Abstract
Amplification bias is a major hurdle in phage display protocols because it imparts additional, unintended selection pressure beyond binding to the desired target. One potential source of amplification bias is the inherent lack of codon optimization that occurs within phage display libraries. Here we present a method that reduces amplification bias by addition of a plasmid that encodes six low abundance tRNAs into K91 Escherichia coli. This new strain, termed K91+, is used to amplify phage during the selection process. We demonstrate the importance of rare codon usage in phage production, and our method produced an overall increase in uniformity of phage production in a random library. Both of these variables are improved in E. coli K91+ compared with the parental K91 strain. This simple solution, requiring only a commercially available plasmid and an additional antibiotic, can reduce amplification bias in phage display protocols.
Keywords: phage display, amplification bias, codon bias, peptide, combinatorial peptide libraries, biopanning
Phage display biopanning is a versatile tool to identify peptides that bind to desired targets. Large libraries of ligands can be generated as fusions to bacteriophage and used to discover binding agents for a variety of targets including inorganic compounds, proteins, cells, and tissues (1,2). However, most phage display experiments suffer from amplification bias, that is, bias that occurs from differential replication of phage inside Escherichia coli (3–7). Small differences in growth rate can have substantial impact on the overall diversity of the library. As such, good binders can be lost in the reiterative process of phage amplification that occurs after each round of binding.
Several methods exist to minimize amplification bias, including growing phage on plates, in emulsion droplets, or microfluidic devices (5,6,8). Deep sequencing in early rounds of panning has been used to identify potential binding phage clones while eliminating or reducing the number of phage amplification steps. However, this results in a high number of false-positive clones that must be sifted out. We sought to develop a simple method to reduce amplification bias while minimally perturbing existing phage display protocols.
One potential source of amplification bias arises from codon bias. Codon bias occurs from nonequivalent expression of tRNAs, which affects translation rates and overall protein levels, potentially impacting the production rate of particular phage clones (9). Peptide phage display libraries consist of random peptides genetically encoded onto one of the coat proteins (2). Thus, it is difficult to generate large random libraries as well as to account for codon bias (10). Pre-assembled trinucleotides can be used instead of single nucleotides during library construction to minimize rare codon use (11). However, co-transformation of plasmids encoding for rare tRNAs as well as the expression plasmid can also minimize the impact of codon bias in nonoptimized protein expression systems (12,13).
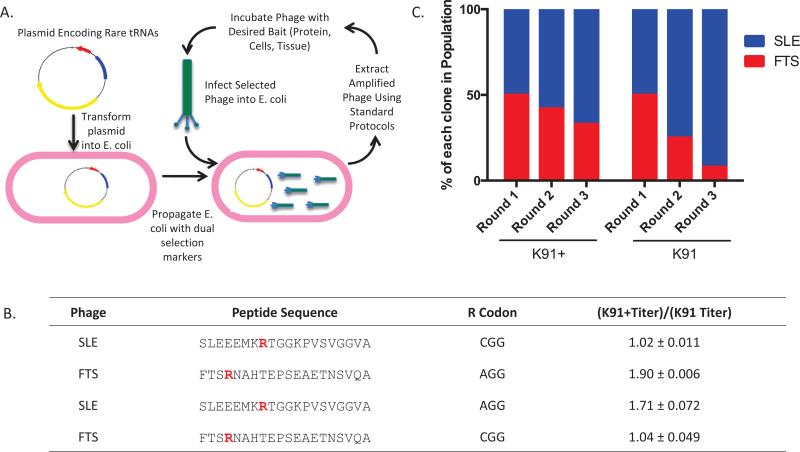
We transformed the pRARE plasmid (Rosetta Competent Cells, 70953; Millipore, San Diego, CA) into chemically competent K91 cells (Hfr-Cavalli thi). Cells recovered in M9 proline dropout media, to maintain F-pilus expression, were plated in M9 proline dropout plates with 30 μg/mL chloramphenicol (Cam) (B20841-14; Alfa Aesar, Heysham, England). The presence of the pRARE plasmid was confirmed using colony PCR (Supplementary Figure S1). These clones retained the ability to internalize phage and thus were termed K91+ cells.
We utilized a library created from the M13-derived, fd-tet construct that encodes a nonlytic phage that imparts tetracycline (Tet) resistance to the host E. coli. A 20-mer peptide is displayed on the pIII coat protein encoded by ligation of random NNK oligonucleotides (14,15). Because of the library's design, it contains the rare AGG codon for arginine (Arg), whose tRNA is encoded by the pRARE plasmid. From this library, we identified a phage clone termed FTS that contained an Arg encoded by the AGG codon. Amplification of FTS in K91+ cells resulted in a 2-fold higher production of phage compared with the parental K91 cells (P < 0.05). The titer of a second phage clone, termed SLE, which contained an Arg encoded by CGG, did not significantly differ between the K91 and K91+ strains. Site-directed mutagenesis was used to change the FTS Arg codon from AGG to CGG and the SLE Arg codon from CGG to AGG (Supplementary Figure S2). The apparent titer of SLE (AGG) was significantly different between K91 and K91+ cells (~1.7-fold increase, P < 0.05). The apparent titer of FTS (CGG) did not significantly differ between K91 and K91+ cells.
Next, we determined the effect of rare codons in iterative amplification. Equal amounts of FTS and SLE were inoculated into either K91 or K91+ cultures and then amplified on YT-Tet plates or YT-Tet+Cam plates, respectively. Twelve colonies from each round of amplification were sequenced to monitor the phage population (McLab, San Francisco, CA). Within 3 rounds in K91, the FTS clone (containing the rare codon AGG) diminished from 6/12 clones (50%) to 1/12 clones (8%). The FTS clone was also lost in the K91+ amplified group, but to a lesser extent; at round 3, 33% clones were FTS compared with 50% initially (Figure 1). In sum, these data support the importance of codon usage in phage amplification and suggest that replication of phage using rare codons is enhanced in the presence of pRARE.
Figure 1. Transformation of K91 cells with the pRARE plasmid increases amplification of a phage clone containing a rare codon.
(A) Scheme outlining the method to reduce phage amplification bias. K91 E. coli cells are transformed with pRARE, which encodes for expression of 6 rare tRNAs and imparts chloramphenicol resistance. Once the cells are transformed, phage amplification occurs by standard protocols using chloramphenicol and tetracycline as section markers. (B) Amino acid sequences of 20-mer phage clones SLE and FTS. Arg residues (R) are highlighted in red to indicate the nucleotide sequence that encodes for the amino acid. The coding sequence for the Arg was switched for each phage clone using site-directed mutagenesis. Presented is quantification of fold changes in titer difference between phage clones amplified in K91+ compared with K91 cells [(K91+ titer)/(K91 titer)]. Titering was performed by inoculating 100-fold phage dilutions into E. coli cultures in log phase growth (0.4–0.6 OD600), incubating at 37°C without shaking, and then plating on YT-tet or YT-tet+cam plates followed by overnight incubation at 37°C. Colonies were counted and CFU/mL was calculated using a standard formula (15). Mean fold change ± SD is presented (n = 3, P < 0.05). (C) Percentage of FTS clones (Arg encoded by AGG) in iterative rounds of amplification in competition with SLE (Arg encoded by CGG) (n = 2). Briefly, 250 phage from each group were incubated with E. coli at 37°C and then plated on 150 cm YT-Tet or YT-Tet+Cam plates. The following day, 12 colonies were picked to monitor phage population, and then phage were extracted using a standard protocol (15). The same procedure was repeated with 500 phage particles from the previous round of amplification.
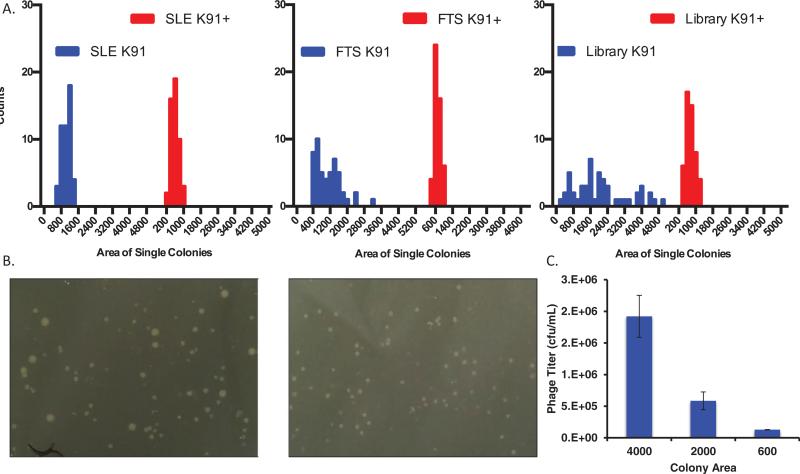
We observed a wide range of colony sizes upon plating of phage-infected K91 cells. However, we noticed that infected K91+ cells demonstrated a significant reduction in colony size variation compared with the K91 group (Figure 2, A and B). In addition, the mean colony size was reduced. This was particularly striking for the FTS phage clone containing AGG codon; the variation in colony size between FTS amplified in K91 versus K91+ was significantly different (P < 0.001), and the mean colony size was reduced by 60% (Supplementary Table S1). SLE colony variation did not significantly differ (P = 0.09), but the colony size was also reduced by 60%. Amplification of the phage library also resulted in significant colony size variation that collapsed in the K91+ cells (P < 0.01). Variation in colony size resulted in significant differences in phage production (Figure 2C); not surprisingly, larger colonies resulted in more phage production. Thus, reducing the variability of colony size can improve the overall diversity of the library and minimize the effects of a phage clone overcoming the population simply due to growth rate. It is unlikely that the vast variability in colony sizes stemmed solely from codon usage; regardless, the K91+ cells mitigated this colony size amplification bias.
Figure 2. K91+ cells demonstrate a significant increase in uniformity of phage amplification.
(A) Histograms quantifying reduced variation in colony size in K91+ cells compared with K91 cells (50 colonies). Reduced variability in colony size indicates reduced overall phage amplification bias. The x-axis represents arbitrary colony area as quantified in ImageJ. A detailed analysis of the data can be found in Supplementary Table S1. From left to right: colony size of K91 and K91+ infected with phage SLE (P = 0.08); colony size of K91 and K91+ infected with FTS (P < 0.01); colony size of K91 and K91+ infected with complete library (P < 0.001). (B) Representative image of a complete 20-mer pIII coat protein–displayed phage library amplified in K91 (left) and K91+ cells (right) shows the range of colony sizes. (C) Quantification of phage production by a single colony was determined to relate colony size to phage production (n = 3). Colonies of the size indicated on the x-axis were picked into 25 μL of PBS and incubated at RT with gentle shaking. The E. coli cells were spun down, and the supernatant containing phage was harvested and then heat inactivated at 65°C for 15 min. The phage were spun again, and supernatant was harvested and titered according to the protocol outlined above.
Our data suggest that transforming pRARE into K91 cells relieves a block in producing phage containing the AGG codon. This is critical since statistically 47% of the phage clones in the initial library should contain at least 1 AGG codon. Poor production of these phage clones will collapse the diversity of the library, and binding phage clones may be lost in the biopanning process. Additionally, each amplification of the starting library for further use will reduce diversity. There are certainly other factors that cause amplification bias, but our approach can mitigate some loss of diversity.
The use of K91+ cells has another unexpected advantage that reduces amplification bias: reduction in colony size variability. Phage production is proportional to colony size; thus, colonies with a growth advantage will produce more phage particles, allowing a clone to dominate the pool over time. Small changes in growth rate between colonies can manifest as large differences in colony size. Phage-infected K91+ cells had fewer super-sized colonies that were larger than the mean. The reason for reduction in colony size in K91+ is not known but may be caused by the expression and/or presence of six additional tRNAs or the addition of the second selection marker.
Antibody phage display protocols utilizing phagemid vectors may also benefit from amplification in E. coli containing pRARE. Typical phagemid vectors derived from the pUC system (pCANTAB or pHEN) utilize ampicillin as a selection marker; thus, pRARE can be utilized directly in these systems by addition of Cam. However, phagemid vectors that utilize Cam as a selection marker would require swapping of antibiotic resistance in pRARE or the phagemid before use.
Here we present a simple method to reduce amplification bias in phage display protocols by the addition of a plasmid coding for rare tRNAs. The protocol presented here requires only commercially available plasmids and the addition of a second antibiotic to decrease amplification bias. Although we transformed the pRARE plasmid into K91 E. coli, it is feasible to transform the pRARE or similar plasmids into ER2738, TG1, or other E. coli strains commonly used in phage display. Future studies into the mechanism of reduced amplification bias as well as creation of new libraries with K91+ are underway.
Supplementary Material
METHOD SUMMARY.
The pRARE plasmid is extracted from Rosetta cells and transformed into Escherichia coli K91 cells to make K91+ cells. These cells are then directly used for phage amplification. The plasmid is maintained by the addition of chloramphenicol during growth and amplification protocols (in addition to markers for selecting pilus- or phage-containing bacteria).
Acknowledgments
Research reported in this work was supported by the National Cancer Institute of the National Institutes of Health under grant number R01CA164447 and the Welch Foundation (I-1622). B.J.U. was supported by a National Science Foundation Graduate Research Fellowship (146339). This paper is subject to the NIH Public Access Policy.
Footnotes
Author contributions
B.J.U. and K.C.B. conceptualized the study and wrote the manuscript. B.J.U., M.J.M., and K.C.B. designed the study. B.J.U. performed the assays presented.
Supplementary material for this article is available at www.BioTechniques.com/article/114256.
Competing interests
The authors declare no competing interests. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
References
- 1.Bassindale AR, Codina-Barrios A, Frascione N, Taylor PG. An improved phage display methodology for inorganic nanoparticle fabrication. Chem. Commun. (Camb.) 2007:2956–2958. doi: 10.1039/b702650a. [DOI] [PubMed] [Google Scholar]
- 2.Smith GP, Petrenko VA. Phage Display. Chem. Rev. 1997;97:391–410. doi: 10.1021/cr960065d. [DOI] [PubMed] [Google Scholar]
- 3.`t Hoen PAC, Jirka SMG, Ten Broeke BR, Schultes EA, Aguilera B, Pang KH, Heemskerk H, Aartsma-Rus A, et al. Phage display screening without repetitious selection rounds. Anal. Biochem. 2012;421:622–631. doi: 10.1016/j.ab.2011.11.005. [DOI] [PubMed] [Google Scholar]
- 4.Kuzmicheva GA, Jayanna PK, Sorokulova IB, Petrenko VA. Diversity and censoring of landscape phage libraries. Protein Eng. Des. Sel. 2009;22:9–18. doi: 10.1093/protein/gzn060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Matochko WL, Ng S, Jafari MR, Romaniuk J, Tang SKY, Derda R. Uniform amplification of phage display libraries in monodisperse emulsions. Methods. 2012;58:18–27. doi: 10.1016/j.ymeth.2012.07.012. [DOI] [PubMed] [Google Scholar]
- 6.Derda R, Tang SKY, Li SC, Ng S, Matochko W, Jafari MR. Diversity of phage-displayed libraries of peptides during panning and amplification. Molecules. 2011;16:1776–1803. doi: 10.3390/molecules16021776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Matochko WL, Cory Li S, Tang SKY, Derda R. Prospective identification of parasitic sequences in phage display screens. Nucleic Acids Res. 2014;42:1784–1798. doi: 10.1093/nar/gkt1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McConnell SJ, Uveges AJ, Spinella DG. Comparison of plate versus liquid amplification of M13 phage display libraries. BioTechniques. 1995;18:803–806. [PubMed] [Google Scholar]
- 9.Tuller T, Waldman YY, Kupiec M, Ruppin E. Translation efficiency is determined by both codon bias and folding energy. Proc. Natl. Acad. Sci. USA. 2010;107:3645–3650. doi: 10.1073/pnas.0909910107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lindner T, Kolmar H, Haberkorn U, Mier W. DNA libraries for the construction of phage libraries: statistical and structural requirements and synthetic methods. Molecules. 2011;16:1625–1641. doi: 10.3390/molecules16021625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Krumpe LRH, Schumacher KM, McMahon JB, Makowski L, Mori T. Trinucleotide cassettes increase diversity of T7 phage-displayed peptide library. BMC Biotechnol. 2007;7:65. doi: 10.1186/1472-6750-7-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Iva nov AV, Korovina AN, Tunitskaya VL, Kostyuk DA, Rechinsky VO, Kukhanova MK, Kochetkov SN. Development of the system ensuring a high-level expression of hepatitis C virus nonstructural NS5B and NS5A proteins. Protein Expr. Purif. 2006;48:14–23. doi: 10.1016/j.pep.2006.02.011. [DOI] [PubMed] [Google Scholar]
- 13.Kirienko NV, Lepikhov KA, Zheleznaya LA, Matvienko NI. Significance of codon usage and irregularities of rare codon distribution in genes for expression of BspLU11III methyltransferases. Biochemistry (Mosc.) 2004;69:527–535. doi: 10.1023/b:biry.0000029851.96180.92. [DOI] [PubMed] [Google Scholar]
- 14.Cwirla SE, Peters EA, Barrett RW, Dower WJ. Peptides on phage: a vast library of peptides for identifying ligands. Proc. Natl. Acad. Sci. USA. 1990;87:6378–6382. doi: 10.1073/pnas.87.16.6378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McGuire MJ, Li S, Brown KC. Biopanning of phage displayed peptide libraries for the isolation of cell-specific ligands. Methods Mol. Biol. 2009;504:291–321. doi: 10.1007/978-1-60327-569-9_18. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.