Abstract
Propionibacterium acnes is the most abundant bacterium on human skin, particularly in sebaceous areas. P. acnes is suggested to be an opportunistic pathogen involved in the development of diverse medical conditions but is also a proven contaminant of human clinical samples and surgical wounds. Its significance as a pathogen is consequently a matter of debate. In the present study, we investigated the presence of P. acnes DNA in 250 next-generation sequencing data sets generated from 180 samples of 20 different sample types, mostly of cancerous origin. The samples were subjected to either microbial enrichment, involving nuclease treatment to reduce the amount of host nucleic acids, or shotgun sequencing. We detected high proportions of P. acnes DNA in enriched samples, particularly skin tissue-derived and other tissue samples, with the levels being higher in enriched samples than in shotgun-sequenced samples. P. acnes reads were detected in most samples analyzed, though the proportions in most shotgun-sequenced samples were low. Our results show that P. acnes can be detected in practically all sample types when molecular methods, such as next-generation sequencing, are employed. The possibility of contamination from the patient or other sources, including laboratory reagents or environment, should therefore always be considered carefully when P. acnes is detected in clinical samples. We advocate that detection of P. acnes always be accompanied by experiments validating the association between this bacterium and any clinical condition.
INTRODUCTION
Propionibacterium acnes is a facultative anaerobic Gram-positive bacterium present on human skin as part of the normal flora, as well as in the oral cavity, large intestine, conjunctiva, and external ear canal (1). P. acnes is the most prevalent bacterium in sebaceous areas of the skin (2, 3) but is also abundant in dry areas (3). P. acnes predominates in the pilosebaceous follicles of the skin (4).
P. acnes is proposed to play a role in the development of acne vulgaris (5) and is considered an opportunistic pathogen causing postoperative infections (6). It has been isolated from patients with endocarditis (7), synovitis, acne, pustulosis, hyperostosis, and osteitis (SAPHO) syndrome (8), and sarcoidosis (9), among other syndromes. Some studies also identify P. acnes to be a contaminant of blood products, tissue cultures, and surgical wounds (10–13), and the significance of this widely abundant skin commensal is consequently debated.
The postulated role of P. acnes in various conditions is often highly speculative and based on the mere detection of the bacterium (8, 9, 14–19). To expand the knowledge of the abundance of P. acnes, we investigated the proportions of P. acnes sequences in samples of a wide variety of cancer types, as well as blood samples. Next-generation sequencing was performed on DNA extracted either after enrichment for intact microbes or directly from the samples. This study is the first large-scale analysis investigating the abundance of P. acnes DNA in such diverse tissue types by next-generation sequencing, and we show that P. acnes can be readily detected in literally all sample types investigated, particularly when samples are subjected to microbial enrichment.
MATERIALS AND METHODS
Ethics statement.
Human sample collection, handling, and analysis were performed under ethical protocol H-2-2012-FSP2 (Regional Committee on Health Research Ethics, Capital Region of Denmark) and case no. 1304226 (National Committee on Health Research Ethics, Denmark). In accordance with Danish national legislation (Sundhedsloven), all human samples were processed anonymously.
Patient samples. (i) Samples collected in Denmark.
Malignant melanoma biopsy specimens and cells from patients with acute myeloid leukemia (AML), B-cell chronic lymphocytic leukemia (B-CLL), and chronic myelogenous leukemia (CML) sorted by fluorescence-activated cell sorting (FACS) were obtained from Aarhus University Hospital. Samples from patients with T-lineage acute lymphoblastic leukemia (T-ALL) were obtained from either Aarhus University Hospital (cells sorted by FACS; n = 9) or Rigshospitalet (Copenhagen University Hospital; bone marrow samples; n = 2). Samples (bone marrow) from patients with B-cell precursor acute lymphoblastic leukemia (BCP-ALL) and biopsy specimens from patients with oropharyngeal or oral cavity head and neck cancer and testicular cancer were obtained from Rigshospitalet. Biopsy specimens from patients with basal cell carcinoma and mycosis fungoides (cutaneous T-cell lymphoma) were obtained from Bispebjerg Hospital. Bladder, breast, and colon cancer biopsy specimens, blood samples from colon cancer patients, and ascitic fluid samples from colon, ovarian, and pancreatic cancer patients were obtained from or collected in collaboration with the Danish Cancer Biobank, Herlev Hospital. Ascitic fluid samples were subjected to low-speed centrifugation upon receipt to pellet intact host cells and cellular debris. The supernatant was used for microbial enrichment, whereas pelleted cells were used for shotgun sequencing. Cryopreserved, fully transformed B-cell lymphoma cell lines (OCI Ly3_M, OCI-Ly7_M, OCI-Ly8, SU-DHL-4, SU-DHL-5, and U698M) and multiple myeloma cell lines (KMS-12-BM, KMS-12-PE, MOLP-2, MOLP-8, RPMI-8226, and U266) were obtained from Aalborg University Hospital.
(ii) Samples collected outside Denmark.
Vulva cancer biopsy specimens were obtained from the National Institute of Oncology, Budapest, Hungary. All samples are listed in Table 1.
TABLE 1.
Samples included in the studya
| Disease | Abbreviation | Sample type | No. of samples subjected to: |
|
|---|---|---|---|---|
| Microbial enrichment | Shotgun sequencing | |||
| Acute myeloid leukemia | AML | Bone marrow (sorted cells) | 9 | |
| B-cell chronic lymphocytic leukemia | B-CLL | Blood/bone marrow (sorted cells) | 9 | |
| B-cell precursor acute lymphoblastic leukemia | BCP-ALL | Bone marrow | 8 | |
| Basal cell carcinoma | BCC | Tumor biopsy specimens | 11 | 11 |
| Bladder cancer | Tumor biopsy specimens | 7 | ||
| Breast cancer (ductal/lobular) | Tumor biopsy specimens | 17 | 20 | |
| Ascitic fluid | 1 | 1 | ||
| Chronic myelogenous leukemia | CML | Bone marrow (sorted cells) | 10 | |
| Colon cancer | Tumor biopsy specimens | 3 | 12 | |
| Blood | 8 | |||
| Ascitic fluid | 1 | |||
| Lymphoma/myeloma | Cell lines | 12 | ||
| Malignant melanoma | Tumor biopsy specimens | 10 | 10 | |
| Mycosis fungoides (cutaneous T-cell lymphoma) | Mycosis | Tumor biopsy specimens | 11 | 11 |
| Oropharyngeal/oral cavity cancer | Oral | Tumor biopsy specimens | 10 | 9 |
| Ovarian cancer | Ascitic fluid | 3 | 5 | |
| Pancreatic cancer | Ascitic fluid | 2 | ||
| T-lineage acute lymphoblastic leukemia | T-ALL | Bone marrow (nonsorted/sorted cells) | 11 | |
| Testicular cancer (sem/non-sem) | Testis | Tumor biopsy specimens | 20 | 5 |
| Vulva cancer | Vulva | Tumor biopsy specimens | 3 | |
| Total no. of sample types and samples | 20 | 143 | 107 | |
The diseases, sample types, and number of samples included are shown.
Laboratory facilities.
All sample processing was performed under conditions designed to diminish contamination through the use of a unidirectional work flow. Separate laboratories were used for reagent preparation, sample extraction and sequencing library preparation, and library amplification. All laboratory work was conducted by individuals who were wearing a lab coat and gloves, as well as sleeves during extraction and library preparation. All sample processing was carried out in laminar-flow cabinets equipped with UV lamps. All benchtops were cleaned with 5% bleach and 70% ethanol before and after any lab work. Only sterile, disposable lab utensils were used. Filter tips were used for all pipetting.
Microbial enrichment, nucleic acid extraction, and library preparation.
Samples used for enrichment were fresh frozen after collection, and no nucleic acid preservatives were added. Each batch of processed samples included a nontemplate control containing only phosphate-buffered saline (PBS), which was run in parallel with the patient samples (n = 19). Samples were kept on ice unless otherwise stated.
Biopsy specimens were thawed and transferred to 400 μl cold PBS. For the samples from leukemia patients, 150 to 400 μl was mixed with cold PBS to a total volume of 400 μl. Two stainless steel beads of 2 to 3 mm in diameter were added to the samples, which were subsequently homogenized using a TissueLyser II tissue disrupter (Qiagen, Hilden, Germany) for 6 min at 30 Hz. All tissue homogenates, samples from leukemia patients, and ascitic fluid specimens (1 ml) devoid of pelleted cells were centrifuged for 2 min at 800 × g to remove tissue debris, and the supernatants were subsequently filtered through 5-μm-pore-size centrifuge filters (Millipore, Darmstadt, Germany). To remove unprotected nucleic acids while retaining the nucleic acids within bacterial cells or viral particles, the filtrates were nuclease digested using 14 μl Turbo DNase (2 U/μl; Ambion, Thermo Fisher Scientific, Waltham, MA, USA), 12 μl Baseline-Zero DNase (1 U/μl; Epicentre, Madison, WI, USA), 16 μl RNase cocktail enzyme mix (Ambion), and 40 μl 10× Turbo DNase buffer in a total volume of 400 μl and incubated at 37°C for 2 h. Nucleic acids from the enriched samples were extracted using the High Pure viral RNA kit (Roche, Basel, Switzerland) according to the manufacturer's instructions with the addition of 10 μg linear acrylamide carrier (Applied Biosystems, Foster City, CA, USA).
DNA libraries were built using the Nextera or Nextera XT DNA sample preparation kit (Illumina, San Diego, CA, USA), according to the manufacturer's instructions. Nextera libraries were amplified by 20 cycles of PCR, and Nextera XT libraries were amplified by 12 cycles of PCR. The amplified and indexed libraries were purified using the Agencourt AMPure XP PCR purification system (Beckman Coulter, Indianapolis, IN, USA). In case of insufficient amplification, libraries were reamplified using AccuPrime Pfx DNA polymerase (Life Technologies, Carlsbad, CA, USA) and P5 and P7 sequence primers.
Total DNA extraction and library preparation.
Total DNA was extracted from samples using the QIAamp DNA minikit (Qiagen) following the manufacturer's instructions. DNA libraries were prepared from 1 μg of DNA using either the TruSeq DNA protocol (catalog number PE-940-2001; Illumina, San Diego, CA, USA) or an in-house protocol (20) using NEBnext reagents (protocol E6070; New England BioLabs, Ipswich, MA, USA).
Sequencing.
Paired-end sequencing (2 reads of 100 bp) was performed on the Illumina HiSeq 2000 platform at BGI-Europe (Copenhagen N, Denmark).
Analysis of sequence data. (i) Preprocessing and depletion of human sequence reads.
Paired-end sequencing reads were trimmed of adapter sequences, and overlapping read pairs were merged using AdapterRemoval software (version 1.5.3) (21). Reads shorter than 30 nucleotides after trimming were excluded from further analysis. For digital subtraction of human sequence reads, the remaining reads were mapped to the human genome (hg38) using the mem algorithm implemented in Burrows-Wheeler Aligner (BWA; version 0.7.7) (22). Reads of a pair were evaluated independently. Reads containing 25 bp or more of low-complexity regions were filtered out using the DustMasker algorithm (version 1.0.0) (23).
(ii) Analysis of reads mapping to P. acnes.
Genomic sequences from 97 P. acnes strains representing complete genomes, scaffolds, or contigs were downloaded from the NCBI genome resource (http://www.ncbi.nlm.nih.gov/genome/genomes/1140) (see Table S1 in the supplemental material). Reads filtered of human and low-complexity sequences were mapped to the P. acnes sequences using the aln algorithm in BWA (22) after duplicate reads had been marked using the SAMBLASTER program (24). Only reads with a perfect match were kept for further analysis. The proportion of P. acnes reads was calculated as the number (parts per million) of unique P. acnes reads relative to the total number of demultiplexed reads.
(iii) Analysis of the P. acnes strains present.
The presence of the different P. acnes strains was assessed on the basis of the mapping to the P. acnes genomic sequences (above). Only reads mapping exclusively to a single strain were considered for this analysis. Information regarding the phylogenetic type of the different strains was obtained from the P. acnes multilocus sequence typing (MLST) database (http://pubmlst.org/pacnes/).
(iv) Analysis of the coverage of the P. acnes genome.
To visualize the genomic coverage of P. acnes, all nonhuman reads were mapped to the 12 full P. acnes genomes currently present in GenBank using the mem algorithm implemented in BWA (version 0.7.7) (22). The resulting alignments are represented as Circos plots (25).
Nucleotide sequence accession numbers.
Sequencing reads mapping to P. acnes have been uploaded to the European Nucleotide Archive (ENA) and are available under accession number PRJEB12636.
RESULTS
P. acnes in microbe-enriched and shotgun-sequenced samples.
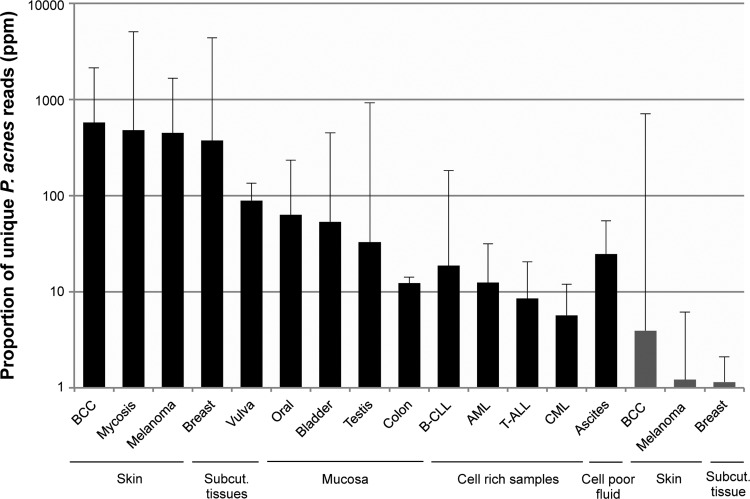
The relative number of P. acnes sequencing reads in 180 samples of 20 different sample types was analyzed (Table 1). Ascitic fluid samples and lymphoma/myeloma cell lines were analyzed as each being one common sample category. One hundred forty-three samples were processed to enrich for intact microbes before extraction of DNA and preparation of sequencing libraries. Sequencing reads from which human reads were removed were mapped to a custom database containing the 97 P. acnes genomes currently present in GenBank as full genomes or genome assemblies. P. acnes levels were assessed as the number (parts per million) of unique P. acnes reads relative to the total number of demultiplexed reads (see Materials and Methods). Using this approach, we detected relative levels of P. acnes reads exceeding 300 ppm in samples from patients with basal cell carcinoma (median, 576 ppm), mycosis fungoides (median, 480 ppm), malignant melanoma (median, 446 ppm), and breast cancer (median, 373 ppm) (Fig. 1, black bars; see also Table S2 in the supplemental material). The proportions of P. acnes reads detected in the remaining enriched samples were below 100 ppm (median, 0.8 to 89 ppm). P. acnes reads were also detected in nontemplate control samples containing PBS run in parallel with the enriched samples (n = 19). In these samples, the relative proportion of P. acnes reads was high (median, 10,377 ppm) (see Discussion).
FIG 1.
Relative P. acnes levels in enriched (black bars) and shotgun-sequenced (gray bars) samples having a median P. acnes proportion of >1 ppm. The proportions of unique P. acnes reads (in parts per million) organized according to tissue type are shown on a logarithmic scale. The error bars indicate 1 standard deviation. BCC, basal cell carcinoma; Subcut., subcutaneous.
Total DNA extracted from 107 samples was subjected to shotgun sequencing (Table 1). Only samples from patients with basal cell carcinoma (median, 3.9 ppm), malignant melanoma (median, 1.2 ppm), and breast cancer (median, 1.1 ppm) contained relative P. acnes levels above 1 ppm (Fig. 1, gray bars; see also Table S2 in the supplemental material). The fold increase in the median number of P. acnes reads (parts per million) in enriched samples relative to the number in shotgun-sequenced samples ranged from 35 in colon cancer biopsy specimens (12 versus 0.3 ppm) to >1,000 in samples from patients with mycosis fungoides (480 versus 0.5 ppm). The blood samples contained the lowest relative P. acnes levels of all samples included in this study (median, 0.05 ppm).
Distribution of P. acnes strains.
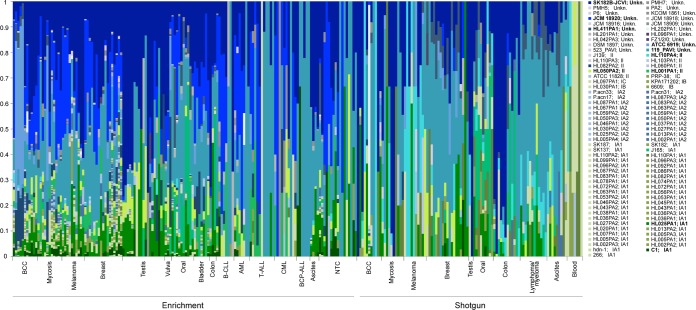
To assess the P. acnes strains and the phylogenetic groups present in our samples, we analyzed the distribution of reads mapping to the 97 P. acnes genomes used in our analysis. Only reads mapping unambiguously to one P. acnes strain were included in the analysis. Using this criterion, an average of 8.6% of the P. acnes reads were included in the analysis (see Table S3 in the supplemental material); however, with this approach we ensured a high stringency when assigning the reads. Figure 2 shows the relative abundance of strains for each sample. The phylogenetic type is indicated next to the individual strain names. For 20 of the strains included, the phylogenetic type was not determined, and these are designated unknown (Unkn.). Overall, two strains predominated across samples for both enriched and shotgun-sequenced samples (Table 2; see also Table S4 in the supplemental material), namely, strains 119_PAVI and SK182B-JCVI (both of unknown phylogenetic type). Strains HL025PA1 and HL050PA2 (type IA1 and type II, respectively) were also among the 10 most abundant strains detected by both methods. Certain strains, such as JCM 18920 (unknown type), HL001PA1 (type II), and C1 (type IA1), were mainly detected in enriched samples, while other strains, such as HL110PA4 (type II) and HL411PA1 (unknown type), were mainly detected in shotgun-sequenced samples.
FIG 2.
Relative distribution of P. acnes strains in the individual samples. The y axis shows the percentage accounted for by each strain, and each vertical line represents a sample. The bottom horizontal lines indicate enriched or shotgun-sequenced samples, and the bottom vertical ticks delimit the sample categories. The phylogenetic type of the P. acnes strains is indicated next to the strain name in the key on the right. The strains mentioned in the main text are highlighted in bold. NTC, nontemplate control; Unkn., unknown.
TABLE 2.
Prevalence of the 20 most frequently detected P. acnes strains in samples investigated by microbial enrichment, shotgun sequencing, and all samples combined
| Strain | Prevalence (%) in: |
||
|---|---|---|---|
| All samples | Microbial enrichment | Shotgun sequencing | |
| 119_PAVI | 82 | 83 | 64 |
| SK182B-JCVI | 78 | 81 | 60 |
| HL025PA1 | 56 | 62 | 36 |
| JCM 18920 | 45 | 66 | 6 |
| HL050PA2 | 44 | 52 | 23 |
| HL001PA1 | 34 | 51 | 4 |
| HL411PA1 | 32 | 26 | 36 |
| C1 | 31 | 46 | 3 |
| HL110PA4 | 29 | 14 | 46 |
| ATCC 6919 | 27 | 32 | 14 |
| JCM 18909 | 26 | 37 | 4 |
| HL082PA2 | 24 | 35 | |
| J165 | 24 | 22 | 22 |
| HL201PA1 | 20 | 29 | |
| HL110PA1 | 19 | 27 | |
| HL030PA1 | 16 | 21 | |
| SK187 | 15 | 22 | |
| HL099PA1 | 15 | 21 | |
| HL078PA1 | 12 | 18 | |
| J139 | 11 | 15 | 4 |
| HL087PA2 | 15 | ||
| KPA171202 | 14 | ||
| HL056PA1 | 7 | ||
| HL110PA3 | 6 | ||
| HL025PA2 | 4 | ||
| HL046PA2 | 3 | ||
| HL074PA1 | 3 | ||
Generally, greater strain diversity was seen in enriched tissue samples (Fig. 2, left, indicated by the striated pattern seen for those samples; see also Fig. S1 in the supplemental material) than in shotgun-sequenced samples, ranging from an average of 10 different strains in samples from patients with testicular cancer to an average of 29 different strains in samples from patients with mycosis fungoides. The remaining samples contained an average of 2 to 9 different strains.
Coverage of the P. acnes genome.
To evaluate the genomic coverage of P. acnes, nonhuman sequencing reads mapping to the 12 full genomes were presented using the Circos program (see Fig. S2 at http://www.cbs.dtu.dk/∼txema/Supplementary/propioni_web/index.html). The plots show evenly distributed sequencing reads leading to almost full coverage of the P. acnes genome in some samples, whereas more sporadic coverage was seen in samples containing lower P. acnes levels. A disproportionately high sequencing depth was seen in two or three regions, depending on which reference genome was used for mapping, corresponding to the regions containing the genes encoding the rRNAs. The high depth of sequencing could be a result of reads originating from bacteria other than P. acnes mapping to the rRNA regions as a result of high sequence similarity between different bacterial rRNAs, as the algorithm used for generating the plots allows some mismatches, unlike the more stringent mapping algorithm used for quantification. To investigate if an excess of reads in the rRNA regions impacted the quantitative proportions of P. acnes, we reanalyzed these data by excluding reads mapping to the rRNA regions. For the majority of the samples, the reanalyzed values of the median relative levels of unique P. acnes reads were <1% lower than the values presented above, except for some sample types containing very low numbers of P. acnes reads (see Table S2 in the supplemental material). These results indicate that an excess of reads mapping to rRNA regions generally was not found in our quantification analysis.
DISCUSSION
During metagenomic analysis of next-generation sequencing data from patient samples, we identified considerable amounts of genetic material from the bacterium P. acnes. P. acnes is known to contaminate human samples, and this initial discovery therefore prompted us to investigate the abundance of P. acnes in metagenomic data from a range of different human clinical samples. In the present study, we investigated the proportions of P. acnes DNA in data from samples subjected to enrichment for intact microbes and from samples subjected to shotgun sequencing of the total pool of DNA. We detected the highest relative levels of P. acnes in samples subjected to enrichment, with relative P. acnes levels being the highest in skin tissue-derived samples and breast cancer biopsy specimens (Fig. 1). Basal cell carcinoma samples contained the highest P. acnes levels among the shotgun-sequenced samples, while most other shotgun-sequenced samples as well as enriched samples from patients with BCP-ALL contained P. acnes at levels below 1 ppm (see Table S2 in the supplemental material).
Prior to surgical procedures or penetration of the skin for sampling of blood, the skin is routinely sterilized using antiseptics. However, antiseptic preparation of the skin does not kill all bacteria or affect bacteria present in the deeper layers of the skin (26, 27), and bacteria from surrounding areas of the skin may rapidly recolonize the antiseptically treated areas (28, 29). Moreover, antiseptics do not remove extracellular bacterial DNA present on the skin. Therefore, detection of P. acnes in skin tissue samples is not unexpected. As the breast tissue lies just beneath the subcutaneous adipose tissue in an area of the skin rich in P. acnes, contamination from the skin during surgery is conceivable.
In our study, microbial enrichment was achieved by reducing the amount of host endogenous DNA by centrifugation and filtration followed by nuclease treatment of fluid samples or tissue homogenates. This procedure is widely used for enrichment of intact viral particles in virus discovery studies (30–33) and results in an increase in the amount of intact viral nucleic acids relative to the amount of host nucleic acids (34). We did not culture bacteria from the samples, and therefore, we do not know if the detected bacterial DNA originated from live bacteria capable of forming colonies. As the DNA of live bacteria is protected from degradation by DNases (35, 36), the up to >1,000-fold increase in the median proportion of P. acnes reads achieved upon enrichment could suggest that the P. acnes DNA detected in our samples at least partially originated from live bacteria.
Over the years, P. acnes has been detected in samples from patients with diverse diseases, which has led to its proposed association with several conditions. Among these, P. acnes has been implicated in the pathogenesis of acne vulgaris, although its role in this condition is still debated (37). Other diseases for which the bacterium is proposed to play a role are sarcoidosis (9), diverse spine conditions (15–17), SAPHO syndrome (8), aortic aneurysms (18), and prostate cancer (19). Common to these studies is that detection of P. acnes is based on culture, PCR, or a combination of the two. In most of the studies, roughly half of the samples are positive, and the identified bacterium is P. acnes in about half of the cases. Besides the proposed role in certain diseases and syndromes, P. acnes is considered a contaminant of blood products (11, 38), surgical wounds (12, 39), and heart valve tissue cultures used in the diagnosis of infection during valve replacement surgery (10). The fact that P. acnes is often detected at similar frequencies in surgical wounds and samples from patients with various diseases could imply that the detection of P. acnes is incidental as a result of its widespread presence. Studies investigating the role of P. acnes or other ubiquitous bacteria in infections or diseases should therefore be designed to allow researchers to distinguish between contamination and true infection. Approaches suggested to help distinguish between these include the performance of multiple cultures (11), histopathological analysis (7), distinguishing of single bacterial cells from biofilm-related infections by immunofluorescence microscopy (12), and investigation of the ability of P. acnes isolates to form biofilm (40). The invasiveness of P. acnes isolates may also hint to a possible disease-causing role of P. acnes in some cases (41). MLST schemes have revealed three major phylogenetic groups of P. acnes, types I, II, and III (42, 43), with type I being further divided into types IA1, IA2, IB, and IC. Clustering analysis found acne vulgaris to be predominantly associated with certain clonal complexes within type IA1, while types IB and II were more frequently recovered from soft tissue and retrieved medical devices. Certain P. acnes sequence types from types II and III were recovered only from healthy skin (44).
In our samples, recurring P. acnes strains predominated across sample types, including in the nontemplate controls (see Table S4 in the supplemental material). Two of these strains were also ubiquitously detected in skin microbiomes (3). These findings could suggest that some strains are widespread among individuals or that they arise from reagent or lab contamination, as P. acnes or propionibacteria are well-known contaminants of next-generation sequencing data (45–47). Our nontemplate controls containing PBS run in parallel with the enriched samples contained high relative levels of P. acnes. This could occur as a result of cross-contamination during sample processing, contamination from lab personnel or the reagents used, or bleeding over from libraries with high P. acnes levels sequenced on the same lanes of the flow cell (34, 48). With the exception of two, all the nontemplate control libraries contained DNA at levels below the detection limit of both the Qubit and Bioanalyzer instruments (data not shown), but the libraries were nevertheless sequenced. Comparison of the nontemplate control data sets to the data sets for the other enriched samples showed that the controls contained an average of 12% human reads, while the biological samples contained an average of 91% human reads (see Table S2 in the supplemental material). This dissimilar composition of the data sets suggests that the sequence reads in the controls, including the P. acnes reads, did not mainly originate from cross contamination or bleeding over. Studies investigating contamination in next-generation sequencing data show that more dilute samples contain higher fractions of contaminating reads (46, 47) and that negative controls contain reads mapping to a variety of bacterial genera, including propionibacteria (47). This could be explained by the more extensive PCR amplification of contaminant templates from samples containing nearly no DNA than of the same amount of contaminant templates from complex samples containing human DNA as well competing for the same quantity of primers and other PCR reagents. Considering this skewed amplification and the circumstance that the biological data sets are diluted with a large amount of endogenous human reads, the relative level of P. acnes reads in the controls becomes less astounding. The detection of P. acnes in the controls can thus be interpreted as further evidence of how easily samples are contaminated with this ubiquitous bacterium, regardless of the source.
We believe that the high relative P. acnes levels detected, particularly in enriched skin tissue and breast cancer biopsy samples, reflect patient-derived contamination. Similar patterns showing traces of P. acnes in most sample types and higher levels in samples from P. acnes-rich body sites have been seen in other microbiome studies (49). Our hypothesis is supported by the high diversity of P. acnes strains seen in these samples compared to the low diversity seen in other samples, including the controls (see Fig. S1 in the supplemental material). The low strain diversity in the controls supports the hypothesis that overamplification occurs in these samples. More adequate negative controls for P. acnes levels in our study were the blood samples from colon cancer patients, as these are, in principle, sterile samples containing high levels of endogenous DNA. In these samples, we detected practically no P. acnes reads (median, 5 in absolute numbers, or 0.05 ppm; see Table S2 in the supplemental material).
Considering our findings, it is possible that contamination derived from the patient, the hospital personnel, or the lab personnel or that arises during sample processing and analysis contributes to the presence of P. acnes DNA in samples processed in clinical and molecular laboratories. The risk of contamination should therefore be considered carefully when interpreting molecular and sequencing data showing the presence of P. acnes, and negative controls should always be included. Typing of P. acnes can be a relevant tool when tracing the origin of P. acnes, but it does not necessarily provide clinically relevant information, as presumed cutaneous P. acnes strains are readily detected in diverse sample types. Efforts should be made to exclude the possibility that P. acnes originates from contamination and, importantly, to prove the causal relationship between this bacterium and the disease in question.
Supplementary Material
ACKNOWLEDGMENTS
We thank Eske Willerslev for support and the use of laboratories. We thank Robert Gniadecki (Bispebjerg Hospital), Christian von Buchwald (Rigshospitalet), Ewa Rajpert-De Meyts (Rigshospitalet), Line Groth-Pedersen (Rigshospitalet), Torben Steinike (Aarhus University Hospital), Hans Johnsen and Karen Dybkaer (Aalborg University Hospital), Estrid Høgdal (The Danish Cancer Biobank), Peter Hokland (Aarhus University Hospital), Ulrik Baandrup (Aalborg University Hospital), Jacob Rosenberg (Herlev Hospital), Zoltan Szallasi (Technical University of Denmark), Ildikó Vereczkey (National Institute of Oncology, Budapest, Hungary), Imre Pete (National Institute of Oncology, Department of Gynecology, Budapest, Hungary), and Zsolt Baranyai (1st Department of Surgery, Semmelweis University, Budapest, Hungary) for providing clinical samples. We thank BGI Europe for sequencing of the samples.
L.P.N., A.J.H., S.M., T.M., L.V., and S.B. conceived the concept and designed the experiments. S.M., L.V., S.R.R., and H.F. performed the experiments. T.M., J.F.-N., J.M.G.I., and T.A.H. conducted the computational analysis. S.M., T.M., A.J.H., L.V., J.F.-N., J.M.G.I., and J.A.R.H. analyzed the computational data. S.M. prepared the manuscript. All authors provided critical revisions to and approved the manuscript.
We declare no conflicts of interest.
The work was supported by the Innovation Fund Denmark (The GenomeDenmark platform, grant no. 019-2011-2), the Danish National Research Foundation (grant no. DNRF94), and the Lundbeck Foundation.
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JCM.02723-15.
REFERENCES
- 1.Brook I, Frazier EH. 1991. Infections caused by Propionibacterium species. Clin Infect Dis 13:819–822. doi: 10.1093/clinids/13.5.819. [DOI] [PubMed] [Google Scholar]
- 2.Grice EA, Kong HH, Conlan S, Deming CB, Davis J, Young AC, NISC Comparative Sequencing Program, Bouffard GG, Blakesley RW, Murray PR, Green ED, Turner ML, Segre JA. 2009. Topographical and temporal diversity of the human skin microbiome. Science 324:1190–1192. doi: 10.1126/science.1171700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Oh J, Byrd AL, Deming C, Conlan S, NISC Comparative Sequencing Program, Kong HH, Segre JA. 2014. Biogeography and individuality shape function in the human skin metagenome. Nature 514:59–64. doi: 10.1038/nature13786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fitz-Gibbon S, Tomida S, Chiu B-H, Nguyen L, Du C, Liu M, Elashoff D, Erfe MC, Loncaric A, Kim J, Modlin RL, Miller JF, Sodergren E, Craft N, Weinstock GM, Li H. 2013. Propionibacterium acnes strain populations in the human skin microbiome associated with acne. J Investig Dermatol 133:2152–2160. doi: 10.1038/jid.2013.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Williams HC, Dellavalle RP, Garner S. 2012. Acne vulgaris. Lancet 379:361–372. doi: 10.1016/S0140-6736(11)60321-8. [DOI] [PubMed] [Google Scholar]
- 6.Achermann Y, Goldstein EJC, Coenye T, Shirtliff ME. 2014. Propionibacterium acnes: from commensal to opportunistic biofilm-associated implant pathogen. Clin Microbiol Rev 27:419–440. doi: 10.1128/CMR.00092-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sohail MR, Gray AL, Baddour LM, Tleyjeh IM, Virk A. 2009. Infective endocarditis due to Propionibacterium species. Clin Microbiol Infect 15:387–394. doi: 10.1111/j.1469-0691.2009.02703.x. [DOI] [PubMed] [Google Scholar]
- 8.Colina M, Monaco AL, Khodeir M. 2007. Propionibacterium acnes and SAPHO syndrome: a case report and literature review. Clin Exp Rheumatol 25:457–460. [PubMed] [Google Scholar]
- 9.Eishi Y, Suga M, Ishige I, Kobayashi D, Yamada T, Takemura T, Takizawa T, Koike M, Kudoh S, Costabel U, Guzman J, Rizzato G, Gambacorta M, du Bois R, Nicholson AG, Sharma OP, Ando M. 2002. Quantitative analysis of mycobacterial and propionibacterial DNA in lymph nodes of Japanese and European patients with sarcoidosis. J Clin Microbiol 40:198–204. doi: 10.1128/JCM.40.1.198-204.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Campbell WN, Tsai W, Mispireta LA. 2000. Evaluation of the practice of routine culturing of native valves during valve replacement surgery. Ann Thorac Surg 69:548–550. doi: 10.1016/S0003-4975(99)01354-5. [DOI] [PubMed] [Google Scholar]
- 11.Kunishima S, Inoue C, Kamiya T, Ozawa K. 2001. Presence of Propionibacterium acnes in blood components. Transfusion 41:1126–1129. doi: 10.1046/j.1537-2995.2001.41091126.x. [DOI] [PubMed] [Google Scholar]
- 12.McLorinan GC, Glenn JV, McMullan MG, Patrick S. 2005. Propionibacterium acnes wound contamination at the time of spinal surgery. Clin Orthop Relat Res 437:67–73. [DOI] [PubMed] [Google Scholar]
- 13.Rood IGH, de Korte D, Savelkoul PHM, Pettersson A. 2011. Molecular relatedness of Propionibacterium species isolated from blood products and on the skin of blood donors. Transfusion 51:2118–2124. doi: 10.1111/j.1537-2995.2011.03139.x. [DOI] [PubMed] [Google Scholar]
- 14.Abe C, Iwai K, Mikami R, Hosoda Y. 1984. Frequent isolation of Propionibacterium acnes from sarcoidosis lymph nodes. Zentralbl Bakteriol Mikrobiol Hyg A 256:541–547. [DOI] [PubMed] [Google Scholar]
- 15.Stirling A, Worthington T, Rafiq M, Lambert PA, Elliott TS. 2001. Association between sciatica and Propionibacterium acnes. Lancet 357:2024–2025. doi: 10.1016/S0140-6736(00)05109-6. [DOI] [PubMed] [Google Scholar]
- 16.Albert HB, Lambert P, Rollason J, Sorensen JS, Worthington T, Pedersen MB, Nørgaard HS, Vernallis A, Busch F, Manniche C, Elliott T. 2013. Does nuclear tissue infected with bacteria following disc herniations lead to Modic changes in the adjacent vertebrae? Eur Spine J 22:690–696. doi: 10.1007/s00586-013-2674-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Arndt J, Charles YP, Koebel C, Bogorin I, Steib J-P. 2012. Bacteriology of degenerated lumbar intervertebral disks. J Spinal Disord Tech 25:E211–E216. doi: 10.1097/BSD.0b013e318269851a. [DOI] [PubMed] [Google Scholar]
- 18.Marques da Silva R, Caugant DA, Eribe ERK, Aas JA, Lingaas PS, Geiran O, Tronstad L, Olsen I. 2006. Bacterial diversity in aortic aneurysms determined by 16S ribosomal RNA gene analysis. J Vasc Surg 44:1055–1060. doi: 10.1016/j.jvs.2006.07.021. [DOI] [PubMed] [Google Scholar]
- 19.Mak TN, Yu S-H, De Marzo AM, Brüggemann H, Sfanos KS. 2013. Multilocus sequence typing (MLST) analysis of Propionibacterium acnes isolates from radical prostatectomy specimens. Prostate 73:770–777. doi: 10.1002/pros.22621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Seguin-Orlando A, Schubert M, Clary J, Stagegaard J, Alberdi MT, Prado JL, Prieto A, Willerslev E, Orlando L. 2013. Ligation bias in Illumina next-generation DNA libraries: implications for sequencing ancient genomes. PLoS One 8:e78575. doi: 10.1371/journal.pone.0078575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lindgreen S. 2012. AdapterRemoval: easy cleaning of next-generation sequencing reads. BMC Res Notes 5:337. doi: 10.1186/1756-0500-5-337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li H, Durbin R. 2010. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 26:589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Morgulis A, Gertz EM, Schäffer AA, Agarwala R. 2006. A fast and symmetric DUST implementation to mask low-complexity DNA sequences. J Comput Biol 13:1028–1040. doi: 10.1089/cmb.2006.13.1028. [DOI] [PubMed] [Google Scholar]
- 24.Faust GG, Hall IM. 2014. SAMBLASTER: fast duplicate marking and structural variant read extraction. Bioinformatics 30:2503–2505. doi: 10.1093/bioinformatics/btu314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, Jones SJ, Marra MA. 2009. Circos: an information aesthetic for comparative genomics. Genome Res 19:1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gårdlund B. 2007. Postoperative surgical site infections in cardiac surgery—an overview of preventive measures. APMIS 115:989–995. doi: 10.1111/j.1600-0463.2007.00845.x. [DOI] [PubMed] [Google Scholar]
- 27.Ostrander RV, Brage ME, Botte MJ. 2003. Bacterial skin contamination after surgical preparation in foot and ankle surgery. Clin Orthop Relat Res 406:246. doi: 10.1097/00003086-200301000-00035. [DOI] [PubMed] [Google Scholar]
- 28.Johnston DH, Fairclough JA, Brown EM, Morris R. 1987. Rate of bacterial recolonization of the skin after preparation: four methods compared. Br J Surg 74:64. doi: 10.1002/bjs.1800740121. [DOI] [PubMed] [Google Scholar]
- 29.Falk-Brynhildsen K, Friberg O, Soderquist B, Nilsson UG. 2013. Bacterial colonization of the skin following aseptic preoperative preparation and impact of the use of plastic adhesive drapes. Biol Res Nurs 15:242–248. doi: 10.1177/1099800411430381. [DOI] [PubMed] [Google Scholar]
- 30.Allander T, Emerson SU, Engle RE, Purcell RH, Bukh J. 2001. A virus discovery method incorporating DNase treatment and its application to the identification of two bovine parvovirus species. Proc Natl Acad Sci U S A 98:11609–11614. doi: 10.1073/pnas.211424698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Victoria JG, Kapoor A, Dupuis K, Schnurr DP, Delwart EL. 2008. Rapid identification of known and new RNA viruses from animal tissues. PLoS Pathog 4:e1000163. doi: 10.1371/journal.ppat.1000163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Daly GM, Bexfield N, Heaney J, Stubbs S, Mayer AP, Palser A, Kellam P, Drou N, Caccamo M, Tiley L, Alexander GJM, Bernal W, Heeney JL. 2011. A viral discovery methodology for clinical biopsy samples utilising massively parallel next generation sequencing. PLoS One 6:e28879. doi: 10.1371/journal.pone.0028879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Alquezar-Planas DE, Mourier T, Bruhn CAW, Hansen AJ, Vitcetz SN, Mørk S, Gorodkin J, Nielsen HA, Guo Y, Sethuraman A, Paxinos EE, Shan T, Delwart EL, Nielsen LP. 2013. Discovery of a divergent HPIV4 from respiratory secretions using second and third generation metagenomic sequencing. Sci Rep 3:2468. doi: 10.1038/srep02468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jensen RH, Mollerup S, Mourier T, Hansen TA, Fridholm H, Nielsen LP, Willerslev E, Hansen AJ, Vinner L. 2015. Target-dependent enrichment of virions determines the reduction of high-throughput sequencing in virus discovery. PLoS One 10:e0122636. doi: 10.1371/journal.pone.0122636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rice KC, Bayles KW. 2008. Molecular control of bacterial death and lysis. Microbiol Mol Biol Rev 72:85–109. doi: 10.1128/MMBR.00030-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pezzulo AA, Kelly PH, Nassar BS, Rutland CJ, Gansemer ND, Dohrn CL, Costello AJ, Stoltz DA, Zabner J. 2013. Abundant DNase I-sensitive bacterial DNA in healthy porcine lungs and its implications for the lung microbiome. Appl Environ Microbiol 79:5936–5941. doi: 10.1128/AEM.01752-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shaheen B, Gonzalez M. 2011. A microbial aetiology of acne: what is the evidence? Br J Dermatol 165:474–485. doi: 10.1111/j.1365-2133.2011.10375.x. [DOI] [PubMed] [Google Scholar]
- 38.Justesen US, Skov MN, Knudsen E, Holt HM, Søgaard P, Justesen T. 2010. 16S rRNA gene sequencing in routine identification of anaerobic bacteria isolated from blood cultures. J Clin Microbiol 48:946–948. doi: 10.1128/JCM.02075-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nandyala SV, Schwend RM. 2013. Prevalence of intraoperative tissue bacterial contamination in posterior pediatric spinal deformity surgery. Spine 38:E482–E486. doi: 10.1097/BRS.0b013e3182893be1. [DOI] [PubMed] [Google Scholar]
- 40.Holmberg A, Lood R, Mörgelin M, Soderquist B, Holst E, Collin M, Christensson B, Rasmussen M. 2009. Biofilm formation by Propionibacterium acnes is a characteristic of invasive isolates. Clin Microbiol Infect 15:787–795. doi: 10.1111/j.1469-0691.2009.02747.x. [DOI] [PubMed] [Google Scholar]
- 41.Furukawa A, Uchida K, Ishige Y, Ishige I, Kobayashi I, Takemura T, Yokoyama T, Iwai K, Watanabe K, Shimizu S, Ishida N, Suzuki Y, Suzuki TG, Yamada T, Ito T, Eishi Y. 2009. Characterization of Propionibacterium acnes isolates from sarcoid and non-sarcoid tissues with special reference to cell invasiveness, serotype, and trigger factor gene polymorphism. Microb Pathog 46:80–87. doi: 10.1016/j.micpath.2008.10.013. [DOI] [PubMed] [Google Scholar]
- 42.Lomholt HB, Kilian M. 2010. Population genetic analysis of Propionibacterium acnes identifies a subpopulation and epidemic clones associated with acne. PLoS One 5:e12277. doi: 10.1371/journal.pone.0012277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McDowell A, Gao A, Barnard E, Fink C, Murray PI, Dowson CG, Nagy I, Lambert PA, Patrick S. 2011. A novel multilocus sequence typing scheme for the opportunistic pathogen Propionibacterium acnes and characterization of type I cell surface-associated antigens. Microbiology 157:1990–2003. doi: 10.1099/mic.0.049676-0. [DOI] [PubMed] [Google Scholar]
- 44.McDowell A, Barnard E, Nagy I, Gao A, Tomida S, Li H, Eady A, Cove J, Nord CE, Patrick S. 2012. An expanded multilocus sequence typing scheme for Propionibacterium acnes: investigation of ‘pathogenic', ‘commensal’ and antibiotic resistant strains. PLoS One 7:e41480. doi: 10.1371/journal.pone.0041480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Strong MJ, Xu G, Morici L, Bon-Durant SS, Baddoo M, Lin Z, Fewell C, Taylor CM, Flemington EK. 2014. Microbial contamination in next generation sequencing: implications for sequence-based analysis of clinical samples. PLoS Pathog 10:e1004437. doi: 10.1371/journal.ppat.1004437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lusk RW. 2014. Diverse and widespread contamination evident in the unmapped depths of high throughput sequencing data. PLoS One 9:e110808. doi: 10.1371/journal.pone.0110808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Salter SJ, Cox MJ, Turek EM, Calus ST, Cookson WO, Moffatt MF, Turner P, Parkhill J, Loman NJ, Walker AW. 2014. Reagent and laboratory contamination can critically impact sequence-based microbiome analyses. BMC Biol 12:87. doi: 10.1186/s12915-014-0087-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kircher M, Sawyer S, Meyer M. 2012. Double indexing overcomes inaccuracies in multiplex sequencing on the Illumina platform. Nucleic Acids Res 40:e3. doi: 10.1093/nar/gkr771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Human Microbiome Project Consortium. 2012. Structure, function and diversity of the healthy human microbiome. Nature 486:207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.