Abstract
The ability to faithfully transmit genetic information across generations via the germ cells is a critical aspect of mammalian reproduction. The process of germ cell development requires a number of large-scale modulations of chromatin within the nucleus. One such occasion arises during meiotic recombination, when hundreds of DNA double-strand breaks are induced and subsequently repaired, enabling the transfer of genetic information between homologous chromosomes. The inability to properly repair DNA damage is known to lead to an arrest in the developing germ cells and sterility within the animal. Chromatin-remodeling activity, and in particular the BRG1 subunit of the SWI/SNF complex, has been shown to be required for successful completion of meiosis. In contrast, remodeling complexes of the ISWI and CHD families are required for postmeiotic processes. Little is known regarding the contribution of the INO80 family of chromatin-remodeling complexes, which is a particularly interesting candidate due to its well described functions during DNA double-strand break repair. Here we show that INO80 is expressed in developing spermatocytes during the early stages of meiotic prophase I. Based on this information, we used a conditional allele to delete the INO80 core ATPase subunit, thereby eliminating INO80 chromatin-remodeling activity in this lineage. The loss of INO80 resulted in an arrest during meiosis associated with a failure to repair DNA damage during meiotic recombination.
Keywords: chromatin remodeling, INO80, meiotic recombination, spermatogenesis
INTRODUCTION
Mammalian spermatogenesis requires chromatin to undergo small- and large-scale changes that involve dynamic epigenetic regulation. Homologous chromosomes synapse, double-strand breaks in DNA are created and resolved, chromosomes segregate, and histones are replaced by protamines [1]. In order to accomplish these tasks, developing germ cells take advantage of a combination of chromatin modulations, including ATPase-dependent chromatin remodeling [2, 3].
ATP-dependent chromatin remodeling utilizes ATP hydrolysis to selectively mobilize nucleosomes leading to localized areas of nucleosome-depleted chromatin [4]. These events are required for a wide variety of cellular and biological processes, including transcription, replication, and DNA repair [5]. Recent reports have shown that chromatin-remodeling events play critical roles during early meiotic stages of germ cell development [6]. As numerous protein complexes harbor ATPase-dependent chromatin-remodeling activity, defining the roles of each is critical to understanding their relevance during gametogenesis.
Chromatin-remodeling complexes have been grouped into four major families [4]. Several are known to have potent roles throughout male germ cell development. The SWI/SNF complex is active early during spermatogenesis. The catalytic subunit, BRG1, is required for homologous recombination (HR), and the ablation of this subunit results in arrest at the pachytene stage of meiosis I [7, 8]. The imitation switch (ISWI) and chromodomain helicase DNA-binding (CHD) complexes function comparatively late in this process, with postmeiotic phenotypes associated with spermiogenesis and fertilization [9–13]. These observations indicate that chromatin-remodeling activity is involved in a wide variety of activities throughout spermatogenesis. One family of chromatin remodelers whose involvement during mammalian germ cell development has yet to be elucidated is INO80. This family includes the INO80, SRCAP, and TIP60 complexes [14]. Importantly, previously described DNA double-strand break repair functions make the INO80 complex a prime candidate for a critical role early in spermatogenesis.
The INO80 chromatin remodeler is a multisubunit chromatin-remodeling complex that is active in a variety of cellular processes, most notably the repair of DNA damage [15, 16]. Yeast INO80 is localized to sites of DNA double-strand breaks by the presence of phosphorylated H2AX (γH2AX) and is responsible for evicting this histone variant [17]. Mammalian cell lines that have been depleted for INO80 demonstrate an increased sensitivity to DNA-damaging agents and an inability to repair double-strand breaks [18]. Like SWI/SNF, complete loss of INO80 is lethal at the earliest embryonic stages [19]. Both complexes share a single subunit in common, BAF53A, an actin-related regulatory protein. In contrast to SWI/SNF, the INO80 complex incorporates three ATPase subunits, INO80 the core ATPase subunit, as well as two additional ATPase-containing helicases, RUVBL1 and RUVBL2 [20]. As the catalytic subunit of the complex, the INO80 core ATPase presents a potent target for addressing the developmental contributions of the complex.
In this study, we present the first tissue-specific knockout of Ino80. Our findings indicate that the INO80 complex is involved in coordinating DNA repair via HR. We show that INO80 expression peaks during early meiosis and that developing spermatocytes lacking the core ATPase subunit die at these stages due to an inability to fully synapse homologous chromosomes and complete meiotic recombination. These defects suggest a potent involvement for INO80 during meiosis that is not compensated for by the SWI/SNF complex. Therefore, meiotic recombination during spermatogenesis requires the presence of both the SWI/SNF and INO80 complexes.
MATERIALS AND METHODS
INO80 Conditional Deletion Mouse Model
Mice harboring an INO80 conditional allele were obtained from the EUCOMM site at Institut Clinique de la Souris (IKMC Project ID: 35678). The allele features a floxed sixth exon inserted with a LacZ gene trap cassette upstream (Supplemental Fig. S1; available online at www.biolreprod.org). The gene trap was flanked by Frt sites and removed through breeding to mice containing a constitutive Flpe transgene. Proper targeting of the construct was determined through long-range PCR assay. Primers used for this assay were a gene-specific forward primer 5′-GTGCCATCTTGCCTGACTCCTTAGATTATG and cassette-universal reverse primer 5′-CACAACGGGTTCTTCTGTTAGTCC.
Ino80 floxed and Stra8Cre [21] mice were maintained on C57BL/6Tac and C57BL/6J backgrounds, respectively, and were intercrossed to obtain Ino80Δ/+; Stra8Cre+ animals. Males of this genotype were crossed to Ino80fl/fl females to obtain wild-type and Ino80cKO (see below) animals for this study. Genotyping primers for Ino80 used in this study were: forward 5′-TGGCACCTTTCCAGTCTTTG and reverse 5′-GCTGTGTGTAGTGGTACATA. Stra8Cre-specific genotyping primers were: forward 5′-GTGCAAGCTGAACAACAGGA and reverse 5′-AGGGACACAGCATTGGAGTC. All the animal work was performed in accordance with Institutional Animal Care and Use Committee protocols at the University of North Carolina-Chapel Hill.
RNA Isolation and RT-PCR
Total RNA from whole individual tissues was extracted from an adult wild-type animal using the TRIzol Reagent method (Invitrogen) followed by cleanup using the RNeasy column (Qiagen) and DNase1 (Ambion) treatment. To analyze transcript levels of Ino80 in individual tissues, quantitative RT-PCR was performed using SsoFast EvaGreen Supermix (Biorad) and analyzed on a CFX96 thermal cycler using CFX Manager Software (Biorad). INO80 quantitative PCR primers used for this assay were: forward 5′-GAAGATGGTGGCTGTAAG and reverse 5′-GATGTCCTGCTGATTGAG.
Testis Histology
Whole testes from adult (8 wk old) wild-type and Ino80cKO animals were dissected and fixed in Bouin fixative solution (11-201; Fisher Scientific Ricca Chemical) at 4°C overnight. Tissues were dehydrated through a graded ethanol series containing lithium carbonate to quickly remove the yellow staining. Following paraffin embedding, 7 μm sections were obtained on a Leica RM2165 microtome. Stains including hematoxylin and eosin and periodic acid-Schiff (PAS) were performed by the Animal Histopathology Core at the University of North Carolina at Chapel Hill. Staging of spermatogonial and spermatocyte cells were identified based on morphology as outlined in [22] and [23], respectively.
Tissue and Spermatocyte Spread Preparation
Whole testes from adult (5 or 8 wk old) wild-type and Ino80cKO animals were dissected and sections obtained as previously reported [7]. In short, after dissection, testes were fixed in 4% paraformaldehyde in 1× PBS (46-013-CM; Cellgro) at 4°C overnight. Samples were washed and dehydrated through a sucrose series and embedded in optimal cutting temperature compound (Sakura Rinetek). Tissue sections were obtained at 8 μm thickness on a Leica CM3050S cryostat. Prior to immunofluorescence staining, slides were washed in 1× PBS and permeabilized in 0.1% Triton in 1× PBS.
Alternatively, some testes were dissected and immediately embedded in optimal cutting temperature compound. Following sectioning, slides were washed in 1× PBS and then fixed for 10 min in ice-cold methanol before continuing with immunofluorescence-staining protocols.
Spermatocyte nuclear spreads were prepared as described previously [24]. In short, testes were dissected and the tunica albuginea removed before the tubules were placed in hypotonic extraction buffer (30 mM Tris, pH 8.2, 50 mM sucrose, 17 mM citrate, 5 mM ethylenediaminetetraacetic acid, 0.5 mM dithiothreitol, and 0.1 mM phenylmethanesulfonyl fluoride) for 10 min. Tubules were then minced in 100 mM sucrose solution and spun down to remove large tissue fragments. The supernatant was spread 1:1 with a 1% paraformaldehyde solution on slides and incubated overnight in a humidified chamber. Dried slides were washed in 1× PBS/Photo-flo and stored at −80°C prior to immunofluorescence staining.
Spermatocyte spreads were staged for prophase I of meiosis I by the immunofluorescent-costaining patterns of several conventional meiotic factors. These include monitoring the condensation of the synaptonemal complex proteins 1 and 3 (SCP1 and SCP3), which mark transverse and axial elements, respectively. These patterns were contrasted with the localization of phosphorylated H2AX, a marker of unrepaired DNA breaks. Representative images for the staging of meiocytes can be seen in Figs. 3, A–C and 4, A–C.
Immunofluorescence Staining
Tissue section or nuclear spread slides were blocked in 5% bovine serum albumin for 1–3 h and incubated in predetermined concentration of primary antibody overnight at 4°C. The following day, slides were washed in PBS/Tween-20 and incubated for 1 h in Alexa-conjugated secondary antibody. Slides were mounted in Prolong Gold antifade medium (P-36931; Life Technologies). Antibodies were used as following: rabbit anti-ATR (1:200, PC538; Calbiochem), rabbit anti-BRCA1 (1:200, 07-434; Millipore), rabbit anti-FANCD2 (1:200, 2986-1; Epitomics), rabbit anti-HORMAD1 (1:200, GTX 119236; GeneTex), mouse anti-H2AX (1:800, 05-636; Millipore), rabbit anti-INO80 (1:200, 18810-1-AP; ProteinTech), rabbit anti-INO80 (1:200, 105451; Abcam), mouse anti-MLH1 (1:500, 51-1327GR; BD BioScience), rabbit anti-replication protein A (RPA) (1:100, IHC-00409; Bethyl Lab), rabbit anti-SCP1 (1:500, GTX15087; GeneTex), mouse anti-SCP3 (1:500, ab97672; Abcam), and rabbit anti-SCP3 (1:500, ab15093; Abcam). Secondary antibodies were highly cross-absorbed goat immunoglobulin G conjugated with fluorescent dyes Alexa Fluor 488, 568, or 647 (1:500, Invitrogen). All imaging in this study was completed using a Zeiss AxioImager-M2 (Carl Zeiss).
RESULTS
INO80 Is an Essential Meiotic Factor
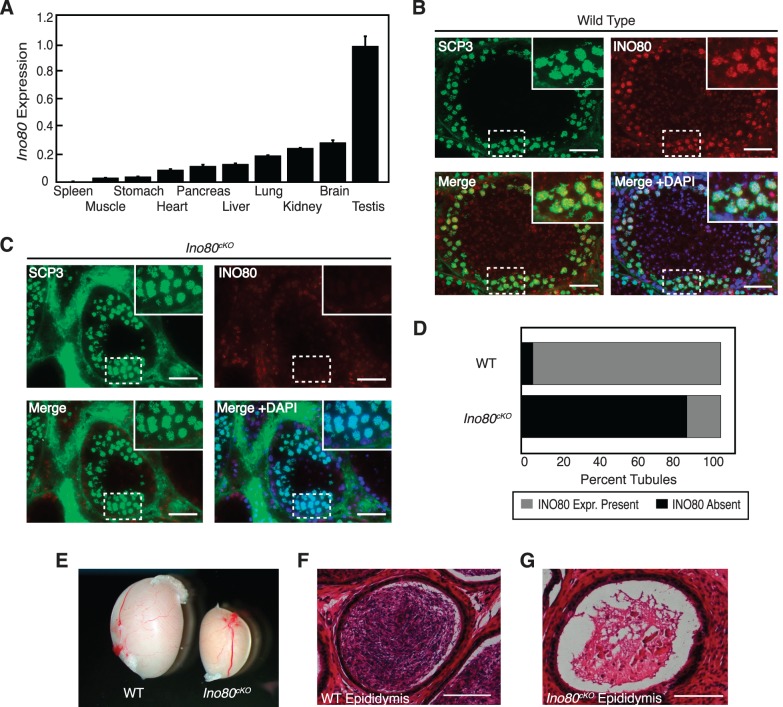
In order to determine a potential in vivo role for the INO80 chromatin-remodeling complex, we analyzed the expression of the core ATPase subunit across a cross section of male adult mouse tissues. INO80 expression was detectable in a majority of these tissues, with the highest expression in the testis (Fig. 1A). Reported roles for INO80 involvement in DNA repair [19, 25–27] made it important to identity the population within the testis that expressed INO80. We detected the presence of INO80 in the spermatocyte population within the seminiferous tubules, colocalizing in developing germ cells with the synaptonemal complex protein SCP3 (Fig. 1B). This stage of meiotic prophase I coincides with timing of the DNA repair associated with the formation of crossovers and recombination.
FIG. 1.
INO80 expression during spermatogenesis. A) RT-PCR analysis of Ino80 mRNA expression from a panel of adult mouse tissues. Error bars: mean ± SEM. B, C) Testis sections showing seminiferous tubules costained for INO80 and SCP3 in wild-type (B) and Ino80cKO (C) animals. Insets show magnified view of the outer edge corresponding to the marked area from the respective section. Bars = 50 μm. D) Quantification of the percentage of seminiferous tubules expressing INO80 protein in wild-type and Ino80cKO animals. Tubules were scored for the presence of INO80 as defined by immunofluorescent staining colocalizing with SCP3 in developing germ cells (grey) or the absence of INO80 signal in the SCP3 population (black). E) Whole mount comparison of testes dissected from wild-type and Ino80cKO 8-wk-old littermate animals. F, G) Hematoxylin and eosin stained histological sections of representative tubules within the cauda epididymis of 8-wk-old wild-type (F) and Ino80cKO (G) littermate animals. Bars = 100 μm.
We obtained a conditional deletion allele for the INO80 core ATPase subunit, which contained a floxed sixth exon. Once exposed to Cre recombinase, the targeted exon was excised, leading to a frameshift and premature stop codon upstream of the previously described Snf2 or helicase functional domains (Supplemental Fig. S1A) [20]. In order to convert this allele into a conditional allele, we utilized FLP-recombinase to excise a gene trap upstream of the floxed exon. Founder animals from this line were tested for correct targeting and orientation of the transgene cassette through a long-range PCR assay. A gene-specific primer binding upstream of the insertion site was paired with a primer internal to the cassette, creating a PCR product if the cassette was inserted in the correct location and orientation. This product was obtained in both male founder animals and was absent in controls, indicating the correct arrangement of the targeting cassette in the conditional allele (Supplemental Fig. S1B).
Initial analysis of the effectiveness of the INO80 conditional allele was performed by intercrossing heterozygous animals carrying the gene trap allele, and no homozygous progeny were obtained from these matings. This was expected based on recent reports showing Ino80 nullizygous animals to be lethal at early embryonic stages [19]. Based on these results, we determined the Ino80 conditional allele to be an effective tool for eliminating the INO80 gene product in specific biological systems.
In order to evaluate the role for INO80 during spermatogenesis, we crossed the Ino80 conditional allele with the Stra8Cre driver [21], ablating Ino80 in the premeiotic spermatogonial population. We confirmed the loss of INO80 by immunofluorescence where spermatocytes expressing the meiotic marker SCP3 costained negatively for INO80, while expression was maintained in cells from the outer portion of the tubule containing both somatic support cells and premeiotic spermatogonia (Fig. 1C). Quantification of fluorescence indicated that exposure to Stra8Cre ablated Ino80 completely in >80% of seminiferous tubules within mutant testes (Fig. 1D).
To determine the effect of ablating INO80 during spermatogenesis, Ino80Δ/fl; Stra8Cre+ (referred to as Ino80cKO) testes were analyzed. These animals had dramatically smaller testes than age-matched control animals (Fig. 1E). In addition, Ino80cKO males were sterile, siring no litters when placed with wild-type CD1 females. The sterility of Ino80cKO animals was consistent with the lack of mature sperm in the epididymis as compared to the dense populations seen in wild-type animals (Fig. 1, F and G).
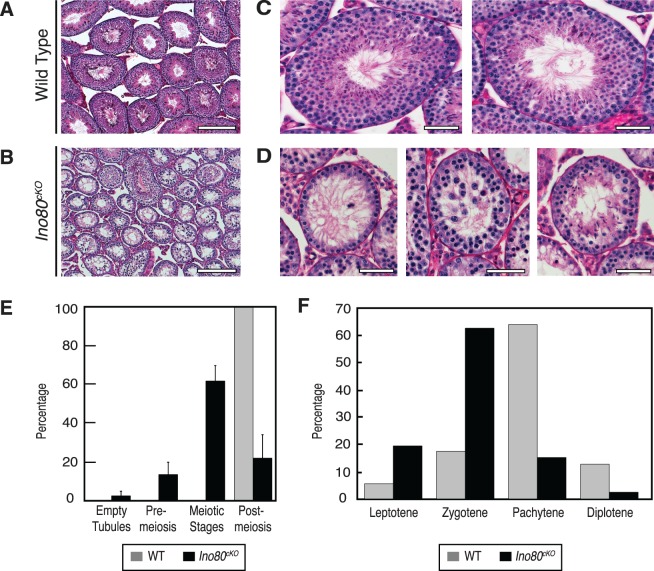
Upon closer inspection, Ino80cKO testes exhibited defects in meiosis. Low magnification images of PAS-stained testis sections taken from 8-wk-old animals demonstrated that the tubules within the Ino80cKO testes were smaller in size and more sparsely populated with developing germ cells (Fig. 2, A and B). When viewed at higher magnifications, the individual tubules showed significant defects in spermatogenesis. In wild-type testes, all tubules contained the full complement of spermatogenic stages, ranging from spermatogonia to elongating spermatids (Fig. 2C). In contrast, the tubules within Ino80cKO testes displayed arrested meiotic progression (Fig. 2D). The majority of tubules (62%) contained a developing germ cell population up to and including meiotic spermatocytes, with no postmeiotic population, indicating a block during meiosis (Fig. 2D, left and center panels, quantified in Fig. 2E). Only 22% of tubules contained later-stage spermatid populations; however, the structure of these tubules appeared compromised (Fig. 2D, right panel, and 2E). This spermatogonial failure and loss of cellularity within the Ino80cKO tubules did not appear to result from a loss of the premeiotic spermatogonial population because <3% of tubules had lost this population leaving empty tubules devoid of germ cells (Fig. 2E). These observations demonstrate that the loss of INO80 during spermatogenesis caused a meiotic defect leading to sterility.
FIG. 2.
Ino80cKO spermatocytes stall in prophase I of meiosis. A, B) Low-magnification PAS-stained histological testis sections from 8-wk-old wild-type (A) and Ino80cKO (B) littermate animals. Bars = 200 μm. C, D) Representative high-magnification PAS-stained tubules from testes sectioned from A and B. Bars = 50 μm. E) Quantification of seminiferous tubules based on the stage of the most advanced cell type in wild-type (grey) and Ino80cKO (black) animals. Error bars: mean ± SD. F) Quantification of the proportion of spermatocytes in individual meiotic prophase I stages from wild-type (grey) and Ino80cKO (black) testes. Staging were determined by SCP1 and SCP3 immunofluorescent staining of spermatocyte spreads.
Synapsis Is Impaired in Ino80cKO Spermatocytes
Given the failure of spermatogenesis in the Ino80cKO mice, we tested how meiotic progression was affected in these animals. The majority of wild-type prophase I meiocytes existed in the pachytene stage (64%). This ratio was skewed in the Ino80cKO spermatocytes (Fig. 2F). Ino80cKO meiocytes displayed a reduced ability to enter the pachytene and diplotene stages (15% and 2%, respectively). The bulk of mutant meiocytes were arrested at the zygotene stage (63%). The presence of spermatocytes that were able to complete meiosis normally was likely due to inefficiencies in Stra8Cre, allowing residual populations to complete critical meiotic functions unperturbed.
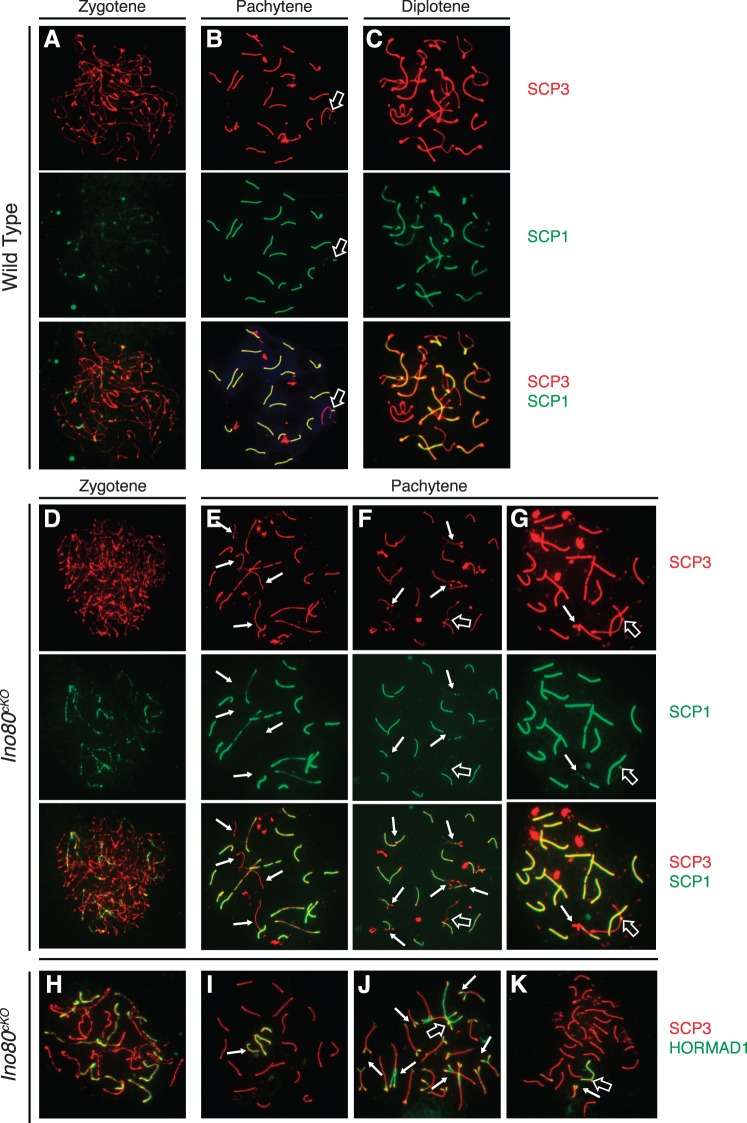
Staging of Ino80cKO spermatocytes with the meiotic markers SCP1 and SCP3 emphasized the meiotic defect. As a cell enters meiosis, the chromosomes begin to condense and SCP3 coats the axial elements. Then as homologous chromosomes synapse, SCP1 forms transverse elements between the individual axes. Therefore, once a spermatocyte enters pachytene, all of the autosomes should be completely coated by both SCP1 and SCP3, and the sex chromosomes should be coated at the pseudoautosomal region [28]. In control populations, this process was completed uniformly, with synapsis completing on all the autosomes (Fig. 3, A–C).
FIG. 3.
Ino80cKO spermatocytes fail to complete synapsis of homologous chromosomes. Comparison of immunofluorsecent staining of SCP3 (lateral elements: red) and SCP1 (transverse elements: green) in wild-type (A–C) and Ino80cKO (D–G) 8-wk-old spermatocyte spreads. H–K) Overlay of SCP3 with HORMAD1 in Ino80cKO spermatocytes. Staging of spermatocytes was determined by the patterns of SCP1 and SCP3 costaining. Open arrows indicate sex chromosomes; closed arrows indicate areas of incomplete synapsis on autosomes.
In contrast Ino80cKO spermatocytes showed aberrations in synapsis. Mutant meiocytes progressed to an abnormal pachytene stage characterized by persistent patterns of asynapsis on either whole or partial portions of chromosomes. These pairing defects were present on the chromosomal axis where SCP3 localized without the presence of transverse element SCP1 (Fig. 3, D–G, closed arrows). While meiosis appeared to initiate properly in Ino80cKO spermatocytes (Fig. 3D), a wide range of synaptic defects quickly became evident. Under normal conditions, the X and Y chromosomes exhibited colocalization of SCP1 and SCP3 only at the pseudoautosomal region (Fig. 3B). Instead, we observed abnormal pachytene spreads where pairing of the sex chromosomes was defective (Fig. 3E). Incomplete autosomal synapsis was apparent on entire chromosomes (Fig. 3, E and F, closed arrows) or restricted to the chromosomal ends (Fig. 3G). The synapsis defects that were observed indicated that INO80 activity was involved in this process. Importantly, it has been shown that defects in synapsis, particularly of the sex chromosomes, are sufficient to activate a pachytene checkpoint, preventing the continued development of defective spermatocytes [29].
Consistent with synaptic defects caused by Ino80 loss, we observed colocalization of SCP3 with HORMAD1 in mutant spermatocytes. HORMAD1 is a meiotic factor that localizes to asynapsed portions of homologous chromosomes in order to prevent HR between sister chromatids [30]. Spermatocytes in prophase I showed localization of HORMAD1 to the unpaired portions of the homologous chromosomes in a pattern opposite to that of SCP1 (Fig. 3, I–K, closed arrows). Together these data suggested that in the absence of INO80 activity, synapsis of homologous chromosomes was impaired.
Repair of DNA Damage in Ino80cKO Spermatocytes Is Defective
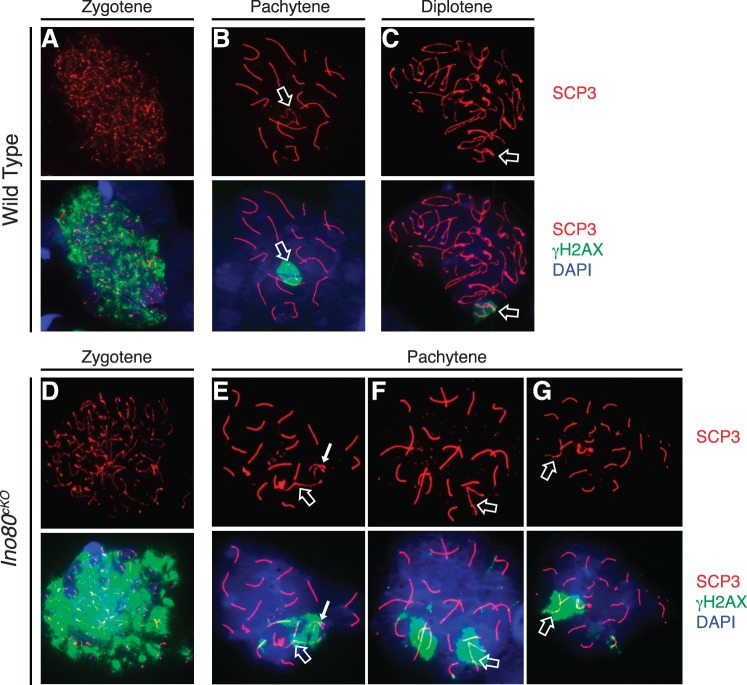
In addition to a defect in synapsis, developing spermatocytes in Ino80cKO animals maintained latent levels of unrepaired DNA breaks. Early in meiosis, SPO11 creates hundreds of DNA double-strand breaks spanning each chromosome [31]. As meiocytes progress through prophase I, the phosphorylated form of histone variant H2AX (γH2AX) marks sites of unrepaired breaks throughout the nucleus, and as DNA repair progresses, it regresses to the sex body [32]. This process occurred normally in wild-type spermatocytes, where a single sex body-specific focus of γH2AX was observed by the pachytene and diplotene stages of prophase I (Fig. 4, A–C). In Ino80cKO spermatocytes, aberrant foci of γH2AX persisted into these later stages (Fig. 4, E–G). We observed latent DNA damage on several of the autosomes regardless of their status of synapsis. As expected, spreads showing extensive asynapsis continued to display hallmarks of DNA damage (Fig. 4E). Alternatively, many Ino80cKO spreads showing complete synapsis continued to have γH2AX localized to one or more of the autosomes (Fig. 4, F and G).
FIG. 4.
Latent unrepaired DNA breaks in Ino80cKO spermatocytes. Distribution of SCP3 (red) with γH2AX (green), a marker of DNA breaks in wild-type (A–C) and Ino80cKO (D–G) spermatocyte spreads. Staging of spermatocytes was determined by the pattern of SCP3 staining. Open arrows indicate the sex chromosomes; closed arrows indicate areas of incomplete synapsis on autosomes.
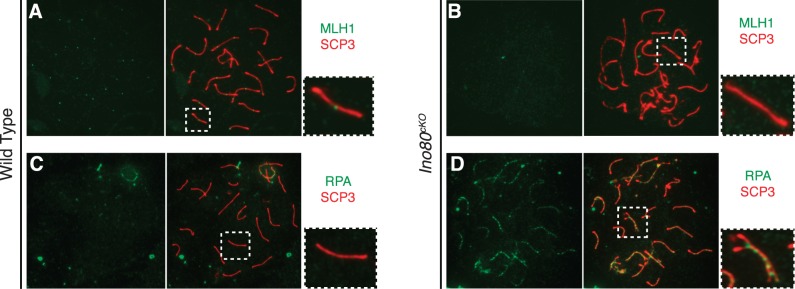
Further analysis of DNA repair factors demonstrated a failure in processing meiotic-associated DNA damage in Ino80cKO spermatocytes. MLH1 is a meiotic factor involved in promoting the repair of DNA breaks into crossovers between the synapsed homologous chromosomes [33]. In wild-type spermatocytes, each pair of homologs formed at least one crossover site, marked by a focus of MLH1 (Fig. 5A). This process was affected in Ino80cKO, where the abnormal pachytene spermatocytes, specifically those harboring synapsis defects on at least one homologous pair, demonstrated a complete absence of MLH1 foci on any of the chromosomes in the nucleus (Fig. 5B). Some spermatocytes displayed normal MLH1 foci, likely corresponding to a population maintaining Ino80 expression. The striking lack of MLH1 foci in the abnormal spermatocyte population is indicative of a failure in the pathway that leads to the formation of meiotic crossovers.
FIG. 5.
Failure to repair DNA damage and form crossovers in Ino80cKO spermatocytes. A, B) Distribution of crossover marker MLH1 (green), costained against SCP3 (red) in pachytene-staged spermatocyte spreads from wild-type (A) and Ino80cKO (B) spermatocytes. C, D) Distribution of ssDNA binding factor RPA in the same populations. Insets show magnified view of a representative autosomal chromosome corresponding to the marked area from the respective spermatocyte spread.
In order to determine the DNA repair pathway affected in Ino80cKO spermatocytes, we surveyed the localization of several DNA repair factors. Single-stranded DNA (ssDNA) binding factor RPA binds ssDNA that is created as a result of DNA damage and is involved in several repair pathways [34]. During meiosis, RPA binds to ssDNA following SPO11-induced DNA damage. Under normal conditions RPA, like γH2AX, localized ubiquitously on the chromosomal axes and regressed to the sex chromosomes as the repair is completed during the pachytene stage (Fig. 5C). However, in many abnormal pachytene-stage spermatocytes from Ino80cKO animals, RPA foci were not resolved and lingered on the axes (Fig. 5D). We observed this pattern on both fully paired chromosomes and those with latent pairing defects, indicating a failure of DNA repair that occurs irrespective of synaptic completion.
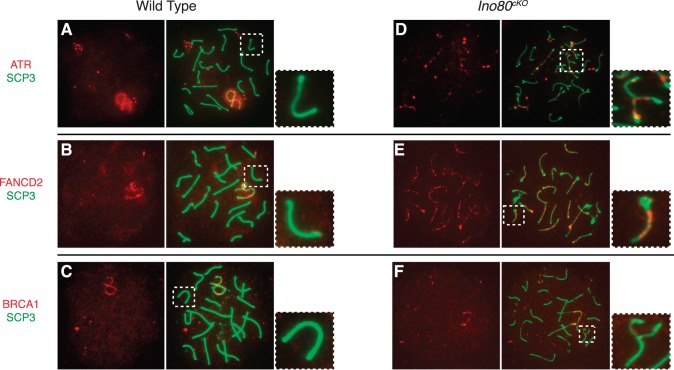
The presence of RPA on the chromosomal axes indicated that early processing events occurred properly, but its continued localization suggested that INO80 might be involved in the completion of later stages of DNA damage repair. To test this hypothesis, we analyzed the expression and localization patterns of ATR, FANCD2, and BRCA1, which are all members of a repair pathway active in HR [35]. ATR senses DNA double-strand breaks and can activate the Fanconi anemia DNA repair pathway [36]. FANCD2 is subsequently ubiquitinated and colocalizes to sites of DNA damage with BRCA1 [37].
We observed an interruption of the Fanconi anemia repair pathway. By the pachytene stage in wild-type spermatocytes, FA-BRCA1 pathway factors were absent from the autosomes, having regressed to the sex chromosomes (Fig. 6, A–C). In Ino80cKO spermatocytes, ATR and FANCD2 remained on the chromosomal axes of both fully and partially synapsed autosomes (Fig. 6, D and E). BRCA1 localized infrequently to asynapsed chromosomes where it would be expected (Fig. 6F). Aberrant localization of repair factors in mutant spermatocytes indicated that INO80 activity was intimately involved in facilitating repair of DNA breaks associated with meiotic recombination.
FIG. 6.
FA-BRCA1 repair pathway factors localize to chromosomal axes. Distribution of factors from the FA-BRCA1 DNA repair pathway in pachytene-stage wild-type (A–C) and Ino80cKO (D–F) spermatocytes. Double staining of SCP3 (green) with repair factors (red), including ATR (A, D), FANCD2 (B, E), and BRCA1 (C, F). Insets show magnified view of a representative autosomal chromosome corresponding to the marked area from the respective spermatocyte spread.
DISCUSSION
Previous studies have determined that SWI/SNF chromatin-remodeling complexes are essential for meiosis [7, 8] and that CHD and ISWI complexes are required for postmeiotic processes [9–13]. In this study we demonstrated that, like the SWI/SNF complex, the INO80 chromatin-remodeling complex is a required participant in meiosis. Ino80cKO spermatocytes were able to initiate the early stages of meiosis properly but began to arrest shortly thereafter. Mutant spermatocytes displayed a striking phenotype resulting from critical defects in DNA repair and synapsis, suggesting that INO80 is critical for the coordination of double-strand break repair and synapsis of homologous chromosomes. These two activities are inextricably linked, with one depending on the other. Disruption of these processes can activate mechanisms that prevent spermatocytes from progressing through meiosis until synapsis is completed [29, 38–40].
Ino80cKO spermatocytes showed severe defects in their ability to complete meiotic recombination. The majority of spermatocytes were arrested at the zygotene stage, while those that made it to later stages displayed disruption of the Fanconi anemia repair pathway, a mechanism of HR. Previous studies in yeast and mammalian cell lines support the involvement of INO80 in both HR, which is involved in forming meiotic crossovers, and nonhomologous end joining pathways [41–43]. In particular, INO80 is involved in preparing the chromatin landscape for HR through the removal of variant histone H2A.Z at sites of DNA damage resulting from irradiation of cells in culture. INO80-mediated depletion of H2A.Z at break sites is crucial for replacement of RPA by RAD51 following resection [44]. Additional support for INO80 involvement in HR is provided by the lack of MLH1 foci in abnormal Ino80cKO spermatocytes, indicating that the specification of crossover sites was disrupted in these cells. Importantly, chromatin architecture contributes to localization of recombination hotspots [45]. Therefore loss of INO80 chromatin-remodeling activity may prevent the establishment of a chromatin structure permissive for crossover formation.
Importantly, the phenotypic consequences of INO80 loss resemble defects caused by experimental manipulation of the SWI/SNF complex. As with INO80, the conditional deletion of the ATPase subunit of the SWI/SNF complex leads to the depletion of spermatocytes during prophase I stages of meiosis, displaying a number of similar defects. These data suggest that these remodelers are unable to compensate for the absence of the other, particularly in their requirement during synapsis and recombination. However, the degree to which these complexes act nonredundantly is unclear. The ablation of both complexes would be required to uncover additional roles that may result in synthetic lethality.
There is precedent for functional interactions between chromatin-remodeling complexes in the regulation of the genome. For example, the yeast ISWI and RSC remodelers act antagonistically to position nucleosomes around the transcription start sites of specific genes. Double mutants for ISWI and RSC suppress the single mutant phenotypes, providing evidence that complexes are in competition [46]. In addition, genomewide localization patterns of ATPase subunits of several remodeling complexes in murine cells suggest that a large portion of genomic targets are shared between complexes [47]. It is unclear whether these patterns represent direct cooperative or antagonistic interactions between complexes at these loci. During spermatogenesis, interactions between remodelers could provide a mechanism for controlling important processes such as recombination. In this context, INO80 and SWI/SNF may be required for the regulation of a distinct set of meiotic functions, leading to similar phenotypic consequences in their absence. On the other hand, the complexes may act on the same targets, but their relationship, whether cooperative or antagonistic, requires the presence of both to complete. Therefore, parsing the relationships between INO80 and SWI/SNF will require further investigation into the mechanistic functions of these complexes during meiosis.
Supplementary Material
ACKNOWLEDGMENT
We would like to thank Andrew Fedoriw, Yuna Kim, Scott Bultman, and the members of the Magnuson laboratory for helpful discussions and comments throughout the duration of this study. We would also like to thank the Mutant Mouse Regional Resource Center at the University of North Carolina at Chapel Hill for their assistance in the acquisition and rederivation of the Ino80 conditional mouse line.
Footnotes
This work was supported by National Institutes of Health (NIH) grants [RO1-HD036655 (T.M.), T32-GM007092 (D.W.S.), and T32-CA071341 (J.S.R.)]. Presented in part at the 2015 Southeastern Regional Society for Developmental Biology Meeting, 11–13 May 2015, Clemson University, South Carolina.
REFERENCES
- Zickler D, Kleckner N. Recombination, pairing, and synapsis of homologs during meiosis. Cold Spring Harb Perspect Biol. 2015;7:a016626. doi: 10.1101/cshperspect.a016626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govin J, Caron C, Lestrat C, Rousseaux S, Khochbin S. The role of histones in chromatin remodelling during mammalian spermiogenesis. Eur J Biochem. 2004;271:3459–3469. doi: 10.1111/j.1432-1033.2004.04266.x. [DOI] [PubMed] [Google Scholar]
- Sasaki H, Matsui Y. Epigenetic events in mammalian germ-cell development: reprogramming and beyond. Nat Rev Genet. 2008;9:129–140. doi: 10.1038/nrg2295. [DOI] [PubMed] [Google Scholar]
- Yodh J. ATP-dependent chromatin remodeling. Adv Exp Med Biol. 2013;767:263–295. doi: 10.1007/978-1-4614-5037-5_13. [DOI] [PubMed] [Google Scholar]
- Lange M, Demajo S, Jain P, Di Croce L. Combinatorial assembly and function of chromatin regulatory complexes. Epigenomics. 2011;3:567–580. doi: 10.2217/epi.11.83. [DOI] [PubMed] [Google Scholar]
- Crichton JH, Playfoot CJ, Adams IR. The role of chromatin modifications in progression through mouse meiotic prophase. J Genet Genomics. 2014;41:97–106. doi: 10.1016/j.jgg.2014.01.003. [DOI] [PubMed] [Google Scholar]
- Kim Y, Fedoriw AM, Magnuson T. An essential role for a mammalian SWI/SNF chromatin-remodeling complex during male meiosis. Development. 2012;139:1133–1140. doi: 10.1242/dev.073478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Gu H, Lin H, Chi T. Essential roles of the chromatin remodeling factor BRG1 in spermatogenesis in mice. Biol Reprod. 2012;86:186. doi: 10.1095/biolreprod.111.097097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowdle JA, Mehta M, Kass EM, Vuong BQ, Inagaki A, Egli D, Jasin M, Keeney S. Mouse BAZ1A (ACF1) is dispensable for double-strand break repair but is essential for averting improper gene expression during spermatogenesis. PLoS Genet. 2013;9:e1003945. doi: 10.1371/journal.pgen.1003945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broering TJ, Wang YL, Pandey RN, Hegde RS, Wang SC, Namekawa SH. BAZ1B is dispensable for H2AX phosphorylation on tyrosine 142 during spermatogenesis. Biol Open. 2015;4:873–884. doi: 10.1242/bio.011734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson PJ, Norton KA, Niri FH, Dawe CE, McDermid HE. CECR2 is involved in spermatogenesis and forms a complex with SNF2H in the testis. J Mol Biol. 2012;415:793–806. doi: 10.1016/j.jmb.2011.11.041. [DOI] [PubMed] [Google Scholar]
- Li W, Wu J, Kim SY, Zhao M, Hearn SA, Zhang MQ, Meistrich ML, Mills AA. Chd5 orchestrates chromatin remodelling during sperm development. Nat Commun. 2014;5:3812. doi: 10.1038/ncomms4812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang T, Hess RA, Kolla V, Higashi M, Raabe TD, Brodeur GM. CHD5 is required for spermiogenesis and chromatin condensation. Mech Dev. 2014;131:35–46. doi: 10.1016/j.mod.2013.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clapier CR, Cairns BR. The biology of chromatin remodeling complexes. Annu Rev Biochem. 2009;78:273–304. doi: 10.1146/annurev.biochem.77.062706.153223. [DOI] [PubMed] [Google Scholar]
- Ebbert R, Birkmann A, Schuller HJ. The product of the SNF2/SWI2 paralogue INO80 of Saccharomyces cerevisiae required for efficient expression of various yeast structural genes is part of a high-molecular-weight protein complex. Mol Microbiol. 1999;32:741–751. doi: 10.1046/j.1365-2958.1999.01390.x. [DOI] [PubMed] [Google Scholar]
- Shen X, Mizuguchi G, Hamiche A, Wu C. A chromatin remodelling complex involved in transcription and DNA processing. Nature. 2000;406:541–544. doi: 10.1038/35020123. [DOI] [PubMed] [Google Scholar]
- van Attikum H, Fritsch O, Gasser SM. Distinct roles for SWR1 and INO80 chromatin remodeling complexes at chromosomal double-strand breaks. EMBO J. 2007;26:4113–4125. doi: 10.1038/sj.emboj.7601835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gospodinov A, Vaissiere T, Krastev DB, Legube G, Anachkova B, Herceg Z. Mammalian Ino80 mediates double-strand break repair through its role in DNA end strand resection. Mol Cell Biol. 2011;31:4735–4745. doi: 10.1128/MCB.06182-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HS, Lee SA, Hur SK, Seo JW, Kwon J. Stabilization and targeting of INO80 to replication forks by BAP1 during normal DNA synthesis. Nat Commun. 2014;5:5128. doi: 10.1038/ncomms6128. [DOI] [PubMed] [Google Scholar]
- Chen L, Cai Y, Jin J, Florens L, Swanson SK, Washburn MP, Conaway JW, Conaway RC. Subunit organization of the human INO80 chromatin remodeling complex: an evolutionarily conserved core complex catalyzes ATP-dependent nucleosome remodeling. J Biol Chem. 2011;286:11283–11289. doi: 10.1074/jbc.M111.222505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadate-Ngatchou PI, Payne CJ, Dearth AT, Braun RE. Cre recombinase activity specific to postnatal, premeiotic male germ cells in transgenic mice. Genesis. 2008;46:738–742. doi: 10.1002/dvg.20437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Rooij DG. Proliferation and differentiation of spermatogonial stem cells. Reproduction. 2001;121:347–354. doi: 10.1530/rep.0.1210347. [DOI] [PubMed] [Google Scholar]
- Meistrich ML, Hess RA. Assessment of spermatogenesis through staging of seminiferous tubules. Methods Mol Biol. 2013;927:299–307. doi: 10.1007/978-1-62703-038-0_27. [DOI] [PubMed] [Google Scholar]
- Peters AH, Plug AW, van Vugt MJ, de Boer P. A drying-down technique for the spreading of mammalian meiocytes from the male and female germline. Chromosom Res. 1997;5:66–68. doi: 10.1023/a:1018445520117. [DOI] [PubMed] [Google Scholar]
- Min JN, Tian Y, Xiao Y, Wu L, Li L, Chang S. The mINO80 chromatin remodeling complex is required for efficient telomere replication and maintenance of genome stability. Cell Res. 2013;23:1396–1413. doi: 10.1038/cr.2013.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Oma Y, Harata M, Kono K, Shima H, Kinomura A, Ikura T, Suzuki H, Mizutani S, Kanaar R, Tashiro S. ATM modulates the loading of recombination proteins onto a chromosomal translocation breakpoint hotspot. PLoS One. 2010;5:e13554. doi: 10.1371/journal.pone.0013554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato D, Waki M, Umezawa M, Aoki Y, Utsugi T, Ohtsu M, Murakami Y. Phosphorylation of human INO80 is involved in DNA damage tolerance. Biochem Biophys Res Commun. 2012;417:433–438. doi: 10.1016/j.bbrc.2011.11.134. [DOI] [PubMed] [Google Scholar]
- Dobson MJ, Pearlman RE, Karaiskakis A, Spyropoulos B, Moens PB. Synaptonemal complex proteins: occurrence, epitope mapping and chromosome disjunction. J Cell Sci. 1994;107(Pt 1):2749–2760. doi: 10.1242/jcs.107.10.2749. [DOI] [PubMed] [Google Scholar]
- Burgoyne PS, Mahadevaiah SK, Turner JM. The consequences of asynapsis for mammalian meiosis. Nat Rev Genet. 2009;10:207–216. doi: 10.1038/nrg2505. [DOI] [PubMed] [Google Scholar]
- Wojtasz L, Daniel K, Roig I, Bolcun-Filas E, Xu H, Boonsanay V, Eckmann CR, Cooke HJ, Jasin M, Keeney S, McKay MJ, Toth A. Mouse HORMAD1 and HORMAD2, two conserved meiotic chromosomal proteins, are depleted from synapsed chromosome axes with the help of TRIP13 AAA-ATPase. PLoS Genet. 2009;5:e1000702. doi: 10.1371/journal.pgen.1000702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeney S, Giroux CN, Kleckner N. Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell. 1997;88:375–384. doi: 10.1016/s0092-8674(00)81876-0. [DOI] [PubMed] [Google Scholar]
- Turner JM, Aprelikova O, Xu X, Wang R, Kim S, Chandramouli GV, Barrett JC, Burgoyne PS, Deng CX. BRCA1, histone H2AX phosphorylation, and male meiotic sex chromosome inactivation. Curr Biol. 2004;14:2135–2142. doi: 10.1016/j.cub.2004.11.032. [DOI] [PubMed] [Google Scholar]
- Moens PB, Marcon E, Shore JS, Kochakpour N, Spyropoulos B. Initiation and resolution of interhomolog connections: crossover and non-crossover sites along mouse synaptonemal complexes. J Cell Sci. 2007;120:1017–1027. doi: 10.1242/jcs.03394. [DOI] [PubMed] [Google Scholar]
- Zou Y, Liu Y, Wu X, Shell SM. Functions of human replication protein A (RPA): from DNA replication to DNA damage and stress responses. J Cell Physiol. 2006;208:267–273. doi: 10.1002/jcp.20622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burdak-Rothkamm S, Rothkamm K, McClelland K, Al Rashid ST, Prise KM. BRCA1, FANCD2 and Chk1 are potential molecular targets for the modulation of a radiation-induced DNA damage response in bystander cells. Cancer Lett. 2015;356:454–461. doi: 10.1016/j.canlet.2014.09.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreassen PR, D'Andrea AD, Taniguchi T. ATR couples FANCD2 monoubiquitination to the DNA-damage response. Genes Dev. 2004;18:1958–1963. doi: 10.1101/gad.1196104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Higuera I, Taniguchi T, Ganesan S, Meyn MS, Timmers C, Hejna J, Grompe M, D'Andrea AD. Interaction of the Fanconi anemia proteins and BRCA1 in a common pathway. Mol Cell. 2001;7:249–262. doi: 10.1016/s1097-2765(01)00173-3. [DOI] [PubMed] [Google Scholar]
- Turner JM. Meiotic sex chromosome inactivation. Development. 2007;134:1823–1831. doi: 10.1242/dev.000018. [DOI] [PubMed] [Google Scholar]
- Mahadevaiah SK, Bourc'his D, de Rooij DG, Bestor TH, Turner JM, Burgoyne PS. Extensive meiotic asynapsis in mice antagonises meiotic silencing of unsynapsed chromatin and consequently disrupts meiotic sex chromosome inactivation. J Cell Biol. 2008;182:263–276. doi: 10.1083/jcb.200710195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian VV, Hochwagen A. The meiotic checkpoint network: step-by-step through meiotic prophase. Cold Spring Harb Perspect Biol. 2014;6:a016675. doi: 10.1101/cshperspect.a016675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukuda T, Fleming AB, Nickoloff JA, Osley MA. Chromatin remodelling at a DNA double-strand break site in Saccharomyces cerevisiae. Nature. 2005;438:379–383. doi: 10.1038/nature04148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison AJ, Highland J, Krogan NJ, Arbel-Eden A, Greenblatt JF, Haber JE, Shen X. INO80 and gamma-H2AX interaction links ATP-dependent chromatin remodeling to DNA damage repair. Cell. 2004;119:767–775. doi: 10.1016/j.cell.2004.11.037. [DOI] [PubMed] [Google Scholar]
- Morrison AJ, Shen X. Chromatin remodelling beyond transcription: the INO80 and SWR1 complexes. Nat Rev Mol Cell Biol. 2009;10:373–384. doi: 10.1038/nrm2693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alatwi HE, Downs JA. Removal of H2A.Z by INO80 promotes homologous recombination EMBO Rep 2015. e201540330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada T, Ohta K. Initiation of meiotic recombination in chromatin structure. J Biochem. 2013;154:107–114. doi: 10.1093/jb/mvt054. [DOI] [PubMed] [Google Scholar]
- Parnell TJ, Schlichter A, Wilson BG, Cairns BR. The chromatin remodelers RSC and ISW1 display functional and chromatin-based promoter antagonism. ELife. 2015;4:e06073. doi: 10.7554/eLife.06073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris SA, Baek S, Sung M-H, John S, Wiench M, Johnson TA, Schiltz RL, Hager GL. Overlapping chromatin-remodeling systems collaborate genome wide at dynamic chromatin transitions. Nat Struct Mol Biol. 2014;21:73–81. doi: 10.1038/nsmb.2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.