Highlight
Semi-Rolled Leaf2 encodes a novel plant-specific protein that modulates leaf rolling by regulating cell development in the abaxial sides of rice leaves.
Key words: Abaxial side, cell differentiation, leaf development, leaf rolling, rice, SRL2.
Abstract
Moderate leaf rolling maintains the erectness of leaves and minimizes the shadowing between leaves which is helpful to establish ideal plant architecture. Here, we describe a srl2 (semi-rolled leaf2) rice mutant, which has incurved leaves due to the presence of defective sclerenchymatous cells on the abaxial side of the leaf and displays narrow leaves and reduced plant height. Map-based cloning revealed that SRL2 encodes a novel plant-specific protein of unknown biochemical function. SRL2 was mainly expressed in the vascular bundles of leaf blades, leaf sheaths, and roots, especially in their sclerenchymatous cells. The transcriptional activities of several leaf development-related YABBY genes were significantly altered in the srl2 mutant. Double mutant analysis suggested that SRL2 and SHALLOT-LIKE1 (SLL1)/ROLLED LEAF9 (RL9) function in distinct pathways that regulate abaxial-side leaf development. Hence, SRL2 plays an important role in regulating leaf development, particularly during sclerenchymatous cell differentiation.
Introduction
Leaf shape has long been considered to be an important agronomic trait in rice. Leaf functions such as photosynthesis, respiration, and transpiration are dependent on leaf shape and its three-dimensional architecture (Govaerts et al., 1996). In rice (Oryza sativa) and other crops cultivated at high density, moderate leaf rolling can help maintain the erectness of leaves which ultimately benefits crops growth by improving light acceptance, delaying leaf senescence, and accelerating the accumulation of dry matter. The last three leaves of super-high-yielding hybrid rice were proposed to be long, erect, narrow, rolled (V-shaped), and thick (Yuan, 1997). Therefore, identifying mutants with moderately rolled leaves and isolating genes that control leaf rolling will be beneficial for breeding crops with the desired architecture and stress tolerance.
As rice is a polymorphic crop, rice varieties and mutants exhibit two different types of transverse leaf rolling, i.e. inward (adaxial) and outward (abaxial) rolling, which are regulated by complicated developmental processes including the control of polarity establishment and cell differentiation (Bowman et al., 2002; Micol and Hake, 2003). Leaf polarity establishment is controlled by both transcription factors and small RNAs (Moon and Hake, 2011). HD-ZIPIII family genes, such as PHABULOSA (PHB), PHAVOLUTA (PHV) (McConnell et al., 2001), REVOLUTE (REV) (Otsuga et al., 2001), and ROLLED1 (RLD1) (Nelson et al., 2002), determine the development of adaxial cells in leaves. By contrast, members of the KANADI family, such as MILK-WEED POD1 (MWP1) (Candela et al., 2008), and genes belonging to the YABBY (YAB) family, such as YAB2 and YAB3 (Eshed et al., 2004), determine the development of abaxial cells. Two small RNAs, trans-acting short interfering RNAs (tasiRNA) and miR165/166, exhibit opposing polar distribution in leaf primordial tissue and are thought to establish the adaxial–abaxial axis during leaf development (Nogueira et al., 2007). Recent reports have demonstrated that cytological architecture can also affect leaf blade morphology. SEMI-ROLLED LEAF1 (SRL1), encoding a putative glycosylphosphatidylinositol-anchored protein, is involved in leaf rolling through inhibiting the formation of bulliform cells (Xiang et al., 2012). In addition, SHALLOT-LIKE1 (SLL1), which encodes a MYB transcription factor of the KANADI family, is involved in leaf rolling through regulating sclerenchyma cell differentiation (Yan et al., 2008; Zhang et al., 2009). Environmental factors such as water deficiency, high temperature, and sunshine may also lead to the inward rolling of leaves (Kadioglu and Terzi, 2007).
Due to its importance, many studies have been performed to characterize the genes controlling leaf rolling in rice. To date, 17 rice mutants with rolled leaves have been characterized, from which six recessive genes (rl1–rl6) were mapped to chromosomes 1, 3, 4, 7, and 12 through classical linkage analysis (Khush and Kinoshita, 1991). However, the other genes were mapped to chromosome 2 (rl (t); Shao et al., 2005b), chromosome 5 (rl7 and rl8; Li, 1998; Shao et al., 2005a), chromosome 7 (rl11, srl1, and sll2; Shi et al., 2009; Xiang et al., 2012; Zhang et al., 2015), chromosome 9 (rl9 and rl10; Luo et al., 2007; Yan et al., 2008; Zhang et al., 2009), and chromosome10 (rl12 and rl14; Fang et al., 2012;Luo et al., 2009) by molecular marker analysis. In addition, overexpression of OsAGO7 results in the upward curling of rice leaves (Shi et al., 2007).
Although many mutants with leaf rolling phenotypes have been characterized and their relevant genes have been identified, the molecular basis of leaf rolling in rice remains fragmented. Here, we report a Semi-Rolled Leaf2 (SRL2) gene which encodes a novel plant-specific protein that functions in regulating rice leaf rolling. SRL2 deficiency leads to abnormal formation of sclerenchymatous cells on the abaxial side of the leaf, resulting in inwardly rolled leaves. Hence, this gene might be involved in the transdifferentiation process from mesophyll cells to sclerenchymatous cells, revealing a possible mechanism underlying leaf rolling in monocot plants.
Materials and methods
Plant materials and growth conditions
The rice (Oryza sativa) semi-rolled leaf mutant srl2 was isolated from the γ-ray-irradiated indica cultivar Zhongxian 3037. The F2 mapping population was generated from a cross between the srl2 mutant and Nipponbare, a typical japonica cultivar. In the F2 population, plants exhibiting rolled leaves were selected for gene mapping. All plants were grown in the paddy fields of the Institute of Genetics and Developmental Biology in Beijing (China) or Sanya (Hainan Province, China) during the natural growing season.
Map-based cloning
SRL2 was mapped and cloned with 2 994 F2 mutant plants using STS molecular markers (see Supplementary Table S1 at JXB online). For the identification of candidate genes the corresponding DNA fragments were amplified from mutants and wild-type plants using LA-Taq (TaKaRa, http://www.takara-bio.com/) and sequenced using an Applied Biosystems 3730 sequencer (http://www.appliedbiosystems.com/).
For functional complementation, a 16.9kb genomic DNA fragment, corresponding to the entire LOC_Os03g19520 gene, was digested from BAC clone OSJNBb0101E03 (Arizona Genomics Institute, http://www.genome.arizona.edu) using EcoRI and inserted into the pCAMBIA1300 vector (Cambia, http://www.cambia.org/) to generate construct pCSRL2. A control construct, pCSRL2-CK, was generated by digesting pCSRL2 with XhoI to remove the whole ORF of SRL2. The two binary plasmids were introduced into Agrobacterium tumefaciens EHA105 by electroporation and transformed into the srl2 mutant plants. For RNAi analysis, a 384bp fragment comprising 316bp of the 5′ UTR and 68bp of the ORF was amplified using primers CTCGAGGTGGAGACGGGTGGTGTGTG and AGATCTGAGCTTGGGCGCAGAGCTGG. After sequencing confirmation, the PCR products were inserted into the vector pCAMBIA23A in both the forward and reverse directions to generate the construct pSRL2RNAi. This binary plasmid was then introduced into the wild-type plants (Zhongxian 3037) by Agrobacterium infection. Rice transformation was performed as described by Hiei et al. (1994).
Microscopy
Culm segments were fixed in formalin-acetic acid-alcohol (FAA) solution (10% formaldehyde, 5% acetic acid, and 47.5% ethanol) at 4 °C overnight. After dehydration in a graded ethanol series, the samples were infiltrated and embedded in Technovit 7100 resin (Heraeus kulzer). Sections, 3 μm thick, were cut, stained with toluidine blue, and viewed under a light microscope (Leica, http://www.leica-microsystems.com). The root cell length was determined by staining the whole roots of 7-d-old seedlings with 20 μg ml−1 of propidium iodide in PBS, followed by examination under a fluorescent microscope (Leica). For scanning electron microscopy (SEM), the 4th leaves of the wild type and srl2 were fixed overnight at 4 °C with 2.5% glutaraldehyde in 0.1M phosphate buffer (pH 7.4). After dehydration in a graded ethanol series and substitution with isoamyl acetate, the samples were critical-point dried, sputter coated with gold, and observed with a S-3000N scanning electron microscope (HITACHI).
Promoter–reporter gene fusion studies
For promoter–GUS fusion studies, a 2.1-kb genomic DNA fragment that contained the promoter region of the SRL2 gene was amplified by PCR and then subcloned into vector pCAMBIA1301 (Cambia), which resulted in a fusion of the SRL2 promoter and the GUS reporter gene. The construct was transformed into wild-type plants as described earlier. Developing leaves of the transgenic plants were harvested and incubated in GUS reaction buffer (1mM 5-bromo-4-chloro-3-indolyl-d-glucuronide, 50mM sodium phosphate (pH 7.0), and 7% (v/v) methanol) at 37 °C overnight. The stained samples were then dehydrated in 95% ethanol until the chlorophyll was removed. For observation of the sclerenchyma in the leaf sheath, samples were hand cut with a sharp razor and mounted for microscopy. For microtome sections (8 μm), samples were prepared according to methods as described by Suzaki et al. (2004).
Phylogenetic analysis
The amino acid sequences of the SRL2 homological proteins were retrieved through a database search using the total amino acid sequences of SRL2. The multiple sequence alignments were performed by Clustal_X 1.83 (Thompson et al., 1997) using the parameters of Weight matrix: Gonnet; Gap opening penalty: 10.0 and Gap extension penalty: 0.10. Phylogenetic trees were constructed with the aligned protein sequences using MEGA 3.1 software (Kumar et al., 2004) based on the Maximum likelihood method with parameters of the Poisson correction model, complete deletion, and bootstrap (1 000 replicates; random seed).
Subcellular localization studies of SRL2
The coding sequence of SRL2 was amplified by PCR and then subcloned into vector pJIT163-GFP to obtain the SRL2–GFP construct. The resultant construct was introduced into rice protoplasts and onion epidermal cells. Transfection of rice protoplasts with SRL–GFP was performed as described by Li et al. (2009). For the transformation of onion cells, those plasmids were bombarded into onion epidermal cells using a PDS-1000 ⁄ He particle gun (Bio-Rad). The green fluorescence was observed by confocal laser-scanning microscopy (Zeiss LSM 510 META) with an argon laser excitation wavelength of 488nm.
Onion epidermis was ground in liquid nitrogen. Total protein was extracted with protein extraction buffer (50mM TRIS–HCl at pH 7.5, 150mM NaCl, 5mM EDTA, 0.2% NP-40, 0.1% Triton X-100, and Complete protease inhibitor cocktail, Roche), usually 600 μl for 0.2g onion epidermis. The extracts were centrifuged at 16 000rpm for 15min at 4 ℃, and the supernatants were collected for Western blot analysis. Western blot analysis was performed with a monoclonal mouse anti-GFP (Abmart) primary antibody at a final dilution of 1:2 000, and with a goat pAb to mouse IgG (Abcam) second antibody at a final dilution of 1:20 000.
In situ hybridization
Leaves were collected from fourth leaf stage of the wild type and fixed in FAA overnight at 4 °C. Samples were then dehydrated through a butanol series and embedded in Paraplast Plus (Sigma–Aldrich, http://www.sigmaaldrich.com/). Sections, 8 μm thick, were obtained using a Leica RM2135 microtome (Leica Biosystems, http://www.leica.com). To prepare the SRL2 probe, a 492-bp fragment of SRL2 cDNA was amplified using primers SRL2 T7 and SRL2 sp6 (SupplementaryTable S2). The probe was synthesized using a DIG RNA labelling kit (SP6/T7; Roche Diagnostics Ltd, http://www.roche.com) in accordance with the manufacturer’s recommendations. Pretreatment of sections, hybridization, and immunological detection were performed as described previously (Xiao et al., 2009).
Real-Time qRT-PCR
Real-time qRT-PCR analysis was performed to examine the expression pattern of SRL2 in various tissues and at different stages of leaf growth in order to identify the transgenic lines in which SRL2 expression was enhanced and complemented. Transcripts from genes related to leaf development that were altered in SRL2 plants compared with the wild type were also identified. Total RNA was extracted using TRIzol solution and reverse transcribed using the Superscript Preamplification System in accordance with the manufacturer’s instructions and then analysed quantitatively on a Bio-Rad CYF96 using the real-time PCR Master Mix (TaKaRa Biotechnology Co. Ltd.). The rice UB gene was amplified and used as an internal standard to normalize the expression of SRL2 and the other genes tested. The primers used to test the expression of SRL2 and the other genes are listed in SupplementaryTable S3.
Results
srl2 plants have semi-rolled leaves and reduced plant height
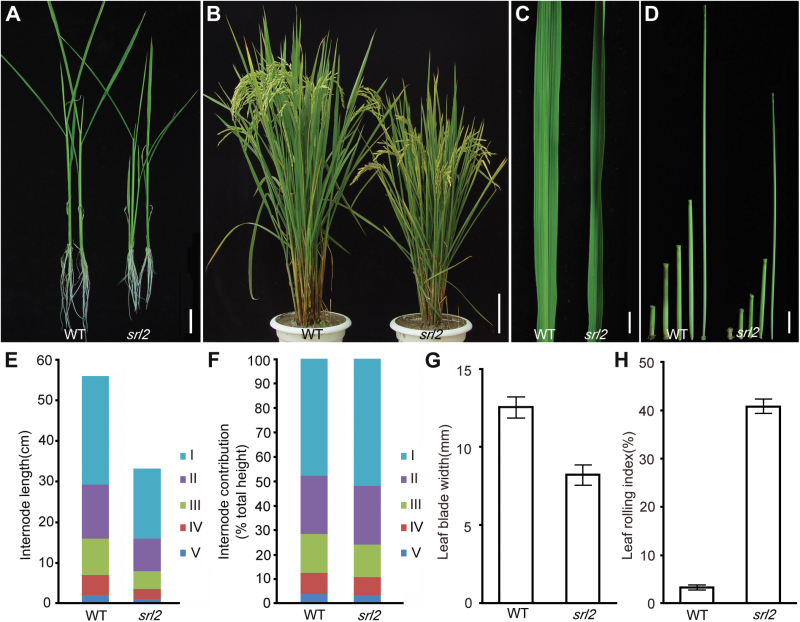
The srl2 mutant was identified in a rice mutant population generated from γ-ray-irradiated indica rice cultivar, Zhongxian 3037. The mutant was designated srl2 based on its semi-rolled leaves. Phenotypic observation revealed that srl2 mutant plants had semi-uprolled leaves (Fig. 1B, C) which appeared during the seedling stage (Fig. 1A) and became more evident during plant growth. Statistical analysis revealed that, compared with wild-type leaves, the leaf rolling index (LRI) of the top second leaves of the srl2 mutant increased by 10-fold (from 4% to 40%) at the heading stage (Fig. 1H;Supplementary Table S4). In addition to rolled leaves, srl2 also displayed narrow leaves (wild type, 12.5±0.67mm, n=29; srl2, 8.2±0.65mm, n=29) (Fig. 1G;Supplementary Table S4) and reduced plant height (Fig. 1B). The dwarfism phenotype of srl2 was exhibited throughout plant growth and development. Two-week-old mutant seedlings had reduced lengths in both the aerial parts and the roots (Fig. 1A). At the flower stage, the height of mutant plants (55.8±1.14cm, n=10) was 70% that of wild-type plants (79.58±1.44cm, n=10) (Fig. 1B;Supplementary Table S5). The reduced height resulted from uniformly shortened internodes in the mutant culms which was confirmed by comparing the length of each internode between the mutant and the wild type (Fig. 1D–F;Supplementary Table S6). In addition, srl2 had thin culms, as well as additional morphological abnormalities including significantly reduced panicle size (SupplementaryFig. S1). Therefore, the mutation in srl2 had multiple effects on plant growth.
Fig. 1.
Phenotypic characterization. (A) Two-week-old wild-type and srl2 seedlings. (B) Mature wild-type and srl2 plants. (C) Wild-type and srl2 leaves. (D) Wild-type and srl2 internodes. (E) Quantification of wild-type and srl2 internode length (mean of 15 culms ±SD). (F) Percentage of contribution of each internode to the total plant height of wild-type and srl2 plants (mean of 15 culms ±SD). I–V, first to fifth internode. (G) Leaf width of the wild type and srl2 (mean of 29 leaves ±SD). (H) LRI of the top two leaves of srl2 at the heading stage. Scale bar 2cm (A, D), 10cm (B), and 1cm (C).
SRL2 encodes a plant-specific protein with unknown biochemical function
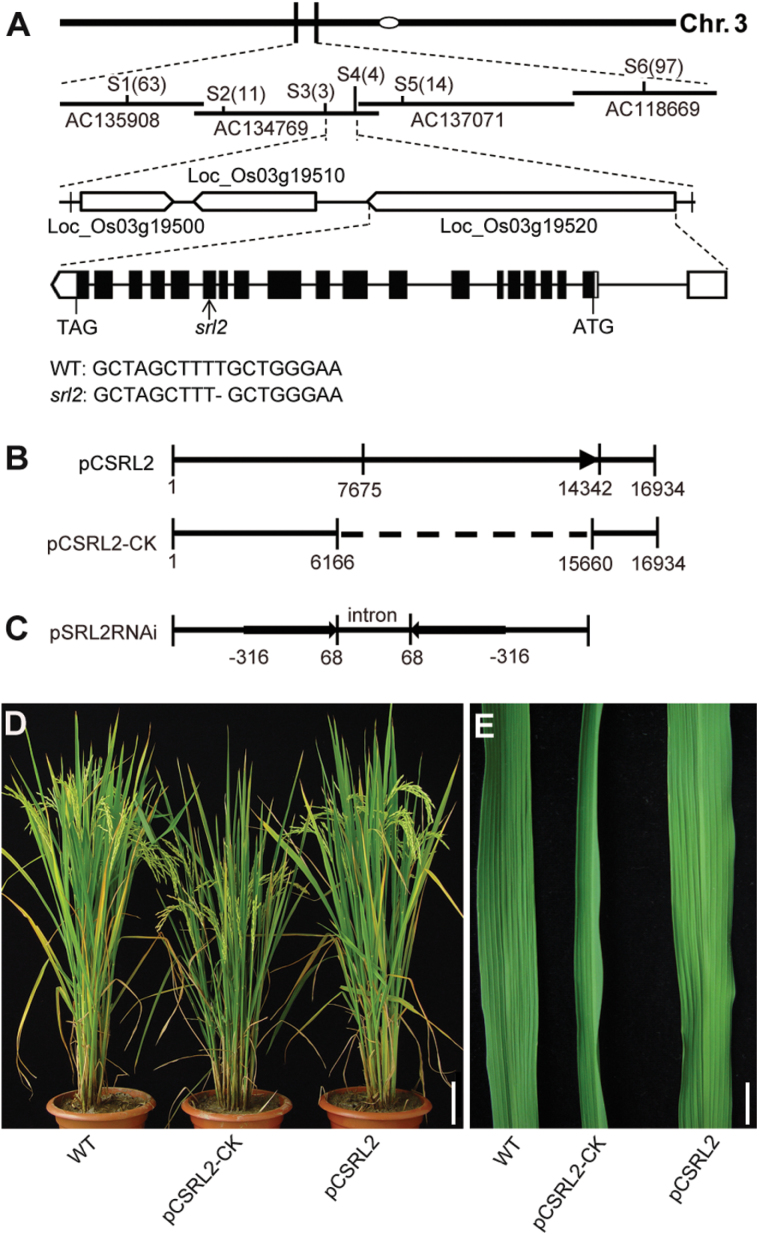
Genetic analysis indicated that the rolled-leaf phenotype is controlled by a single recessive gene, with one-quarter of the F2 progeny displaying leaf rolling (110: 34, χ2 (1)=0.15, P >0.10). To unravel the molecular basis of this phenotype, we performed map-based cloning to isolate this gene. From the F2 population generated by crossing the srl2 mutant with a japonica variety Nipponbare, 2 994 homozygous mutant plants were obtained and used for positional cloning. The SRL2 locus was preliminarily mapped to the long arm of chromosome 3 between sequence-tagged site (STS) markers S1 and S6. Further mapping using adjacent STS markers indicated that the SRL2 locus is present in BAC clone AC134769 (Supplementary Table S1); the candidate gene region was ultimately narrowed down to a 56-kb region. According to the annotations of the rice genome database (Rice Genome Annotation Project, http://rice.plantbiology.msu. edu/cgi-bin/gbrowse/rice/), this region comprises three putative genes with annotated functions. DNA sequencing revealed that one of these genes contains a mutation within the open reading frame (ORF) that co-segregated with the srl2 phenotype. The mutation in SRL2 in the srl2 mutant is a deletion of a T residue in the 15th exon of LOC_Os03g19520 (Fig. 2A), which causes a reading frame shift, resulting in the production of a predicted truncated protein. Compared with the wild type, in the deduced mutant protein, 14 amino acid residues (aa) are altered, with the exception of a 270 aa deletion (Supplementary Fig. S2).
Fig. 2.
Map-based cloning of SRL2. (A) The srl2 locus was mapped to a 56-kb DNA region on chromosome 3. The SRL2 gene comprises 20 exons (boxes) and 19 introns (line). (B) Constructs used for complementary analysis; pCSRL2 and pCSRL2-CK were used to transform srl2 plants. The dotted line in pCSRL2-CK indicates the deleted genomic region. (C) Constructs used for RNAi analysis; pSRL2RNAi was used to transform wild-type plants. (D, E) Results of complementary analysis. Scale bars=10cm (D) and 1cm (E).
To confirm that LOC_Os03g19520 corresponds to the SRL2 locus, we performed a functional complementation assay. Specifically, 16.9kb or 7.44kb of genomic DNA (with or without the entire ORF) was inserted into vector pCAMBIA 1300 to generate plasmids pCSRL2 and pCSRL2-CK, respectively (Fig. 2B). These plasmids were then transformed into the srl2 mutant, and 46 and 34 independent transgenic lines were obtained, respectively. The mutant phenotypes were fully rescued in lines transformed with pCSRL2, but not in lines transformed with pCSRL2-CK (Fig. 2D, E). Moreover, SRL2 knockdown transgenic plants generated by transforming an RNAi construct (Fig. 2C) into wild-type plants mimicked the srl2 phenotypes (Supplementary Fig. S3). Thus, we conclude that SRL2 corresponds to LOC_Os03g19520.
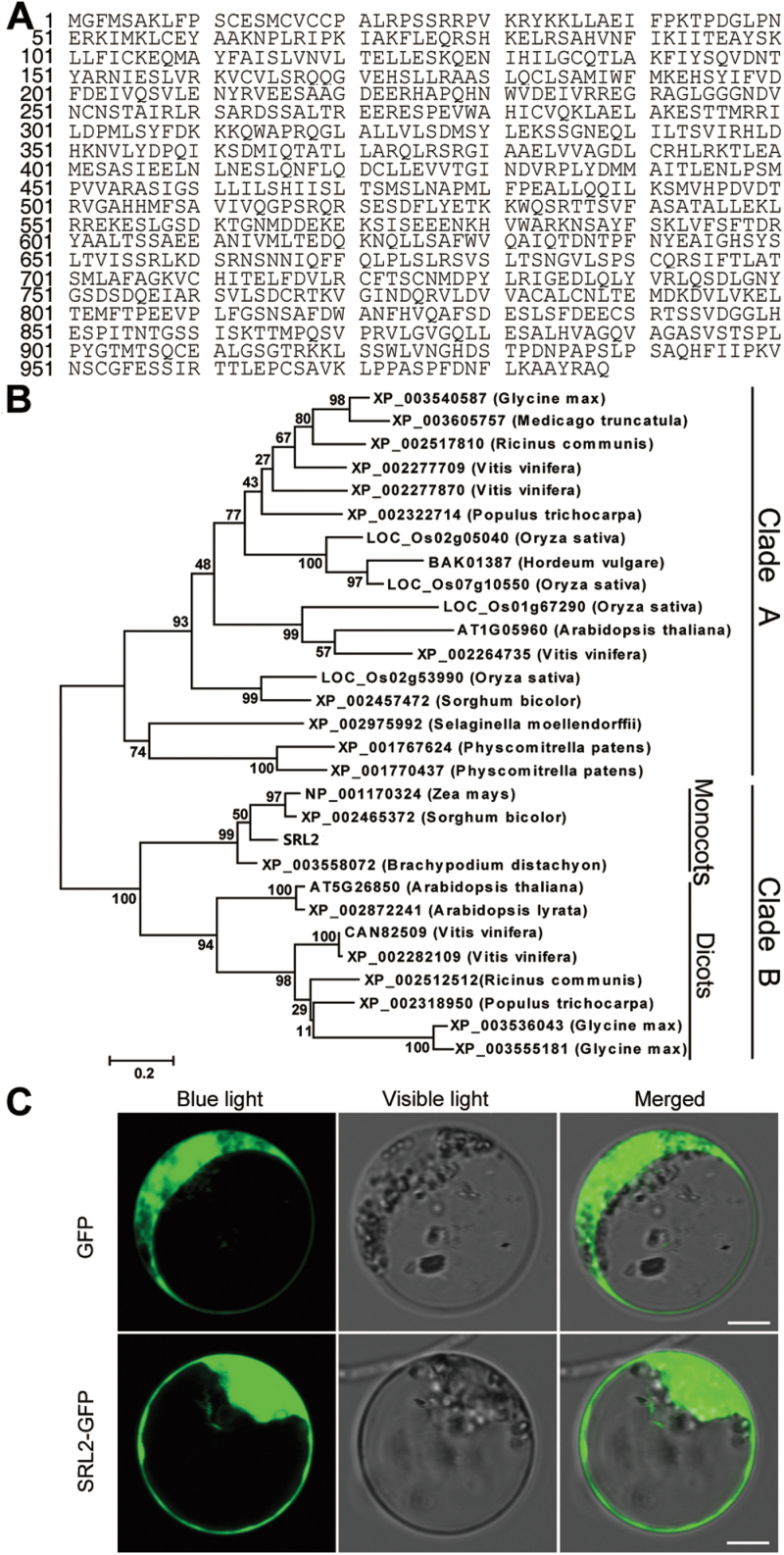
A KOME cDNA database search (http://cdna01.dna.affrc.go.jp/cDNA/) revealed the presence of a full-length cDNA (AK072741) corresponding to SRL2 (LOC_Os03g19520). We performed RT-PCR and confirmed the sequence validation of the cDNA (AK072741). Sequence comparison between genomic DNA and cDNA revealed that SRL2 consists of 20 exons and 19 introns (Fig. 2A) and encodes a protein with 988 amino acids (Fig. 3A). A survey of the published rice genome database (http://www.gramene.org/) revealed the existence of four additional genes designated SRL2-like genes (LOC_Os02g05040, LOC_Os01g67290, LOC_Os02g53990, and LOC_Os07g10550), which were predicted to encode proteins that share various levels of amino acid sequence identity with SRL2. We performed a BLASTP search against the NCBI non-redundant protein database using the SRL2 protein sequence as the query (http://blast.ncbi.nlm.nih.gov), revealing 24 SRL-like proteins from other plant species with significant homology (≥50% identity at the amino acid level) to SRL2, including monocots and dicots. Intriguingly, the existence of SRL2-like genes in different plant species suggests that SRL2 family members share conserved biochemical functions. However, the SRL2 sequence did not match any protein of known biochemical function in the public databases. Therefore, we propose that SRL2 encodes a novel plant-specific protein.
Fig. 3.
Phylogenetic analysis and subcellular localization of SRL2 protein. (A) Amino acid sequence of SRL2. (B) Phylogenetic relationships among the SRL2-like proteins. The numbers at each node represent the bootstrap support (percentage). The scale bar indicates the genetic distance based on branch length. (C) Subcellular location of the SRL2–GFP fusion protein in rice protoplasts. Scale bars=50 μm.
To elucidate the evolutionary relationships among these SRL2-like proteins, we constructed a phylogenetic tree based on the full-length protein sequences of SRL2 and the 28 SRL2-like proteins using the Maximum likelihood method in MEGA3.1. The 29 proteins were classified into two major clades (Clade A and Clade B; Fig. 3B). While the four SRL2 paralogous genes belong to Clade A, SRL2 belongs to Clade B, indicating the functional diversification between SRL2 and its paralogues in rice.
To determine the subcellular localization of SRL2, we fused the green fluorescent protein (GFP) gene to the ORF of SRL2 driven by the 35S promoter and transformed this construct into rice protoplasts. SRL2–GFP chimeric protein, like the control, displayed a ubiquitous distribution pattern throughout the cell (Fig. 3C). We also certified the result in onion epidermis cell (Supplementary Fig. S4) and the Western blot analysis showed that the SRL–GFP was expressed normally (Supplementary Fig. S5).
srl2 plants have defective sclerenchymatous cells in the abaxial sides of leaves
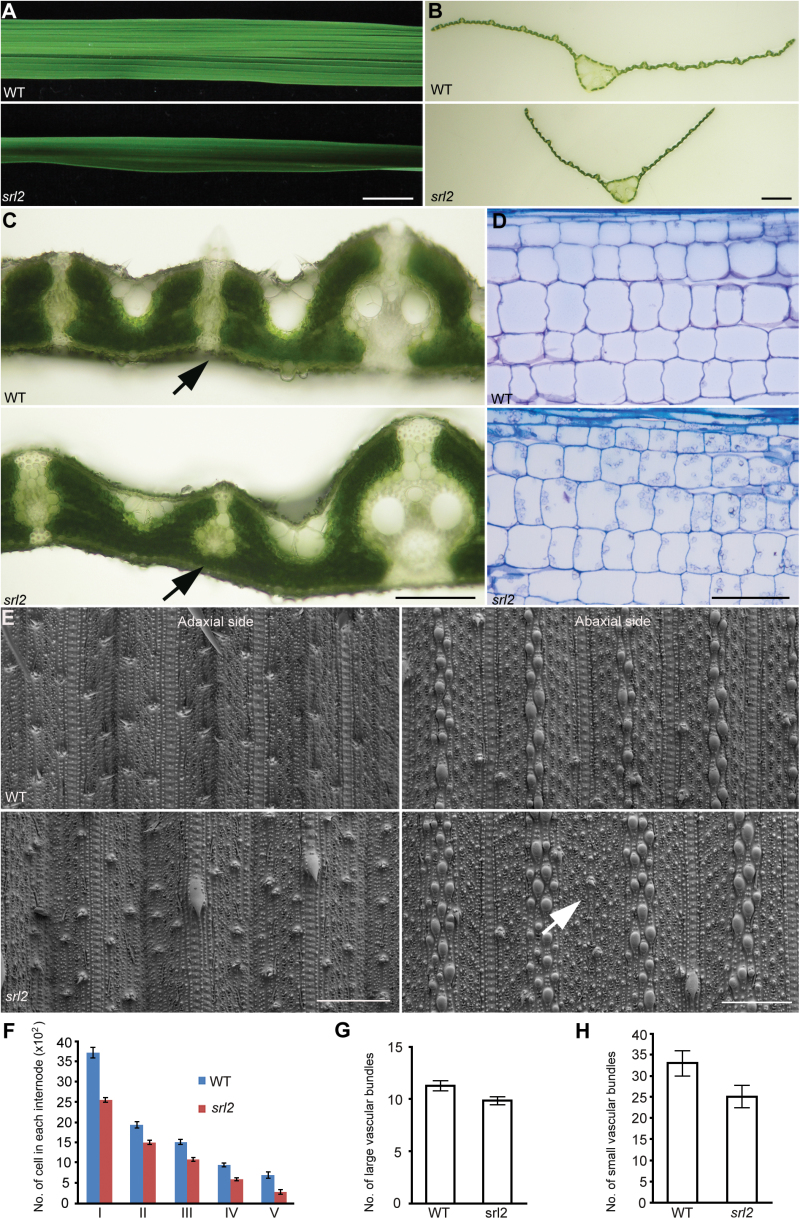
As mentioned above, the srl2 mutant had semi-rolled leaves. To determine the cause of the rolled leaf phenotype, we performed anatomical observations of leaves. In contrast to the wild type, cross-sections of mature srl2 leaves displayed altered mesophyll cell differentiation and distribution. The mutant did not form abaxial sclerenchymatous cells in the small veins of the lateral region (Fig. 4A–C) where the leaf began to roll; however, the cellular organization in the midrib region and large veins was similar to that of the wild type. In addition, not all small veins lacked sclerenchymatous cells in the abaxial side, statistical analysis indicated that approximate 20% of small veins per leaf (20.37±2%, n=7) lacked sclerenchymatous cells (Supplementary Table S7).
Fig. 4.
The srl2 mutant has incurved leaves, with a deficiency of sclerenchymatous cells on the abaxial side. (A–C) Defective sclerenchymatous cells on the abaxial side of srl2 leaves. The defect is mainly present in the small veins where the curl occurs. (D) Longitudinal sections of wild-type and srl2 internodes. (E) Scanning electron micrograph of the adaxial and abaxial surfaces of wild-type and srl2 leaves. (F) Number of cells in each internode (mean of four culms ±SD). Quantification of the large veins (LV) (G) and small veins (SV) (H) in leaves of wild-type and srl2 plants (mean of six leaves ±SD). The arrows in (C) indicate the difference in abaxial sclerenchymatous cells between the wild type and srl2. The arrow in (E) indicates the absence of dumb-bell-shaped silica cells in the abaxial side of srl2 leaves. Scale bars=1cm (A), 1mm (B), 100 μm (C, D) and 150 μm (E).
To confirm our results further, we performed an ultrastructural observation of the leaf epidermis by scanning electron microscopy (SEM). Epidermal cells in wild-type rice leaves are arranged regularly in rows that are parallel to the midrib. At the centre of the leaf surface (on both the adaxial and abaxial sides) where a vascular bundle occurs, dumb-bell-shaped silica cells lay side-by-side. These dumb-bell-shaped silica cells form one file directly over or under a small vascular bundle. The adaxial side of the leaf epidermis in srl2 was comparable with that of the wild type. In the abaxial side of srl2 leaf epidermis, however, the file formed by dumb-bell-shaped silica cells was absent under some small vascular bundles (Fig. 4E). These results further confirm that the differentiation of some perivascular sclerenchyma cells in the abaxial sides of srl2 leaves was defective, indicating that the absence of sclerenchymatous cells in the abaxial side of leaves may account for the rolled leaf phenotype. A similar defect in sclerenchymatous cell development in the abaxial sides of leaves was also observed in the sll1/rl9 mutant (Yan et al., 2008; Zhang et al., 2009), and this defect was also considered to produce rolled leaves in sll1/rl9.
In addition, another major phenotype of srl2 is moderate dwarfism at all stages of growth and development. At the mature stage, the height of mutant plants was reduced by more than 30% (Fig. 1B). To determine the cause of the dwarf phenotype in srl2 plants, we examined the anatomical features of cells in mutant and wild-type culms. First, we compared the internode cell size in both the transverse and longitudinal directions in mutant versus wild-type plants and found that these values were not significantly different (Fig. 4D). We also compared the cell number in the longitudinal direction (which contributes to the length of each internode) between srl2 and wild-type plants. The cell numbers in each internode of srl2 were only 40–70% those of the wild type (Fig. 4F; Supplementary Table S8). Furthermore, examining cross-sections of leaves and leaf sheaths helped confirm that the thin culms and narrow leaves resulted from reduced cell numbers, as determined by counting the number of vascular bundles (Fig. 4G, 4H; Supplementary Table S9). Therefore, reduced cell numbers might be the main factor contributing to the small size of the mutant plants.
Expression patterns of SRL2
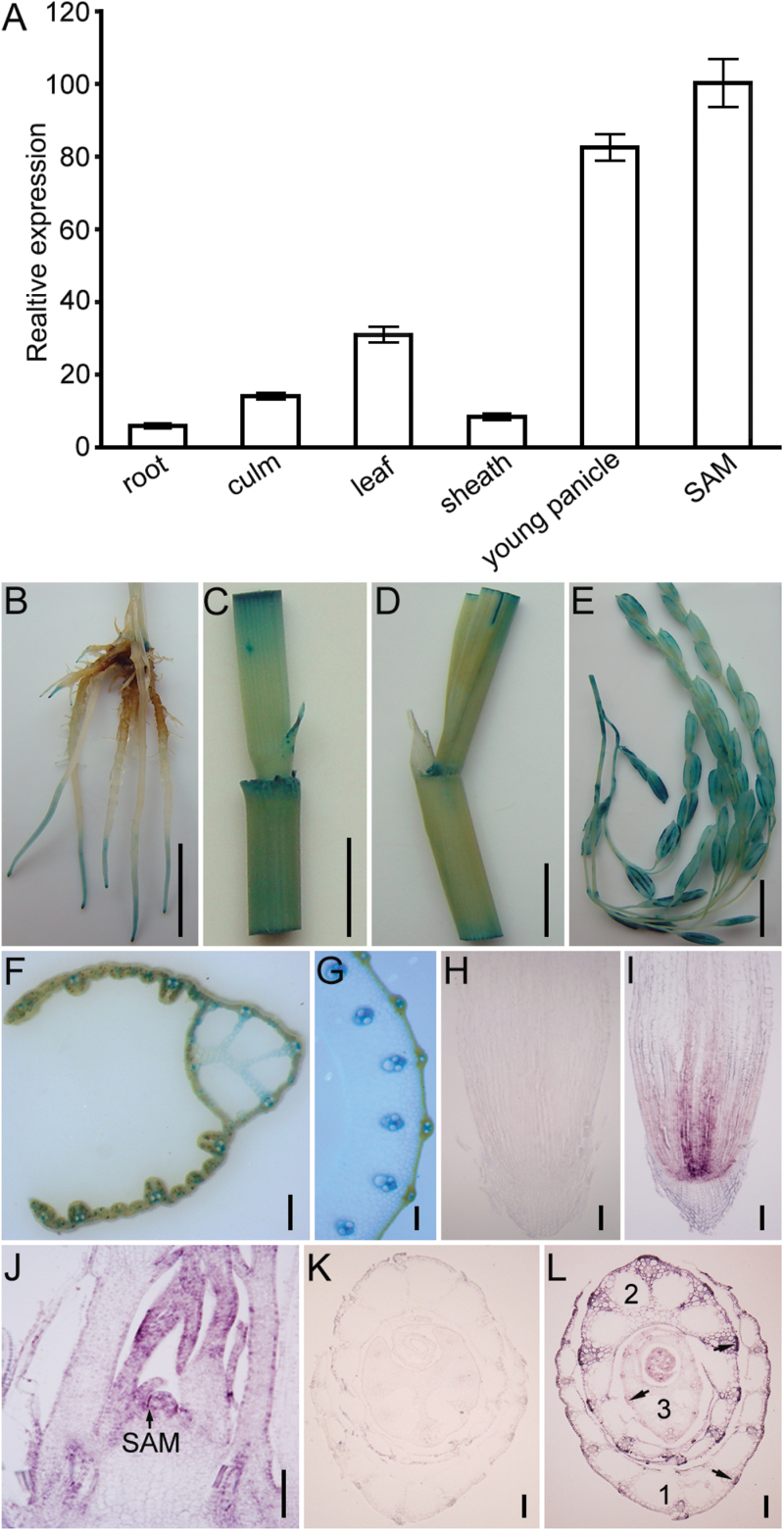
We used quantitative real-time RT-PCR (qRT-PCR) analysis to investigate the expression of SRL2 in various tissues, including root, mature leaf, internode (culm), and leaf sheath tissue (Fig. 5A). Strong expression of SRL2 was detected in the shoot apical meristem (SAM) and young panicles before heading (Fig. 5A), suggesting that SRL2 is highly expressed in rapidly growing tissues.
Fig. 5.
Expression pattern of SRL2. (A) qRT-PCR analysis of SRL2 in various rice tissues. The expression levels are percentages of that of Ubiquitin. Values are means ±SD of triplicate assays. (B–G) GUS patterns in various organs of Pro SRL2 -GUS transgenic plants, showing GUS signals in root tips (cell division zone) (B), culm (C), leaf sheath (D), and young panicle (E). Hand-cut section of leaf (F) and culm (G) tissue. (H–L) In situ hybridization analysis of SRL2 expression in root and SAM tissue. (H, K) In situ hybridization of control tissue using a sense probe. SRL2 transcript was detected in root tips (cell division zone) (I), SAM (J), and the leaf sheath abaxial cell layer and vascular bundles (L). The arrows in (J) and (L) indicate the in situ hybridization signal. Scale bars=1cm (B–E), 100 μm (F–L).
To assess the expression patterns comprehensively, we generated transgenic rice plants expressing the GUS reporter gene driven by the SRL2 putative promoter (Pro SRL2 -GUS). β-glucuronidase (GUS) histochemical staining of transgenic plants demonstrated that SRL2 was preferentially expressed in root tips (cell division zone) (Fig. 5B). In the aerial parts of plants, GUS staining was also visualized in all tissues, including culms (Fig. 5C), leaf sheaths (Fig. 5D), and young panicles (Fig. 5E) which is consistent with the results of qRT-PCR. Hand-cut sectioning of mature leaf blades and leaf sheaths further demonstrated that GUS was mainly expressed in vascular bundles (Fig. 5F, G; Supplementary Fig. S6).
To confirm that SRL2 is preferentially expressed in rapidly growing tissues, we performed a more detailed characterization of SRL2 transcript levels in root and SAM tissues by mRNA in situ hybridization. Hybridization signals were clearly expressed in root tip cells undergoing cell division (Fig. 5I), especially in the region that differentiates into the stele or vascular cylinder. Similar results were obtained for SAM tissue (Fig. 5J). Hybridization signals were not detected in sections probed with sense-strand SRL2 RNA, which was used as a negative control (Fig. 5H).
We also examined the spatial and temporal localization of SRL2 during leaf development by in situ hybridization analysis. Cross-section analysis of the shoot apex showed that SRL2 was highly expressed in the leaf sheath abaxial cell layer and in vascular bundles, especially in sclerenchymatous cells that had undergone a certain degree of development. SRL2 was dynamically expressed in the sclerenchymatous cell zone. As shown in Fig. 5L, the leaf sheath comprises three layers (layer 1–3, from the outside to the inside). SRL2 mRNA signals were relatively weak in layer 1, significantly stronger in layer 2, and weak in layer 3 (Fig. 5L). However, very weak hybridization signals were detected in sections probed with sense-strand SRL2 RNA (Fig. 5K).
In summary, SRL2 appears to be ubiquitously expressed, although it is preferentially expressed in rapidly growing tissues and at specific stages of leaf development.
YAB4 and YAB5 are dramatically down-regulated in srl2
Leaf development is regulated by several groups of genes such as auxin synthesis-related genes including YUCCA genes (Fujino et al., 2008), auxin transport-associated genes (Benkova et al., 2003), and YABBY genes (Dai et al., 2007; Stahle et al., 2009; Sarojam et al., 2010; Tanaka et al., 2012), as well as other leaf development-related genes. To gain insights into the SRL2-mediated mechanism in leaf organogenesis, we measured the mRNA levels of leaf development-associated genes in young leaves by qRT-PCR. We first examined the expression of auxin signal transduction, transport, and biosynthesis-related genes, such as AUXIN RESPONSE FACTOR (ARF), PIN-FORMED (PIN), and YUC family genes, revealing no obvious change in expression in srl2.
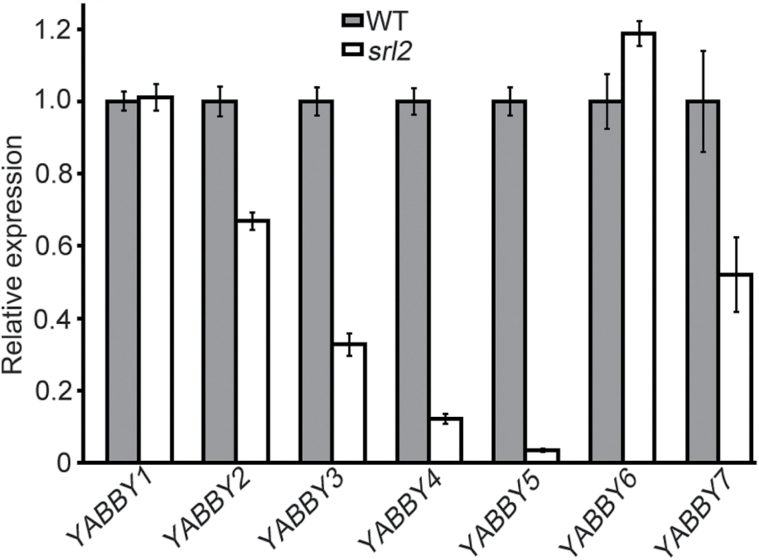
In rice, SLL1/RL9 encodes a KANADI family protein involved in leaf abaxial cell development. Like SRL2, mutation of SLL1/RL9 leads to defective development of sclerenchymatous cells in the abaxial sides of leaves (Yan et al., 2008; Zhang et al., 2009). These similar mutant phenotypes prompted us to investigate changes in SLL1/RL9 expression in srl2, revealing that SLL1/RL9 expression was unaffected in the mutant. Moreover, when we crossed srl2 and sll1 plants, srl2 sll1 double mutant plants were obtained. Statistical analysis revealed that, compared with srl2 and sll1 single mutants, the srl2 sll1 double mutant had more severely defective development of sclerenchymatous cells in the leaf abaxial side, together with a much more narrow leaf blade phenotype (Supplementary Fig. S7). These results suggest that SRL2 and SLL1/RL9 function in distinct pathways regulating the development of the abaxial side of the leaf. Finally, we examined the expression of other leaf development-related genes, such as YABBY family genes (Stahle et al., 2009; Sarojam et al., 2010; Tanaka et al., 2012), NAL1 (Qi et al., 2008), ND1 (Li et al., 2009), and RL14 (Fang et al., 2012). Compared with the wild type, YAB4 and YAB5 were significantly down-regulated in srl2 (5-fold and 27-fold, respectively), and YAB2, YAB3, and YAB7 were also slightly down-regulated (Fig. 6). However, the expression levels of other YABBY genes, as well as NAL1, ND1, and RL14 were not altered. These results suggest that the phenotypes of srl2 may be related to the altered transcriptional activity of leaf development-related YABBY genes.
Fig. 6.
Expression of YABBYs in the wild type and srl2, using the ubiquitin gene as the internal control. Values are means ±SD of triplicate assays.
Discussion
Abnormal sclerenchymatous cells account for rolled leaves in srl2
Leaf rolling is a complex trait affected by multiple genes and even the same degree of leaf rolling can be caused by different genes (Hibara et al., 2009; Zhang et al., 2009; Li et al., 2010; Zou et al., 2011). Furthermore, differences in the expression of a single gene have been associated with differences in leaf rolling index (Zou et al., 2011). In addition, the leaf rolling phenotype may be caused by more than one alteration in the structure of a particular cell type and may even be caused by changes in different or multiple cell types (Zou et al., 2014). Bulliform cells, which are located between two vascular bundle ridges in parallel with the more adaxial-localized veins of grass leaves, play an important role in regulating leaf rolling in rice (Zou et al., 2011; Xiang et al., 2012). Sclerenchyma cells are important for maintaining the proper morphology in rice leaves. Sclerenchymatous cells, which originate from mesophyll cells through transdifferentiation, are lignified dead cells with thickened secondary cell walls. A deficiency in abaxial sclerenchyma cells results in adaxially rolled leaves due to the loss of force provided by sclerenchyma cells, whereas improper formation of sclerenchyma cells on the adaxial side of the leaf is associated with abaxial leaf rolling. SHALLOT-LIKE1 (SLL1) controls the formation of abaxial sclerenchyma cells, and SLL1 deficiency results in tight inwardly rolled leaves (Zhang et al., 2009). Deficient adaxial sclerenchyma cells associated with increased parenchyma cells in vascular bundles in the pci mutants lead to outwardly rolled leaves (Zou et al., 2014). Using SEM and histological analysis of srl2 leaves, we found that srl2 mutant leaves had abnormal sclerenchymatous cells. Since sclerenchymatous cells contribute continuous mechanical strength to maintain normal leaf shape, the lack of sclerenchymatous cells on the abaxial surface in curved regions of srl2 leaves indicates that sclerenchyma plays a critical role in controlling leaf rolling in monocots and that SRL2 functions in sclerenchyma cells formation.
SRL2 and SLL1 may function in distinct pathways regulating abaxial-side development in leaves
SLL1 encodes an SHAQKYF class MYB transcription factor involved in the transdifferentiation of mesophyll cells to sclerenchymatous cells through the modulation of programmed cell death and its deficiency leads to impaired formation of sclerenchymatous cells on the abaxial side of the leaf, resulting in extremely inwardly rolled leaves (Zhang et al., 2009). In addition, SLL1 is crucial to polarity formation and helps direct the development of the leaf abaxial cell layer (Zhang et al., 2009). The defective leaf development in the srl2 mutant is quite similar to that of sll1 although they have varying degrees of leaf rolling (perhaps because not all small veins lacked sclerenchymatous cells in the abaxial sides of srl2 leaves, and approximately 20% of small veins per leaf lacked sclerenchymatous cells). However, we did not find any obvious alteration in leaf polarity in srl2. Interestingly, the srl2 sll1 double mutant exhibited a more severe defect in the development of sclerenchymatous cells in the abaxial side of the leaf, together with a more narrow leaf blade phenotype. Meanwhile, qRT-PCR revealed that the expression of SLL1 was not affected in the srl2 mutant. We propose that SLL1 and SRL2 act in two different ways to modulate abaxial development in leaves.
SRL2 is involved in multiple developmental processes
The expression of YABBY genes was altered in srl2. Genes in this small, plant-specific family encode putative transcription factors containing two conserved domains: a zinc-finger domain in the N-terminal region and a YABBY domain (helix-loop-helix motif) in the C-terminal region (Siegfried et al., 1999). YABBY genes play important roles in regulating diverse developmental processes such as the establishment of adaxial–abaxial polarity, lamina expansion, and floral organ development in eudicots (Tanaka et al., 2012). Arabidopsis thaliana contains only six YABBY genes, namely, FILAMENTOUS FLOWER (FIL), CRABS CLAW (CRC), INNER NO OUTER (INO), YAB2, YAB3, and YAB5, which play distinct roles in development (Bowman and Smyth, 1999; Sawa et al., 1999; Siegfried et al., 1999; Villanueva et al., 1999). Rice contains eight YABBY genes, including OsYABBY1, OsYABBY2, OsYABBY3, OsYABBY4, OsYABBY5, OsYABBY6, OsYABBY7, and DROOPING LEAF (DL) (Toriba et al., 2007). DL, a rice orthologue of CRC has been well characterized. Loss-of-function of DL causes the homeotic transformation of carpels into stamens in the flower and loss of the midrib in the leaf, suggesting that DL regulates carpel specification and midrib formation in rice (Nagasawa et al., 2003; Yamaguchi et al., 2004; Ohmori et al., 2008; Ohmori et al., 2011). OsYABBY1, which is similar to Arabidopsis YAB2, is expressed in putative precursor cells of a specific cell type, such as sclerenchyma (Juarez et al., 2004; Toriba et al., 2007). OsYABBY5 (TOB1) is involved in lateral organ development and maintaining meristem organization in the rice spikelet. Mutation of this gene leads to pleiotropic phenotypes in spikelets, such as the formation of a cone-shaped organ instead of a lemma or palea, the development of two florets in a spikelet, and premature termination of the floret meristem, in addition to reduced growth of the lemma or palea and elongation of the awn (Tanaka et al., 2012). In the srl2 mutant, the expression of some YABBY genes was altered. For example, YAB2, YAB3, YAB4, YAB5, and YAB7 were down-regulated, but YAB6 was up-regulated. In particular, the expression of YAB5 was significantly down-regulated (27-fold) in srl2, but this mutant did not exhibit the same phenotype as the tob1 mutant which might be due to the effect of the altered expression of more than one YABBY gene. These results suggest that the phenotype of srl2 may be related to the altered transcriptional activity of YABBY genes. All of these results demonstrate that SRL2 has multiple effects on plant development.
Mutants represent an excellent genetic resource for ideal plant architecture breeding in rice
Improving grain yield is the ultimate goal of rice breeding. As leaves represent the major photosynthetic organ of plants, optimal leaf structure is important for maximizing light capture and for efficient gas exchange. The moderately rolled leaf is regarded as a critical component of the ideal rice phenotype as it is important for improving photosynthetic efficiency and grain yields by maintaining the erectness of leaves and minimizing shadowing between leaves (Yuan, 1997; Wu, 2009). The shortage of new germplasm resources with important agronomic features is a major barrier to the development of cultivars with higher yields (Wang and Li, 2005). Therefore, it is critical to identify new mutants with moderately rolled leaves and to isolate genes related to these features. In the present study, we performed an in-depth characterization of a semi-rolled leaf mutant which may be useful for breeding rice with an ideal plant architecture.
Supplementary data
Supplementary data can be found at JXB online.
Table S1. Primers used in map-based cloning of SRL2.
Table S2. Primers used for in situ hybridization analysis.
Table S3. Primers used in real-time qRT-PCR analysis.
Table S4. The width of the leaf blade in both the wild type and srl2.
Table S5. The height of plants in both the wild type and srl2.
Table S6. The length of internode in both the wild type and srl2.
Table S7. The number of abnormal small vascular bundles in srl2.
Table S8. The number of cells in each internode in both the wild type and srl2.
Table S9. The number of large vascular bundles and small vascular bundles in both the wild type and srl2.
Figure S1. Morphological comparison of wild-type and srl2 panicles.
Figure S2. Deduced amino acid sequences of SRL2 and the changes in amino acids caused by mutations.
Figure S3. RNAi analyses.
Figure S4. Subcellular location of the SRL2–GFP fusion protein in onion epidermal cells.
Figure S5. Western blot assays.
Figure S6. Hand-cut section of a Pro SRL2 -GUS transgenic rice leaf.
Figure S7. Characteristics of the srl2 sll1 double mutant.
Acknowledgements
This work was supported by grants from the Ministry of Sciences and Technology of China (2014ZX0800939B and 2013ZX08009-003), the National Natural Science Foundation of China (31371589), and the Jiangsu Agricultural Science and Technology Innovation Fund (CX12-5072).
References
- Benkova E, Michniewicz M, Sauer M, Teichmann T, Seifertova D, Jurgens G, Friml J. 2003. Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell 115, 591–602. [DOI] [PubMed] [Google Scholar]
- Bowman JL, Eshed Y, Baum SF. 2002. Establishment of polarity in angiosperm lateral organs. Trends in Genetics 18, 134–141. [DOI] [PubMed] [Google Scholar]
- Bowman JL, Smyth DR. 1999. CRABS CLAW, a gene that regulates carpel and nectary development in Arabidopsis, encodes a novel protein with zinc finger and helix-loop-helix domains. Development 126, 2387–2396. [DOI] [PubMed] [Google Scholar]
- Candela H, Johnston R, Gerhold A, Foster T, Hake S. 2008. The milkweed pod1 gene encodes a KANADI protein that is required for abaxial/adaxial patterning in maize leaves. The Plant Cell 20, 2073–2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai MQ, Hu YF, Zhao Y, Liu HF, Zhou DX. 2007. A WUSCHEL-LIKE HOMEOBOX gene represses a YABBY gene expression required for rice leaf development. Plant Physiology 144, 380–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eshed Y, Izhaki A, Baum SF, Floyd SK, Bowman JL. 2004. Asymmetric leaf development and blade expansion in Arabidopsis are mediated by KANADI and YABBY activities. Development 131, 2997–3006. [DOI] [PubMed] [Google Scholar]
- Fang LK, Zhao FM, Cong YF, Sang XC, Du Q, Wang DZ, Li Y, Ling YH, Yang ZL, He GH. 2012. Rolling-leaf14 is a 2OG-Fe (II) oxygenase family protein that modulates rice leaf rolling by affecting secondary cell wall formation in leaves. Plant Biotechnology Journal 10, 524–532. [DOI] [PubMed] [Google Scholar]
- Fujino K, Matsuda Y, Ozawa K, Nishimura T, Koshiba T, Fraaije MW, Sekiguchi H. 2008. NARROW LEAF 7 controls leaf shape mediated by auxin in rice. Molecular Genetics and Genomics 279, 499–507. [DOI] [PubMed] [Google Scholar]
- Govaerts YM, Jacquemoud S, Verstraete MM, Ustin SL. 1996. Three-dimensional radiation transfer modeling in a dicotyledon leaf. Applied Optics 35, 6585–6598. [DOI] [PubMed] [Google Scholar]
- Hibara K, Obara M, Hayashida E, Abe M, Ishimaru T, Satoh H, Itoh J, Nagato Y. 2009. The ADAXIALIZED LEAF1 gene functions in leaf and embryonic pattern formation in rice. Developmental Biology 334, 345–354. [DOI] [PubMed] [Google Scholar]
- Hiei Y, Ohta S, Komari T, Kumashiro T. 1994. Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. The Plant Journal 6, 271–282. [DOI] [PubMed] [Google Scholar]
- Juarez MT, Twigg RW, Timmermans MCP. 2004. Specification of adaxial cell fate during maize leaf development. Development 131, 4533–4544. [DOI] [PubMed] [Google Scholar]
- Kadioglu A, Terzi R. 2007. A dehydration avoidance mechanism: leaf rolling. The Botanical Review 73, 290–302. [Google Scholar]
- Khush GS, Kinoshita T. 1991. Rice karyotype, marker genes, and linkage groups. Rice Biotechnology 83–108. [Google Scholar]
- Kumar S, Tamura K, Nei M. 2004. MEGA3: Integrated software for Molecular Evolutionary Genetics Analysis and sequence alignment. Briefings in Bioinformatics 5, 150–163. [DOI] [PubMed] [Google Scholar]
- Li L, Shi ZY, Li L, Shen GZ, Wang XQ, An LS, Zhang JL. 2010. Overexpression of ACL1 (abaxially curled leaf 1) increased bulliform cells and induced abaxial curling of leaf blades in rice. Molecular Plant 3, 807–817. [DOI] [PubMed] [Google Scholar]
- Li M, Xiong GY, Li R, Cui JJ, Tang D, Zhang BC, Pauly M, Cheng Z, Zhou YH. 2009. Rice cellulose synthase-like D4 is essential for normal cell-wall biosynthesis and plant growth.The Plant Journal 60, 1055–1069. [DOI] [PubMed] [Google Scholar]
- Li SG, Ma YQ, He P, Li HY, Chen Y, Zhou KD, Zhu LH. 1998. Genetics analysis and mapping the flag leaf roll in rice (Oryza sativa L.). Journal of Sichuan Agricultural University 16, 391–393. [Google Scholar]
- Luo Y, Zhao FM, Sang XC, Ling YH, Yang ZL, He GH. 2009. Genetic analysis and gene mapping of a novel rolled-leaf mutant rl12 (t) in rice. Acta Agronomica Sinica 35, 1967–1972. [Google Scholar]
- Luo ZK, Yang ZL, Zhong BQ, Li YF, Xie R, Zhao FM, Ling YH, He GH. 2007. Genetic analysis and fine mapping of a dynamic rolled leaf gene, RL10(t), in rice (Oryza sativa L.). Genome/Conseil national de recherches Canada 50, 811–817. [DOI] [PubMed] [Google Scholar]
- McConnell JR, Emery J, Eshed Y, Bao N, Bowman J, Barton MK. 2001. Role of PHABULOSA and PHAVOLUTA in determining radial patterning in shoots. Nature 411, 709–713. [DOI] [PubMed] [Google Scholar]
- Micol JL, Hake S. 2003. The development of plant leaves. Plant Physiology 131, 389–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon J, Hake S. 2011. How a leaf gets its shape. Current Opinion in Plant Biology 14, 24–30. [DOI] [PubMed] [Google Scholar]
- Nagasawa N, Miyoshi M, Sano Y, Satoh H, Hirano H, Sakai H, Nagato Y. 2003. SUPERWOMAN1 and DROOPING LEAF genes control floral organ identity in rice. Development 130, 705–718. [DOI] [PubMed] [Google Scholar]
- Nelson JM, Lane B, Freeling M. 2002. Expression of a mutant maize gene in the ventral leaf epidermis is sufficient to signal a switch of the leaf’s dorsoventral axis. Development 129, 4581–4589. [DOI] [PubMed] [Google Scholar]
- Nogueira FTS, Madi S, Chitwood DH, Juarez MT, Timmermans MCP. 2007. Two small regulatory RNAs establish opposing fates of a developmental axis. Genes & Development 21, 750–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohmori Y, Abiko M, Horibata A, Hirano HY. 2008. A transposon, Ping, is integrated into intron 4 of the DROOPING LEAF gene of rice, weakly reducing its expression and causing a mild drooping leaf phenotype. Plant and Cell Physiology 49, 1176–1184. [DOI] [PubMed] [Google Scholar]
- Ohmori Y, Toriba T, Nakamura H, Ichikawa H, Hirano HY. 2011. Temporal and spatial regulation of DROOPING LEAF gene expression that promotes midrib formation in rice. The Plant Journal 65, 77–86. [DOI] [PubMed] [Google Scholar]
- Otsuga D, DeGuzman B, Prigge MJ, Drews GN, Clark SE. 2001. REVOLUTA regulates meristem initiation at lateral positions. The Plant Journal 25, 223–236. [DOI] [PubMed] [Google Scholar]
- Qi J, Qian Q, Bu QY, et al. 2008. Mutation of the rice Narrow leaf1 gene, which encodes a novel protein, affects vein patterning and polar auxin transport. Plant Physiology 147, 1947–1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarojam R, Sappl PG, Goldshmidt A, Efroni I, Floyd SK, Eshed Y, Bowman JL. 2010. Differentiating Arabidopsis shoots from leaves by combined YABBY activities. The Plant Cell 22, 2113–2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawa S, Watanabe K, Goto K, Liu YG, Shibata D, Kanaya E, Morita EH, Okada K. 1999. FILAMENTOUS FLOWER, a meristem and organ identity gene of Arabidopsis, encodes a protein with a zinc finger and HMG-related domains. Genes & Development 13, 1079–1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao YJ, Chen ZX, Zhang YF, Chen EH, Qi DC, Miao J, Pan XB. 2005. a One major QTL mapping and physical map construction for rolled leaf in rice. Acta Genetica Sinica 32, 501–506. [PubMed] [Google Scholar]
- Shao YJ, Pan CH, Chen ZX, Zuo SM, Zhang YF, Pan XB. 2005. b Fine mapping of an incomplete recessive gene for leaf rolling in rice (Oryza sativa L.). Chinese Science Bulletin 50, 2466–2472. [Google Scholar]
- Shi YF, Chen J, Liu WQ, Huang QN, Shen B, Leung H, Wu JL. 2009. Genetic analysis and gene mapping of a new rolled-leaf mutant in rice (Oryza sativa L.). Science in China, Series C, Life Sciences 52, 885–890. [DOI] [PubMed] [Google Scholar]
- Shi ZY, Wang J, Wan XS, Shen GZ, Wang XQ, Zhang JL. 2007. Over-expression of rice OsAGO7 gene induces upward curling of the leaf blade that enhanced erect-leaf habit. Planta 226, 99–108. [DOI] [PubMed] [Google Scholar]
- Siegfried KR, Eshed Y, Baum SF, Otsuga D, Drews GN, Bowman JL. 1999. Members of the YABBY gene family specify abaxial cell fate in Arabidopsis . Development 126, 4117–4128. [DOI] [PubMed] [Google Scholar]
- Stahle MI, Kuehlich J, Staron L, von Arnim AG, Golz JF. 2009. YABBYs and the transcriptional corepressors LEUNIG and LEUNIG_HOMOLOG maintain leaf polarity and meristem activity in Arabidopsis . The Plant Cell 21, 3105–3118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzaki T, Sato M, Ashikari M, Miyoshi M, Nagato Y, Hirano HY. 2004. The gene FLORAL ORGAN NUMBER1 regulates floral meristem size in rice and encodes a leucine-rich repeat receptor kinase orthologous to Arabidopsis CLAVATA1. Development 131, 5649–5657. [DOI] [PubMed] [Google Scholar]
- Tanaka W, Toriba T, Ohmori Y, Yoshida A, Kawai A, Mayama-Tsuchida T, Ichikawa H, Mitsuda N, Ohme-Takagi M, Hirano HY. 2012. The YABBY gene TONGARI-BOUSHI1 is involved in lateral organ development and maintenance of meristem organization in the rice spikelet. The Plant Cell 24, 80–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Research 25, 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toriba T, Harada K, Takamura A, Nakamura H, Ichikawa H, Suzaki T, Hirano HY. 2007. Molecular characterization the YABBY gene family in Oryza sativa and expression analysis of OsYABBY1 . Molecular Genetics and Genomics 277, 457–468. [DOI] [PubMed] [Google Scholar]
- Villanueva JM, Broadhvest J, Hauser BA, Meister RJ, Schneitz K, Gasser CS. 1999. INNER NO OUTER regulates abaxial–adaxial patterning in Arabidopsis ovules. Genes & Development 13, 3160–3169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YH, Li JY. 2005. The plant architecture of rice (Oryza sativa). Plant Molecular Biology 59, 75–84. [DOI] [PubMed] [Google Scholar]
- Wu XJ. 2009. Prospects of developing hybrid rice with super high yield. Agronomy Journal 101, 688–695. [Google Scholar]
- Xiang JJ, Zhang GH, Qian Q, Xue HW. 2012. Semi-rolled leaf1 encodes a putative glycosylphosphatidylinositol-anchored protein and modulates rice leaf rolling by regulating the formation of bulliform cells. Plant Physiology 159, 1488–1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao H, Tang J, Li YF, et al. 2009. STAMENLESS 1, encoding a single C2H2 zinc finger protein, regulates floral organ identity in rice. The Plant Journal 59, 789–801. [DOI] [PubMed] [Google Scholar]
- Yamaguchi T, Nagasawa N, Kawasaki S, Matsuoka M, Nagato Y, Hirano HY. 2004. The YABBY gene DROOPING LEAF regulates carpel specification and midrib development in Oryza sativa . The Plant Cell 16, 500–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan S, Yan CJ, Zeng XH, Yang YC, Fang YW, Tian CY, Sun YW, Cheng ZK, Gu MH. 2008. ROLLED LEAF 9, encoding a GARP protein, regulates the leaf abaxial cell fate in rice. Plant Molecular Biology 68, 239–250. [DOI] [PubMed] [Google Scholar]
- Yuan L. 1997. Super-high yield hybrid rice breeding. Hybrid Rice 12, 1–6. [Google Scholar]
- Zhang GH, Xu Q, Zhu XD, Qian Q, Xue HW. 2009. SHALLOT-LIKE1 is a KANADI transcription factor that modulates rice leaf rolling by regulating leaf abaxial cell development. The Plant Cell 21, 719–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang JJ, Wu SY, Jiang L, Wang JL, Zhang X, Guo XP, Wu CY, Wan JM. 2015. A detailed analysis of the leaf rolling mutant sll2 reveals complex nature in regulation of bulliform cell development in rice (Oryza sativa L.). Plant Biology 17, 437–448. [DOI] [PubMed] [Google Scholar]
- Zou LP, Sun XH, Zhang ZG, Liu P, Wu JX, Tian CJ, Qiu JL, Lu TG. 2011. Leaf rolling controlled by the homeodomain leucine zipper class IV gene Roc5 in rice. Plant Physiology 156, 1589–1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou LP, Zhang ZG, Qi DF, Peng M, Lu TG. 2014. Cytological mechanisms of leaf rolling in rice. Crop Science 54, 198–209. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.