinctive phenotypes for screening. APMIS 119:543–550. doi: 10.1111/j.1600-0463.2011.02772.x. [DOI] [PubMed] [Google Scholar]
- 23.Yan JJ, Ko WC, Chuang CL, Wu JJ. 2002. Metallo-beta-lactamase-producing Enterobacteriaceae isolates in a university hospital in Taiwan: prevalence of IMP-8 in Enterobacter cloacae and first identification of VIM-2 in Citrobacter freundii. J Antimicrob Chemother 50:503–511. doi: 10.1093/jac/dkf170. [DOI] [PubMed] [Google Scholar]
- 24.Dortet L, Poirel L, Abbas S, Oueslati S, Nordmann P. 2015. Genetic and biochemical characterization of FRI-1, a carbapenem-hydrolyzing class A β-lactamase from Enterobacter cloacae. Antimicrob Agents Chemother 59:7420–7425. doi: 10.1128/AAC.01636-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Doumith M, Dhanji H, Ellington MJ, Hawkey P, Woodford N. 2012. Characterization of plasmids encoding extended-spectrum beta-lactamases and their addiction systems circulating among Escherichia coli clinical isolates in the UK. J Antimicrob Chemother 67:878–885. doi: 10.1093/jac/dkr553. [DOI] [PubMed] [Google Scholar]
- 26.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 28.Hernandez-Alles S, Alberti S, Alvarez D, Domenech-Sanchez A, Martinez-Martinez L, Gil J, Tomas JM, Benedi VJ. 1999. Porin expression in clinical isolates of Klebsiella pneumoniae. Microbiology 145:673–679. [DOI] [PubMed] [Google Scholar]
- 29.Carlone GM, Thomas ML, Rumschlag HS, Sottnek FO. 1986. Rapid microprocedure for isolating detergent-insoluble outer membrane proteins from Haemophilus species. J Clin Microbiol 24:330–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Naas T, Aubert D, Fortineau N, Nordmann P. 2002. Cloning and sequencing of the beta-lactamase gene and surrounding DNA sequences of Citrobacter braakii, Citrobacter murliniae, Citrobacter werkmanii, Escherichia fergusonii and Enterobacter cancerogenus. FEMS Microbiol Lett 215:81–87. doi: 10.1111/j.1574-6968.2002.tb11374.x. [DOI] [PubMed] [Google Scholar]
- 31.Thiolas A, Bornet C, Davin-Regli A, Pages JM, Bollet C. 2004. Resistance to imipenem, cefepime, and cefpirome associated with mutation in Omp36 osmoporin of Enterobacter aerogenes. Biochem Biophys Res Commun 317:851–856. doi: 10.1016/j.bbrc.2004.03.130. [DOI] [PubMed] [Google Scholar]
- 32.Izdebski R, Baraniak A, Herda M, Fiett J, Bonten MJ, Carmeli Y, Goossens H, Hryniewicz W, Brun-Buisson C, Gniadkowski M, MOSAR WP2, WP3, and WP5 Study Groups. 2015. MLST reveals potentially high-risk international clones of Enterobacter cloacae. J Antimicrob Chemother 70:48–56. doi: 10.1093/jac/dku359. [DOI] [PubMed] [Google Scholar]
- 33.Tenover FC, Gay EA, Frye S, Eells SJ, Healy M, McGowan JE Jr. 2009. Comparison of typing results obtained for methicillin-resistant Staphylococcus aureus isolates with the DiversiLab system and pulsed-field gel electrophoresis. J Clin Microbiol 47:2452–2457. doi: 10.1128/JCM.00476-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guérin F, Isnard C, Cattoir V, Giard JC. 2015. Complex regulation pathways of AmpC-mediated beta-lactam resistance in Enterobacter cloacae complex. Antimicrob Agents Chemother 59:7753–7761. doi: 10.1128/AAC.01729-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hanson ND, Sanders CC. 1999. Regulation of inducible AmpC beta-lactamase expression among Enterobacteriaceae. Curr Pharm Des 5:881–894. [PubMed] [Google Scholar]
- 36.Genereux C, Dehareng D, Devreese B, Van Beeumen J, Frere JM, Joris B. 2004. Mutational analysis of the catalytic centre of the Citrobacter freundii AmpD N-acetylmuramyl-l-alanine amidase. Biochem J 377:111–120. doi: 10.1042/bj20030862. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.