Abstract
In mouse mammary epithelial cells, CPEB1 mediates the apical localization of ZO-1 mRNA, which encodes a critical tight junction component. In mice lacking CPEB1 and in cultured cells from which CPEB has been depleted, randomly distributed ZO-1 mRNA leads to the loss of cell polarity. We have investigated whether this diminution of polarity results in an epithelial-to-mesenchyme (EMT) transition and possible increased metastatic potential. Here, we show that CPEB1-depleted mammary epithelial cells alter their gene expression profile in a manner consistent with an EMT and also become motile, which are made particularly robust when cells are treated with TGF-β, an enhancer of EMT. CPEB1-depleted mammary cells become metastatic to the lung following injection into mouse fat pads while ectopically-expressed CPEB1 prevents metastasis. Surprisingly, CPEB1 depletion causes some EMT/metastasis-related mRNAs to have shorter poly(A) tails while other mRNAs to have longer poly(A) tails. Matrix metalloproteinase 9 (MMP9) mRNA, which encodes a metastasis-promoting factor, undergoes poly(A) lengthening and enhanced translation upon CPEB reduction. Moreover, in human breast cancer cells that become progressively more metastatic, CPEB1 is reduced while MMP9 becomes more abundant. These data suggest that at least in part, CPEB1 regulation of MMP9 mRNA expression mediates metastasis of breast cancer cells.
Keywords: CPEB1, metastasis, polyadenylation, mRNA translation
Introduction
The Cytoplasmic Polyadenylation Element Binding Protein 1 (CPEB1) is a sequence-specific RNA binding protein that regulates mRNA translation by dynamically controlling poly(A) tail length. CPEB1 anchors the non-canonical poly(A) polymerases Gld2 or Gld4, as well as the deadenylating enzyme PARN (poly(A) ribonuclease), to mRNA 3′ untranslated regions (UTRs) that contain a cytoplasmic polyadenylation element (CPE). By doing so and in response to phosphorylation/dephoshorylation events on serine 174 or threonine 171 (species-dependent), CPEB1 regulates poly(A) tail growth or removal, which consequently promotes or represses translation, respectively.1–4 The biological manifestations of CPEB1 regulation include control of gametogenesis, cell cycle progression, cellular senescence, glucose homeostasis, inflammation, and higher cognitive function and neurologic disease.1,4–11
In addition to mediating the levels of specific protein synthesis, CPEB1 also coordinates where mRNAs are translated by regulating their subcellular localization. In oocytes, for example, CPE-containing RNAs are localized to the centrosome and mitotic spindles by CPEB1, where they are translated into proteins that promote cell division.12–14 In neurons, CPEB1 is involved in RNA transport into dendrites where they are translated in synapto-dendritic compartments in response to synaptic stimulation.15–19 In mouse mammary epithelial cells, CPEB1-regulated RNA localization is important for cell polarity. This phenomenon was first detected in CPEB1-deficient mice where mammary epithelial cells exhibit disrupted polarity as indicated by impaired co-localization of the tight junction proteins ZO-1 and apical protein syntaxin-3 and increased improper co-localization of ZO-1 and the baso-lateral protein E-cadherin.20 In cultured mouse EpH4 mouse mammary cells, epithelial sheets are formed with ZO-1 localized apically and E-cadherin localized baso-laterally; when CPEB1 is depleted, this two-dimensional polarity is lost and ZO-1 is aberrantly distributed baso-laterally. Three-dimensional cultures of EpH4 cells, which when grown in an anchorage-independent manner form hollow lumen much like mammary gland terminal end buds (TEBs),21 also lose their polarity following CPEB1 depletion and do not contain a hollow cavity. Under normal conditions, CPEB1 and the CPE-containing ZO-1 mRNA are co-localized apically where translation takes place. When CPEB1 is depleted, ZO-1 mRNA translation occurs without spatial specification, thereby resulting in loss of cell polarity.20
Epithelial cells that lose their strong apposition and adhesiveness through tight junctions can experience an epithelial-to-mesenchyme transition (EMT), a state of dedifferentiation often linked to increased metastatic potential.22,23 EMTs are characterized by altered morphology of cells from, for example, planar epithelial sheets to loosely dispersed cells with a spindle-shaped mesenchymal phenotype that is conducive to cell migration. A number of molecular markers are used to define an EMT, most prominently the down-regulation of E-cadherin and the up-regulation of N-cadherin, fibronectin, vimentin, twist, snail, slug and others.23,24 Cells that have experienced an EMT can colonize distant tissues via the bloodstream; if such cells are derived from a malignant tumor, the colonization represents the metastatic spread of cancer.23,25
Because CPEB1 depletion results in the loss of apical-basal polarity of mammary epithelial cells, we suspected it might presage an EMT and as result possibly enhance their metastatic potential. Indeed, CPEB1 depletion from mouse mammary epithelial NMuMG cells, when combined with administration of TGF-β, an enhancer of EMT,26 causes molecular alterations that are consistent with EMT. CPEB1-depleted cells also become more motile, another trait that characterizes EMT. Mouse mammary 4T1 epithelial cells, which are highly metastatic, express little detectable CPEB1. However, ectopic expression of CPEB1 in these cells significantly reduces metastasis to the lung following injection into the fat pads of mice. Conversely, MCF7 human mammary epithelial cells are not metastatic and express high levels of CPEB1. When depleted of CPEB1, they become metastatic to the lung. An analysis of EMT-related RNAs that undergo changes in poly(A) tail size in mouse mammary cells following CPEB1 depletion revealed several that had reduced poly(A) and very surprisingly, others that gained poly(A). One mRNA that gained poly(A) and whose translation was elevated following CPEB1 reduction encodes matrix metalloproteinase 9 (MMP9), a metastasis-stimulatory enzyme whose levels are inverse to that of CPEB1 in human breast metastases. These data suggest that depletion of CPEB1 not only causes loss of mammary epithelial cell polarity, but that it also leads to EMT and increased metastasis possibly by altering the translational efficiency of MMP9 mRNA.
Results
CPEB1 Regulates Cell Motility
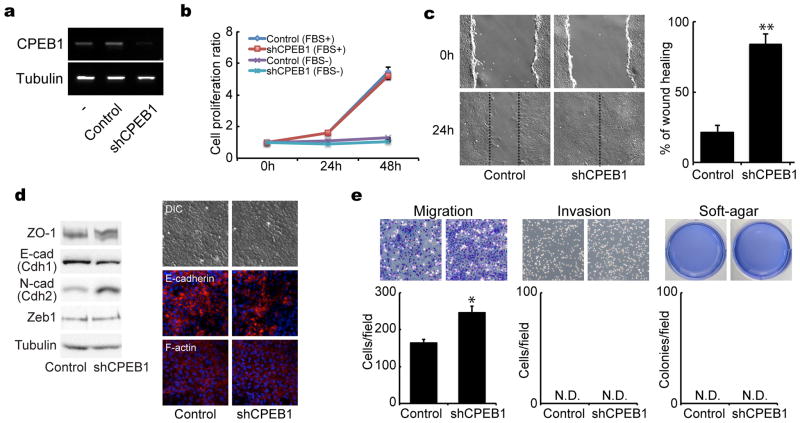
To determine whether reduction of CPEB1 results in characteristics that resemble EMT or tumorigenesis, NMuMG mouse mammary epithelial cells were infected with lentivirus expressing an shRNA for CPEB1, which we have previously shown specifically depletes CPEB1.20 Although the shRNA effectively reduced CPEB1, it had no effect on cell proliferation irrespective of whether growth-promoting fetal bovine serum (FBS) was added to the culture medium (Figure 1a and b). In a cell motility assay a score was made through confluent cells and the rate at which the border cells infiltrated the scratch was determined. Figure 1c shows that depletion of CPEB1 stimulated the rate of repopulation of the score compared to control cells in this 2 dimensional migration assay. CPEB1 depletion also elicited an increase in N-cadherin (Cdh2) levels and a decrease in E-cadherin levels (Cdh1), which are consistent with induction of an EMT (Figure 1d, western blots and immunocytochemistry). However, Zeb1, another EMT marker was unaffected by CPEB1 knockdown. CPEB1 depletion also enhanced cell migration in a Transwell assay, but had no effect on cell invasiveness in matrigel or a tumorigenesis-related phenotype based on growth in soft agar (Figure 1e). These data indicate that CPEB1 controls cell motility and is likely to be involved in EMT, but probably does not influence oncogenic transformation.
Figure 1.
CPEB1 regulates migration of mammary epithelial cells. (a) NMuMG cells were infected with lentiviruses expressing GFP only (control) or GFP plus shRNA against CPEB1 (shCPEB1-1). The relative levels of CPEB1 were determined by RT-PCR; tubulin RNA served as a control. (b) Proliferation rate of cells infected with control of shCPEB1 in the absence or presence of FBS. (c) Control and CPEB1-depleted cells were analyzed for cell motility in a wound-healing scratch assay. The dotted lines define the areas lacking cells. The histogram displays the ratio of wound healing from time 0h to time 24h by measuring the area of each scratch closure. (d) NMuMG cells infected with control or CPEB1 knockdown virus were western blotted for ZO-1, E-cadherin, N-cadherin, Zeb1, and tubulin, and immunostained for E-cadherin and F-actin. (e) Migration, invasion, and soft-agar colony formation assays following CPEB1 knockdown. In this and all subsequent figures, one asterisk refers to p<0.05 and two asterisks refer to p<0.01 (two-sided Tukey’s multiple range test). All experiments were performed three times (biologic replicates). All histograms in all figures refer to mean +/− sem. Refer to Materials and Methods for further details. N.D. refers to not detected.
TGF-β Potentiates CPEB1-Mediation of EMT
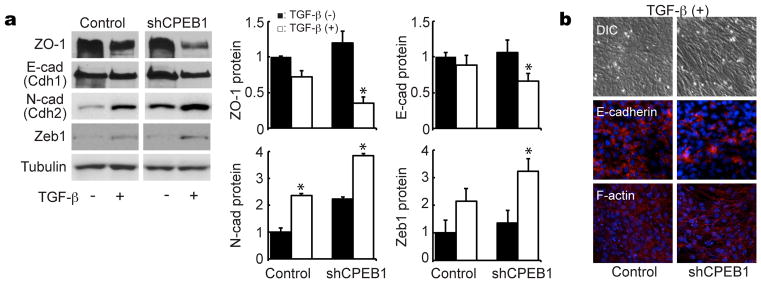
Although the loss of CPEB1 in mouse mammary epithelial cells prevented the establishment of apical-basal polarity,20 it did not obviously induce an EMT, at least based on cell morphology and notwithstanding its stimulation of cell migration, it did not stimulate cell invasiveness (Figure 1). Because TGF-β potentiates EMT,26 we surmised that simultaneous treatment of NMuMG cells with CPEB1 shRNA and this growth factor might induce EMT (TGF-β treatment reduces CPEB mRNA, Supplementary Figure S1a). Consequently, we determined whether TGF-β enhances the expression of EMT markers in the presence or absence of CPEB1. TGF-β treatment elicited a more profound decrease of both ZO-1 and E-cadherin when CPEB1 was depleted. Conversely, this same regimen evoked enhanced expression of N-cadherin as well as Zeb1 (Figure 2a), all of which are consistent with the induction of EMT. Immunocytochemical analysis also showed reduction of E-cadherin and fiber structure of F-actin upon TGF-β treatment and CPEB1 depletion (Figure 2b). To gain a greater sense of the degree of EMT induction as noted above, RNA from control or CPEB1 knockdown cells treated with TGF-β were use to screen a microarray specific for EMT marker sequences. Supplementary Figure S2 shows that in a manner consistent with an EMT, many markers were up or down-regulated in CPEB1 knockdown cells. Similar to protein levels, RNAs that encode N-cadherin (Cdh2) and Zeb1 increased, conversely, E-cadherin (Cdh1) decreased. Taken together, these data indicate that TGF-β enhances EMT when CPEB1 is depleted.
Figure 2.
CPEB1-depletion enhances EMT induction by TGF-β. (a) NMuMG cells infected with a control or CPEB1 shRNA-expressing virus were treated with TGF-β and western blotted for ZO-1, E-cadherin, N-cadherin, Zeb1, and tubulin. (b) NMuMG cells infected with control or CPEB1 knockdown virus were treated with TGF-β and immunostained for E-cadherin and F-actin.
CPEB1 Regulates Mammary Epithelial Cell Metastasis
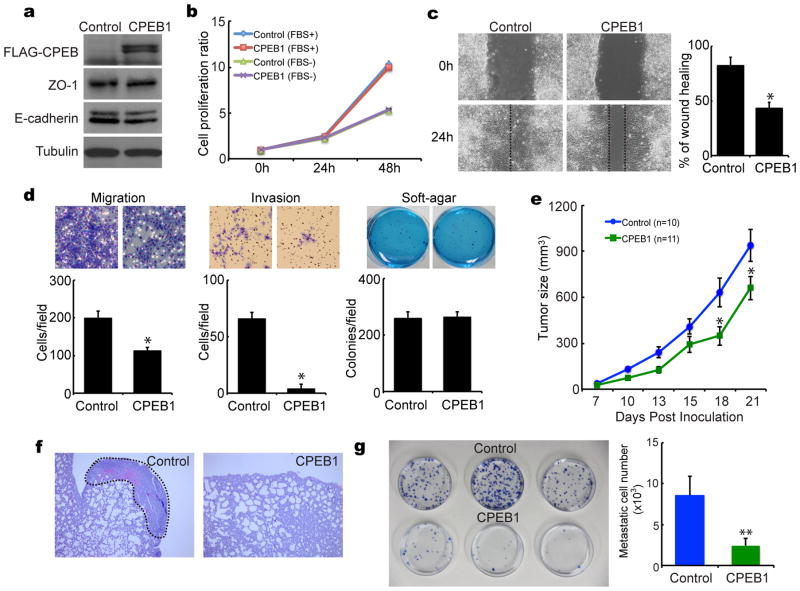
Mouse mammary Eph4 and NMUMG cells, which express relatively high levels of CPEB1 have low metastatic potential, Conversely, 4T1 cells, which have nearly undetectable levels of CPEB1, are highly metastatic (Supplementary Figure S1b). This correlation between CPEB1 and metastatic potential is also conserved in human cells. In the relatively non-metastatic mammary epithelial cells MCF10A and MCF7, CPEB1 levels are high; in contrast the highly metastatic MDA-MB-453 and MDA-MB-231 cells CPEB1 levels are low (Supplementary Figure S1b). These data suggest a strong inverse correlation between CPEB1 and metastasis. To begin to assess whether this relationship is causal, we ectopically expressed CPEB1 in 4T1 cells that are normally deficient for this protein, did not alter ZO-1 or E-cadherin levels (Figure 3a) (N-cadherin and Zeb1analysis was ambiguous) or affect cell proliferation (Figure 3b). However, ectopic CPEB1 significantly reduced cell migration when analyzed by a scratch assay (Figure 3c) and repressed cell migration and invasiveness while having no effect on cell growth in soft agar (Figure 3d).
Figure 3.
CPEB1 suppresses mammary tumor metastasis. (a) Mouse mammary 4T1 cells were infected with a retrovirus expressing CPEB1 and western blotted for FLAG-CPEB, ZO-1, E-cadherin, and tubulin. (b) Control and ectopic CPEB1-expressing cells cultured in the absence or presence of 10% fetal bovine serum (FBS) were examined for cell proliferation. (c) Control and ectopic CPEB1-expressing cells were examined for motility in a wound-healing scratch assay. The dotted lines define the areas lacking cells. The histogram displays the ratio of wound healing from time 0h to time 24h by measuring the area of each scratch closure. (d) Migration, invasion, and soft agar colony formation were assayed cells harboring ectopically-expressed CPEB1. (e) Control and ectopic CPEB1-expressing 4T1 cells were inoculated into the fat pads of 8 week old female Balb/c mice. Tumor sizes were measured with calipers along the two main axes and volumes calculated by the long axis x (short axis)2 × 0.5. (f) Hematoxylin and eosin staining of lung tissue after control and ectopic CPEB1-expressing 4T1 cells were injected into mammary fat pad. (g) Quantification of metastatic cells in the lung. Three weeks after fat pad inoculation, dissociated lung cells were cultured with 6-thioguanine; the 4T1 cell metastases were stained with methylene blue and quantified.
To investigate whether ectopic CPEB1 regulates tumor growth and/or metastasis in vivo, 4T1 cells infected with a retrovirus expressing CPEB1 were injected into the fatpads of mice. Several days after inoculation, tumor size was determined as was the incidence of metastasis to the lung. Figure 3e shows that by 3 weeks, ectopic CPEB1 retarded tumor cell growth by ~25%, which is surprising given that CPEB1 had no effect on cell proliferation in vitro (panel b). Perhaps this reflects an intrinsic difference between in vitro and in vivo growth control. In the control lung, metastasized 4T1 cells formed a solid mass with heightened vasculature, which was not the case with CPEB1-expressign cells (Figure 3f). When the 4T1 tumor reached 1000 mm3 it was isolated from the lung and the disaggregated cells were cultured with 6-thioguanine (6-TGN), which is toxic only to normal cells. Figure 3g shows that the metastatic 4T1 cells were strongly resistant to 6-TGN and formed many colonies in vitro, which was not the case when CPEB1 was ectopically expressed. These data show an inverse correlation between CPEB1 expression and mammary epithelial cell metastasis.
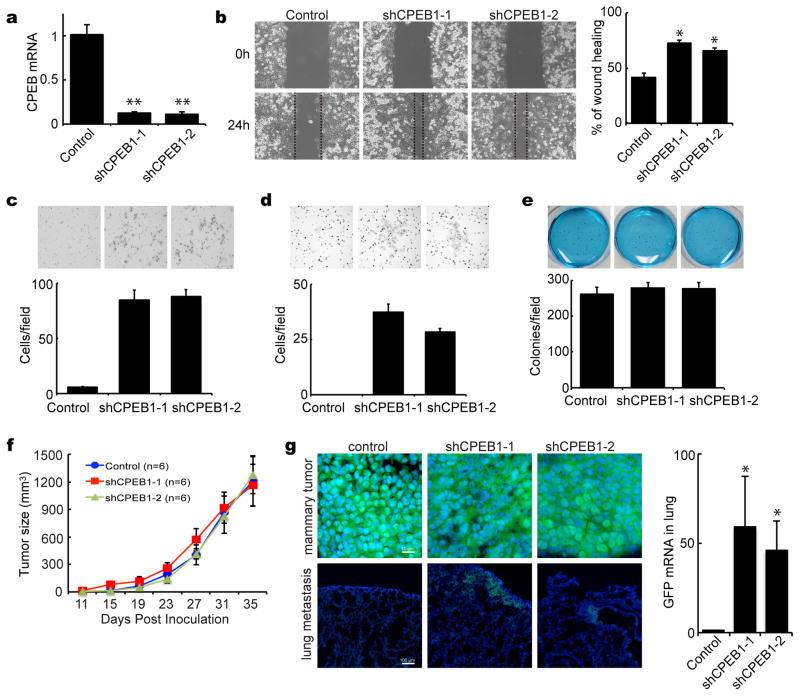
To assess whether the high level of CPEB1 in human mammary epithelial MCF7 cells is at least partially causative for their inability to metastasize, we infected them with lentiviruses expressing GFP and 2 different shRNAs for CPEB1 or a control shRNA. CPEB1 RNA was depleted by >90% with specific shRNAs (Figure 4a), which dramatically stimulated cell motility as revealed by a wound-healing scratch assay (Figure 4b). CPEB1 depletion also stimulated migration (Figure 4d) and invasiveness (Figure 4d) in anchorage-independent assays but had no effect on growth in soft agar (Figure 4e). CPEB1-depletion also had no effect on tumor size when the transduced MCF-7 cells were injected into the fatpad of nude (athymic) mice (Figure 4f). Most importantly, however, CPEB1 knockdown with two different shRNAs produced cell metastasis to the lung. Compared to controls, there was detectable colonization of GFP-positive and CPEB1-depleted MCF7 cells in the lung (Figure 4g). Moreover, RT-PCR analysis showed that GFP RNA levels in the lung were high when CPEB1 was depleted (Figure 4g). Therefore, the data in Figures 3 and 4 demonstrate that high levels of CPEB1 are refractory to mammary cell metastasis to the lung.
Figure 4.
CPEB1 depletion stimulates mammary tumor metastasis. (a) MCF7 cells were infected with lentiviruses expressing GFP only or GFP plus two different shRNAs against CPEB1. Real-time RT-PCR was used to assess CPEB1 depletion. (b) Wound-healing scratch in MCF-7 cells, some of which express CPEB1 shRNA. Migration assay (c), invasion assay (d) and soft-agar colony formation assay (e) following CPEB1 knockdown. (f) MCF7 cells infected with control or CPEB1 knockdown virus were inoculated into the fat pads of 8 week old female nude (nu/nu) mice. Calipers were used to measure tumor sizes along the two main axes. Tumor volumes were estimated by the relationship of the long axis × (short axis)2 × 0.5. (g) Metastatic MCF-7 cells in the lung were detected by immuno-fluorescence for GFP in tissue that was snap frozen and sectioned. Quantification of metastatic cells in the lung was determined by real-time RT-PCR for GFP.
CPEB1 Regulates mRNA Poly(A) tail Growth and Removal
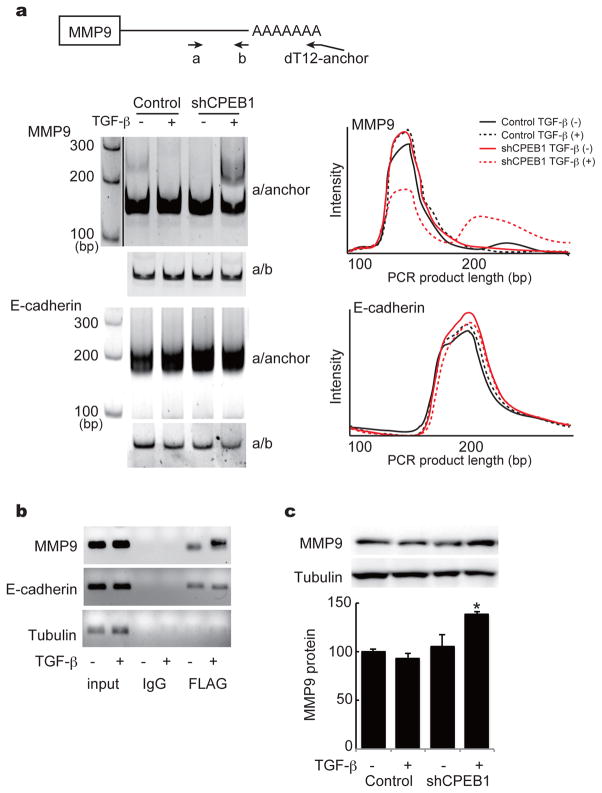
Because many mRNAs contain 3′ UTR CPE-like sequences, it seems implausible that the mis-regulation of a single or even a few mRNAs upon CPEB1 depletion promotes metastasis. Even so, we have sought to identify mRNA targets of CPEB1 that we surmise are likely to be involved in metastasis. Although CPEB1 controls RNA metabolism through alternative splicing,27 RNA localization,16,21 translational repression,e.g.,2,8 and translational activation,4 the most prevalent form of regulation is translational activation via cytoplasmic polyadenylation. Consequently, we thought changes in poly(A) tail length might occur during EMT and thus promote metastasis at least in part by this mechanism. To test this possibility, NMuMG cells were depleted of CPEB1 and treated with TGF-β; extracted RNA was then subjected to chromatography on poly(U) agarose followed by thermal elution.18,28 The RNAs eluting at 60°C were then used to screen an EMT PCR microarray. Supplementary Figure S3 shows that of the 84 RNAs on the array, 14 underwent detectable poly(A) tail size changes. Surprisingly, CPEB1 depletion caused some mRNAs to gain tails while causing others to lose tails. The growth in poly(A) was unexpected because in all cases so far examined, the loss of CPEB1 leads to poly(A) tail reduction, not elongation.e.g.,5,29 We were particularly intrigued by the potential increase in MMP9 mRNA polyadenylation because high levels of the encoded protein are known to promote metastasis.30 A validation of the polyadenylation by a PCR-based assay shows that indeed MMP9 mRNA, but not E-cadherin mRNA, underwent poly(A) tail lengthening upon CPEB1 depletion (Figure 5a) even though both are co-immunoprecipitated with CPEB (Figure 5b). Moreover, MMP9 protein increased by nearly 50% following CPEB1 knockdown (Figure 5c). Thus, the loss of CPEB1 results in increased polyadenylation and translation of MMP9 mRNA.
Figure 5.
CPEB1 regulation of MMP9 mRNA expression. (a) Illustration of PAT (poly(A) tail) assay (top). Cells transduced with control or shCPEB1 and incubated in the absence or presence of TGF-β were subjected to a PAT assays for MMP9 and E-cadherin. The right portion of the panel depicts a scan of the gels that are portrayed at left. (b) RT-PCR analysis of the indicated mRNAs co-immunoprecipitated with IgG or FLAG-CPEB antibody. Input represents 10% of the starting material. (c) Western blots of MMP9 and tubulin in control or shCPEB1 transduced cells cultured in the absence or presence of TGF-β.
CPEB1 Levels are Inversely Correlated with Human Breast Cancer Metastasis
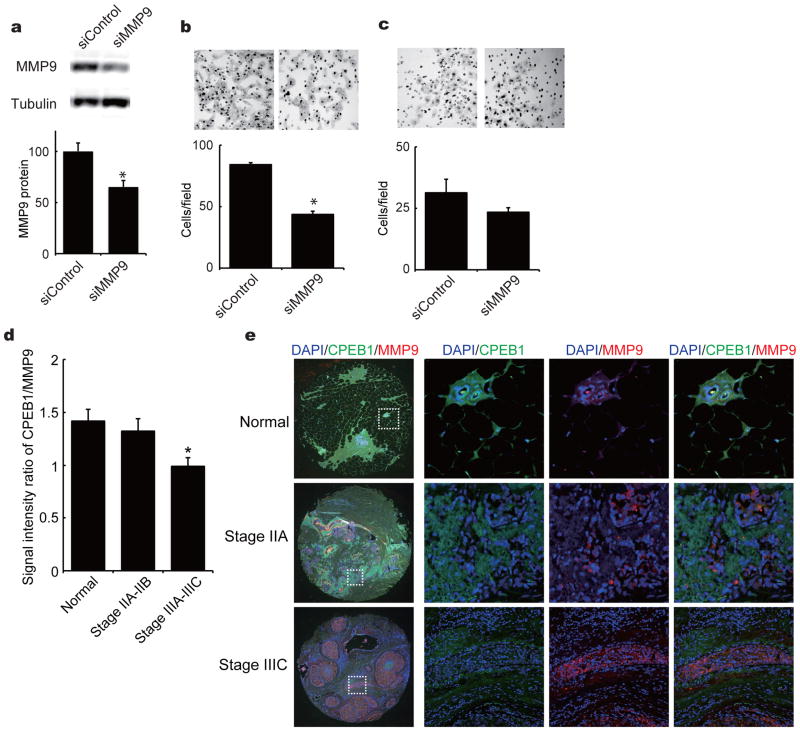
To assess whether loss of MMP9 recovered high cell invasiveness by CPEB1 knockdown, we transduced CPEB1-depleted MCF7 cells with a siRNA for MMP9. The siRNA, which caused a reduction of MMP9 protein by ~40%, significantly reduced cell migration in vitro (Figures 6b). We also injected MMP9 siRNA-transduced cells into mouse fatpads to assess metastasis to the lung in vivo, however, because the siRNA was not retained at a high level in the cells due to both instability and dilution as the cells divided, there was little effect on metastasis.
Figure 6.
CPEB1 and MMP9 in human breast metastases. (a) CPEB1-depleted MCF7 cells were transfected with 20nM siRNA against MMP9. After 48 hours, the relative levels of MMP9 were determined by western blot. Migration assay (b) and invasion assay (c) following MMP9 knockdown. Forty-nine samples of normal (9), stage IIA–IIB (24), or stage IIIA–IIIC (16) human breast cancer metastatic tissue were immunostained for CPEB1 and MMP9. The ratio of these two proteins was calculated based on fluorescence intensity. (b) Immunostaining of normal, stage IIA and stage IIIC human metastatic breast tissue with antibody for CPEB1 (green) and MMP9 (red). DAPI was used as the counterstain.
We screened 49 human tissue samples corresponding to normal breast, stage IIA-IIB breast cancer, and stage IIIA-IIIC metastatic breast cancer tissue samples with antibodies for CPEB1 and MMP9. Based on the fluorescence signal intensities, the ratio of CPEB1 to MMP9 decreased about 40% between normal and stage IIIA-IIIC metastatic breast cancer, which reflects both a decrease in CPEB1 and an increase in MMP9 (Figure 6c). The inverse relationship between these two proteins during metastasis is evident in Figure 6d. Immunocytochemistry for CPEB1 and MMP9 shows that as the degree of metastasis increased from normal to stage IIIC, CPEB1 fell dramatically while MMP9 exhibited an equally robust increase. These data, together with those outlined above, indicate that the reduction of CPEB1 leads to increased MMP9 mRNA polyadenylation and translation and resulting enhanced metastasis of breast cancer cells.
Discussion
CPEB1 depletion from primary mouse or human cells makes them refractory to cellular as well as oncogene-induced senescence, which is caused by reduced p53 mRNA translation.2,8,9,31,32 However, with the exception of an acceleration of cutaneous papillomas in CPEB1 knockout mice treated with 7,12-dimethylbenzanthracene (DMBA) and the phorbol ester 12-O-tetradecanoylphorbol-13-acetate (TPA),9 little evidence suggests that the lack of CPEB1 promotes tumor growth. Indeed, although MEFs derived from CPEB1 KO mice are immortal, they do not form tumors in nude mice.8 On the other hand, CPEB1 depletion disrupts polarity in mammary epithelial cells,21 and although this would not be predicted to induce tumor formation, it could initiate an EMT and potentiate metastasis. Moreover, other emerging evidence has linked CPEB1 levels to EMT, primarily through translational control of TWIST mRNA.33,34 Indeed, our study confirms a number of conclusions reached by Grudzien-Nogalska et al34, who performed an extensive in vitro study of the relationship between CPEB1 and EMT. In our case, we find that CPEB1 depletion not only induces an EMT in vitro (as previously shown by Grudzien-Nogalska et al34), but indeed also promotes metastasis of mammary cells to the lung in vivo. Although we not observe that CPEB1 modifies TWIST mRNA poly(A) tail length or translation, we surprisingly found that CPEB1 depletion stimulates MMP9 mRNA polyadenylation and translation. Moreover, we found that at least in vitro, depletion of MMP9, a key metastasis-promoting factor, decreases cell migration in CPEB1-depleted cells. An interesting corollary is our observation that as human breast cancer cells become progressively more metastatic, CPEB1 decreases and MMP9 increases. These data suggest that the diminution of CPEB1 during EMT gives rise to an increase in MMP9, which in turn promotes metastasis.
CPEB1 alters gene expression in three different ways as cells become metastatic. First, the loss of mammary epithelial polarity by CPEB1 depletion is due to mis-localization of ZO-1 mRNA with no detectable effect on polyadenylation or translation.20 Second, when CPEB1-depleted cells are treated with TGF-β to potentiate EMT,35,36 there are substantial alterations in the levels of mRNAs that encode EMT-relevant proteins. Third, CPEB1 depletion increases the polyadenylation and translation of MMP9 mRNA, which promotes metastasis. How can CPEB1 affect these processes by different mechanisms? CPEB1 is already known to regulate mRNA localization by virtue of its binding to the molecular motors kinesin and dynein,16 and it could mediate ZO-1 mRNA localization to the apical region of mammary epithelial cells by these or other molecular motors. CPEB1 control of polyadenylation is regulated by its association with non-canonical poly(A) polymerases and deadenylases.2,13,37 Therefore, it is likely that in some circumstances or cell types, CPEB1 interacts with factors to regulate polyadenylation while in other cell types, such interactions may not occur and thus CPEB1 affects RNA metabolism in other ways.
We found that CPEB1 depletion stimulated polyadenylation of MMP9 mRNA, which was surprising because the loss of CPEB1 would be expected to reduce polyadenylation. How this could occur is unclear, but one possibility is that CPEB1 association with MMP9 RNA might preclude the binding of a different protein (e.g., FMRP)38 that in turn recruits the non-canonical poly(A) polymerases Gld2 or Gld4. Such a situation could be analogous to control of alternative splicing, where an RNA binding protein prevents the association of the splicing machinery for some exons but not others.39
Our results show that in vivo, reduced CPEB1 levels promote metastasis of breast cancer cells to the lung whereas ectopic expression of CPEB1 strongly inhibits it, suggesting that CPEB1 could be a good prognosticator of human breast cancer metastasis. The extent to which CPEB1 mediates metastasis of other cell types is unclear. The loss of CPEB1 has no effect on polarity in intestinal or kidney epithelial cells,20 and if one assumes this is an essential initiating event, then CPEB1 would not be expected to have much of an influence on metastasis in these cells. Moreover, in glioblastoma cells, ectopic expression of a dominant negative CPEB1 that cannot stimulate polyadenylation blocks expression of metadherin (MRDH), a metastasis-promoting factor and reduces cell migration and tumor growth.40 Because these observations are seemingly the opposite of what we observe in breast cancer cells, it appears that CPEB1 can have diametrically opposed effects on metastasis depending on the cell type.
Materials and Methods
Plasmid Construction, siRNA and Antibodies
A lentiviral shRNA targeting mouse CPEB1 was cloned into pLentiLox 3.7 vector; shCPEB1-1: 5′-GTCGTGTGACTTTCAATAA-3′, shCPEB1-2: 5′-GTCCCAGAGACCCTCTAAA. Mouse CPEB1 tagged with an amino terminal three rimes reiterated Flag epitope was cloned into the pBABE retrovirus vector. Mission esiRNA for target mouse MMP9 mRNA and siRNA universal negative control were purchased from Sigma. The following antibodies were used: anti-ZO-1 (Invitrogen, catalogue #40-2200), anti-E-cadherin (BD Transduction, catalogue #610181), anti-N-cadherin (Cell Signaling, catalogue #4061), anti-ZEB1 (Cell Signaling, catalogue #3396), anti-FLAG (Cell Signaling, catalogue #2368), anti-MMP9 (Sigma, catalogue #AV33090), and anti-γ-tubulin (Sigma, catalogue #T6557). Anti-CPEB1 is described in Udagawa et al (2013).
Cell Culture, Transfection, Virus Production and Infection
Mouse mammary gland epithelial cells (NMuMG), mouse mammary tumor cells (4T1), and human breast cancer cells (MCF7) were purchased from ATCC (not tested further for mycoplasma) and maintained in DMEM containing 5 μg/ml insulin, 10% FBS and antibiotic-antimycotic reagent (Invitrogen). The lentiviral and retroviral constructs were co-transfected with their respective packaging plasmids into 293T cells using lipofectamine-2000 (Invitrogen). Virus-containing supernatants were collected 48 hours after transfection and filtered; the target cells were infected in the presence of 6 μg/ml polybrene. Infected cells were selected with puromycin or single clones strongly expressing GFP were identified by the limited dilution method, isolated, and expanded.
PCR Array and Quantitative PCR
For PCR array, RNA was extracted from TGF-β–treated control and shCPEB1 knockdown NMuMG using Trizol (Invitrogen), followed by, reverse transcribed using RT2 First Strand kit (QIAGEN, Hilden, Germany), and applied to an Epithelial to Mesenchymal Transition (EMT) PCR array as detailed by the manufacturer (QIAGEN). The expression levels of the mRNA of each gene were normalized using the expression of Actb, B2m, Gapdh, Gusb, and Hap90ab1, which are considered to be housekeeping genes.
For real-time PCR, complementary DNA (cDNA) was synthesized using PrimeScript reverse transcriptase (TaKaRa) according to the manufacturer’s instructions. The amount of cDNA targets was determined based on real-time PCR results. Oligonucleotide primers were selected using a web-based Primer3 software and are listed in Supplementary Table S1. PCR reactions were run using SYBR Premix Ex Taq II (TaKaRa), and the expression of each target mRNA relative to tubulin mRNA was determined using the 2−ΔΔCT method.
Western Blot Analysis
Whole-cell lysates were prepared in RIPA buffer (50 mM Tris-HCl, 150 mM NaCl, 1 mM EDTA, 1 mM NaVO4, 50 mM NaF, 0.1% SDS, 1% Triton-100, and Protease Inhibitor Cocktail). Protein samples (15 μg) were separated by electrophoresis on SDS-PAGE gels and transferred onto nitrocellulose membranes (Immobilon; Millipore, Bedford, MA). Membranes were blocked with 5% skim milk at room temperature for 1 h before incubation with primary antibodies at 4°C for 12 h. The membranes were washed 4 times in PBS containing Tween-20, incubated with horseradish peroxidase-conjugated anti-mouse or anti-rabbit IgG (GE Healthcare UK Ltd., Buckinghamshire, UK) at room temperature for 1 h, and again washed 4 times in PBS-Tween 20. Protein bands were detected using the ECL Plus Western Blotting Detection System (GE Healthcare).
Immunofluorescence
Cells were cultured on glass coverslips and fixed in 4% paraformaldehyde for 30 min. They were permeabilized using 0.5% Triton X-100/PBS for 10 min, blocked with 2% BSA/PBS for 1 h, treated with primary antibodies for 1 h, and incubated with Alexa 488-conjugated anti-mouse IgG or Alexa 568-conjugated anti-rabbit IgG for 1 h. Slides were mounted with ProLong Gold with DAPI (Invitrogen). Images were captured using an inverted confocal microscope (LSM710: Carl Zeiss, Oberkochen, Germany).
Cell Proliferation, Motility, Migration, and Invasion
Cells were plated in 96-well plates at a concentration of 2000 cells/well in complete medium. At 24h and 48h time points, cell proliferation was measured using the Cell Counting Kit-8 (DOJINDO, Kumamoto, Japan) according to the manufacturer’s protocol. For the wound-healing scratch assays, cells were grown in monolayers on 6-well plates and scored with a pipette tip. After 24h, cell migration was assessed.
For the migration and invasion assays, 24well-Transwells (Costar) coated without or with Matrigel (BD Bioscience) were used. Cells at concentrations of 50000 cells/well were seeded in the top well with DMEM containing 5 ng/ml TGF-β and were allowed to migrate and invade for 24h. The membranes were removed, washed with PBS, fixed and stained with Dif-Quick (Kokusai Shiyaku, Kobe, Japan). The number of cells that passed to the lower surface was microscopically counted at six randomly chosen high power fields.
Anchorage-independent growth, an indicator of cell transformation was evaluated in soft agar assays. Briefly, 1 × 104 cells were plated in complete DMEM containing 0.4% agarose in 6-cm plates over a layer of solidified DMEM containing 0.75% agarose. After 5 weeks, colonies were stained with crystal violet for 3 h and counted.
In vivo Transplantation Assay
Cells (1 × 106) were injected into the no. 4 or no. 9 fatpad of 8 week old female BALB/c mice for 4T1 cells (n=21) and BALB/c nude mice for MCF7 cells (n=18) using a 30-gauge needle. After 1 week, tumor sizes were subsequently measured every 3 days and the tumor volumes were estimated as long axis × (short axis)2 × 0.5. For 4T1 cells, when tumor size reached ~1000 mm3, lung samples were collected from animals, minced into small pieces with scissors and digested in Hank’s balanced salt solution (HBSS) containing 1 mg/ml collagenase-A (Roche) for 75 min at 4 °C. After the enzyme digestion, samples were filtered through 70-μm nylon cell strainers (BD Bioscience), washed with HBSS and then suspended in DMEM supplemented with 10% FBS, 1% antibiotic-antimycotic reagent, and 60 μM 6-thioguanine. After culturing for 2 weeks, the cells were fixed by 10% formalin and stained with 0.03% methylene blue, and blue colonies were counted. For MCF7 cells, when tumors were ~1500 mm3, lung samples were snap frozen in liquid nitrogen and used for GFP detection. All experiments with mice were performed according to the guidelines of the Institutional Animal Care and Use Committee of Tokyo University of Agriculture and Technology.
Poly(A) Analysis
RNA was extracted from NMuMG control or shCPEB1-expressing cells treated with or without TGF-β (Supplementary Figure S2). Polyadenylated RNA was fractionated on poly(U) Sepharose, which washed at 50°C and the RNA eluted at 65°C. Total RNA and RNA from 65°C eluates were used as templates for EMT PCR array. For the poly(A) tail (PAT) assay, cDNA was synthesized with oligo(dT) anchor primer (5′-GCG AGC TCC GCG GCC GCG-T12-3′), and subsequent PCR was conducted with anchor primer (5′-GCG AGC TCC GCG GCC GCG-3′) and specific primer for MMP9 (5′-ACT AGG GCT CCT TCT TTG CTT CAA C-3′) or E-cadherin (5′-TGT ATG TGT GTG GGT GCT GA-3′) RNA in the presence of SYBR Green Dye.
CPEB1 and MMP9 in Human Breast Cancer Cells
Human breast tissue array slides containing normal, cancer, and metastatic biopsies were purchased from SuperBioChips Laboratories (Korea). The slides were deparaffinized and subjected to antigen retrieval by boiling for 5 min in 0.01M sodium citrate buffer (pH 6.0). The slides were then blocked with 2% BSA/PBS for 1 h, treated with primary antibodies for 1 h, and incubated with Alexa 488-conjugated anti-mouse IgG or Alexa 568-conjugated anti-rabbit IgG for 1 h. Slides were then mounted with ProLong Gold with DAPI (Invitrogen). Images were captured using an immunofluorescence microscope (BZ-X700an: Keyence, Osaka, Japan).
Statistical Analysis
The data are presented as mean ± SEM of 3 independent experiments (biological replicates), each performed in triplicate (technical replicates). The level of significance was determined using one-way analysis of variance, followed by two-sided Tukey’s multiple range tests (GraphPad Prism5). Sample size was chosen for statistical power. Differences were considered statistically significant when p<0.05 (center values are the mean). All experiments were replicated three times, which were sufficient for statistical analysis. All error bars refer to s.e.m. For the animal studies (i.e., in vivo transplantation), all data were included in the analysis – none were excluded. The experimenter was not blinded to the studies, and the data were not randomized.
Supplementary Material
(a) NMuMG cells were treated with TGF-β and real-time PCR for CPEB1. (b)CPEB1 mRNA levels (assessed by RT-qPCR) in Eph4, NMuMG, and 4T1 mouse breast epithelial cells as well as MCF10S, MCF7, MDA-MB-453, and MDA-MB-231 human breast epithelial cells. There is an inverse correlation between CPEB1 mRNA levels and metastatic potential.
NMuMG cells were infected with lentiviruses expressing GFP or GFP plus an shRNA against CPEB1. The cells were then treated with TGF-β and the extracted RNA analyzed for their gene expression by EMT PCR Array. Of the 84 RNAs on the array, 26 were up-regulated while 21 were down-regulated upon CPEB1 depletion.
Flow chart illustration of mRNAs with altered poly(A tail length in control and shCPEB1 transduced Eph4 cells cultured in the presence or absence of TGF-β.
Migration of assayed cells harboring ectopically-expressed CPEB1 and cultured in the absence or presence of TGF-β were determined.
Acknowledgments
This work was supported by NIH grant GM46779 (to JDR).
Footnotes
Author contributions: KN and JDR designed the research; KN, HZ, KF, and GW performed research; MI made antibody; KN and JDR analyzed data; and KN and JDR wrote the manuscript
References
- 1.Groisman I, Jung MY, Sarkissian M, Cao Q, Richter JD. Translational control of the embryonic cell cycle. Cell. 2002;109:473–483. doi: 10.1016/s0092-8674(02)00733-x. [DOI] [PubMed] [Google Scholar]
- 2.Burns DM, D’Ambrogio A, Nottrott S, Richter JD. CPEB and two poly(A) polymerases control miR-122 stability and p53 mRNA translation. Nature. 2011;473:105–108. doi: 10.1038/nature09908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.D’Ambrogio A, Nagaoka K, Richter JD. Translational control of cell growth and malignancy by the CPEB1s. Nat Rev Cancer. 2013;13:283–290. doi: 10.1038/nrc3485. [DOI] [PubMed] [Google Scholar]
- 4.Ivshina M, Lasko P, Richter JD. Cytoplasmic polyadenylation element binding proteins in development, health, and disease. Annu Rev Cell Dev Biol. 2014;30:393–415. doi: 10.1146/annurev-cellbio-101011-155831. [DOI] [PubMed] [Google Scholar]
- 5.Tay J, Richter JD. Germ cell differentiation and synaptonemal complex formation are disrupted in CPEB1 knockout mice. Dev Cell. 2001;1:201–213. doi: 10.1016/s1534-5807(01)00025-9. [DOI] [PubMed] [Google Scholar]
- 6.Alarcon JM, Hodgman R, Theis M, Huang YS, Kandel ER, Richter JD. Selective modulation of some forms of schaffer collateral-CA1 synaptic plasticity in mice with a disruption of the CPEB1-1 gene. Learn Mem. 2004;11:318–327. doi: 10.1101/lm.72704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berger-Sweeney J, Zearfoss NR, Richter JD. Reduced extinction of hippocampal-dependent memories in CPEB1 knockout mice. Learn Mem. 2006;13:4–7. doi: 10.1101/lm.73706. [DOI] [PubMed] [Google Scholar]
- 8.Groisman I, Ivshina M, Marin V, Kennedy NJ, Davis RJ, Richter JD. Control of cellular senescence by CPEB1. Genes Dev. 2006;20:2701–2712. doi: 10.1101/gad.1438906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Burns DM, Richter JD. CPEB1 regulation of human cellular senescence, energy metabolism, and p53 mRNA translation. Genes Dev. 2008;22:3449–3460. doi: 10.1101/gad.1697808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alexandrov IM, Ivshina M, Jung DY, Friedline R, Ko HJ, Xu M, et al. Cytoplasmic polyadenylation element binding protein deficiency stimulates PTEN and Stat3 mRNA translation and induces hepatic insulin resistance. PLoS Genet. 2012;8:e1002457. doi: 10.1371/journal.pgen.1002457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Udagawa T, Farny NG, Jakovcevski M, Kaphzan H, Alarcon JM, Anilkumar S, et al. Genetic and acute CPEB1 depletion ameliorate fragile X pathophysiology. Nat Med. 2013;19:1473–1477. doi: 10.1038/nm.3353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Groisman I, Huang YS, Mendez R, Cao Q, Theurkauf W, Richter JD. CPEB1, maskin, and cyclin B1 mRNA at the mitotic apparatus: implications for local translational control of cell division. Cell. 2000;103:435–447. doi: 10.1016/s0092-8674(00)00135-5. [DOI] [PubMed] [Google Scholar]
- 13.Barnard DC, Ryan K, Manley JL, Richter JD. Symplekin and xGLD-2 are required for CPEB1-mediated cytoplasmic polyadenylation. Cell. 2004;119:641–651. doi: 10.1016/j.cell.2004.10.029. [DOI] [PubMed] [Google Scholar]
- 14.Eliscovich C, Peset I, Vernos I, Méndez R. Spindle-localized CPE-mediated translation controls meiotic chromosome segregation. Nat Cell Biol. 2008;10:858–865. doi: 10.1038/ncb1746. [DOI] [PubMed] [Google Scholar]
- 15.Huang YS, Jung MY, Sarkissian M, Richter JD. N-methyl-D-aspartate receptor signaling results in Aurora kinase-catalyzed CPEB1 phosphorylation and alpha CaMKII mRNA polyadenylation at synapses. EMBO J. 2002;21:2139–2148. doi: 10.1093/emboj/21.9.2139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang YS, Carson JH, Barbarese E, Richter JD. Facilitation of dendritic mRNA transport by CPEB1. Genes Dev. 2003;17:638–653. doi: 10.1101/gad.1053003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bestman JE, Cline HT. The RNA binding protein CPEB1 regulates dendrite morphogenesis and neuronal circuit assembly in vivo. Proc Natl Acad Sci USA. 2008;105:20494–20499. doi: 10.1073/pnas.0806296105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Udagawa T, Swanger SA, Takeuchi K, Kim JH, Nalavadi V, Shin J, et al. Bidirectional control of mRNA translation and synaptic plasticity by the cytoplasmic polyadenylation complex. Mol Cell. 2012;47:253–266. doi: 10.1016/j.molcel.2012.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shen W, Liu HH, Schiapparelli L, McClatchy D, He HY, Yates JR, 3rd, et al. Acute synthesis of CPEB is required for plasticity of visual avoidance behavior in Xenopus. Cell Rep. 2014;6:737–747. doi: 10.1016/j.celrep.2014.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nagaoka K, Udagawa T, Richter JD. CPEB1-mediated ZO-1 mRNA localization is required for epithelial tight-junction assembly and cell polarity. Nat Commun. 2012;3:675. doi: 10.1038/ncomms1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mailleux AA, Overholtzer M, Brugge JS. Lumen formation during mammary epithelial morphogenesis: insights from in vitro and in vivo models. Cell Cycle. 2008;7:57–62. doi: 10.4161/cc.7.1.5150. [DOI] [PubMed] [Google Scholar]
- 22.Knights AJ, Funnell AP, Crossley M, Pearson RC. Holding Tight: Cell Junctions and Cancer Spread. Trends Cancer Res. 2012;8:61–69. [PMC free article] [PubMed] [Google Scholar]
- 23.Scheel C, Weinberg RA. Cancer stem cells and epithelial-mesenchymal transition: concepts and molecular links. Semin Cancer Biol. 2012;22:396–403. doi: 10.1016/j.semcancer.2012.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Voulgari A, Pintzas A. Epithelial-mesenchymal transition in cancer metastasis: mechanisms, markers and strategies to overcome drug resistance in the clinic. Biochim Biophys Acta. 2009;1796:75–90. doi: 10.1016/j.bbcan.2009.03.002. [DOI] [PubMed] [Google Scholar]
- 25.Chaffer CL, Weinberg RA. A perspective on cancer cell metastasis. Science. 2011;331:1559–1564. doi: 10.1126/science.1203543. [DOI] [PubMed] [Google Scholar]
- 26.Zavadil J, Bitzer M, Liang D, Yang YC, Massimi A, Kneitz S, et al. Genetic programs of epithelial cell plasticity directed by transforming growth factor-beta. Proc Natl Acad Sci USA. 2001;98:6686–6691. doi: 10.1073/pnas.111614398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lin CL, Evans V, Shen S, Xing Y, Richter JD. The nuclear experience of CPEB: implications for RNA processing and translational control. RNA. 2010;16:338–348. doi: 10.1261/rna.1779810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Du L, Richter JD. Activity-dependent polyadenylation in neurons. RNA. 2005;11:1340–1347. doi: 10.1261/rna.2870505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Racki WJ, Richter JD. CPEB controls oocyte growth and follicle development in the moue. Development. 2006;133:4527–4537. doi: 10.1242/dev.02651. [DOI] [PubMed] [Google Scholar]
- 30.Hiratsuka S, Nakamura K, Iwai S, Murakami M, Itoh T, Kijima H, et al. MMP9 induction by vascular endothelial growth factor receptor-1 is involved in lung-specific metastasis. Cancer Cell. 2002;2:289–300. doi: 10.1016/s1535-6108(02)00153-8. [DOI] [PubMed] [Google Scholar]
- 31.Groppo R, Richter JD. CPEB control of NF-kappaB nuclear localization and interleukin-6 production mediates cellular senescence. Mol Cell Biol. 2011;31:2707–2714. doi: 10.1128/MCB.05133-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.D’Ambrogio A, Gu W, Udagawa T, Mello CC, Richter JD. Specific miRNA stabilization by Gld2-catalyzed monoadenylation. Cell Rep. 2012;2:1537–1545. doi: 10.1016/j.celrep.2012.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nairismägi ML, Vislovukh A, Meng Q, Kratassiouk G, Beldiman C, Petretich M, et al. Translational control of TWIST1 expression in MCF-10A cell lines recapitulating breast cancer progression. Oncogene. 2012;31:4960–4966. doi: 10.1038/onc.2011.650. [DOI] [PubMed] [Google Scholar]
- 34.Grudzien-Nogalska E, Reed BC, Rhoads RE. CPEB1 promotes differentiation and suppresses EMT in mammary epithelial cells. J Cell Sci. 2014;127:2326–2338. doi: 10.1242/jcs.144956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gonzalez DM, Medici D. Signaling mechanisms of the epithelial-mesenchymal transition. Sci Signal. 2014;7:re8. doi: 10.1126/scisignal.2005189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang J, Tian XJ, Zhang H, Teng Y, Li R, Bai F, et al. TGF-β-induced epithelial-to-mesenchymal transition proceeds through stepwise activation of multiple feedback loops. Sci Signal. 2014;7:ra91. doi: 10.1126/scisignal.2005304. [DOI] [PubMed] [Google Scholar]
- 37.Kim JH, Richter JD. Opposing polymerase-deadenylase activities regulate cytoplasmic polyadenylation. Mol Cell. 2006;24:173–83. doi: 10.1016/j.molcel.2006.08.016. [DOI] [PubMed] [Google Scholar]
- 38.Janusz A, Milek J, Perycz M, Pacini L, Bagni C, Kaczmarek L, et al. The Fragile X mental retardation protein regulates matrix metalloproteinase 9 mRNA at synapses. J Neurosci. 2013;33:18234–18241. doi: 10.1523/JNEUROSCI.2207-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Licatalosi DD, Mele A, Fak JJ, Ule J, Kayikci M, Chi SW, et al. HITS-CLIP yields genome-wide insights into brain alternative RNA processing. Nature. 2008;456:464–469. doi: 10.1038/nature07488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kochanek DM, Wells DG. CPEB1 regulates the expression of MTDH/AEG-1 and glioblastoma cell migration. Mol Cancer Res. 2013;11:149–160. doi: 10.1158/1541-7786.MCR-12-0498. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(a) NMuMG cells were treated with TGF-β and real-time PCR for CPEB1. (b)CPEB1 mRNA levels (assessed by RT-qPCR) in Eph4, NMuMG, and 4T1 mouse breast epithelial cells as well as MCF10S, MCF7, MDA-MB-453, and MDA-MB-231 human breast epithelial cells. There is an inverse correlation between CPEB1 mRNA levels and metastatic potential.
NMuMG cells were infected with lentiviruses expressing GFP or GFP plus an shRNA against CPEB1. The cells were then treated with TGF-β and the extracted RNA analyzed for their gene expression by EMT PCR Array. Of the 84 RNAs on the array, 26 were up-regulated while 21 were down-regulated upon CPEB1 depletion.
Flow chart illustration of mRNAs with altered poly(A tail length in control and shCPEB1 transduced Eph4 cells cultured in the presence or absence of TGF-β.
Migration of assayed cells harboring ectopically-expressed CPEB1 and cultured in the absence or presence of TGF-β were determined.