Abstract
mRNA positioning in the cell is important for diverse cellular functions and proper development of multicellular organisms. Single-molecule RNA FISH (smFISH) enables quantitative investigation of mRNA localization and abundance at the level of individual molecules in the context of cellular features. Details about spatial mRNA patterning at various times, in different genetic backgrounds, at different developmental stages, and under varied environmental conditions provide invaluable insights into the mechanisms and functions of spatial regulation. Here, we describe detailed methods for performing smFISH along with immunofluorescence for two large, multinucleate cell types: the fungus Ashbya gossypii and cultured mouse myotubes. We also put forward a semi-automated image processing tool that systematically detects mRNAs from smFISH data and statistically analyzes the spatial pattern of mRNAs using a customized MATLAB code. These protocols and image analysis tools can be adapted to a wide variety of transcripts and cell types for systematically and quantitatively analyzing mRNA distribution in three-dimensional space.
Keywords: Single molecule RNA FISH, spatial analysis, Ripley’s H, A. gossypii, C2C12 myotubes
1. Introduction
Subcellular localization of mRNAs is known to be crucial to a variety of cellular functions including cell cycle regulation and proper development in many organisms [1–12]. To unravel the mechanisms controlling this positioning and the role such patterning plays in cellular processes, conventional In Situ Hybridization has long been used to localize mRNAs [13]. However, its low sensitivity and technically challenging experimental procedures hinder the acceleration of mRNA positioning studies. Its incompatibility with other visualization methods such as DAPI staining or immunofluorescence (IF) also limits further research on the role and mechanism of spatial distribution of mRNAs using this technique. The emergence and recent advances in fluorescence in situ hybridization (FISH) have largely alleviated these limitations. The resolution and sensitivity of detection was greatly improved by using haptenated antisense probes and matching anti-hapten antibodies conjugated with bright fluorophores (e.g. Alexa Fluor dyes) [14–17]. Employing various tags and fluorophores in FISH enabled the multiplex detection of several RNA species and global assessments of RNA intracellular localization features in detail [18–20]. However, indirect signal amplification resulting from the FISH technique may complicate the experimental procedures and limit a systematic quantitation of transcription.
Emergence of single-molecule RNA Fluorescence In Situ Hybridization (smFISH), which enables detection of individual RNAs using small oligonucleotide probes directly tagged with a fluorophore, was a major turning point for many researches interested in mRNA positioning and regulation [2,12,21–23]. The smFISH technique also opened a new era of visualizing unprocessed transcripts and single molecule-level quantitation of transcription because of its sensitivity, which is sufficient to distinguish different classes of RNAs at the same time such as primary transcripts and spliced mRNAs [24,25]. By utilizing the recent advances in imaging tools and probe development, it becomes possible to use multiple probes for visualizing several different mRNA species as well as proteins using immunofluorescence (IF) in the same cell.
Large 3D data sets are readily generated with smFISH protocols, however tools to systematically and quantitatively analyze the spatial patterns of mRNA in these images are not broadly available. The various shapes and sizes of cells in three-dimensional space make it especially difficult to consistently analyze the spatial pattern of mRNAs. Variable intensities between transcripts in cells or between cells are also problematic for estimating the number of RNA molecules in a single diffraction-limited spot. Finally, it is difficult to quantitatively compare mRNA spatial distributions amongst cells that have differences in mRNA abundance either due to differences in genetic background or stochastic differences between genetically identical cells.
Here, we describe smFISH protocols with and without IF and recommended imaging conditions for both wide-field deconvolution microscopy followed and confocal microscopy. We discuss generation of complete spatial randomness (CSR) models, which are crucial for statistically evaluating spatial distributions, as well as the Ripley’s H function, which is widely used for assessing the clustering or dispersion of objects [26–29]. This workflow can be adapted to analyze subcellular positioning and protein-mRNA colocalization for a wide variety of transcripts and cell types.
2. Visualization of RNA
2.1 Designing the smFISH probes
The smFISH probes (Stellaris RNA probes) can be purchased from Biosearch Technologies (www.biosearchtech.com). Before ordering, the fluorophores conjugated to the probes should be carefully chosen based on their compatibility with the microscope setup (e.g. filter and dichroic mirror options for the wide-field microscope or excitation laser options for the confocal microscope) and their use (e.g. multiplexing or combining with immunofluorescence). For multiplexing the smFISH probe sets, the fluorophores should be chosen in a way to maximize the difference of excitation/emission wavelengths between two fluorophores. Fluorophores excited at lower energy wavelengths (450–500 nm) are generally not recommended due to the high autofluorescence at these wavelengths in many cells, including A. gossypii. In our lab, the fluorophores Quasar 570, TAMRA, and Quasar 670 have been utilized successfully with filter sets CZ915, 41002B, and 41008, respectively (all from Chroma Technology Corp) equipped on an AxioImager-M1 upright light microscope (Carl Zeiss). The commercial smFISH probes from Biosearch Technologies are comprised of up to 48 20-nucleotide oligonucleotides complementary to the RNA of interest. We obtained a good signal-to-noise ratio (>5) even with as few as 23 oligonucleotides [12]. It has been reported that single mRNAs could be visualized with only five oligonucleotides [30], but this is dependent on a variety of factors including the species examined and imaging considerations such as the excitation source, the sensitivity of the detector, and the background fluorescence. Probes designed to target exons or the complete RNA of interest allow visualization of both nuclear primary transcripts (enriched at transcription sites) and cytoplasmic mRNA. 3′ and 5′ untranslated regions (UTRs) can also be included in the probe target for such studies [31,32]. To specifically detect primary transcripts, only intronic sequences should be used for designing smFISH probes.
2.2 smFISH with immunofluorescence (IF) using A. gossypii
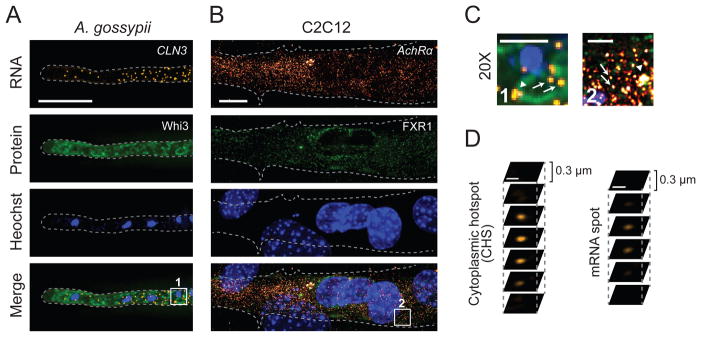
Visualization of an mRNA along with specific proteins may be of interest in different contexts. Our lab has previously shown that mRNA of the G1 cyclin CLN3 as well as polarisome components BNI1 and SPA2 are heterogeneously localized in A. gossypii using smFISH [2,12]. The RNA-binding protein Whi3 directly binds to these transcripts and affects their localization and stability. However, the spatial pattern of CLN3 is different from that of BNI1 and SPA2. We simultaneously visualized CLN3 mRNAs and Whi3 protein in cells using smFISH and IF, respectively (Figure 1A; 1C, left). Combining this technique with genetic approaches to manipulate the CLN3-Whi3 interaction would allow dissection of the role of CLN3 transcript spatial patterning in cellular processes and its regulation in A. gossypii. For example, the fraction of CLN3 mRNA colocalized with Whi3 may change when Whi3, especially its RNA-binding domain, is mutated. In our hands, the following procedure for smFISH staining has also been successfully adapted to another filamentous fungus, Neurospora crassa. Others have modified the protocol for Cryptococcus neoformans, and we are confident that it can be optimized for other species with cell walls [33]. Application of this protocol to N. crassa only required modification of the cell wall digestion step in order to allow probes access to the cell interior. Specifically, longer digestion times were required. Therefore, if you seek to adapt smFISH to another organism, we recommend investigating permeabilization methods for that (or related) organism developed for immunofluorescence and adapting them to this procedure.
Figure 1.
smFISH with immunofluorescence (IF) (A) CLN3 mRNA (orange; smFISH) and Whi3-GFP (green; IF) are visualized by smFISH and IF, respectively in A. gossypii. Rabbit anti-GFP IgG (1:100) and rat antirabbit IgG conjugated with Alexa-488 (1:500; Santa Cruz Biotech, TX) are used for IF. (B) AchRα transcripts (orange; smFISH) and FXR1 (green; IF) in differentiated C2C12 mouse myotubes. FXR1 was detected using 1:5,000 rabbit α-FXR1 ML14 antiserum (a generous gift from Edward Khandjian at Universite Laval), and 1:200 goat α-rabbit Alexa 488 (Molecular probes). (A,B) Nuclei are visualized by Hoechst staining. The gray dashed line indicates cell outline. Scale bar: 10 μm. (C) Magnification (20X) of a region within boxes in Figure 1A and 1B, bottom panels. Arrowheads indicate the cytoplasmic hotspots and arrows indicate single mRNA foci. Scale bar: 2 μm. (D) Z-sections of a cytoplasmic hotspot and mRNA spot. Scale bar: 500 nm.
2.2.1 RNase decontamination
Before performing smFISH, make sure to wipe the bench, pipette handles, tip boxes, and the bottles containing RNase-free reagents with a nuclease decontaminant such as RNase Zap (Ambion) to minimize RNase contamination. Wipe down bench and instruments with decontaminant at least 5 min. prior to working. Make all solutions in RNase-free water. Triple-distilled water was adequate for RNase-free experiments on our hands. Wear clean lab coat, gloves, and facemask at all times. Minimize talking or breathing directly on the RNase-free samples or reagents to get the best signal for smFISH.
2.2.2 Probe preparation
The probes from Biosearch Technologies are diluted in TE buffer (10 mM TrisCl, 1 mM EDTA, pH 8.0) to a stock concentration of 250 μM. These probe stocks can be stored in the dark at −20°C for at least 1 year. Then probes can be further diluted by 1:10, 1:20, 1:50 or 1:100 in TE buffer as a working stock. These working stocks can be stored at least six months at −20°C. 1:10 dilution generally works well but different dilutions may be required for specific probe sets to minimize non-specific binding, which results in high background.
2.2.3 Growth and preparation of fungal cells for smFISH
Fungal cells should be prepared after growth to a suitable stage or density. While the steps below are detailed for A. gossypii, slight modifications to the type of enzyme used for digestion and digestion times should make it adaptable to any fungal cells. For A. gossypii, “clean” spores (i.e. isolated by hydrophobicity with minimal mycelial contamination) are germinated and grown in Ashbya Full Media (AFM: 10 g/L yeast extract, 10 g/L peptone, 20 g/L glucose, 1 g/L myo-inositol) shaking at 30°C in a baffled flask. Generally, cells are grown for 14–16 h. The quantity of cells can be very important in the smFISH procedure and will likely need to be optimized in adapting the protocol for another species. Loss of material during enzymatic digestion (in the case of fungi) and wash steps can result in very few or no cells remaining if the procedure is started with insufficient biomass. However, too many cells present during digestion can lead to heterogeneous cell permeability and considerable variability in smFISH signal. The automated analysis independently determines the signal intensity for a single mRNA for each hypha, which compensates for the majority of heterogeneity caused by variable digestion. However, excessive digestion heterogeneity can result from attempted digestion of too many cells and this can cause problems during automated image processing steps. Typically, for wild-type fungal cells, a 50 mL culture inoculated with 50 μl spores is sufficient. Directly add 37% formaldehyde or freshly-made paraformaldehyde to the cell culture (final concentration: 3.7% v/v) to fix cells. Incubate cells shaking at 30°C for 1 h. Wash cells twice with ice-cold RNase-free Buffer B (1.2 M sorbitol, 0.1 M potassium phosphate, pH 7.5, RNase-free). Resuspend the cells in 1 mL RNase-free spheroplasting buffer (10 mL Buffer B, 2 mM vanadyl ribonucleoside complex (NEB) and transfer cells to a new RNase-free microcentrifuge tube.
To permeabilize cells, add 1.5 mg Zymolyase (MP Biomedicals) to the cells and incubate them at 37°C with gentle mixing for 35 – 40 min. for wild-type A. gossypii cells. The incubation time varies from strain to strain and can depend upon the Zymolyase preparation. Check the cell wall every 5–10 min. using a phase microscope to ensure proper digestion. Presence of the cell wall results in a phase-bright halo around the cells under the phase microscope with halogen light source. Both the interior and the periphery of the cells turn phase-dark during cell wall digestion and cells are physically broken down to smaller hyphal fragments during Zymolyase treatment. If cells are over-digested, they begin to burst. Roughly 80% of hyphae should appear phase-dark before the digestion is stopped. Once the digestion is complete, wash cells twice with cold Buffer B by spinning at low speed (<700 g). Spinning at too high a speed can cause digested cells to burst, therefore all centrifugation steps after digestion should be at low speed. Resuspend cells in RNase-free 70% EtOH and incubate cells at 4°C for at least 4 h. The cells may be stored in EtOH at 4°C for up to a week.
Aspirate EtOH and wash cells with wash buffer (20X SSC, 10% v/v deionized formamide). If the cells stick to the microcentrifuge tubes, add 0.1 % Tween-20 to the wash buffer to limit cell loss. Resuspend the cells in 100 μL hybridization buffer (1 g dextran sulfate, 10 mg E. Coli tRNA, 2 mM vanadyl ribonucleoside complex, 2 mg BSA, 20X SSC, 10% v/v deionized formamide in 50 mL RNase-free water) containing the 1–3 μL working stock of RNA FISH probe (start with 1 μL and modify if necessary) and incubate cells in the dark overnight at 37°C. Wash cells with wash buffer, resuspend the cells in wash buffer, and incubate them at 37°C for 30 min.
2.2.4 Completion of staining without IF
Wash cells with 1X PBS and incubate the cells in 1X PBS with 5 μg/mL Hoechst or DAPI at room temperature for 10 min. to counterstain DNA. Wash cells twice with 1X PBS and mount the cells on the RNase-free slide with an RNase free coverslip (glass slide baked at 250 °C for at least 8 h) with 5–15 μL mounting media (Prolong Gold antifade reagent, Molecular Probes). Seal the slide with transparent nail polish and proceed to image. The slides can be stored in the dark at −20°C.
2.2.5 Completion of staining with IF
Wash cells with wash buffer and resuspend the cells in RNase-free 1X PBS containing 1 mg/mL BSA (globulin-free). Incubate the cells at room temperature for 30–60 min. to block. Wash cells twice with 1X PBS+BSA and resuspend the cells in the 1X PBS containing BSA and primary antibody. In case of GFP immunofluorescence, 1:100 diluted rabbit anti-GFP antibody (Life technologies) was used. Incubate the cells for 1–2 h. at room temperature or overnight at 4°C. Wash cells 3 times with 1X PBS+BSA and incubate the cells for 1 h. at room temperature in 1X PBS+BSA with the secondary antibody. For GFP, 1:500 diluted mouse anti-rabbit IgG antibody (Santa Cruz Biotechnology) was used. Wash cells with 1X PBS and incubate the cells in 1X PBS with 5 μg/mL Hoechst or DAPI at room temperature for 10 min. to counterstain DNA (optionally this can instead be included in the secondary antibody incubation). Wash cells twice with 1X PBS and remove as much supernatant as possible without removing cells. Add a roughly equivalent amount of mounting medium (Prolong Gold antifade reagent, Molecular Probes) to cellular material. Mount the cells on an RNase-free slide with an RNase-free coverslip (glass slide baked at 250°C for at least 8 h.) by spotting a total of 5–15 μL mounting media/cell solution around the slide, depending on the coverslip used. We typically use 22 x 50 mm coverslips (Corning). Seal the slide with transparent nail polish and proceed to image. The slides can be stored in the dark at 4°C.
2.3 smFISH using C2C12 Mouse Myotubes
A. gossypii has proven an excellent model system for the study of functional cellular organization in examining the outputs of asynchronous nuclear division and specialized cellular regions such as the growing hyphal tips. More recently, our lab has adapted some of the approaches we have used in A. gossypii to other multinucleate cell types, including differentiated C2C12 mouse myotubes (Figure 1B; 1C, right). C2C12 myoblasts fuse to form large, multinucleate cells with different functional regions including the neuromuscular junction. The following smFISH procedure, as presented, has also been successfully utilized for visualization of mRNA in mammalian neurons. We have found the step that requires the most alteration in both muscle and neuron is the detergent permeabilization step: Insufficient permeabilization does not allow smFISH probes to enter the cell, while excessive permeabilization allows cytoplasmic RNA to leak out of the cell, causing signal loss and negatively impacting spatial analyses. For most transcripts, in both muscle and neuron, a two-minute permeabilization time was optimal. However, this may need to be altered for visualization of different transcripts or different cell types. For testing, we typically permeabilized cells for 30 seconds, one, two, five, seven, or ten minutes. Longer permeabilization times (with cytoplasmic RNA loss) may be required to visualize nuclear mRNAs.
2.3.1 RNase decontamination
The RNase decontamination should be conducted as described above in 2.2.1.
2.3.2 Probe preparation
Prepare the probes as described above in 2.2.2.
2.3.3 Cell preparation and smFISH hybridization
Start with myotubes grown and differentiated on coated coverslips (e.g. poly-lysine or laminin) in multi-well tissue culture plates. For simplicity in later steps, after sterilization of coverslips and before growing cells, one side of the coverslip can be labeled “UP” to indicate the cell side with a solvent-resistant marker. We typically hybridize 3–6 coverslips at a time to ensure a sufficient number of cells can be imaged and processed. Rinse the coverslips twice with room-temperature Hank’s Buffered Salt Solution (HBSS, stored at 4°C). For all steps, add wash solution by pipetting down the side of the container and aspirate gently to minimize cell loss off the coverslips. Fix cells with 1 mL 4% paraformaldehyde in HBSS at room temperature for 10 min. Wash cells twice with HBSS.
To permeabilize the cells, add 1 mL RNase-free CSK buffer (100 mM NaCl, 30 mM sucrose, 10 mM PIPES, 3 mM MgCl2; store at 4°C), supplemented with 0.5 % Triton X-100 and 10 mM vanadyl ribonucleoside complex (NEB) to the cells and incubate for 2 min. on ice. Incubation time can vary from 30 sec. to 10 min., and may need to be optimized depending on probe set and cell type, we have found that 2 min. is generally best. Rinse coverslips with 2 mL CSK buffer (without Triton) and wash cells with RNase-free 70% EtOH. The cells may be stored in a parafilm-sealed container in EtOH at 4°C for up to a week.
Make 20 μL probe hybridization mixture per coverslip: Mix 6.8 μL RNase-free water containing and 3μL (30% v/v) deionized formamide with 0.2 μL (5 pmol) of probe working solution. Add 10 μL of hybridization buffer (20% w/v dextran sulfate, 4 mg/mL BSA, 4X SSC, store at 4°C for up to three months) and mix (final 15% formamide). Generally 15% formamide works well but the concentration may be adjusted between 10–25% for different probe sets (up to 50%). Prepare hybridization chamber: Cover the bottom of a petri dish with parafilm. Decontaminate parafilm by wiping with RNase Zap and rinsing with RNase-free 90% EtOH, and let dry. Spot 20 μL of the probe hybridization mixture onto parafilm in hybridization chamber for each coverslip. Using forceps, place coverslip (cell side down) on top of hybridization solution. Gently press down so that the solution contacts the entire cell surface of the coverslip, but avoid bubbles and avoid pressing firmly enough so that probe solution leaks out from the edges of the coverslip. Cover all coverslips with a second piece of decontaminated parafilm. Seal around edges to prevent the probe solution from drying out. Close petri dish and seal with parafilm. Incubate the cells at 37°C in the dark overnight. Transfer coverslips to decontaminated coverslip holder in beaker. To equilibrate cells for imaging, wash with RNase-free 2X SSC containing 15% formamide for 20 min. at 37°C, with 2X SSC for 20 min. at 37°C, with 1X SSC for 20 min. at room temperature with gentle mixing, and with 4X SSC for 2 min. at 37°C.
2.3.4 Completion of staining without IF
For nuclear counterstaining, incubate cells in 1X PBS with 5 μg/mL Hoechst (or DAPI) for 30 min. at room temperature in the dark. Wash the cells twice with 1X PBS and mount the coverslips on RNase-free glass slides (glass slide baked at 250 °C for at least 8 h, ensure cell side down) with 5–7 μL of Prolong Gold mounting medium (Molecular Probes). Seal edges with nail polish and proceed to image. The slides can be stored in the dark at −20°C.
2.3.5 Completion of staining with IF
Wash cells in 1X PBS. Incubate in 1X PBS + 10 mg/mL BSA at room temperature for 1 h. Incubate with primary antibody diluted in 1X PBS + 10 mg/mL BSA at 4°C overnight. Rinse three times with 1X PBS for 5 min. Incubate with secondary antibody diluted in 1X PBS + 10 mg/mL BSA with 5 μg/mL Hoechst at room temperature for 1 h. in the dark. Rinse three times with 1X PBS for 5 min. Mount coverslips on RNase-free glass slides (glass slide baked at 250 °C for at least 8 h, ensure cell side is down) onto 5–7 μL of Prolong Gold mounting medium (Molecular Probes). Seal edges with nail polish, and store slides in the dark at −20°C.
2.4 Image acquisition
Any suitable fluorescence microscope can be used for image acquisition. Both wide-field and confocal microscopy have been used for imaging smFISH depending on their purpose and RNA species [12,34–36]. Often, after wide-field image acquisition, a step for deconvolution or image enhancement (e.g. Laplacian of Gaussian filtering) is included to reduce out of focus light, which causes a haze around emitting particles (e.g. mRNA foci). The number of iterations for image deconvolution should be carefully determined not to over- or under-process the images which may result in image manipulation (e.g. generating random foci from the background or saturating pixel values at certain regions). However, the mRNA spots may not be resolved into single spots but appear as erroneous blobs or tubules even after careful deconvolution if mRNA is highly abundant. Confocal microscopes (e.g. spinning disk, laser scanner) can be used in this case to minimize manipulating the mRNA signal. On our hands, smFISH images acquired using laser scanner (Leica LSM SP8) achieved a comparable image quality compared to wide-field microscope (Supplementary figure 1). Since, the confocal microscopy sacrifices a significant amount of emitted light, bright and photo-stable fluorophores such as TAMRA (TMR) or CAL fluor Red 610 are recommended.
A Z-stack should be taken thoroughly from the bottom to the top of the cell. Missing a part of the cell may lead to an error in spatial pattern analyses. The Z-step size has to be carefully determined so that the same mRNA spot can be captured over multiple Z-slices. In our hands, 0.3 μm Z-spacing has worked very well but this will vary depending on the microscopic setup specifics such as the optics, pinhole size, and detector pixel size. Our typical imaging (detecting TAMRA-conjugated probes) used a wide-field microscope (AxioImager-M1; Zeiss) equipped with 63x 1.4 NA objective (Zeiss), TRITC filter cube with a dichroic mirror/beamsplitter (41002B; Chroma Technology corp) and Hamamatsu Orca-ER CCD camera (C4742-80-12AG) driven by Volocity 4.4 (PerkinElmer) or μManager [37,38]. Typically, the smFISH samples were excited for 30–100 msec. with 100% neutral density using an Exfo X-Cite 120 lamp. With this setup and Z-step size, mRNAs and hotspots are seen in two to four Z-planes and up to seven Z-planes, respectively (Figure 1D). It is hard to distinguish true mRNA signal from background if they are detected only in one Z-plane. Some background spots that look similar to actual mRNA can be easily excluded during image analysis by scoring only mRNAs seen in at least two consecutive Z-planes. The exposure time (wide-field microscopy) or laser power (confocal microscopy) should be carefully chosen to minimize photo-bleaching but maximize the signal-to-noise ratio. Supplementary Figure 2 summarizes the smFISH procedures explained above.
3. Image preparation and 3-D detection of nuclei, mRNAs and cell outline
We designed the ImageJ macros and MATLAB code to process single-cell images for the fast and systematic image analysis. We mainly use these codes to process smFISH images of the multinucleate filamentous fungus A. gossypii, but they can be adapted to essentially any cell type and even multi-cellular samples. After acquiring Z-stack images we rotate and crop the original field using ImageJ (Fiji 1.46a) so that each cropped image contains only one hyphal fragment, horizontally oriented with the center of the image being inside the cell. For A. gossypii and for other filamentous cells, it is recommended that hyphae are aligned in a way that their tips consistently point to left (or right) side of the image. This image preparation process is not critical for general spatial pattern analysis of nuclei or mRNA, but is important for some analyses in which relative position of mRNAs from the hyphal tip is taken into account (e.g. comparison of local mRNA density relative to the distance from the hyphal tip). For the Ripley’s H analysis (described in detail in Section 5), we recommend cropped images contain >4 transcripts (for both univariate and bivariate) and nuclei (for bivariate). Images with very few objects can promote artifacts in the statistical analysis. These cropped image stacks are automatically categorized by labels from their image file names, which may contain description of their strains or conditions. The same categorization is used in further image processing and analysis.
The customized ImageJ macro script is used to detect nuclei, mRNAs, and cell outline, and record their coordinates in 3D (Material 1: 3D_OC_mask.ijm or Material 2: 3D_OC_multiThreshold.ijm). The core function of 3-D detection is the ImageJ plugin “3D-OC (3D Object Counter 2.0),” which uses one (or multiple) threshold for pixel intensity values to distinguish objects (nuclei or mRNAs) from background. The threshold value needs to be manually determined by testing various values on a set of images (>15 images as a training set). The customized MATLAB code “DetectionTest.m” generates a reconstruction of detected mRNA and nucleus spots, which can be used to find the best threshold by comparing detected spots to the actual micrographs (Material 3). To easily find the appropriate threshold and automate further processes, we recommend using cells processed at the same time and images acquired on the same day with the same settings to minimize signal variability that may increase detection errors. In the event that there is substantial signal variability between images, the 3D_OC_multiThreshold.ijm code can be used to apply different thresholds to each image and ensure appropriate mRNA detection (see Section 6.2). In the case of low signal-to-noise in smFISH images, it is recommended that the threshold be set to a low value in order to detect as many transcripts as possible even along with some background spots. Most background spots captured are removed in the process of mRNA detection in MATLAB by scoring their size, shape, and intensity and comparing them to average mRNA spot parameters. The cell outline mask is obtained from the phase channel. In case of using A. gossypii, the mask for middle–widest–region of the cell is recorded in ImageJ and used to reconstitute the hypha as a cylinder with changing radii in MATLAB. Thus, this cell reconstitution method should be modified depending on the shape of the cell to accurately reconstruct the cell area. Defining the cell area is critical for several analyses including mRNA density measurement. The output files contain various data such as X, Y, and Z-coordinates of each detected mRNA and its volume.
4. Reconstruction of smFISH data
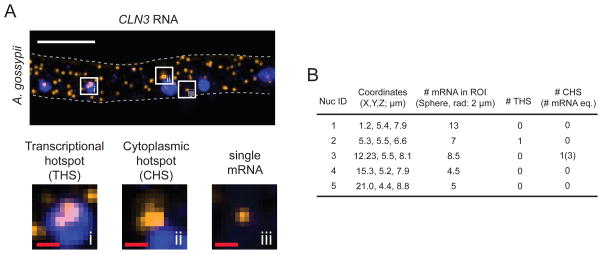
The MATLAB code <smFISH3D_pro1.m> reads all data files generated by ImageJ processing in Section 3 and reconstructs the cell with detected objects in hypothetical 3D space for each image stack (Material 4). First, the code collects the 3D position, size, and signal intensity of nuclei or mRNA spots and calculates the mean values for each image stack. Any nuclei or mRNA too big (3D size >10X mean), too small (<0.5X Mean) or too dim (<0.5X Mean) are removed before any further analyses because these are likely background signal or particles on the slide or coverslip. Although these criteria (e.g. <0.5X mean) efficiently remove most background signals in general, they should be modified if the smFISH signal is highly variable from spot to spot or cell-to-cell. In most cases in our hands, the signal intensity of an individual mRNA had very low variability (CV < 0.5 in the histogram for mRNA signal intensity), and the mean intensity for detected objects was a reasonable approximation of the average intensity of a single mRNA [2,12]. We recommend examining the histogram of intensities of objects detected by ImageJ in Materials 1 or 2 and comparing the mean value to the peak of the histogram. Frequent detection of small background objects or presence of a significant number of diffraction-limited clusters of transcripts may skew the average. In this case, the approximation of the intensity of a single mRNA may need to be adjusted (See section 6.4). The number of mRNAs can be over- or under-estimated if the signal from single mRNAs are too variable or if the mean value does not approximate a single mRNA because the MATLAB code may exclude true mRNAs or include background signal. The edge of the 3D-reconstructed cell is used to remove any objects detected outside of the cell during execution of the MATLAB code. In many cases, we observed that some RNA spots are significantly larger and brighter than the average. We termed spots with these characteristics “hotspots.” They are categorized into two classes: transcriptional hotspot (THS) or cytoplasmic hotspot (CHS), depending on their location (Figure 2A): THS indicating nuclear and CHS indicating cytosolic. THSs are thought to consist of primary transcripts at sites of active transcription at the gene locus [2,39], thus THSs are useful for localizing and counting the nuclei that are transcriptionally active for the gene being visualized and estimating their degree of transcriptional activity by scoring THS intensity. In contrast, CHS implies a concentration of mRNAs in the cytoplasm in a diffraction-limited spot that could be either because the mRNAs are in the same complex or are by chance less than ~200 nm apart.
Figure 2.
Analysis of smFISH data (A) A transcriptional hotspot (THS), cytoplasmic hotspot (CHS), and single mRNA spot are shown as a maximum-intensity Z-projection. Each white square box is enlarged in the bottom panel. The gray dashed line indicates the cell outline. White scale bar: 5 μm. Red scale bars: 500 nm. (B) An example of output data (Material 5) generated by the customized MATLAB code (Materials 4,6). The coordinates in the table refer to the positions of nuclei in the smFISH image shown in A. The top-left corner of the image represents 0 μm for X and Y coordinates. In the Z-stack of the smFISH image, the bottom Z-slice represents 0 μm for Z coordinate. The MATLAB data includes the following: the number of mRNAs within 2 μm of the center point of each nucleus (i.e. within a spherical ROI with a 2 μm radius), the number of THSs detected within each nucleus, and the number of CHSs detected within 2 μm of the center point of the nucleus. The number of mRNAs within a CHS is indicated in parentheses, and was calculated from the intensity value of a single mRNA determined for this hypha.
CHSs need further consideration during analysis because there are multiple mRNAs at the same apparent X, Y, Z coordinate which could confound later statistical analysis. We estimated the number of mRNAs likely in each CHS by dividing its total signal intensity by the average mRNA intensity. Each predicted mRNA can be assigned its own coordinates within the CHS for spatial statistical analysis. To generate the positions of transcripts within a diffraction-limited CHS, we developed a method using complete spatial random (CSR) simulation to assign distinct positions to each mRNA within the CHS. Spots that are significantly brighter (>3X) than the average are tested for multiple signal peaks by the signal integrated density. If multiple mRNAs are estimated in one detected spot, then random X, Y, Z coordinates are generated for each mRNA based on Gaussian probability in the space of corresponding CHS and the generated hypothetical mRNAs replace the single object detected by thresholding for more accurate assessment of their spatial clustering. Our final mRNA/nucleus detection results from the smFISH image shown in Figure 2A were very similar to the detection results obtained with FISH-quant [40] which is a widely-used software for detection/analysis of smFISH data (Supplementary Figure 3).
The final output is a record of the data about objects estimated to be real mRNA signals such as X, Y, Z coordinates, volume, integrated signal intensity, and nearest-neighbor distance in an output file (Figure 2B; Material 5: MatlabOutputExample.xls). Information for each image is recorded in a spreadsheet in the output files, which are generated for each strain or condition. With the current settings, image files with the same name except for numbering are treated as the same category. The MATLAB code <smFISH3D_pro2.m> summarizes the data obtained from multiple strains or conditions by <smFISH3D_pro1.m> and generates an Excel file with multiple spreadsheets, where all data are sorted as tables for convenience in performing statistical tests between strains or conditions all at once (Material 6: smFISH3D_pro2.m; Material 7: SummaryExample.xls).
4. Random simulation of mRNA positions
A random simulation is essential for statistically testing if a localization pattern is different from what would be expected by chance and to compare cells under different conditions [41,42]. We developed code to generate randomly-distributed mRNAs using Poisson distribution with the same mRNA density and cell shape as the experimental data. Our customized MATLAB code automatically creates 100 such simulations per image of mRNA locations using CSR. Regions outside of the cell and within nuclear area are excluded from the space where random mRNAs are generated. The nuclear area was defined based on the average size of A. gossypii nuclei (i.e. a sphere with 1-μm radius). The nuclear size should be modified depending on the species. For more sophisticated analyses, a 95% confidence interval is estimated for each image using the 100 CSR simulations by calculating parametric or non-parametric confidence intervals [43–45] depending on the shape of the random distribution. Random simulations enable assessment of the degree of mRNAs are clustering or dispersal compared to the distribution expected by chance. The data for random mRNAs are useful for many statistical tests (e.g. nearest-neighbor distance) because they can serve as the null hypothesis.
5. Spatial point pattern analysis
5.1 mRNA abundance and simple analyses
Once the 3-D information for mRNAs is collected, it needs to be systematically analyzed to examine the structure and scale of an mRNA localization pattern. One simple way to analyze this spatial information is to calculate the global or local mRNA density. The global mRNA density is defined as [total number of mRNAs in the cell] divided by [total cytoplasmic volume of the cell in the image] and is used to compare the mRNA expression level (or stability) between strains or conditions. The local mRNA density is defined as [number of mRNAs in the region of interest (ROI)] divided by [volume of ROI] and can provide insights into the regional regulation of transcripts. For example, mRNA of G1 cyclin CLN1/2 was found to be highly enriched at the hyphal tip by comparing the local mRNA density at the hyphal tips versus non-tip regions [2]. The enrichment of CLN1/2 at the tip implies active mRNA transport to the tip or mRNA retention at tips. To calculate how many mRNAs are around each nucleus, a spherical ROI with a user-defined radius (we frequently use 2 μm) is centered on each nucleus. The number of mRNAs in each ROI is recorded along with the position and volume of the ROI, which is adjusted if any of the ROI falls outside of the cell. The local mRNA density near each nucleus can be used to estimate how heterogeneous the number of mRNAs is between neighboring nuclei. Following are some examples of spatial analyses using local mRNA density: correlation between inter-nuclear distance and number of mRNAs around each nucleus, correlation between transcriptional activity of individual nuclei (signal intensity of THS) and number of surrounding mRNAs, and heterogeneity in mRNA abundance compared to CSR to test if mRNAs are significantly clustered or dispersed. Several simple spatial analyses are automatically done and recorded in the output file generated by the MATLAB code.
5.2 Ripley’s H function
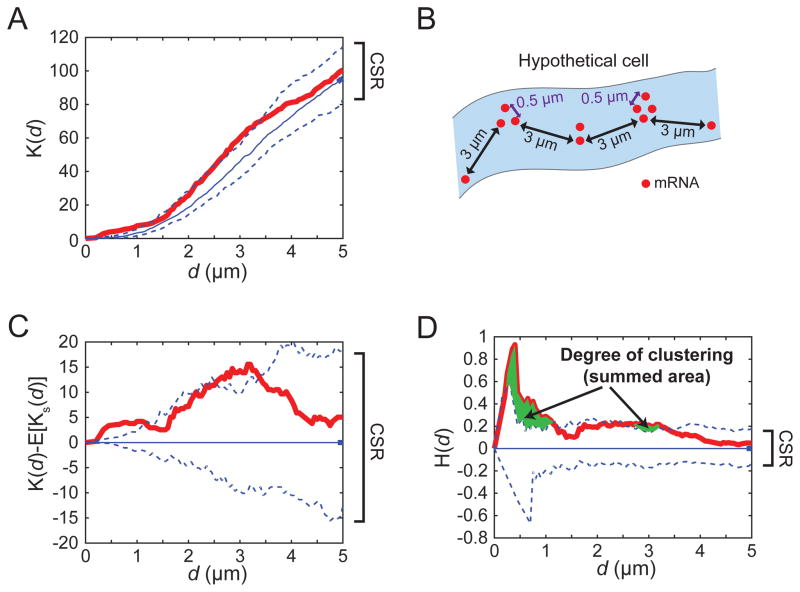
One of the biggest challenges in analyzing the spatial pattern of mRNAs is the fact that mRNA abundance is different between individual cells, strains (e.g. mutants in which mRNA abundance greatly changes) and between mRNA species due to differences in mRNA synthesis and degradation rates. We use the 3-D Ripley’s H function to quantitatively assess the spatial patterns in mRNA localization [2,12]. 3-D Ripley’s H function is derived from Ripley’s K function (Eq. 1), which calculates the ratio of the average number of mRNAs within a distance d of each mRNA to the overall mRNA density of the cell [29].
| Eq.1 |
where, n is the number of mRNA spots, V is the total volume of the cytoplasm, w is an edge correction function, I is a (0,1) indicator function, and D is the Euclidean distance. The procedure to calculate Ripley’s K(d) can conceptualized as follows: a sphere with a radius d is centered on each mRNA, and the total number of mRNAs within each sphere is determined. Then the sphere radius is increased (larger d), and the number of mRNAs is determined within each larger sphere. This process repeats up to a user-specified maximum d. These raw mRNA counts are converted to ratios to the total number of mRNA. These ratios estimate the portion of mRNA localized within a certain distance (d) of each other.
The value of Ripley’s K(d) is independent from the mRNA abundance because it is internally normalized as a ratio to the total mRNA. The Ripley’s K(d) output is a curve changing its ratio value from 0 on the Y-axis as the distance d increases on the X-axis (Figure 3A). The value of Ripley’s K(d) is independent from the mRNA abundance because it is internally normalized as a ratio to the total mRNA. The Ripley’s K(d) output is a curve changing its ratio value from 0 on the Y-axis as the distance d increases on the X-axis (Figure 3A). The curves obtained from the biological data and the random simulations must be compared to understand the spatial distribution of the transcripts. If mRNAs are randomly distributed, the Ripley’s K(d) value will increase at the same rate as the curve for randomly positioned mRNAs (i.e. if this were the case in Figure 3A, the red, data curve and blue, CSR curve would overlap). If mRNAs are clustered at a certain distance from each other, the ratio will increase at d greater than that for randomly positioned mRNA. Therefore, at d, the Ripley’s K curve for the biological data (red) will have values above the 95% CI for the the CSR (blue dotted line) (see red mRNA curve at d=0.5 and 3 μm in Figure 3A). Figure 3B illustrates hypothetical mRNA localization in a cell that could result in the Ripley’s K(d) output in Figure 3A. Conversely, experimental ratios that drop below the 95% CI indicate significant dispersal of the transcripts. Although the conventional Ripley’s H(d) (Figure 3C) takes the CSR into consideration, it only accounts for a single dimension, which results in an underestimation of the clustering at a closer distance (small d) in 3-D space [2,46]. The 3-D Ripley’s H(d) (Eq. 2; Figure 3D) resolves this issue by converting Ripley’s K(d) values obtained from 3-D space into 1 dimension. This conversion enables the direct comparison of the degree of clustering at a small d to that at a larger d. The 3-D Ripley’s H(d) is defined as Ripley’s K(d) normalized by the complete spatial randomness (CSR).
Figure 3.
Ripley’s spatial analysis (A) One example of Ripley’s K function plot. Red line: Ripley’s K function for actual mRNA. Blue line: median Ripley’s K function for 100 Poisson-distributed (CSR) simulations. Blue, dashed lines indicate the 95% confidence interval (CI) of 100 CSR simulations. (B) A hypothetical cell with mRNA clusters that could generate the Ripley’s K(d) result in A. (C) Ripley’s K(d)-E[Ks(d)] plot is derived from Figure 3A. E[Ks(d)] is Ripley’s K function for the simulations representing CSR. (D) Ripley’s H(d) is derived from Figure 3B to take account 3D space. The area where mRNA curve (Ripley’s H(d) value; red) breaches the upper 95% CI of CSR population (upper blue dashed line) is summed to estimate the degree of clustering (green regions).
| Eq.2 |
where, Ks(d) is the Ripley’s K function for the simulations representing CSR. This analysis is also called univariate Ripley’s H(d) since it analyzes just one object class (mRNA). A Ripley’s H(d) value breaching over the 95% confidence interval (CI) calculated from the CSR simluations indicates the statistically significant clustering of mRNAs at that d value. Conversely, values below the lower boundary of 95% CI indicate the dispersion of mRNA. We invented the term “degree of clustering” which is unit-less and can be used to compare clustering or dispersion between strains or conditions (Figure 3D). The degree of clustering is obtained by summing the area where Ripley’s H(d) deviates from the 95% CI of the random distribution. Similarly, Ripley’s H(d) can be modified to assess if mRNAs are clustered/dispersed at specific positions relative to nuclei. This is called the bivariate Ripley’s H(d) because two classes of objects (nucleus and mRNA) are involved in the analysis. Essentially the same principles as the univariate Ripley’s H(d) are applied to bivariate pattern analysis. The graphical output and values of both univariate and bivariate Ripley’s H(d) analysis are generated as output files for each image. In the output files, the Ripley’s H(d) is normalized by upper-(e.g. ‘bSummary4_strainName_H(t).jpg’) or lower-boundary (e.g. ‘cSummary4_strainName_H(t).jpg’) of the random population. The output plots of Ripley’s H(d) normalized by upper random boundary are useful to determine at what distance and to what degree clustering of mRNA occurs because any Ripley’s H(d) curve breach over the boundary line indicates significant clustering. Conversely, the plots normalized by the lower random boundary are useful for examining dispersed mRNA species. The median and 95% CI of each population are highlighted as described in previous studies in the lab [2,12]. Additionally, the Ripley’s H(d) curves from multiple images with the same strain and condition can be examined together and compared to other populations (e.g. Ripley’s H(d) for mRNAs in a mutant) using t-test or Wilcoxon test for parametric or non-parametric distributions, respectively.
6. Code execution
6.1 < 3D_OC_mask.ijm > (Material 1)
Before running this code, put all image files that are to be processed at the same time in one folder. This macro is designed to process 5-channel image stacks exported from Volocity 4.4 (PerkinElmer), which open in ImageJ as single-channel images with each channel concatenated in the Z-series. If using a multi-channel image file, or a file with a different number of channels, the usage “nSlices/5” (5 channels are concatenated) in this code must be modified properly as well as the “Channel” information in <smFISH3d_pro1.m>. In ImageJ, open the text editor (File>New>Script or press ‘[’) and open the “3D_OC_mask.ijm” macro. Open one image file (any of them) and select “Run” at the bottom of the text editor. The macro detects objects above the threshold designated in lines 33 (nuclei) and 51 (mRNA) and records all information in D:\3D-OC results\. If the folder path needs to be changed, especially in Mac OS for proper syntax, change line 23 as desired in “3D_OC_mask.ijm.” Once the macro has completed processing the image currently open, it will automatically close the current image, open the next image and process it until all of the images in the working folder are processed. There may be a pop-up message reporting an error during the image processing. In our experience, this typically occurs when the image being processed has very dim signal and no objects above threshold are detected. These images can be excluded from the image pool or a lower threshold for object detection can be used. In the event that smFISH signal is sufficiently variable between samples that different thresholds are required for different images, set the threshold for this macro to the lowest threshold for the set of images and proceed to using 3D_OC_multiThreshold.ijm (Section 6.2). After the object detection is done, there should be five files per image. Each result file has the same name as the image file with an additional suffix. ‘Filename_rna.xls’ contains information for RNA (X, Y, Z coordinates, volume, signal intensity, etc.) and ‘filename_nuc.xls’ contains the same information for nuclei. ‘Filename_mask.txt’ contains X and Y coordinates for the cell outline mask and ‘filename_mask.tif’ is an image of the mask. Finally, ‘filename_info.txt’ contains information for the image such as image size, Z-step size, pixel size, and so on. To make sure cell outline was properly detected, ‘filename_mask.tif’ should be compared to the actual phase image. Only the outline of the object contacting the centermost point is considered for masking and this is detected with the “Magic Wand” tool in ImageJ, this area should be bordered with a thin yellow line. If manual modification of the mask is necessary, detect the edge of the object with the wand tool after modification and overwrite both the filename_mask.tif and filename_mask.txt (File > Save As > XY Coordinates) results files. All these files are used in MATLAB code for further analyses and do not need to be edited.
6.2 < 3D_OC_multiThreshold.ijm > (Material 2)
We strongly recommend optimizing experimental and imaging conditions to result in the most homogeneous smFISH signal possible. However, if individual thresholds for each image remain necessary, save the threshold values in column format as a tab-delimited text file. Threshold values only must be in this file (no text), or ImageJ will not be able to process these values correctly. Thresholds must be listed in order of the images as the computer will process them. Zero-padding the numbered image files greatly simplifies predicting the order of image processing. Run the “3D_OC_mask.ijm” as described above and then open “3D_OC_multiThreshold.ijm” (Material 2). The path for input and output files should be specified on lines 10, 13, and 20. New ‘filename_rna.xls’ and ‘filename_info.txt’ result files are generated in the output folder after running the “3D_OC_multiThreshold.ijm.” Replace the files with these names generated by “3D_OC_mask.ijm” with the files output by 3D_OC_multiThreshold.ijm and then they are ready to be read by the MATLAB code.
6.3 < DetectionTest.m > (Material 3)
“DetectionTest.m” processes each image in the exact same way as “smFISH3D_pro1.m” only until the final list of nuclei and mRNAs is generated (after elimination of background objects and accounting for multiple transcripts in CHSs). In MATLAB, open “DetectionTest.m” and modify line 59, which designates the location of the input folder containing detection result files, which are generated by ImageJ macro “3D_OC_mask.ijm.” The preset is ‘D:\3D-OC results\’ which is consistent with the output folder preset for the ImageJ macro (Section 6.1). Modify the output folder as desired at lines 65, 67, and 68. The preset is ‘D:\smFISH Analysis Results\.’ Select “Run” in the top ribbon menu under “EDITOR” tab. The image files depicting nuclei as blue circles and mRNA as red dots are saved in the output folder with names consistent with the input files. These images should be compared to the original smFISH images to validate the precision of object detection. The threshold can be adjusted based on this comparison and the ImageJ image processing can be redone for better detection.
6.4 < smFISH3D_pro1.m > (Material 4)
Before running the code, modify the preset parameters for smFISH analyses if needed. Following are the parameters that can be modified: ‘radius (line 38)’ defines the size of the region of interest (ROI) within which the number of mRNAs are counted around each nucleus; ‘TRITC (line 44)’ is the order the mRNA (e.g. if mRNA signal is the 4th channel in the concatenated stack this value should be 4, be sure to use the correct channel if you have both a raw and deconvolved channel included in the image); ‘DAPI (line 45)’ specifies the order of the nuclear channel; ‘NSc (line 46)’ indicates the total number of channels the images have. Specify the input folder at line 68 and output folder at line 73. This code uses the ‘xlswrite1’ function. In order for it to run properly, ensure the xlswrite1 file (available through MATLAB Central File Exchange) is saved in your working path. The output Excel files which contain the analysis results for each image (one spreadsheet for each image) are saved in the output folder. The results for images with the same label except for numbering suffix will be together in one Excel file (Material 5). The ‘smFISH workspace 3D.mat’ file in the output folder contains all results in MATLAB form.
There are 3 subfolders in the output folder. The folder “Coor_reconstruct” has MATLAB-generated images of nuclei and mRNAs. The folder “K(d)” has plots for Ripley’s K(d) or H(d) for each images. The plots are separated in univariate or bivariate analysis folders within the “K(d)” folder. To calculate the degree of clustering, use the function code “cluHarea.m” (Material 8). The usage example: Harea_CLN3_WT = cluHarea (1, 1, ori_range_plot), where first parameter 1 indicates univariate analysis, the second parameter 1 indicates n-th strain (or condition; when multiple strains are processed at the same time), and the last parameter is the input variable in MATLAB, which should stay the same.
6.5 < smFISH3D_pro2.m > (Material 6)
This code can be run once “smFISH3D_pro1.m” has been run. Specify the input folder (the output folder from smFISH3D_pro2.m) at line 15 (preset: ‘D:\smFISH Analysis Results’) in “smFISH3D_pro2.m,” which will read and summarize the result files generated by “smFISH3D_pro1.m. This code uses the ‘xlswrite1’ function. In order for it to run properly, ensure the xlswrite1 file (available through MATLAB Central File Exchange) is saved in your working path. The Excel file for each strain (or condition) will be saved in the “Summary” subfolder (Material 7). Each spreadsheet contains a different type of analysis such as the shortest inter-transcript distance (ITD). There are labels at the top of each spreadsheet denoting the variable in each column.
7. Concluding remarks
Here we describe smFISH protocols to visualize individual transcripts, and customized ImageJ/MATLAB codes that systematically record smFISH data and analyze the spatial pattern of transcripts and nuclei. The smFISH technique can be performed with multiplexed probe sets for visualizing multiple mRNA species in the same cell. This technique can also be coupled with protein visualization using immunofluorescence. smFISH data can be systematically and quantitatively analyzed using spatial pattern analysis, including the Ripley’s H function discussed here. The Ripley’s H function enables systematic spatial pattern analysis even despite obstacles such as irregular cell shape and differences in mRNA abundance. By utilizing internal normalization and Poisson random simulations, the degree of clustering or dispersion can be compared between different strains or conditions. These analytical pipelines and protocols can potentially be adapted to other systems to provide insights into spatial control of gene expression.
Supplementary Material
Supplementary Figure 1. BNI1 smFISH in A. gossypii acquired using a confocal laser scanner (Leica LSM SP8). Images are shown as a maximum-intensity Z-projection. Gray dashed lines mark cell outlines. Scale bar: 5 μm.
Supplementary Figure 2. A flow chart schematic of the smFISH procedure for A. gossypii and C2C12 in parallel.
Supplementary Figure 3. Comparison of the transcript detection using FISH-quant or our customized MATLAB code (bottom). The image shown in Figure 2A was analyzed. Top: FISH-quant v2e was used. The threshold for mRNA signal intensity was 26 (max: 255); Bottom: Blue dots mark nuclei, red dots mark mRNA and gray dashed lines mark cell outline. The threshold for mRNA and nuclei were 120 and 40 (max: 255), respectively.
Highlights.
smFISH allows examination of the spatial distribution of individual transcripts.
smFISH can be combined with other cytological techniques such as immunofluorescence.
Protocols are provided with considerations for adaptation to other species.
An analysis pipeline and tools (ImagJ/MATLAB codes) are provided with example outputs.
Acknowledgments
We thank the Gladfelter Lab for useful discussions in the development of these tools. This work was funded in part by NIH grant R01GM081506 (A.S.G.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Weil TT, Forrest KM, Gavis ER. Localization of bicoid mRNA in Late Oocytes Is Maintained by Continual Active Transport. Dev Cell. 2006;11:251–262. doi: 10.1016/j.devcel.2006.06.006. [DOI] [PubMed] [Google Scholar]
- 2.Lee C, Zhang H, Baker AE, Occhipinti P, Borsuk ME, Gladfelter AS. Protein aggregation behavior regulates cyclin transcript localization and cell-cycle control. Dev Cell. 2013;25:572–84. doi: 10.1016/j.devcel.2013.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kershner AM, Shin H, Hansen TJ, Kimble J. Discovery of two GLP-1/Notch target genes that account for the role of GLP-1/Notch signaling in stem cell maintenance. Proc Natl Acad Sci U S A. 2014;111:3739–44. doi: 10.1073/pnas.1401861111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Roper M, Lee C, Hickey PC, Gladfelter AS. Life as a moving fluid: fate of cytoplasmic macromolecules in dynamic fungal syncytia. Curr Opin Microbiol. 2015;26:116–122. doi: 10.1016/j.mib.2015.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Long RM, Singer RH, Meng X, Gonzalez I, Nasmyth K, Jansen RP. Mating type switching in yeast controlled by asymmetric localization of ASH1 mRNA. Science. 1997;277:383–387. doi: 10.1126/science.277.5324.383. [DOI] [PubMed] [Google Scholar]
- 6.Roegiers F. Insights into mRNA transport in neurons. Proc Natl Acad Sci U S A. 2003;100:1465–6. doi: 10.1073/pnas.0630376100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buxbaum AR, Haimovich G, Singer RH. In the right place at the right time: visualizing and understanding mRNA localization. Nat Rev Mol Cell Biol. 2014;16:95–109. doi: 10.1038/nrm3918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jansen RP. mRNA localization: message on the move. Nat Rev Mol Cell Biol. 2001;2:247–256. doi: 10.1038/35067016. [DOI] [PubMed] [Google Scholar]
- 9.Baumann S, Konig J, Koepke J, Feldbrugge M. Endosomal transport of septin mRNA and protein indicates local translation on endosomes and is required for correct septin filamentation. EMBO Rep. 2014;15:94–102. doi: 10.1002/embr.201338037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jansen RP, Niessing D, Baumann S, Feldbrugge M. mRNA transport meets membrane traffic. Trends Genet. 2014;30:408–417. doi: 10.1016/j.tig.2014.07.002. [DOI] [PubMed] [Google Scholar]
- 11.Palacios IM. Hop-on hop-off: polysomes take a tour of the cell on endosomes. J Cell Biol. 2014;204:287–9. doi: 10.1083/jcb.201401019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee C, Occhipinti P, Gladfelter AS. PolyQ-dependent RNA-protein assemblies control symmetry breaking. J Cell Biol. 2015;208:533–44. doi: 10.1083/jcb.201407105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zeller R, Rogers M, Haramis AG, Carrasceo AS. In situ hybridization to cellular RNA. Curr Protoc Mol Biol. 2001;Chapter 14(Unit 14.3) doi: 10.1002/0471142727.mb1403s55. [DOI] [PubMed] [Google Scholar]
- 14.Brown JM, Buckle VJ. Detection of nascent RNA transcripts by fluorescence in situ hybridization. Methods Mol Biol. 2010;659:33–50. doi: 10.1007/978-1-60761-789-1_3. [DOI] [PubMed] [Google Scholar]
- 15.van Raamsdonk CD. Optimizing the detection of nascent transcripts by RNA fluorescence in situ hybridization. Nucleic Acids Res. 2001;29:42e–42. doi: 10.1093/nar/29.8.e42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Arvey A, Hermann A, Hsia CC, Ie E, Freund Y, McGinnis W. Minimizing off-target signals in RNA fluorescent in situ hybridization. Nucleic Acids Res. 2010;38:e115. doi: 10.1093/nar/gkq042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mattsson G, Tan SY, Ferguson DJP, Erber W, Turner SH, Marafioti T, et al. Detection of genetic alterations by immunoFISH analysis of whole cells extracted from routine biopsy material. J Mol Diagn. 2007;9:479–89. doi: 10.2353/jmoldx.2007.070041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kosman D, Mizutani CM, Lemons D, Cox WG, McGinnis W, Bier E. Multiplex detection of RNA expression in Drosophila embryos. Science. 2004;305:846. doi: 10.1126/science.1099247. [DOI] [PubMed] [Google Scholar]
- 19.Lecuyer E, Yoshida H, Parthasarathy N, Alm C, Babak T, Cerovina T, et al. Global analysis of mRNA localization reveals a prominent role in organizing cellular architecture and function. Cell. 2007;131:174–87. doi: 10.1016/j.cell.2007.08.003. [DOI] [PubMed] [Google Scholar]
- 20.Jambor H, Surendranath V, Kalinka AT, Mejstrik P, Saalfeld S, Tomancak P. Systematic imaging reveals features and changing localization of mRNAs in Drosophila development. Elife. 2015;4:e05003. doi: 10.7554/eLife.05003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Raj A, van Oudenaarden A. Nature, Nurture, or Chance: Stochastic Gene Expression and Its Consequences. Cell. 2008;135:216–226. doi: 10.1016/j.cell.2008.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Raj A, Rifkin SA, Andersen E, van Oudenaarden A. Variability in gene expression underlies incomplete penetrance. Nature. 2010;463:913–918. doi: 10.1038/nature08781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Raj A, van den Bogaard P, Rifkin SA, van Oudenaarden A, Tyagi S. Imaging individual mRNA molecules using multiple singly labeled probes. Nat Methods. 2008;5:877–9. doi: 10.1038/nmeth.1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Raj A, Tyagi S. Detection of individual endogenous RNA transcripts in situ using multiple singly labeled probes. 1. Elsevier Inc; 2010. [DOI] [PubMed] [Google Scholar]
- 25.Shaffer SM, Wu MT, Levesque MJ, Raj A. Turbo FISH: a method for rapid single molecule RNA FISH. PLoS One. 2013;8:e75120. doi: 10.1371/journal.pone.0075120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jafari-Mamaghani M, Andersson M, Krieger P. Spatial Point Pattern Analysis of Neurons Using Ripley’s K-Function in 3D. Front Neuroinform. 2010;4:9. doi: 10.3389/fninf.2010.00009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kiskowski MA, Hancock JF, Kenworthy AK. On the use of Ripley’s K-function and its derivatives to analyze domain size. Biophys J. 2009;97:1095–1103. doi: 10.1016/j.bpj.2009.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dixon PM. Ripley’s K function Theoretical K (t) function. Statistics (Ber) 2001:1–16. [Google Scholar]
- 29.Ripley BD. Spatial Statistics. 2004 doi: 10.1002/0471725218. [DOI] [Google Scholar]
- 30.Trcek T, Sato H, Singer RH, Maquat LE. Temporal and spatial characterization of nonsense-mediated mRNA decay. Genes Dev. 2013;27:541–51. doi: 10.1101/gad.209635.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Raj A, van Oudenaarden A. Single-molecule approaches to stochastic gene expression. Annu Rev Biophys. 2009;38:255–70. doi: 10.1146/annurev.biophys.37.032807.125928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Padovan-Merhar O, Nair GP, Biaesch AG, Mayer A, Scarfone S, Foley SW, et al. Single Mammalian Cells Compensate for Differences in Cellular Volume and DNA Copy Number through Independent Global Transcriptional Mechanisms. Mol Cell. 2015;58:339–52. doi: 10.1016/j.molcel.2015.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chacko N, Zhao Y, Yang E, Wang L, Cai JJ, Lin X. The lncRNA RZE1 Controls Cryptococcal Morphological Transition. PLoS Genet. 2015;11:e1005692. doi: 10.1371/journal.pgen.1005692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kwon S. Single-molecule fluorescence in situ hybridization: quantitative imaging of single RNA molecules. [accessed October 23, 2015];BMB Rep. 2013 46:65–72. doi: 10.5483/BMBRep.2013.46.2.016. http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=4133856&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Trcek T, Grosch M, York A, Shroff H, Lionnet T, Lehmann R. Drosophila germ granules are structured and contain homotypic mRNA clusters. Nat Commun. 2015;6:7962. doi: 10.1038/ncomms8962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Oka Y, Sato TN. Whole-mount single molecule FISH method for zebrafish embryo. Sci Rep. 2015;5:8571. doi: 10.1038/srep08571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Edelstein AD, Tsuchida MA, Amodaj N, Pinkard H, Vale RD, Stuurman N. Advanced methods of microscope control using μManager software. J Biol Methods. 2014;1:10. doi: 10.14440/jbm.2014.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Edelstein A, Amodaj N, Hoover K, Vale R, Stuurman N. Computer control of microscopes using μManager. Curr Protoc Mol Biol. 2010;Chapter 14(Unit14.20) doi: 10.1002/0471142727.mb1420s92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Senecal A, Munsky B, Proux F, Ly N, Braye FE, Zimmer C, et al. Transcription factors modulate c-Fos transcriptional bursts. Cell Rep. 2014;8:75–83. doi: 10.1016/j.celrep.2014.05.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mueller F, Senecal A, Tantale K, Marie-Nelly H, Ly N, Collin O, et al. FISH-quant: automatic counting of transcripts in 3D FISH images. Nat Methods. 2013;10:277–8. doi: 10.1038/nmeth.2406. [DOI] [PubMed] [Google Scholar]
- 41.Ullah M, Wolkenhauer O. Stochastic approaches in systems biology. Wiley Interdiscip Rev Syst Biol Med. 2010;2:385–397. doi: 10.1002/wsbm.78. [DOI] [PubMed] [Google Scholar]
- 42.Alfonsi A, Cances E, Turinici G, Di Ventura B, Huisinga W. Adaptive simulation of hybrid stochastic and deterministic models for biochemical systems. ESAIM Proc. 2005;14:1–13. doi: 10.1051/proc:2005001. [DOI] [Google Scholar]
- 43.Zhou XH, Dinh P. Nonparametric confidence intervals for the one- And two-sample problems. Biostatistics. 2005;6:187–200. doi: 10.1093/biostatistics/kxi002. [DOI] [PubMed] [Google Scholar]
- 44.Beaulieu-Prevost D. Confidence Intervals: From tests of statistical signifigance to confidence intervals, range hypotheses and substantial effects. Tutor Quant Methods Psychol. 2006;2:11–19. [Google Scholar]
- 45.Efron B. Nonparametric Standard Errors and Confidence Intervals. Can J Stat / La Rev Can Stat. 1981;9:139–158. doi: 10.2307/3314608. [DOI] [Google Scholar]
- 46.Ehrlich M, Boll W, Van Oijen A, Hariharan R, Chandran K, Nibert ML, et al. Endocytosis by random initiation and stabilization of clathrin-coated pits. Cell. 2004;118:591–605. doi: 10.1016/j.cell.2004.08.017. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. BNI1 smFISH in A. gossypii acquired using a confocal laser scanner (Leica LSM SP8). Images are shown as a maximum-intensity Z-projection. Gray dashed lines mark cell outlines. Scale bar: 5 μm.
Supplementary Figure 2. A flow chart schematic of the smFISH procedure for A. gossypii and C2C12 in parallel.
Supplementary Figure 3. Comparison of the transcript detection using FISH-quant or our customized MATLAB code (bottom). The image shown in Figure 2A was analyzed. Top: FISH-quant v2e was used. The threshold for mRNA signal intensity was 26 (max: 255); Bottom: Blue dots mark nuclei, red dots mark mRNA and gray dashed lines mark cell outline. The threshold for mRNA and nuclei were 120 and 40 (max: 255), respectively.