Abstract
Overexpression of HOXA/MEIS1/PBX3 homeobox genes is the hallmark of mixed lineage leukemia (MLL)-rearranged acute myeloid leukemia (AML). HOXA9 and MEIS1 are considered to be the most critical targets of MLL fusions and their co-expression rapidly induces AML. MEIS1 and PBX3 are not individually able to transform cells and were therefore hypothesized to function as cofactors of HOXA9. However, in this study we demonstrate that co-expression of PBX3 and MEIS1 (PBX3/MEIS1), without ectopic expression of a HOX gene, is sufficient for transformation of normal mouse hematopoietic stem/progenitor cells in vitro. Moreover, PBX3/MEIS1 overexpression also caused AML in vivo, with a leukemic latency similar to that caused by forced expression of MLL-AF9, the most common form of MLL fusions. Furthermore, gene expression profiling of hematopoietic cells demonstrated that PBX3/MEIS1 overexpression, but not HOXA9/MEIS1, HOXA9/PBX3 or HOXA9 overexpression, recapitulated the MLL-fusion-mediated core transcriptome, particularly upregulation of the endogenous Hoxa genes. Disruption of the binding between MEIS1 and PBX3 diminished PBX3/MEIS1-mediated cell transformation and HOX gene upregulation. Collectively, our studies strongly implicate the PBX3/MEIS1 interaction as a driver of cell transformation and leukemogenesis, and suggest that this axis may play a critical role in the regulation of the core transcriptional programs activated in MLL-rearranged and HOX-overexpressing AML. Therefore, targeting the MEIS1/PBX3 interaction may represent a promising therapeutic strategy to treat these AML subtypes.
Keywords: PBX3, MEIS1, HOXA, homeobox gene, MLL-rearranged leukemia, core transcriptome, gene expression profiles, cell transformation, leukemogenesis
INTRODUCTION
The mixed lineage leukemia (MLL) gene, located on human chromosome 11 band q23 (11q23), is a common target of chromosomal translocations in acute leukemia. MLL-rearranged leukemia accounts for 5-10% of patients with acute myeloid leukemia (AML) and 7-10% of patients with acute lymphoblastic leukemia (ALL), ~80% of infant acute leukemia, and the majority of patients with therapy-related AML/ALL secondary to therapy that targets topoisomerase II (like etoposide) (1-7). The critical feature of MLL-rearrangements is the generation of a chimeric transcript consisting of 5' MLL and 3' sequences of one of more than 60 different partner genes (2, 3, 6, 7). MLL-AF9, resulting from t(9;11)(p22;q23), is the most common form of MLL-fusion genes in AML (2, 6).
Aberrant expression of a group of homeobox genes including the Cluster A HOX genes and genes encoding HOX cofactors, e.g., MEIS1 (but not MEIS2 or MEIS3) and PBX3 (but not PBX1 or PBX2), is the hallmark of MLL-rearranged leukemias (8-16). HOXA9 and MEIS1 are the two most well-studied downstream target genes of MLL-fusion proteins; their aberrant overexpression has been considered to be required for the induction and maintenance of MLL-rearranged leukemia (11, 17-19), and their coexpression is sufficient to transform cells and induce rapid leukemia (12, 20-23). In contrast, although PBX3 is also significantly up-regulated in MLL-rearranged AML (11-15). the role of PBX3 in leukemogenesis was largely unappreciated. Pbx proteins have been shown to be required for linking Hoxa and Meis1 proteins together (24, 25). However, the previous studies of Pbx genes were focusing on Pbx1, the founding member of the Pbx family, which was shown to have no synergistic effect with Hoxa9 in cell transformation and leukemogenesis (12, 20, 23). Instead, we recently showed that PBX3 and three HOXA genes (HOXA7, HOXA9 and HOXA11) composed of an independent predictor of unfavorable survival of patients with cytogenetically abnormal AML (CA-AML) (16). We showed further that PBX3, rather than PBX1 or PBX2, is an important cofactor of HOXA9 in cytogenetically abnormal AML and their co-expression can cause development of rapid AML in mice (26). The prognostic impact and essential oncogenic role of PBX3 have also been observed in cytogenetic normal AML (27).
Thus far, both MEIS1 and PBX3 have been shown to be essential co-factors of HOXA9 and co-expression of HOXA9 with either MEIS1 or PBX3 can induce rapid AML (12, 20-23, 26). However, unlike HOXA9, neither MEIS1 nor PBX3 alone can transform normal hematopoietic stem/progenitor cells (HSPCs) (20, 21, 23, 26). Thus, MEIS1 and PBX3 were simply recognized as cofactors of HOX proteins (especially HOXA9) and it was believed that they exert their function solely or mainly through cooperating with HOX proteins (18, 24-28). Surprisingly, here we show that without ectopic expression of a HOX gene, co-expression of PBX3 and MEIS1 is sufficient to transform normal HSPCs and induce rapid AML in mice. More interestingly, our genome-wide gene expression profiling analysis shows that forced expression of PBX3/MEIS1, but not that of HOXA9/MEIS1, HOXA9/PBX3 or HOXA9 alone, can induce the core transcriptome (especially, the up-regulation of endogenous Hoxa genes, Pbx3 and Meis1) of MLL-rearranged AML in mice.
MATERIALS AND METHODS
Retroviral constructs
MSCVneo-HOXA9 and MSCV-PIG-PBX3 plasmids with human gene HOXA9 and PBX3 coding regions, respectively, were constructed previously by us (26). MSCVneo-MLL-AF9 is a kind gift from Dr. Scott Armstrong (29). MSCV-PIG vector containing a PGK-puromycin-IRES-GFP (PIG) cassette was kindly provided by Drs. Hannon and He (30). MEIS1 coding region sequence was PCR amplified from human normal BM mononuclear cells with primers, forward 5’- ATAGAATTCATGGCGCAAAGGTAC-3’, and reverse 5’- GGCCTCGAGTAGATGAAGGTTACA -3’, was then cloned into MSCVneo (Clontech, Mountain View, CA) as MSCVneo-MEIS1, or cloned into MSCV-PIG as MSCV-PIG-MEIS1. Mouse Pbx3 coding region sequence was PCR amplified from mouse normal BM mononuclear cells with primers, forward 5’- AATAGATCTACCACCATGGACGATCAATCCAGGATG-3’, and reverse 5’- ACTCTCGAGTTAGTTAGAG GTATCCGAGTGC-3’, was then cloned into MSCV-PIG, as MSCV-PIG-Pbx3. Wild-type and mutant mouse Meis1 coding regions were PCR amplified from MSCVpuro-VP16-Meis1 (WT), MSCVpuro-VP16-Meis1-M2ΔLRF/ΔLLEL, and MSCVpuro-VP16-Meis1-Δ64-202, which were kindly provided by Dr. Kamps (24, 25), and then subcloned into MSCVneo, as MSCVneo-Meis1, MSCVneo-Meis1-M2ΔLRF/ΔLLEL, and MSCVneo-Meis1-Δ64-202, respectively. All inserts have been confirmed by Sanger sequencing.
Mouse bone marrow transplantation (BMT; in vivo reconstitution) assays
For primary BMT assays, mouse BM progenitor cells from 4- to 6-week-old wild-type (C57BL/6J CD45.2) mice were co-transduced with different retroviral combinations, and were then cultured in methylcellulose medium to select double-transduction positive cells. Seven days later, colony cells were collected and washed, and were then injected by tail vein into lethally irradiated (960 rads) 8- to 10-week-old B6.SJL (CD45.1) recipient mice with 0.5×106 donor cells plus a radioprotective dose of whole BM cells (1×106; freshly harvested from a B6.SJL mouse) per recipient mouse.
For secondary BMT assays, primary leukemic BM cells were obtained from the primary BMT recipients of the HOXA9+MEIS1, HOXA9+PBX3, PBX3+MEIS1 and MLL-AF9 groups, when the mice developed with full-blown AML. The CD45.1+ leukemic BM cells were sorted by flow cytometry and then injected through tail vein into lethally irradiated (850 rads) 8- to 10-week-old B6.SJL (CD45.1) secondary recipient mice with 1×105 donor cells plus 1.8×106 whole BM cells (from a B6.SJL mouse) per recipient mouse. Leukemic BM cells from two independent primary leukemic mice were used as the donors for the secondary BMT of each group, with 5-6 recipients per donor. Another secondary transplantation assay was conducted with leukemic spleen cells (CD45.1+) obtained from the spleen of primary HOXA9+MEIS1, PBX3+MEIS1 and MLL-AF9 AML mice, with 1×105 donor cells (CD45.1) plus 1.8×106 radioprotective whole BM cells (CD45.2) per recipient mouse.
Affymetrix gene arrays of mouse samples
A total of 20 mouse BM samples from the Control (NC; n=5), HOXA9+MEIS1 (n=3), HOXA9+PBX3 (n=3), HOXA9 (n=3), PBX3+MEIS1 (n=3), and MLL-AF9 (n=3) AML groups were analyzed with Affymetrix GeneChip Mouse Gene 1.0 ST Array (Affymetirx). For each sample, the CD45.1+ BM cells (i.e., transplanted donor cells) were sorted with flow cytometry from whole BM cells collected from BMT recipients at the end point. 1 μg total RNA was used for each sample. The QC test and array assays were done in the core facility of National Human Genome Research Institute, NIH (Bethesda, MD). The quantified signals were normalized using Robust Multi-array Average (RMA) (31), after hybridization and background correction according to the standard protocols. All genes with a standard deviation of expression values over 0.3 across the 20 samples were used for the unsupervised hierarchical clustering and followed statistic analyses. The microarray data has been deposited into the GEO database (GSE68643; Reviewer access: http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token=clorwkcodjuxzyl&acc=GSE68643).
Gene set analyses
Gene Set Enrichment Analysis (GSEA) (32) was used to analyze gene sets enriched in different groups of samples. KEGG (Kyoto Encyclopedia of Genes and Genomes Pathway Database) gene set (33) and “chemical and genetic perturbation” gene set obtained from MsigDB (The Molecular Signatures Database) (32) were used as the gene set input for GSEA. The Database for Annotation, Visualization and Integrated Discovery (DAVID; v6.7) (34, 35) was used to analyze gene sets enriched in genes significantly differentially expressed between different groups of samples.
RNA extraction, quantitative RT-PCR (qPCR), flow cytometry, cytospin, histological hematoxylin and eosin (H&E) staining, and Western blotting
These assays were conducted as described previously (16, 36-39) with some modifications. Antibodies for Pbx3 (sc-891), Hoxa9 (sc-17155), Hoxa7 (sc-17152) and Meis1 (sc-10599) were purchased from Santa Cruz Biotechnology, Inc (Dallas, TX). Anti-Hoxa5 antibody (ab140636) was purchased from Abcam (Cambridge, MA). Anti-Hoxa4 antibody (PA1603) was purchased from Boster Biological Technology (Pleasanton, CA). Anti- β-Actin antibody (#3700) was purchased from Cell Signaling Technology, LTD (Danvers, MA).
Statistical analyses
Overall Survival was estimated according to the method of Kaplan and Meier, and the log rank test was used to assess statistical significance. The Kaplan-Meier method, log-rank test, and t-test were performed with Partek Genomics Suite (Partek Inc, St. Louis, MI), WinSTAT (R. Fitch Software), and/or Bioconductor R packages. Any p values less than 0.05 were considered statistically significant.
Human AML cell line and siRNA transfection
MonoMac-6 AML cell line, carrying t(9;11)/MLL-AF9, was maintained in the Chen laboratory and cultured as described previously (36-38). Continued testing to authenticate this cell line was done using qPCR and Western blot to validate the existence of MLL-AF9 when this line was used in this project. Anti-PBX3 or anti-MEIS1 siRNA oligos and control siRNA oligos were purchased from Dharmacon (Catalog #: L-020121-00-0005, L-011726-00-0005 and D-001810-10-05; GE Healthcare Bio-Sciences, Pittsburgh, PA), and transfected into MonoMac-6 cells Amaxa® Nucleofector® Technology (Amaxa Biosystems, Berlin, Germany) at a concentration of 100 nM as described previously (16, 36-38).
RESULTS
Co-expression of PBX3 and MEIS1 is sufficient to transform normal HSPCs
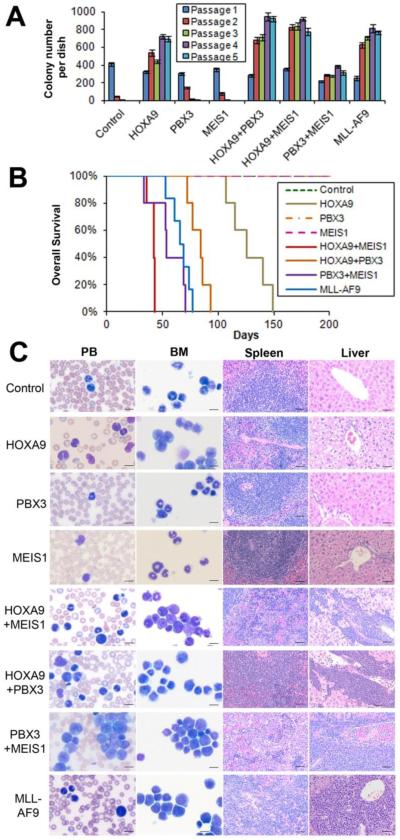
Based on previous studies from us and others showing crucial roles of both PBX3 and MEIS1 in AML (16, 24-28), we query whether co-expression of PBX3/MEIS1 can mimic that of HOXA9/MEIS1 or HOXA9/PBX3 in inducing cell transformation. To this end, we conducted in vitro colony-forming/replating assays. As shown in Figure 1A, PBX3/MEIS1 co-expression (PBX3+MEIS1) was sufficient to transform and immortalize normal mouse bone marrow (BM) progenitor cells, although with relatively fewer colonies compared to co-expression of HOXA9/MEIS1 (HOXA9+MEIS1) or HOXA9/PBX3 (HOXA9+PBX3), or forced expression of MLL-AF9. As expected, HOXA9 alone, but not MEIS1 or PBX3 alone, also transformed mouse BM progenitor cells (Figure 1A).
Figure 1. Co-expression of PBX3 and MEIS1 can transform normal mouse bone marrow (BM) progenitor cells in vitro and induce rapid AML in vivo.
(A) In vitro colony-forming/replating assays. Briefly, mouse normal BM progenitor (lineage negative; Lin-) cells collected from 4- to 6-week-old B6.SJL (CD45.1) mice were retrovirally co-transduced with MSCVneo+MSCV-PIG (Control), MSCVneo-HOXA9+MSCV-PIG (HOXA9), MSCVneo+MSCV-PIG-MEIS1 (MEIS1), MSCVneo+MSCV-PIG-PBX3 (PBX3), MSCVneo-HOXA9+MSCV-PIG-MEIS1 (HOXA9+MEIS1), MSCVneo-HOXA9+MSCV-PIG-PBX3 (HOXA9+PBX3), MSCVneo-MEIS1+MSCV-PIG-PBX3 (PBX3+MEIS1), or MSCVneo-MLL-AF9+MSCV-PIG (MLL-AF9). The colony cells were replated every 7 days for up to 5 passages and colony numbers were counted for each passage. Mean±SD values of colony counts are shown. (B) Mouse BM transplantation (BMT) assays were conducted for the above 8 groups with the first-passage colony cells (CD45.1) as donors, which were transplanted into lethally irradiated 8- to 10-week-old C57BL/6 (CD45.2) recipient mice. Kaplan-Meier curves are shown. Five mice were studied in each group, except for the MLL-AF9 group in which 6 mice were studied. (C) Cell/tissue morphologies of the 8 groups. Peripheral blood (PB) and BM cells were stained with Wright-Giemsa. The spleen and liver tissues were stained with hematoxylin and eosin (H&E). The length of bars represents 10 μm for PB and BM, and 100 μm for spleen and liver.
Co-expression of PBX3 and MEIS1 induces rapid AML in mice
To determine whether co-expression of PBX3/MEIS1 is also sufficient to induce AML in mice, we conducted in vivo mouse BM transplantation (BMT) assays. As shown in Figure 1B and Supplemental Figure 1, similar to the co-expression of HOXA9 with MEIS1 or PBX3, co-expression of PBX3 and MEIS1 also caused a rapid AML in mice. The leukemic latency of the PBX3+MEIS1 group was comparable to that of the MLL-AF9 group (p=0.24; log-rank test) and slightly longer than that of the HOXA9+MEIS1 group (p=0.054), but significantly shorter than that of the HOXA9+PBX3 group (p=0.021) or HOXA9 alone group (p=0.0018). Neither MEIS1 nor PBX3 alone caused leukemia within 200 days (Figure 1B). Notably, mice of the PBX3+MEIS1 group exhibited similar, if not more aggressive, leukemic phenotypes in peripheral blood (PB), BM, spleen and liver tissues compared to the HOXA9+MEIS1, HOXA9+PBX3 and MLL-AF9 groups (Figure 1C). Our flow cytometry analysis showed that leukemic BM cells of the PBX3+MEIS1 group had largely similar proportions of Mac-1+ (a myeloid lineage cell marker) and/or c-Kit+ (a stem/progenitor cell marker) cells compared to those of the other AML groups (Supplemental Figure 1).
PBX3/MEIS1-induced AML is transplantable
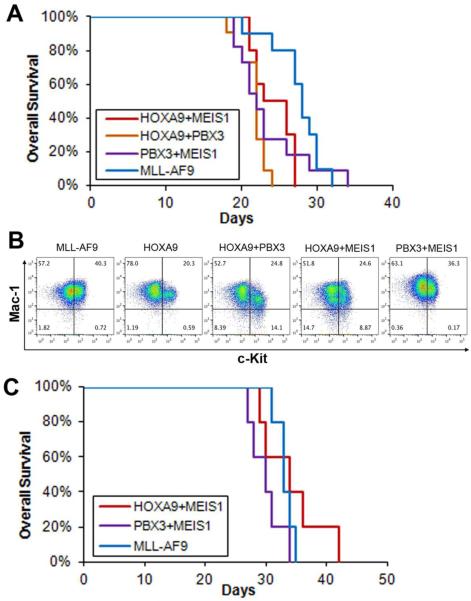
To determine whether PBX3/MEIS1-induced AML is transplantable, we conducted secondary transplantation assays. First, we collected leukemic BM cells from primary BMT recipient mice of the HOXA9+MEIS1, HOXA9+PBX3, PBX3+MEIS1 and MLL-AF9 groups and then transplanted into lethally irradiated secondary recipient mice. Leukemic BM cells from two independent primary recipient mice were used as donor cells for the secondary BMT of each group. All secondary BMT recipient mice transplanted with PBX3+MEIS1 primary AML cells developed full-blown AML within 35 days, with similar leukemic latency to those transplanted with HOXA9+MEIS1, HOXA9+PBX3, or MLL-AF9 AML cells (Figures 2A and B). The two independent donors of each group caused full-blown leukemia with similar latency in secondary recipients (Supplemental Figure 2). In addition, we also conducted secondary transplantation assays using primary leukemic spleen cells as donor cells, and also found that PBX3+MEIS1 leukemic spleen cells caused full-blown AML in all secondary BMT recipients within 35 days, in a manner similar to HOXA9+MEIS1 or MLL-AF9 leukemic spleen cells (Figure 2C).
Figure 2. PBX3/MEIS1-induced AML is transmissible in secondary transplantation recipients.
(A) Kaplan-Meier survival curves of secondary transplantation recipient (CD45.2+) mice transplanted with primary leukemic BM cells (CD45.1+) of the HOXA9+MEIS1 (recipient mouse number: n=10), HOXA9+PBX3 (n=11), PBX3+MEIS1 (n=11) and MLL-AF9 (n=10) groups. Primary AML BM cells from two donor mice were used for each group. There is no significant difference (p>0.1) between survival of the PBX3+MEIS1 group and that of any other three groups. (B) Flow cytometry analysis of leukemic BM cells from the above secondary BMT recipient mice. Antibodies against Mac-1 and c-Kit were used. Flow data of leukemic BM samples from one recipient mouse is shown as representative for each group. (C) Kaplan-Meier survival curves of secondary transplantation recipient (CD45.2+) mice transplanted with primary leukemic spleen cells (CD45.1+) of the HOXA9+MEIS1 (n=5), PBX3+MEIS1 (n=5) and MLL-AF9 (n=5) groups. Primary AML spleen cells from one donor mouse were used for each group. There is no significant difference (p>0.1) between survival of the PBX3+MEIS1 group and that of any other two groups.
Genome-wide gene expression profiles of PBX3/MEIS1 AML are more similar to those of MLL-AF9 AML than to those of HOXA9/MEIS1, HOXA9/PBX3 or HOXA9 AML
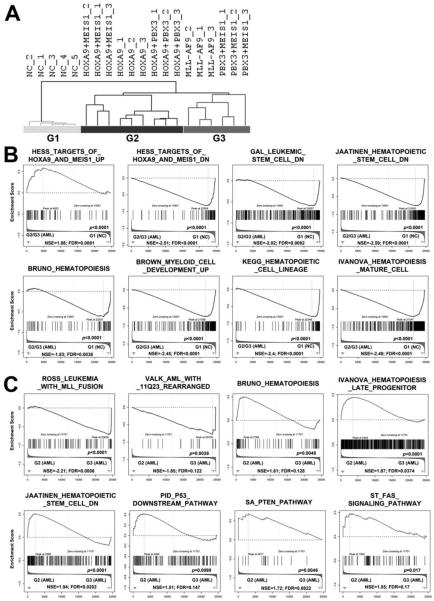
We then conducted microarray-based genome-wide gene expression profiling assays with leukemic or control BM cells collected from primary BMT recipients. Surprisingly, in an unsupervised hierarchical clustering analysis, we found that PBX3+MEIS1 AML samples clustered together with MLL-AF9 AML samples, whereas HOXA9+MEIS1, HOXA9+PBX3 and HOXA9 AML samples formed a separate cluster and so did the normal control samples (see Figure 3A). Thus, there are three separate groups, including Group 1/G1 (normal controls; NC); Group 2/G2 (HOXA9+MEIS1, HOXA9+PBX3 and HOXA9) and Group 3/G3 (MLL-AF9 and PBX3+MEIS1). We then conducted gene set enrichment analysis (GSEA) (32). No surprise, as both Groups 2 and 3 samples are leukemia samples with ectopic and/or induced endogenous expression of homeobox genes, they share a series of gene sets that distinguish them from normal control samples. Notably, target genes up-regulated by HOXA9 and MEIS1, and genes related to DNA replication, mRNA processing, microRNA biogenesis, RNA metabolism or translation, are enriched in genes expressed at a higher level in Groups 2 and 3 samples than in Group 1 samples (Figure 3B and Supplemental Figure 3A). In contrast, target genes down-regulated by HOXA9 and MEIS1, genes down-regulated in leukemic stem cells or hematopoietic stem cells, genes related to hematopoiesis, hematopoietic cell lineage, apoptosis or p53 signaling pathway, and genes up-regulated in myeloid cell development or in hematopoietic mature cells are enriched in genes expressed at a higher level in Group1 samples than in Groups 2 and 3 samples (Figure 3B and Supplemental Figure 3A).
Figure 3. Comparison of gene expression profiles between different groups of leukemic or normal control cells.
(A) Unsupervised hierarchical clustering analysis of the Control (n=5), HOXA9+MEIS1 (n=3), HOXA9+PBX3 (n=3), HOXA9 (n=3), PBX3+MEIS1 (n=3), and MLL-AF9 (n=3) groups. The hierarchical clustering tree is shown. (B) Gene sets that are shared by Group 2 (G2; including HOXA9+MEIS1, HOXA9+PBX3, and HOXA9) and Group 3 (G3; including PBX3+MEIS1 and MLL-AF9 (MA9)) samples, with a significantly different pattern in Group 1 (G1; normal control (NC)) samples, as detected by GSEA (32). (C) Gene sets that are differentially enriched between Group 2 and Group 3 samples. NSE, normalized enrichment score; FDR, false discovery rate.
On the other hand, Groups 2 and 3 samples also have significantly different enrichments of a series of gene sets. In particular, genes are highly expressed in MLL-rearranged leukemia or AML with NPM1 mutations, and target genes up-regulated by NUP98-HOXA9 fusion gene, are enriched in genes expressed at a higher level in Group 3 samples than in Group 2 samples (Figure 3C and Supplemental Figure 3B). Indeed, besides MLL-rearranged leukemia, previous studies have reported that the HOXA/MEIS1/PBX3 genes are also highly expressed in AML with NPM1 mutations or NUP98-HOXA9 fusion (40-43). In contrast, genes related to hematopoiesis, p53 pathway, PTEN pathway, FAS signaling pathway, genes highly expressed in hematopoietic late progenitor cells, and genes down-regulated in hematopoietic stem cells or in NPM1-mutated AML, are enriched in genes expressed at a higher level in Group 2 samples than in Group 3 samples (Figure 3C and Supplemental Figure 3B). Thus, genes highly expressed in Group 2 are more enriched with genes belonging to the pathways related to apoptosis and cell differentiation, while genes highly expressed in Group 3 are enriched with gene sets up-regulated in MLL-rearranged, NPM1 mutated or NUP98-HOXA9 leukemic cells. Most of these gene sets are also significantly (p<0.05) enriched in the group of PBX3+MEIS1 samples relative to Group 2 samples (see Supplemental Figure 4), further suggesting that the PBX3+MEIS1 samples share the core gene sets with the MLL-AF9 samples.
PBX3/MEIS1 AML recapitulates the core transcriptome of MLL-AF9 AML
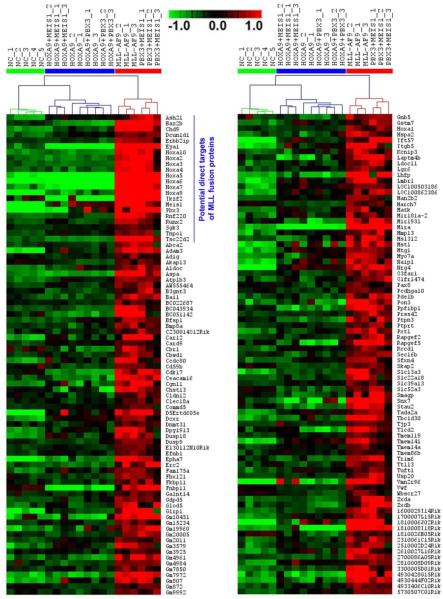
We then performed ANOVA analysis and identified 2,562 genes that are significantly differentially expressed (false discover rate (FDR) <0.01) between the three groups of samples (Supplemental Figure 5A). Through Significance Analysis of Microarrays (SAM) (44), we found that 166 and 211 genes are expressed at a significantly higher and lower level (q<0.05; FDR<0.01), respectively, in Group 3 compared to both Groups 1 and 2 (Figure 4; Supplemental Figures 5B and 6). A total of 279 and 85 genes are expressed at a significantly higher and lower level (q<0.05; FDR<0.05), respectively, in Group 2 compared to both Groups 1 and 3 (Supplemental Figures 5B, 7 and 8). A total of 1,190 and 1,022 genes are expressed at a significantly higher and lower level (q<0.05; FDR<0.05), respectively, in Group 1 compared to both Groups 2 and 3 (Supplemental Figure 5B; Supplemental Tables 1 and 2).
Figure 4. Heatmap of expression profiles of the 116 genes that are expressed at a significantly higher level in Group 3 (i.e., PBX3+MEIS1 and MLL-AF9 samples) than in both Groups 1 and 2.
Of them, 22 genes are potential direct target genes of MLL-fusion proteins. Expression data was mean centered. Red represents a high expression (scale shown in the upper middle).
Through searching for the Database for Annotation, Visualization and Integrated Discovery (DAVID; v6.7) (34, 35), we found that gene sets associated with ‘homobox transcription factor’, ‘embryonic development’, ‘DNA binding’ and ‘transcription’/‘transcription regulator activity’ are significantly enriched (FDR<0.05) in genes highly expressed in Group 3 (see Supplemental Figure 5C). Gene sets associated with ‘transferase activity’, ‘G-protein modulator’, ‘lysosome’ and ‘cell surface’ are significantly enriched in genes highly expressed in Group 2 (Supplemental Figure 5C). Gene sets associated with ‘immune response’, ‘hematopoiesis’, ‘leukocyte/ lymphocyte/ erythrocyte/ myeloid cell differentiation’ and ‘regulation of cell death/apoptosis’ are significantly enriched in genes highly expressed in Group 1 (Supplemental Figure 5C). Consistent with our GSEA analysis results (Figure 3 B and Supplemental Figure 3A), genes associated with cell death/apoptosis and hematopoietic cell differentiation are significantly down-regulated in both Groups 2 and 3 AML samples relative to normal control (Group 1) samples (see Supplemental Figure 9). Such data suggests that the repression of those genes is highly likely critical for the development of leukemia of both Groups 2 and 3. In genes that are expressed at a significantly lower level in Group 3 than in both Groups 1 and 2, gene sets related to ‘hematopoiesis’, ‘regulation of cell differentiation’, ‘stem cell differentiation’, ‘apoptosis’, ‘immune system process’, and ‘positive regulation of cell activation’ are significantly enriched (Supplemental Figure 5C); notably, those genes are expressed at the lowest level in Group 3 and at the highest level in Group 1, while at the middle level in Group 2 samples (see Supplemental Figures 10).
Notably, 22 (i.e., Ash2l, Baz2b, Chd9, Dcun1d1, Erbb2ip, Eya1, Hoxa2, Hoxa3, Hoxa4, Hoxa5, Hoxa6, Hoxa7, Hoxa9, Hoxa10, Ikzf2, Meis1, Pbx3, Rnf220, Runx2, Sgk3, Tnpo1 and Tsc22d2; see Figure 4) out of the 166 highly expressed genes in Group 3 (13.3%) are potential direct targets of MLL-fusion proteins as detected by at least one of the three published genome-wide ChIPseq or ChIP-chip assays of MLL-fusion targets (45-47). Such proportion (13.3%) is significantly greater (p<0.0001; χ2-test) than that (4/279; 1.4%) of the genes highly expressed in Group 2 (Supplemental Table 3). In contrast, none of the 211 genes under-expressed in Group 3 and none of the 1,190 genes highly expressed in Group 1 is potential direct target of MLL-fusions (Supplemental Table 3). These data indicate that MLL-fusion proteins often up-regulate, rather than down-regulate, expression of their target genes, and that the PBX3/MEIS1 combination sufficiently resembles MLL-fusion proteins in promoting expression of a cohort of core target genes of MLL fusions.
Unlike the PBX3+MEIS1 AML cells, the AML cells with HOXA9+PBX3 or HOXA9+MEIS1 exhibited a very low level of endogenous expression of the Hoxa genes (see Figure 4). To investigate whether ectopic HOXA9 exerted a suppressive effect on the expression of endogenous Hoxa genes, we ectopically expressed HOXA9 in PBX3+MEIS1 AML cells. As expected, forced expression of HOXA9 did not suppress endogenous Hoxa expression (Supplemental Figure 11). Thus, our data indicate that the incapability of HOXA9+PBX3 or HOXA9+MEIS1 to upregulate endogenous expression of Hoxa genes is not due to a transcriptional suppression mediated by exogenous expression of HOXA9.
Knockdown of PBX3 and/or MEIS1 leads to a significant down-regulation of endogenous HOXA expression in MLL-rearranged AML cells
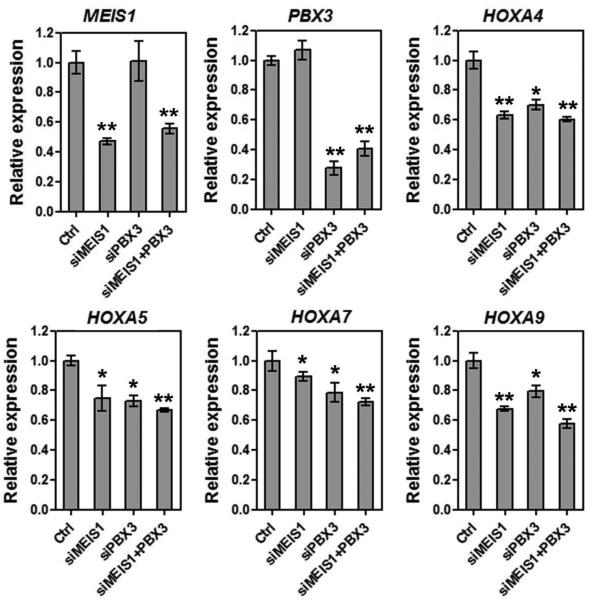
To investigate whether a high level of PBX3 or MEIS1 expression is also required to maintain the high level of endogenous HOXA expression in MLL-rearranged AML cells, we transfected anti-PBX3 and/or anti-MEIS1 siRNA oligos into MonoMac-6 cells. As expected, knockdown of PBX3 and MEIS1 each alone or together resulted in a significant down-regulation of expression of HOXA genes (Figure 5), further demonstrating the importance of PBX3/MEIS1 in regulating expression of the HOXA genes.
Figure 5. Effects of knockdown of PBX3 and/or MEIS1 on the expression of endogenous HOXA genes in MLL-rearranged AML cells.
Endogenous expression levels of MEIS1, PBX3, HOXA4, HOXA5, HOXA7 and HOXA9 in MonoMac-6/t(9;11) AML cells were detected by qPCR 48 hours post transfection of anti-MEIS1 siRNA oligos (siMEIS1) and/or anti-PBX3 siRNA oligos (siPBX3), or of scrambled control oligos (Ctrl). Mean±SD values are shown. *, p<0.05; **, p<0.01.
The binding between Meis1 and Pbx3 is essential for their synergistic effects on cell transformation and up-regulation of endogenous homeobox genes
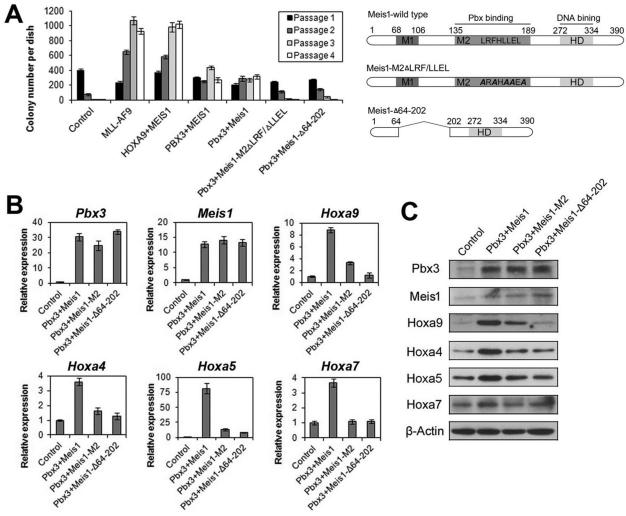
Previous studies have identified a functional domain in Meis1 protein that is critical for its binding with Pbx proteins (24, 25). To determine whether the binding between PBX3 and MEIS1 is also critical for the biological effects caused by co-expression of PBX3 and MEIS1, we cloned mouse Pbx3 gene, and also subcloned the wild-type mouse Meis1 gene and two mutants with point mutations or a regional deletion in the Pbx-binding domain (i.e., Meis1-M2ΔLRF/ΔLLEL and Meis1-Δ64-202) based on the constructs reported previously (24, 25). We then conducted colony-forming/replating assays with mouse BM progenitor cells retrovirally transduced with Pbx3+Meis1 (wilt-type), Pbx3+Meis1-M2ΔLRF/ΔLLEL, Pbx3+Meis1-Δ64-202, along with PBX3+MEIS1, HOXA9+MEIS1, MLL-AF9, and control vectors. As shown in Figure 6A, co-expression of mouse Pbx3 and Meis1 genes had a similar effect to co-expression of human PBX3 and MEIS1 genes on cell transformation/immortalization, with more than 200 colonies per dish after series of replating. In contrast, co-expression of Pbx3 with mutated Meis1 (Meis1-M2ΔLRF/ΔLLEL or Meis1-Δ64-202) lost the capacity to form a significant number of colonies after series of replating (Figure 6A), indicating that disruption of the binding between Meis1 and Pbx3 substantially inhibits their synergistic effect on cell transformation.
Figure 6. The binding between Meis1 and Pbx3 is critical for their synergistic effects on cell transformation and up-regulation of homeobox genes.
(A) In vitro colony-forming/replating assays. Briefly, mouse normal BM progenitor (lineage negative; Lin-) cells collected from 4- to 6-week-old B6.SJL (CD45.1) mice were retrovirally co-transduced with MSCVneo+MSCV-PIG (Control), MSCVneo-MLL-AF9+MSCV-PIG (MLL-AF9), MSCVneo-HOXA9+MSCV-PIG-MEIS1 (HOXA9+MEIS1), MSCVneo-MEIS1+MSCV-PIG-PBX3 (PBX3+MEIS1), MSCVneo-Meis1+MSCV-PIG-Pbx3 (Pbx3+Meis1), MSCVneo-Meis1-M2ΔLRF/ΔLLEL+MSCV-PIG-Pbx3 (Pbx3+Meis1-M2ΔLRF/ΔLLEL) or MSCVneo-Meis1-Δ64-202+MSCV-PIG-Pbx3 (Pbx3+ Meis1-Δ64-202). The colony cells were replated every 7 days for up to 4 passages and colony numbers were counted for each passage. Mean±SD values of colony counts are shown (left panel). The domain structure of the wild-type and mutant Meis1 proteins are also shown (right panel). (B,C) qPCR (B) and Western blotting (C) analyses of expression levels of a series of representative Homeobox genes in colony cells from the first passage of four groups (samples generated from colony-forming/replating assays shown in Figure 6A). Mean±SD values are shown in Figure 6B.
Not surprisingly, our further qPCR assays showed that, despite that Pbx3 and Meis1, wild-type or mutants, show the comparable level, co-expression of Pbx3 with Meis1 mutants (Meis1-M2ΔLRF/ΔLLEL or Meis1-Δ64-202) could not up-regulate expression of endogenous Hoxa genes to a level as high as that induced by co-expression of Pbx3 with wild-type Meis1, a phenomenon verified by immunoblotting (Figures 6B and C). Thus, disruption of the binding between Meis1 and Pbx3 also substantially abrogates their synergistic effect on up-regulation of expression of endogenous homeobox genes.
DISCUSSION
Here we show, for the first time, that forced expression of both PBX3 and MEIS1 can transform/immortalize normal HSPCs in vitro and induce a rapid AML in vivo. Although the binding ability of Meis1 with Pbx proteins has been reported previously to be essential for the synergistic effect between Meis1 and Hoxa9 and for the function of MLL-fusion proteins (18, 24, 25), no efforts have been exerted to investigate whether forced expression of both MEIS1 and PBX3 is sufficient to transform cells and induce leukemia, without forced expression of a HOXA gene. Indeed, because neither Meis1 nor Pbx3 alone can induce cell transformation and leukemogenesis (20, 21, 23, 26), they have been thought to mainly play supportive roles in facilitating HOXA proteins in regulating their downstream targets (18, 24-28). Therefore, our new finding reveals the functional importance of PBX3 and MEIS1 in cell transformation and leukemogenesis.
More strikingly, we further show that it is the co-expression of PBX3 and MEIS1, not co-expression of HOXA9 and MEIS1 (or HOXA9 and PBX3), that can recapitulate the core transcriptome of MLL-rearranged AML, especially the significant up-regulation of a set of homeobox transcriptional factors (in particular, the Hoxa genes, Meis1 and Pbx3). In fact, a total of more than 160 genes (including the Homeobox genes) are significantly up-regulated by both co-expression of PBX3 and MEIS1 and forced expression of MLL-AF9. Notably, forced expression of HOXA9 alone or together with MEIS1 or PBX3 cannot up-regulate expression of most of those genes at all. Indeed, consistent with our observation, previous studies also showed that forced expression of Hoxa9 and Meis1 each alone or together cannot significantly up-regulate expression of endogenous Pbx3 and Hoxa genes (25, 48). In addition, more than 210 genes are significantly repressed by both MLL-fusion proteins and the PBX3/MEIS1 combination; although these genes are also repressed by HOXA9 alone or together with MEIS1 or PBX3, the repression degrees are much more moderate. Therefore, despite the close relationship between HOXA9, MEIS1 and PBX3 (18, 24-27), only the PBX3/MEIS1 combination, but not the HOXA9/MEIS1 or HOXA9/PBX3 combination (or HOXA9 alone), can resemble MLL-fusion proteins in inducing critical transcriptional programs. Further systematic studies are warranted in the future to decipher the underlying molecular mechanism(s). It is possible that a certain threshold of abundance of both PBX3 and MEIS1 is required for them to initiate their cooperation in inducing expression of endogenous HOXA genes, and the induced HOXA proteins can further cooperate with PBX3 and MEIS1 and enhance their downstream transcriptional programs, which in turn leads to the full establishment of the core transcriptome of MLL-rearranged AML. In contrast, because forced expression of HOXA9 alone or together with MEIS1 or PBX3 cannot achieve sufficiently high abundance of both PBX3 and MEIS1, and thereby cannot induce the core transcriptome of MLL-rearranged AML.
Besides MLL-rearranged AML, AMLs with NPM1 mutations or NUP98-HOXA9 fusion, which together account for more than 30% of total AML cases, are also featured with aberrant overexpression of a set of homeobox genes, including the HOXA/MEIS1/PBX3 genes (40-43, 49). No surprise, gene sets up-regulated in AML with NPM1 mutations or NUP98-HOXA9 fusion are significantly enriched in genes highly expressed in both PBX3/MEIS1 and MLL-AF9 AML cells. Thus, it is highly likely that the cooperation between PBX3 and MEIS1 is critical for the establishment of the core transcriptomes of a wide range of AMLs, not limited to MLL-rearranged AML.
Furthermore, our present study together with previously studies (18, 24, 25) suggest that the binding between MEIS1 and PBX proteins is critical for the biological functions of the PBX3/MEIS1 combination and MLL-fusion proteins in cell immortalization, leukemogenesis and regulation of transcriptional programs. Therefore, targeting the binding between MEIS1 and PBX3 may represent a promising therapeutic strategy to cure leukemia with aberrant overexpression of homeobox genes, such as AMLs with MLL-rearrangements, NPM1 mutations or NUP98-HOXA9 fusion. We propose that small-molecule compounds or small peptides can be developed to specially inhibit the binding between MEIS1 and PBX3 and they can be applied in clinic, alone or together with other therapeutic agents, to treat leukemia.
In summary, we demonstrate that forced expression of PBX3 and MEIS1 is sufficient to induce cell transformation and leukemogenesis, and to resemble MLL-fusion proteins in inducing the core transcriptional programs, especially the up-regulation of a set of homeobox genes. The biological functions of the PBX3/MEIS1 combination are dependent on their binding ability. Thus, our finding is of great significance as it shows that two previously assumed “supporting actors” (i.e., PBX3 and MEIS1) highly likely act as “leading actors” in the establishment of the core transcriptome of AML with MLL-rearrangements, and probably also in that of AML with NPM1 mutations or NUP98-HOXA9 fusion. Targeting the binding between PBX3 and MEIS1 represents an attractive therapeutic strategy to treat a wide-range of AML with aberrant overexpression of homeobox genes.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Drs. Mark P. Kamps, Scott Armstrong, Gregory Hannon, and Lin He for providing retroviral constructs. This work was supported in part by the National Institutes of Health (NIH) R01 Grants CA178454, CA182528 and CA127277 (J.C.), Leukemia & Lymphoma Society (LLS) Special Fellowship (Z.L.), LLS Translational Research Grant (J.C.), American Cancer Society (ACS) Research Scholar grant (J.C.), ACS-IL Research Scholar grant (Z.L.), Gabrielle’s Angel Foundation for Cancer Research (J.C., Z.L., X.J. and H.H.), and Intramural Research Program of National Human Genome Research Institute, NIH (A.G.E. and P.P.L.).
Abbreviations
- AML
acute myeloid leukemia
- BMT
bone marrow transplantation
- HSPC
hematopoietic stem/progenitor cell
- GSEA
Gene Set Enrichment Analysis
- KEGG
Kyoto Encyclopedia of Genes and Genomes Pathway Database
- DAVID
The Database for Annotation, Visualization and Integrated Discovery
Footnotes
Conflict of Interest: The authors declare no conflict of interest.
AUTHOR CONTRIBUTIONS
Z.L. and J.C. conceived the project and designed the research. Z.L., P.C., R.S., C.H., Y.L., A.G.E., Z.Z., S.G., S.A., H.W., Y.W., S.L., H.H., M.B.N., X.J. and J.C. performed experiments and/or data analyses; Z.L., A.G.E., H.H., G.G.W., X.J., P.P.L., J.J. and J.C. contributed reagents/analytic tools and/or grant support; Z.L. and J.C. wrote the paper. All authors discussed the results and commented on the manuscript.
REFERENCES
- 1.Pui CH, Chessells JM, Camitta B, Baruchel A, Biondi A, Boyett JM, et al. Clinical heterogeneity in childhood acute lymphoblastic leukemia with 11q23 rearrangements. Leukemia. 2003;17:700–6. doi: 10.1038/sj.leu.2402883. [DOI] [PubMed] [Google Scholar]
- 2.Krivtsov AV, Armstrong SA. MLL translocations, histone modifications and leukaemia stem-cell development. Nat Rev Cancer. 2007;7:823–33. doi: 10.1038/nrc2253. [DOI] [PubMed] [Google Scholar]
- 3.Popovic R, Zeleznik-Le NJ. MLL: how complex does it get? J Cell Biochem. 2005;95:234–42. doi: 10.1002/jcb.20430. [DOI] [PubMed] [Google Scholar]
- 4.Rowley JD, Olney HJ. International workshop on the relationship of prior therapy to balanced chromosome aberrations in therapy-related myelodysplastic syndromes and acute leukemia: overview report. Genes Chromosomes Cancer. 2002;33:331–45. doi: 10.1002/gcc.10040. [DOI] [PubMed] [Google Scholar]
- 5.Krivtsov AV, Feng Z, Lemieux ME, Faber J, Vempati S, Sinha AU, et al. H3K79 methylation profiles define murine and human MLL-AF4 leukemias. Cancer Cell. 2008;14:355–68. doi: 10.1016/j.ccr.2008.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Muntean AG, Hess JL. The pathogenesis of mixed-lineage leukemia. Annu Rev Pathol. 2012;7:283–301. doi: 10.1146/annurev-pathol-011811-132434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Slany RK. The molecular biology of mixed lineage leukemia. Haematologica. 2009;94:984–93. doi: 10.3324/haematol.2008.002436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Armstrong SA, Staunton JE, Silverman LB, Pieters R, den Boer ML, Minden MD, et al. MLL translocations specify a distinct gene expression profile that distinguishes a unique leukemia. Nat Genet. 2002;30:41–7. doi: 10.1038/ng765. [DOI] [PubMed] [Google Scholar]
- 9.Yeoh EJ, Ross ME, Shurtleff SA, Williams WK, Patel D, Mahfouz R, et al. Classification, subtype discovery, and prediction of outcome in pediatric acute lymphoblastic leukemia by gene expression profiling. Cancer Cell. 2002;1:133–43. doi: 10.1016/s1535-6108(02)00032-6. [DOI] [PubMed] [Google Scholar]
- 10.Lawrence HJ, Rozenfeld S, Cruz C, Matsukuma K, Kwong A, Komuves L, et al. Frequent co-expression of the HOXA9 and MEIS1 homeobox genes in human myeloid leukemias. Leukemia. 1999;13:1993–9. doi: 10.1038/sj.leu.2401578. [DOI] [PubMed] [Google Scholar]
- 11.Faber J, Krivtsov AV, Stubbs MC, Wright R, Davis TN, van den Heuvel-Eibrink M, et al. HOXA9 is required for survival in human MLL-rearranged acute leukemias. Blood. 2009;113:2375–85. doi: 10.1182/blood-2007-09-113597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zeisig BB, Milne T, Garcia-Cuellar MP, Schreiner S, Martin ME, Fuchs U, et al. Hoxa9 and Meis1 are key targets for MLL-ENL-mediated cellular immortalization. Mol Cell Biol. 2004;24:617–28. doi: 10.1128/MCB.24.2.617-628.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ross ME, Mahfouz R, Onciu M, Liu HC, Zhou X, Song G, et al. Gene expression profiling of pediatric acute myelogenous leukemia. Blood. 2004;104:3679–87. doi: 10.1182/blood-2004-03-1154. [DOI] [PubMed] [Google Scholar]
- 14.Andersson A, Olofsson T, Lindgren D, Nilsson B, Ritz C, Eden P, et al. Molecular signatures in childhood acute leukemia and their correlations to expression patterns in normal hematopoietic subpopulations. Proc Natl Acad Sci U S A. 2005;102:19069–74. doi: 10.1073/pnas.0506637102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Andersson A, Eden P, Lindgren D, Nilsson J, Lassen C, Heldrup J, et al. Gene expression profiling of leukemic cell lines reveals conserved molecular signatures among subtypes with specific genetic aberrations. Leukemia. 2005;19:1042–50. doi: 10.1038/sj.leu.2403749. [DOI] [PubMed] [Google Scholar]
- 16.Li Z, Huang H, Li Y, Jiang X, Chen P, Arnovitz S, et al. Up-regulation of a HOXA-PBX3 homeobox-gene signature following down-regulation of miR-181 is associated with adverse prognosis in patients with cytogenetically abnormal AML. Blood. 2012;119:2314–24. doi: 10.1182/blood-2011-10-386235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ayton PM, Cleary ML. Transformation of myeloid progenitors by MLL oncoproteins is dependent on Hoxa7 and Hoxa9. Genes Dev. 2003;17:2298–307. doi: 10.1101/gad.1111603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wong P, Iwasaki M, Somervaille TC, So CW, Cleary ML. Meis1 is an essential and rate-limiting regulator of MLL leukemia stem cell potential. Genes Dev. 2007;21:2762–74. doi: 10.1101/gad.1602107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kumar AR, Li Q, Hudson WA, Chen W, Sam T, Yao Q, et al. A role for MEIS1 in MLL-fusion gene leukemia. Blood. 2009;113:1756–8. doi: 10.1182/blood-2008-06-163287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kroon E, Krosl J, Thorsteinsdottir U, Baban S, Buchberg AM, Sauvageau G. Hoxa9 transforms primary bone marrow cells through specific collaboration with Meis1a but not Pbx1b. Embo J. 1998;17:3714–25. doi: 10.1093/emboj/17.13.3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bach C, Buhl S, Mueller D, Garcia-Cuellar MP, Maethner E, Slany RK. Leukemogenic transformation by HOXA cluster genes. Blood. 2010;115:2910–8. doi: 10.1182/blood-2009-04-216606. [DOI] [PubMed] [Google Scholar]
- 22.Jin G, Yamazaki Y, Takuwa M, Takahara T, Kaneko K, Kuwata T, et al. Trib1 and Evi1 cooperate with Hoxa and Meis1 in myeloid leukemogenesis. Blood. 2007;109:3998–4005. doi: 10.1182/blood-2006-08-041202. [DOI] [PubMed] [Google Scholar]
- 23.Thorsteinsdottir U, Kroon E, Jerome L, Blasi F, Sauvageau G. Defining roles for HOX and MEIS1 genes in induction of acute myeloid leukemia. Mol Cell Biol. 2001;21:224–34. doi: 10.1128/MCB.21.1.224-234.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang GG, Pasillas MP, Kamps MP. Meis1 programs transcription of FLT3 and cancer stem cell character, using a mechanism that requires interaction with Pbx and a novel function of the Meis1 C-terminus. Blood. 2005;106:254–64. doi: 10.1182/blood-2004-12-4664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang GG, Pasillas MP, Kamps MP. Persistent transactivation by meis1 replaces hox function in myeloid leukemogenesis models: evidence for co-occupancy of meis1-pbx and hox-pbx complexes on promoters of leukemia-associated genes. Mol Cell Biol. 2006;26:3902–16. doi: 10.1128/MCB.26.10.3902-3916.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li Z, Zhang Z, Li Y, Arnovitz S, Chen P, Huang H, et al. PBX3 is an important cofactor of HOXA9 in leukemogenesis. Blood. 2013;121:1422–31. doi: 10.1182/blood-2012-07-442004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dickson GJ, Liberante FG, Kettyle LM, O'Hagan KA, Finnegan DP, Bullinger L, et al. HOXA/PBX3 knockdown impairs growth and sensitizes cytogenetically normal acute myeloid leukemia cells to chemotherapy. Haematologica. 2013;98:1216–25. doi: 10.3324/haematol.2012.079012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Garcia-Cuellar MP, Steger J, Fuller E, Hetzner K, Slany RK. Pbx3 and Meis1 cooperate through multiple mechanisms to support Hox-induced murine leukemia. Haematologica. 2015 doi: 10.3324/haematol.2015.124032. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Krivtsov AV, Twomey D, Feng Z, Stubbs MC, Wang Y, Faber J, et al. Transformation from committed progenitor to leukaemia stem cell initiated by MLL-AF9. Nature. 2006;442:818–22. doi: 10.1038/nature04980. [DOI] [PubMed] [Google Scholar]
- 30.He L, Thomson JM, Hemann MT, Hernando-Monge E, Mu D, Goodson S, et al. A microRNA polycistron as a potential human oncogene. Nature. 2005;435:828–33. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 2003;31:e15. doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–50. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 35.Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li Z, Huang H, Chen P, He M, Li Y, Arnovitz S, et al. miR-196b directly targets both HOXA9/MEIS1 oncogenes and FAS tumour suppressor in MLL-rearranged leukaemia. Nat Commun. 2012;2:688. doi: 10.1038/ncomms1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jiang X, Huang H, Li Z, Li Y, Wang X, Gurbuxani S, et al. Blockade of miR-150 maturation by MLL-fusion/MYC/LIN-28 is required for MLL-associated leukemia. Cancer Cell. 2012;22:524–35. doi: 10.1016/j.ccr.2012.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huang H, Jiang X, Li Z, Li Y, Song CX, He C, et al. TET1 plays an essential oncogenic role in MLL-rearranged leukemia. Proc Natl Acad Sci USA. 2013;110:11994–9. doi: 10.1073/pnas.1310656110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li Z, Lu J, Sun M, Mi S, Zhang H, Luo RT, et al. Distinct microRNA expression profiles in acute myeloid leukemia with common translocations. Proc Natl Acad Sci USA. 2008;105:15535–40. doi: 10.1073/pnas.0808266105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Haferlach C, Mecucci C, Schnittger S, Kohlmann A, Mancini M, Cuneo A, et al. AML with mutated NPM1 carrying a normal or aberrant karyotype show overlapping biologic, pathologic, immunophenotypic, and prognostic features. Blood. 2009;114:3024–32. doi: 10.1182/blood-2009-01-197871. [DOI] [PubMed] [Google Scholar]
- 41.Becker H, Marcucci G, Maharry K, Radmacher MD, Mrozek K, Margeson D, et al. Favorable prognostic impact of NPM1 mutations in older patients with cytogenetically normal de novo acute myeloid leukemia and associated gene- and microRNA-expression signatures: a Cancer and Leukemia Group B study. J Clin Oncol. 2010;28:596–604. doi: 10.1200/JCO.2009.25.1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mullighan CG, Kennedy A, Zhou X, Radtke I, Phillips LA, Shurtleff SA, et al. Pediatric acute myeloid leukemia with NPM1 mutations is characterized by a gene expression profile with dysregulated HOX gene expression distinct from MLL-rearranged leukemias. Leukemia. 2007;21:2000–9. doi: 10.1038/sj.leu.2404808. [DOI] [PubMed] [Google Scholar]
- 43.Calvo KR, Sykes DB, Pasillas MP, Kamps MP. Nup98-HoxA9 immortalizes myeloid progenitors, enforces expression of Hoxa9, Hoxa7 and Meis1, and alters cytokine-specific responses in a manner similar to that induced by retroviral co-expression of Hoxa9 and Meis1. Oncogene. 2002;21:4247–56. doi: 10.1038/sj.onc.1205516. [DOI] [PubMed] [Google Scholar]
- 44.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA. 2001;98:5116–21. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Guenther MG, Lawton LN, Rozovskaia T, Frampton GM, Levine SS, Volkert TL, et al. Aberrant chromatin at genes encoding stem cell regulators in human mixed-lineage leukemia. Genes Dev. 2008;22:3403–8. doi: 10.1101/gad.1741408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bernt KM, Zhu N, Sinha AU, Vempati S, Faber J, Krivtsov AV, et al. MLL-rearranged leukemia is dependent on aberrant H3K79 methylation by DOT1L. Cancer Cell. 2011;20:66–78. doi: 10.1016/j.ccr.2011.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang QF, Wu G, Mi S, He F, Wu J, Dong J, et al. MLL fusion proteins preferentially regulate a subset of wild-type MLL target genes in the leukemic genome. Blood. 2011;117:6895–905. doi: 10.1182/blood-2010-12-324699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mulgrew NM, Kettyle LM, Ramsey JM, Cull S, Smyth LJ, Mervyn DM, et al. c-Met inhibition in a HOXA9/Meis1 model of CN-AML. Dev Dyn. 2014;243:172–81. doi: 10.1002/dvdy.24070. [DOI] [PubMed] [Google Scholar]
- 49.Falini B, Sportoletti P, Martelli MP. Acute myeloid leukemia with mutated NPM1: diagnosis, prognosis and therapeutic perspectives. Curr Opin Oncol. 2009;21:573–81. doi: 10.1097/CCO.0b013e3283313dfa. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.