Abstract
MLL (for mixed-lineage leukemia) is a proto-oncogene that is mutated in a variety of human leukemias. Its product, a homolog of Drosophila melanogaster trithorax, displays intrinsic histone methyltransferase activity and functions genetically to maintain embryonic Hox gene expression. Here we report the biochemical purification of MLL and demonstrate that it associates with a cohort of proteins shared with the yeast and human SET1 histone methyltransferase complexes, including a homolog of Ash2, another Trx-G group protein. Two other members of the novel MLL complex identified here are host cell factor 1 (HCF-1), a transcriptional coregulator, and the related HCF-2, both of which specifically interact with a conserved binding motif in the MLLN (p300) subunit of MLL and provide a potential mechanism for regulating its antagonistic transcriptional properties. Menin, a product of the MEN1 tumor suppressor gene, is also a component of the 1-MDa MLL complex. Abrogation of menin expression phenocopies loss of MLL and reveals a critical role for menin in the maintenance of Hox gene expression. Oncogenic mutant forms of MLL retain an ability to interact with menin but not other identified complex components. These studies link the menin tumor suppressor protein with the MLL histone methyltransferase machinery, with implications for Hox gene expression in development and leukemia pathogenesis.
MLL is a proto-oncogene that was originally discovered at the site of chromosomal translocations in human leukemias (15, 22, 56). It is mutated in a subset of acute leukemias that display distinctive biological, genetic, and clinical features underscoring a critical role for MLL in dictating disease pathogenesis (14). As a result of chromosomal translocations, MLL is fused with 1 of more than 40 different partner proteins to yield a diverse collection of chimeric fusion proteins (6, 12, 28). Despite their substantial diversity, protein fusions appear to activate MLL through two alternative mechanisms by either conferring constitutive transcriptional effector activity or inducing forced MLL dimerization and oligomerization (26, 40, 52, 54, 68). Both mechanisms result in the inappropriate expression of a subset of Hox genes, particularly HoxA9, whose consistent expression is a characteristic, albeit not exclusive, feature of human MLL leukemias (5, 7, 48, 63, 67). Furthermore, genetic studies in mouse models have shown that HoxA9 makes critical contributions to the incidence and/or phenotypes of MLL leukemias (7, 34, 53), likely reflecting its role as a direct target gene for MLL oncoproteins.
Despite these advances in understanding the oncogenic contributions of MLL, little is known about the biochemical properties and roles of wild-type MLL, a large (431 kDa) and structurally complex protein with conserved motifs often found in chromatin-associated transcriptional regulators (1, 50). Genetic studies have shown that MLL is a functional homolog of Drosophila melanogaster trithorax, which is required for the maintenance but not initiation of Hox gene expression to establish proper segment identity throughout embryonic development (23, 65, 66). This cellular memory role is likely to be mediated in part through epigenetic mechanisms. MLL displays intrinsic histone methyltransferase (HMT) activity, which is conferred by a SET domain active site that specifically methylates lysine 4 of histone H3, an epigenetic mark typically associated with transcriptionally active chromatin (42, 44). A more extensive transcriptional role for MLL is suggested by its proteolytic processing by the endopeptidase taspase I into two portions (MLLN and MLLC) that have antagonistic transcriptional effector properties but reassociate with and potentially stabilize each other (24, 25, 64). MLLN displays transcriptional repression activity and, under experimental conditions, is capable of interacting with corepressor proteins and members of the PcG family of silencing proteins (62, 64). Conversely, MLLC displays features of a transcriptional activation module containing, in addition to the HMT active site, a strong transactivation domain that recruits the coactivator CBP (16, 64, 69). An even more diverse transcriptional role was suggested by immunopurification of MLL, which resulted in the copurification of at least 29 associated factors unrelated to those cited above and implicated in basal transcription, corepression, chromatin remodeling, and RNA processing (44). Taken together, these studies suggest a multifaceted transcriptional role for MLL in several aspects of gene expression.
We demonstrate here that MLL assembles a novel multimember complex whose composition is highly similar to that of the SET1 HMT complexes of yeast and humans, thereby establishing a conserved and ancient biochemical machinery for histone H3 lysine 4 methylation. Furthermore, host cell factor 1 (HCF-1) (33, 58), a transcriptional coregulator that associates with human SET1 (61), and the related but functionally distinct HCF-2 (31) both specifically interact with MLL, suggesting a potential mechanism for differentially regulating its antagonistic transcriptional properties (60). We also demonstrate that the tumor suppressor protein menin (10, 11) is an essential component of the MLL complex, is required for maintenance of Hox gene expression, and is the only identified component to also interact with oncogenic MLL fusion proteins. Because the tumor suppressor menin functions together with the proto-oncoprotein MLL to regulate Hox target gene expression, our studies provide new insights into the mechanism of MLL-mediated leukemogenesis and establish a functional link between menin and the HMT epigenetic machinery.
MATERIALS AND METHODS
Cell culture.
293 and HeLa cells were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal calf serum (FCS). K562, REH, and HB cells were cultured in RPMI 1640 medium supplemented with 10% FCS. HeLa cells stably expressing Flag-tagged HCF-1N (residues 2 to 1011; f-HCF-1N) or HCF-1Kelch (residues 2 to 380; f-HCF-1Kelch) or transduced with empty pBabeFlag-Puro vector were described elsewhere (61).
Purification of MLL complexes.
K562 cells (1010) were harvested by centrifugation and were washed once with phosphate-buffered saline. Nuclei were prepared by suspension of washed cells in isotonic buffer (150 mM NaCl, 1.5 mM MgCl2, 10 mM Tris-HCl at pH 7.5, 0.5% NP-40, EDTA-complete protease inhibitor cocktail [Roche]) on ice for 5 min, followed by sedimentation at 300 × g for 5 min. The nuclei were suspended in lysis buffer (250 mM NaCl, 20 mM sodium phosphate at pH 7.0, 30 mM sodium pyrophosphate, 5 mM EDTA, 10 mM NaF, 0.1% NP-40, 10% glycerol, 1 mM dithiothreitol, EDTA-complete protease inhibitor cocktail), and the resulting extract was clarified by ultracentrifugation at 30,000 rpm (SW 32Ti; Beckman Coulter) for 1 h at 4°C. The clarified extract was passed through a 0.45-μm-pore-size filter, mixed with 4 volumes of 50 mM KCl in buffer A (20 mM HEPES at pH 7.9, 1 mM EDTA, 0.2% NP-40, 10% glycerol, 2 mM dithiothreitol, 2 μg of aprotinin/ml, 2 μg of leupeptin/ml, 2 μg of pepstatin/ml, 1 mM phenylmethylsulfonyl fluoride), and applied to a 16/40 Q-Sepharose FF column (Amersham Pharmacia Biotech). Fractions were eluted stepwise at 100, 200, 300, 400, 500, and 600 mM KCl in buffer A. The 300 mM KCl fraction, which contained the majority of MLL by immunoblotting, was applied to a 5-ml HiTrap heparin Sepharose column (Amersham Pharmacia Biotech). Following stepwise elution in buffer A, the 500 mM KCl fraction containing most of MLL (Hep500 fraction) was used for subsequent affinity purification.
Monoclonal antibodies (3 mg) specific for either MLLN (mmN4.4), MLLC (mmC2.1), or SUV39H1 (negative control) were incubated with a 0.5-ml bed volume of protein G-Sepharose beads (Amersham Pharmacia Biotech) in 10 ml of lysis buffer at 4°C for 2 h with rotation. The beads were then washed three times with 10 ml of 0.2 M sodium borate at pH 9.0 and were suspended in 10 ml of 0.2 M sodium borate at pH 9.0. Following addition of dimethyl pimelimidate-2HCl (Pierce) to a final concentration of 20 mM, the beads were incubated at room temperature for 30 min with rotation. The cross-linking reaction was stopped by washing the beads once in 0.2 M ethanolamine at pH 8.0 (EA buffer) followed by incubation for 2 h at room temperature in 10 ml of 0.2 M EA buffer with rotation. The beads were again washed with 10 ml of 0.2 M EA buffer and then were suspended in phosphate-buffered saline with 0.01% sodium azide. For affinity purification, a 1/100 volume of antibody-conjugated beads was added to the Hep500 fraction or nuclear extract and was incubated at 4°C for 4 h with gentle rotation. The beads were then washed six times with lysis buffer and were suspended in sodium dodecyl sulfate-polyacrylamide gel electrophoresis sample buffer (62.5 mM Tris-HCl at pH 6.8, 2% SDS, 5% β-mercaptoethanol, 0.01% bromphenol blue). A 1/50 volume of sample was separated in denaturing gel and visualized by silver stain (Silver stain plus; Bio-Rad).
Protein identification by mass spectrometry.
The purified MLL complex was subjected to SDS-PAGE fractionation, and complex components were visualized by Coomassie brilliant blue staining (Colloidal Blue Stain kit; Invitrogen). Stained bands were excised and processed for in-gel trypsin digestion following standard protocols. Resulting peptides were extracted, purified, and analyzed by LC-MS/MS at the Stanford Mass Spectroscopy Laboratory, and spectra were analyzed with Mascot software (Matrix Science).
Construction of expression vectors.
Expression vectors of MLL for His- and Flag-tagged MLL Δ3820, MLL Δ2254, MLL Δ1406, and MLL-p300 [named pLNCX(HF)MLL 34/3820, pLNCX(HF)MLL34/2254, pLNCX(HF)MLL34/1406, and pLNCX(HF)MLL-p300, respectively] were generated by restriction enzyme digestion and ligation of pLNCX(HF)MLL vector and various deletion mutants as reported previously (64). The expression vector for His- and Flag-tagged MLL ΔHBM (Δ1799/1803) was generated by PCR-mediated mutagenesis using primers 5′-GACAGCCAGAAATTAAAAAA-3′, 5′-TCCTCTCGCTCCTGCCACTGAAGTGAAGGTGGAAGCACTG-3′, 5′-CAGTGCTTCCACCTTCACTTCAGTGGCAGGAGCGAGAGGA-3′, and 5′-CTTCCTGCAGAAGGCAACGG-3′. The amplified fragment containing an HCF-1 binding motif (HBM) deletion was cloned into pLNCX(HF)MLL vector by restriction enzyme digestion and ligation [named pLNCX(HF)MLL ΔHBM].
Transfections.
293 cells were seeded at 10% confluency in 175-cm2 flasks, and 2 days later (∼60 to 80% confluency) they were transfected with various expression plasmids by using Lipofectamine 2000 (Invitrogen). At 4 h posttransfection, cells were washed and placed in fresh DMEM growth medium. Cells were harvested 36 h posttransfection. Nuclear extract (5 ml) was prepared as described above for K562 cells and was used for immunoprecipitations and Western blotting.
Coimmunoprecipitation and immunoblot analysis.
Nuclear extract (5 ml) was incubated with primary antibodies at a concentration of approximately 1 μg/200 μl of extract or 50 μl of half the slurry of FLAG M2-agarose affinity gel and was used for immunoprecipitations and Western blotting as described elsewhere (64). Primary antibodies consisted of monoclonal anti-MLLN (mmN4.4), anti-MLLC (mmC2.1), and anti-SUV39H1 as described previously (17, 64). Mouse monoclonal anti-actin antibody (C4) was purchased from Chemicon International. Polyclonal anti-MLLN (rpN1), anti-HCF-1N (N18), anti-ENL, and anti-HCF-1C (H12) antibodies were reported previously (9, 21, 58, 59, 64). Polyclonal anti-ASH2L, anti-WDR5, and anti-HCF-2 antibodies were raised against synthetic peptides (61). Anti-menin (H-300 or C19), anti-mSin3A (K-20), anti-SNF2H (H-300), anti-HIS (D-8), and anti-BRM (N-19) antibodies were purchased from Santa Cruz Biotechnology, Inc. Anti-RBBP5 (BL766) was purchased from Bethyl Laboratories, Inc. Anti-Flag (M2) agarose affinity gel was purchased from Sigma.
Gel filtration chromatography.
The Hep500 fraction was passed through a 0.45-μm-pore-size filter and was applied to a 16/60 Superose 6 column (Amersham Pharmacia Biotech) at a flow rate of 0.75 ml/min in gel filtration running buffer (200 mM KCl, 20 mM HEPES at pH 7.9, 1 mM EDTA, 0.2% NP-40, 10% glycerol, 2 mM dithiothreitol, 2 μg of aprotinin/ml, 2 μg of leupeptin/ml, 2 μg of pepstatin/ml, 1 mM phenylmethylsulfonyl fluoride). Fractions (2.5 ml) were collected and proteins were precipitated by addition of ice-cold acetone (10 ml) and incubation at −20°C overnight. Following centrifugation at 3,220 × g for 30 min at 4°C, protein pellets were suspended in SDS-PAGE sample buffer and were subjected to Western blot analysis. The molecular size corresponding to each fraction was estimated by elution profiles of Blue Dextran (2 MDa), Thyroglobulin (669 kDa), and Apoferritin (443 kDa) (Molecular Weight Marker kit; Sigma).
RNA interference.
RNA oligonucleotides were purchased as duplexes from Dharmacon Research. HeLa cells were transfected with siRNA duplexes (200 pmol) by using Oligofectamine (Invitrogen) according to the manufacturer's instructions. The medium was changed on the following day. Two additional rounds of transfection were performed using identical conditions 24 and 48 h after the initial transfection. Cells were harvested 72 h after initial transfection and were either lysed in SDS-PAGE sample buffer or subjected to RNA preparation with an RNeasy mini kit (QIAGEN).
The sequences of small interfering RNAs (siRNAs) employed were the following: GL2, 5′-CGUACGCGGAAUACUUCGAdTdT-3′ and 5-UCGAAGUAUUCCGCGUACGdTdT-3′; MLL, 5′-GAAGUCAGAGUGCGAAGUCdTdT-3′ and 5′-GACUUCGCACUCUGACUUCdTdT-3′; ASH2L, 5′-GGCAAACUUGGUCGAUGUAdTdT-3′ and 5′-UACAUCGACCAAGUUUGCCdTdT-3′; WDR5, 5′-CACCUGUGAAGCCAAACUAdTdT-3′ and 5′-UAGUUUGGCUUCACAGGUGdTdT-3′; HCF-1, 5′-GGAGCUCAUCGUGGUGUUUdTdT-3′ and 5′-AAACACCACGAUGAGCUCCdTdT-3′; HCF-2, 5′-GCAAGUCGUUGGUUAUGGAdTdT-3′ and 5′-UCCAUAACCAACGACUUGCdTdT-3′; menin, 5′-GUCGCAAGUGCAGAUGAAGdTdT-3′ and 5′-CUUCAUCUGCACUUGCGACdTdT-3′.
Real-time quantitative PCR analysis of HoxA9 expression.
Total RNA (1 μg) was reverse transcribed by using an oligo(dT) primer and Superscript First-Strand Synthesis System for reverse transcription (RT)-PCR (Invitrogen) according to the manufacturer's instructions. The reaction products were diluted (50×) with Tris-EDTA buffer, and 5 μl was subjected to real-time PCR, which was performed in triplicate using Taqman probes and the ABI prism 7700 sequence detection system. Taqman probes for HoxA9 (Hs00365956_m1) and GAPDH (Hs99999905_m1) were purchased from Applied Biosystems. Relative expression levels of HoxA9 were calculated using a standard curve, and the relative quantitation method used was that described in ABI User Bulletin no. 2.
RESULTS
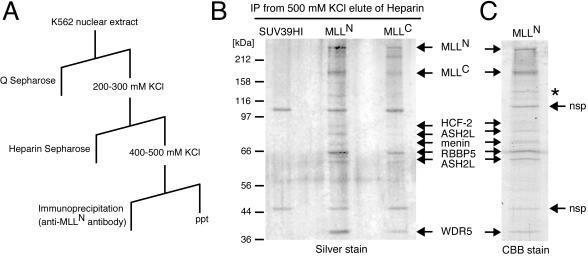
MLL associates with a cohort of proteins shared with the yeast and human SET1 HMT complexes. MLL and associated factors were biochemically purified by using conventional and affinity chromatography. Nuclear extract prepared from the K562 erythroleukemia cell line was subjected to Q-Sepharose (anion exchange) and heparin Sepharose chromatography and then was immunoabsorbed by using a monoclonal antibody specific for the p300 subunit of MLL (MLLN) (Fig. 1A). Denaturing gel electrophoretic analysis of the MLLN immunopurification product revealed the presence of eight polypeptides that were not present in control purifications with an unrelated monoclonal antibody (anti-SUV39H1) (Fig. 1B). A similar polypeptide profile was observed when the purification was performed with a monoclonal antibody specific for the p180 MLLC subunit (Fig. 1B). The eight polypeptides were identified by mass spectrometry to be the processed portions of MLL (MLLN and MLLC) and six previously known but mostly uncharacterized proteins, including HCF-2, ASH2L1 and -2 (human homologs of Drosophila Ash2) (29, 57), menin (gene product of MEN1), RBBP5 (also called RBQ-3) (49), and WDR5 (also called BIG-3) (20).
FIG. 1.
Purification of MLL and identification of associated proteins. (A) The scheme for purification of MLL complex indicates the chromatography and elution conditions employed. ppt, precipitate. (B) Purification of MLL complex. The protein eluted at 500 mM KCl from heparin Sepharose (Hep500 fraction) was subjected to immunopurification (IP) using protein G beads conjugated with anti-MLLN, anti-MLLC, or anti-SUV39H1 (negative control) monoclonal antibodies as indicated at the top of the gel. Bound proteins were eluted, subjected to SDS-PAGE, and visualized by silver staining. Molecular sizes of protein markers are shown on the left. (C) A large-scale purification sample was eluted from anti-MLLN beads, fractionated in SDS-7% PAGE, and stained with Coomassie brilliant blue. Visualized bands were excised and analyzed by LC-MS/MS. Arrows indicate protein identities. *, MLL degradation product; nsp, nonspecific products that bind to beads alone.
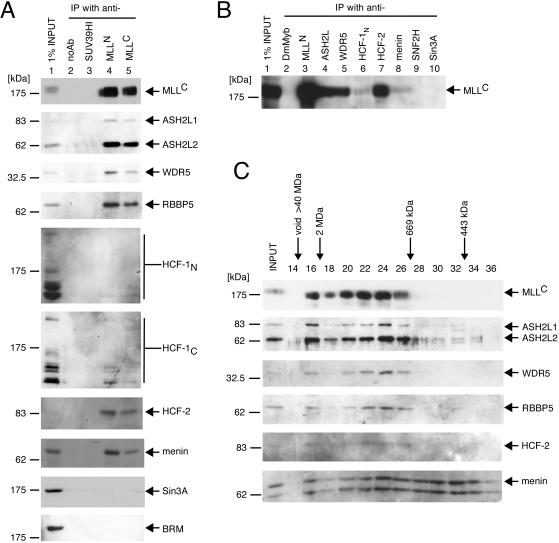
The composition of the MLL multiprotein complex in K562 cells was further confirmed by immunoprecipitation analysis of nuclear extracts. ASH2L1 and -2, WDR5, RBBP5, menin, and HCF-2 were each detected by specific antisera on Western blot analysis of the immunoprecipitates generated with anti-MLLN or -MLLC antibodies (Fig. 2A). HCF-1 (a homolog of HCF-2) was also detected in the MLL immunoprecipitates (Fig. 2A). Multiple HCF-1 bands on Western blot analysis reflect its known proteolytic processing into several different-sized polypeptides (58, 59). Its coprecipitation efficiency was much less than that of HCF-2, suggesting that although MLL associates with HCF-1, HCF-2 is a preferred component of the MLL complex in K562 cells. In contrast to a previous report (44), we were unable to coprecipitate either the corepressor Sin3A or chromatin remodeling protein BRM with MLLN or MLLC. Immunoprecipitation analysis was also performed with antibodies specific for individual components of the MLL complex, which clearly demonstrated the presence of MLLC in immunoprecipitates of ASH2L, WDR5, HCF-1, HCF-2, and menin, respectively (Fig. 2B). However, MLLC was not detected in the immunoprecipitates of Sin3A or hSNF2H, previously reported as components of an MLL supercomplex (44).
FIG. 2.
Coprecipitation of MLL with associated factors. (A) A nuclear extract of K562 cells (lane 1 input) was subjected to immunoprecipitation (IP) using antibodies specific for MLLN (lane 4) or MLLC (lane 5). As negative controls, precipitations were also performed with an SUV39H1 antibody (lane 3) or no antibody (noAb; lane 2). The immunoprecipitates were fractionated in SDS-PAGE and then immunoblotted with the antibodies indicated to the right of the panels (anti-MLLC [mmC2.1], anti-ASH2L, anti-WDR5, anti-RBBP5 [BL766], anti-HCF-1N [N18], anti-HCF-1C [H12], anti-HCF-2, anti-menin [C19], anti-Sin3A [K-20], and anti-BRM [N-19], respectively). Molecular sizes of marker proteins are shown on the left. (B) A nuclear extract of K562 cells was subjected to immunoprecipitation analysis using antibodies specific for various components of the MLL complex as indicated at the tops of the respective lanes (anti-MLLN [rpN1], anti-ASH2L, anti-WDR5, anti-HCF-1N [N18], anti-HCF-2, and anti-menin [H-300], respectively). Precipitations were also performed with antibodies specific for SNF2H (H-300) and Sin3A (K-20) or Drosophila Myb as a negative control. The immune precipitates were separated in SDS-PAGE and then immunoblotted with a monoclonal antibody specific for MLLC (mmC2.1). (C) The protein eluate from heparin Sepharose chromatography (Hep500 fraction) obtained according to the purification scheme shown in Fig. 1 was subjected to Superose 6 gel filtration chromatography. Each fraction was concentrated by acetone precipitation, fractionated in SDS-PAGE, and immunoblotted with antibodies indicated to the right of the respective panels (anti-MLLC [mmC2.1], anti-ASH2L, anti-WDR5, anti-RBBP5 [BL766], anti-HCF-2, and anti-menin [C19], respectively). MLL-associated factors preferentially cofractionated with MLL, which peaked in fraction 24 (approximately 1 MDa). Elution of standard molecular size markers is indicated at the top.
Further evidence in support of the observed MLL complex composition was obtained by gel filtration chromatography. The fraction eluted at 500 mM KCl from heparin Sepharose (Hep500) purified according to the scheme depicted in Fig. 1 was subjected to Superose 6 gel filtration chromatography. Fractions were concentrated by acetone precipitation and then were immunoblotted with antibodies specific for components of the MLL complex. Elution of MLLC and other complex components overlapped extensively and was broadly distributed from the void to fraction 26, peaking at fraction 24 (approximately 1 MDa), consistent with the predicted size of the MLL complex. The elution profile of menin was broader, indicating that not all menin forms a complex with MLL (Fig. 2C). A minor MLL peak at fraction 16 suggested the presence of another form of MLL sizing at more than 2 MDa, which is reminiscent of the MLL supercomplex (Fig. 2C). However, immunoprecipitates from either nuclear extract or the Hep500 fraction did not contain MLL supercomplex components, such as Sin3A or BRM (Fig. 2A and unpublished data). In conclusion, our data demonstrate that MLL forms a multiprotein complex with ASH2L, WDR5, RBBP5, menin, and HCF proteins. We hereafter refer to this complex as MLL/HCF to distinguish it from other possible forms of MLL.
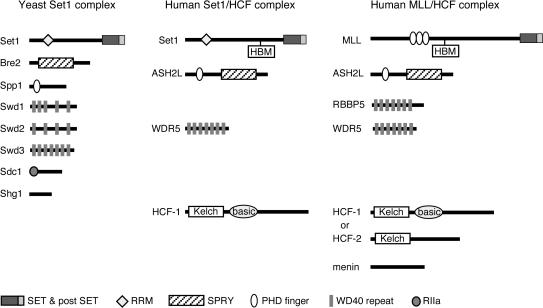
The composition of MLL/HCF shares considerable similarity with the previously characterized SET1 HMT complexes of Saccharomyces cerevisiae and humans (41, 43, 47, 61) (Fig. 3). All three complexes contain homologs of Drosophila Ash2, a trxG gene product required for imaginal disk pattern formation (36). The human homolog, ASH2L, is expressed as two isoforms (denoted 1 and 2) by alternative splicing (57). In yeast, two gene products (Bre2 and Spp1) together appear to constitute a bipartite functional homolog of Ash2 as described previously (43). All three complexes also contain highly similar WD repeat-containing proteins. WDR5 is a mammalian homolog of two proteins (Swd2 or Swd3) which are components of the yeast SET1 complex and are required for its histone methylation (43). RBBP5, another WD repeat-containing protein, also has a homolog (Swd1) in the yeast SET1 complex but has not yet been reported in mammalian SET1. Conversely, S. cerevisiae lacks a homolog of HCF-1, which is a common component of the mammalian SET1 and MLL complexes (61). It was originally identified as a host cell factor targeted by the herpes simplex virus VP16 protein (33, 58). Menin, a tumor suppressor protein with multiple known roles and interactions (11), has not been reported to be present in the human SET1 complex and thus appears to be a unique component of MLL/HCF. Our data indicate that MLL forms a multimember complex whose composition is conserved in part with the SET1 complexes associated with histone H3 lysine 4 methylation, suggesting that they all employ similar enzymatic mechanisms.
FIG. 3.
MLL complex composition is conserved with yeast and human SET1 methyltransferase complexes. Similarities between components of the yeast SET1, human SET1/HCF-1, and human MLL/HCF complexes are shown. The protein components of each complex are shown schematically with structural domains and motifs as defined in the key at the bottom. RRM, RNA recognition motif; SET, Set domain; postSET, Set domain-associated cysteine-rich motif; PHD, PHD zinc-finger motif; SPRY, domain in SP1a and the Ryanodine receptor; RIIa, protein kinase A regulatory subunit dimerization domain motif.
The SET domain of MLLC assembles methyltransferase-associated cofactors.
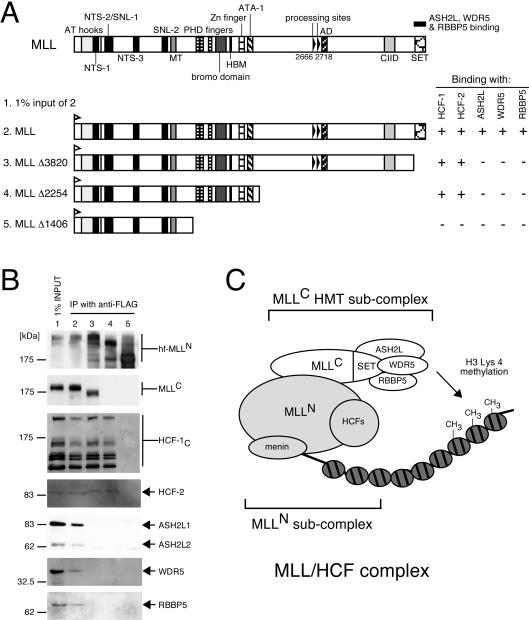
To define heterologous protein interactions within the MLL/HCF complex, various mutant forms of MLL were transiently expressed in 293 cells and were assessed for their ability to interact with and coprecipitate endogenous members of the complex. Under these conditions, endogenous HCF1, HCF2, ASH2L, RBBP5, and WDR5 readily coprecipitated with transfected wild-type MLL as assessed by immunoprecipitation-Western blot analysis (Fig. 4). A mutant MLL lacking C-terminal sequences spanning the SET domain (Δ3820), however, lost the ability to interact and coprecipitate with ASH2L, WDR5, and RBBP5 but retained an ability to coprecipitate HCF proteins. Therefore, ASH2L, WDR5, and RBBP5 interact with the p180 MLLC subunit through its SET domain, which is highly conserved with the mammalian and yeast SET1 proteins (4, 47, 61). The homologous proteins Bre2, Swd1, Swd2, and Swd3 in the yeast SET1 complex are necessary for its full HMT activity in vivo (43). Hence, it is likely that ASH2L, WDR5, and RBBP5 form an MLLC subcomplex to effect HMT catalytic activity (Fig. 4C).
FIG. 4.
Conserved methyltransferase components associate with MLLC. (A) 293 cells were transiently transfected with expression vectors encoding various MLL deletion mutants (shown schematically) containing HIS and FLAG epitope tags at their N termini. Nuclear extracts prepared from transfectants were subjected to immunoprecipitation (IP) with anti-FLAG antibody (M2). (B) Immunoprecipitates were analyzed by SDS-PAGE and were immunoblotted with antibodies indicated to the right of the respective panels (anti-His [D-8] for various MLL mutants, anti-MLLC [mmC2.1], anti-HCF-1C [H12], anti-HCF-2, anti-ASH2L, anti-WDR5, and anti-RBBP5 [BL766], respectively). Positions of molecular size markers are indicated on the left. Coprecipitation of endogenous proteins with exogenous MLL (+ or −) is indicated to the right of the schematic. (C) Composition of subcomplexes in MLL/HCF. MLL/HCF complex and chromatin with or without methylation are illustrated.
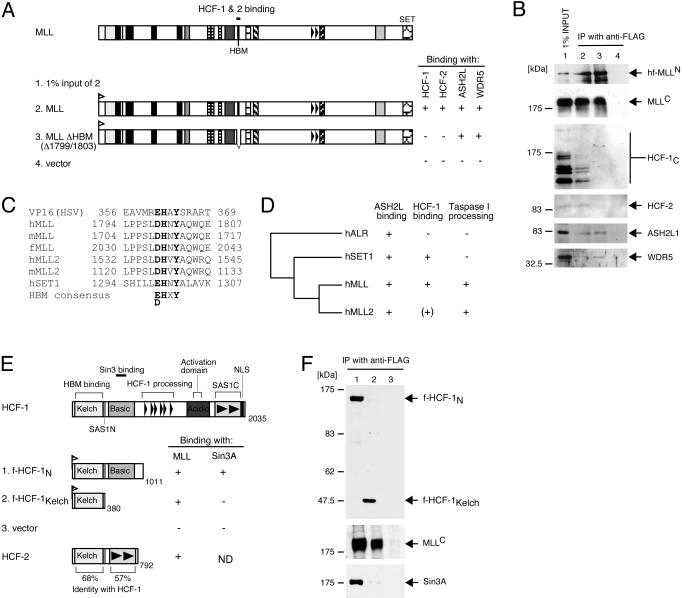
Conserved motif in MLLN mediates interactions with HCF proteins.
More refined mutation analysis indicated that interactions with HCF-1 and -2 required amino acids 1799 to 1802 of MLL, whereas interactions with ASH2L and WDR5 did not (Fig. 5A and B). This sequence fits a consensus (D/EHXY) for the previously defined HBM (Fig. 5C) found in all proteins known to physically associate with HCF-1 through its amino-terminal Kelch domain, including VP16 (18, 38). In addition to MLL and its paralogs, HBMs are conserved in mammalian MLL-related proteins MLL2 and SET1 but not in ALR (Fig. 5D). However, characterization of a menin-MLL2 complex didn't detect HCF-1 or HCF-2 (27). Therefore, the possibility of MLL2 interaction with HCFs [signified by (+) in Fig. 5D] remains to be investigated. Previous studies have shown that the HCF-1 Kelch domain binds human SET1 and that the amino-terminal portion of HCF-1 (HCF-1N) tethers SET1 with the corepressor Sin3A (61). When Flag-tagged HCF-1 mutants were stably expressed in HeLa cells, immunoprecipitation-Western blot analysis showed that the Kelch domain of HCF-1N was sufficient for interaction with MLL (Fig. 5E and F), whereas interaction with Sin3A was dependent on the adjacent basic region (Fig. 5F) as previously reported (61). Therefore, MLL interacts with HCF proteins through its HBM, consistent with the proposed mechanism of Kelch-HBM interaction.
FIG. 5.
Conserved binding motif in MLLN mediates interactions with HCF-1 and HCF-2. (A and B) 293 cells were transiently transfected with expression vectors encoding wild-type MLL or a mutant lacking the HBM (shown schematically in panel A). Nuclear extracts prepared from transfectants were subjected to immunoprecipitation (IP) with anti-FLAG antibody (M2). Immune precipitates were analyzed by SDS-PAGE and were immunoblotted with antibodies indicated to the right of the respective gels in panel B (anti-His [D-8] for various MLL mutants, anti-MLLC [mmC2.1], anti-HCF-1C [H12], anti-HCF-2, anti-ASH2L, and anti-WDR5, respectively). Coprecipitation of endogenous proteins associated with MLL (+ or −) is summarized to the right of the schematic. (C) Amino acid sequence alignment of HBMs present in HSV VP16, human, mouse, and fugu MLL, human and mouse MLL2, and human SET1. The HBM consensus (D/EHXY) is highlighted in bold. (D) Dendrogram of MLL superfamily proteins summarizing their known or predicted abilities to interact with ASH2 and HCF-1 or to undergo proteolytic processing [summarized by +, −, or (+)]. + indicates that HBM is present in MLL2 but HCF-1 and -2 interactions are yet to be identified. (E) Schematic representation of HCF-1 and HCF-2 constructs assessed for their abilities to interact with MLL or Sin3A (summarized by +, −, or ND [not determined] to right of schematics). (F) Nuclear extracts were prepared from cells transduced with f-HCF-1N (lane 1), f-HCF-1Kelch (lane 2), or empty vector (lane 3). Extracts were diluted to a concentration of 100 mM KCl and were directly incubated with FLAG antibody beads (M2). Immunoprecipitates were washed and eluted from the beads with peptide and were analyzed by immunoblotting with antibodies specific for HCF-1N (top gel, N18), MLLC (middle gel, mmC2.1), or Sin3A (bottom gel, K-20). Samples were normalized to cell equivalents. NLS, nuclear localization signal.
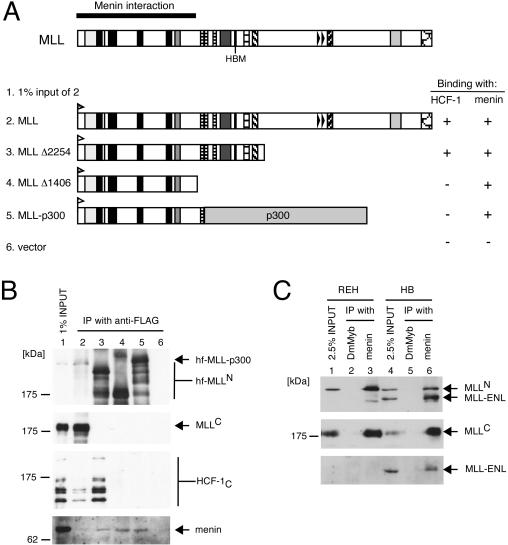
Menin interacts with wild-type MLL and leukemic MLL fusion proteins.
Domain mapping revealed that menin interaction occurs within the first 1,406 amino acids of MLLN (Fig. 6A and B). This portion is consistently retained in all MLL fusion proteins associated with human leukemias. Immunoprecipitation-Western blot analysis showed that endogenous menin coprecipitated with the MLL-p300 fusion protein in transfected 293 cells (Fig. 6B). To further confirm that menin interacts with MLL fusion proteins, coimmunoprecipitation assays were performed with the HB cell line, which expresses endogenous MLL-ENL fusion protein due to chromosomal translocation. Immunoprecipitation of menin resulted in the coprecipitation of both wild-type and fusion MLL proteins from HB cells (Fig. 6C, lane 6). A similar analysis using REH cells, which lack an MLL chromosomal translocation, resulted in coprecipitation of only wild-type MLL with menin. Therefore, it is likely that all MLL fusion proteins interact with menin, which is the only identified component of the MLL/HCF complex that associates with mutant MLL proteins and thus is potentially implicated in leukemia pathogenesis.
FIG. 6.
Menin interacts with wild-type and oncogenic MLL proteins. (A and B) 293 cells were transiently transfected with expression vectors encoding various MLL deletion or fusion mutants (shown schematically in panel A) containing HIS and FLAG epitope tags at their N termini. Nuclear extracts prepared from transfectants were subjected to immunoprecipitation (IP) with anti-FLAG antibody (M2). Immune precipitates were analyzed by SDS-PAGE and were immunoblotted with antibodies indicated to the right of the respective panels (for panel B, anti-His [D-8] for various MLL mutants, anti-MLLC [mmC2.1], anti-HCF-1C [H12], and antimenin [C19], respectively). Positions of molecular size markers are indicated on the left. Coprecipitation of endogenous menin with exogenous MLL (+ or −) is indicated to the right of the schematic. (C) Nuclear extracts of REH and HB cells were subjected to immunoprecipitation using antimenin antibody (C19) or a negative control antibody (anti-DmMyb). Immune precipitates were analyzed by SDS-PAGE and were immunoblotted with anti-MLLN (mmN4.4, top gel), anti-MLLC (mmC2.1, middle gel), and anti-ENL antibodies (bottom gel) as indicated to the right of the panels. Wild-type MLL coprecipitated with menin in REH cells (lane 3). Both wild-type and fusion (MLL-ENL) MLL proteins coprecipitated with menin in HB cells (lane 6).
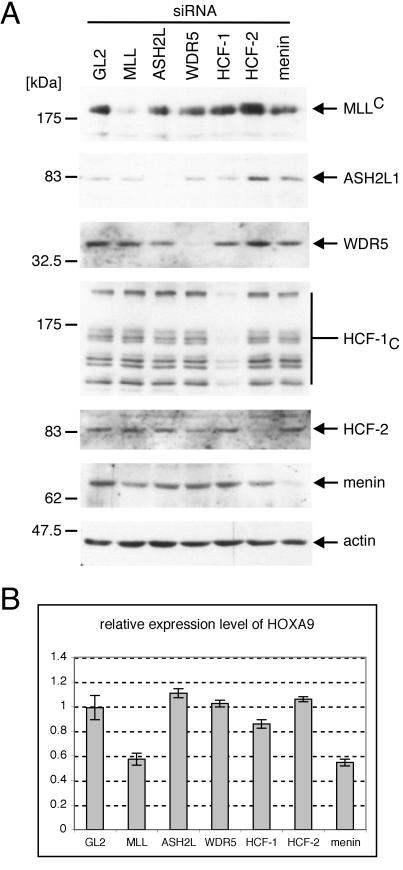
Menin, but not other components, is required for maintenance of HoxA9 gene expression.
The requirement for each component of the MLL/HCF complex in the maintenance of Hox gene expression was investigated in HeLa cells. The expression of complex components was specifically eliminated by siRNA techniques, and knockdown efficiencies were estimated by Western blotting, which revealed more than 80% reduction in expression for each component (Fig. 7A). Quantitative real-time RT-PCR analysis showed that knockdown of MLL expression resulted in a significant downregulation (50% decrease) in HoxA9 mRNA levels relative to that of GAPDH, whereas knockdown of the control GL2 protein showed no effect on HoxA9 (Fig. 7B). Despite more than 80% knockdown efficiency of ASH2L, WDR5, HCF-1, and HCF-2, their reductions had no measurable effect on HoxA9 expression, suggesting that low levels of these components may be sufficient for maintenance of HoxA9 transcription in HeLa cells. Conversely, menin knockdown caused downregulation of HoxA9 transcripts comparable to that observed for MLL knockdown (Fig. 7B). Therefore, we conclude that menin is an essential component that is required for maintenance of HoxA9 gene expression.
FIG. 7.
Menin is required for maintenance of Hox gene expression. (A and B) HeLa cells were subjected to three rounds of transfection with siRNAs specific for transcripts encoding components of MLL/HCF complex (indicated at the tops of gel lanes). RNA and protein samples were prepared 3 days after the initial transfection. Proteins were subjected to Western blot analysis with the antibodies indicated to the right of the panels (anti-MLLC [mmC2.1], anti-ASH2L, anti-WDR5, anti-HCF-1C [H12], anti-HCF-2, antimenin [C19], and antiactin [C4], respectively), which showed specific and efficient knockdown of protein expression. RNA samples were reverse transcribed and used for quantitative real-time PCR analysis for HoxA9 and GAPDH expression determined in triplicate. (B) Relative expression of HoxA9 to GAPDH is shown with error bars indicating standard deviations.
DISCUSSION
MLL forms a conserved SET1-like HMT complex. Using a biochemical approach, we have established that MLL forms a higher order protein complex that shares several notable features with the yeast and human SET1 HMT complexes (Fig. 3). All three complexes contain HMT enzymes with carboxy-terminal SET domains that are highly conserved with each other and define a specific branch of the larger SET domain protein superfamily. All three complexes also contain homologs of Drosophila Ash2, a Trx-G protein whose presence in the MLL complex provides a biochemical rationale for the similar genetic roles of these two proteins in Hox gene regulation in the fly embryo (8, 36, 51). As noted previously, the S. cerevisiae homolog of Ash2 appears to comprise two polypeptides (Bre2 and Spp1) separately containing the highly conserved SPRY and PHD finger motifs present in the single polypeptides of Drosophila Ash2 and mammalian ASH2L. All three HMT complexes also contain conserved WD40 repeat-containing proteins (WDR5 and/or RBBP5). A recently reported menin-MLL2 (the closest homolog of MLL) complex also contains ASH2L and RBBP5 (27). The yeast homologs of WDR5 and RBBP5 (Swd2/3 and Swd1, respectively) are necessary for methylation of histone H3 on lysine 4, possibly mediating histone binding (43). Detailed domain mapping revealed that WDR5, RBBP5, and ASH2L interact with MLL through its SET domain, suggesting that MLLC specifically assembles an HMT subcomplex (Fig. 4). The distantly related ALR, whose homology with MLL and SET1 is confined to the SET domain, also has been shown to associate with ASH2L and RBBP5 (19). Conversely, more remotely related SET domain proteins, such as SUV39H1 and EZH, apparently do not associate with ASH2L and RBBP5 (Fig. 2 and unpublished data) (35). Therefore, association with ASH2L, WDR5, and RBBP5 is a characteristic feature of the MLL-SET1-ALR subgroup of SET domain proteins and is likely to dictate features of their highly specific HMT activities.
The composition of the MLL/HCF complex reported here differs substantially from that reported for previous purifications of MLL and trithorax. Purification of trithorax from Drosophila embryos revealed its stable association with the transcriptional coactivator CBP and the pseudophosphatase Sbf1, both of which are capable of interacting in vitro with MLL (13, 16, 45). However, neither protein was detected here in MLL/HCF or previously in the MLL super complex. The latter contains WDR5 and RBBP5 but lacks ASH2L and shares no other components with MLL/HCF. Furthermore, we were unable to coprecipitate MLL with supercomplex components such as Sin3a, hSNF2H, and BRM. Although technical factors could account for these significant differences in composition, it is also possible that MLL complex composition varies with different cellular conditions and/or subcellular localizations (44). Nevertheless, the composition of MLL/HCF reported here indicates an evolutionary conservation of specific cofactors for a defined subclass of HMTs that mediate histone H3 lysine 4 methylation. In this context, studies of yeast SET1 should continue to provide a tractable model for understanding the function of MLL.
MLL is a candidate target for regulation by HCF proteins.
A novel feature of the mammalian SET1 and MLL complexes that distinguishes them from yeast SET1 is the presence of HCF proteins, which have no recognizable homologs in S. cerevisiae. HCF-1 was originally identified as a key cellular target of the herpes simplex virus VP16 transactivator protein (33, 58). HCF-1 binds VP16 and also several cellular proteins through its Kelch domain, which recognizes a conserved sequence motif known as the HBM (18, 38). MLL contains a short sequence that matches the HBM consensus (39), and this motif is required for its association with HCF-1 and the related HCF-2 protein. Because the Kelch domains of HCF-1 and HCF-2 are 68% identical, they likely bind MLL by similar mechanisms. An HBM is present in human SET1 but not in the more distantly related ALR (Fig. 5D). MLL2 contains an HBM in addition to endoprotease processing sites, suggesting that MLL2 may have functions and heterologous protein interactions similar to those of MLL. Taken together, our biochemical data and the conserved presence of HBM sequences provide strong evidence that MLL and its close relatives are partners for HCF proteins in vivo.
The physiological significance of MLL-HCF association is presently unclear. HCF-1 has features of a transcriptional coregulator because it can selectively tether together the Sin3 deacetylase and SET1 methyltransferase complexes, whose enzymatic activities are associated with opposing effects on the transcriptional state of chromatin. Despite this role for HCF-1 and previous reports of Sin3 and deacetylase interactions with MLL, we were unable to demonstrate the association or coprecipitation of Sin3 or histone deacetylases (HDACs) in MLL/HCF. One possibility is that their interactions are highly transient and target gene specific. Furthermore, the vast majority of MLL in K562 cells is associated with HCF-2, which lacks a basic region motif that mediates the interaction of HCF-1 with Sin3-HDAC (Fig. 5E). Thus, HCF-2 should be incapable of tethering MLL with Sin3-HDAC, which may account for their absence in the MLL/HCF complex identified in our studies. This would be consistent with the proposed model that switching between HCF-1 and HCF-2 partners may mediate interconversion between on and off transcriptional states (60). Taken together, these data raise the interesting possibility that HCF proteins function as transcriptional switches for the actions of MLL on select target genes, and future studies are warranted to address this possibility.
Menin, unlike other components, is essential for MLL target gene expression.
Our studies identify MLL as a new protein partner for menin, which has not been reported in the SET1 or other HMT complexes, thereby providing a link between this tumor suppressor protein and the HMT machinery. Initially identified as a product of the MEN1 tumor suppressor gene, menin loss of function plays a significant role in human neoplasms of multiple endocrine organs (10). Despite extensive genetic and biochemical analyses, the biological functions of menin remain unclear. Its tumor suppressor role may be mediated in part through an ability to tether JunD with the Sin3-HDAC complex, thereby converting it from a growth promoter to a growth suppressor (2, 3, 32). However, menin reportedly interacts with a wide variety of additional transcriptional proteins and with intermediate filaments (46). It has also been implicated in telomere biology and DNA replication and repair (30, 37, 55). Thus, its biological and tumor suppressor functions may be broad and pleiotropic. The potential physiological importance of menin-MLL interactions is underscored by our results showing that menin is required for proper maintenance of HoxA9 gene expression in a model system that reads out MLL transcriptional function in HeLa cells (24, 44). Recently, menin has also been shown to be required for maintenance of HoxC8 expression in mouse embryo fibroblasts (27). Menin was the only MLL/HCF complex component whose knockdown phenocopied loss of MLL, an unexpected result considering the contributions of the homologs of WDR5 and ASH2L for full HMT activity of yeast SET1 (43). Because their knockdown was not complete, the results suggest that even low levels of these SET domain-interacting components may be sufficient for maintenance of HoxA9 expression by wild-type MLL in a cell line model system, in contrast to a more sensitive dependence on the levels of menin.
The requirement for menin in maintenance of HoxA9 gene expression by wild-type MLL raises the possibility that menin may contribute to MLL-mediated leukemogenesis. HoxA9 is a critical target gene of MLL fusion proteins and is consistently expressed in leukemic cells carrying MLL translocations, serving as a molecular signature for this subtype of acute leukemia (5, 7, 48, 63). Our studies indicate that menin contacts the amino-terminal third of MLL, a region that is retained in all leukemic MLL fusion proteins, and coprecipitates with the latter from leukemia cell lines. Thus, menin is distinguished as the only identified MLL/HCF component that interacts with both wild-type and fusion MLL proteins, both of which are implicated in maintenance of HoxA9 gene expression. Genetic studies with appropriate model systems are necessary to establish if menin is required for MLL-mediated leukemogenesis and potentially constitutes a common therapeutic target shared by the diverse array of MLL fusion proteins. Our studies also raise the possibility that the developmental and tumor suppressor roles of menin could be mediated in part through effects on MLL function. In this regard, it will be of interest to assess mouse embryos deficient for MEN1 and tumors resulting from MEN1-inactivating mutations for aberrations in Hox gene expression or histone methylation.
Acknowledgments
We thank J. Lipsick and J. Manak for generously providing a rabbit polyclonal anti-Drosophila Myb antibody, P. Nagy for comments on the manuscript, and Bich-Tien Rouse for technical assistance.
A.Y. was supported by the Uehara Memorial Foundation and an ASH Scholar Award from the American Society of Hematology. We acknowledge support from the Children's Health Initiative; PHS grants CA55029 (M.L.C.), CA13106 (W.H.), and GM54598 (W.H.); and the Program for Promotion of Fundamental Studies in Health Sciences of the Organization for Pharmaceutical Safety and Research of Japan (I.K.).
REFERENCES
- 1.Aasland, R., T. J. Gibson, and A. F. Stewart. 1995. The PHD finger: implications for chromatin-mediated transcriptional regulation. Trends Biochem. Sci. 20:56-59. [DOI] [PubMed] [Google Scholar]
- 2.Agarwal, S. K., S. C. Guru, C. Heppner, M. R. Erdos, R. M. Collins, S. Y. Park, S. Saggar, S. C. Chandrasekharappa, F. S. Collins, A. M. Spiegel, S. J. Marx, and A. L. Burns. 1999. Menin interacts with the AP1 transcription factor JunD and represses JunD-activated transcription. Cell 96:143-152. [DOI] [PubMed] [Google Scholar]
- 3.Agarwal, S. K., E. A. Novotny, J. S. Crabtree, J. B. Weitzman, M. Yaniv, A. L. Burns, S. C. Chandrasekharappa, F. S. Collins, A. M. Spiegel, and S. J. Marx. 2003. Transcription factor JunD, deprived of menin, switches from growth suppressor to growth promoter. Proc. Natl. Acad. Sci. USA 100:10770-10775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alvarez-Venegas, R., and Z. Avramova. 2002. SET-domain proteins of the Su(var)3-9, E(z) and trithorax families. Gene 285:25-37. [DOI] [PubMed] [Google Scholar]
- 5.Armstrong, S. A., J. E. Staunton, L. B. Silverman, R. Pieters, M. L. den Boer, M. D. Minden, S. E. Sallan, E. S. Lander, T. R. Golub, and S. J. Korsmeyer. 2002. MLL translocations specify a distinct gene expression profile that distinguishes a unique leukemia. Nat. Genet. 30:41-47. [DOI] [PubMed] [Google Scholar]
- 6.Ayton, P. M., and M. L. Cleary. 2001. Molecular mechanisms of leukemogenesis mediated by MLL fusion proteins. Oncogene 20:5695-5707. [DOI] [PubMed] [Google Scholar]
- 7.Ayton, P. M., and M. L. Cleary. 2003. Transformation of myeloid progenitors by MLL oncoproteins is dependent on Hoxa7 and Hoxa9. Genes Dev. 17:2298-2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Breen, T. R., and P. J. Harte. 1993. Trithorax regulates multiple homeotic genes in the bithorax and antennapedia complexes and exerts different tissue-specific, parasegment-specific and promoter-specific effects on each. Development 117:119-134. [DOI] [PubMed] [Google Scholar]
- 9.Butler, L. H., R. Slany, X. Cui, M. L. Cleary, and D. Y. Mason. 1997. The HRX proto-oncogene product is widely expressed in human tissues and localizes to nuclear structures. Blood 89:3361-3370. [PubMed] [Google Scholar]
- 10.Chandrasekharappa, S. C., S. C. Guru, P. Manickam, S. E. Olufemi, F. S. Collins, M. R. Emmert-Buck, L. V. Debelenko, Z. Zhuang, I. A. Lubensky, L. A. Liotta, J. S. Crabtree, Y. Wang, B. A. Roe, J. Weisemann, M. S. Boguski, S. K. Agarwal, M. B. Kester, Y. S. Kim, C. Heppner, Q. Dong, A. M. Spiegel, A. L. Burns, and S. J. Marx. 1997. Positional cloning of the gene for multiple endocrine neoplasia-type 1. Science 276:404-407. [DOI] [PubMed] [Google Scholar]
- 11.Chandrasekharappa, S. C., and B. T. Teh. 2003. Functional studies of the MEN1 gene. J. Intern. Med. 253:606-615. [DOI] [PubMed] [Google Scholar]
- 12.Collins, E. C., and T. H. Rabbitts. 2002. The promiscuous MLL gene links chromosomal translocations to cellular differentiation and tumour tropism. Trends Mol. Med. 8:436-442. [DOI] [PubMed] [Google Scholar]
- 13.Cui, X., I. De Vivo, R. Slany, A. Miyamoto, R. Firestein, and M. L. Cleary. 1998. Association of SET domain and myotubularin-related proteins modulates growth control. Nat. Genet. 18:331-337. [DOI] [PubMed] [Google Scholar]
- 14.Dimartino, J. F., and M. L. Cleary. 1999. Mll rearrangements in haematological malignancies: lessons from clinical and biological studies. Br. J. Haematol. 106:614-626. [DOI] [PubMed] [Google Scholar]
- 15.Djabali, M., L. Selleri, P. Parry, M. Bower, B. D. Young, and G. A. Evans. 1992. A trithorax-like gene is interrupted by chromosome 11q23 translocations in acute leukaemias. Nat. Genet. 2:113-118. [DOI] [PubMed] [Google Scholar]
- 16.Ernst, P., J. Wang, M. Huang, R. H. Goodman, and S. J. Korsmeyer. 2001. MLL and CREB bind cooperatively to the nuclear coactivator CREB-binding protein. Mol. Cell. Biol. 21:2249-2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Firestein, R., X. Cui, P. Huie, and M. L. Cleary. 2000. SET domain-dependent regulation of transcriptional silencing and growth control by SUV39H1, a mammalian ortholog of Drosophila Su(var)3-9. Mol. Cell. Biol. 20:4900-4909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Freiman, R. N., and W. Herr. 1997. Viral mimicry: common mode of association with HCF by VP16 and the cellular protein LZIP. Genes Dev. 11:3122-3127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goo, Y. H., Y. C. Sohn, D. H. Kim, S. W. Kim, M. J. Kang, D. J. Jung, E. Kwak, N. A. Barlev, S. L. Berger, V. T. Chow, R. G. Roeder, D. O. Azorsa, P. S. Meltzer, P. G. Suh, E. J. Song, K. J. Lee, Y. C. Lee, and J. W. Lee. 2003. Activating signal cointegrator 2 belongs to a novel steady-state complex that contains a subset of trithorax group proteins. Mol. Cell. Biol. 23:140-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gori, F., and M. B. Demay. 2004. BIG-3, a novel WD-40 repeat protein, is expressed in the developing growth plate and accelerates chondrocyte differentiation in vitro. Endocrinology 145:1050-1054. [DOI] [PubMed] [Google Scholar]
- 21.Goto, H., S. Motomura, A. C. Wilson, R. N. Freiman, Y. Nakabeppu, K. Fukushima, M. Fujishima, W. Herr, and T. Nishimoto. 1997. A single-point mutation in HCF causes temperature-sensitive cell-cycle arrest and disrupts VP16 function. Genes Dev. 11:726-737. [DOI] [PubMed] [Google Scholar]
- 22.Gu, Y., T. Nakamura, H. Alder, R. Prasad, O. Canaani, G. Cimino, C. M. Croce, and E. Canaani. 1992. The t(4;11) chromosome translocation of human acute leukemias fuses the ALL-1 gene, related to Drosophila trithorax, to the AF-4 gene. Cell 71:701-708. [DOI] [PubMed] [Google Scholar]
- 23.Hanson, R. D., J. L. Hess, B. D. Yu, P. Ernst, M. van Lohuizen, A. Berns, N. M. van der Lugt, C. S. Shashikant, F. H. Ruddle, M. Seto, and S. J. Korsmeyer. 1999. Mammalian trithorax and polycomb-group homologues are antagonistic regulators of homeotic development. Proc. Natl. Acad. Sci. USA 96:14372-14377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hsieh, J. J., E. H. Cheng, and S. J. Korsmeyer. 2003. Taspase1: a threonine aspartase required for cleavage of MLL and proper HOX gene expression. Cell 115:293-303. [DOI] [PubMed] [Google Scholar]
- 25.Hsieh, J. J., P. Ernst, H. Erdjument-Bromage, P. Tempst, and S. J. Korsmeyer. 2003. Proteolytic cleavage of MLL generates a complex of N- and C-terminal fragments that confers protein stability and subnuclear localization. Mol. Cell. Biol. 23:186-194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hsu, K., and A. T. Look. 2003. Turning on a dimer: new insights into MLL chimeras. Cancer Cell 4:81-83. [DOI] [PubMed] [Google Scholar]
- 27.Hughes, C. M., O. Rozenblatt-Rosen, T. A. Milne, T. D. Copeland, S. S. Levine, J. C. Lee, D. N. Hayes, K. S. Shanmugam, A. Bhattacharjee, C. A. Biondi, G. F. Kay, N. K. Hayward, J. L. Hess, and M. Meyerson. 2004. Menin associates with a trithorax family histone methyltransferase complex and with the hoxc8 locus. Mol. Cell 13:587-597. [DOI] [PubMed] [Google Scholar]
- 28.Huret, J. L., P. Dessen, and A. Bernheim. 2001. An atlas of chromosomes in hematological malignancies. Example: 11q23 and MLL partners. Leukemia 15:987-989. [DOI] [PubMed] [Google Scholar]
- 29.Ikegawa, S., M. Isomura, Y. Koshizuka, and Y. Nakamura. 1999. Cloning and characterization of ASH2L and Ash2l, human and mouse homologs of the Drosophila ash2 gene. Cytogenet. Cell Genet. 84:167-172. [DOI] [PubMed] [Google Scholar]
- 30.Jin, S., H. Mao, R. W. Schnepp, S. M. Sykes, A. C. Silva, A. D. D'Andrea, and X. Hua. 2003. Menin associates with FANCD2, a protein involved in repair of DNA damage. Cancer Res. 63:4204-4210. [PubMed] [Google Scholar]
- 31.Johnson, K. M., S. S. Mahajan, and A. C. Wilson. 1999. Herpes simplex virus transactivator VP16 discriminates between HCF-1 and a novel family member, HCF-2. J. Virol. 73:3930-3940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim, H., J. E. Lee, E. J. Cho, J. O. Liu, and H. D. Youn. 2003. Menin, a tumor suppressor, represses JunD-mediated transcriptional activity by association with an mSin3A-histone deacetylase complex. Cancer Res. 63:6135-6139. [PubMed] [Google Scholar]
- 33.Kristie, T. M., J. L. Pomerantz, T. C. Twomey, S. A. Parent, and P. A. Sharp. 1995. The cellular C1 factor of the herpes simplex virus enhancer complex is a family of polypeptides. J. Biol. Chem. 270:4387-4394. [DOI] [PubMed] [Google Scholar]
- 34.Kumar, A. R., W. A. Hudson, W. Chen, R. Nishiuchi, Q. Yao, and J. H. Kersey. 2003. Hoxa9 influences the phenotype but not the incidence of Mll-AF9 fusion gene leukemia. Blood 100:1823-1828. [DOI] [PubMed] [Google Scholar]
- 35.Kuzmichev, A., K. Nishioka, H. Erdjument-Bromage, P. Tempst, and D. Reinberg. 2002. Histone methyltransferase activity associated with a human multiprotein complex containing the Enhancer of Zeste protein. Genes Dev. 16:2893-2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.LaJeunesse, D., and A. Shearn. 1995. Trans-regulation of thoracic homeotic selector genes of the antennapedia and bithorax complexes by the trithorax group genes: absent, small, and homeotic discs 1 and 2. Mech. Dev. 53:123-139. [DOI] [PubMed] [Google Scholar]
- 37.Lin, S. Y., and S. J. Elledge. 2003. Multiple tumor suppressor pathways negatively regulate telomerase. Cell 113:881-889. [DOI] [PubMed] [Google Scholar]
- 38.Lu, R., P. Yang, S. Padmakumar, and V. Misra. 1998. The herpesvirus transactivator VP16 mimics a human basic domain leucine zipper protein, luman, in its interaction with HCF. J. Virol. 72:6291-6297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Luciano, R. L., and A. C. Wilson. 2003. HCF-1 functions as a coactivator for the zinc finger protein Krox20. J. Biol. Chem. 278:51116-51124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Martin, M. E., T. A. Milne, S. Bloyer, K. Galoian, W. Shen, D. Gibbs, H. W. Brock, R. Slany, and J. L. Hess. 2003. Dimerization of MLL fusion proteins immortalizes hematopoietic cells. Cancer Cell 4:197-207. [DOI] [PubMed] [Google Scholar]
- 41.Miller, T., N. J. Krogan, J. Dover, H. Erdjument-Bromage, P. Tempst, M. Johnston, J. F. Greenblatt, and A. Shilatifard. 2001. COMPASS: a complex of proteins associated with a trithorax-related SET domain protein. Proc. Natl. Acad. Sci. USA 98:12902-12907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Milne, T. A., S. D. Briggs, H. W. Brock, M. E. Martin, D. Gibbs, C. D. Allis, and J. L. Hess. 2002. MLL targets SET domain methyltransferase activity to Hox gene promoters. Mol. Cell 10:1107-1117. [DOI] [PubMed] [Google Scholar]
- 43.Nagy, P. L., J. Griesenbeck, R. D. Kornberg, and M. L. Cleary. 2002. A trithorax-group complex purified from Saccharomyces cerevisiae is required for methylation of histone H3. Proc. Natl. Acad. Sci. USA 99:90-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nakamura, T., T. Mori, S. Tada, W. Krajewski, T. Rozovskaia, R. Wassell, G. Dubois, A. Mazo, C. M. Croce, and E. Canaani. 2002. ALL-1 is a histone methyltransferase that assembles a supercomplex of proteins involved in transcriptional regulation. Mol. Cell 10:1119-1128. [DOI] [PubMed] [Google Scholar]
- 45.Petruk, S., Y. Sedkov, S. Smith, S. Tillib, V. Kraevski, T. Nakamura, E. Canaani, C. M. Croce, and A. Mazo. 2001. Trithorax and dCBP acting in a complex to maintain expression of a homeotic gene. Science 294:1331-1334. [DOI] [PubMed] [Google Scholar]
- 46.Poisson, A., B. Zablewska, and P. Gaudray. 2003. Menin interacting proteins as clues toward the understanding of multiple endocrine neoplasia type 1. Cancer Lett. 189:1-10. [DOI] [PubMed] [Google Scholar]
- 47.Roguev, A., D. Schaft, A. Shevchenko, W. W. Pijnappel, M. Wilm, R. Aasland, and A. F. Stewart. 2001. The Saccharomyces cerevisiae Set1 complex includes an Ash2 homologue and methylates histone 3 lysine 4. EMBO J. 20:7137-7148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rozovskaia, T., E. Feinstein, O. Mor, R. Foa, J. Blechman, T. Nakamura, C. M. Croce, G. Cimino, and E. Canaani. 2001. Upregulation of Meis1 and HoxA9 in acute lymphocytic leukemias with the t(4:11) abnormality. Oncogene 20:874-878. [DOI] [PubMed] [Google Scholar]
- 49.Saijo, M., Y. Sakai, T. Kishino, N. Niikawa, Y. Matsuura, K. Morino, K. Tamai, and Y. Taya. 1995. Molecular cloning of a human protein that binds to the retinoblastoma protein and chromosomal mapping. Genomics 27:511-519. [DOI] [PubMed] [Google Scholar]
- 50.Schneider, R., A. J. Bannister, and T. Kouzarides. 2002. Unsafe SETs: histone lysine methyltransferases and cancer. Trends Biochem. Sci. 27:396-402. [DOI] [PubMed] [Google Scholar]
- 51.Sedkov, Y., S. Tillib, L. Mizrokhi, and A. Mazo. 1994. The bithorax complex is regulated by trithorax earlier during Drosophila embryogenesis than is the antennapedia complex, correlating with a bithorax-like expression pattern of distinct early trithorax transcripts. Development 120:1907-1917. [DOI] [PubMed] [Google Scholar]
- 52.Slany, R. K., C. Lavau, and M. L. Cleary. 1998. The oncogenic capacity of HRX-ENL requires the transcriptional transactivation activity of ENL and the DNA binding motifs of HRX. Mol. Cell. Biol. 18:122-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.So, C. W., H. Karsunky, P. Wong, I. L. Weissman, and M. L. Cleary. 2004. Leukemic transformation of hematopietic progenitors by MLL-GAS7 in the absence of Hoxa7 or Hoxa9. Blood 103:3192-3199. [DOI] [PubMed] [Google Scholar]
- 54.So, C. W., M. Lin, P. M. Ayton, E. H. Chen, and M. L. Cleary. 2003. Dimerization contributes to oncogenic activation of MLL chimeras in acute leukemias. Cancer Cell 4:99-110. [DOI] [PubMed] [Google Scholar]
- 55.Sukhodolets, K. E., A. B. Hickman, S. K. Agarwal, M. V. Sukhodolets, V. H. Obungu, E. A. Novotny, J. S. Crabtree, S. C. Chandrasekharappa, F. S. Collins, A. M. Spiegel, A. L. Burns, and S. J. Marx. 2003. The 32-kilodalton subunit of replication protein A interacts with menin, the product of the MEN1 tumor suppressor gene. Mol. Cell. Biol. 23:493-509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tkachuk, D. C., S. Kohler, and M. L. Cleary. 1992. Involvement of a homolog of Drosophila trithorax by 11q23 chromosomal translocations in acute leukemias. Cell 71:691-700. [DOI] [PubMed] [Google Scholar]
- 57.Wang, J., Y. Zhou, B. Yin, G. Du, X. Huang, G. Li, Y. Shen, J. Yuan, and B. Qiang. 2001. ASH2L: alternative splicing and downregulation during induced megakaryocytic differentiation of multipotential leukemia cell lines. J. Mol. Med. 79:399-405. [DOI] [PubMed] [Google Scholar]
- 58.Wilson, A. C., K. LaMarco, M. G. Peterson, and W. Herr. 1993. The VP16 accessory protein HCF is a family of polypeptides processed from a large precursor protein. Cell 74:115-125. [DOI] [PubMed] [Google Scholar]
- 59.Wilson, A. C., M. G. Peterson, and W. Herr. 1995. The HCF repeat is an unusual proteolytic cleavage signal. Genes Dev. 9:2445-2458. [DOI] [PubMed] [Google Scholar]
- 60.Wysocka, J., and W. Herr. 2003. The herpes simplex virus VP16-induced complex: the makings of a regulatory switch. Trends Biochem. Sci. 28:294-304. [DOI] [PubMed] [Google Scholar]
- 61.Wysocka, J., M. P. Myers, C. D. Laherty, R. N. Eisenman, and W. Herr. 2003. Human Sin3 deacetylase and trithorax-related Set1/Ash2 histone H3-K4 methyltransferase are tethered together selectively by the cell-proliferation factor HCF-1. Genes Dev. 17:896-911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Xia, Z. B., M. Anderson, M. O. Diaz, and N. J. Zeleznik-Le. 2003. MLL repression domain interacts with histone deacetylases, the polycomb group proteins HPC2 and BMI-1, and the corepressor C-terminal-binding protein. Proc. Natl. Acad. Sci. USA 100:8342-8347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yeoh, E. J., M. E. Ross, S. A. Shurtleff, W. K. Williams, D. Patel, R. Mahfouz, F. G. Behm, S. C. Raimondi, M. V. Relling, A. Patel, C. Cheng, D. Campana, D. Wilkins, X. Zhou, J. Li, H. Liu, C. H. Pui, W. E. Evans, C. Naeve, L. Wong, and J. R. Downing. 2002. Classification, subtype discovery, and prediction of outcome in pediatric acute lymphoblastic leukemia by gene expression profiling. Cancer Cell 1:133-143. [DOI] [PubMed] [Google Scholar]
- 64.Yokoyama, A., I. Kitabayashi, P. M. Ayton, M. L. Cleary, and M. Ohki. 2002. Leukemia proto-oncoprotein MLL is proteolytically processed into 2 fragments with opposite transcriptional properties. Blood 100:3710-3718. [DOI] [PubMed] [Google Scholar]
- 65.Yu, B. D., R. D. Hanson, J. L. Hess, S. E. Horning, and S. J. Korsmeyer. 1998. MLL, a mammalian trithorax-group gene, functions as a transcriptional maintenance factor in morphogenesis. Proc. Natl. Acad. Sci. USA 95:10632-10636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yu, B. D., J. L. Hess, S. E. Horning, G. A. Brown, and S. J. Korsmeyer. 1995. Altered Hox expression and segmental identity in Mll-mutant mice. Nature 378:505-508. [DOI] [PubMed] [Google Scholar]
- 67.Zeisig, B. B., T. Milne, M. P. Garcia-Cuellar, S. Schreiner, M. E. Martin, U. Fuchs, A. Borkhardt, S. K. Chanda, J. Walker, R. Soden, J. L. Hess, and R. K. Slany. 2004. Hoxa9 and Meis1 are key targets for MLL-ENL-mediated cellular immortalization. Mol. Cell. Biol. 24:617-628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zeisig, B. B., S. Schreiner, M. P. Garcia-Cuellar, and R. K. Slany. 2003. Transcriptional activation is a key function encoded by MLL fusion partners. Leukemia 17:359-365. [DOI] [PubMed] [Google Scholar]
- 69.Zeleznik-Le, N. J., A. M. Harden, and J. D. Rowley. 1994. 11q23 translocations split the “AT-hook” cruciform DNA-binding region and the transcriptional repression domain from the activation domain of the mixed-lineage leukemia (MLL) gene. Proc. Natl. Acad. Sci. USA 91:10610-10614. [DOI] [PMC free article] [PubMed] [Google Scholar]