Summary
Dysregulated metabolism is a hallmark of cancer, manifested through alterations in metabolites. We performed metabolomic profiling on 138 matched clear cell renal cell carcinoma (ccRCC)/normal tissue pairs and found that ccRCC is characterized by broad shifts in central carbon metabolism, one-carbon metabolism and anti-oxidant response. Tumor progression and metastasis were associated with metabolite increases in glutathione and cysteine/methionine metabolism pathways. We develop an analytic pipeline and visualization tool (“metabolograms”) to bridge the gap between TCGA transcriptomic profiling and our metabolomic data, which enables us to assemble an integrated pathway-level metabolic atlas and to demonstrate discordance between transcriptome and metabolome. Lastly, expression profiling was performed on a high-glutathione cluster, which correspond to a poor-survival subgroup in the ccRCC TCGA cohort.
Graphical Abstract
Introduction
Dysregulated metabolism is a cancer hallmark and presents opportunities for cancer diagnostics, prognostics, and therapeutics (DeBerardinis et al., 2008; Hanahan and Weinberg, 2011; Hsu and Sabatini, 2008; Vander Heiden et al., 2009). Tumors reorganize their metabolism to produce sufficient energy and biosynthetic building blocks, such as nucleotides, lipids, and amino acids, for malignant cellular proliferation. Moreover, recent studies have demonstrated that a pathological accumulation of metabolic intermediates, such as fumarate and 2-hydroxyglutarate, can contribute to tumorigenesis (Kaelin and McKnight, 2013; Raimundo et al., 2011).
Clear cell renal cell carcinoma (ccRCC) is the most common (~75%), lethal subtype of kidney cancer (Funakoshi et al., 2014; Hakimi et al., 2013b; Wei and Hsieh, 2015). Morphologically, ccRCC cells are lipid- and glycogen- laden (Gebhard et al., 1987), implicating altered fatty acid and glucose metabolism in the development of ccRCC. Genetically, ccRCC is characterized by a biallelic loss of the Von Hippel-Lindau (VHL) tumor suppressor gene which encodes an E3 ubiquitin ligase that degrades hypoxia inducible factors (HIF) 1α and HIF2 α (Kaelin, 2004). Loss of VHL leads to aberrant accumulation of HIFα despite an adequately oxygenated tissue microenvironment, which in turn results in uncontrolled activation of HIFα-target genes that regulate angiogenesis, glycolysis, and apoptosis (Majmundar et al., 2010; Semenza, 2013). Interestingly, the landmark TCGA analysis of ccRCC highlighted a key role for metabolic alteration in ccRCC progression (The Cancer Genome Atlas Research et al., 2013). In that study and subsequent analysis, worse patient survival was shown to correlate with upregulation of pentose phosphate pathway and fatty acid synthesis pathway genes, and downregulation of TCA cycle genes (Hakimi et al., 2013a; The Cancer Genome Atlas Research et al., 2013). Separately, a cross-cancer study of metabolic gene expression profiles further characterized ccRCC with concerted down-regulation of most metabolic genes in comparison with other malignancies (Anders et al., 2013; Gatto et al., 2014).
The fundamental unit in studying metabolism is the activity (“flux”) of a metabolic reaction. However, the vast majority of large cancer profiling studies, including the TCGA, have studied cancer metabolism using transcriptomics data (Gatto et al., 2014; Hu et al., 2013; The Cancer Genome Atlas Research et al., 2013). While it is well established that gene expression changes of particular metabolic pathways correlate with clinical aggressiveness in ccRCC, limited large-scale metabolomics data exists to support prior findings connecting metabolism to kidney cancer pathogenesis and/or progression (Gatto et al., 2014; The Cancer Genome Atlas Research et al., 2013).
Results
Metabolic Profiling on 138 Human ccRCC Tumor-Normal Pairs
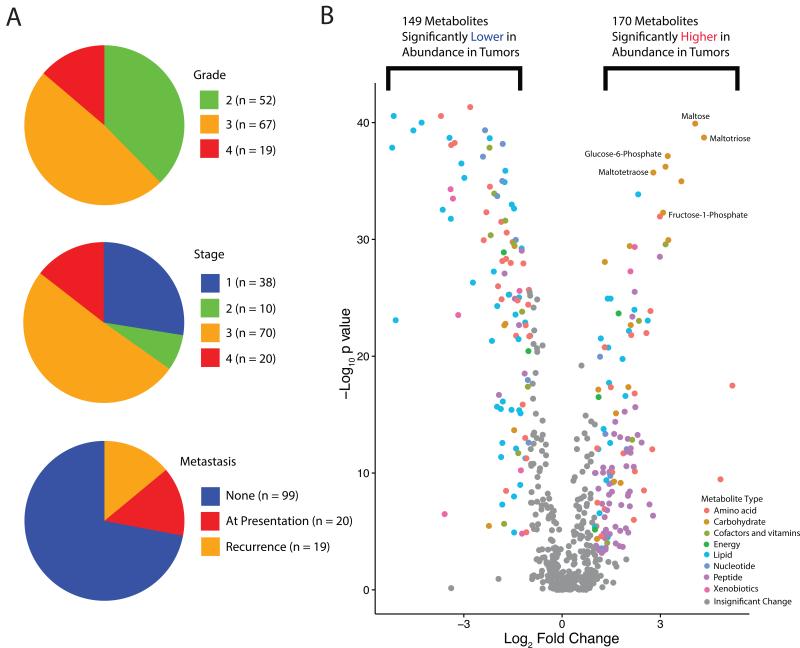
To enable comprehensive metabolomic profiling of ccRCC, we assembled a human ccRCC sample set containing sufficient quantities of fresh frozen high-quality matched tumor/adjacent normal tissue materials. This cohort included 138 ccRCC tumor-normal (T/N) pairs encompassing tumors of different Fuhrman nuclear grades and American Joint Committee on Cancer (AJCC) clinical stages (Figure 1A and Table S1). Mass spectrometry detected 877 (577 named and 300 unnamed) metabolites in these samples (Table S2). Principal component analysis showed clear separation between tumor and normal samples (Figure S1A). FDR-corrected Mann Whitney U tests identified 319 metabolites (170 higher and 149 lower) that display differential abundance between tumor and normal tissue samples (FDR-corrected p value ≤0.001) (Figure 1B). Interestingly, carbohydrates were overrepresented and highly abundant in tumors, e.g. maltotriose, maltose, maltotetraose, fructose-1-phosphate, and glucose-6-phosphate (Figure 1B). These results correlated with a prior metabolomics analysis of 20 ccRCC tumor/normal pairs (Figure S1B) (Li et al., 2014).
Figure 1. Clinical and metabolic features of the MSK ccRCC Metabolomics Cohort.
(A) Clinical characteristics of the patient cohort at presentation. Among the 118 patients who presented with Stage I-III diseases, 19 (16.1%) developed a new recurrence by the end of 2014. (B) A volcano plot of the 577 named metabolites profiled. 319 exhibited significant differential abundance (p value <0.001, absolute fold change > 2) when comparing ccRCC tumors to adjacent normal kidney tissues. Mann-Whitney U tests were used to calculate statistical significance, and p values were corrected using the Benjamini-Hochberg procedure. Differentially abundant metabolites of different categories were individually color-coded. See also Table S1 and S2, Figure S1.
Metabolic Landscape of ccRCC
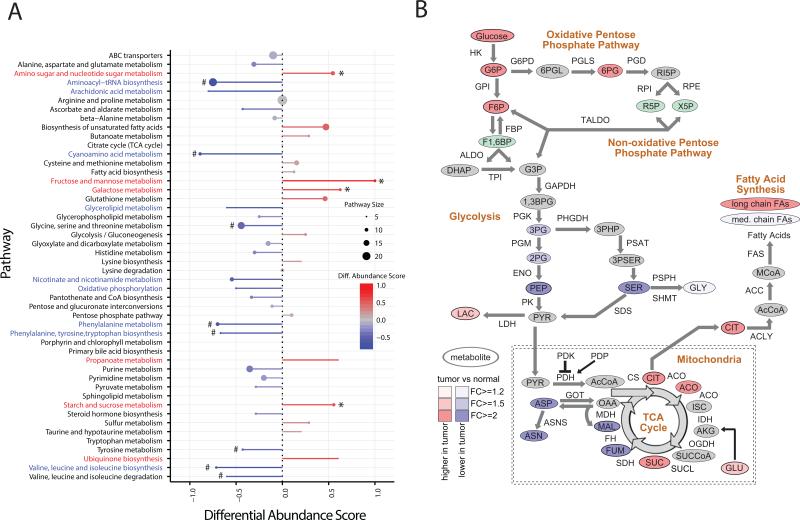
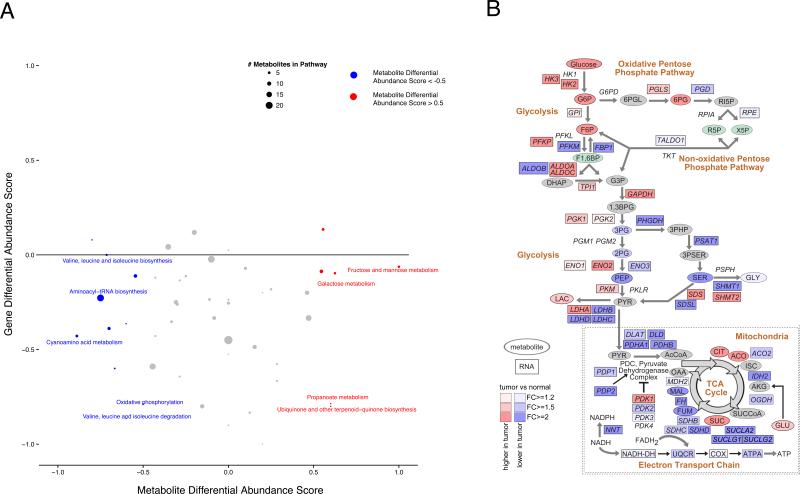
To systematically investigate the metabolic alterations associated with ccRCC pathogenesis, we performed KEGG pathway-based analysis utilizing metabolites that were present at differential abundance between tumor and normal kidney tissues. Differential abundance scores were calculated, which captured the tendency for metabolites in a pathway to be increased/decreased relative to normal samples. Among the 48 metabolic pathways for which at least five metabolites were captured in our profiling, 6 were up (>0.5 differential abundance score, red) and 9 were down (<−0.5, blue) (Figure 2A). Interestingly, most of the pathways elevated in tumor tissues were involved in carbohydrate metabolism (*), whereas most of the decreased pathways were involved in amino acid metabolism (#) (Figure 2A). Of note, all amino acids except cysteine, glutamate, and glutamine were significantly decreased in tumor samples (FDR-corrected p value < 0.05) (Table S3).
Figure 2. Pathway-based analysis of ccRCC metabolomics.
(A) A pathway-based analysis of metabolic changes upon comparing primary ccRCC to adjacent normal kidney tissues. The differential abundance score captures the average, gross changes for all metabolites in a pathway. A score of 1 indicates all measured metabolites in the pathway increase, and −1 indicates all measured metabolites in a pathway decrease. # the amino acid pathways. * glucose metabolism. (B) Metabolic changes of central carbon metabolism in ccRCC. Metabolites are labeled as color-coded ovals. Color corresponds to the log2 fold changes between tumor and normal tissues. Red, increase; Blue, decrease; Green, isomers; Gray, not measured. Enzymes for individual chemical reactions were denoted next to arrows connecting two metabolites. See also Table S3, and Figure S2.
Metabolic Alterations of Glycolysis and the TCA Cycle in ccRCC
The fundamental tumor-initiating event during ccRCC pathogenesis is the loss of VHL, which results in the accumulation of HIF1α and the ensuing persistent activation of HIF1 transcriptional programs. HIF1 normally functions as a master transcriptional regulator that organizes cellular adaptation to low oxygen microenvironment, e.g. ischemic tissues and growing solid tumors (Kaelin, 2008; Semenza, 2013). Hence, the two main transcriptional endpoints of HIF1 are (1) to reduce oxygen demand by increasing glycolytic flux and reducing oxidative phosphorylation, and (2) to increase oxygen supply through activating new blood vessel formation. One of the key HIF1 targets in glycolysis is pyruvate dehydrogenase kinase (PDK) that phosphorylates and inhibits pyruvate dehydrogenase (PDH), thereby impeding fueling of pyruvate-derived carbons to the TCA cycle (Gordan et al., 2007; Kaelin, 2008; Semenza, 2013). We anticipated that ccRCC would exhibit elevated levels of metabolites in glycolysis and decreased levels of metabolites involved in oxidative phosphorylation, which was partially supported by the metabolic pathway analysis based on RNA sequencing (The Cancer Genome Atlas Research et al., 2013). To this end, we constructed a metabolic map detailing the shift in abundance of central carbon metabolites from normal kidney to ccRCC (Figure 2B). Consistent with increased glucose uptake, we observed that metabolites in upper glycolysis, including glucose, glucose-6-phosphate (G6P), and fructose-6-phosphate (F6P), showed > 2-fold increases in abundance (Figure 2B). Notably, increased levels of G6P and F6P correlated with increases in 6-phosphogluconate (6PG: p<1e-6, F6P: p<1e-6, Figure S2), an intermediate of the pentose-phosphate pathway (PPP), suggesting increased shunting into the pentose phosphate pathway to produce ribose-5-phosphate (R5P) and NADPH. On the contrary, we observed that the majority (3 of 4) of quantified metabolites in lower glycolysis, i.e. downstream of F6P, were reduced in abundance. The upper part of glycolysis diverts hexose-phosphates to the pentose phosphate pathway, and the lower part diverts triose-phosphates to either the TCA cycle or one carbon metabolism. The distinct behavior of metabolites in the upper and lower parts of glycolysis is intriguing, and suggests that flux through glycolysis may be differentially partitioned (e.g. overflow into serine biosynthesis vs. PPP).
We next investigated the TCA cycle, which plays central roles in both ATP production through oxidation of acetyl-CoA to CO2, as well as the production of biosynthetic precursors (Owen et al., 2002). Five TCA cycle metabolites were measured, which revealed a dichotomous pattern of changes. In comparison to normal kidney tissues, citrate, cis-aconitate and succinate levels were markedly elevated (> 2 fold) whereas fumarate and malate were markedly reduced (>2 fold) in ccRCC (Figure 2B). The conversion of succinate to fumarate is catalyzed by succinate dehydrogenase (SDH), which also acts as an electron carrier in the inner membrane of the mitochondrial electron transport chain (Gottlieb and Tomlinson, 2005). The reduced levels of malate and fumarate, in tandem with an increase in succinate levels, suggest that the rate of oxidative phosphorylation may be reduced in tumors, which is consistent with the negative effect of HIF1 on oxidative phosphorylation in ccRCC. Notably, citrate can produce cytosolic acetyl-CoA through the action of ATP citrate lyase (ACL) for the synthesis of fatty acids in support of lipid membrane formation, and ccRCC tumors are exceptionally lipid-laden, giving rise to the clear cell morphology (Gebhard et al., 1987). Cell-based assays demonstrated that reductive carboxylation of glutamine generates citrate needed for the growth of mitochondrion-defective tumor cells (Metallo et al., 2012; Mullen et al., 2012), and reductive carboxylation of α-ketoglutarate to citrate for cell growth can be promoted by hypoxia and HIF1 (Wise et al., 2011). The splitting discordance in levels of TCA cycle metabolites with increased glutamine abundance in tumors implicates reductive carboxylation in ccRCC.
Metabolic Changes upon ccRCC Cancer Progression
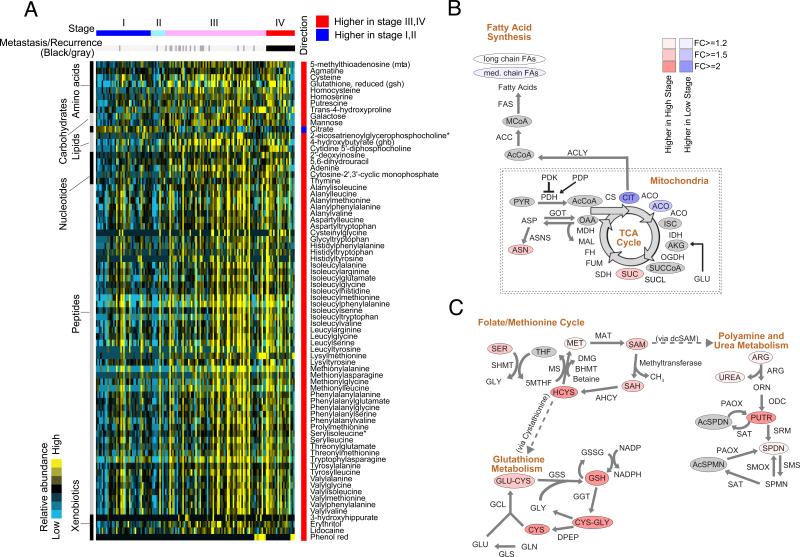
The large number of samples in our stage-distributed ccRCC cohort allowed us to investigate the association between metabolic shifts and ccRCC progression using AJCC clinical stage. There were 208 identifiable metabolites exhibiting statistically significant differential abundance between early- (I, II) and late-stage (III, IV) tumors (Mann-Whitney U-test FDR corrected p value < 0.05) (Table S4). Of these, 73 metabolites showed a greater than 2-fold increase in late-stage tumors, while one metabolite, citrate, showed a greater than 2-fold decrease in late-stage tumors (Figure 3A, Mann-Whitney U test FDR-corrected p value < 0.05, absolute log2 fold change > 1). Of note, with >2-year clinical followup of our cohort, 17 (24%) of the 70 patients who presented with stage III cancer eventually developed disease recurrence (Figure 3A, dark gray bars).
Figure 3. Metabolites associated with ccRCC progression.
(A) Mann-Whitney U tests were used to identify metabolites significantly higher or lower in Stage III/IV tumors, compared to Stage I/II tumors (Benjamini-Hochberg corrected p value < 0.05, absolute log2 fold change > 2). Metabolites were grouped, labeled on the left and detailed on the right. Clinical stages at presentation were color-labeled. Recurrences (n = 19) in the original Stage I to III patients (n = 118) were marked as darker gray bars. (B) Metabolic shifts in the TCA cycle and fatty acids during the progression. (C) Depicted are metabolic shifts of several interconnected metabolic programs upon ccRCC progression, including folate/methionine cycle, glutathione metabolism, and polyamine/urea metabolism. (B, C) Color corresponds to the log2 fold change between high stage (III/IV) and low stage (I/II) disease. Red, increase; Blue, decrease; Gray, not measured. Metabolites are labeled as color-coded ovals. Enzymes for individual chemical reactions were denoted next to arrows connecting two metabolites. See also Table S4.
Among these 74 metabolites, we identified several distinct groups of metabolites displaying synchronous patterns of variation with respect to clinical stages. Late-stage ccRCC is characterized by significant increases in galactose and mannose (Figure 3A). Notably, the decrease in citrate and smaller-magnitude decreases in cis-aconitate, and medium chain fatty acids in late-stage tumors (Figure 3B and Table S4) are consistent with the known observation that high-grade ccRCC is associated with decreased lipid content (Gebhard et al., 1987). We also noted increases in a large number of dipeptides (Figure 3A) that may be produced through protein degradation/reutilization processes, such as lysosomal degradation, phagocytosis, endocytosis, pinocytosis, and autophagy (Commisso et al., 2013; Kimmelman, 2015; Mizushima and Komatsu, 2011; Rubinsztein et al., 2012; Settembre and Ballabio, 2014; Son et al., 2013).
Elevated One Carbon Metabolic Network in Aggressive ccRCC: Folate/Methionine, Urea/Polyamine, and Glutathione Pathways
Of note, there were a number of metabolites, concentrated in interconnected metabolic pathways, exhibiting statistically significant but smaller-magnitude shifts that correlate with AJCC stages (Table S4). Accordingly, we assembled a metabolic map depicting the distribution of these changes in three interconnected pathways: the folate/methionine cycle, glutathione metabolism, and polyamine/urea metabolism (Figure 3C). The increases with respect to low stage tumors in serine, homocysteine, methionine, S-adenosyl methionine (SAM), and S-adenosyl homocysteine (SAH) supports a metabolic scenario in which serine feeds a methyl group (one carbon) into the tetrahydrofolate cycle for nucleotide synthesis and methionine replenishment which in turn supports protein/DNA/RNA methylation and polyamine synthesis (Figure 3C). The synthesis of polyamines requires decarboxylated SAM (dcSAM), a derivative of SAM, as a cofactor, and releases MTA (methylthioadenosine) which was significantly increased with stage progression (Figure 3C and Table S4). Of note, polyamines, including spermine (SPMN) and spermidine (SPDN), are essential for cell proliferation (Casero and Marton, 2007). We also observed an increase in late-stage tumors of a large number of metabolites related to the biosynthesis of glutathione, including cysteine, γ-glutamyl cysteine (GLU-CYS), and glutathione (GSH) (Figure 3C). Glutathione is the primary cellular antioxidant that buffers reactive oxygen species. Interestingly, the observed increases of methionine metabolism in late-stage tumors may be connected to increases in glutathione metabolism, via conversion of homocysteine (resulting from degradation of SAH) to cysteine through cystathionine for glutathione biogenesis (Lu, 1999). The association of increased buffering capacity for oxidative stress and aggressiveness is not unique to ccRCC, and has been observed in several cancers (Fan et al., 2014).
Unsupervised Clustering Reveals Four Metabolic Clusters (mClusters) with Prognostic Value
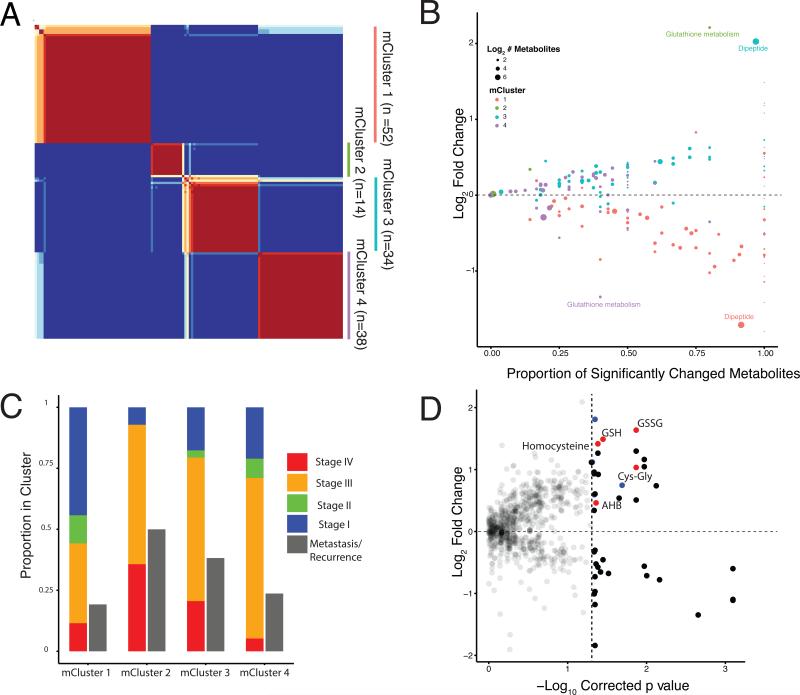
To evaluate whether ccRCC tumors could be partitioned into clusters with distinct metabolic phenotypes, we performed unsupervised Non-negative Matrix Factorization (NMF) consensus clustering (Lee and Seung, 1999). This approach yielded 4 distinct clusters (Figure 4A). Mann-Whitney U-tests were then used to calculate which metabolites were significantly increased or decreased in each cluster relative to all other tumors (Benjamini-Hochberg corrected p value < 0.05). To explore the metabolic underpinnings of each cluster, metabolic pathway-based analysis was performed to identify representative metabolites that distinguish each cluster against the remainder of the cohort. For each pair of metabolite cluster (mCluster) and pathway, we plotted the proportion of metabolites that changed significantly in that cluster, relative to all other tumors (X-axis), versus the average log-fold changes of these metabolites (Y-axis) (Figure 4B and Table S5). The clinical stages of each cluster at presentation were summarized in Figure 4C. Interestingly, mCluster 1 has the highest percentage (56%) of early stage (I & II) tumors and is characterized by the low abundances of dipeptides; mCluster 2 has the highest percentage (93%) of late stage (III & IV) tumors and displays exceptionally high levels of glutathione-related metabolites; mCluster 3, characterized by the highest abundance of dipeptides, has 79% late stage tumors; and mCluster 4 has of 71% of late stage tumors (Figure 4B and 4C). We stratified our mClusters into low-risk (mCluster 1) or high-risk (mClusters 2/3/4) groups, based on a 50% threshold of stage III/IV tumors, since ~80% of Stage I and Stage II ccRCC patients at presentation are cured by surgery alone and 30-50% of Stage III patients are expected to recur after surgery. We then examined relapse free survival on non-metastatic patients (Stage I, II, and III at presentation), which reveals a separation between low- and high-risk groups (Figure S3A), which does not reach statistical significance likely due to the relatively small number of events during follow-up (log-rank p value 0.12).
Figure 4. Unsupervised clustering of ccRCC based on metabolite signatures.
(A) Nonnegative matrix factorization (NMF) clustering of metabolomics data. Note that consensus results show consistency for k=4. (B) Mann-Whitney U-tests were used to calculate which metabolites were significantly increased or decreased in each cluster, relative to all other tumors (Benjamini-Hochberg corrected p value < 0.05). X-axis indicates, for a given cluster of patient samples, the proportion of metabolites in a pathway that are significantly changed (both increased and decreased) in a cluster. Y-axis plots the average log2 fold change of these metabolites. mCluser 2 & mCluser 4 are enriched in either increase or decrease of metabolites concerning glutathione metabolism, respectively. mCluster 1 & 3 show large differences in dipeptide levels, relative to other tumor samples. (C) The clinical stages at sample collection and the eventual metastasis of each individual metabolic cluster are presented. mCluster 1 is particularly enriched with Stage 1 tumors (p<0.0001 Chi-Square). (D) Comparison of metabolite abundances in tumors developing metastases versus those not developing metastases at preparation of this report. Red, the metabolites in the glutathione biosynthetic pathway that are increased in tumors that developed metastases. Blue dots correspond to dipeptides. See also Table S5 and S6 and Figure S3.
High Glutathione Pathway and Dipeptide Metabolites Associate with Stage IV ccRCC
With >2-year clinical follow-up of our cohort, 17 of the 70 patients who presented with stage III cancer eventually developed disease recurrence either locally or distantly, whereas only 2 of the 48 original early-stage (Stage I, II) patients developed recurrence (Figure 3A). With this dataset, we were particularly interested in identifying metabolites that correlate with the potential of disease recurrence among Stage III patients. Interestingly, only one metabolite, α-hydroxybutyrate (AHB), significantly distinguished patients who eventually recurred from those did not at preparation of this manuscript (FDR-corrected p value 0.05), Figure S3B). This is noteworthy because high AHB level is a surrogate of high α-ketobutyrate (AKB) which is produced when cystathionine is hydrolyzed to cysteine–a critical precursor for glutathione synthesis (Gall et al., 2010). We next compared tumor metabolic profiles patients either presenting with metastasis or later recurring (eventual Stage IV disease, n=39) versus others (n=99) patients (Figures 1C and 4E). There were 16 identifiable metabolites that increased significantly with stage IV disease (right upper quadrant, Figure 4D), of which 5, including AHB, involve glutathione metabolism and 4 are dipeptides (Table S6). These data again support a association between two distinct classes of metabolites, i.e. glutathione-related and dipeptides, and stage IV kidney cancers.
Relationship between Metabolic Genes and Metabolites in ccRCC
To this point, we have focused on analyzing ccRCC metabolism using our own metabolomic profiling. However, the proliferation of transcriptomic analysis of cancers, including ccRCC, gives us an orthogonal perspective (the expression of metabolic genes) from which to investigate ccRCC metabolism (Gatto et al., 2014; Hu et al., 2013). In this regard, it is interesting to note that a prior analysis of metabolic gene expression across many cancer types identified ccRCC as an outlier (Gatto et al., 2014). In that study, metabolic genes in ccRCC displayed reduced expression across nearly all metabolic pathways compared to adjacent normal kidney tissues, a pattern that was not apparent in other tumor types. To explore the degree to which transcriptomic and metabolomic changes were consistent with each other in ccRCC, we analyzed RNA-Seq data from the KIRC TCGA project consisting of 480 ccRCC tumors and 71 adjacent normal kidney tissues. Differential expressions of all genes was calculated using the limma voom R package (Law et al., 2014), and genes were mapped to KEGG metabolic pathways. Differential abundance scores indicating the tendency for genes in a pathway to go up/down in tumors relative to normal tissues were obtained and subsequently compared to metabolite scores (Figure 2A) for the same pathway. The results enabled a comparison of changes in gene expression to changes in metabolite abundance between tumors and adjacent normal tissues (Figure 5A). Interestingly, such analysis revealed a lack of linear correlation between transcriptomics (KIRC TCGA) and metabolomics (MSK) (Figure 5) (Spearman rho - 0.02, p value 0.89). It was intriguing to find that a large number of metabolic pathways exhibited reduced levels of gene expression, but increased levels of metabolites (Figure 5A, right lower quadrant). To address the possibility that this heterogeneity arose from population differences between the TCGA gene expression cohort and the MSK metabolomics cohort, we repeated this analysis using RNA-Seq data from 10 MSK metabolomics samples from rCluster 2. Because normal kidney tissue was not available for RNA-sequencing, we compared these 10 samples to all normal kidney tissue from the TCGA. We again found no correlation between differential abundance scores for metabolites and genes (Spearman rho 0.06, p value 0.7) (Figure S4A).
Figure 5. Correlation between the KIRC TCGA transcriptomics and the MSK ccRCC global metabolomics.
(A) For each KEGG pathway, the average fold changes of all genes were calculated. A differential abundance score was subsequently calculated for each pathway, which equals to the proportion of genes significantly increased in abundance in the pathway (FDR p value < 0.05) minus the proportion of genes significantly decreased in the pathway. The process was repeated for all metabolites in each pathway, and the two scores were plotted against each other. Size of dots indicates number of quantified metabolites in the pathway. (B) Detailed network map of central carbon highlights discordant behavior of metabolite and gene expression levels in ccRCC tumors. Coloring corresponds to the log2 fold change between tumor and normal tissue. Ovals represent metabolites (TKCRP ccRCC Metabolomics Cohort) and rectangles represent mRNA levels (KIRC TCGA RNA-Seq). Red, increase; Blue, decrease; Green, isomers; Gray, not measured. See also Figure S4.
Lack of Correlation between Enzymes Expression Level and Metabolites Abundance in ccRCC
We reasoned that this heterogeneity might be partially explained by examining metabolic genes at a detailed network level. To do so, we incorporated enzyme transcript levels into our corresponding central carbon metabolic network pathway map (Figure 5B). The result highlighted the inherent difficulty of inferring metabolic pathway activity from measurements of RNA: the highly interconnected nature of central carbon metabolism made it difficult to identify regions displaying consistent changes in gene and metabolite levels (Figure 5B). An example of the magnitude of heterogeneity between our metabolomics data and existing expression data was in the genes and metabolites constituting KEGG TCA cycle pathway. There, we observed that nearly all (28/30) genes were downregulated in tumor vs. normal tissue, while changes in metabolite levels were evenly split between increases and decreases in abundance (Figure 5B). Furthermore, using the Recon2 human metabolic reconstruction, we examined the heterogeneity between metabolomic and transcriptomic data at individual reaction level. We extracted pairs of genes/metabolites reported to interact in irreversible metabolic reactions, and separated these pairs into two sets based on whether the metabolite was used as a substrate, or produced as a product of the reaction. We again observed no correlation between gene expression data and metabolomics data in either comparison (substrate: Spearman p value 0.64; product: Spearman p value 0.69) (Figure S4B).
While the heterogeneity between transcriptomic and metabolomics data can be striking (e.g. in the TCA cycle), it is critical to remember that transcriptomic data is not a reliable surrogate for metabolic flux. A metabolic reaction can be catalyzed by any one of potentially many distinct isoforms of an enzyme, and such isoenzymes are characterized by distinct kinetic parameters, and are subject to different regulatory mechanisms, e.g. allosteric cofactors and protein modifications. Furthermore, the relationship between transcript level and protein abundance is not always linear. Nevertheless, our findings here suggest that changes in metabolite levels and enzyme transcript levels are often not synchronous in ccRCC, pointing to a complex paradigm for metabolic regulation.
Metabologram: A Web-based Application Integrates Large-scale Transcriptomics and Metabolomics
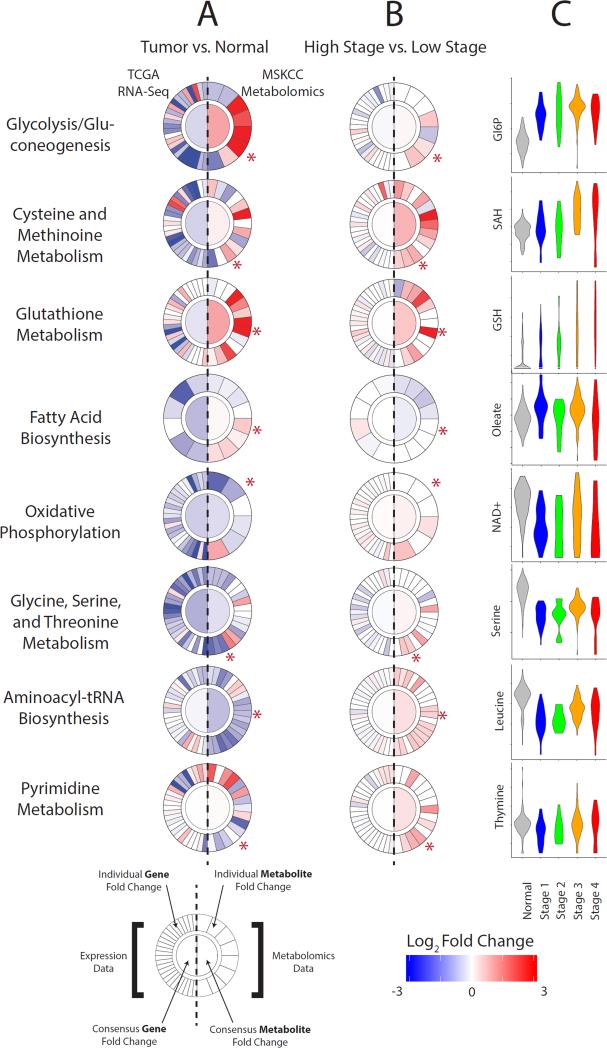
The difficulty in examining detailed metabolic networks when simultaneously layered with metabolomic and transcriptomic data motivated us to develop an integrative approach to study metabolism at pathway level. We developed a tool, called “Metabologram,” to integrate the MSK metabolomics and the KIRC TCGA transcriptomics data. Metabolograms enable concurrent visualization of metabolic pathway-level data on gene expression (plotted on the left) and metabolomics (plotted on the right) (Figure 6A). The ability to visualize data at a pathway level enables a quick, systematic comparison of (1) the average changes of genes and metabolites in a pathway (plotted in the center) and (2) the change of each individual component of the pathway (plotted as slices at the periphery) (Figure 6A and Figure S5). Metabolograms can be used as a rapid assessment of cancer metabolism when comparing changes between tumor and normal tissues (Figure 6A), as well as between late- and early-stage tumors (Figure 6B). The Metabologram tool is one part of a public, interactive data portal (http://kidneymetab.chenghsiehlab.org). The portal also enables users to interactively explore associations between metabolite abundances and 24 clinical parameters such as age, gender and tumor grade.
Figure 6. Metabolic pathway-based integration of transcriptomics and metabolomics with a novel web-based analytic tool, “Metabologram”.
(A, B) Each circular metabologram corresponds to a metabolic pathway. The left half circle corresponds to transcriptomics and the right half circle corresponds to metabolomics. The inner round center corresponds to the average fold change among all constituents of the pathway. The outer circle displays the fold change for each individual gene (left) and metabolite (right). 66 metabolograms from KEGG metabolic pathways are accessible through the web data portal, where interactive features enable detailed exploration by users. Metabolograms illustrate the metabolic differences between kidney tumors and adjacent normal tissue (A) and between late- and early-stage tumors (B). (C) Metabolites indicated by asterisk in (A, B) are displayed in violin plots as a function of normal kidney tissues and tumors at different stages. See also Figure S5.
Using metabolograms, we first interrogated pathway-level metabolic shifts between tumor and normal tissues for eight critical metabolic pathways that contained large number of metabolites recurrently arising as significant in our analyses to this point (Figure 6A). The patterns evident in the metabolograms in Figure 6A highlighted two distinct types of heterogeneity. First, it was rare to find a pathway where all metabolites showed a synchronous, large change in metabolite levels. For example, in the peripheral slices of glycolysis in Figure 6A, we observed again that approximately half of the metabolites showed exceptionally strong increases in tumors (e.g. F6P, G6P), while others (e.g. PEP) showed exceptionally strong decreases. These discordant patterns likely reflect the fact that metabolic flux patterns do not follow the canonical flow delineated by curated metabolic pathways, but rather branch, link, and shunt flux into neighboring pathways. A second heterogeneity was the evident contrast between the left half of each circle (gene expression) and the right half (metabolomics). The effect was particularly striking for glutathione metabolism. From the perspective of gene transcription, the majority of genes involved in glutathione metabolism showed decreases in expression in tumor relative to normal tissues (13 increased, 23 decreased), whereas from the perspective of metabolites, the majority of metabolites significantly increased in tumors relative to normal tissues, e.g. GSH, GSSG, cysteinylglycine, and putrescine (all > 4-fold) (Figure 6A).
Metabolic Shifts During Kidney Cancer Progression Assessed with Metabolograms
We next evaluated metabolic changes upon kidney cancer progression of these same metabolic pathways (Figure 6B). From the perspective of gene expression changes (left half centers), these pathways showed weak to no significant changes with cancer progression. On the other hand, there were significant consensus changes in a number of pathways from the perspective of metabolomics data (right half centers) (Figure 6B). The heterogeneous patterns when comparing changes between (1) tumor/normal samples, and (2) late-stage/early-stage tumors (i.e. comparing Figure 6B with 6A) was informative. For example, there were increases in metabolite levels in fatty acid biosynthesis and decreases in oxidative phosphorylation at pathogenesis (Figure 6A) and a reversal of these patterns during progression (Figure 6B). Among the most striking observations was the contrast between the initial decreases in amino acid metabolite levels at tumor initiation (Figure 6A) and the subsequent increases at progression (Figure 6B, serine and leucine inset in Figure 6C). Captured in the KEGG pathway “Amino-acyl tRNA biosynthesis”, 17 of 20 amino acids were decreased at tumor initiation (Figure 6A) whereas 13 of 20 amino acids were increased at progression (Figure 6B).
Mapping the MSK High Glutathione mCluster to the KIRC TCGA Dataset
Our supervised and unsupervised ccRCC metabolomics analyses identified two groups of high-risk patients whose tumors were characterized by either high glutathione metabolism (mCluster 2) or dipeptides (mCluster3) (Figure 4). Given the highest overall incidence (50%) of stage IV disease in the high-glutathione cluster, and the potential predictive value of high AHB with the later development of metastasis in Stage III patients, mCluster 2 was further investigated. Like most metabolic pathways, there is no clear correlation between transcriptomic and metabolomic signatures in the glutathione metabolism pathway (Figure 6). However, given the highly interconnected nature of the metabolic network, we envisioned that a metabolic gene expression signature might exist for the mCluster 2 tumors, which could offer molecular insights overlooked by our univariate analyses. To this end, we leveraged data from the KIRC TCGA dataset. Our goal was to identify a set of patients in the TCGA with similar expression profiles to our high-glutathione tumors, and then interrogate the genomic data in the TCGA to uncover possible genetic causes underlying elevated glutathione levels.
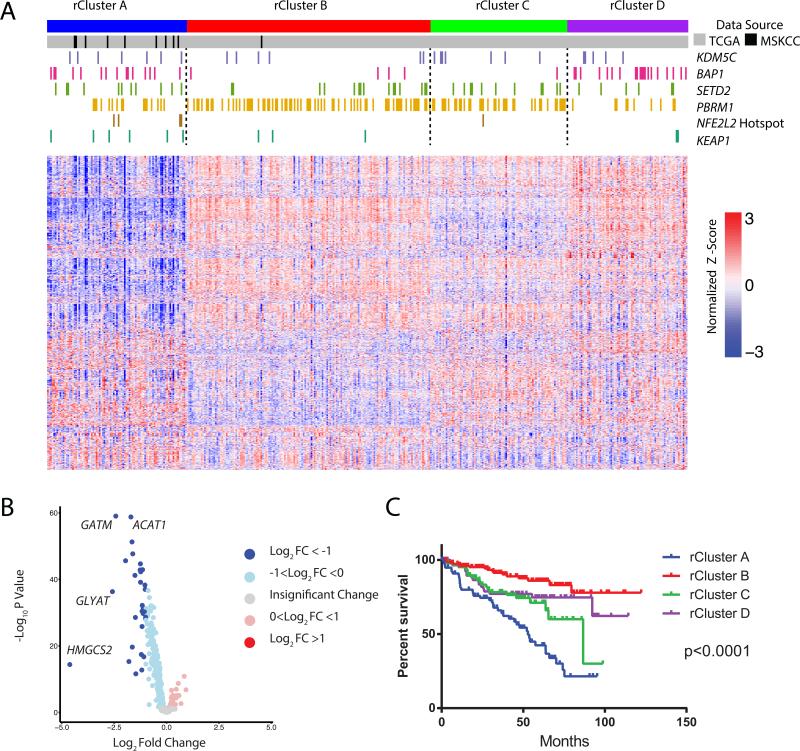
To enable a potential link of these two datasets, we performed RNA-Seq expression profiling on all available materials (n=10) of our mCluster 2 tumor samples. Reads from our high-glutathione cohort and the KIRC TCGA project were jointly aligned using a common pipeline. To identify a transcriptomic metabolic signature, we reasoned that we should restrict ourselves to comparison of 1,506 metabolic genes from the Recon2 human metabolic network (Thiele et al., 2013). Consensus hierarchical clustering was performed on the RNA-Seq counts from the joint TCGA/high-glutathione dataset, and yielded four robust and stable metabolic RNA clusters (rClusters) (Figure 7A), which showed association with the four reported TCGA RNA clusters (The Cancer Genome Atlas Research et al., 2013) (Chi-squared p value < 2e-16, Table S6). Remarkably, 9 of our 10 mCluster 2 tumors mapped to rCluster A (Figure 7A). To learn what metabolic gene patterns distinguished rCluster A from the other rClusters, pathway analysis was performed based on functionally related groups of genes that were significantly over- or under-expressed in rCluster A, relative to other rClusters. Notably, we found that mitochondrial genes were significantly under-expressed in rCluster A (Figure 7B and Table S7).
Figure 7. Mapping the MSK high glutathione ccRCC cluster with the KIRC-TCGA mutli-platform omics dataset.
(A) Consensus clustering was performed on 1,506 metabolic genes from the Recon2 human metabolic network reconstruction using RNA-Seq data from the KIRC TCGA (n = 398, gray bars), and the MSK TKCRP ccRCC Metabolomics high-glutathione tumors (n = 10, black bars). Mutations of indicated genes were marked by color bars. Depicted are the top 1000 most variable metabolic genes across the cohort, using log-normalized counts from limma voom. (B) Volcano plots of differentially expressed metabolic genes among four rClusters. HMGCS2, GLYAT, GATM, and ACAT1 are nuclear DNA-encoded mitochondrial genes. (C) Kaplan-Meier curves of cancer specific survival of individual rClusters (p value <0.0001, log-rank test). See also Table S7 and S8 and Figure S6.
Given that mCluster2 patients had worst clinical outcomes among four mClusters, we assessed the survival of patients in rCluster A patients, and found that they exhibited the worst clinical outcome among the four rClusters (log-rank test, p value <0.0001) (Figure 7C). We next interrogated if any specific mutations distinguished rCluster A from the other rClusters (Figure 7A) by analyzing all significantly mutated genes (SMG) reported by the KIRC TCGA project. ccRCC has high frequency mutations in chromatin modulators including PBRM1, BAP1, SETD2 and KDM5C, however, no specific mutations of these genes distinguish rCluster A from B, C or D (Figure 7A). Interestingly, rCluster A did show enrichment for NFE2L2 hotspot mutations (p value 0.008 chi-square) and KEAP1 mutations and copy number loss (p value 0.02 chi-square), two genes involved in the transcriptional control of glutathione metabolism genes. NFE2L2 encodes the transcription factor NRF2 whose function is negatively regulated by KEAP1 (Wakabayashi et al., 2003). Mutations in the KEAP1/NRF2 axis have been shown to promote tumor invasiveness and resistance to oxidative stress in pancreatic cancers (DeNicola et al., 2011). Consistently, the majority of KIRC TCGA patients whose tumors harbored NFE2L2 hotspot mutations or KEAP1 loss showed a propensity for metastasis (Figure S6). Altogether, our analyses identify a poor prognostic cluster in the KIRC TCGA cohort that is enriched for mutations in NFE2L2 or KEAP1, supporting the feasibility in integrating global metabolomics with published multi-omics datasets.
Discussion
Cancer metabolism has recently attracted renewed interest, in part due to the identification of tumorigenic mutations in the metabolic genes IDH1, IDH2, FH, and SDHB (Kaelin and McKnight, 2013; Linehan et al., 2010). Furthermore, it is now established that key oncogenes such as MYC and KRAS can reorganize cellular metabolism to support tumor fitness (Commisso et al., 2013; Dang, 2012). This interest in cancer metabolism has run parallel to the proliferation of genomic, transcriptomic, proteomic, and epigenomic profiling of tumors, and has fueled enthusiasm to study cancer metabolism from an integrative “multi-omics” perspective. However, due to technical challenges in identifying, measuring and tracing individual metabolites, metabolic flux profiling studies have largely focused on individual metabolic pathways and mainly employed in vitro cell-based assays.
In addition to targeted approaches, global metabolomic profiling has been performed on a comparatively smaller number cancer types, including prostate, breast, brain, liver and kidney cancers, at modest scale (n ≤ 65) (Budhu et al., 2013; Huang et al., 2013; Jaramillo and Zhang, 2013; Prabhu et al., 2014; Sreekumar et al., 2009). Based on such studies, metabolites such as sarcosine, 2-hydroxyglutarate, cysteine, lipids, and fatty acids have been implicated in the disease processes of prostate, breast, brain, liver and pancreas cancer, respectively. However, due to the lack of a common analytical pipeline to interrogate metabolomics data at a large, genome-wide scale, and the rarity of large sample sets (n ≥ 100 tumors), the feasibility and the value of routine inclusion of metabolomics in cancer research remained to be determined.
To address these translational issues concerning cancer metabolism as a whole and provide metabolic insights into kidney cancer, we performed global metabolic profiling on 138 kidney cancer/adjacent normal tissue pairs. Our study interrogated metabolic shifts changes during kidney cancer pathogenesis and highlighted a network of metabolic shifts associated with the genesis and progression of ccRCC tumors. In addition to analyzing metabolomic data in isolation, we proposed a pathway-based pipeline for studying metabolomics data in tandem with transcriptomic measurements of enzyme abundances. The results highlighted heterogeneity between gene expression changes and metabolomics changes in the same pathway and underscored the importance of both data types in generating a complete atlas of ccRCC metabolism. This heterogeneity was apparent both at the level of gross pathways and individual reactions, and has been reported in other systems (Chubukov et al., 2013). Nevertheless, it will be important to determine in future studies if such heterogeneity may arise from other confounding factors, e.g. incomplete coverage of the metabolome or ambiguity of metabolite abundances due to compartmentalization, or whether it is a bona fide feature of eukaryotic metabolic systems. Importantly, these analyses, and the visualization tools we have created are made publically available.
The large sample size and a relatively long clinical follow-up time of our study presented an opportunity to understand metabolic changes underlying kidney cancer aggressiveness. Interestingly, both supervised and unsupervised analyses identified both high glutathione and high dipeptide levels as aggressive metabolic signatures and as key features of distinctive metabolic clusters (mClusters) 2 and 3, respectively. Within our ccRCC cohort, the high-glutathione mCluster 2 was most aggressive (> 50% of stage IV patients) and AHB, a byproduct of metabolic flux from methionine metabolism to glutathione biosynthesis, was correlated with the disease recurrence in stage III patients. The role of glutathione metabolism in ccRCC was recently reported (Li et al., 2014). RNA-Seq profiling of the high-glutathione mCluster 2 led to the identification of an analogous cluster of tumors profiled in the KIRC TCGA project. This transcriptomic cluster (rCluster A) was not only associated with poor clinical outcome but also enriched in somatic alterations of central redox regulators KEAP1 and NRF2 (Jaramillo and Zhang, 2013), supporting the proposed roles of reactive oxygen species (ROS) and altered redox state in certain cancers (Trachootham et al., 2009). These data suggest that targeting ROS-buffering capacity could be exploited as a therapeutic strategy in an aggressive subset of kidney cancer (Nogueira and Hay, 2013).
Overall, our study demonstrated the value of large-scale tumor metabolomics. By establishing analytical pipelines for integrating metabolomics data with the large amount of transcriptomic profiling now available through projects such as the TCGA, we hope to provide an example for future studies embarking on metabolomic profiling of human tissue. Furthermore, our data here has provided pathologic insights that could have future prognostic and/or therapeutic value, and offer evidence for the incorporation of tumor metabolomics into the future study of human cancer biology.
Experimental Procedures
Tumor Samples and Patient Characteristics
After acquiring written informed consent and Memorial Sloan-Kettering Cancer Center institutional review board approval, 138 matched/pairs of RCC tumor and adjacent normal kidney tissue from partial or radical nephrectomies performed at Memorial Sloan Kettering Cancer Center (New York, NY, USA) were obtained by and stored at the MSK Translational Kidney Research Program (TKCRP). Samples were fresh frozen and stored at −80°C prior to metabolomic characterization. Disease and patient associated metadata including tumor pathologic and clinical stage, nuclear grade, metastatic status, and patient characteristics were also obtained. (See Figure 1 and Table S1). All samples were reviewed by two expert genitourinary pathologists.
The median age of the cohort was 63 (range 36-82). The median tumor size was 4.5 cm (range 2-13.5). Median follow up was 60 months. 28% of patients had metastatic disease at last follow up with 14% of patients presenting with metastatic disease at the time of nephrectomy and 14% of patients developing recurrence by October, 2014.
Sample Preparation and Metabolic Profiling
Metabolomic profiling was performed in collaboration with Metabolon Inc. Details were included in the Supplemental Methods.
Differential Abundance Score
The differential abundance (DA) score captures the tendency for a pathway to have increased levels of metabolites, relative to a control group. The score is calculating by first applying a non-parametric differential abundance test (in this study, Benjamini-Hochberg corrected Mann-Whitney U-tests) to all metabolites in a pathway. Then, after determining which metabolites are significantly increased/decreased in abundance, the differential abundance score is defined as:
Thus, the DA score varies from −1 to 1. A score of −1 indicates that all metabolites in a pathway decreased in abundance, while a score of 1 indicates that all metabolites increased.
Metabolic Clustering
Variance of metabolites across samples was calculated and the top 20% of metabolites (176 metabolites) were retained. This data was then clustered using NMF(Gaujoux and Seoighe, 2010). Primary clustering of the tumors was run in NMF for 200 iterations of ranks 2-5, with default settings of method brunet and seed random. Rank estimation was calculated using 50 iterations of ranks 2-8. All above analyses were performed in R. Four clusters were chosen as the stable breakdown based on the major inflection point of the cophentic estimate and the second largest peak (after rank 2) of the dispersion estimate. A consensus labeling was then determined based on cluster inclusion in ranks 2-4.
Differential Expression Analysis and Metabolic Gene Expression Clustering
For all RNA-Seq analysis except that in Figure 7, TCGA RSEM unnormalized counts were downloaded from the Broad Firehose. The limma voom package was used to calculate fold changes and statistical significance between tumor/normal and high/low stage samples. Details of metabolic gene expression clustering were included in the supplementary methods.
Supplementary Material
Significance.
Changes in cellular metabolism contribute to the development and progression of tumors, and can render tumors vulnerable to interventions. However, studies of human cancer metabolism remain limited due to technical challenges of detecting and quantifying small molecules, the highly interconnected nature of metabolic pathways, and the lack of designated tools to analyze and integrate metabolomics with other –omics data. Our study generates a comprehensive metabolomics dataset on a single cancer type and enables integration of metabolomics with sequencing data. Our results highlight the massive re-organization of cellular metabolism as tumors progress and acquire more aggressive features. The results of our work are made available through an interactive public data portal for cancer research community.
Acknowledgement
We thank our patients for donating their tumors and the team members of the MSK TKCRP for the advancement of kidney cancer research. We also thank Drs. Craig Thompson and Lydia Finley for their critical comments on the manuscript. This investigation was supported by: AAH and PR were supported by the Sidney Kimmel Center for Prostate and Urologic Cancers and by the MSK Cancer Center Support Grant/Core Grant (P30 CA008748). CJ was supported by the NIH P30 CA125123. ARB was supported by the NCI 5T32CA160001. CS was supported by MSK Center for Translational Cancer Genomic Analysis; and U24 CA143840; JJH was supported by The Jill and Jeffrey Weiss Fund to the Cure of Kidney Cancer; J. Randall & Kathleen L. MacDonald Kidney Cancer Research Fund.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Accession Number: RNA sequencing from glutathione-high cluster is deposited under GEO accession number GSE74734.
Author Contributions: AAH, ER and JJH – designed the study, analyzed the data and drafted the manuscript. CHL and EC – analyzed the data and edited the manuscript. CJC and ARB – analyzed the data and assisted with figure generation. ER, AL, BAA, and EML – analyzed the data and developed the online metabologram application. RS and WL – analyzed the data and performed statistical analysis. YC and SMS – performed metabolic quantification and analysis. PR – provided surgical samples and edited the manuscript. YBC, SKT and VER – performed pathologic analysis and edited the manuscript. CS – performed study supervision analyzed the data and edited the manuscript.
Reference
- Anders S, McCarthy DJ, Chen Y, Okoniewski M, Smyth GK, Huber W, Robinson MD. Count-based differential expression analysis of RNA sequencing data using R and Bioconductor. Nature protocols. 2013;8:1765–1786. doi: 10.1038/nprot.2013.099. [DOI] [PubMed] [Google Scholar]
- Budhu A, Roessler S, Zhao X, Yu Z, Forgues M, Ji J, Karoly E, Qin LX, Ye QH, Jia HL, et al. Integrated metabolite and gene expression profiles identify lipid biomarkers associated with progression of hepatocellular carcinoma and patient outcomes. Gastroenterology. 2013;144:1066–1075. e1061. doi: 10.1053/j.gastro.2013.01.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casero RA, Jr., Marton LJ. Targeting polyamine metabolism and function in cancer and other hyperproliferative diseases. Nature reviews Drug discovery. 2007;6:373–390. doi: 10.1038/nrd2243. [DOI] [PubMed] [Google Scholar]
- Chubukov V, Uhr M, Le Chat L, Kleijn RJ, Jules M, Link H, Aymerich S, Stelling J, Sauer U. Transcriptional regulation is insufficient to explain substrate-induced flux changes in Bacillus subtilis. Molecular systems biology. 2013;9:709. doi: 10.1038/msb.2013.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Commisso C, Davidson SM, Soydaner-Azeloglu RG, Parker SJ, Kamphorst JJ, Hackett S, Grabocka E, Nofal M, Drebin JA, Thompson CB, et al. Macropinocytosis of protein is an amino acid supply route in Ras-transformed cells. Nature. 2013;497:633–637. doi: 10.1038/nature12138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang CV. MYC on the path to cancer. Cell. 2012;149:22–35. doi: 10.1016/j.cell.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBerardinis RJ, Lum JJ, Hatzivassiliou G, Thompson CB. The biology of cancer: metabolic reprogramming fuels cell growth and proliferation. Cell metabolism. 2008;7:11–20. doi: 10.1016/j.cmet.2007.10.002. [DOI] [PubMed] [Google Scholar]
- DeNicola GM, Karreth FA, Humpton TJ, Gopinathan A, Wei C, Frese K, Mangal D, Yu KH, Yeo CJ, Calhoun ES, et al. Oncogene-induced Nrf2 transcription promotes ROS detoxification and tumorigenesis. Nature. 2011;475:106–109. doi: 10.1038/nature10189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan J, Ye J, Kamphorst JJ, Shlomi T, Thompson CB, Rabinowitz JD. Quantitative flux analysis reveals folate-dependent NADPH production. Nature. 2014;510:298–302. doi: 10.1038/nature13236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funakoshi T, Lee CH, Hsieh JJ. A systematic review of predictive and prognostic biomarkers for VEGF-targeted therapy in renal cell carcinoma. Cancer treatment reviews. 2014;40:533–547. doi: 10.1016/j.ctrv.2013.11.008. [DOI] [PubMed] [Google Scholar]
- Gall WE, Beebe K, Lawton KA, Adam KP, Mitchell MW, Nakhle PJ, Ryals JA, Milburn MV, Nannipieri M, Camastra S, et al. alpha-hydroxybutyrate is an early biomarker of insulin resistance and glucose intolerance in a nondiabetic population. PloS one. 2010;5:e10883. doi: 10.1371/journal.pone.0010883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatto F, Nookaew I, Nielsen J. Chromosome 3p loss of heterozygosity is associated with a unique metabolic network in clear cell renal carcinoma. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:E866–875. doi: 10.1073/pnas.1319196111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaujoux R, Seoighe C. A flexible R package for nonnegative matrix factorization. BMC bioinformatics. 2010;11:367. doi: 10.1186/1471-2105-11-367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebhard RL, Clayman RV, Prigge WF, Figenshau R, Staley NA, Reesey C, Bear A. Abnormal cholesterol metabolism in renal clear cell carcinoma. Journal of lipid research. 1987;28:1177–1184. [PubMed] [Google Scholar]
- Gordan JD, Thompson CB, Simon MC. HIF and c-Myc: sibling rivals for control of cancer cell metabolism and proliferation. Cancer cell. 2007;12:108–113. doi: 10.1016/j.ccr.2007.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottlieb E, Tomlinson IP. Mitochondrial tumour suppressors: a genetic and biochemical update. Nature reviews Cancer. 2005;5:857–866. doi: 10.1038/nrc1737. [DOI] [PubMed] [Google Scholar]
- Hakimi AA, Furberg H, Zabor EC, Jacobsen A, Schultz N, Ciriello G, Mikklineni N, Fiegoli B, Kim PH, Voss MH, et al. An epidemiologic and genomic investigation into the obesity paradox in renal cell carcinoma. Journal of the National Cancer Institute. 2013a;105:1862–1870. doi: 10.1093/jnci/djt310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakimi AA, Pham CG, Hsieh JJ. A clear picture of renal cell carcinoma. Nature genetics. 2013b;45:849–850. doi: 10.1038/ng.2708. [DOI] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- Hsu PP, Sabatini DM. Cancer cell metabolism: Warburg and beyond. Cell. 2008;134:703–707. doi: 10.1016/j.cell.2008.08.021. [DOI] [PubMed] [Google Scholar]
- Hu J, Locasale JW, Bielas JH, O'Sullivan J, Sheahan K, Cantley LC, Vander Heiden MG, Vitkup D. Heterogeneity of tumor-induced gene expression changes in the human metabolic network. Nature biotechnology. 2013;31:522–529. doi: 10.1038/nbt.2530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Q, Tan Y, Yin P, Ye G, Gao P, Lu X, Wang H, Xu G. Metabolic characterization of hepatocellular carcinoma using nontargeted tissue metabolomics. Cancer research. 2013;73:4992–5002. doi: 10.1158/0008-5472.CAN-13-0308. [DOI] [PubMed] [Google Scholar]
- Jaramillo MC, Zhang DD. The emerging role of the Nrf2-Keap1 signaling pathway in cancer. Genes & development. 2013;27:2179–2191. doi: 10.1101/gad.225680.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaelin WG., Jr. The von Hippel-Lindau tumor suppressor gene and kidney cancer. Clin Cancer Res. 2004;10:6290S–6295S. doi: 10.1158/1078-0432.CCR-sup-040025. [DOI] [PubMed] [Google Scholar]
- Kaelin WG., Jr. The von Hippel-Lindau tumour suppressor protein: O2 sensing and cancer. Nature reviews Cancer. 2008;8:865–873. doi: 10.1038/nrc2502. [DOI] [PubMed] [Google Scholar]
- Kaelin WG, Jr., McKnight SL. Influence of metabolism on epigenetics and disease. Cell. 2013;153:56–69. doi: 10.1016/j.cell.2013.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmelman AC. Metabolic Dependencies in RAS-Driven Cancers. Clin Cancer Res. 2015;21:1828–1834. doi: 10.1158/1078-0432.CCR-14-2425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law CW, Chen Y, Shi W, Smyth GK. voom: Precision weights unlock linear model analysis tools for RNA-seq read counts. Genome biology. 2014;15:R29. doi: 10.1186/gb-2014-15-2-r29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee DD, Seung HS. Learning the parts of objects by non-negative matrix factorization. Nature. 1999;401:788–791. doi: 10.1038/44565. [DOI] [PubMed] [Google Scholar]
- Li B, Qiu B, Lee DS, Walton ZE, Ochocki JD, Mathew LK, Mancuso A, Gade TP, Keith B, Nissim I, et al. Fructose-1,6-bisphosphatase opposes renal carcinoma progression. Nature. 2014;513:251–255. doi: 10.1038/nature13557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linehan WM, Srinivasan R, Schmidt LS. The genetic basis of kidney cancer: a metabolic disease. Nature reviews Urology. 2010;7:277–285. doi: 10.1038/nrurol.2010.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu SC. Regulation of hepatic glutathione synthesis: current concepts and controversies. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 1999;13:1169–1183. [PubMed] [Google Scholar]
- Majmundar AJ, Wong WJ, Simon MC. Hypoxia-inducible factors and the response to hypoxic stress. Molecular cell. 2010;40:294–309. doi: 10.1016/j.molcel.2010.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metallo CM, Gameiro PA, Bell EL, Mattaini KR, Yang J, Hiller K, Jewell CM, Johnson ZR, Irvine DJ, Guarente L, et al. Reductive glutamine metabolism by IDH1 mediates lipogenesis under hypoxia. Nature. 2012;481:380–384. doi: 10.1038/nature10602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N, Komatsu M. Autophagy: renovation of cells and tissues. Cell. 2011;147:728–741. doi: 10.1016/j.cell.2011.10.026. [DOI] [PubMed] [Google Scholar]
- Mullen AR, Wheaton WW, Jin ES, Chen PH, Sullivan LB, Cheng T, Yang Y, Linehan WM, Chandel NS, DeBerardinis RJ. Reductive carboxylation supports growth in tumour cells with defective mitochondria. Nature. 2012;481:385–388. doi: 10.1038/nature10642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogueira V, Hay N. Molecular pathways: reactive oxygen species homeostasis in cancer cells and implications for cancer therapy. Clin Cancer Res. 2013;19:4309–4314. doi: 10.1158/1078-0432.CCR-12-1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owen OE, Kalhan SC, Hanson RW. The key role of anaplerosis and cataplerosis for citric acid cycle function. The Journal of biological chemistry. 2002;277:30409–30412. doi: 10.1074/jbc.R200006200. [DOI] [PubMed] [Google Scholar]
- Prabhu A, Sarcar B, Kahali S, Yuan Z, Johnson JJ, Adam KP, Kensicki E, Chinnaiyan P. Cysteine catabolism: a novel metabolic pathway contributing to glioblastoma growth. Cancer research. 2014;74:787–796. doi: 10.1158/0008-5472.CAN-13-1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raimundo N, Baysal BE, Shadel GS. Revisiting the TCA cycle: signaling to tumor formation. Trends in molecular medicine. 2011;17:641–649. doi: 10.1016/j.molmed.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinsztein DC, Codogno P, Levine B. Autophagy modulation as a potential therapeutic target for diverse diseases. Nature reviews Drug discovery. 2012;11:709–730. doi: 10.1038/nrd3802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semenza GL. HIF-1 mediates metabolic responses to intratumoral hypoxia and oncogenic mutations. The Journal of clinical investigation. 2013;123:3664–3671. doi: 10.1172/JCI67230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Settembre C, Ballabio A. Lysosome: regulator of lipid degradation pathways. Trends in cell biology. 2014;24:743–750. doi: 10.1016/j.tcb.2014.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Son J, Lyssiotis CA, Ying H, Wang X, Hua S, Ligorio M, Perera RM, Ferrone CR, Mullarky E, Shyh-Chang N, et al. Glutamine supports pancreatic cancer growth through a KRAS-regulated metabolic pathway. Nature. 2013;496:101–105. doi: 10.1038/nature12040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sreekumar A, Poisson LM, Rajendiran TM, Khan AP, Cao Q, Yu J, Laxman B, Mehra R, Lonigro RJ, Li Y, et al. Metabolomic profiles delineate potential role for sarcosine in prostate cancer progression. Nature. 2009;457:910–914. doi: 10.1038/nature07762. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- The Cancer Genome Atlas Research, N. Analysis working group: Baylor College of, M. Creighton CJ, Morgan M, Gunaratne PH, Wheeler DA, Gibbs RA, Agency BCC, Gordon Robertson A, Chu A, et al. Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature. 2013 [Google Scholar]
- Thiele I, Swainston N, Fleming RM, Hoppe A, Sahoo S, Aurich MK, Haraldsdottir H, Mo ML, Rolfsson O, Stobbe MD, et al. A community-driven global reconstruction of human metabolism. Nature biotechnology. 2013;31:419–425. doi: 10.1038/nbt.2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trachootham D, Alexandre J, Huang P. Targeting cancer cells by ROS-mediated mechanisms: a radical therapeutic approach? Nature reviews Drug discovery. 2009;8:579–591. doi: 10.1038/nrd2803. [DOI] [PubMed] [Google Scholar]
- Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakabayashi N, Itoh K, Wakabayashi J, Motohashi H, Noda S, Takahashi S, Imakado S, Kotsuji T, Otsuka F, Roop DR, et al. Keap1-null mutation leads to postnatal lethality due to constitutive Nrf2 activation. Nature genetics. 2003;35:238–245. doi: 10.1038/ng1248. [DOI] [PubMed] [Google Scholar]
- Wei EY, Hsieh JJ. A river model to map convergent cancer evolution and guide therapy in RCC. Nature reviews Urology. 2015 doi: 10.1038/nrurol.2015.260. [DOI] [PubMed] [Google Scholar]
- Wise DR, Ward PS, Shay JE, Cross JR, Gruber JJ, Sachdeva UM, Platt JM, DeMatteo RG, Simon MC, Thompson CB. Hypoxia promotes isocitrate dehydrogenase-dependent carboxylation of alpha-ketoglutarate to citrate to support cell growth and viability. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:19611–19616. doi: 10.1073/pnas.1117773108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.