Centriole elimination is essential for sexual reproduction. Borrego-Pinto et al. show that in starfish oocytes elimination proceeds by extrusion of mother centrioles to polar bodies, whereas the single remaining daughter centriole is eliminated in the cytoplasm, preparing the egg for fertilization.
Abstract
Centriole elimination is an essential process that occurs in female meiosis of metazoa to reset centriole number in the zygote at fertilization. How centrioles are eliminated remains poorly understood. Here we visualize the entire elimination process live in starfish oocytes. Using specific fluorescent markers, we demonstrate that the two older, mother centrioles are selectively removed from the oocyte by extrusion into polar bodies. We show that this requires specific positioning of the second meiotic spindle, achieved by dynein-driven transport, and anchorage of the mother centriole to the plasma membrane via mother-specific appendages. In contrast, the single daughter centriole remaining in the egg is eliminated before the first embryonic cleavage. We demonstrate that these distinct elimination mechanisms are necessary because if mother centrioles are artificially retained, they cannot be inactivated, resulting in multipolar zygotic spindles. Thus, our findings reveal a dual mechanism to eliminate centrioles: mothers are physically removed, whereas daughters are eliminated in the cytoplasm, preparing the egg for fertilization.
Introduction
The centrosome functions as the primary microtubule-organizing center of animal cells, with key roles in polarity, migration, division, and cilium formation (Arquint et al., 2014; Chavali et al., 2014; Reina and Gonzalez, 2014; Stinchcombe and Griffiths, 2014). The centrosome is typically composed of two microtubule-based cylindrical centrioles surrounded by pericentriolar material (PCM).
Cycling somatic cells are born with one mother centriole and one daughter centriole. The mother centriole acquired additional protein components and posttranslational modifications during the previous cell cycle and is fully able to organize PCM (Brito et al., 2012; Kong et al., 2014; Winey and O’Toole, 2014). The mother centriole features a set of distal and subdistal appendages apparent by EM that enable attachment to the plasma membrane during cilium formation, as well as to anchor microtubules during interphase (Vorobjev and Chentsov YuS, 1982; Paintrand et al., 1992; Winey and O’Toole, 2014). Starting at the G1/S-phase transition, both mother and daughter centrioles seed the formation of a new procentriole orthogonal to its proximal end. The two procentrioles then grow and mature, so that each spindle pole at the G2/M transition harbors two centrioles, an older one (the mother centriole) and a younger one (the daughter centriole). These two centrioles disengage from one another during mitosis, thus completing the centriole duplication cycle (Fırat-Karalar and Stearns, 2014; Fu et al., 2015). Defects in this canonical centriole cycle lead to errors in cell division and development and are thought to contribute to tumorigenesis (Godinho and Pellman, 2014; Gönczy, 2015).
Importantly, the centriole cycle needs to be altered when the zygote forms: if egg and sperm each contributed a pair of centrioles, the zygote would contain double of the normal centriole number. This would have severe adverse consequences such as multipolar spindles and aneuploidy during early development (Sathananthan et al., 2006; Scheer, 2014). Therefore, centriole elimination from oocytes is thought to be an essential mechanism in metazoa to ensure the exclusive contribution of sperm-derived centrioles to the zygote (Delattre and Gönczy, 2004; Manandhar et al., 2005).
Although centriole elimination has been studied in many species, the underlying mechanisms are far from being understood. In many of the common model systems, including Caenorhabditis elegans, Drosophila melanogaster, Xenopus laevis, and the mouse, centrioles are eliminated during the long meiotic prophase, and thus meiotic spindles are anastral (Szollosi et al., 1972; Dävring and Sunner, 1973; Albertson and Thomson, 1993; Gard, 1994). Meiotic prophase is particularly difficult to access experimentally, and therefore mechanisms of centriole elimination remain largely unclear. However, it has been shown that elimination is likely to be a rapid process occurring during the diplotene stage and that it is promoted by the CGH-1 helicase in C. elegans (Mikeladze-Dvali et al., 2012).
In contrast, in several other species, centrioles are eliminated during the actual meiotic divisions, and consequently meiotic spindles are astral. Although this was considered initially to be a specific feature of echinoderms (sea urchins, starfish, and sea cucumbers; Kato et al., 1990; Nakashima and Kato, 2001; Miyazaki et al., 2005), classic as well as recent evidence suggest that centrosomal meiotic spindles are widely spread across metazoan groups. In addition to echinoderms, meiotic spindles are astral in annelids, nemertea, and mollusks (Longo and Anderson, 1969; Crowder et al., 2015), indicating that centriole elimination during the meiotic divisions is a widespread phenomenon.
From an experimental point of view, this second group of organisms is much better suited to study centriole elimination, because meiotic divisions are rapid and synchronous, allowing the whole process to be imaged in live specimens. In particular, centriole elimination has been investigated in the oocytes of the starfish Patiria (previously known as Asterina) pectinifera, using serial section EM to detect centrioles and transmitted light microscopy to visualize microtubule asters (Kato et al., 1990; Tamura and Nemoto, 2001; Uetake et al., 2002; Zhang et al., 2004; Shirato et al., 2006). These experiments established that of the four centrioles contained in the oocyte, two are extruded into the first polar body (PBI) and one in the second polar body (PBII). The single centriole that remains in the egg is inactivated before the first embryonic division (Kato et al., 1990; Saiki and Hamaguchi, 1998). It was further proposed that an intrinsic difference must exist between centrioles extruded into the polar bodies and the one remaining in the mature egg cytoplasm. This centriole was referred to as “nonreplicative” and those extruded into the polar bodies as “replicative,” because the latter maintained aster-forming capacity if reintroduced into the mature egg by polar body transplantation, retention, or oocyte fusion (Tamura and Nemoto, 2001; Uetake et al., 2002; Zhang et al., 2004; Shirato et al., 2006). Moreover, it was proposed that the replicative potential may be associated with the maturation state of the centrioles (Washitani-Nemoto et al., 1994; Saiki and Hamaguchi, 1998; Tamura and Nemoto, 2001). However, whether this is the case has not been tested experimentally. Furthermore, studies in other starfish species, Asterias forbesi and Pisaster ocraceus, arrived at a different conclusion and suggested instead that the reproductive capacity of all four centrioles is degraded during first meiosis (MI; Sluder et al., 1989, 1993).
Here, we set out to address the mechanisms of centriole elimination in starfish oocytes. By following the entire process by live imaging of molecular markers specific to mother or daughter centrioles, we demonstrate that mother centrioles are selectively extruded into the polar bodies. By tracking centriole motion, we further show that extrusion of mother centrioles results from dynein-driven transport and subsequent anchoring to the plasma membrane. Ultrastructural analysis indicates that the mother centriole is anchored at the site of polar body extrusion by centriolar appendages. We further establish that selective extrusion is essential for centriole elimination: if mother centrioles are retained in the egg, they remain active and interfere with zygotic spindle assembly.
Results
Identification of centriolar proteins and fluorescent centriole markers in starfish
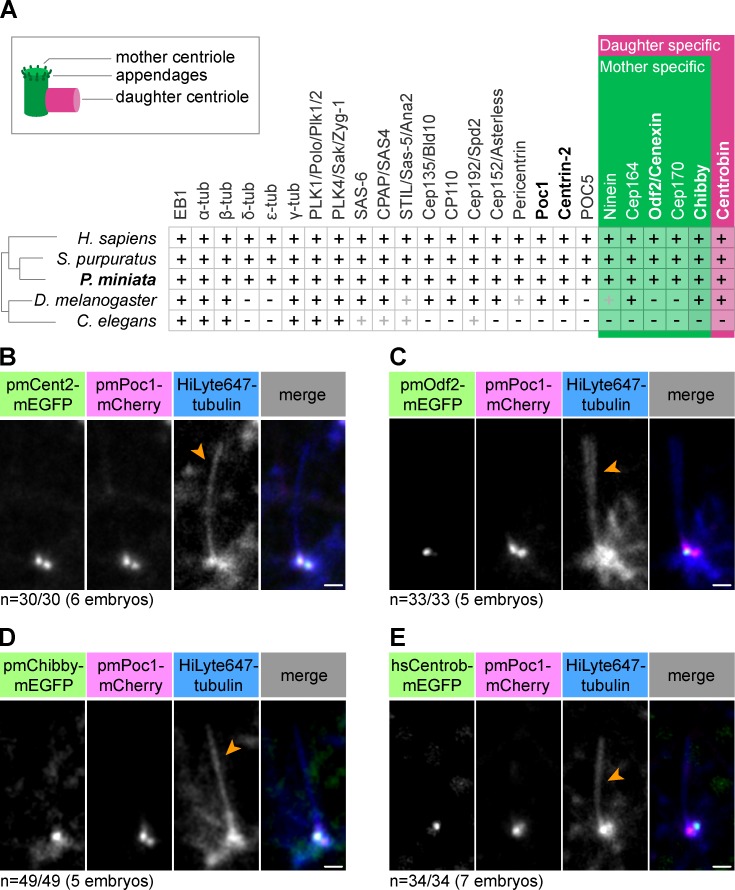
Centriole composition is well conserved across metazoan evolution, and the phylogeny of centrosomal proteins has already been established for multiple organisms (Hodges et al., 2010; Carvalho-Santos et al., 2011). Using this information and a transcriptome-based sequence database, we identified starfish (Patiria miniata) homologues of all 25 centrosomal proteins queried (Table S1). Sequence comparisons reveal that, consistent with the phylogenetic position of starfish in the deuterostome lineage, starfish centrosome components resemble most those of sea urchin (Strongylocentrotus purpuratus) and vertebrates (Homo sapiens), whereas they are more distinct from the protostome centrosomes of the fly (D. melanogaster) and nematodes (C. elegans; Fig. 1 A).
Figure 1.
Identification and characterization of centriole markers in starfish. (A) Overview of identified starfish (P. miniata) homologues of centriolar and PCM proteins compared with those of select model organisms. Black plus sign indicates the presence of a clear homologue; gray plus sign indicates functional homology with divergent amino acid sequence. Mother centriole–specific markers are shown in green and daughter centriole–specific markers are shown in pink (see also schematic representation on the top left). Genes coding for proteins shown in bold were used in this study. (B–E) Fluorescent protein fusions of indicated centriolar proteins were validated as live cell markers in the ciliated epithelium of starfish embryos. HiLyte647 tubulin labels the cilium (orange arrowheads). pmPoc1-mCherry and mEGFP-pmCentrin-2 label both mother and daughter centrioles; pmOdf2-mEGFP and pmChibby-mEGFP are mother-specific markers; hsCentrobin-mEGFP is a daughter-specific marker. Maximum intensity projections of 2–5 confocal sections are shown. Bars, 1 µm.
The localization of most of these proteins within centrosomes has been described in other species (Brito et al., 2012; Gönczy, 2012). Many localize to both centrioles, hereafter referred to as general markers, whereas others are specific to the mother or daughter centriole. In particular, binding partners and constituents of the two sets of appendages, including Chibby and Odf2, are mother centriole specific, whereas Centrobin specifically localizes to the daughter centriole (Lange and Gull, 1995; Ishikawa et al., 2005; Zou et al., 2005; Voronina et al., 2009; Steere et al., 2012; Burke et al., 2014; Winey and O’Toole, 2014; Fig. 1 A).
Based on these data, we constructed fluorescent protein (FP) fusions to several centriolar proteins and tested them as live cell markers by expressing them in starfish embryos (Fig. 1 A, names in bold). Starfish embryos develop a ciliated epithelial cell layer at the early gastrula stage that proved useful for this analysis. In ciliated cells, the mother centriole acts as a platform from which the axoneme of the cilium grows, whereas the daughter centriole is located more laterally from this structure (Reiter et al., 2012). We found that the general centriole markers mEGFP-pmCentrin-2 and pmPoc1-mCherry colocalized at two foci at the base of the cilium (Fig. 1 B). In contrast, the mother centriole–specific pmOdf2-mEGFP and pmChibby-mEGFP labeled only one of these foci, the one connected to the cilium (Fig. 1, C and D; and Fig. S1 A). Conversely, hsCentrobin-mEGFP colocalized with the pmPoc1-mCherry focus distal to the cilium base, the expected localization for the daughter centriole (Fig. 1 E).
Collectively, by homology searches in a transcriptome dataset assembled de novo, we identified all major centriole components in starfish, a species previously uncharacterized in this regard. Moreover, by using FP fusions of these proteins, we were able to establish live cell markers that allow reliable detection of centrioles and distinction of mother versus daughter centrioles.
Live imaging of centrioles in meiotic starfish oocytes
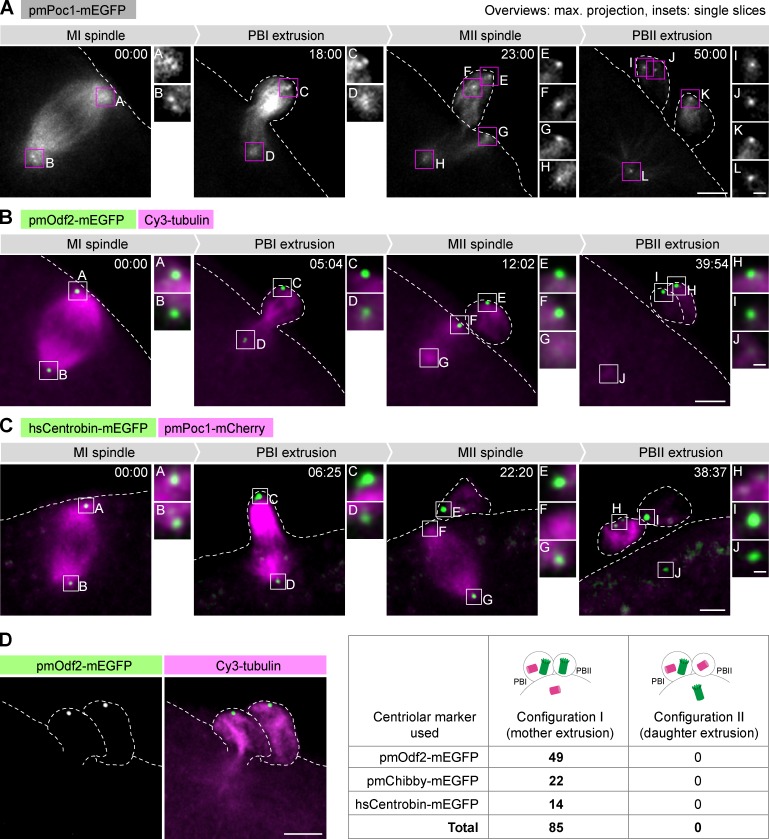
We next used high-resolution confocal 3D time-lapse imaging of these markers to follow the fate of centrioles throughout meiosis; previously, this behavior had merely been inferred from imaging of microtubule asters and analysis of fixed specimens (Kato et al., 1990; Uetake et al., 2002; Zhang et al., 2004; Shirato et al., 2006; Ucar et al., 2013). Of the two general centriole markers, pmPoc1-mEGFP (and to a lesser extent pmPoc1-mCherry) was preferred for its reliability over a wide range of expression levels, and because it conveniently colabels microtubules and thus simultaneously gives information on spindle dynamics (Fig. 2 A and Video 1). Identical results were obtained using mEGFP-pmCentrin-2 (Fig. S2 A). Using these markers, we detected a constant number of four centrioles from meiosis onset to meiosis completion. The poles of the MI spindle are organized by two centrosomes containing two centrioles each (Figs. 2 A and S2 A, MI spindle). The centrosome localized at the outer spindle pole, i.e., the pole facing the plasma membrane, is extruded into the PBI (Figs. 2 A and S2 A, PBI extrusion). The two centrioles contained in the other centrosome separate thereafter, and in the absence of centriole duplication, the resulting two single centrioles organize the poles of the MII spindle (Figs. 2 A and S2 A, MII spindle). One of these centrioles is extruded into the PBII, leaving a single centriole in the mature egg at the end of meiosis (Figs. 2 A and S2 A, PBII extrusion).
Figure 2.
Mother centrioles are invariably extruded into the polar bodies, leaving a single daughter centriole in the mature egg. (A–C) Oocytes injected with and expressing the indicated fluorescent markers were imaged by 3D confocal microscopy throughout meiosis, starting at metaphase I. Overview images are maximum intensity projections of the entire z-stacks; insets are single confocal sections of the regions marked on the overviews. Here and in other figures, unless otherwise indicated, the dashed line indicates the oocyte contour based on the transmitted light image (not depicted). z-Stacks were acquired every 30–60 s, and time is shown in mm:ss. Bars: (overview images) 5 µm; (insets) 1 µm. (A) pmPoc1-mEGFP labels centrioles and microtubules (see Video 1). (B) pmOdf2-mEGFP specifically labels mother centrioles; Cy3-tubulin labels microtubules (see Video 2). (C) hsCentrobin-mEGFP labels daughter centrioles; pmPoc1-mCherry labels centrioles and microtubules (see Video 3). (D) Distribution of mother and daughter centrioles after PBII extrusion determined using the indicated centriolar markers. In all oocytes examined, the two mother centrioles were extruded into the polar bodies (configuration I); extrusion of the daughter centriole into PBII (configuration II) was never observed. An Odf2-mEGFP–expressing oocyte, coinjected with Cy3-Tubulin, is shown as an example on the left (maximum intensity projection of a z-stack; processed to remove autofluorescence as described in Materials and methods). Bar, 5 µm.
Overall, the fact that four centrioles were detected throughout meiosis in live oocytes allows us to unequivocally conclude that centrioles are neither eliminated nor duplicated during meiosis in starfish oocytes.
Mother centrioles are extruded into the polar bodies, leaving a single daughter centriole in the mature egg
Next, we used mother- or daughter-specific centriole markers to monitor the maturation state of centrioles during meiosis. As expected, the MI spindle contains a mother–daughter pair at each pole: one centriole is labeled with pmOdf2-mEGFP and one with hsCentrobin-mEGFP (Fig. 2, B and C, MI spindle; and Videos 2 and 3). One such pair is extruded into PBI (Fig. 2, B and C, PBI extrusion), whereas the other pair separates, forming the poles of the MII spindle (Fig. 2, B and C, MII spindle). We conclude that centriole maturation from daughter to mother, measured by a transition from hsCentrobin-mEGFP to pmOdf2-mEGFP, does not occur during starfish oocyte meiosis. Thus, starfish oocyte meiosis starts and ends with the same number of centrioles, two mothers and two daughters, which do not change their state of maturation during this process.
An important consequence of this altered centriole cycle is that single centrioles organize the poles of the MII spindle: a mother centriole at one pole and a daughter centriole at the other (Fig. 2, B and C, MII spindle). Strikingly, we found that in the MII spindle, the pmOdf2-mEGFP–labeled mother centriole is reproducibly located at the pole facing the plasma membrane (Fig. 2 B, MII spindle). Consequently, the mother centriole is invariably extruded into PBII (Fig. 2, B [PBII extrusion] and D; and Fig. S2 B). The same conclusion was reached using pmChibby-mEGFP, another mother-specific marker (Fig. S2, C and D). In contrast, we found that in all oocytes imaged, the centriole bearing the daughter-specific hsCentrobin-mEGFP marker is facing inward of the MII spindle and is thus retained in the mature egg (Fig. 2, C [PBII extrusion] and D).
Collectively, our data imply that a specific mechanism must exist to position the MII spindle so that the mother centriole faces outward. As a result, the two mother centrioles are extruded into the polar bodies, along with one daughter centriole extruded into PBI, whereas a single daughter centriole remains in the mature egg.
The MII spindle is positioned by directed transport and subsequent anchoring of the mother centriole to the plasma membrane
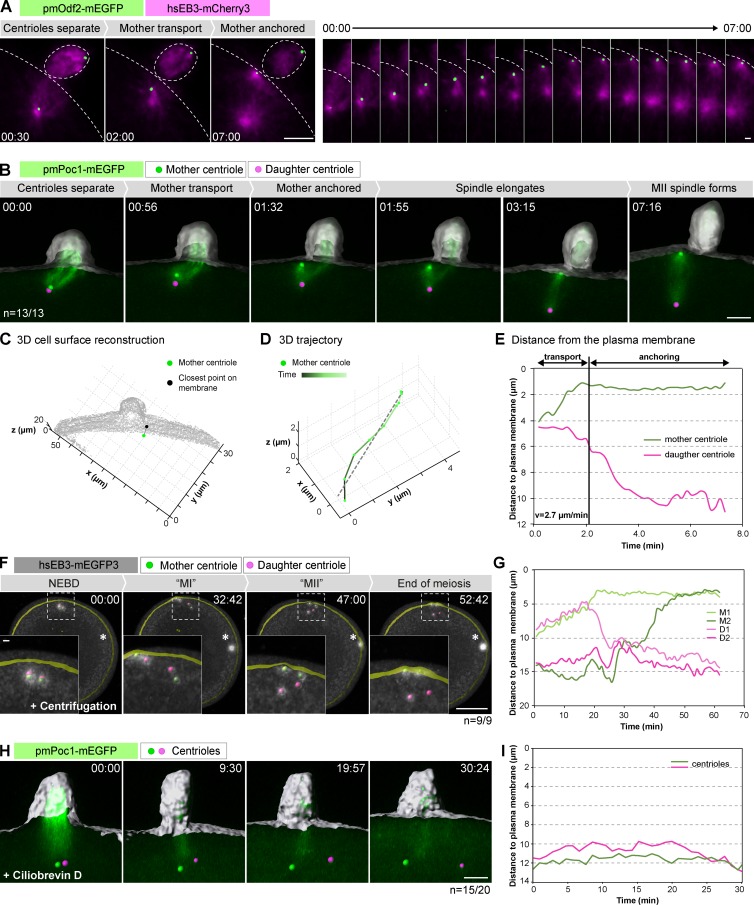
To address the mechanism of mother centriole–specific positioning, we imaged oocytes expressing the microtubule plus tip marker hsEB3-mCherry3 and pmOdf2-mEGFP from anaphase I onwards to visualize the mother centriole in the context of spindle assembly. These recordings revealed that the mother centriole moves progressively to the cell membrane starting almost immediately after anaphase I; once at the cell membrane, the centriole remains stably attached while the MII spindle continues to elongate and does not reorient (Fig. 3 A).
Figure 3.
The mother centriole is transported and anchored to the cell membrane shortly after PBI extrusion. (A) pmOdf2-mEGFP labels mother centrioles; EB3-mCherry3 labels microtubule plus ends. The left panel shows maximum intensity projections for selected time points, the right panel shows the region around the centriole for all frames. Bars: (left) 5 µm; (right) 1 µm. (B) pmPoc1-mEGFP imaged in 3D at high temporal resolution (see Video 4). 3D volume rendering of the data overlaid with an isosurface reconstruction of the cell outline (gray). Identified positions of centrioles are shown as green and pink spheres for mother and daughter centrioles, respectively. z-Stacks were acquired every 12 s. Bar, 5 µm. (C) Example of retrieved 3D coordinates of a mother centriole and the closest point on the plasma membrane. Time point selected from the dataset in B. (D) Example of a 3D mother centriole trajectory during the transport phase showing the linear fit (dashed line) to estimate the speed of motion. (E) Plot of centriole distance from the plasma membrane over time for the oocyte shown in B. (F) Dashed squares indicate the area shown in insets (see Video 5). Asterisk in the first frame shows nuclear position after centrifugation. t = 0 is shortly after NEBD. 3D rendering as in B. Bars: (main) 40; (inset) 5 µm. (G) Distance measurements of mother and daughter centrioles to the plasma membrane over time for the oocyte shown in F. M1 and M2 are mother centrioles; D1 and D2 are daughter centrioles identified based on their microtubule nucleating activity at the end of meiosis. (H) Centrioles were tracked in an oocyte expressing pmPoc1-mEGFP and hsEB3-mCherry3 starting from anaphase I onset (only pmPoc1-mEGFP is shown). 3D rendering as in B. Bar, 5 µm (see Video 6). (I) Plot of centriole distance from the plasma membrane over time for the oocyte shown in H.
To characterize this behavior in a quantitative manner, we imaged this process at higher temporal resolution using pmPoc1-mEGFP (Fig. 3 B and Video 4). In these datasets, centriole positions were detected automatically at every time point, and the cell outline was segmented using the soluble pmPoc1-mEGFP signal (Fig. 3 C). Together, this allowed us to calculate the minimum distance between centrioles and the plasma membrane over time and in 3D. Analyses of the resulting tracks reveal a two-step mechanism of mother centriole positioning (Figs. 3 E and S3). In the first ∼2–4 min after anaphase I, the mother centriole is transported to the plasma membrane, even before the complete disassembly of the MI spindle and completion of cytokinesis I (i.e., PBI extrusion). This transport is directed toward the plasma membrane and has a relatively constant speed of 2.0 ± 0.8 µm/min (Figs. 3 D and S3). Second, the mother centriole remains stably anchored to the plasma membrane, maintaining a constant distance until the end of the recording at cytokinesis II (i.e., PBII extrusion; Fig. 3, A, B, and E; and Fig. S3; note that the distance to the plasma membrane is a relative value derived by the segmentation algorithm; for the absolute distance, see Fig. 4). The daughter centriole shows a very different behavior: no directional motion is observed in the first phase, after which the centriole is pushed deeper in the cytoplasm by the elongating MII spindle (Fig. 3, A, B, and E; and Fig. S3).
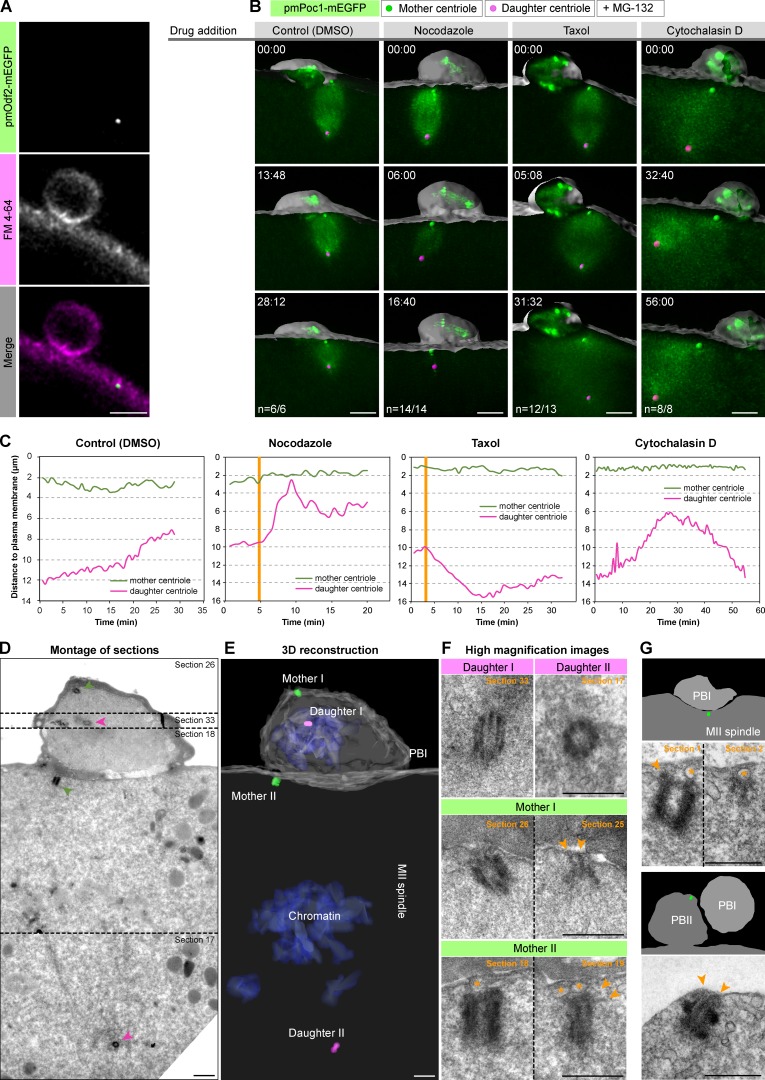
Figure 4.
The mother centriole is anchored to the cell membrane via appendages. (A) The mother centriole labeled by pmOdf2-mEGFP colocalizes with the cell membrane labeled by FM4-64 in metaphase II. Bar, 5 µm. (B) Oocytes expressing pmPoc1-mEGFP were arrested in metaphase II by MG-132. Mother centriole anchoring was assessed in control cells (DMSO) and in oocytes treated with nocodazole, taxol, or cytochalasin D. 3D volume renderings are shown as in Fig. 3 B. t = 0 corresponds to the time of drug addition, time is in mm:ss. (C) Distance of mother and daughter centrioles to the plasma membrane over time for each oocyte shown in B. Orange vertical lines indicate when the drug effect becomes visible. (D) Serial semi-thin section (130 nm) electron micrographs showing the area encompassing the MII spindle and PBI. The panel shows a montage of selected serial sections containing the four centrioles. (E) 3D reconstruction of the serially sectioned volume. (F) High-magnification images of the four centrioles, showing the relevant sections for each of them. Vesicles are identified by an orange asterisk. Orange arrowheads indicate electron-dense connections to the plasma membrane, reminiscent of mother centriole appendages. (G) Top panel shows another example of a mother centriole attached to vesicles (orange asterisk) via electron dense connections reminiscent of appendages during metaphase II (arrowheads). Bottom panel shows a mother centriole still anchored to the plasma membrane after PBII extrusion; arrowheads indicate attachment points to the plasma membrane. Schematic overviews of the whole area and position of the mother centriole within (green sphere) are depicted. Bars: (D and E) 1 µm; (F and G) 0.5 µm.
The directed transport of the mother centriole is reminiscent of the movement of the mother centriole toward the midbody observed in mammalian somatic cells at the end of cytokinesis (Piel et al., 2001). To address a potential similarity in mechanisms, we tested whether mother centriole transport depends on midbody formation of the preceding cytokinesis I in starfish oocytes. Gentle centrifugation of starfish oocytes can be used to relocate the nucleus, separating it from the centrosomes, which remain attached at the cell cortex (Matsuura and Chiba, 2004). These oocytes undergo nuclear envelope breakdown (NEBD), but polar body extrusion does not take place (Barakat et al., 1994; Matsuura and Chiba, 2004). Thus, if centriole transport depended on PBI cytokinesis and midbody formation, it would be prevented in these centrifuged oocytes. We tracked centrosomes in such centrifuged oocytes using hsEB3-mEGFP3, and found that the two mother centrioles—identified by the persistence of microtubule asters after the end of meiosis (see Fig. 5)—are transported to the plasma membrane despite the complete block of PBI extrusion and absence of cytokinetic structures (Fig. 3, F and G; and Video 5).
Figure 5.
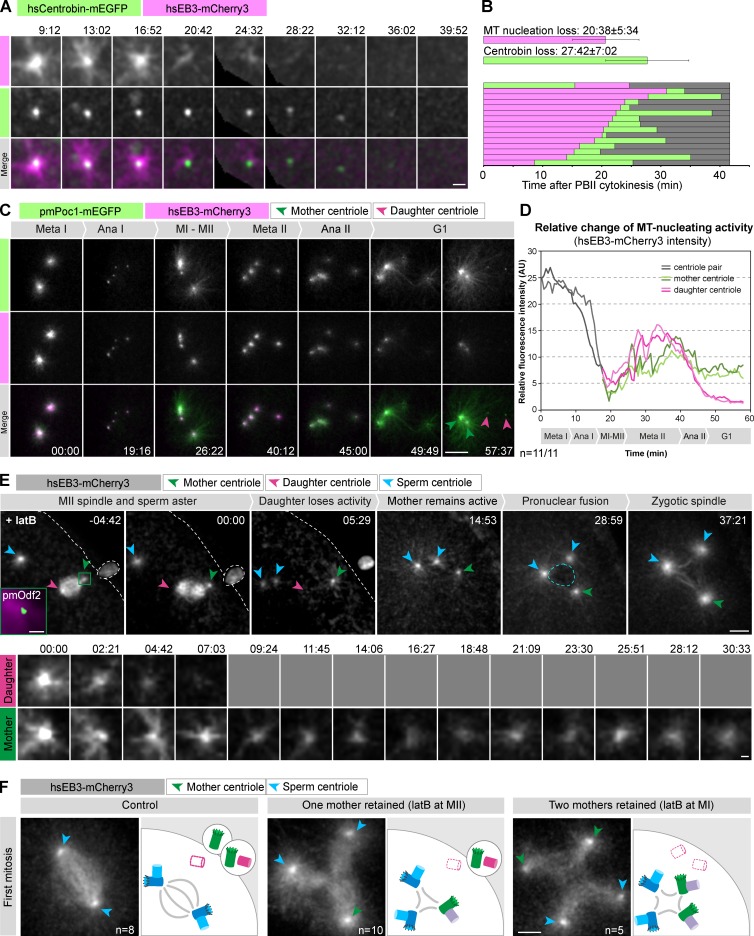
Mother and daughter centrioles follow different fates at the end of meiosis. (A) High-magnification view of a daughter centriole losing microtubule nucleating activity (here visualized by hsEB3-mCherry3), followed by centriole elimination as indicated by loss of the hsCentrobin-mEGFP focus. t = 0 (mm:ss) is set at PBII cytokinesis; panels show single confocal sections. Bar, 2 µm. (B) Quantification of the timing of microtubule nucleating activity loss and of centriole elimination. Top bars indicate the mean times and SDs; bottom bars show data for all individual oocytes imaged. (C) An oocyte expressing pmPoc1-mEGFP and hsEB3-mCherry3 was centrifuged to spatially separate spindle and centrioles. Centrioles were then imaged from MI to the end of meiosis (see Video 7). Maximum intensity z-projections of selected frames are shown. Time in mm:ss; scale bar: 10 µm. (D) Quantification of hsEB3-mCherry3 intensities of individual centrioles. Background-corrected mean fluorescence intensities were measured in a sphere 3 µm in diameter and plotted over time for the oocyte shown in (C). Mother centrioles were identified by their attachment to the plasma membrane. (E) Oocytes expressing hsEB3-mCherry3 and pmOdf2-mEGFP were fertilized and then treated with latB at MII (one mother centriole retained; see Video 9). Top panels show the major events of the process (maximum intensity projections). The centrioles are monitored by hsEB3-mCherry3. The green square in the first frame shows the initial position of the mother centriole identified by pmOdf2-mEGFP (green) and hsEB3-mCherry3 (magenta) at the time of latB addition. t = 0 is set at anaphase II onset. Blue arrowheads indicate sperm, green and pink arrowheads the maternal mother and daughter asters, respectively. The blue and white dashed lines delineate the zygotic nucleus after pronuclear fusion and the cell membrane, respectively. Bottom panels show a higher magnification of a single confocal section (every 2.35 min, the daughter centriole lost activity and therefore could not be followed after the fourth time point as indicated by gray squares). Time is in mm:ss. Bars: (top) 10 µm; (bottom) 5 µm. (F) The left panel shows a control oocyte treated with DMSO and fertilized (see Video 8); the middle panel shows a fertilized oocyte treated with latB at MII (one mother retained, see Video 9); the right panel shows a fertilized oocyte after latB treatment at MI (two mothers retained, see Video 10). For each case, a frame at metaphase of the first zygotic mitosis is shown (maximum intensity projections). On the corresponding schematics on the right, maternal mother and daughter centrioles are shown in green and in pink, respectively. Maternal daughter centrioles after elimination are shown with dashed pink lines. Sperm centrioles are shown in blue and the new generation of zygotic daughter centrioles in violet. The same color code is used for the arrowheads marking centrioles on the images. Bar, 10 µm.
To address the mechanism of mother centriole transport, we next attempted to impair cytoskeletal elements that might be involved in this process. These experiments have proven challenging because transport takes place minutes after anaphase I and concomitantly with cytokinesis I, both of which depend on actin and microtubules. Thus, although these experiments suggested a dependency on microtubules but not on actin, their outcome was too variable to allow a firm conclusion to be reached (unpublished data). In contrast, acute treatment of oocytes with ciliobrevin D, an inhibitor of the minus end–directed motor protein dynein (Firestone et al., 2012), invariably and effectively blocked transport of the mother centriole to the plasma membrane (Fig. 3, H and I; and Video 6). Ciliobrevin D treatment in MI or MII resulted in spindle phenotypes expected for dynein inhibition, confirming the specificity of the treatment (Firestone et al., 2012). Together, these data indicate that centriole transport is dynein driven and imply that it is microtubule dependent.
Collectively, our experiments show that the mother centriole is transported and subsequently anchored to the plasma membrane at early steps of MII spindle assembly. Transport is mediated by dynein and is specific to the mother centriole. Transport is independent of PBI cytokinesis and the meiotic spindle, as it also occurs in oocytes in which centrioles have been spatially separated from the spindle by centrifugation.
The mother centriole stably anchors to the plasma membrane via its appendages
After its transport to the cell cortex, the mother centriole associates stably and tightly with the plasma membrane in the MII spindle (Fig. 4 A; Ucar et al., 2013). This is very different from the configuration in mitotic spindles, where long astral microtubules connect centrosomes to the cell cortex (Grill et al., 2001; von Dassow et al., 2009). To address the mechanism underlying the tight association in the starfish MII oocyte, we first tested whether astral microtubules or actin dynamics are involved. Therefore, we arrested oocytes by MG-132 at metaphase II, in which case the spindle remains stably anchored to the plasma membrane (Fig. 4, B and C, Control). When such arrested oocytes are treated with nocodazole, spindle microtubules rapidly depolymerize, leading to spindle collapse and erratic movement of the daughter centriole (Fig. 4 C, nocodazole). Despite this, the tight association of the mother centriole with the plasma membrane is not affected (Fig. 4, B and C, nocodazole). Similarly, upon microtubule stabilization by taxol, whereas the MII spindle extends to double its normal length, pushing the daughter centriole deeper in the cytoplasm (Fig. 4 C, taxol), the mother centriole remains stably anchored (Fig. 4, B and C, taxol). To test whether actin is involved, we treated arrested oocytes with either cytochalasin D or latrunculin B (latB) to block actin dynamics and found that these treatments also did not affect the tight anchoring of the mother centriole to the plasma membrane (Fig. 4, B and C, cytochalasin D; and Fig. S4, A and B, latrunculin B). Collectively, our results show that after transport, the mother centriole is anchored to the plasma membrane, establishing a tight connection that is stable for hours in oocytes arrested in MII and that does not require the microtubule or the actin cytoskeleton.
To our knowledge, such close and stable association between centriole and plasma membrane was not yet documented in cell division. However, a similar close association of centrioles and the plasma membrane occurs upon formation of the cilium as well as the immunological synapse in T cells. In these cases, the mother centriole anchors to the plasma membrane through its appendages (Reiter et al., 2012; Sung and Leroux, 2013; Jana et al., 2014; Stinchcombe and Griffiths, 2014; Stinchcombe et al., 2015).
We thus performed EM to visualize the details of the tight association between mother centriole and plasma membrane in the MII starfish oocyte to address whether it likewise involves appendages. We used live imaging to identify oocytes in metaphase II or shortly after PBII extrusion, and then immobilized them by chemical fixation. The oocytes were processed for EM, and serial sections were cut around the polar body region using microscopic x-ray computed tomography (microCT) targeting (n = 10 oocytes; see Materials and methods and Karreman et al., 2016, for details). This analysis identified four individual centrioles in total, fully consistent with the light microscopy data and earlier EM studies (Kato et al., 1990; Fig. 4, D and E; compare with Fig. 2). Whereas two of these centrioles are located in PBI, a single centriole is found at each pole of the MII spindle (Fig. 4, D and E). Strikingly, two of the four centrioles are tightly associated with the plasma membrane in a perpendicular configuration: one in PBI and one at the outer spindle pole of the MII spindle. Importantly, analysis at higher magnification revealed electron-dense connections between these mother centrioles and the plasma membrane or membrane vesicles located beneath the plasma membrane, which are typical of mother centriole appendages (Fig. 4 F, arrowheads). Similar ultrastructural features were found in oocytes fixed at later stages of MII or even after formation of PBII, consistent with the high stability of this association (Fig. 4 G). In clear contrast, the centrioles identified as daughters do not feature structures reminiscent of appendages and do not establish connections with the plasma membrane (Fig. 4 F).
Collectively, the association of the mother centriole with the plasma membrane through characteristic electron densities indicates that the mother centriole anchors to the plasma membrane via its appendages. This explains the observations that neither microtubules nor actin are required for anchoring of the mother centriole to the plasma membrane, and that pmOdf2-mEGFP, a component of mother centriole appendages, colocalizes with the plasma membrane at the resolution of light microscopy (Fig. 4 A). Because appendages are specific to the mother centriole, these data also provide an explanation for why only the mother centriole anchors to the plasma membrane.
The daughter centriole is inactivated and eliminated in the egg cytoplasm, but artificially retained mother centrioles remain active
Our data show that mother centrioles are selectively removed from the oocyte into the polar bodies, leaving a single daughter centriole in the mature egg. We next aimed to address how this remaining centriole is eliminated and whether extrusion of mother centrioles is necessary for elimination of centrioles originally contained in the oocyte.
We used the microtubule plus tip marker hsEB3-mCherry3 to assay the microtubule nucleating activity of the remaining daughter centriole. Time-lapse imaging revealed a loss of activity down to undetectable levels 20.6 ± 5.6 min after anaphase II (Fig. 5, A and B). This was accompanied by gradual reduction of the focus of hsCentrobin-mEGFP, which reached background levels 27.7 ± 7.0 min after anaphase II, a mean of 7.1 min after the loss of microtubule nucleating activity (Fig. 5, A and B). Thus, the daughter centriole is first inactivated, as indicated by loss of microtubule nucleating activity, and then eliminated, as indicated by disappearance of a daughter centriole protein. Interestingly, elimination thus proceeds in the same order as during C. elegans oogenesis, where loss of microtubule nucleating activity precedes dissociation of core centriolar components (Mikeladze-Dvali et al., 2012).
Next, we set out to investigate whether elimination is specific to the daughter centriole or whether all maternal centrioles that are contained in the cytoplasm are inactivated and eliminated. Therefore, we spatially separated chromatin and centrioles before meiosis by centrifugation to avoid potentially confounding contributions of spindle microtubules and cytokinetic structures. We then quantified hsEB3-mCherry3 levels at individual centrioles to dynamically assay microtubule-nucleating activity through meiosis. This assay clearly visualized the expected cell cycle–driven changes in nucleation activity but did not reveal any systematic difference between centrioles before meiosis II (Fig. 5, C and D; and Video 7). Interestingly, we found that pairs of centrioles in MI nucleate roughly twice as many microtubules as individual centrioles in MII, suggesting that the microtubule-nucleating activity of all four centrioles is similar in MI and MII (Fig. 5, C and D). Importantly, a dramatic difference emerges just after anaphase II, when the two daughter centrioles abruptly and synchronously lose microtubule nucleating activity, whereas mothers retain it (Fig. 5, C and D).
Next we wanted to address the functional importance of this difference in microtubule nucleating activity between mother and daughter centrioles emerging after anaphase II. For this purpose, we artificially retained mother centrioles in the egg and followed effects on zygotic spindle formation after fertilization. As expected, in untreated oocytes fertilized at MI, after inactivation of the last remaining daughter centriole, the two sperm-derived centrioles directed assembly of the bipolar zygotic spindle (Fig. 5 F, left; and Video 8).
To retain one mother centriole in the egg, oocytes were treated with latB at metaphase II to prevent PBII cytokinesis (Fig. 5 E and Video 9). This results in the reabsorption of PBII contents but does not interfere with further meiotic progression, pronuclear migration, fusion, or assembly of the zygotic spindle (Fig. 5 E). As shown in Fig. 5 E, we found that after such a treatment, the daughter centriole is inactivated as in control cells. In contrast, the mother centriole retains microtubule nucleating activity and contributes, together with the sperm-derived centrioles, to the assembly of a tripolar spindle (Fig. 5, E and F, middle panel).
Next, to test the consequences of retaining all four maternal centrioles in the oocyte, we prevented extrusion of both polar bodies by treating cells with latB at metaphase I. In this case, the two mother centrioles remain active and contribute to the assembly of a tetrapolar spindle, together with the sperm-derived centrioles (Fig. 5 F, right; and Video 10). This additionally implies that the first daughter centriole, which normally would have been extruded into PBI, is equally inactivated and does not contribute to zygotic spindle assembly. These behaviors were independent of fertilization, as mother centrioles persisted in unfertilized eggs for several hours after preventing extrusion of one or both polar bodies (Fig. S5).
In summary, we conclude that mother and daughter centrioles exhibit intrinsic differences emerging at the end of meiosis: mother centrioles retain microtubule nucleating activity, whereas daughter centrioles lose it after the completion of meiosis and are eliminated shortly thereafter. This difference in fate explains why mother centrioles have to be physically removed from the egg by extrusion into polar bodies to prevent assembly of multipolar zygotic spindles leading to aneuploidy and impaired embryonic development (Zhang et al., 2004; Maderspacher, 2008; Scheer, 2014).
Discussion
In a manner that is conceptually similar to the reduction of DNA content in meiosis, centriole number also needs to be reset before fusion of the gametes. This is achieved in most species by elimination of the maternal centrioles, whereas the sperm provides active paternal centrioles at fertilization (Delattre and Gönczy, 2004; Manandhar et al., 2005). Therefore, centriole elimination from the oocyte is essential for sexual reproduction of animal species, but the underlying mechanisms are very poorly understood.
Here, we took advantage of starfish oocytes, which are exceptionally well suited to image and manipulate this process in living cells. We show that, of the four total centrioles contained in the oocyte, two (one mother and one daughter) are extruded into PBI. The two centrioles that remain in the egg, one mother and one daughter, organize the MII spindle. Importantly, we show that the MII spindle is invariably positioned with the mother centriole facing the plasma membrane at the site of PBII extrusion. This positioning is achieved by dynein-driven directed transport and subsequent stable anchoring of the mother centriole to the plasma membrane via mother-specific appendages. As a result, the mother centriole is extruded into PBII, and a single daughter centriole remains in the mature egg. This daughter centriole loses microtubule-nucleating activity and is eliminated shortly after the completion of meiosis.
Our findings are consistent with and explain several previous observations in which the “aster forming capacity” of centrioles retained or transplanted from polar bodies were tested. We can principally equate “centrioles with replicative potential” in these earlier studies with the mother centrioles, and “non-replicative centrioles” with daughter centrioles (Tamura and Nemoto, 2001; Uetake et al., 2002; Zhang et al., 2004; Shirato et al., 2006). However, in our system, we did not find support for the model in which all maternal centrioles would be eliminated in the specific environment provided by the egg’s cytoplasm (Sluder et al., 1989, 1993).
A key element of the model unveiled here is that mechanisms of centriole elimination differ depending on centriole age. Mother centrioles cannot be eliminated in the cytoplasm, as shown here and by previous polar body retention experiments (Zhang et al., 2004) and therefore need to be physically removed into the polar bodies. Intriguingly, our data suggest that the anchoring, and possibly the transport, that are required for extrusion of the mother centriole are mediated by mother centriole–specific appendages. Thus, in our model the mother centriole uses its specific appendages to ensure its own extrusion.
To our knowledge, this is the first case in which mother appendages are involved in spindle positioning. However, mother appendages have been demonstrated to form morphologically similar tight membrane attachments in ciliogenesis and immunological synapse formation (Reiter et al., 2012; Sung and Leroux, 2013; Jana et al., 2014; Stinchcombe and Griffiths, 2014; Stinchcombe et al., 2015). Intriguingly, we observe vesicles between the plasma membrane and appendages that are morphologically similar to those observed during ciliogenesis, where they have been proposed to deliver the centriole to the cell membrane (Reiter et al., 2012; Sung and Leroux, 2013; Jana et al., 2014). Furthermore, during T cell polarization, the mother centriole is transported toward the plasma membrane with a speed and trajectory remarkably similar to what we observed in starfish oocytes, in a process that has been proposed to require dynein (Kuhn and Poenie, 2002; Yi et al., 2013). Furthermore, the attachment to the plasma membrane via appendages shows striking morphological similarities in the two cases (Stinchcombe et al., 2015). It will be interesting in the future to test a possible conservation of molecular mechanisms between these processes.
In contrast to mother centrioles, daughter centrioles are eliminated in the egg’s cytoplasm at the end of meiosis. We propose that this might be the consequence of the disengagement of the two centrioles at the end of MI, combined with the lack of centriole duplication between MI and MII, which results in a meiosis-specific configuration that does not occur in somatic cells. In the somatic cycle, PCM is recruited by the mother centriole, with the daughter centriole being automatically coembedded because of its engagement with the mother (Wang et al., 2011). Thus, the single daughter centriole, resulting from the meiosis-specific centriole cycle described here, may not be stable owing to its inability to maintain a PCM, as it is no longer associated with a mother centriole. Indeed, at the end of meiosis, we observed an abrupt loss of microtubule nucleating activity that may reflect PCM dispersion, which in turn could cause the destabilization and collapse of the core centriolar structure.
In summary, our data lead us to propose a comprehensive model for centriole elimination in oocytes. In this model, known features of centrioles are combined in a meiosis-specific sequence to physically remove replicative mother centrioles, leaving an unstable nonreplicative daughter centriole in the mature egg. Although it is likely that mechanisms of centriole elimination differ between species, our data suggest that mother and daughter centrioles rely on distinct mechanisms for elimination.
Materials and methods
Oocyte injection, maturation, and fertilization
Starfish (P. miniata, also known as Asterina miniata) were obtained from Southern California Sea Urchin Co., Marinus Scientific, South Coast Bio-Marine, or Monterey Abalone Co. and maintained in seawater tanks at 16°C at the European Molecular Biology Laboratory (EMBL) Marine Facility or at the Marine Resource Center of the Marine Biological Laboratory (Woods Hole, MA). Oocytes were isolated and injected using mercury-filled needles as described in Terasaki (1994). Protein markers were injected shortly before initiation of meiosis, whereas mRNAs encoding fluorescent markers were injected the day before and incubated overnight at 14°C to obtain sufficient levels of protein expression. Meiotic maturation was triggered by addition of 10 µM 1-methyladenine (Acros Organics). Oocyte centrifugation was performed at 2,400 rpm for 1 h (Multifuge 3 liter-R; Heraeus) at 4°C, as detailed in Matsuura and Chiba (2004) and Mori et al. (2011).
Testes excised from starfish were kept dry at 4°C. Just before fertilization, a minced piece of testis was diluted to ∼1:8,000 in seawater. Successful fertilization was confirmed under a dissecting microscope, and thereafter embryos were allowed to develop for ∼18 h at 14°C, by which time they reached the early gastrula stage with a ciliated epithelium. For latB experiments, oocytes were first fertilized and then treated with latB, as fertilization is known to be actin dependent.
Identification of centriolar proteins in starfish
Starfish homologs were identified in a transcriptome dataset prepared from mature P. miniata eggs as follows. Total RNA was extracted using the TRIzol LS reagent following the manufacturer’s instructions (Life Technologies), polyA+ RNA was isolated, and mRNAs were chemically fragmented (Wery et al., 2013) and sequenced on an Illumina HiSeq2000 platform using 50-bp paired-end reads at EMBL’s Genomics Core Facility. The resulting reads were assembled using Trinity (Grabherr et al., 2011) and are available at http://www.lenartlab.embl.de:4567.
Using a list of human centriole proteins (Hodges et al., 2010; Carvalho-Santos et al., 2011), we first identified sea urchin homologs in the S. purpuratus genome (http://sugp.caltech.edu/SpBase/wwwblast/blast.php). Sea urchin sequences were then used to search for homologs in the P. miniata transcriptome by BLASTP (Altschul et al., 1990). Hits were considered homologs when the e-value was <10−20 and the human protein was the best reciprocal hit in a reverse BLAST search. Identified starfish proteins and the corresponding sea urchin and human homologs are listed in Table S1.
Live cell fluorescent markers
A cDNA library was made from polyA+ total RNA extracted from mature starfish eggs as described in the previous section and reverse-transcribed with the GeneRacer kit (Invitrogen). Full-length cDNAs for pmCentrin-2, pmOdf2, and pmPoc1 were isolated using specific primers designed based on the transcriptome data. pmChibby cDNA was synthetized by GENEWIZ. hsCentrobin was a gift from G. Lukinavičius (Swiss Federal Institute of Technology, Lausanne, Switzerland; Lukinavičius et al., 2013). These sequences were typically N-terminally (C-terminally, in the case of pmCentrin-2) fused to mEGFP or mCherry and subcloned into pGEMHE for in vitro transcription as described (Lénárt et al., 2003). Capped mRNAs were synthesized from linearized templates using the AmpliCap-Max T7 High Yield Message Maker kit and extended with poly(A) tails at the 3′ termini using the Poly(A) Tailing kit (both CellScript). hsEB3-mEGFP3 and -mCherry3 mRNAs were synthesized using the same protocol. mRNAs were dissolved in 11 µl RNase-free water (typically at 6–12 µg/µl) and injected to 1–5% of the oocyte volume. Cy3-tubulin (gift from the Nédélec laboratory, EMBL, Heidelberg, Germany) and HiLyte 647-tubulin (Cytoskeleton, reconstituted according to the manufacturer’s instructions) were injected to oocytes. FM 4–64 (Thermo Fisher Scientific) was reconstituted according to the manufacturer’s instructions and added to the oocytes at a final concentration of 5 µg/ml. Labeling of the plasma membrane occurred progressively 2–5 min after FM4-64 addition.
Drug treatments
To provoke metaphase II arrest, oocytes were treated with a final concentration of 250 µM MG-132 (Calbiochem) added 45 min after inducing maturation. To confirm MG-132–induced arrest, oocytes were monitored for at least 45 min in metaphase II arrest before additional drug treatments. After this period, oocytes were treated with given cytoskeletal inhibitors and imaged for 20–45 min. Cytochalasin D (Sigma-Aldrich) was used at a final concentration of 10 µM, latrunculin B (EMD Biosciences) at 250 nM, taxol (Sigma-Aldrich) at 11 nM, and nocodazole (EMD Biosciences) at 3.3 µM. Ciliobrevin D (Merck Millipore) was used at 100 µM final concentration and was added 2 min after anaphase I.
Light microscopy and image processing
Microscopy was done on a Leica SP5 or SP8 confocal microscope equipped with a fast Z-focusing device (SuperZ Galvo stage) and using a 40× HCX PL APO 1.10 NA water immersion objective lens (all Leica Microsystems). Starfish oocytes were imaged in 3D (with a z-step of 1–2 µm; typically 1.5 µm) over time (typical time step 20–40 s) using a square frame of 304–512 pixels (typically 304) at a pixel size of 130–500 nm (typically 220 nm). In a typical experiment, three to five oocytes were imaged using the multilocation mode, except for the centriole transport experiments (Fig. 3), in which a single oocyte was imaged at a faster speed (typically 11–20 s). All imaging was performed at RT (20–22°C).
Image data are shown after brightness and contrast adjustment and application of a 2D or 3D Gaussian blur filter (sigma value: 0.4–0.8). Panels show either single Z-slices, maximum intensity projections, or 3D renderings as indicated in the figure legends. Image processing was done using Fiji (Schindelin et al., 2012) or Imaris (Bitplane). Figures were assembled in Adobe Illustrator.
Additionally, where indicated, the strong autofluorescence of cortical granules was removed post-acquisition. For this purpose an additional red-shifted channel, which detects autofluorescence but no mEGFP emission, was recorded. This autofluorescence channel was then subtracted from the mEGFP channel to remove the contribution of the autofluorescence.
Centriole detection and tracking
A Fiji macro was used to automatically segment the cell outline based on the cytoplasmic pmPoc1-mEGFP fluorescence to obtain a set of 3D surface coordinates for each time point. This pipeline included an anisotropic diffusion filtering step, followed by interpolation to isotropic XYZ resolution, a 3D Gaussian blur filtering step, and automatic thresholding using the “Mean” algorithm. The macro code is available on request. Centriole tracking was performed in Imaris using the automatic spot detection and tracking functions. Tracks were manually validated and corrected when necessary. XYZ coordinates were then exported to an Excel file. The minimum distance between centrioles and 3D surface coordinates was calculated using a script written in Matlab (MathWorks) by searching for the minimum Euclidian distance among all possible combinations of cell outline and centriole coordinates. The velocity of the mother centriole was calculated by fitting a 3D line to the tracks. The transport phase was manually defined. For quantifying hsEB3-mCherry3 associated with centrioles, centrioles were similarly tracked in Imaris using the spot detection and tracking functions; the same function was then used to extract the mean background-corrected intensities contained in a 3-µm diameter spot.
For Fig. S1, centrioles were automatically detected by the spot detection function in Imaris. Pairwise distances between all pairs labeled by the two markers were calculated by solving the linear sum assignment problem (Papadimitriou and Steiglitz, 1982) using the “clue” package (Hornik, 2005). A fixed threshold based on cell diameter was used to assign the corresponding pairs.
EM
Oocytes labeled with pmPoc1-mEGFP or Cy3-tublin were followed by live imaging up to metaphase II or shortly after PBII extrusion. Oocytes were then fixed for 1 h at RT with 1% glutaraldehyde and 2% formaldehyde (both Electron Microscopy Sciences) in PHEM buffer (60 mM Pipes, 25 mM Hepes, 10 mM EGTA, and 2 mM MgCl2, pH adjusted to 6.9 with KOH), and stored at 4°C. Samples were then mounted in cellulose capillaries or agar blocks and further processed for EM in a PELCO Biowave Pro microwave (Ted Pella, Inc.). The oocytes were first washed in cacodylate buffer, pH 7.4, and primary postfixed with 1% OsO4 (Electron Microscopy Sciences) and 1.5% K4Fe(CN)6 (Merck) in 0.1 M cacodylate buffer. A second postfixation was then performed with 1% OsO4 in 0.1 M cacodylate buffer. The samples were stained with 1% aqueous uranyl acetate (UA) and gradually dehydrated in ethanol and embedded in epon. Oocytes were then mounted in resin molds. After polymerization and trimming, we performed microCT scanning in a Phoenix Nanotom (GE Sensing & Inspection Technologies GmbH; Karreman et al., 2016). The microCT volume was reconstructed using Phoenix datos|x reconstruction software (GE Sensing & Inspection Technologies GmbH), and the volume was then processed using VGStudio MAX software (Volume Graphics). The microCT datasets were then loaded in Amira software (FEI company), and semiautomatically segmented using the “Labels” module. 3D surface models were generated to target the area around the PBI, which was approached by targeted ultramicrotomy. At the position of the PBI, a series of thin sections (typically 130 nm) were obtained. These sections were poststained with 2% UA in 70% methanol and lead citrate, followed by imaging using a transmission electron microscope at 120 kV (FEI company).
Online supplemental material
Table S1 lists names and IDs of all starfish centriolar proteins as well as those of sea urchin and human homologs. Fig. S1 shows automated counting of centriole configuration in a large number of basal bodies. Fig. S2 shows additional centriole markers, mEGFP-pmCentrin-2 and pmChibby-mEGFP, confirming findings shown in Fig. 2. Fig. S3 shows tracking data from 12 individual oocytes complementing the single example shown in Fig. 3. Fig. S4 confirms the independence of mother centriole anchoring from actin dynamics by latB treatment. Fig. S5 shows that persistence of mother centrioles is independent of fertilization. Videos show complete time-lapse series of which selected frames are displayed in Figs. 2 A (Video 1), 2 B (Video 2), 2 C (Video 3), 3 B (Video 4), 3 F (Video 5), 3 H (Video 6), 5 C (Video 7), and 5 F (Videos 8, 9, and 10). Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201510083/DC1.
Supplementary Material
Acknowledgments
We thank Konrad Rudolph, Serge Dimitrieff, and Philippe Bun for helping with implementing scripts for centriole detection and tracking. We also thank Kasia Tarnawska and Francois Nédélec for the gift of Cy3-tubulin, as well as Grazvydas Lukinavičius and Benita Wolf for providing the hsCentrobin clone. We thank Edgar Gomes and Jan Ellenberg for comments on the manuscript. We thank Gabriele Marcassa for his help with the EM sample preparation and Bernhard Ruthensteiner for his assistance with the microCT. We thank Francesca Peri, Pedro Machado, and Inês Bento for helpful discussions. We thank Mark Terasaki for his constant support throughout the project.
The work was supported by the European Molecular Biology Laboratory (EMBL) and specifically the EMBL International PhD Program to J. Borrego-Pinto. We acknowledge the essential support of EMBL’s Genomics, Advanced Light Microscopy, and Electron Microscopy Core Facilities as well as Laboratory Animal Resources, to K. Crnokic in particular. Part of this work was performed at the Marine Biological Laboratory (Woods Hole, MA), supported by the Laura and Arthur Colwin Endowed Summer Research Fellowship to P. Gönczy. T. Müller-Reichert received funding from the Deutsche Forschungsgemeinschaft grant MU1423/4-1.
The authors declare no competing financial interests.
Footnotes
Abbreviations used in this paper:
- FP
- fluorescent protein
- MI
- first meiotic division
- MII
- second meiotic division
- NEBD
- nuclear envelope breakdown
- PBI
- first polar body
- PBII
- second polar body
- PCM
- pericentriolar material
References
- Albertson D.G., and Thomson J.N.. 1993. Segregation of holocentric chromosomes at meiosis in the nematode, Caenorhabditis elegans. Chromosome Res. 1:15–26. 10.1007/BF00710603 [DOI] [PubMed] [Google Scholar]
- Altschul S.F., Gish W., Miller W., Myers E.W., and Lipman D.J.. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403–410. 10.1016/S0022-2836(05)80360-2 [DOI] [PubMed] [Google Scholar]
- Arquint C., Gabryjonczyk A.-M., and Nigg E.A.. 2014. Centrosomes as signalling centres. Philos. Trans. R. Soc. B Biol. Sci. 369:20130464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barakat H., Geneviere-Garrigues A.-M., Schatt P., and Picard A.. 1994. Subcellular distribution of aster-nucleated microtubule length: A more or less mitotic status of cytoplasmic areas during meiosis I of starfish oocytes. Biol. Cell. 81:205–213. 10.1016/0248-4900(94)90002-7 [DOI] [Google Scholar]
- Brito D.A., Gouveia S.M., and Bettencourt-Dias M.. 2012. Deconstructing the centriole: Structure and number control. Curr. Opin. Cell Biol. 24:4–13. 10.1016/j.ceb.2012.01.003 [DOI] [PubMed] [Google Scholar]
- Burke M.C., Li F.-Q., Cyge B., Arashiro T., Brechbuhl H.M., Chen X., Siller S.S., Weiss M.A., O’Connell C.B., Love D., et al. 2014. Chibby promotes ciliary vesicle formation and basal body docking during airway cell differentiation. J. Cell Biol. 207:123–137. 10.1083/jcb.201406140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho-Santos Z., Azimzadeh J., Pereira-Leal J.B., and Bettencourt-Dias M.. 2011. Evolution: Tracing the origins of centrioles, cilia, and flagella. J. Cell Biol. 194:165–175. 10.1083/jcb.201011152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavali P.L., Pütz M., and Gergely F.. 2014. Small organelle, big responsibility: The role of centrosomes in development and disease. Philos. Trans. R. Soc. B Biol. Sci. 369:20130468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowder M.E., Strzelecka M., Wilbur J.D., Good M.C., von Dassow G., and Heald R.. 2015. A comparative analysis of spindle morphometrics across metazoans. Curr. Biol. 25:1542–1550. 10.1016/j.cub.2015.04.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dävring L., and Sunner M.. 1973. Female meiosis and embryonic mitosis in Drosophila melanogaster. I. Meiosis and fertilization. Hereditas. 73:51–64. 10.1111/j.1601-5223.1973.tb01067.x [DOI] [PubMed] [Google Scholar]
- Delattre M., and Gönczy P.. 2004. The arithmetic of centrosome biogenesis. J. Cell Sci. 117:1619–1630. 10.1242/jcs.01128 [DOI] [PubMed] [Google Scholar]
- Fırat-Karalar E.N., and Stearns T.. 2014. The centriole duplication cycle. Philos. Trans. R. Soc. B Biol. Sci. 369:20130460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firestone A.J., Weinger J.S., Maldonado M., Barlan K., Langston L.D., O’Donnell M., Gelfand V.I., Kapoor T.M., and Chen J.K.. 2012. Small-molecule inhibitors of the AAA+ ATPase motor cytoplasmic dynein. Nature. 484:125–129. 10.1038/nature10936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu J., Hagan I.M., and Glover D.M.. 2015. The centrosome and its duplication cycle. Cold Spring Harb. Perspect. Biol. 7:a015800 10.1101/cshperspect.a015800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gard D.L. 1994. γ-tubulin is asymmetrically distributed in the cortex of Xenopus oocytes. Dev. Biol. 161:131–140. 10.1006/dbio.1994.1015 [DOI] [PubMed] [Google Scholar]
- Godinho S.A., and Pellman D.. 2014. Causes and consequences of centrosome abnormalities in cancer. Philos. Trans. R. Soc. B Biol. Sci. 369:20130467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gönczy P. 2012. Towards a molecular architecture of centriole assembly. Nat. Rev. Mol. Cell Biol. 13:425–435. 10.1038/nrm3373 [DOI] [PubMed] [Google Scholar]
- Gönczy P. 2015. Centrosomes and cancer: Revisiting a long-standing relationship. Nat. Rev. Cancer. 15:639–652. 10.1038/nrc3995 [DOI] [PubMed] [Google Scholar]
- Grabherr M.G., Haas B.J., Yassour M., Levin J.Z., Thompson D.A., Amit I., Adiconis X., Fan L., Raychowdhury R., Zeng Q., et al. 2011. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 29:644–652. 10.1038/nbt.1883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grill S.W., Gönczy P., Stelzer E.H., and Hyman A.A.. 2001. Polarity controls forces governing asymmetric spindle positioning in the Caenorhabditis elegans embryo. Nature. 409:630–633. 10.1038/35054572 [DOI] [PubMed] [Google Scholar]
- Hodges M.E., Scheumann N., Wickstead B., Langdale J.A., and Gull K.. 2010. Reconstructing the evolutionary history of the centriole from protein components. J. Cell Sci. 123:1407–1413. 10.1242/jcs.064873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornik K. 2005. A CLUE for CLUster Ensembles. J. Stat. Softw. 14:65–72. 10.18637/jss.v014.i12 [DOI] [Google Scholar]
- Ishikawa H., Kubo A., Tsukita S., and Tsukita S.. 2005. Odf2-deficient mother centrioles lack distal/subdistal appendages and the ability to generate primary cilia. Nat. Cell Biol. 7:517–524. 10.1038/ncb1251 [DOI] [PubMed] [Google Scholar]
- Jana S.C., Marteil G., and Bettencourt-Dias M.. 2014. Mapping molecules to structure: unveiling secrets of centriole and cilia assembly with near-atomic resolution. Curr. Opin. Cell Biol. 26:96–106. 10.1016/j.ceb.2013.12.001 [DOI] [PubMed] [Google Scholar]
- Karreman M.A., Mercier L., Schieber N.L., Solecki G., Allio G., Winkler F., Ruthensteiner B., Goetz J.G., and Schwab Y.. 2016. Fast and precise targeting of single tumor cells in vivo by multimodal correlative microscopy. J. Cell Sci. 16:144–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato K.H., Washitani-Nemoto S., Hino A., and Nemoto S.. 1990. Ultrastructural studies on the behavior of centrioles during meiosis of starfish oocytes. Dev. Growth Differ. 32:41–49. 10.1111/j.1440-169X.1990.00041.x [DOI] [PubMed] [Google Scholar]
- Kong D., Farmer V., Shukla A., James J., Gruskin R., Kiriyama S., and Loncarek J.. 2014. Centriole maturation requires regulated Plk1 activity during two consecutive cell cycles. J. Cell Biol. 206:855–865. 10.1083/jcb.201407087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn J.R., and Poenie M.. 2002. Dynamic polarization of the microtubule cytoskeleton during CTL-mediated killing. Immunity. 16:111–121. 10.1016/S1074-7613(02)00262-5 [DOI] [PubMed] [Google Scholar]
- Lange B.M., and Gull K.. 1995. A molecular marker for centriole maturation in the mammalian cell cycle. J. Cell Biol. 130:919–927. 10.1083/jcb.130.4.919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lénárt P., Rabut G., Daigle N., Hand A.R., Terasaki M., and Ellenberg J.. 2003. Nuclear envelope breakdown in starfish oocytes proceeds by partial NPC disassembly followed by a rapidly spreading fenestration of nuclear membranes. J. Cell Biol. 160:1055–1068. 10.1083/jcb.200211076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longo F.J., and Anderson E.. 1969. Cytological aspects of fertilization in the lamellibranch, Mytilus edulis. I. Polar body formation and development of the female pronucleus. J. Exp. Zool. 172:69–95. 10.1002/jez.1401720107 [DOI] [PubMed] [Google Scholar]
- Lukinavičius G., Lavogina D., Orpinell M., Umezawa K., Reymond L., Garin N., Gönczy P., and Johnsson K.. 2013. Selective chemical crosslinking reveals a Cep57-Cep63-Cep152 centrosomal complex. Curr. Biol. 23:265–270. 10.1016/j.cub.2012.12.030 [DOI] [PubMed] [Google Scholar]
- Maderspacher F. 2008. Theodor Boveri and the natural experiment. Curr. Biol. 18:R279–R286. 10.1016/j.cub.2008.02.061 [DOI] [PubMed] [Google Scholar]
- Manandhar G., Schatten H., and Sutovsky P.. 2005. Centrosome reduction during gametogenesis and its significance. Biol. Reprod. 72:2–13. 10.1095/biolreprod.104.031245 [DOI] [PubMed] [Google Scholar]
- Matsuura R.K., and Chiba K.. 2004. Unequal cell division regulated by the contents of germinal vesicles. Dev. Biol. 273:76–86. 10.1016/j.ydbio.2004.04.038 [DOI] [PubMed] [Google Scholar]
- Mikeladze-Dvali T., von Tobel L., Strnad P., Knott G., Leonhardt H., Schermelleh L., and Gönczy P.. 2012. Analysis of centriole elimination during C. elegans oogenesis. Development. 139:1670–1679. 10.1242/dev.075440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazaki A., Kato K.H., and Nemoto S.. 2005. Role of microtubules and centrosomes in the eccentric relocation of the germinal vesicle upon meiosis reinitiation in sea-cucumber oocytes. Dev. Biol. 280:237–247. 10.1016/j.ydbio.2005.01.026 [DOI] [PubMed] [Google Scholar]
- Mori M., Monnier N., Daigle N., Bathe M., Ellenberg J., and Lénárt P.. 2011. Intracellular transport by an anchored homogeneously contracting F-actin meshwork. Curr. Biol. 21:606–611. 10.1016/j.cub.2011.03.002 [DOI] [PubMed] [Google Scholar]
- Nakashima S., and Kato K.H.. 2001. Centriole behavior during meiosis in oocytes of the sea urchin Hemicentrotus pulcherrimus. Dev. Growth Differ. 43:437–445. 10.1046/j.1440-169x.2001.00580.x [DOI] [PubMed] [Google Scholar]
- Paintrand M., Moudjou M., Delacroix H., and Bornens M.. 1992. Centrosome organization and centriole architecture: their sensitivity to divalent cations. J. Struct. Biol. 108:107–128. 10.1016/1047-8477(92)90011-X [DOI] [PubMed] [Google Scholar]
- Papadimitriou C.H., and Steiglitz K.. 1982. Combinatorial Optimization: Algorithms and Complexity. Prentice Hall, Upper Saddle River, NJ. 528 pp. [Google Scholar]
- Piel M., Nordberg J., Euteneuer U., and Bornens M.. 2001. Centrosome-dependent exit of cytokinesis in animal cells. Science. 291:1550–1553. 10.1126/science.1057330 [DOI] [PubMed] [Google Scholar]
- Reina J., and Gonzalez C.. 2014. When fate follows age: Unequal centrosomes in asymmetric cell division. Philos. Trans. R. Soc. B Biol. Sci. 369:20130466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiter J.F., Blacque O.E., and Leroux M.R.. 2012. The base of the cilium: Roles for transition fibres and the transition zone in ciliary formation, maintenance and compartmentalization. EMBO Rep. 13:608–618. 10.1038/embor.2012.73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saiki T., and Hamaguchi Y.. 1998. Aster-forming abilities of the egg, polar body, and sperm centrosomes in early starfish development. Dev. Biol. 203:62–74. 10.1006/dbio.1998.9045 [DOI] [PubMed] [Google Scholar]
- Sathananthan A.H., Ratnasooriya W.D., de Silva A., and Randeniya P.. 2006. Rediscovering Boveri’s centrosome in Ascaris (1888): its impact on human fertility and development. Reprod. Biomed. Online. 12:254–270. 10.1016/S1472-6483(10)60867-X [DOI] [PubMed] [Google Scholar]
- Scheer U. 2014. Historical roots of centrosome research: discovery of Boveri’s microscope slides in Würzburg. Philos. Trans. R. Soc. B Biol. Sci. 369:20130469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., et al. 2012. Fiji: An open-source platform for biological-image analysis. Nat. Methods. 9:676–682. 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirato Y., Tamura M., Yoneda M., and Nemoto S.. 2006. Centrosome destined to decay in starfish oocytes. Development. 133:343–350. 10.1242/dev.02193 [DOI] [PubMed] [Google Scholar]
- Sluder G., Miller F.J., Lewis K., Davison E.D., and Rieder C.L.. 1989. Centrosome inheritance in starfish zygotes: Selective loss of the maternal centrosome after fertilization. Dev. Biol. 131:567–579. 10.1016/S0012-1606(89)80027-2 [DOI] [PubMed] [Google Scholar]
- Sluder G., Miller F.J., and Lewis K.. 1993. Centrosome inheritance in starfish zygotes. II: Selective suppression of the maternal centrosome during meiosis. Dev. Biol. 155:58–67. 10.1006/dbio.1993.1006 [DOI] [PubMed] [Google Scholar]
- Steere N., Chae V., Burke M., Li F.-Q., Takemaru K., and Kuriyama R.. 2012. A Wnt/beta-catenin pathway antagonist Chibby binds Cenexin at the distal end of mother centrioles and functions in primary cilia formation. PLoS One. 7:e41077 10.1371/journal.pone.0041077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stinchcombe J.C., and Griffiths G.M.. 2014. Communication, the centrosome and the immunological synapse. Philos. Trans. R. Soc. B Biol. Sci. 369:20130463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stinchcombe J.C., Randzavola L.O., Angus K.L., Mantell J.M., Verkade P., and Griffiths G.M.. 2015. Mother centriole distal appendages mediate centrosome docking at the immunological synapse and reveal mechanistic parallels with ciliogenesis. Curr. Biol. 25:3239–3244. 10.1016/j.cub.2015.10.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung C.-H., and Leroux M.R.. 2013. The roles of evolutionarily conserved functional modules in cilia-related trafficking. Nat. Cell Biol. 15:1387–1397. 10.1038/ncb2888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szollosi D., Calarco P., and Donahue R.P.. 1972. Absence of centrioles in the first and second meiotic spindles of mouse oocytes. J. Cell Sci. 11:521–541. [DOI] [PubMed] [Google Scholar]
- Tamura M., and Nemoto S.. 2001. Reproductive maternal centrosomes are cast off into polar bodies during maturation division in starfish oocytes. Exp. Cell Res. 269:130–139. 10.1006/excr.2001.5305 [DOI] [PubMed] [Google Scholar]
- Terasaki M. 1994. Redistribution of cytoplasmic components during germinal vesicle breakdown in starfish oocytes. J. Cell Sci. 107:1797–1805. [DOI] [PubMed] [Google Scholar]
- Ucar H., Tachibana K., and Kishimoto T.. 2013. The Mos-MAPK pathway regulates Diaphanous-related formin activity to drive cleavage furrow closure during polar body extrusion in starfish oocytes. J. Cell Sci. 126:5153–5165. 10.1242/jcs.130476 [DOI] [PubMed] [Google Scholar]
- Uetake Y., Kato K.H., Washitani-Nemoto S., and Nemoto Si S.. 2002. Nonequivalence of maternal centrosomes/centrioles in starfish oocytes: Selective casting-off of reproductive centrioles into polar bodies. Dev. Biol. 247:149–164. 10.1006/dbio.2002.0682 [DOI] [PubMed] [Google Scholar]
- von Dassow G., Verbrugghe K.J.C., Miller A.L., Sider J.R., and Bement W.M.. 2009. Action at a distance during cytokinesis. J. Cell Biol. 187:831–845. 10.1083/jcb.200907090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vorobjev I.A., and Chentsov YuS. 1982. Centrioles in the cell cycle. I. Epithelial cells. J. Cell Biol. 93:938–949. 10.1083/jcb.93.3.938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voronina V.A., Takemaru K., Treuting P., Love D., Grubb B.R., Hajjar A.M., Adams A., Li F.-Q., and Moon R.T.. 2009. Inactivation of Chibby affects function of motile airway cilia. J. Cell Biol. 185:225–233. 10.1083/jcb.200809144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W.-J., Soni R.K., Uryu K., and Tsou M.-F.B.. 2011. The conversion of centrioles to centrosomes: Essential coupling of duplication with segregation. J. Cell Biol. 193:727–739. 10.1083/jcb.201101109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Washitani-Nemoto S., Saitoh C., and Nemoto S.. 1994. Artificial parthenogenesis in starfish eggs: Behavior of nuclei and chromosomes resulting in tetraploidy of parthenogenotes produced by the suppression of polar body extrusion. Dev. Biol. 163:293–301. 10.1006/dbio.1994.1148 [DOI] [PubMed] [Google Scholar]
- Wery M., Descrimes M., Thermes C., Gautheret D., and Morillon A.. 2013. Zinc-mediated RNA fragmentation allows robust transcript reassembly upon whole transcriptome RNA-Seq. Methods. 63:25–31. 10.1016/j.ymeth.2013.03.009 [DOI] [PubMed] [Google Scholar]
- Winey M., and O’Toole E.. 2014. Centriole structure. Philos. Trans. R. Soc. B Biol. Sci. 369:20130457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi J., Wu X., Chung A.H., Chen J.K., Kapoor T.M., and Hammer J.A.. 2013. Centrosome repositioning in T cells is biphasic and driven by microtubule end-on capture-shrinkage. J. Cell Biol. 202:779–792. 10.1083/jcb.201301004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q.Y., Tamura M., Uetake Y., Washitani-Nemoto S., and Nemoto S.. 2004. Regulation of the paternal inheritance of centrosomes in starfish zygotes. Dev. Biol. 266:190–200. 10.1016/j.ydbio.2003.10.027 [DOI] [PubMed] [Google Scholar]
- Zou C., Li J., Bai Y., Gunning W.T., Wazer D.E., Band V., and Gao Q.. 2005. Centrobin: a novel daughter centriole-associated protein that is required for centriole duplication. J. Cell Biol. 171:437–445. 10.1083/jcb.200506185 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.