Abstract
β1–3-N-Acetylglucosaminyltransferases (β3GlcNAcTs) and β1–4-galactosyltransferases (β4GalTs) have been broadly used in enzymatic synthesis of N-acetyllactosamine (LacNAc)-containing oligosaccharides and glycoconjugates including poly-LacNAc, and lacto-N-neotetraose (LNnT) found in the milk of human and other mammals. In order to explore oligosaccharides and derivatives that can be synthesized by the combination of β3GlcNAcTs and β4GalTs, donor substrate specificity studies of two bacterial β3GlcNAcTs from Helicobacter pylori (Hpβ3GlcNAcT) and Neisseria meningitidis (NmLgtA), respectively, using a library of 39 sugar nucleotides were carried out. The two β3GlcNAcTs have complementary donor substrate promiscuity and 13 different trisaccharides were produced. They were used to investigate the acceptor substrate specificities of three β4GalTs from Neisseria meningitidis (NmLgtB), Helicobacter pylori (Hpβ4GalT), and bovine (Bβ4GalT), respectively. Ten of the 13 trisaccharides were shown to be tolerable acceptors for at least one of these β4GalTs. The application of NmLgtA in one-pot multienzyme (OPME) synthesis of two trisaccharides including GalNAcβ1–3Galβ1–4GlcβProN3 and Galβ1–3Galβ1–4Glc was demonstrated. The study provides important information for using these glycosyltransferases as powerful catalysts in enzymatic and chemoenzymatic syntheses of oligosaccharides and derivatives which can be useful probes and reagents.
Keywords: Carbohydrate, Glycosyltransferase, Donor substrate promiscuity, Acceptor substrate specificity, Enzymatic synthesis, One-pot multienzyme
Graphical abstract
1. Introduction
β1–3-N-Acetylglucosaminyltransferases (β3GlcNAcTs) and β1–4-galactosyltransferases (β4GalTs) have been broadly used in enzymatic synthesis of Galβ1–4GlcNAcβ1–3Gal-containing oligosaccharides and glycoconjugates such as poly-N-acetyllactosamine [poly-LacNAc, (-3Galβ1–4GlcNAcβ1-)n]-containing structures1–3 with or without additional fucosylation and/or sialylation4–10 as well as lacto-N-neotetraose (LNnT, Galβ1–4GlcNAcβ1–3Galβ1–4Glc)11–13 and longer LacNAc-containing oligosaccharides13 found in the milk of human and other mammals. Both mammalian and bacterial glycosyltransferases have been used in the synthesis of these structures.6, 8, 13–15
For β3GlcNAcTs, mammalian enzymes were found to be quite specific on the acceptor substrates.1, 15, 16 In comparison, recombinant β3GlcNAcT from Neisseria meningitidis (NmLgtA) was shown to be more promiscuous and was able to use both α- and β-galactosides as acceptors.15, 17 It was found to have low15 or no activity towards type 1 (Galβ1–3GlcNAc) or type 3 (Galβ1–3GalNAcα) galactoside acceptors.1, 6 Branched glycans or longer LacNAc repeats were less efficient acceptors for NmLgtA. More recently, a β3GlcNAcT from Helicobacter pylori (Hpβ3GlcNAcT) was cloned and characterized.1, 18 Hpβ3GlcNAcT was shown to be specific for the β-linked galactoside acceptors14 and prefer type 2 (Galβ1–4GlcNAc) acceptors but can also use type 1 (Galβ1–3GlcNAc), type 3 (Galβ1–3GalNAcα), and type 4 (Galβ1–3GalNAcβ) acceptors on both linear and branched glycan structures.1 Regarding donor substrate specificity, only a limited number of UDP-sugars have been tested for NmLgtA which was shown to tolerate uridine 5'-diphosphate-N-acetylglucosamine (UDP-GlcNAc), uridine 5'-diphosphate-N-acetylgalactosamine (UDP-GalNAc), UDP-4-deoxy-GalNAc, UDP-6-deoxy-GalNAc/GlcNAc, and UDP-N-propanoylglucosamine.15, 19 The donor substrate specificity of Hpβ3GlcNAcT has not been explored in detail.
Bovine β4GalT 1 (Bβ4GalT) is the most well studied mammalian glycosyltransferase that has been used for biosynthesis.20, 21 In addition, β4GalTs have been cloned from Neisseria meningitidis, Neisseria gonorrhoeae, and Helicobacter pylori.21–25 Neisseria meningitidis β4GalT (NmLgtB) and Helicobacter pylori β4GalT (Hpβ4GalT), and Bβ4GalT have been used broadly for enzymatic and chemoenzymatic synthesis of β1–4-linked galactosides.13, 21, 26–30 NmLgtB has been fused with a Streptococcus thermophilus UDP-galactose 4-epimerase (NmLgtB-StGalE) to allow the use of a less expensive sugar nucleotide uridine 5'-disphosphate glucose (UDP-Glc) as the starting material which can be converted to uridine 5'-disphosphate galactose (UDP-Gal) by StGalE for NmLgtB-catalyzed synthesis of β1–4-linked galactosides.31 Although the tolerance of the modifications on the acceptor substrates of β4GalTs has been tested using monosaccharide derivatives,21, 32 their tolerance of modifications on longer oligosaccharides such as trisaccharides is less clear.
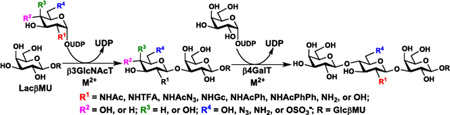
In order to explore the scope of oligosaccharide derivatives that can be synthesized by the combination of β3GlcNAcTs and β4GalTs, herein we report the donor substrate specificities of NmLgtA and Hpβ3GlcNAcT using 4-methylumbelliferyl (MU) β-lactoside Galβ1–4GlcβMU (or LacβMU) as an acceptor and a library of 39 sugar nucleotides including 37 UDP-sugars, cytidine 5'-monophosphate N-acetylneuraminic acid (CMP-Neu5Ac), and guanosine 5'-diphosphate L-fucose (GDP-Fuc) as potential donors. Thirteen trisaccharides have been successfully obtained by NmLgtA or Hpβ3GlcNAcT-catalyzed reactions and have been used to investigate the acceptor substrate specificities of NmLgtB, Hpβ4GalT, and Bβ4GalT (Scheme 1). We have also demonstrated the application of NmLgtA in one-pot multienzyme (OPME) preparative-scale synthesis of two trisaccharides including GalNAcβ1–3Galβ1–4GlcβProN3 and Galβ1–3Galβ1–4Glc from simple monosaccharides and suitable acceptors. The study provides important information for choosing suitable catalysts in enzymatic and chemoenzymatic synthesis of desired glycans and glycoconjugates that can be useful probes or reagents.
Scheme 1.
Donor substrate specificity and acceptor substrate specificity studies of β3GlcNAcTs and β4GalTs, respectively. LacβMU was used as an acceptor for β3GlcNAcTs (NmLgtA or Hpβ3GlcNAcT) and the resulting trisaccharide products were used as acceptor substrates for β4GalTs (NmLgtB, Hpβ4GalT, or Bβ4GalT).
2. Results and discussion
2.1. Cloning and expression of β3GlcNAcTs and β4GalTs
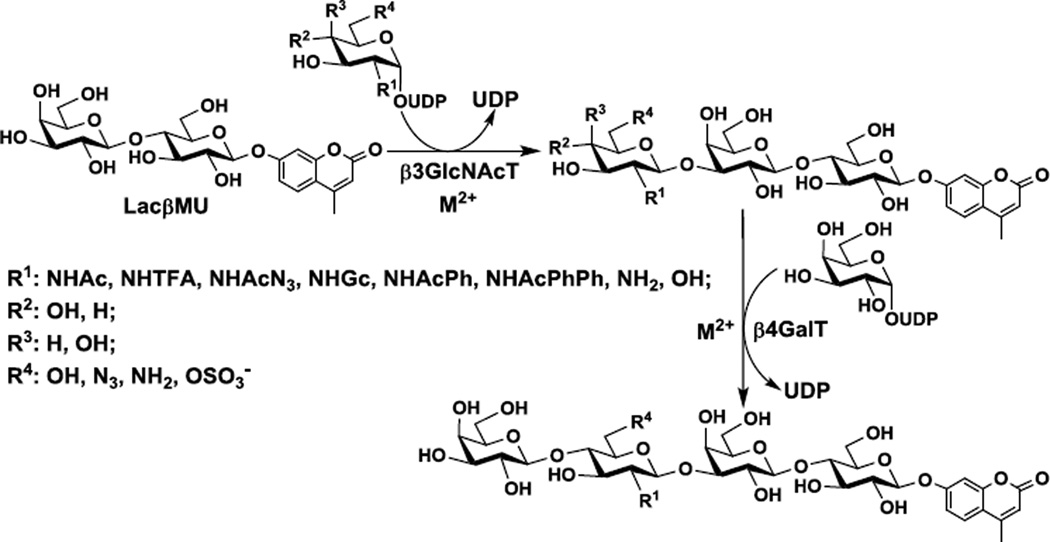
Both NmLgtA15 and Hpβ3GlcNAcT1 have been cloned and expressed in Escherichia coli previously. In our attempts, initial cloning into pET15b vectors led to a low expression of soluble and active enzymes in Escherichia coli BL21(DE3) cells. In order to improve the protein expression, NmLgtA and Hpβ3GlcNAcT were cloned into pMAL-c2X and pMAL-c4X vectors, respectively, both as N-terminal maltose-binding protein (MBP)-fused and C-terminal hexa-histidine (His6)-tagged recombinant proteins. Soluble and active enzymes could be obtained by inducing Escherichia coli BL21(DE3) cells with 0.1 mM of isopropyl β-D-1-thiogalactopyranoside (IPTG) followed by incubation at 20 °C for 20 hours. Purification was achieved by one-step nickel-nitrilotriacetic acid (Ni2+-NTA) affinity chromatography. About 10 mg of NmLgtA and only about 1 mg of Hpβ3GlcNAcT could be obtained from 1 liter of Escherichia coli cell culture. The differences of their expression levels could be contributed by their protein sequence differences as the two share only 16% protein sequence similarity (http://imed.med.ucm.es/Tools/sias.html) (Fig. 1A). Sodium dodecylsulfate-polyacrylamide gel electrophoresis (SDS-PAGE) analysis indicated that the apparent molecular weights of purified Hpβ3GlcNAcT (Fig. 1B) and NmLgtA (Fig. 1C) were about 88 kDa and 76 kDa respectively, which were consistent with their calculated molecular weights of 88.9 kDa and 76.6 kDa. Recombinant Bβ4GalT was usually expressed in Escherichia coli as inclusion bodies and active soluble enzyme was obtained after proper refolding procedures.33, 34 We found that when cloned in vector plasmid pET15b as an N-His6-tagged fusion protein with an N-terminal 128 amino acid truncation, soluble and active Bβ4GalT was readily obtained without refolding although its expression level (< 1 mg L−1 culture) was low (Fig. S1). In comparison, NmLgtB and Hpβ4GalT (previously named as Hp1–4GalT) were obtained at expression levels of 12.5 mg and 5 mg per liter of culture, respectively, as reported previously.21
Figure 1.
Protein sequence alignment (A) and SDS-PAGE analysis (8% Tris-Glycine gel) of Hpβ3GlcNAcT (B) and NmLgtA (C). Lanes: BI, whole cell extract before induction; AI, whole cell extract after induction; L, lysate after induction; P, Ni2+-NTA column purified protein; M, protein markers (Bio-Rad precision Plus Protein Standards, 10–250 kDa).
2.2. pH Profile and metal effect of β3GlcNAcTs
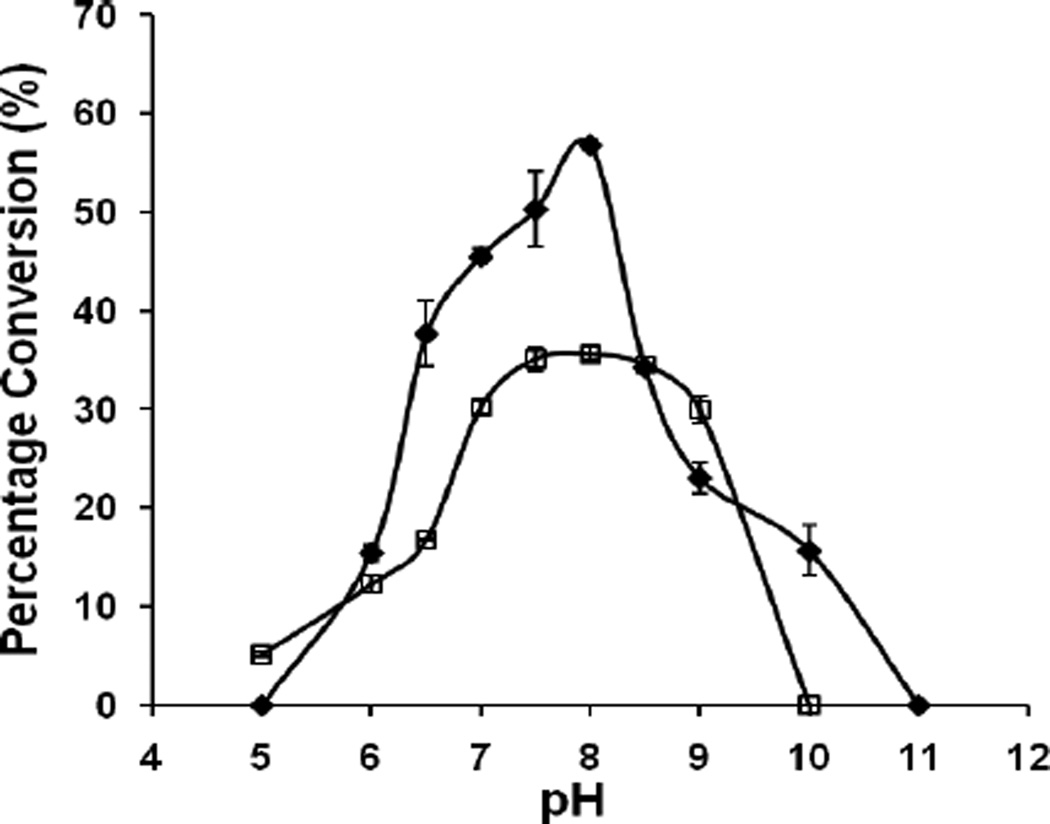
When 4-methylumbelliferyl β-lactoside (LacβMU) was used as the acceptor, the observed optimal pH for both NmLgtA and Hpβ3GlcNAcT was 8.0 (Fig. 2). The slight different optimal pH observed for NmLgtA compared to the previously reported pH 7.515 may be caused by the differences of the buffers (Tris-HCl buffer for this study versus sodium cacodylate buffer used before) used. Both enzymes retain activities in a pH range of 6.0–9.0 although at pH 10.0, only NmLgtA was active.
Figure 2.
pH profiles of NmLgtA (filled diamond) and Hpβ3GlcNAcT (open square). Buffers used were: MES, pH 5.0–6.5; Tris-HCl, pH 7.0–9.0; CAPS, pH 10.0–11.0.
The effects of several divalent metal ions including Mn2+, Mg2+, and Ca2+, as well as ethylenediaminetetraacetic acid (EDTA) on the activity of both β3GlcNAcTs were investigated. As shown in Fig. S2, both Hpβ3GlcNAcT (Fig. S2A) NmLgtA (Fig. S2B) required a divalent metal ion for activity as adding 10 mM of EDTA completed abolished their activities. Hpβ3GlcNAcT exhibited optimal activity in the presence of Mg2+. Increasing the concentration of Mg2+ from 1 mM to 10 mM did not affect the activity of Hpβ3GlcNAcT significantly, while its activity dropped slightly when the concentration of Mg2+ increased from 10 mM to 20 mM. Mn2+ was shown to be a less efficient cofactor for Hpβ3GlcNAcT and increasing the concentration of Mn2+ from 1 mM to 20 mM caused gradual decrease of Hpβ3GlcNAcT activity. Ca2+ was shown to be a less efficient cofactor for Hpβ3GlcNAcT compared to Mg2+ and its concentration ranging from 1–20 mM did not affect the Hpβ3GlcNAcT activity significantly. Unlike Hpβ3GlcNAcT, NmLgtA exhibited the best activity in the presence of Mn2+ with an optimal activity observed in the presence of 10 mM of Mn2+. Increasing or decreasing the concentrations of Mn2+ led to the decrease of NmLgtA activity. Although a less effective cofactor compared to Mn2+ at concentrations at or lower than 10 mM, increasing the concentration of Mg2+ from 1 mM to 20 mM led to a gradual increase of NmLgtA activity. Ca2+ was not a suitable cofactor for NmLgtA. The results of the metal effect of NmLgtA were consistent to those reported previously.15
2.3. Donor Substrate Specificities of β3GlcNAcTs
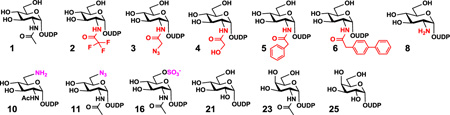
Once the optimal conditions were identified for Hpβ3GlcNAcT and NmLgtA, donor substrate specificity studies were carried out using a library of 39 sugar nucleotides (Fig. S3) including 37 UDP-sugars as well as CMP-Neu5Ac and GDP-Fuc. UDP-sugars used in the assays included UDP-GlcNAc (1) and its 2'-, 3'-, and/or 6'-modified derivatives (2–20), UDP-glucose (UDP-Glc) (21), UDP-glucuronic acid (UDP-GlcA) (22), UDP-N-acetylgalactosamine (UDP-GalNAc) (23) and its 6'-modified derivative (24), UDP-galactose (UDP-Gal) (25) and its 6'-modified derivative (26), UDP-galacturonic acid (UDP-GalA) (27), UDP-N-acetylmannosamine (UDP-ManNAc) (28) and its 2'-modified derivatives (29–35), UDP-mannose (36) and its 2'-modified derivative (37), as well as CMP-Neu5Ac (38), and GDP-Fuc (39) (Fig. S3). The reactions were analyzed by capillary electrophoresis (CE) equipped with a photodiode array (PDA) detector and were monitored at 315 nm to determine the ratio of the UV active fluorescent trisaccharide product formed and the fluorescent LacβMU acceptor. High performance liquid chromatography (HPLC) was also used to assist the analysis for some of the substrates. The presence of each trisaccharide product was further confirmed by high resolution mass spectrometry (HRMS).
As shown in Table 1, both β3GlcNAcTs exhibited promiscuous and somewhat complementary substrate specificities. NmLgtA was able to use 11 UDP-sugars and Hpβ3GlcNAcT could take 7 UDP-sugars tested as donor substrates. The estimated yields based on high-resolution mass spectrometry (HRMS) matched well with the capillary electrophoresis and/or high performance liquid chromatography (HPLC) results. For the reaction using UDP-Gal (25), the product and the acceptor peaks had the same retention time and could not be separated by CE using sodium tetraborate (25 mM, pH 9.4) as a running buffer but could be resolved by CE using sodium tetraborate (50 mM, pH 11.0) as a running buffer. For reactions using compounds 8, 10, 16, and 23, neither CE nor HPLC could separate the acceptor and the product peaks. However, the product formation was verified by HRMS analysis. In addition to UDP-GlcNAc (1), both β3GlcNAcTs could use 2'-modified UDP-GlcNAc derivatives including UDP-GlcNTFA (2), UDP-GlcNAcN3 (3), and UDP-GlcNGc (4) as well as a 6'-modified UDP-GlcNAc derivative UDP-GlcNAc6NH2 (10) as donor substrates. UDP-GlcNTFA (2) containing an N-trifluoroacetyl group was an excellent donor substrate for both β3GlcNAcTs. In contrary to a previous report indicating UDP-GlcNAcN3 (3) containing an N-azidoacetyl group as an intolerable donor for NmLgtA,19 the compound was confirmed by both HRMS and CE assays to be an acceptable donor substrate for both β3GlcNAcTs although the yields were not high.
Table 1.
Donor substrate specificities of Hpβ3GlcNAcT and NmLgtA analyzed by HRMS and CE or HPLC. LacβMU was used as the acceptor. Reactions were carried out at 37 °C for 24 hours.
 | |||||
|---|---|---|---|---|---|
| # | Donors | NmLgtA | Hpβ3GlcNAcT | ||
| HRMSa | CEb,c or HPLCd | HRMSa | CEb or HPLCd | ||
| 1 | UDP-GlcNAc | 90–95% | 81.2±0.8%b | 55–60% | 66.2±1.3%b |
| 2 | UDP-GlcNTFA | 100% | 100±0%b | 90–95% | 92.9±0.3%b |
| 3 | UDP-GlcNAcN3 | 25–35% | 29.1±0.1%b | 20–30% | 26.3±0.4 %b |
| 4 | UDP-GlcNGc | 30–40% | 36.1±0.8%b | 55–60% | 77.5±0.1%b |
| 5 | UDP-GlcNAcPh | 20–25% | 12.9±0.5%b | 0 | 0 |
| 6 | UDP-GlcNAcPhPh | <5% | N.D. | 0 | 0 |
| 8 | UDP-GlcNH2 | 20–25% | peaks togetherd | 0 | 0 |
| 10 | UDP-GlcNAc6NH2 | 20–25% | peaks togetherd | 70–75% | peaks togetherd |
| 11 | UDP-GlcNAc6N3 | 0 | 0 | 30–35% | 27.8±1.1%b |
| 16 | UDP-GlcNAc6S | 0 | 0 | 65–70% | peaks togetherd |
| 21 | UDP-Glc | 20–25% | 23.2±2.6%b | 0 | 0 |
| 23 | UDP-GalNAc | 80–90% | peaks togetherd | 0 | 0 |
| 25 | UDP-Gal | 40–45% | 39.6±0.6%c | 0 | 0 |
Yields estimated by HRMS.
Quantitative yields obtained by CE using sodium tetraborate (25 mM, pH 9.4) as the running buffer.
Quantitative yields obtained by CE using sodium tetraborate (50 mM, pH 11.0) as the running buffer.
Results obtained by HPLC. The acceptor and the product peaks had the same retention time and could not be separated.
N.D: not detected by CE or HPLC.;
Quite interestingly, two β3GlcNAcTs showed some complementary donor substrate specificities. For example, NmLgtA could tolerate UDP-GlcNAc derivatives with a small or a large substituent at C-2 of the GlcNAc including UDP-GlcNAcPh (5) with an N-phenylacetyl group, UDP-GlcNAcPhPh (6) with an N-diphenylacetyl group, and UDP-GlcNH2 (8) with an amino group. However, these UDP-sugars were not acceptable donor substrates for Hpβ3GlcNAcT. On the other hand, 6'-modifications of UDP-GlcNAc were more tolerable by Hpβ3GlcNAcT compared to NmLgtA. For example, UDP-GlcNAc6NH2 (10) containing a 6'-amino group was a better donor substrate for Hpβ3GlcNAcT and the reaction was achieved with a higher yield than that with NmLgtA. In addition, UDP-GlcNAc6N3 (11) containing an 6'-azido group and UDP-GlcNAc6S (16) containing an 6'-O-sulfate group were suitable donor substrates for Hpβ3GlcNAcT, but not for NmLgtA. It was quite exciting to notice the well tolerance of UDP-GlcNAc6S (16) which contains a relatively large and negatively charged O-sulfate group at C-6 of GlcNAc as the donor substrate by Hpβ3GlcNAcT. In consistent with previous reports,15, 19 UDP-GalNAc (23) was an excellent donor substrate for NmLgtA. The compound, however, was not a donor for Hpβ3GlcNAcT. To our surprise, unlike a previous report indicating both UDP-Glc (21) and UDP-Gal (25) were not donor substrate for NmLgtA,15 our results showed that both of these UDP-sugars were suitable donor substrates for NmLgtA but not for Hpβ3GlcNAcT.
Neither β3GlcNAcTs could use UDP-GlcA (22), UDP-GalNAc (24), UDP-Gal (26), UDP-GalA (27), UDP-ManNAc (28) or its derivatives (29–35), UDP-mannose (36), UDP-mannose (37), CMP-Neu5Ac (38), and GDP-Fuc (39) (Fig. S3) as donor substrates under the reaction conditions used.
2.4. Kinetic studies of β3GlcNAcTs
Due to their potential in synthetic applications and chemical biological studies, three UDP-sugars including UDP-GlcNAc (1), UDP-GlcNTFA (2), and UDP-GlcNAcN3 (3) that could be used by both β3GlcNAcTs were chosen for kinetic studies. As shown in Table 2, in general the catalytic efficiency (kcat/KM) of NmLgtA was higher than that of Hpβ3GlcNAcT no matter which substrate among the three was used. Compared to UDP-GlcNTFA (2), the catalytic efficiencies of both β3GlcNAcTs were higher (~3-fold) when UDP-GlcNAc (1) was used as the donor substrate mainly due to the higher kcat values observed for UDP-GlcNAc (1). Compared to UDP-GlcNTFA (2), the catalytic efficiencies of both β3GlcNAcTs were lower (3–4-fold) when UDP-GlcNAcN3 (3) was used as the donor substrate mainly due to the higher KM values observed for UDP-GlcNAcN3 (3).
Table 2.
Apparent kinetic data for Hpβ3GlcNAcT and NmLgtA.
| NmLgtA | Hpβ3GlcNAcT | |||||
|---|---|---|---|---|---|---|
| Substrate | kcat (s−1) | KM (mM) | kcat/KM (s−1 mM−1) | kcat (s−1) | KM (mM) | kcat/KM (s−1 mM−1) |
| UDP-GlcNAc (1) | 0.55 ± 0.13 | 0.47 ± 0.11 | 1.16 | 0.46 ± 0.08 | 1.66 ± 0.28 | 0.68 |
| UDP-GlcNTFA (2) | 0.19 ± 0.06 | 0.59 ± 0.20 | 0.32 | 0.32 ± 0.08 | 1.23 ± 0.31 | 0.26 |
| UDP-GlcNAcN3 (3) | 0.15 ± 0.03 | 1.34 ± 0.24 | 0.11 | 0.29 ± 0.07 | 4.74 ± 1.09 | 0.062 |
2.5. Acceptor substrate specificities of β4GalTs
Thirteen trisaccharides could be obtained by reactions catalyzed by NmLgtA and/or Hpβ3GlcNAcT. They were used to test the acceptor substrate specificities of three β4GalTs including NmLgtB, Hpβ4GalT, and Bβ4GalT with UDP-Gal as the donor substrate. Protein sequence alignment (Fig. S4) showed that these enzymes share very low sequence identity. For example, 7.3% sequence identity was found for Hpβ4GalT and NmLgtB, 6.2% for Bβ4GalT and Hpβ4GalT, and 2.9% for NmLgtB and Bβ4GalT. The overall sequence similarity among the three was less than 16%.
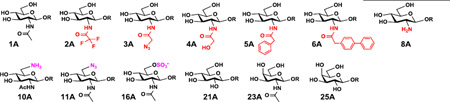
Despite their low sequence homology, NmLgtB, Hpβ4GalT, and Bβ4GalT are effective β4GalTs and can use several trisaccharides and derivatives produced by β3GlcNAcT-catalyzed reactions as acceptors. As shown in Table 3, ten of the 13 trisaccharides could be used as acceptors by either NmLgtB or Hpβ4GalT or both. Bβ4GalT could use only four as acceptors (HRMS data are shown in Fig. S5). Besides GlcNAc-containing trisaccharide GlcNAcβ1–3LacβMU (1A), all three β4GalTs could use 2'-modified GlcNAc-containing trisaccharides GlcNTFAβ1–3LacβMU (2A), GlcNAcN3β1–3LacβMU (3A), and GlcNAcPhβ1–3LacβMU (5A) as acceptors. While GlcNTFA-containing trisaccharide GlcNTFAβ1–3LacβMU (2A) was well tolerated by Bβ4GalT, other 2'-modifications on GlcNAc decrease its activity more significantly. 6'-Modification completely abolished its activity. While not acceptable by Bβ4GalT, GlcNGcβ1–3LacβMU (4A) was an excellent acceptor for both NmLgtB and Hpβ4GalT.
Table 3.
Acceptor substrate specificities of β4GalTs using a sequential two-step process.a
 | ||||||
|---|---|---|---|---|---|---|
| # | Acceptor | β3GlcNAcT* | NmLgtB | Hpβ4GalT | Bβ4GalT | |
| 1A | GlcNAcβ1–3LacβMU | 90–95% | NmLgtA | 100% | 100% | 90–95% |
| 2A | GlcNTFAβ1–3LacβMU | 100% | NmLgtA | 95–100% | 100% | 60–65% |
| 3A | GlcNAcN3β1–3LacβMU | 25–35% | NmLgtA | 100% | 100% | 35–40% |
| 4A | GlcNGcβ1–3LacβMU | 55–60% | Hpβ3GlcNAcT | 90–95% | 75–80% | 0 |
| 5A | GlcNAcPhβ1–3LacβMU | 20–25% | NmLgtA | 100% | 50% | 20–25% |
| 6A | GlcNAcPhPhβ1–3LacβMU | <5% | NmLgtA | 100% | 0 | 0 |
| 8A | GlcNH2β1–3LacβMU | 20–25% | NmLgtA | 0 | 0 | 0 |
| 10A | GlcNAc6NH2β1–3LacβMU | 70–75% | Hpβ3GlcNAcT | 0 | 20% | 0 |
| 11A | GlcNAc6N3β1–3LacβMU | 30–35% | Hpβ3GlcNAcT | 5–10% | 100% | 0 |
| 16A | GlcNAc6Sβ1–3LacβMU | 65–70% | Hpβ3GlcNAcT | 0 | 100% | 0 |
| 21A | Glcβ1–3LacβMU | 20–25% | NmLgtA | 95–100% | 0 | 0 |
| 23A | GalNAcβ1–3LacβMU | 80–90% | NmLgtA | 0 | 0 | 0 |
| 25A | Galβ1–3LacβMU | 40–45% | NmLgtA | 0 | 0 | 0 |
Yields estimated by HRMS. R= LacβMU.
LacβMU was used as the acceptor for β3GlcNAcT-catalyzed reactions. The trisaccharide producing reactions with a higher yield (catalyzed by either Hpβ3GlcNAcT or NmLgtA) were used for the acceptor substrate specificity studies of β-4GalTs.
In consistent with those observed previously using monosaccharide derivatives as acceptors,21 NmLgtB was more promiscuous towards acceptors containing a terminal 2'-modified GlcNAc, while Hpβ4GalT was more tolerable towards acceptors containing a terminal 6'-modified GlcNAc. For example, trisaccharide GlcNAcPhPhβ1–3Galβ1–4GlcβMU (6A) containing a terminal N-diphenylacetylglucosmine (GlcNAcPhPh) was a suitable acceptor for NmLgtB but not for Hpβ4GalT. On the other hand, GlcNAc6NH2β1–3LacβMU (10A) and GlcNAc6Sβ1–3LacβMU (16A) could be used as acceptors by Hpβ4GalT but not NmLgtB. In addition, a high yield (100%) was achieved when GlcNAc6N3β1–3LacβMU (11A) was used as an acceptor for Hpβ4GalT compared to a low yield (5–10%) for NmLgtB. Quite excitingly, the Glcβ1–3LacβMU (21A) obtained by NmLgtA-catalyzed reaction can be extended further by NmLgtB for the formation of presumably Lacβ1–3LacβMU tetrasaccharide. It is interesting to notice that the trisaccharide derivatives formed by NmLgtA-catalyzed reaction were more tolerable acceptors by NmLgtB. Similarly, Hpβ3GlcNAcT and Hpβ4GalT seemed to also work as a good pair in tolerating modified glycan substrates.
2.6. Preparative-scale synthesis of GalNAcβ1–3Galβ1–4GlcβProN3 and Galβ1–3Galβ1–4Glc
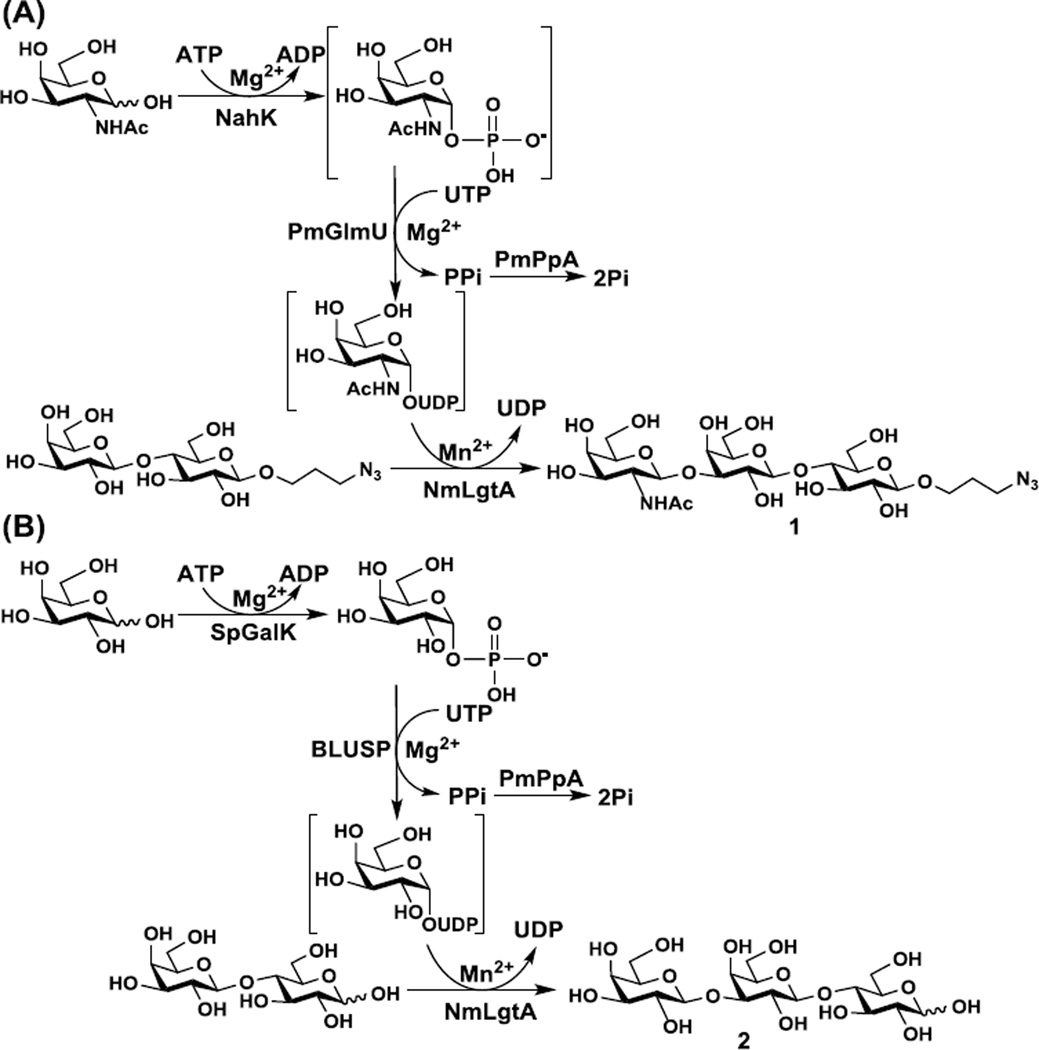
Taking advantage of the interesting tolerance of UDP-GalNAc and UDP-Gal as the donor substrates by NmLgtA, preparative-scale synthesis of trisaccharides GalNAcβ1–3Galβ1–4GlcβProN3 (1) (Scheme 2A) and Galβ1–3Galβ1–4Glc (2) (Scheme 2B) were carried out.
Scheme 2.
One-pot four-enzyme synthesis of GalNAcβ1–3Galβ1–4GlcβProN3 (23A) (A) and Galβ1–3Galβ1–4Glc (25A) (B). Enzyme abbreviations: NahK, Bifidobacterium longum ATCC55813 N-acetylhexosamine 1-kinase;37 PmGlmU, Pasteurella multocida N-acetylglucosamine-1-phosphate uridylyltransferase;38 PmPpA, Pasteurella multocida inorganic pyrophosphatase;21 SpGalK, Streptococcus pneumoniae TIGR4 galactokinase;39 BLUSP, Bifidobacterium longum UDP-sugar pyrophosphorylase.40
GalNAcβ1–3Galβ1–4GlcβOR component has been found in glycosphingolipids identified in rat granuloma, rat abdominal macrophage,35 starfish Asterias amurensis versicolor Sladen.36 As shown in Scheme 2A, GalNAcβ1–3Galβ1–4GlcβProN3 (1) was synthesized from Galβ1–4GlcβProN3 and simple free monosaccharide GalNAc using a one-pot four-enzyme (OP4E) system containing Bifidobacterium longum ATCC55813 N-acetylhexosamine 1-kinase (NahK),37 Pasteurella multocida N-acetylglucosamine- 1-phosphate uridylyltransferase (PmGlmU),38 Pasteurella multocida inorganic pyrophosphatase (PmPpA),21 and NmLgtA. In this system, NahK was responsible for the formation of GalNAc-1-phosphate from GalNAc and adenosine 5'-triphosphate (ATP), PmGlmU catalyzed the formation of UDP-GalNAc which was used by NmLgtA to catalyze the transfer of GalNAc to the acceptor for the formation of the trisaccharide product. PmPpA was used to break down the pyrophosphate formed in the UDP-GalNAc production step to drive the reaction towards the formation of the sugar nucleotide. This OP4E system was quite efficient in synthesizing GalNAcβ1–3Galβ1–4GlcβProN3 (1) and the yield was quantitative. Compared to a previous report,15 the OP4E process here avoided the use of relatively expensive UDP-GalNAc and allowed the production of the desired glycan in a high-yield more economic process. As shown in Table S1, comparing the 13C nuclear magnetic resonance (NMR) chemical shifts of the disaccharide LacβProN3 and the trisaccharide product indicated that significant changes occurred at C-3 of internal Gal residue (a downfield shift of 9.26 ppm from 72.37 ppm in the disaccharide starting material to 81.63 ppm in the trisaccharide product). This confirmed that the glycosylation took place at C-3 of the Gal residue of LacβProN3. The anomeric proton of terminal GalNAc in the trisaccharide product had a chemical shift at 4.58 ppm as a doublet with a coupling constant J1,2= 8.8 Hz indicates the formation of a β-linkage. These results confirmed the structure of the trisaccharide product as GalNAcβ1–3Galβ1–4GlcβProN3 (1).
Galβ1–3Galβ1–4Glc (3'-galactosyllactose) has been found in human milk41 and eastern quoll (Dasyurus viverrinus) milk42 as well as in the colostrums of bovine,43 ovine,44 primate,45 camel,46 and horse.47 It is also an abundant oligosaccharide in the milk of elephant48 and marsupials such as brushtail possum (Trichosurus vulpecula),49 tammar wallaby (Macropus eugenii) and grey kangaroo,50 and koala (Phascolarctos cinereus).51 As shown in Scheme 2B, Galβ1–3Galβ1–4Glc (2) was synthesized from lactose and galactose (Gal) using a one-pot four-enzyme (OP4E) system containing a Streptococcus pneumoniae TIGR4 galactokinase (SpGalK),39 a Bifidobacterium longum ATCC55813 UDP-sugar pyrophosphorylase (BLUSP),40 PmPpA,21 and NmLgtA. SpGalK, BLUSP, and PmPpA were used previously for high efficient (85% yield) one-pot multienzyme (OPME) synthesis of UDP-Gal.40 As UDP-Gal was a relatively weak substrate for NmLgtA, the synthetic yield (10%) of Galβ1–3Galβ1–4Glc (25A) was lower compared to the yield (40–45%) shown in Table 1, which may be caused by lactose as a less effective acceptor than the fluorophore-tagged beta-linked lactoside (LacβMU) similar to that observed previously.15 Nevertheless, this is a tailor-made process of synthesizing 3'-galactosyllactose which is not readily available by reactions catalyzed by other glycosyltransferases. The chemical structure of the trisaccharide product was confirmed by NMR. As shown in Table S2, comparing the 13C NMR chemical shifts of the disaccharide lactose and the trisaccharide product indicated that significant changes occurred at C-3 of internal Gal residue (a downfield shift of 9.37 ppm from 72.38 ppm in the disaccharide starting material to 81.75 ppm in the trisaccharide product). This confirmed that the glycosylation took place at C-3 of the Gal residue of lactose. The anomeric proton of terminal Gal in the trisaccharide product had a chemical shift at 4.59 ppm as a doublet with a coupling constant J1,2= 8.0 Hz indicates the formation of a β-linkage. These results confirmed the structure of the trisaccharide product as Galβ1–3Galβ1–4Glc (2).
3. Conclusions
The donor substrate specificity of two β1–3-N-acetylglucosaminyltransferases (Hpβ3GlcNAcT and NmLgtA) towards 39 sugar nucleotides and the acceptor substrate specificity of three β1–4-galactosyltransferases (Hpβ4GalT, NmLgtB, and Bβ4GalT) towards 13 trisaccharide acceptors formed by Hpβ3GlcNAcT and/or NmLgtA-catalyzed reactions were investigated for the synthesis of diverse LacNAc-containing glycans or derivatives. Compared to Hpβ3GlcNAcT, NmLgtA could tolerate more 2'-modified UDP-GlcNAc derivatives including those with large groups such as phenyl group and even diphenyl group or small groups such as NH2. Besides UDP-GlcNAc and its derivatives, UDP-GalNAc, UDP-Glc, and UDP-Gal were also tolerable donor substrates for NmLgtA but not for Hpβ3GlcNAcT. However, for 6'-modified UDP-GlcNAc derivatives, Hpβ3GlcNAcT was a better catalyst than NmLgtA. Interestingly, enzymes from the same species can tolerate a similar set of modified substrates. For example, similarly to the tolerance of 6'-modified UDP-GlcNAc derivatives by Hpβ3GlcNAcT, β4GalT from H. pylori (Hpβ4GalT) was the best catalyst in using 6'-modified GlcNAc derivatives as acceptors. Also, GlcNAcPhPh- and Glc-containing trisaccharides were only acceptable substrates for NmLgtB but not for Hpβ4GalT. In addition, preparative-scale enzymatic synthesis of trisaccharides GalNAcβ1–3Galβ1–4GlcβProN3 and Galβ1–3Galβ1–4Glc from simple monosaccharides GalNAc and Gal, respectively, were successfully achieved using NmLgtA-containing one-pot four-enzyme (OP4E) systems. The substrate promiscuity of β3GlcNAcTs and β4GalTs will allow efficient chemoenzymatic synthesis of diverse derivatives of LacNAc-containing structures as probes for protein binding study and for developing potential therapeutics.
4. Experimental section
4.1. Materials
Vector plasmids pMAL-c4X and pMAL-c2X were purchased from Amersham Biosciences (Piscataway, NJ, USA). Restriction enzymes EcoRI and SalI were purchased from New England Biolabs, Inc. (Ipswich, MA, USA). Herculase II DNA polymerase was purchased from Stratagene (La Jolla, CA, USA). T4 DNA ligase and 1 kb DNA ladder were bought from Promega (Madison, WI. USA). QIAprep spin miniprep kit, QIAEX II gel extraction kit, and Nickel-nitrilotriacetic acid agarose (Ni2+-NTA agarose) were from Qiagen (Valencia, CA, USA). Escherichia coli electrocompetent DH5α and chemically competent BL21 (DE3) cells were purchased from Invitrogen (Carlsbad, CA, USA). Sugar nucleotides and derivatives were enzymatically or chemoenzymatically synthesized previously.38, 40, 52–55
4.2. Cloning of MBP-Hpβ3GlcNAcT-His6 and MBP-NmLgtA-His6
MBP-Hpβ3GlcNAcT-His6 (GenBank accession number: NP_223749) was cloned from Helicobacter pylori J99 genomic DNA into pMAL-c4X vector as a fusion protein with N-terminal maltose-binding protein (MBP) and C-terminal His6-tag. Primers used were: forward: 5'-CCGGAATTCATGACTTCAGCTTCAAGCCATTCTTTTAAA G-3' (EcoRI restriction site is underlined) and reverse: 5'-ACGCGTCGACTCAGTGGTGGTGGTGGTGGTGGCCCTTTTTCAAGAGCTTGAGTT-3' (SalI restriction site is underlined). MBP-NmLgtA-His6 (GenBank accession number: U25839) was cloned from Neisseria meningitidis MC58 genomic DNA into pMAL-c2X vector as a fusion protein with N-terminal maltose-binding protein (MBP) and C-terminal His6-tag. Primers used for polymerase chain reaction (PCR) are: Forward: 5'-GACCGAATTCATGCAGCCTTTAGTCAGCGTATTG-3' (EcoRI restriction site is underlined) and reverse: 5'-CAGCAAGCTTTTAGTGGTGGTGGTGGTGGTGACGGTTTTTCAGCAATCGGTGCAAAATG-3' (HindIII restriction site is underlined). Polymer chain reactions (PCRs) were carried out in a reaction mixture (50 µL) containing 10× Herculase buffer (5 µL), the genomic DNA (1 µg), forward and reverse primers (1 µM each), dNTP mixture (1 mM), and 5 U (1 µL) of Herculase II DNA polymerase. The reaction mixture was subjected to 30 cycles of normal PCR amplification as followed: denaturation at 95 °C for 30 s, annealing at 50 °C for 45 s, elongation at 72 °C for 2 min. Followed by a final elongation at 72 °C for 10 min. The PCR product was subsequently purified, digested, and ligated with pre-digested pMAL-c4X vector (for MBP-Hpβ3GlcNAcT-His6) or pMAL-c2X vector (for MBP-NmLgtA-His6). The ligation products were transformed into electrocompetent E. coli DH5α cells. The positive clones were confirmed by restriction digestion analysis and DNA sequencing (performed by UC-Davis Sequencing Facility). The correct plasmids with the target genes were subsequently transformed into expression host, E. coli BL21 (DE3) chemically competent cells.
4.3. Expression and purification
E. coli BL21 (DE3) strains harboring the recombinant plasmids with target genes were cultured in 50 mL Luria-Bertani (LB) rich medium (10 g L−1 tryptone, 5 g L−1 yeast extract, and 10 g L−1 NaCl) containing 0.1 mg mL−1 ampicillin with rapid shaking (220 rpm) at 37 °C overnight. Then 15 mL of the overnight cell culture was transferred into 1 L of LB rich medium with 0.1 mg mL−1 ampicillin and incubated at 37 °C. When the OD600 nm of the cell culture reached 0.8–1.0, isopropyl-1-thio-β-d-galactopyranoside (IPTG) was added to a final concentration of 0.1 mM to induce the over-expression of Hpβ3GlcNAcT, which was followed by incubation at 20 °C with slow shaking (190 rpm) for 20 h. Over-expression of NmLgtA was achieved by the same method.
All the cells were collected by centrifugation at 4 °C, 4000 rpm for 2 h. Harvested cells were resuspended with lysis buffer (100 mM, pH 8.0, Tris-HCl buffer containing 0.1 % Triton X-100). Lysis buffer (20 mL) was used to resuspend collected cells from one liter cell culture. The cells were broken by sonication and the method was as followed: amplitude at 68% for big tip, 3 s pulse on and 2 s pulse off for 60 cycles. Then the cell lysate was centrifuged at 4 °C, 12,000 rpm for 15 min, the supernatant was collected and loaded onto a Ni2+-NTA affinity column that was pre-equilibrated with 6 column volumes of binding buffer (50 mM, pH 7.5, Tris-HCl buffer, 10 mM imidazole, 0.5 M NaCl). The column was washed with 10 column volumes of binding buffer and 10 column volumes of washing buffer (Tris-HCl buffer, 50 mM, pH7.5, 40 mM imidazole for NmLgtA and 60 mM imidazole for Hpβ3GlcNAcT, 0.5 M NaCl) sequentially to wash away the nonspecific binding protein. The target protein was eluted using Tris-HCl buffer (50 mM, pH 7.5) containing 200 mM of imidazole and 0.5 M of NaCl. Fractions containing the purified protein were combined and dialyzed against Tris-HCl buffer (20 mM, pH 7.5). The enzyme solution was stored at 4°C with 10% glycerol. All procedures were performed at 4 °C.
4.4. pH profile by capillary electrophoresis (CE)
Standard enzymatic assays were carried out in reactions (10 µL) each containing a buffer (100 mM) with a pH in the range of 5.0–11.0, UDP-GlcNAc (1 mM), LacβMU (1 mM), MnCl2 (10 mM), and Hpβ3GlcNAcT (3 µg) or NmLgtA (2 µg). Buffers used here were: MES, pH 5.0–6.5; Tris-HCl, pH 7.0–9.0; and CAPS, pH 10.0–11.0. Reactions were incubated at 30 °C for 10 min and quenched by heating at 98 °C for 2 min. Samples were centrifuged at 13,000 rpm for 5 min, the supernatants were analyzed by a P/ACETM Capillary Electrophoresis (CE) system equipped with a Photodiode Array (PDA) detector (Bechman Coulter, Inc., Fullerton, CA, USA). CE conditions were as follows: 75 µm i.d. capillary, 25 KV/80 µA, 6 s vacuum injections, monitored at 315 nm, the running buffer used was sodium tetraborate (25 mM, pH 9.4).
4.5. Effect of metal ions and EDTA
In order to investigate the effect of metal ions of Hpβ3GlcNAcT and NmLgtA, the reactions were carried out with the addition of different concentrations (1, 5, 10, and 20 mM) of different metal ions (MgCl2, MnCl2, CaCl2) or EDTA (10 mM) in Tris-HCl buffer (100 mM, pH 8.0) containing UDP-GlcNAc (1 mM), LacβMU (1 mM), and Hpβ3GlcNAcT (3 µg) or NmLgtA(2 µg). Reactions were performed at 30 °C for 10 min. The reaction without metal ion nor EDTA was used as a control.
4.6. Donor substrate specificity of Hpβ3GlcNAcT and NmLgtA
All reactions were performed in duplicates. The enzymatic assay was carried out in 40 µL reaction system for both β3GlcNAcTs. For Hpβ3GlcNAcT, the reactions were carried out in Tris-HCl (100 mM, pH 8.0) containing LacβMU (2 mM), UDP-GlcNAc or its derivative or other sugar nucleotide (2 mM), a metal ion (10 mM OF MgCl2 was used for Hpβ3GlcNAcT and 10 mM of MnCl2 was used for NmLgtA), and Hpβ3GlcNAcT or NmLgtA (10 µg). The reaction mixtures were incubated at 30 °C for 24 h. The supernatants obtained after centrifugation were analyzed by CE as described for the pH profile. For reactions using UDP-Gal as the donor, the supernatants of the reaction mixtures after centrifugation were analyzed by CE using sodium tetraborate (50 mM, pH 11.0) as the running buffer to allow the separation of LacβMU and the trisaccharide product.
For HPLC analysis, aliquots of 1.0 µL from above supernatants were withdrawn and added to ice-cold 18% acetonitrile (99 µL) to make 100-fold dilution. The samples were kept on ice until aliquots of 10 µL were injected and analyzed by a Shimadzu LC-2010A system equipped with a membrane online degasser, a temperature control unit, and a fluorescence detector. A reverse-phase Premier C18 column (250 × 4.6 mm i.d., 5 µm particle size, Shimadzu) protected with a C18 guard column cartridge was used. The mobile phase was 18% (v/v) acetonitrile. The fluorescent 4-methylumbelliferyl compounds were detected with excitation at 325 nm and emission at 372 nm.
4.7. Acceptor substrate specificity of Hpβ4GalT, NmLgtB and Bβ4GalT
These assays were performed in duplicate in two steps using a sequential two-step process. An aliquot of 10 µL of the supernatant of the reaction mixture for the donor substrate specificity of β3GlcNAcT was drawn and added to a solution to give a final mixture of 20 µL containing Tris-HCl buffer (pH 7.5, 200 mM), MgCl2 (10 mM), UDP-Gal (1 mM), and Hpβ4GalT (~2.0 µg), NmLgtB (~2.0 µg), or Bβ4GalT (~1.0 µg). The reaction mixtures were allowed to proceed for 24 h at 37 °C.
4.8. Kinetic study of Hpβ3GlcNAcT and NmLgtA by capillary electrophoresis (CE)
Kinetic assays were carried out in duplicates in a 10 µL system containing Tris-HCl buffer (100 mM, pH 8.0), MgCl2 or MnCl2 (10 mM), LacβMU (1 mM), UDP-GlcNAc or its derivatives at different concentration ranging from 0.1–10 mM (0.1 mM, 0.2 mM, 0.4 mM, 1 mM, 2 mM, 4 mM, 5 mM, 10 mM), and Hpβ3GlcNAcT (0.7 µg µL−1) or NmLgtA (1.0 µg µL−1). Reactions were incubated at 30 °C for 10 min, followed by heating at 98 °C for 2 min. The reaction mixtures were subsequently centrifuged at 13,000 rpm for 5 min to remove the protein precipitate. The supernatants were analyzed by CE as described for the pH profile.
4.9. One-pot four-enzyme preparative-scale synthesis of trisaccharide GalNAcβ1–3Galβ1–4GlcβProN3
To prepare the trisaccharide, a reaction mixture in Tris-HCl buffer (100 mM, pH 7.5) in a total volume of 5 mL containing LacβProN3 (40 mg, 0.094 mmol), N–acetylgalactosamine (GalNAc, 27 mg, 0.122 mmol), adenosine 5'-triphosphate (ATP) (67 mg, 0.122 mmol), uridine 5'-triphosphate (UTP, 65 g, 0.122 mmol), MgCl2 (20 mM). NahK (1.5 mg), PmGlmU (1.5 mg), NmLgtA (2.0 mg), and PmPpA (1.5 mg) were incubated in a shaker at 37 °C for 24 hrs. The product formation was monitored by TLC (EtOAc:MeOH:H2O:HOAc = 4:2:1:0.2, by volume and detected by p-anisaldehyde sugar stain) and mass spectrometry. To stop the reaction, the reaction mixture was added with same volume (10 mL) of ethanol and incubated at 4 °C for 30 min. After centrifugation, the supernatant was concentrated and passed through a Bio Gel P-2 gel filtration column (water was used as an eluent). The fractions containing the product were collected, concentrated, further purified by silica gel column (EtOAc:MeOH:H2O = 5:2:1, by volume). Further purification by preparative HPLC using C18 column provided the trisaccharide GalNAcβ1–3Galβ1–4GlcβProN3 as a white powder (58 mg, 98%). 1H NMR (800 MHz, D2O) δ 4.59 (d, J = 8.8 Hz, 1H), 4.46 (d, J = 8.0 Hz, 1H), 4.40 (d, J = 8.0 Hz, 1H), 4.13 (d, J = 3.6 Hz, 1H), 3.99–3.27 (m, 21H), 2.01 (s, 3H), 1.88 (m, 2H). 13C NMR (200 MHz, D2O) δ 175.03, 103.20, 102.81, 101.97, 81.63, 78.20, 74.84, 74.74, 74.63, 74.22, 72.65, 70.59, 69.93 (2C), 68.34, 67.61, 67.23, 60.85, 60.82, 59.92, 47.73, 28.10, 22.09. HRMS (ESI) m/z calculated for C23H40N4NaO16 (M+Na) 651.2337, found 651.2335.
4.10. One-pot four-enzyme preparative-scale synthesis of trisaccharide Galβ1–3Galβ1–4Glc
To prepare the trisaccharide, a reaction mixture in Tris-HCl buffer (100 mM, pH 8.0) in a total volume of 10 mL containing lactose (Galβ1–4Glc, 102 mg, 0.3 mmol), galactose (Gal, 65 mg, 0.36 mmol), adenosine 5'-triphosphate (ATP) (198 mg, 0.36 mmol), uridine 5'-triphosphate (UTP, 190 g, 0.36 mmol), MgCl2 (10 mM). SpGalK (2.0 mg), BLUSP (2.0 mg), NmLgtA (2.0 mg), and PmPpA (1.5 mg) were incubated in a shaker at 37 °C for 24 hrs. The product formation was monitored by LC-MS. To stop the reaction, the reaction mixture was added with same volume (10 mL) of ethanol and incubated at 4 °C for 30 min. After centrifugation, the supernatant was concentrated and passed through a Bio Gel P-2 gel filtration column (water was used as an eluent). The fractions containing the product were collected, concentrated, further purified by C18 column (H2O, 0–7 min; 0–50% ACN in H2O, 7–35 min; 50% ACN in H2O, 35–40min). The pure trisaccharide Galβ1–3Galβ1–4Glc was obtained by H2O wash. The synthesis yield of this trisaccharide Galβ1–3Galβ1–4Glc was 10% (15 mg). 1H NMR (800 MHz, D2O) δ 5.20 (d, J = 3.2 Hz, 0.4H), 4.65 (d, J = 8.0 Hz, 0.6H), 4.46 (d, J = 7.2 Hz, 1H), 4.59 (d, J = 8.0 Hz, 1H), 4.49 (d, J = 8.0 Hz, 1H), 4.17 (d, J = 3.2 Hz, 1H), 3.94–3.26 (m, 17H); 13C NMR (200 MHz, D2O) α-isomer: δ 104.21, 102.39, 91.70, 81.75, 78.08, 74.85, 74.65, 72.36, 71.27, 71.03, 70.07, 69.95, 68.43, 68.30, 68.28, 60.87, 59.94, 59.80. HRMS (ESI) m/z calculated for C18H32NaO16 (M+Na) 527.1588, found 527.1584.
Supplementary Material
Highlights.
N. meningitidis and H. pylori β3GlcNAcTs were actively expressed in E. coli.
Bovine β4GalT 1 was expressed in E. coli as a soluble active enzyme.
Thirteen UDP-sugars could be used as donor substrates for NmLgtA and Hpβ3GlcNAcT.
Ten trisaccharides could be used as acceptors for one of the three β4GalTs.
GalNAcβ1–3LacβProN3 and Galβ1–3Lac were produced by OPME reaction containing NmLgtA.
Acknowledgments
This work was supported by National Science Foundation grant CHE-1300449 as well as, National Institutes of Health grants R01HD065122 and R01GM094523, National Basic Research Program of China 973 Program No. 2012CB822102, and the Key Grant Project of Chinese Ministry of Education No. 313033.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and notes
- 1.Peng W, Pranskevich J, Nycholat C, Gilbert M, Wakarchuk W, Paulson JC, Razi N. Glycobiology. 2012;22:1453. doi: 10.1093/glycob/cws101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Di Virgilio S, Glushka J, Moremen K, Pierce M. Glycobiology. 1999;9:353. doi: 10.1093/glycob/9.4.353. [DOI] [PubMed] [Google Scholar]
- 3.Naruchi K, Hamamoto T, Kurogochi M, Hinou H, Shimizu H, Matsushita T, Fujitani N, Kondo H, Nishimura S. J. Org. Chem. 2006;71:9609. doi: 10.1021/jo0617161. [DOI] [PubMed] [Google Scholar]
- 4.Nycholat CM, Peng W, McBride R, Antonopoulos A, de Vries RP, Polonskaya Z, Finn MG, Dell A, Haslam SM, Paulson JC. J. Am. Chem. Soc. 2013;135:18280. doi: 10.1021/ja409781c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Renkonen O. Cell. Mol. Life Sci. 2000;57:1423. doi: 10.1007/PL00000627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vasiliu D, Razi N, Zhang Y, Jacobsen N, Allin K, Liu X, Hoffmann J, Bohorov O, Blixt O. Carbohydr. Res. 2006;341:1447. doi: 10.1016/j.carres.2006.03.043. [DOI] [PubMed] [Google Scholar]
- 7.Sauerzapfe B, Krenek K, Schmiedel J, Wakarchuk WW, Pelantova H, Kren V, Elling L. Glycoconj. J. 2009;26:141. doi: 10.1007/s10719-008-9172-2. [DOI] [PubMed] [Google Scholar]
- 8.Blixt O, Razi N. Methods Enzymol. 2006;415:137. doi: 10.1016/S0076-6879(06)15009-0. [DOI] [PubMed] [Google Scholar]
- 9.Nycholat CM, McBride R, Ekiert DC, Xu R, Rangarajan J, Peng W, Razi N, Gilbert M, Wakarchuk W, Wilson IA, Paulson JC. Angew. Chem. Int. Ed. 2012;51:4860. doi: 10.1002/anie.201200596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chien WT, Liang CF, Yu CC, Lin CH, Li SP, Primadona I, Chen YJ, Mong KK, Lin CC. Chem. Commun. 2014;50:5786. doi: 10.1039/c4cc01227e. [DOI] [PubMed] [Google Scholar]
- 11.Johnson KF. Glycoconj. J. 1999;16:141. doi: 10.1023/a:1026440509859. [DOI] [PubMed] [Google Scholar]
- 12.Yu H, Lau K, Thon V, Autran CA, Jantscher-Krenn E, Xue M, Li Y, Sugiarto G, Qu J, Mu S, Ding L, Bode L, Chen X. Angew. Chem. Int. Ed. 2014;53:6687. doi: 10.1002/anie.201403588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Priem B, Gilbert M, Wakarchuk WW, Heyraud A, Samain E. Glycobiology. 2002;12:235. doi: 10.1093/glycob/12.4.235. [DOI] [PubMed] [Google Scholar]
- 14.Gebus C, Cottin C, Randriantsoa M, Drouillard S, Samain E. Carbohydr. Res. 2012;361:83. doi: 10.1016/j.carres.2012.05.015. [DOI] [PubMed] [Google Scholar]
- 15.Blixt O, van Die I, Norberg T, van den Eijnden DH. Glycobiology. 1999;9:1061. doi: 10.1093/glycob/9.10.1061. [DOI] [PubMed] [Google Scholar]
- 16.Togayachi A, Kozono Y, Ishida H, Abe S, Suzuki N, Tsunoda Y, Hagiwara K, Kuno A, Ohkura T, Sato N, Sato T, Hirabayashi J, Ikehara Y, Tachibana K, Narimatsu H. Proc. Natl. Acad. Sci. U. S. A. 2007;104:15829. doi: 10.1073/pnas.0707426104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wakarchuk W, Martin A, Jennings MP, Moxon ER, Richards JC. J. Biol. Chem. 1996;271:19166. doi: 10.1074/jbc.271.32.19166. [DOI] [PubMed] [Google Scholar]
- 18.Logan SM, Altman E, Mykytczuk O, Brisson JR, Chandan V, Schur MJ, St Michael F, Masson A, Leclerc S, Hiratsuka K, Smirnova N, Li J, Wu Y, Wakarchuk WW. Glycobiology. 2005;15:721. doi: 10.1093/glycob/cwi057. [DOI] [PubMed] [Google Scholar]
- 19.Guan W, Ban L, Cai L, Li L, Chen W, Liu X, Mrksich M, Wang PG. Bioorg. Med. Chem. Lett. 2011;21:5025. doi: 10.1016/j.bmcl.2011.04.100. [DOI] [PubMed] [Google Scholar]
- 20.Palcic MM. Curr. Opin. Chem. Biol. 2011;15:226. doi: 10.1016/j.cbpa.2010.11.022. [DOI] [PubMed] [Google Scholar]
- 21.Lau K, Thon V, Yu H, Ding L, Chen Y, Muthana MM, Wong D, Huang R, Chen X. Chem. Commun. 2010;46:6066. doi: 10.1039/c0cc01381a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wakarchuk WW, Cunningham A, Watson DC, Young NM. Protein Eng. 1998;11:295. doi: 10.1093/protein/11.4.295. [DOI] [PubMed] [Google Scholar]
- 23.Park JE, Lee KY, Do SI, Lee SS. J. Biochem. Mol. Biol. 2002;35:330. doi: 10.5483/bmbrep.2002.35.3.330. [DOI] [PubMed] [Google Scholar]
- 24.Logan SM, Conlan JW, Monteiro MA, Wakarchuk WW, Altman E. Mol. Microbiol. 2000;35:1156. doi: 10.1046/j.1365-2958.2000.01784.x. [DOI] [PubMed] [Google Scholar]
- 25.Endo T, Koizumi S, Tabata K, Ozaki A. Glycobiology. 2000;10:809. doi: 10.1093/glycob/10.8.809. [DOI] [PubMed] [Google Scholar]
- 26.Namdjou DJ, Chen HM, Vinogradov E, Brochu D, Withers SG, Wakarchuk WW. Chembiochem. 2008;9:1632. doi: 10.1002/cbic.200700775. [DOI] [PubMed] [Google Scholar]
- 27.Dumon C, Bosso C, Utille JP, Heyraud A, Samain E. Chembiochem. 2006;7:359. doi: 10.1002/cbic.200500293. [DOI] [PubMed] [Google Scholar]
- 28.Wong CH, Wang R, Ichikawa Y. J. Org. Chem. 1992;57:4343. [Google Scholar]
- 29.Palcic MM, Srivastava OP, Hindsgaul O. Carbohydr. Res. 1987;159:315. doi: 10.1016/s0008-6215(00)90224-6. [DOI] [PubMed] [Google Scholar]
- 30.Berliner LJ, Davis ME, Ebner KE, Beyer TA, Bell JE. Mol. Cell. Biochem. 1984;62:37. doi: 10.1007/BF00230075. [DOI] [PubMed] [Google Scholar]
- 31.Blixt O, Brown J, Schur MJ, Wakarchuk W, Paulson JC. J. Org. Chem. 2001;66:2442. doi: 10.1021/jo0057809. [DOI] [PubMed] [Google Scholar]
- 32.Brockhausen I, Benn M, Bhat S, Marone S, Riley JG, Montoya-Peleaz P, Vlahakis JZ, Paulsen H, Schutzbach JS, Szarek WA. Glycoconj. J. 2006;23:525. doi: 10.1007/s10719-006-7153-x. [DOI] [PubMed] [Google Scholar]
- 33.Boeggeman EE, Balaji PV, Sethi N, Masibay AS, Qasba PK. Protein Eng. 1993;6:779. doi: 10.1093/protein/6.7.779. [DOI] [PubMed] [Google Scholar]
- 34.Ramakrishnan B, Shah PS, Qasba PK. J. Biol. Chem. 2001;276:37665. doi: 10.1074/jbc.M102458200. [DOI] [PubMed] [Google Scholar]
- 35.Hanada E, Handa S, Konno K, Yamakawa T. J. Biochem. 1978;83:85. doi: 10.1093/oxfordjournals.jbchem.a131915. [DOI] [PubMed] [Google Scholar]
- 36.Higuchi R, Inukai K, Jhou JX, Honda M, Komori T, Tsuji S, Nagai Y. Liebigs Ann. Chem. 1993:359. [Google Scholar]
- 37.Li Y, Yu H, Chen Y, Lau K, Cai L, Cao H, Tiwari VK, Qu J, Thon V, Wang PG, Chen X. Molecules. 2011;16:6396. doi: 10.3390/molecules16086396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen Y, Thon V, Li Y, Yu H, Ding L, Lau K, Qu J, Hie L, Chen X. Chem. Commun. 2011;47:10815. doi: 10.1039/c1cc14034e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen M, Chen LL, Zou Y, Xue M, Liang M, Jin L, Guan WY, Shen J, Wang W, Wang L, Liu J, Wang PG. Carbohydr. Res. 2011;346:2421. doi: 10.1016/j.carres.2011.08.014. [DOI] [PubMed] [Google Scholar]
- 40.Muthana MM, Qu J, Li Y, Zhang L, Yu H, Ding L, Malekan H, Chen X. Chem. Commun. 2012;48:2728. doi: 10.1039/c2cc17577k. [DOI] [PubMed] [Google Scholar]
- 41.Donald AS, Feeney J. Carbohydr. Res. 1988;178:79. doi: 10.1016/0008-6215(88)80103-4. [DOI] [PubMed] [Google Scholar]
- 42.Urashima T, Sun Y, Fukuda K, Hirayama K, Taufik E, Nakamura T, Saito T, Merchant J, Green B, Messer M. Glycoconj. J. 2015;32:361. doi: 10.1007/s10719-015-9600-z. [DOI] [PubMed] [Google Scholar]
- 43.Saito T, Itoh T, Adachi S. Carbohydr. Res. 1987;165:43. doi: 10.1016/0008-6215(87)80076-9. [DOI] [PubMed] [Google Scholar]
- 44.Urashima T, Saito T, Nishimura J, Ariga H. Bioch. Biophys. Acta. 1989;992:375. doi: 10.1016/0304-4165(89)90099-8. [DOI] [PubMed] [Google Scholar]
- 45.Urashima T, Kawai Y, Nakamura T, Arai I, Saito T, Namiki M, Yamaoka K, Kawahawa K, Messer M. Comp. Biochem. Physiol. C Pharmacol. Toxicol. Endocrinol. 1999;124:295. doi: 10.1016/s0742-8413(99)00080-8. [DOI] [PubMed] [Google Scholar]
- 46.Fukuda K, Yamamoto A, Ganzorig K, Khuukhenbaatar J, Senda A, Saito T, Urashima T. J. Dairy Sci. 2010;93:5572. doi: 10.3168/jds.2010-3151. [DOI] [PubMed] [Google Scholar]
- 47.Difilippo E, Willems HA, Vendrig JC, Fink-Gremmels J, Gruppen H, Schols HA. J. Agric. Food Chem. 2015;63:4805. doi: 10.1021/acs.jafc.5b01127. [DOI] [PubMed] [Google Scholar]
- 48.Kunz C, Rudloff S, Schad W, Braun D. Br. J. Nutr. 1999;82:391. doi: 10.1017/s0007114599001798. [DOI] [PubMed] [Google Scholar]
- 49.Urashima T, Fujita S, Fukuda K, Nakamura T, Saito T, Cowan P, Messer M. Glycoconj. J. 2014;31:387. doi: 10.1007/s10719-014-9533-y. [DOI] [PubMed] [Google Scholar]
- 50.Messer M, Trifonoff E, Stern W, Collins JG, Bradbury JH. Carbohydr. Res. 1980;83:327. doi: 10.1016/s0008-6215(00)84545-0. [DOI] [PubMed] [Google Scholar]
- 51.Urashima T, Taufik E, Fukuda R, Nakamura T, Fukuda K, Saito T, Messer M. Glycoconj. J. 2013;30:801. doi: 10.1007/s10719-013-9484-8. [DOI] [PubMed] [Google Scholar]
- 52.Zhang L, Muthana MM, Yu H, McArthur JB, Qu J, Chen X. Carbohydr. Res. 2016;419:18. doi: 10.1016/j.carres.2015.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Muthana MM, Qu J, Xue M, Klyuchnik T, Siu A, Li Y, Zhang L, Yu H, Li L, Wang PG, Chen X. Chem. Commun. 2015;51:4595. doi: 10.1039/c4cc10306h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yu H, Yu H, Karpel R, Chen X. Bioorg. Med. Chem. 2004;12:6427. doi: 10.1016/j.bmc.2004.09.030. [DOI] [PubMed] [Google Scholar]
- 55.Zhang L, Lau K, Cheng J, Yu H, Li Y, Sugiarto G, Huang S, Ding L, Thon V, Wang PG, Chen X. Glycobiology. 2010;20:1077. doi: 10.1093/glycob/cwq068. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.