Summary
Fluoroquinolone resistance can be conferred through chromosomal mutations or acquisition of plasmids that carry genes such as qnr. In this study 3,201 strains of commensal Escherichia coli were isolated from: 1) 2,374 (74%) strains from humans and chickens from rural northern coastal Ecuador, 2) 827 (26%) strains from chickens from an Ecuadorian industrial poultry operation. We also analyzed 114 fluoroquinolone resistant strains from patients with urinary infection attending three urban hospitals in Quito Ecuador. Isolates were subjected to antibiotic susceptibility screening, and fluoroquinolone resistant isolates (FRIs) were screened for the presence of qnrB genes. We found significantly higher phenotypic resistance to fluoroquinolones in E. coli from chickens in both the rural area (22%) and from the industrial operation (10%) than those from humans in the rural communities (3%). However E. coli isolates from healthy humans in the rural communities had higher rates of qnrB genes (31%, 11 of 35 isolates) compared to chickens from industrial operations (6%, 3 of 81 isolates) and chickens from rural communities (2.8%, 7 of 251 isolates). In addition, human FRIs from urban hospitals had low occurrence of qnrB genes (0.87%, 1 of 114 isolates). These results suggest that the qnrB gene is more widely distributed in rural settings, where antibiotic usage is low, compared to urban hospital and industrial poultry settings. The role of qnrB genes in clinical resistance to fluoroquinolones is still unclear and requires more research.
Keywords: Escherichia coli, qnrB, quinolone resistance
Introduction
Antibiotic resistance is an increasingly serious public health issue. Every year in the United States alone, more than two million people are infected with antibiotic resistant bacteria, resulting in more than 23,000 deaths [3]. Both medical and agricultural usage of antibiotics has been linked to antibiotic resistance in pathogens [6, 7, 15, 16]. Resistance to fluoroquinolones is particularly troubling because they are broad-spectrum antibiotics used to treat serious bacterial infections, especially those acquired in hospitals [2, 3, 5]. Resistance to fluoroquinolones can be caused by mutations in chromosomal genes [2] and by the presence of conjugative or non-conjugative plasmids carrying qnr or other genes [10, 21, 27]. Plasmid-mediated quinolone resistance in human pathogens has been associated with food-producing animals [8, 28], although this claim has been challenged by other studies [20]. Additionally, qnrB genes have been found widely distributed in South American countries [17, 18, 20]. In Ecuador, where there is no restriction on the use of fluoroquinolones in food animals, the prevalence of fluoroquinolone resistance in community-acquired urinary tract E. coli isolates is reported to be 41% [22]. In this study, we assess fluoroquinolone resistance and the presence of qnrB genes in E. coli isolates obtained from fecal samples of chickens and humans in a rural, low antibiotic use setting to fecal samples obtained in two higher antibiotic use settings. Specifically, we compare fluoroquinolone resistant isolates (FRIs) from 1) healthy human individuals from rural communities, 2) chickens (broiler and free range) from rural communities, 3) humans from urban hospitals, and 4) chickens from an industrial poultry operation in Ecuador.
Materials and methods
Samples and bacterial isolates
E. coli was isolated from 1,167 human fecal samples and 1,207 chicken cloacal swabs (from 1,134 chickens) cultured on MacConkey agar. Five lactose-fermenting colonies were selected from each sample and tested for glucuronidase activity on Chromocult agar; glucuronidase positive colonies were subjected to antibiotic susceptibility tests and 1 FRI was selected from each sample. FRIs showed an inhibition zone ≤ to 20 mm using discs with 5 μg ciprofloxacin and Kirby-Bauer antibiotic susceptibility testing, in accordance with the Clinical and Laboratory Standards Institute guidelines.
Isolates from chickens in rural communities
We obtained 1,207 E. coli isolates from chickens in a rural community involved with small-scale poultry farming operations in northwestern Ecuador from January to March 2009. The majority of these isolates (961; 80%) were from broiler chickens raised in small-scale commercial poultry operations. These birds were purchased from a local distributor and fed with commercial poultry feed. Of these 961 isolates from broiler chickens, 831 came from 30 chickens on three farms in one community sampled weekly for six weeks, and 124 came from 25 chickens sampled once cross-sectionally from another farm in the same community. In addition, another 246 (20%) isolates were from household varietals (other breeds of chickens also purchased from a local distributor). Isolates from all chickens in rural communities were labeled RemCHK. In addition, one isolate (Rem6) was isolated from water extracted from a well in the same community where we collected chicken isolates.
Isolates from humans in rural communities
We obtained 1,167 commensal E. coli isolates from healthy humans (controls) participating in a case-control study of diarrheal diseases from 24 communities in northwestern Ecuador between February 2009 to February 2010. Details about the region, study design and sampling strategy were described previously [4]. Human isolates were labeled RemHUM. All interaction with human subjects was approved by the University of Michigan’s Institutional Review Board and Universidad San Francisco de Quito’s bioethics committee.
Isolates from chickens in an industrial operation
We obtained 827 E. coli isolates from an industrial poultry operation (located in the Ecuadorian Coast approximately 300 kilometers from the study region), from broiler chickens sampled between March to November 2010. Isolates from these industrial chickens were labeled IndCHK. These animals were kept in coops and received oxytetracycline in the drinking water (10mg/litre).
Isolates from humans in an urban hospitals
We obtained 114 clinical FRI E. coli from patients with urinary infections collected in three hospitals in urban Quito from May to July 2010. Of these isolates, 61 were from Hospital Vozandes (kindly provided by Jeannette Zurita), 42 were from Hospital Carlos Andrade Marín (kindly provided by Isabel Narváez), and 11 were from the Institute of Microbiology at Universidad San Francisco de Quito. These clinical human isolates were labeled E. coli Quito.
Polymerase chain reaction amplification and DNA sequence analysis
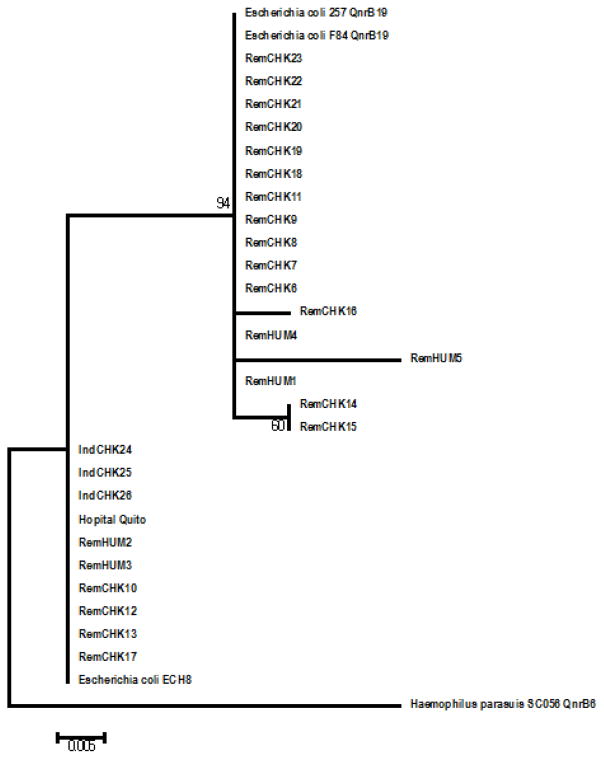
Bacterial DNA from a single FRI colony was released from cells using a boiling technique [25]. A PCR reaction for qnrB genes was carried out following [8], using primers to amplify internal fragments of the target genes: qnrB F 5’-GGMATHGAAATTCGCCACTG-3’ and qnrB R 5′-TTTGCYGYYCGCCAGTCGAA-3′. PCR conditions were as follows: 95 °C for 5 min, 35 cycles of 94 °C for 30 sec, 56 °C for 40 sec and 72 °C for 1 min, and a final incubation at 72 °C for 10 min. Amplicons were sequenced by the University of Michigan DNA Sequencing Core. The qnrB gene sequences described in this report comprise accession numbers JN714812 to JN714838 (Table 2). A subset of these amplicons (IndCHK24, IndCHK25, IndCHK26 and E. coli Quito Hospital) were sequenced a second time at Functional Biosciences, Inc, Madison, WI to rule out mistakes and they were used for phylogenetic analysis using Mega5.1 program (Figure 1). Two qnrB sequences from broiler chicken FRIs came from the same animal, from two colonies collected two weeks apart (RemCHKb7 y RemCHKb9).
Table 2.
Nucleotide sequences of qnrB genes from fluoroquinolone resistant isolates (FRIs) from humans and chickens inhabiting rural communities in the northern coast of Ecuador, FRIs from chicken industrial operation and a FRI from an urban hospital in Quito.
| Amplicon | Gen Bank Accession number |
|---|---|
| RemHUM1 | JN714812 |
| RemHUM2 | JN714813 |
| RemHUM3 | JN714814 |
| RemHUM4 | JN714815 |
| RemHUM5 | JN714816 |
| RemCHK6 | JN714817 |
| RemCHK7 | JN714818 |
| RemCHK8 | JN714819 |
| RemCHK9 | JN714820 |
| RemCHK10 | JN714821 |
| RemCHK11 | JN714822 |
| RemCHK12 | JN714823 |
| RemCHK13 | JN714824 |
| RemCHK14 | JN714825 |
| RemCHK15 | JN714826 |
| RemCHK17 | JN714828 |
| RemCHK18 | JN714829 |
| RemCHK19 | JN714830 |
| RemCHK20 | JN714831 |
| RemCHK21 | JN714832 |
| RemCHK22 | JN714833 |
| RemCHK23 | JN714834 |
| IndCHK24 | JN714835 |
| IndCHK25 | JN714836 |
| IndCHK26 | JN714837 |
| Quito Hospital | JN714838 |
Fig. 1.
Maximum likelihood analysis of qnrB genes: Bacterial sequences obtained from humans in Ecuadorian rural communities are named RemHUM; sequences from chickens in rural communities are RemCHK; qnrB from hospital isolates in Quito: E. coli Quito Hospital hospital and sequences from isolates from a poultry industrial operation are IndCHK. The rest of the sequences were obtained from GenBank: E. coli strain F84, accession number KM094204.1; E. coli strain F257 accession number KM094205.1; E. coli strain ECH8, accession number KP268825.1; H parasuis strain SC056, accession number HQ117877.1. Numbers are bootstrap values obtained after 500 pseudoreplicates. Asterisk indicates sequences analysed in this paper.
Statistical analysis
A χ2 test was used to analyze the differences in fluoroquinolone resistance and qnrB gene frequency between the sample types.
Results and Discussion
Rates of resistance
Significantly higher rates of resistance to fluoroquinolones (p<0.001) were observed in isolates from chickens in rural communities (broilers 214 of 961 isolates, 22.3% and household varietals: 37 of 246 isolates, 15%) when compared with isolates from chickens in industrial operations (81of 827 isolates, 9.8%) or with human isolates from rural communities (35 of 1,167 isolates, 3%).
Presence of qnrB gene in fluoroquinolone-resistant isolates
There was a substantial difference in the percentage of human FRIs carrying qnrB genes depending on the origin of the isolates: 31% (11 of 35 isolates) of human FRIs from rural communities carried the qnrB gene versus 0.87% (1 of 114 isolates) of FRIs from hospitals in Quito (p<0.0001) (Table 1). Differences were less evident in chicken isolates: 1.87% (4 of 214 isolates) of FRIs from broiler chickens in rural communities and 8% (3 of 37 isolates) from household varietals carried the qnrB gene, versus 6% (3 of 81 isolates) in FRIs from broiler chickens from the industrial operation (p=0.4). No significant differences were observed between FRIs from broiler chickens and household varietals in the rural community (p = 0.069).
Table 1.
Fluoroquinolone resistant isolates FRIs of E. coli with and without qnrB. Five E. coli isolates were analyzed from each fecal sample from either humans or chickens inhabiting rural communities in the northern coast of Ecuador.
| Source of isolates | Number of isolates | FRIs % | FRIs with qnrB % | p-value |
|---|---|---|---|---|
| Human isolates from rural area | 1,167 | 35 3.0% | 11 31% | |
| Human isolates from urban hospitals | ND** | 114 | 1 0.87% | p<0.0001 |
| Broiler chicken isolates from rural area | 961 | 214 22.3% | 4 1.87 % | |
| Household varietals from rural area | 246 | 37 15% | 3 8.1% | |
| Broiler chicken isolates from an industrial poultry operation | 827 | 81 9.8% | 3 6.0%*** |
ND= not determined;
Only 50 isolates were analyzed for qnrB genes. P value obtained from comparison of human isolates from the rural community and the hospital in Quito
Phylogenetic analysis of qnrB genes
Nucleotide sequences of all the amplicons showed high similarity to previously described qnrB genes Figure 1.
Conclusions
Based on the published literature [1] and the high prevalence of fluoroquinolone resistance reported in Ecuador [22], we expected to encounter a greater presence of qnrB genes in pathogenic FRIs from hospitals in Quito where antibiotic use is high, compared to rural settings where antibiotic use is low. On the contrary, where we found a low overall prevalence of FRI, in commensal E. coli isolated from humans in rural communities 3%, a high proportion of these carried qnrB genes 31%. Where we found a high prevalence of FRIs, chickens from these rural communities, a low proportion of these carried qnrB genes 2.7%. The lowest proportion of qnrB gene carriage was found in FRI from urban hospitals in Quito 0.9%, where antibiotic use is high. Although qnr genes cause low-level resistance to quinolones [9], they are thought to be important in the development of the highly resistant phenotype [14]. Therefore it was surprising to find a lack of association of the qnrB gene with clinical fluoroquinolone resistance in this study.
Other studies have found that qnrB genes are widely distributed in commensal E. coli isolated from healthy humans including children in urban settings in Peru and Bolivia [17, 18], humans in remote Peruvian Amazon communities [19], and from domestic and farm animals in Germany [23]. We found a high rate of qnrB carriage in healthy humans from rural, but not urban areas of Ecuador. Despite high rates of fluoroquinolone resistance in chickens, we did not find high rates of qnrB carriage in the FRIs from these animals.
Although our findings may suggest that qnrB genes are not linked to fluoroquinolone clinical resistance, they have been associated with the development high resistance to quinolones [14, 24] and qnrB genes are one of the most common plasmid-mediated quinolone resistance genes [13]. It is also a concern the presence of qnrB genes in isolates from domestic animals as these genes have been found in food pathogens, such as Salmonella [11]. Additional research about the role of qnr genes in fluoroquinolone resistance is necessary. The main limitation of this study was the segment of DNA amplified which did not allow us to determine the diversity of qnr genes detected.
Acknowledgments
The authors would like to thank the Ecologia, Desarrollo, Salud, y Sociedad EcoDSS field team for their invaluable contribution collecting the data, especially Nadia Lopez. This study was supported by grant numbers R01-AI050038 and K01-AI103544 from the National Institute of Allergy and Infectious Diseases NIAID, and grant number 0811934 from the Ecology of Infectious Diseases program, Fogarty International Centre FIC of the National Institutes of Health NIH and the National Science Foundation NSF.
Footnotes
Competing interests
The authors declare that they have no competing interest
References
- 1.Anssour L, Messai Y, Derkaoui M, Alouache S, Estepa V, Somalo S, Torres C, Bakour R. ESBL, plasmidic AmpC, and associated quinolone resistance determinants in coliforms isolated from hospital effluent: first report of qnrB2, qnrB9, qnrB19, and bla CMY-4 in Algeria. J Chemother. 2014;26:74–79. doi: 10.1179/1973947813Y.0000000115. [DOI] [PubMed] [Google Scholar]
- 2.Cavaco LM, Aarestrup FM. Evaluation of quinolones for use in detection of determinants of acquired quinolone resistance, including the new transmissible resistance mechanisms qnrA, qnrB, qnrS, and aac(6′)Ib-cr, in Escherichia coli and Salmonella enterica and determinations of wild-type distributions. J Clin Microbiol. 2009;47:2751–2758. doi: 10.1128/JCM.00456-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Centers for Disease Control and Prevention. Antibiotic resistance threats in the United States. 2013 http://www.cdc.gov/drugresistance/threatreport-2013.
- 4.Eisenberg JN, Cevallos W, Ponce K, Levy K, Bates SJ, Scott JC, Hubbard A, Vieira N, Endara P, Espinel M, Trueba G, Riley LW, Trostle J. Environmental change and infectious disease: how new roads affect the transmission of diarrheal pathogens in rural Ecuador. Proc Natl Acad Sci U S A. 2006;103:19460–19465. doi: 10.1073/pnas.0609431104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goossens H, Ferech M, Vander Stichele R, Elseviers M. Outpatient antibiotic use in Europe and association with resistance: a cross-national database study. Lancet. 2005;365:579–587. doi: 10.1016/S0140-6736(05)17907-0. [DOI] [PubMed] [Google Scholar]
- 6.Harada K, Asai T. Role of antimicrobial selective pressure and secondary factors on antimicrobial resistance prevalence in Escherichia coli from food-producing animals in Japan. J Biomed Biotechnol. 2010;2010:180682. doi: 10.1155/2010/180682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hawkey PM, Jones AM. The changing epidemiology of resistance. J Antimicrob Chemother. 2009;64:13–20. doi: 10.1093/jac/dkp256. [DOI] [PubMed] [Google Scholar]
- 8.Huang SY, Dai L, Xia LN, Du XD, Qi YH, Liu HB, Wu CM, Shen JZ. Increased prevalence of plasmid-mediated quinolone resistance determinants in chicken Escherichia coli isolates from 2001 to 2007. Foodborne Pathog Dis. 2009;6:1203–1209. doi: 10.1089/fpd.2009.0348. [DOI] [PubMed] [Google Scholar]
- 9.Jacoby GA, Walsh KE, Mills DM, Walker VJ, Oh H, Robicsek A, Hooper DC. qnrB, another plasmid-mediated gene for quinolone resistance. Antimicrob Agents Chemother. 2006;50:1178–1182. doi: 10.1128/AAC.50.4.1178-1182.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jacoby GA, Strahilevitz J, Hooper DC. Plasmid-mediated quinolone resistance. Microbiol Spectr. 2014;2(2) doi: 10.1128/microbiolspec.PLAS-0006-2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Karczmarczyk M, Martins M, McCusker M, Mattar S, Amaral L, Leonard N, Aarestrup FM, Fanning S. Characterization of antimicrobial resistance in Salmonella enterica food and animal isolates from Colombia: identification of a qnrB19-mediated quinolone resistance marker in two novel serovars. FEMS Microbiol Lett. 2010;313:10–19. doi: 10.1111/j.1574-6968.2010.02119.x. [DOI] [PubMed] [Google Scholar]
- 12.Kim HB, Park CH, Kim CJ, Kim EC, Jacoby GA, Hooper DC. Prevalence of plasmid-mediated quinolone resistance determinants over a 9-year period. Antimicrob Agents Chemother. 2009;53:639–645. doi: 10.1128/AAC.01051-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ma J, Zeng Z, Chen Z, Xu X, Wang X, Deng Y, Lü D, Huang L, Zhang Y, Liu J, Wang M. High prevalence of plasmid-mediated quinolone resistance determinants qnr, aac(6′)-Ib-cr, and qepA among ceftiofur- resistant Enterobacteriaceae isolates from companion and food-producing animals. Antimicrob Agents Chemother. 2009;53:519–524. doi: 10.1128/AAC.00886-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Martínez-Martínez L, Pascual A, Jacoby GA. Quinolone resistance from a transferable plasmid. Lancet. 1998;351:797–799. doi: 10.1016/S0140-6736(97)07322-4. [DOI] [PubMed] [Google Scholar]
- 15.Mathew AG, Cissell R, Liamthong S. Antibiotic resistance in bacteria associated with food animals: A United States perspective of livestock production. Foodborne Pathog Dis. 2007;4:115–133. doi: 10.1089/fpd.2006.0066. [DOI] [PubMed] [Google Scholar]
- 16.McGowan JE. Antimicrobial resistance in hospital organisms and its relation to antibiotic use. Rev Infect Dis. 1983;5:1033–1048. doi: 10.1093/clinids/5.6.1033. [DOI] [PubMed] [Google Scholar]
- 17.Pallecchi L, Riccobono E, Mantella A, Bartalesi F, Sennati S, Gamboa H, Gotuzzo E, Bartoloni A, Rossolini GM. High prevalence of qnr genes in commensal enterobacteria from healthy children in Peru and Bolivia. Antimicrob Agents Chemother. 2009;53:2632–2635. doi: 10.1128/AAC.01722-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pallecchi L, Riccobono E, Mantella A, Fernandez C, Bartalesi F, Rodriguez H, Gotuzzo E, Bartoloni A, Rossolini GM. Small qnrB-harbouring ColE-like plasmids widespread in commensal enterobacteria from a remote Amazonas population not exposed to antibiotics. Antimicrob Chemother. 2011;66:1176–1178. doi: 10.1093/jac/dkr026. [DOI] [PubMed] [Google Scholar]
- 19.Pallecchi L, Riccobono E, Sennati S, Mantella A, Bartalesi F, Trigoso C, Gotuzzo E, Bartoloni A, Rossolini GM. Characterization of small ColE-like plasmids mediating widespread dissemination of the qnrB19 gene in commensal enterobacteria. Antimicrob Agents Chemother. 2010;54:678–682. doi: 10.1128/AAC.01160-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Riccobono E, Pallecchi L, Mantella A, Bartalesi F, Chavez I, Trigoso C, Villagran AL, Bartoloni A, Rossolini GM. Carriage of antibiotic-resistant Escherichia coli among healthy children and home-raised chickens: a household study in a resource-limited setting. Microb Drug Resist. 2012;18:83–87. doi: 10.1089/mdr.2011.0003. [DOI] [PubMed] [Google Scholar]
- 21.Robicsek A, Jacoby GA, Hooper DC. The worldwide emergence of plasmid-mediated quinolone resistance. Lancet Infect Dis. 2006;6:629–640. doi: 10.1016/S1473-3099(06)70599-0. [DOI] [PubMed] [Google Scholar]
- 22.Salles MJ, Zurita J, Mejía C, Villegas MV. Resistant Gram-negative infections in the outpatient setting in Latin America. Epidemiol Infect. 2013;141:2459–2472. doi: 10.1017/S095026881300191X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schink AK, Kadlec K, Schwarz S. Detection of qnr genes among Escherichia coli isolates of animal origin and complete sequence of the conjugative qnrB19-carrying plasmid pQNR2078. J Antimicrob Chemother. 2012;67:1099–1102. doi: 10.1093/jac/dks024. [DOI] [PubMed] [Google Scholar]
- 24.Strahilevitz J, Jacoby GA, Hooper DC, Robicsek A. Plasmid-mediated quinolone resistance: a multifaceted threat. Clin Microbiol Rev. 2009;22:664–689. doi: 10.1128/CMR.00016-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vieira N, Bates SJ, Solberg OD, Ponce K, Howsmon R, Cevallos W, Trueba G, Riley L, Eisenberg JN. High prevalence of enteroinvasive Escherichia coli isolated in a remote region of northern coastal Ecuador. Am J Trop Med Hyg. 2007;76:528–533. [PMC free article] [PubMed] [Google Scholar]
- 26.Wang M, Guo Q, Xu X, Wang X, Ye X, Wu S, Hooper DC, Wang M. New plasmid-mediated quinolone resistance gene, qnrC1, found in a clinical isolate of Proteus mirabilis. Antimicrob Agents Chemother. 2009;53:1892–1897. doi: 10.1128/AAC.01400-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu JJ, Ko WC, Tsai SH, Yan JJ. Prevalence of plasmid-mediated quinolone resistance determinants QnrA, QnrB, and QnrS among clinical isolates of Enterobacter cloacae in a Taiwanese hospital. Antimicrob Agents Chemother. 2007;51:1223–1227. doi: 10.1128/AAC.01195-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yue L, Jiang HX, Liao XP, Liu JH, Li SJ, Chen XY, Chen CX, Lü DH, Liu YH. Prevalence of plasmid-mediated quinolone resistance qnr genes in poultry and swine clinical isolates of Escherichia coli. Vet Microbiol. 2008;132:414–420. doi: 10.1016/j.vetmic.2008.05.009. [DOI] [PubMed] [Google Scholar]