Abstract
ATP-dependent chromatin remodeling complexes are essential for transcription regulation, and yet it is unclear how these multisubunit complexes coordinate their activities to facilitate diverse transcriptional responses. In this study, we found that the conserved Arp5 and Ies6 subunits of the Saccharomyces cerevisiae INO80 chromatin-remodeler form an abundant and distinct subcomplex in vivo and stimulate INO80-mediated activity in vitro. Moreover, our genomic studies reveal that the relative occupancy of Arp5-Ies6 correlates with nucleosome positioning at transcriptional start sites and expression levels of >1,000 INO80-regulated genes. Notably, these genes are significantly enriched in energy metabolism pathways. Specifically, arp5Δ, ies6Δ, and ino80Δ mutants demonstrate decreased expression of genes involved in glycolysis and increased expression of genes in the oxidative phosphorylation pathway. Deregulation of these metabolic pathways results in constitutively elevated mitochondrial potential and oxygen consumption. Our results illustrate the dynamic nature of the INO80 complex assembly and demonstrate for the first time that a chromatin remodeler regulates glycolytic and respiratory capacity, thereby maintaining metabolic stability.
INTRODUCTION
Eukaryotic genomic DNA is assembled with histones to form chromatin, a complex structure that undergoes constant dynamic reorganization in coordination with DNA-templated processes. Chromatin remodeling, an ATP-dependent mechanism by which nucleosomes are repositioned and reconstructed, is a fundamental component of chromatin manipulation and influences numerous DNA-templated pathways. Because chromatin remodelers are involved in essential cellular processes, defects in remodeling activity directly result in fitness deficiencies in lower eukaryotes, as well as developmental defects and disease in higher eukaryotes (1, 2).
In particular, disruption of INO80, an evolutionarily conserved chromatin remodeling complex, results in pluripotency defects and carcinogenesis (3–6). The INO80 complex has demonstrated roles in transcription (7–9), replication (10–12), DNA damage responses (13–16), telomere regulation (17), and mitotic stability (18, 19). These studies exemplify the functional diversity of the INO80 complex in different pathways (20). Moreover, they highlight the need for regulatory mechanisms that direct its activity among, and within, these processes.
Ample opportunities for regulation of chromatin remodeling exist at the level of individual remodeler complex subunits. For example, different subunits of the INO80 complex are involved in DNA repair and cell cycle checkpoint responses (13, 16). Structural studies demonstrate that these subunits are components of different modules that interact with distinct domains of the S. cerevisiae Ino80 ATPase subunit (21) and thus may impart regulatory functions on ATP-dependent activities of the INO80 complex.
For example, a module consisting of actin and actin-related proteins (Arps) Arp8 and Arp4 interacts with the helicase-associated SANT domain of the Ino80 ATPase subunit (22). This Arp4/Arp8/actin module is important for nucleosome recognition, which stimulates ATP hydrolysis and nucleosome sliding in vitro (21, 23–27). In addition, the Arp5 and INO80 subunit 6 (Ies6) subunits comprise a separate module that is particularly interesting because of its structural placement at the “enzymatic center” of the complex, between the ATPase domain of Ino80 and the large Rvb1-Rvb2 helicase module (21). Indeed, assembly of the Arp5-Ies6 module into the INO80 complex requires the Ies2 subunit (28), Rvb1-Rvb2 (29), and the Ino80 ATPase domain (28, 30). Deletion of ARP5 or IES6 diminishes INO80 activity in vitro (21, 23, 31), although their precise role in nucleosome positioning, particularly in vivo, has not been well established.
Nevertheless, chromatin remodelers have demonstrated roles in the positioning of nucleosomes in vivo, a process that is influenced by DNA sequence, epigenetic modifications, and other chromatin-modifying factors (32). Indeed, ATP-dependent chromatin remodelers have been implicated as primary determinants of in vivo nucleosome positioning (33–35). Small deviations in nucleosome positioning can alter accessibility of transcription factors at transcriptional start sites (TSSs) (36–38). Although the INO80 complex influences transcription (8) and is abundantly localized to nucleosomes proximal to TSSs (39), very little is known regarding its chromatin-remodeling activity at target genes. Moreover, the biological relevance of INO80-regulated gene programs has not been reported.
We show here that the evolutionarily conserved S. cerevisiae Arp5-Ies6 subcomplex regulates INO80-mediated gene expression in vivo and nucleosome positioning in vitro through dynamic association with the INO80 complex. Thus, the assembly and composition of chromatin remodeler complexes directly regulate diverse gene expression programs. For the INO80 complex, this may provide mechanisms to maintain metabolic homeostasis, since arp5Δ, ies6Δ, and ino80Δ mutants have altered expression of genes involved in glycolysis and oxidative phosphorylation, concomitant with deviations in energy metabolism and respiration.
MATERIALS AND METHODS
Protein purifications and fractionations.
The yeast strains are listed in Table 1. FLAG tags were integrated genomically at endogenous loci. Proteins were purified using anti-FLAG affinity beads as previously described (13, 16). For gradient separation, purified complexes were loaded onto a 10 to 30% (wt/wt) sucrose gradient and centrifuged at 130,000 × g for 20 h. Proteins in collected fractions were precipitated by trichloroacetic acid prior to electrophoresis and silver staining. Chromatin fractionations were conducted as described previously (40). Western blots utilize anti-FLAG M2 (Sigma), anti-Arp5 (Abcam), antihexokinase (Novus), or anti-H3 C-terminal (Active Motif) antibodies.
TABLE 1.
Yeast strains used in this study
| Straina | Description |
|---|---|
| BY4741* | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 |
| WYY0193 | BY4741 ino80::LEU2 |
| WYY0194 | BY4741 arp5::LEU2 |
| WYY0195 | BY4741 ies6::LEU2 |
| WYY0196 | BY4741 arp8::LEU2 |
| Ino80-FLAG** | BY4733 MATa his3Δ200 leu2Δ0 met15Δ0 trp1Δ63 ura3Δ0 INO80-2-FLAG |
| WYY0197 | Ino80-FLAG arp5::URA3 |
| WYY0198 | Ino80-FLAG ies6::URA3 |
| WYY0199 | Ino80-FLAG arp8::URA3 |
| WYY0200 | BY4741 ARP5-2-FLAG::URA3 |
| WYY0201 | WYY0200 ino80::LEU2 |
| WYY0202 | WYY0200 ies6::LEU2 |
| WYY0203 | WYY0200 arp8::LEU2 |
| WYY0204 | BY4741 IES6-2-FLAG::URA3 |
| WYY0205 | WYY0204 ino80::LEU2 |
| WYY0206 | WYY0204 arp5::LEU2 |
| WYY0207 | BY4741 ARP8-2-FLAG::URA3 |
| BY4743* | MATa/α his3Δ1/his3Δ1 leu2Δ0/leu2Δ0 LYS2/lys2Δ0 met15Δ0/MET15 ura3Δ0/ura3Δ0 |
| WYY0208 | BY4743 arp5::LEU2/ARP5 ies6::HIS3/IES6 |
| WYY0209 | BY4741 arp5::HIS3 arp8::KANMX4(pRS415) |
| WYY0211 | WYY0209(pRS415-ARP5) |
| WYY0212 | WYY0209(pRS415-ARP8) |
| WYY0213 | WYY0209(pRS415-ARP5-ARP8) |
| WYY0210 | BY4741 ies6::HIS3 arp8::KANMX4(pRS415) |
| WYY0214 | WYY0210(pRS415-IES6) |
| WYY0215 | WYY0210(pRS415-ARP8) |
| WYY0216 | WYY0210(pRS415-IES6-ARP8) |
All strains are from an S288C background and were constructed for this study, except as indicated. *, obtained from Open Biosystems; **, obtained from Shen et al. (8).
In vitro biochemical assays. Cy5-tagged mononucleosomes with 601 DNA sequence and 60 bp of linker DNA were prepared as previously described (41). Remodeling reaction mixtures contained 2 nM INO80 or INO80d (INO80-deficient, lacking Arp5-Ies6) and Arp5-Ies6 complexes purified by Arp5-Flag, and 2 nM mononucleosomes in reaction buffer (25 mM HEPES-KOH [pH 7.6], 70 mM KCl, 3.6 mM MgCl2, 0.37 mM EDTA, 0.37 mM EGTA, 0.017% NP-40, 10% glycerol, 1 mM dithiothreitol, 100 μg of bovine serum albumin/ml, and 1× protease inhibitors). After incubation at 30°C for 30 min, reactions were initiated by addition of 2 mM ATP-Mg2+ and stopped with 2× stop buffer (42 mM ADP, 20% glycerol, and 0.3 mg of nonspecific plasmid DNA/ml). Electrophoretic mobility shift assays were performed with 2 nM Cy5-tagged 601 DNA fragment or mononucleosomes incubated with the indicated amounts of INO80 complex or Arp5-Ies6 subcomplex in reaction buffer as described above. Samples were electrophoresed on native 6% PAGE gel in 0.5× Tris-borate-EDTA and visualized using a Typhoon 9210 imager (GE Healthcare Life Sciences).
RNA sequencing.
Poly(A) mRNA enrichment was performed on RNA extracted from mid-log-phase yeast cultures (two biological replicates) via the Illumina TruSeq RNA sample preparation v2 low-throughput protocol. Illumina sequencing services (10 million single-end reads per sample) were performed at the Stanford Center for Genomics and Personalized Medicine on an Illumina HiSeq2000 platform. Fragments per kilobase of transcript per million mapped reads (FPKM) were processed using the Tuxedo software suite (Bowtie2 [v2.2.2], TopHat2 [v2.0.11], and Cufflinks [v2.1.1]) according to the “quantification of reference annotation only protocol,” as previously described (42). Significant transcriptional changes between wild-type and deletion strains were selected at a false discovery rate-adjusted P value of 0.05. The distribution of log-transformed expression values was bimodal, and statistical outliers (∼4% of transcripts) largely involved in ribosomal function were excluded from further analysis.
Gene expression analysis.
Gene expression plots were ordered by Arp5 chromatin immunoprecipitation (ChIP) occupancy (43) and smoothed by fitting a spline function selected by ordinary cross-validation in R using smooth.spline (…, cv = TRUE). Verification of the validated spline fit was performed by bootstrap resampling and is represented as 95% confidence bands. The Wilcoxon rank-sum test was used to compare distribution location shift in FPKM expression values from two populations. Gene ontology (GO) enrichment was computed using GORILLA (44), and network illustration was generated using REVIGO (45). All significance results are based on P values from the hypergeometric distribution after false-discovery-rate correction for multiple hypothesis testing. Dimensionality reduction and visualization for principal component analysis (PCA) was performed using the “PCAplot()” function in the cummeRbund R package (Bioconductor).
MNase-ChIP occupancy and chromatin analysis.
Micrococcal nuclease (MNase) ChIP data of TAP-tagged Ino80, Arp5, and Hht1 (H3) was obtained and processed as previously described (43). The MNase-ChIP data were generated using SOLiD sequencing of two biological replicates with an average of 2.3 million spots and 109 million bases and are available at Sequence Read Archive SRA051347. Nucleosome fuzziness was defined as before (46) and indicates the standard deviation of a nucleosome's genomic location from a population of cells around that nucleosome's consensus location. The nucleosome-free-region (NFR) length is defined as the distance (in base pairs) from the −1 nucleosome dyad to the +1 nucleosome dyad. After ordering genes by Arp5 occupancy at +1 nucleosomes, cross-validated spline functions were fit to nucleosome fuzziness data, and confidence bands were estimated by bootstrapping as described above.
Metabolic assays.
For oxygen consumption assays, cells were first grown to mid-log phase in yeast extract-peptone-dextrose (YPD). Cells (5 × 105per well) were then washed, resuspended in assay medium (0.167% yeast nitrogen base, 0.5% ammonium sulfate, and 2% ethanol or glucose), and plated in poly-l-lysine-coated XF96 plates via centrifugation (500 rpm for 3 min). Oxygen consumption was determined using an XF96 extracellular flux analyzer (Seahorse Biosciences) after 1 h of incubation. Mitochondrial potential was determined in mid-log-phase cultures in YPD that were washed and resuspended in YP plus 3% glucose or 3% ethanol for 1 h. Then, 20 ml of cells at an optical density at 660 nm (OD660) of 0.65 were incubated with 0.15 μM tetramethylrhodamine ethyl ester (TMRE) for 30 min at 30°C with constant aeration. Cells were spun down, washed, and resuspended in phosphate-buffered saline before fluorescent measurement (excitation, 560 nm; emission, 610 nm). Fitness assays were performed as previously described (47). Briefly, cells were grown to saturation in YPD and diluted to pre-log phase (OD660 of 0.1) in YP plus 3% glucose, YP plus 3% glucose and 1.5 mM 2-deoxyglucose (2-DG), or YP plus 3% ethanol. The cells were then plated in 96-well plate with clear breathable film sealant and grown in a Tecan Sunrise microplate reader with constant shaking. OD measurements were used to calculate population doubling times in mid-log phase between an OD660 of 0.4 and an OD660 of 0.6.
Accession number.
Gene list and associated FPKM values, as well as Arp5 and Ino80 occupancies, nucleosome fuzziness, and NFR length, can be found in the Gene Expression Omnibus (GEO) database under accession number GSE77257.
RESULTS
Arp5 and Ies6 are critical components of both the INO80 complex and a separate subcomplex.
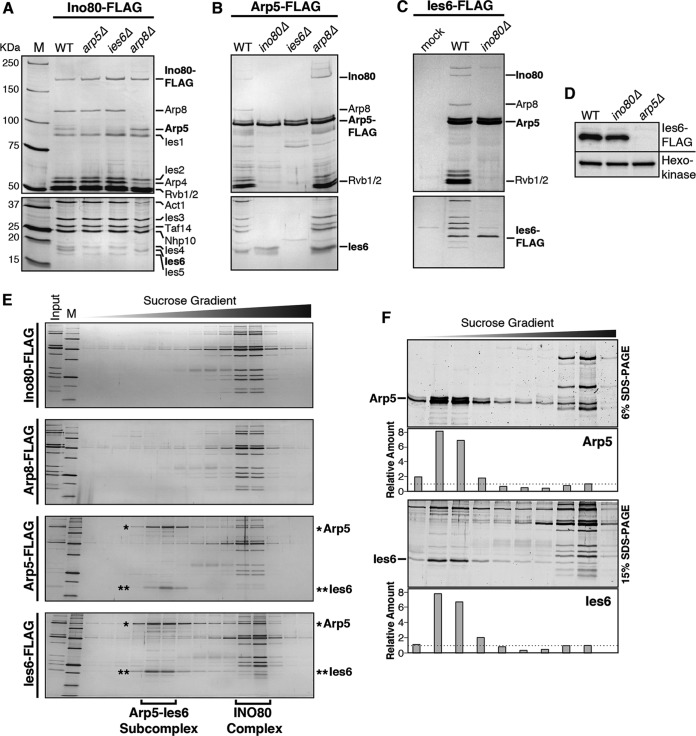
Deletion of either IES6 or ARP5 results in loss of both subunits from the purified INO80 complex, demonstrating that these subunits are dependent on each other for association with the complex (Fig. 1A). Deletion of ARP8, which abrogates Arp4 and actin association with the complex (23), does not alter Arp5-Ies6 association (Fig. 1A and B). These results confirm previous findings (21, 31) and demonstrate that S. cerevisiae Arp5-Ies6 occupy a subunit module separate from the Arp8 module. Interestingly, Arp5 and Ies6 copurify even in the absence of the Ino80 ATPase subunit, the main subunit scaffold for the INO80 complex (Fig. 1B and C). Unexpectedly, deletion of ARP5 results in an undetectable amount of FLAG-tagged Ies6 cellular protein (Fig. 1D). However, deletion of IES6 does not substantial change Arp5 protein levels in purified protein (Fig. 1B) and whole-cell extracts (data not shown). The precise reason for decreased Ies6 protein in arp5Δ cells is currently unknown; however, it does not appear to originate from a significant loss of IES6 gene expression in arp5Δ cells, as determined by transcriptome sequencing (RNA-seq) (data not shown). Thus, it is likely that Arp5 stabilizes Ies6 at the posttranscriptional level. This finding further strengthens the functional dependence between both subunits. Interestingly, unlike Arp8, sucrose gradient separation reveals that Arp5 and Ies6 form a separate subcomplex (Fig. 1E), which is approximately in 8-fold excess of that which is present within the INO80 complex (Fig. 1F). This separate subcomplex is also observed in sucrose gradient fractions from whole-cell extracts (data not shown).
FIG 1.
Arp5 and Ies6 form an independent subcomplex. (A to C) Silver-stained 6% (top) and 15% (bottom) SDS-PAGE gels of FLAG-purified proteins. Subunits are designated on the right of each gel. M, molecular mass markers. (A to C) Ino80-FLAG, Arp5-FLAG, and Ies6-FLAG purified from the wild type (WT) and the indicated deletion strains and mock purification from cell lysate lacking FLAG-tagged protein. (D) Anti-Flag Western blot of Ies6-FLAG in WT and the indicated deletion strain lysates. An antihexokinase Western blot was used as a loading control. (E) Ten to 25% sucrose gradient sedimentation of complexes purified from Ino80-FLAG, Arp8-FLAG, Arp5-FLAG, and Ies6-FLAG. (F) Fifteen to 20% sucrose gradient sedimentation of complexes purified from Ies6-FLAG-expressing cells. Samples were electrophoresed on 6% and 15% SDS-PAGE gels to resolve Arp5 and Ies6, respectively. The relative protein amount was quantified using Oriole fluorescent protein stain.
The INO80 complex regulates many critical DNA-templated processes. Accordingly, yeast strains that lack the Ino80 ATPase or Arp8 subunit have decreased fitness (8, 23). In addition, arp5Δ and ies6Δ mutants exhibit decreased fitness in the absence of inositol and at elevated temperature, which cause metabolic stress and protein instability, respectively (Fig. 2A). In fitness assays, an epistatic relationship between ARP5 and IES6 is observed, since the growth of the double mutant is similar to that of single mutants. This differs from the additive fitness defect observed in arp5Δ arp8Δ and ies6Δ arp8Δ double-mutant cells (Fig. 2B), demonstrating that the in vivo functions of Arp5-Ies6 module are distinct from that of the Arp8-containing module.
FIG 2.
Arp5-Ies6 function together to maintain cellular fitness. (A) Serial dilution fitness assays of wild-type (WT) and ino80Δ, arp5Δ, ies6Δ, and arp8Δ mutant strains grown at 30 or 37°C or on medium lacking inositol. (B) Serial dilutions of WT, arp5Δ, ies6Δ, and arp5Δ ies6Δ strains. Tetrad segregation of arp5Δ and ies6Δ heterozygous diploids, indicating an epistatic genetic interaction between ARP5 and IES6, as cells with either single deletion or the double deletion exhibit similar growth. (C) Fitness assays of WT and indicated deletion strains show nonepistatic genetic interactions between ARP8 and ARP5 or IES6, as the growth defect is additive of single mutants.
Thus, Arp5 and Ies6 function as two crucial subunits of the INO80 complex, and together as an abundant, distinct subcomplex. These results illustrate the divergent activities among Arp subunits of the INO80 complex and highlight the unique cooperative function between Arp5 and Ies6.
Arp5-Ies6 subcomplex influences INO80-mediated nucleosome positioning.
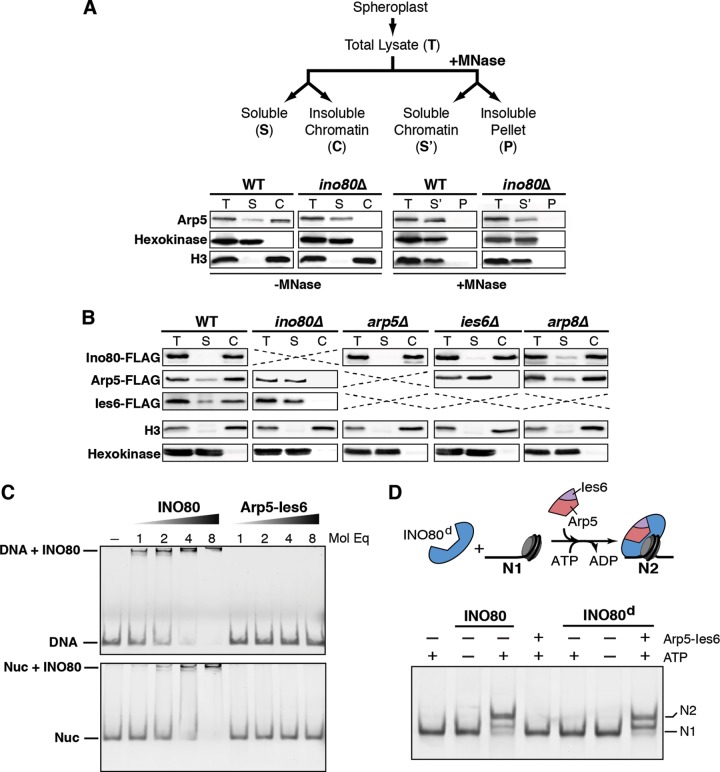
Arp5-Ies6 forms a subcomplex independent of the complete INO80 complex. However, this subcomplex requires Ino80 for chromatin association, as previously demonstrated (28). Specifically, chromatin fractionation assays demonstrate that deletion of INO80 results in a dramatic decrease in Arp5 chromatin association (Fig. 3A). Nuclease digestion of the chromatin fraction efficiently solubilizes both histone H3 and Arp5, thus confirming that Arp5 is dependent on chromatin to remain within the insoluble fraction. Similarly, deletion of IES6 results in comparable defects in Arp5 chromatin association (Fig. 3B). Furthermore, deletion of INO80 results in an undetectable amount of both Ies6 and Arp5 chromatin association. Ino80, Arp5, and Ies6 chromatin association does not substantially change in arp8Δ cells. In addition, the in vivo chromatin association of Ino80 is not significantly altered by deletion of ARP5 or IES6 (Fig. 3B). In agreement with the in vivo chromatin fractionation assay, in vitro binding assays demonstrate that the Arp5-Ies6 subcomplex does not independently bind to nucleosomes or DNA at concentrations that confer association of the INO80 complex to these chromatin substrates (Fig. 3C).
FIG 3.
Arp5-Ies6 subcomplex associates with chromatin in an INO80-dependent manner and modulates nucleosome sliding. (A) The illustration at the top shows the experimental design for in vivo chromatin fractionation with micrococcal nuclease (MNase). Below is the chromatin fractionation of wild-type (WT) and ino80Δ cells. Arp5 was detected by anti-Arp5 antibody. Hexokinase and histone H3 are indicative of cytoplasmic and chromatin fractions, respectively. MNase treatment solubilizes H3 chromatin fraction. (B) In vivo chromatin fractionation of WT and indicated deletion strains expressing Ino80-FLAG, Arp5-FLAG, or Ies6-FLAG. Arp5 and Ies6 are predominantly present in the chromatin fraction in WT cells and the soluble fraction in ino80Δ cells. Ies6-FLAG protein is undetectable in arp5Δ strains, as shown in Fig. 1D. A strain with both Ies6-FLAG and arp8Δ is nonviable. (C) Native PAGE gels of DNA (left panel) and nucleosome containing 60 bp extranucleosomal DNA (Nuc, right panel). A 2 nM concentration of DNA or nucleosome was incubated with the indicated molar equivalents (Mol Eq) of INO80 complex or Arp5-Ies6 subcomplex. (D) The illustration at the top depicts the experimental setup, wherein INO80 complex deficient of the Arp5-Ies6 subcomplex (INO80d) is preincubated with end-positioned (N1) nucleosomal substrate. Upon addition of the Arp5-Ies6 subcomplex and ATP, INO80d activity is stimulated, and the nucleosome is moved to the center position (N2).
In order to examine the functional relationship between the abundant Arp5-Ies6 subcomplex and the INO80 complex on chromatin, in vitro chromatin-remodeling assays were performed. The INO80 complex exhibits nucleosome remodeling activity that “slides” an end-positioned nucleosome to the center of a DNA fragment (N1 to N2, Fig. 3D, top panel) (23). Nucleosome substrates that stimulate INO80 complex sliding activity contain a minimum of 30-bp extranucleosomal DNA, with optimal activity exhibited with at least 53 bp (48). Using this well-defined system, the INO80 complex lacking Arp5-Ies6 (INO80d) does not exhibit nucleosome sliding on substrates with 60 bp of extranucleosomal DNA (Fig. 3D). These results are in agreement with previous findings regarding the requirement for yeast Arp5 in chromatin remodeling (21, 23, 28, 31). Surprisingly, INO80d-mediated sliding activity is stimulated by the addition of ectopic Arp5-Ies6 subcomplex, whereas Arp5-Ies6 alone does not affect nucleosome sliding (Fig. 3D). In these assays, an INO80 complex deficient of Arp5-Ies6 (INO80d) is preincubated with nucleosomes, and the sliding reaction is stimulated with the addition of Arp5-Ies6 and ATP. Taken together, these results demonstrate that Arp5-Ies6 is able to assemble into the INO80 complex to stimulate sliding activity and reveal that the assembly of a chromatin remodeler can occur as a dynamic process to influence in vitro catalytic activity.
Arp5 abundance at promoters influences expression of >1,000 genes.
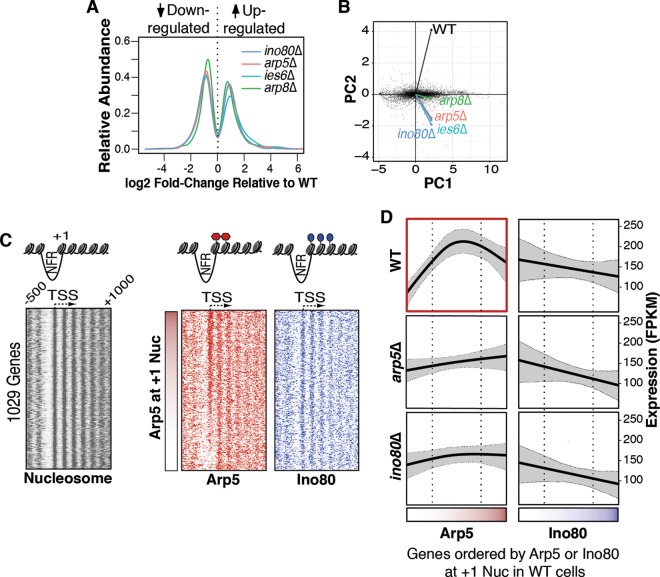
In order to gain insight into the in vivo mechanisms by which Arp5-Ies6 regulates the INO80 complex, RNA-seq was performed in individual ino80Δ, arp5Δ, ies6Δ, and arp8Δ mutant cells. More than 15% (n = 1,029) of yeast genes are misregulated upon deletion of INO80 subunits, with roughly half of the significantly differentially expressed (SDE) genes upregulated in the mutants and the other half downregulated (Fig. 4A), indicative of transcriptional activation and repression functions. PCA, which reduces dimensionality and variance in large data sets (49), was performed on genome-wide expression profiles, revealing striking similarities between the arp5Δ and ies6Δ mutants (Fig. 4B). Furthermore, arp5Δ and ies6Δ mutants are more similar to ino80Δ mutants than they are to arp8Δ mutants, suggesting that the Arp5-Ies6 subcomplex plays a key role in governing Ino80 ATPase's involvement in global transcription regulation.
FIG 4.
Arp5 occupancy at promoters influences the transcription of >1,000 yeast genes. (A) Distribution of significant gene expression changes determined by RNA-seq, shown on a log2 scale, in the indicated deletion strains relative to the wild type (WT). (B) PCA, log-transformed gene expression values from ino80Δ, arp5Δ, ies6Δ, and arp8Δ samples were projected onto primary (PC1) and secondary (PC2) principal components. Vectors represent projections of the indicated sample onto principal components, whereas black dots are log-transformed expression values of individual genes transformed into principal components. Note the close proximity of arp5Δ, ies6Δ, and ino80Δ vectors. (C) Micrococcal nuclease-ChIP (MNase-ChIP) maps of nucleosomes, Arp5, and Ino80 on 1,029 Arp5 and Ino80-regulated genes aligned by their TSSs. Genes are ordered by the total Arp5 occupancy at the +1 nucleosome (+1 Nuc) and are illustrated by the vertical red gradient strip. The NFR and promoter proximal nucleosomes are illustrated. (D) RNA-seq gene expression profiles in WT, ino80Δ, and arp5Δ strains are ordered by wild-type Arp5 occupancy (horizontal red gradient strip) and Ino80 occupancy (horizontal blue gradient strip) at the +1 Nuc. Note, in the ino80Δ and arp5Δ plots, that the gene expression in mutant cells is plotted according to Arp5 or Ino80 occupancy in wild-type cells. The fragments per kilobase of transcript per million (FPKM) mapped reads are shown. The cross-validated (CV) spline function (solid, black line) and 95% confidence bands for the spline fit (dashed, gray lines) are shown. Genes within the interquartile range of Arp5 occupancy (dotted, black lines) have relatively higher expression levels. The red box outline highlights the asymmetric trend line in wild-type (WT) expression that is not observed in deletion strains.
In order to focus in vivo analyses on genomic loci where Arp5-Ies6 is likely to directly participate in transcription, the relative subunit occupancy of Ino80 and Arp5-Ies6 at SDE gene promoters was assessed by ChIP (43). The chromatin architecture adopted by the vast majority of genes in the S. cerevisiae genome is characterized by a +1 nucleosome adjacent to the TSS and an NFR upstream of the TSS. This well-defined architecture plays critical roles in gene expression, in part due to its influence on accessibility of transcription factors to promoter regions (32). Micrococcal nuclease (MNase) digestion, which digests DNA between nucleosomes, followed by ChIP, has been successful in mapping chromatin-associated factors at single-nucleosome resolution and effectively captures chromatin remodelers with nucleosome substrates at individual promoters (43).
Analysis of MNase-ChIP data reveals a relative enrichment of Arp5 and Ino80 found at the transcriptionally relevant +1 nucleosome when 1029 SDE genes are aligned according to their TSSs (Fig. 4C). Interestingly, we observe a divergent pattern of Arp5 occupancy at the +1 nucleosome of individual genes, with levels varying considerably between different promoters (Fig. 4C). Given the ability of Arp5-Ies6 to stimulate INO80-mediated nucleosome sliding in vitro (Fig. 3D) and the differing patterns of Arp5 abundance in vivo (Fig. 4C), we investigated the possibility that differences in gene expression are associated with the relative abundance of Arp5 at the +1 nucleosome of specific promoters. Indeed, when genes are ordered by Arp5 promoter occupancy (as indicated by the horizontal red gradient in the bottom panel of Fig. 4D), we observe a distinguishable pattern in the expression profile of wild-type cells. Promoter loci with undetectable or reduced levels of Arp5 are relatively lowly expressed, whereas genes within the interquartile range of Arp5 occupancy are associated with significantly higher transcription levels (P = 4.4 × 10−9).
More precisely, among INO80-occupied promoters, genes with relatively low Arp5 occupancy (within the bottom quartile) tend to be lowly expressed in wild-type cells and upregulated in arp5Δ mutants (note the higher expression levels of genes in the left quadrant of Fig. 4D in the arp5Δ mutant relative to the wild type). This suggests that the INO80 complex with relatively low levels of Arp5 at promoter nucleosomes is generally repressive to transcription. Conversely, INO80-occupied promoters with relatively higher levels of Arp5 are highly expressed in wild-type cells (Fig. 4D, top panel, middle quadrant). These genes tend to be significantly downregulated in arp5Δ mutants. This implies that Arp5-Ies6 association with the INO80 complex, which can stimulate nucleosome repositioning activity in vitro (Fig. 3D), actively upregulates transcription of these genes in vivo. Genes with the highest quartile of Arp5 occupancy show no significant bias in up- or downregulation in arp5Δ mutants, although the downward trend observed in Fig. 4D may indicate a slight, yet not statistically significant, repressive role at these loci. These data support a model that Arp5-Ies6 assembly into the INO80 complex repositions promoter nucleosomes to facilitate gene expression activation at these genes. It should be noted that these results present a generalized model of INO80 and Arp5-Ies6-regulated transcription at the +1 nucleosome and may not be indicative of specialized Arp5 function. For example, Arp5 has been shown to prevent euchromatic spreading into transcriptionally silent chromatin (9) and repress long noncoding RNA expression (7).
The observed pattern of INO80-mediated gene expression is dependent on the presence of Ino80, but irrespective of its relative abundance, since ordering genes by Ino80 occupancy levels does not produce an distinguishable expression trend (Fig. 4D, right panels). As such, the relative level of Arp5-Ies6 at the +1 nucleosome is a significant determinant of expression levels at these loci.
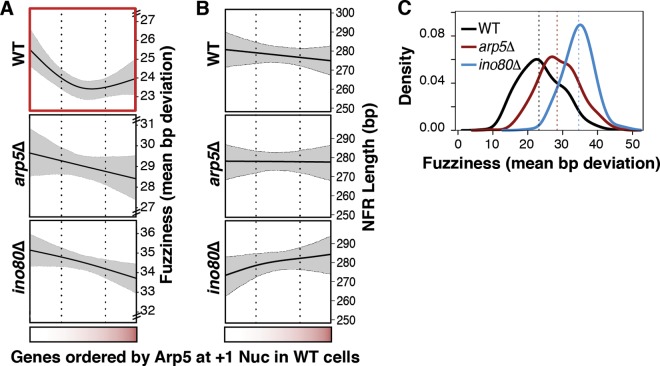
We then investigated the chromatin architecture of INO80-occupied promoters to gain insight into the specific mechanism by which Arp5-Ies6 regulates gene expression. Genes were again ordered by Arp5 occupancy and corresponding profiles of NFR length and +1 nucleosome fuzziness were generated. Nucleosome fuzziness depicts variable nucleosome positioning in a cell population, which as previously described, can be regulated by trans-acting ATP-dependent chromatin remodelers (33–35). Importantly, +1 nucleosome positioning deviations as small as 2 bp can alter accessibility of transcription factors at the TSS (36–38), and TSSs close to the nucleosome midpoint are found to be repressed (50, 51). Thus, correct positioning of nucleosomes is critical for the proper control of gene expression.
When SDE genes are ordered by Arp5 abundance, +1 fuzziness inversely parallels the Arp5-Ies6-regulated gene expression profile previously shown in Fig. 4D (Fig. 5A). However, there is no substantial relationship between Arp5 occupancy and NFR length (Fig. 5B), +2 or +3 nucleosomes fuzziness, or linker DNA length between +1, +2, and + 3 nucleosomes (data not shown). Strikingly, in ino80Δ and arp5Δ cells, the mean nucleosome positioning pattern was dramatically disrupted, as evidenced by an overall increase in fuzziness of +1 nucleosomes (Fig. 5C). These results suggest an Arp5-Ies6-regulated mode of gene expression that depends on the positioning of the +1 nucleosome.
FIG 5.
Arp5 abundance at +1 nucleosomes regulates promoter architecture. (A) As in Fig. 4D, genes are ordered by Arp5 occupancy at +1 nucleosomes, as indicated by the horizontal red gradient strip. Nucleosome fuzziness, which is the base pair standard deviation from the mean position (mean bp deviation) in wild-type (WT), ino80Δ, and arp5Δ strains are shown. The cross-validated spline function (solid, black line) and 95% confidence bands for the spline fit (dashed, gray lines) are shown. Horizontal dotted lines indicate the interquartile range. The red box outline highlights the asymmetric trend line in WT nucleosome fuzziness that is not observed in deletion strains. (B) NFR length profiles of Arp5-regulated genes are shown as in panel A. (C) Distribution of +1 nucleosome fuzziness values of all Arp5-regulated genes in WT, arp5Δ, and ino80Δ cells.
Arp5-Ies6 and the INO80 complex regulate the expression of metabolic gene networks.
In order to characterize the in vivo implications of Arp5-Ies6 regulation of INO80 activity, functional enrichment analysis was performed on SDE genes introduced in Fig. 4. This analysis reveals that in the set of Arp5-Ies6-dependent genes, all significantly enriched biological processes pertain to cellular metabolism (Fig. 6).
FIG 6.
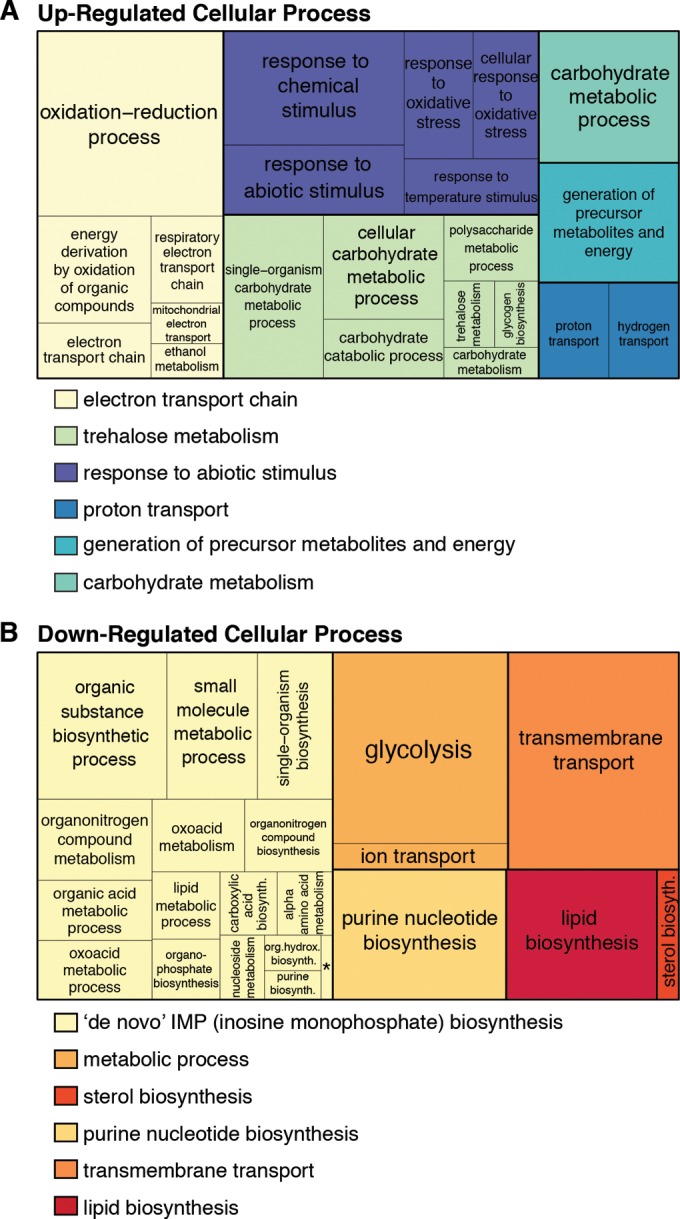
Arp5-Ies6-dependent genes are enriched in metabolic pathways. A TreeMap of enriched gene ontology (GO) categories (P < 10−3) grouped by biological process for significantly differentially expressed upregulated (A) or downregulated (B) genes in arp5Δ mutants (according to the statistical analysis described in Materials and Methods) is shown. GO terms are hierarchically clustered into REVIGO-determined parent categories indicated by color key below TreeMap. Ambiguous parents categories include “single organism cellular process,” “single organism metabolism,” and “biosynthesis,” which were manually curated to indicate the largest constituent group. The size of the box is scaled to represent the number of genes in the indicated category. *, “de novo” IMP biosynthetic process.
It is important to note that S. cerevisiae have a unique adaptive metabolism that can utilize diverse carbon sources in both aerobic and anaerobic conditions. In oxygen and glucose-rich conditions, S. cerevisiae preferentially utilize glycolysis to produce pyruvate, followed by fermentation to produce carbon dioxide and ethanol. However, under oxygen-rich, glucose-poor conditions, cells undergo a respiratory shift to aerobic ethanol metabolism. Ethanol is converted to acetyl coenzyme A and is metabolized by the citric acid cycle, which is coupled to the electron transport chain for ATP production (52). The evolution of these diverse metabolic capabilities ensures survival during fluctuating nutrient conditions. Growth in high glucose results in “glucose repression,” which is characterized by transcriptional repression of genes involved in alternate carbon source metabolism, including those in respiration (53).
arp5Δ mutants appear to have lost this glucose repression, since genes upregulated in these mutants are enriched in mitochondrial energy generation processes, including cellular respiration and storage carbon utilization (Fig. 6A). In addition, downregulated genes are enriched in metabolic pathways, such as glycolysis and RNA and protein metabolism (Fig. 6B). Previous microarray studies confirm these results through the identification of several metabolic genes regulated by the INO80 complex (29). Moreover, multiple published ChIP data sets confirm significant enrichment of Arp5 occupancy on genes of the mitochondrion gene ontology cellular component (P = 4.4 × 10e−7) in wild-type cells grown in glucose-rich media (39, 43).
Transcriptional deregulation of these metabolic gene pathways is not believed to originate from cell cycle disruptions, since fluorescence-activated cell sorting analysis demonstrates comparable cell cycle distributions in asynchronously growing cells: wild-type cells exhibit 27% G1, 30% S, and 43% G2/M phases; arp5Δ cells exhibit 30% G1, 27% S, and 45% G2/M phases; and ino80Δ cells exhibit 32% G1, 24% S, and 44% G2/M phases. Although mutant cells have altered DNA content that resembles that of diploids (18), variations in ploidy associated with cell size do not alter expression of metabolic genes regulated by INO80 (54). This strongly suggests that the INO80 complex plays a critical and direct role in regulating metabolic gene networks.
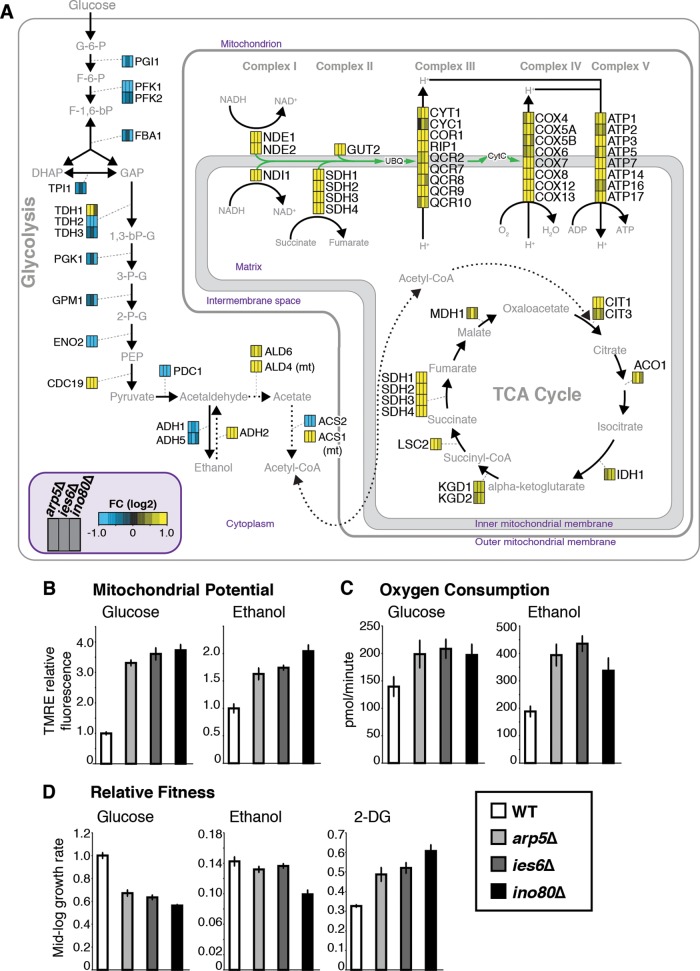
A closer inspection of the Arp5-Ies6-regulated genes reveals that the expression of nearly every gene involved in glycolysis is decreased while every gene in the mitochondrial electron transport chain is increased (Fig. 7A). Accordingly, mitochondrial potential, indicative of active respiration, is increased in arp5Δ, ies6Δ, and ino80Δ mutants (Fig. 7B). Oxygen consumption is also increased in these mutants relative to wild-type cells, demonstrating that the entire respiration capacity is elevated (Fig. 7C). These alterations of metabolic activity in the mutants are observed in both glucose-containing media, when oxidative phosphorylation is normally repressed, and ethanol-containing media, when oxidative phosphorylation is active. Thus, arp5Δ, ies6Δ, and ino80Δ mutants have constitutively increased rates of respiration, independent of carbon source availability.
FIG 7.
Arp5-Ies6 regulates INO80-mediated glycolytic function and respiration capacity. (A) Genes with ≥1.5-fold change (FC) in arp5Δ, ies6Δ, and ino80Δ mutants compared to the wild type are shown in glycolysis, citric acid cycle, ethanol fermentation (dashed lines), and the electron transport chain. Gene expression in each mutant is color coded as indicated. (B) Mitochondrial potential, in the wild type (WT) and the indicated mutant cells grown in glucose or ethanol-containing media, measured as TMRE fluorescence that accumulates in active mitochondria. Values are given relative to the WT signal within each growth condition. (C) Oxygen consumption (pmol/min/million cells) in wild-type (WT) and the indicated mutant cells grown in glucose or ethanol-containing media, measured by using an XF96 extracellular flux analyzer (Seahorse Biosciences). (D) Doubling time in the mid-log-phase growth of WT and indicated mutant cells in glucose-, ethanol-, and 2-DG-containing media. Fitness is given as relative to the WT growth in glucose-containing media.
Interestingly, the fitness defects of the mutants relative to the wild type can be partially suppressed by replacing glucose with ethanol in the culture media, which results in glycolytic inhibition and increased respiration (Fig. 7D). In these assays, mid-log-phase growth was monitored using OD measurements in order to quantitatively calculate relative growth rates indicative of fitness. The previously described fitness defects of arp5Δ, ies6Δ, and ino80Δ mutants (Fig. 2) are recapitulated using this approach (Fig. 7D). Furthermore, when the glycolytic inhibitor 2-DG is added to the media, growth is inhibited in wild-type cells to a greater extent than in mutant cells. 2-DG is a glucose analogue that cannot be fully metabolized, effectively inhibiting glycolytic enzymes (55). Thus, the relative fitness defects observed in the arp5Δ, ies6Δ, and ino80Δ mutants during normal growth in glucose can be substantially suppressed under conditions that inhibit glycolysis and support aerobic respiration. Again, these differences do not appear to be a direct consequence of differences in DNA content in the mutants, as haploid and diploid wild-type cells have similar mitochondrial potential and growth in the presence of 2-DG (data not shown). Collectively, these results demonstrate that Arp5-Ies6 regulates the INO80 complex to optimize metabolic capacity and corresponding fitness of cells through regulation of genes involved in glycolysis and respiration.
DISCUSSION
The Arp5-Ies6 subcomplex regulates INO80-mediated function.
Although individual chromatin remodelers can exhibit both transcriptional activation and repression properties (56, 57), it is largely unknown how these varied activities are regulated. Moreover, the composition of chromatin-remodeling complexes is often considered static. In this study, we demonstrate that the composition of the INO80 complex is fluid, with dynamic association of the Arp5-Ies6 subcomplex and subsequent modulation of in vitro chromatin-remodeling function.
Specifically, our results suggest that an INO80 complex lacking the Arp5-Ies6 module is unable to reposition nucleosomes in vitro and is correlated with repressed gene expression in vivo. Furthermore, Arp5-Ies6 can assemble into the subcomplex-deficient INO80 complex and modify nucleosome positioning and gene expression. These results illustrate the various ways in which one chromatin-remodeling complex can contribute to diverse modes of transcription through the dynamic composition of its subunits. Indeed, an emerging theme of chromatin-remodeling control is the regulation of distinct chromatin functions through the varying composition of individual complexes (58, 59). Recent genome-wide mapping analyses reveal nonoverlapping genomic occupancy of INO80 subunits (39). In addition, targeting of individual subunits to synthetic reporter genes confirms diverse subunit function in transcriptional regulation (57). These observations have important implications for gene expression, since varied compositions of the SWI/SNF, ISW1, and SWR1 complexes at specific target genes have been linked to varied rates of transcription (40, 60–62). Moreover, subcellular localization of chromatin-remodeling subunits can change in response to stress, further indicating the adaptable functionality of chromatin remodelers in coordination with environmental conditions (63).
Structural analyses of these multisubunit chromatin-remodeling complexes have aided in developing combinatorial models of chromatin-remodeling function. As previously mentioned, the Arp8-Arp4 and Arp5-Ies6 modules associate with distinct regions within the Ino80 ATPase subunit (21). Results presented in this study demonstrate that these two Arp-containing modules can participate in different INO80-mediated functions. For example, the Arp8-Arp4-actin module enhances nucleosome binding affinity (23, 26, 31). In addition, we recently demonstrated that Arp5-Ies6 is needed to couple ATP hydrolysis with nucleosome sliding (28). The results presented here reveal that the Arp5-Ies6 subcomplex is in excess abundance compared to the INO80 complex (Fig. 1). Interestingly, our results further reveal that the excess Arp5-Ies6 is chromatin associated in an INO80-dependent manner (Fig. 3). Thus, one intriguing model is that Arp5-Ies6 transiently associates with the INO80 complex before its autonomous association with chromatin. Because the Arp5-Ies6 subcomplex has much lower in vitro nucleosome affinity than the INO80 complex, it may be that the INO80 complex alters Arp5-Ies6 or chromatin conformation to one that is more conducive to association and/or retention or that Arp5-Ies6 is indirectly associating to chromatin via other unknown factors. Nevertheless, full understanding of the mechanisms by which Arp5-Ies6 regulates dynamic activities of the INO80 complex will require future examination.
It should be noted that previous ChIP experiments revealed Arp5 and Ino80 occupancy at many genic locations throughout the genome (39, 43). However, not all of these genes are significantly differentially expressed in arp5Δ and ino80Δ mutant cells. The contribution of Arp5-Ies6 and INO80 complex function to the expression of individual genes is likely dependent on myriad factors, including regulatory DNA elements, epigenetic modifications, and interaction with other trans-acting chromatin factors. Thus, although our data suggest that Arp5-Ies6 and INO80 may be a dominant mode of transcriptional regulation at a subset of metabolic genes, they may have more subtle influences on gene expression of other occupied loci. Alternatively, INO80 subunits may adopt “poised” chromatin-bound states that are responsive to cellular signaling pathways (such as inositol and stress signaling, as previously demonstrated [16, 64]) to elicit gene expression programs in response to changing cellular contexts. Indeed, the directed recruitment of Arp5-Ies6 to the INO80 complex may be dependent the metabolic state of the cell.
Arp5-Ies6 are primary determinants of INO80-mediated metabolic gene expression.
S. cerevisiae have evolved unique metabolic diversity in carbon catabolism pathways. Similar metabolic plasticity has been observed in cancer cells that endure and thrive in environments of varying oxygen and nutrient availability (65). These data illustrate the importance of chromatin-remodeling in balancing metabolic processes, which are commonly disrupted in developmental disorders and disease. For example, the SWI/SNF (SWItch/Sucrose Non-Fermentable) remodeler, first identified as a regulator of carbon metabolism in yeast (66), is known to be an important regulator of development (58) and is commonly disrupted in cancers (67). However, the link between SWI/SNF-regulated metabolic function and development and cancer is currently underexplored.
The results presented here suggest that the INO80 complex regulates glycolytic specification under nutrient-rich conditions. It is noteworthy that recent genome sequencing studies identify INO80 subunits as amplified in several cancer types (68), including amplification in more than 40% of lung squamous cell carcinomas (69), 43% of ovarian adenocarcinoma (70), and 65% of xenografts from breast cancer patients (71). Future research investigating the significance of INO80 subunit amplification in carcinogenesis is warranted, since cancer cells often exhibit a preference for glycolytic metabolism (65) that could be facilitated by an increase in INO80-mediated expression of genes involved in glycolysis.
A rapidly growing theme in the field of chromatin biology is that the chromatin template and chromatin-associated proteins respond to and integrate changing environmental and metabolic signals to enact homeostatic transcriptional responses (72). However, the extent to which chromatin-remodeling complexes are involved in these processes is largely unknown. Our analyses highlight Arp5-Ies6 and INO80 as critical transcriptional regulators of gene networks involved in cellular energy balance and protein synthesis and demonstrate the importance of epigenetic regulation in metabolic gene expression.
ACKNOWLEDGMENTS
We thank Rafael Herrera, Geeta Narlikar, and Frank Pugh for helpful advice in preparation of the manuscript.
We declare that no competing interests exist.
REFERENCES
- 1.Clapier CR, Cairns BR. 2009. The biology of chromatin remodeling complexes. Annu Rev Biochem 78:273–304. doi: 10.1146/annurev.biochem.77.062706.153223. [DOI] [PubMed] [Google Scholar]
- 2.Narlikar GJ, Sundaramoorthy R, Owen-Hughes T. 2013. Mechanisms and functions of ATP-dependent chromatin-remodeling enzymes. Cell 154:490–503. doi: 10.1016/j.cell.2013.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chia N-Y, Chan Y-S, Feng B, Lu X, Orlov YL, Moreau D, Kumar P, Yang L, Jiang J, Lau M-S, Huss M, Soh B-S, Kraus P, Li P, Lufkin T, Lim B, Clarke ND, Bard F, Ng HH. 2010. A genome-wide RNAi screen reveals determinants of human embryonic stem cell identity. Nature 468:316–320. doi: 10.1038/nature09531. [DOI] [PubMed] [Google Scholar]
- 4.Kim JH, Lee JM, Nam HJ, Choi HJ, Yang JW, Lee JS, Kim MH, Kim S-I, Chung CH, Kim KI, Baek SH. 2007. SUMOylation of pontin chromatin-remodeling complex reveals a signal integration code in prostate cancer cells. Proc Natl Acad Sci U S A 104:20793–20798. doi: 10.1073/pnas.0710343105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang L, Du Y, Ward JM, Shimbo T, Lackford B, Zheng X, Miao Y-L, Zhou B, Han L, Fargo DC, Jothi R, Williams CJ, Wade PA, Hu G. 2014. INO80 facilitates pluripotency gene activation in embryonic stem cell self-renewal, reprogramming, and blastocyst development. Cell Stem Cell 14:575–591. doi: 10.1016/j.stem.2014.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim JH, Choi HJ, Kim B, Kim MH, Lee JM, Kim IS, Lee MH, Choi SJ, Kim KI, Kim S-I, Chung CH, Baek SH. 2006. Roles of sumoylation of a reptin chromatin-remodelling complex in cancer metastasis. Nat Cell Biol 8:631–639. doi: 10.1038/ncb1415. [DOI] [PubMed] [Google Scholar]
- 7.Alcid EA, Tsukiyama T. 2014. ATP-dependent chromatin remodeling shapes the long noncoding RNA landscape. Genes Dev 28:2348–2360. doi: 10.1101/gad.250902.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shen X, Mizuguchi G, Hamiche A, Wu C. 2000. A chromatin remodelling complex involved in transcription and DNA processing. Nature 406:541–544. doi: 10.1038/35020123. [DOI] [PubMed] [Google Scholar]
- 9.Xue Y, Van C, Pradhan SK, Su T, Gehrke J, Kuryan BG, Kitada T, Vashisht A, Tran N, Wohlschlegel J, Peterson CL, Kurdistani SK, Carey MF. 2015. The Ino80 complex prevents invasion of euchromatin into silent chromatin. Genes Dev 29:350–355. doi: 10.1101/gad.256255.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vincent JA, Kwong TJ, Tsukiyama T. 2008. ATP-dependent chromatin remodeling shapes the DNA replication landscape. Nat Struct Mol Biol 15:477–484. doi: 10.1038/nsmb.1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shimada K, Oma Y, Schleker T, Kugou K, Ohta K, Harata M, Gasser SM. 2008. Ino80 chromatin remodeling complex promotes recovery of stalled replication forks. Curr Biol 18:566–575. doi: 10.1016/j.cub.2008.03.049. [DOI] [PubMed] [Google Scholar]
- 12.Papamichos-Chronakis M, Peterson CL. 2008. The Ino80 chromatin-remodeling enzyme regulates replisome function and stability. Nat Struct Mol Biol 15:338–345. doi: 10.1038/nsmb.1413. [DOI] [PubMed] [Google Scholar]
- 13.Morrison AJ, Highland J, Krogan NJ, Arbel-Eden A, Greenblatt JF, Haber JE, Shen X. 2004. INO80 and γ-H2AX interaction links ATP-dependent chromatin remodeling to DNA damage repair. Cell 119:767–775. doi: 10.1016/j.cell.2004.11.037. [DOI] [PubMed] [Google Scholar]
- 14.Van Attikum H, Fritsch O, Hohn B, Gasser SM. 2004. Recruitment of the INO80 complex by H2A phosphorylation links ATP-dependent chromatin remodeling with DNA double-strand break repair. Cell 119:777–788. doi: 10.1016/j.cell.2004.11.033. [DOI] [PubMed] [Google Scholar]
- 15.Falbo KB, Alabert C, Katou Y, Wu S, Han J, Wehr T, Xiao J, He X, Zhang Z, Shi Y, Shirahige K, Pasero P, Shen X. 2009. Involvement of a chromatin remodeling complex in damage tolerance during DNA replication. Nat Struct Mol Biol 16:1167–1172. doi: 10.1038/nsmb.1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Morrison AJ, Kim J-A, Person MD, Highland J, Xiao J, Wehr TS, Hensley S, Bao Y, Shen J, Collins SR, Weissman JS, Delrow J, Krogan NJ, Haber JE, Shen X. 2007. Mec1/Tel1 phosphorylation of the INO80 chromatin remodeling complex influences DNA damage checkpoint responses. Cell 130:499–511. doi: 10.1016/j.cell.2007.06.010. [DOI] [PubMed] [Google Scholar]
- 17.Yu EY, Steinberg-Neifach O, Dandjinou AT, Kang F, Morrison AJ, Shen X, Lue NF. 2007. Regulation of telomere structure and functions by subunits of the INO80 chromatin remodeling complex. Mol Cell Biol 27:5639–5649. doi: 10.1128/MCB.00418-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chambers AL, Ormerod G, Durley SC, Sing TL, Brown GW, Kent NA, Downs JA. 2012. The INO80 chromatin remodeling complex prevents polyploidy and maintains normal chromatin structure at centromeres. Genes Dev 26:2590–2603. doi: 10.1101/gad.199976.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ogiwara H, Enomoto T, Seki M. 2007. The INO80 chromatin remodeling complex functions in sister chromatid cohesion. Cell Cycle 6:1090–1095. doi: 10.4161/cc.6.9.4130. [DOI] [PubMed] [Google Scholar]
- 20.Morrison AJ, Shen X. 2009. Chromatin remodelling beyond transcription: the INO80 and SWR1 complexes. Nat Rev Mol Cell Biol 10:373–384. doi: 10.1038/nrm2693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tosi A, Haas C, Herzog F, Gilmozzi A, Berninghausen O, Ungewickell C, Gerhold CB, Lakomek K, Aebersold R, Beckmann R, Hopfner K-P. 2013. Structure and subunit topology of the INO80 chromatin remodeler and its nucleosome complex. Cell 154:1207–1219. doi: 10.1016/j.cell.2013.08.016. [DOI] [PubMed] [Google Scholar]
- 22.Szerlong H, Hinata K, Viswanathan R, Erdjument-Bromage H, Tempst P, Cairns BR. 2008. The HSA domain binds nuclear actin-related proteins to regulate chromatin-remodeling ATPases. Nat Struct Mol Biol 15:469–476. doi: 10.1038/nsmb.1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shen X, Ranallo R, Choi E, Wu C. 2003. Involvement of actin-related proteins in ATP-dependent chromatin remodeling. Mol Cell 12:147–155. doi: 10.1016/S1097-2765(03)00264-8. [DOI] [PubMed] [Google Scholar]
- 24.Kapoor P, Chen M, Winkler DD, Luger K, Shen X. 2013. Evidence for monomeric actin function in INO80 chromatin remodeling. Nat Struct Mol Biol 20:426–432. doi: 10.1038/nsmb.2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Harata M, Oma Y, Mizuno S, Jiang YW, Stillman DJ, Wintersberger U. 1999. The nuclear actin-related protein of Saccharomyces cerevisiae, Act3p/Arp4, interacts with core histones. Mol Biol Cell 10:2595–2605. doi: 10.1091/mbc.10.8.2595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Saravanan M, Wuerges J, Bose D, McCormack EA, Cook NJ, Zhang X, Wigley DB. 2012. Interactions between the nucleosome histone core and Arp8 in the INO80 chromatin remodeling complex. Proc Natl Acad Sci U S A 109:20883–20888. doi: 10.1073/pnas.1214735109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gerhold CB, Winkler DD, Lakomek K, Seifert FU, Fenn S, Kessler B, Witte G, Luger K, Hopfner K-P. 2012. Structure of actin-related protein 8 and its contribution to nucleosome binding. Nucleic Acids Res 40:11036–11046. doi: 10.1093/nar/gks842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yao W, Beckwith SL, Zheng T, Young T, Dinh VT, Ranjan A, Morrison AJ. 2015. Assembly of the Arp5 (actin-related protein) subunit involved in distinct INO80 chromatin remodeling activities. J Biol Chem 290:25700–25709. doi: 10.1074/jbc.M115.674887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jónsson ZO, Jha S, Wohlschlegel JA, Dutta A. 2004. Rvb1p/Rvb2p recruit Arp5p and assemble a functional Ino80 chromatin remodeling complex. Mol Cell 16:465–477. doi: 10.1016/j.molcel.2004.09.033. [DOI] [PubMed] [Google Scholar]
- 30.Chen L, Conaway RC, Conaway JW. 2013. Multiple modes of regulation of the human Ino80 SNF2 ATPase by subunits of the INO80 chromatin-remodeling complex. Proc Natl Acad Sci U S A 110:20497–20502. doi: 10.1073/pnas.1317092110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Watanabe S, Tan D, Lakshminarasimhan M, Washburn MP, Erica Hong E-J, Walz T, Peterson CL. 2015. Structural analyses of the chromatin remodelling enzymes INO80-C and SWR-C. Nat Commun 6:7108. doi: 10.1038/ncomms8108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sadeh R, Allis CD. 2011. Genome-wide “re-”modeling of nucleosome positions. Cell 147:263–266. doi: 10.1016/j.cell.2011.09.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gkikopoulos T, Schofield P, Singh V, Pinskaya M, Mellor J, Smolle M, Workman JL, Barton GJ, Owen-Hughes T. 2011. A role for Snf2-related nucleosome-spacing enzymes in genome-wide nucleosome organization. Science 333:1758–1760. doi: 10.1126/science.1206097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang Y, Moqtaderi Z, Rattner BP, Euskirchen G, Snyder M, Kadonaga JT, Liu XS, Struhl K. 2009. Intrinsic histone-DNA interactions are not the major determinant of nucleosome positions in vivo. Nat Struct Mol Biol 16:847–852. doi: 10.1038/nsmb.1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang Z, Wippo CJ, Wal M, Ward E, Korber P, Pugh BF. 2011. A packing mechanism for nucleosome organization reconstituted across a eukaryotic genome. Science 332:977–980. doi: 10.1126/science.1200508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rhee HS, Pugh BF. 2012. Genome-wide structure and organization of eukaryotic pre-initiation complexes. Nature 483:295–301. doi: 10.1038/nature10799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lomvardas S, Thanos D. 2001. Nucleosome sliding via TBP DNA binding in vivo. Cell 106:685–696. doi: 10.1016/S0092-8674(01)00490-1. [DOI] [PubMed] [Google Scholar]
- 38.Martinez-Campa C, Politis P, Moreau J-L, Kent N, Goodall J, Mellor J, Goding CR. 2004. Precise nucleosome positioning and the TATA box dictate requirements for the histone H4 tail and the bromodomain factor Bdf1. Mol Cell 15:69–81. doi: 10.1016/j.molcel.2004.05.022. [DOI] [PubMed] [Google Scholar]
- 39.Yen K, Vinayachandran V, Pugh BF. 2013. SWR-C and INO80 chromatin remodelers recognize nucleosome-free regions near +1 nucleosomes. Cell 154:1246–1256. doi: 10.1016/j.cell.2013.08.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yoshida T, Shimada K, Oma Y, Kalck V, Akimura K, Taddei A, Iwahashi H, Kugou K, Ohta K, Gasser SM, Harata M. 2010. Actin-related protein Arp6 influences H2A.Z-dependent and -independent gene expression and links ribosomal protein genes to nuclear pores. PLoS Genet 6:e1000910. doi: 10.1371/journal.pgen.1000910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xiao H, Mizuguchi G, Wisniewski J, Huang Y, Wei D, Wu C. 2011. Nonhistone Scm3 binds to AT-rich DNA to organize atypical centromeric nucleosome of budding yeast. Mol Cell 43:369–380. doi: 10.1016/j.molcel.2011.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L. 2012. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc 7:562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yen K, Vinayachandran V, Batta K, Koerber RT, Pugh BF. 2012. Genome-wide nucleosome specificity and directionality of chromatin remodelers. Cell 149:1461–1473. doi: 10.1016/j.cell.2012.04.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Eden E, Navon R, Steinfeld I, Lipson D, Yakhini Z. 2009. GOrilla: a tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinformatics 10:48. doi: 10.1186/1471-2105-10-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Supek F, Bošnjak M, Škunca N, Šmuc T. 2011. REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS One 6:e21800. doi: 10.1371/journal.pone.0021800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jiang C, Pugh BF. 2009. A compiled and systematic reference map of nucleosome positions across the Saccharomyces cerevisiae genome. Genome Biol 10:R109. doi: 10.1186/gb-2009-10-10-r109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Onge RPS, Mani R, Oh J, Proctor M, Fung E, Davis RW, Nislow C, Roth FP, Giaever G. 2007. Systematic pathway analysis using high-resolution fitness profiling of combinatorial gene deletions. Nat Genet 39:199–206. doi: 10.1038/ng1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Udugama M, Sabri A, Bartholomew B. 2011. The INO80 ATP-dependent chromatin remodeling complex is a nucleosome spacing factor. Mol Cell Biol 31:662–673. doi: 10.1128/MCB.01035-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ringnér M. 2008. What is principal component analysis? Nat Biotechnol 26:303–304. doi: 10.1038/nbt0308-303. [DOI] [PubMed] [Google Scholar]
- 50.Schmid A, Fascher KD, Hörz W. 1992. Nucleosome disruption at the yeast PHO5 promoter upon PHO5 induction occurs in the absence of DNA replication. Cell 71:853–864. doi: 10.1016/0092-8674(92)90560-Y. [DOI] [PubMed] [Google Scholar]
- 51.Whitehouse I, Tsukiyama T. 2006. Antagonistic forces that position nucleosomes in vivo. Nat Struct Mol Biol 13:633-640. [DOI] [PubMed] [Google Scholar]
- 52.Fiechter A, Fuhrmann GF, Käppeli O. 1981. Regulation of glucose metabolism in growing yeast cells. Adv Microb Physiol 22:123–183. doi: 10.1016/S0065-2911(08)60327-6. [DOI] [PubMed] [Google Scholar]
- 53.Gancedo JM. 1998. Yeast carbon catabolite repression. Microbiol Mol Biol Rev 62:334–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wu C-Y, Rolfe PA, Gifford DK, Fink GR. 2010. Control of transcription by cell size. PLoS Biol 8:e1000523. doi: 10.1371/journal.pbio.1000523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.McCartney RR, Chandrashekarappa DG, Zhang BB, Schmidt MC. 2014. Genetic analysis of resistance and sensitivity to 2-deoxyglucose in Saccharomyces cerevisiae. Genetics 198:635–646. doi: 10.1534/genetics.114.169060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lenstra TL, Benschop JJ, Kim T, Schulze JM, Brabers NACH, Margaritis T, Van De Pasch LAL, Van Heesch SAAC, Brok MO, Groot Koerkamp MJA, Ko CW, Van Leenen D, Sameith K, Van Hooff SR, Lijnzaad P, Kemmeren P, Hentrich T, Kobor MS, Buratowski S, Holstege FCP. 2011. The specificity and topology of chromatin interaction pathways in yeast. Mol Cell 42:536–549. doi: 10.1016/j.molcel.2011.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Keung AJ, Bashor CJ, Kiriakov S, Collins JJ, Khalil AS. 2014. Using targeted chromatin regulators to engineer combinatorial and spatial transcriptional regulation. Cell 158:110–120. doi: 10.1016/j.cell.2014.04.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ho L, Crabtree GR. 2010. Chromatin remodelling during development. Nature 463:474–484. doi: 10.1038/nature08911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gerhold CB, Gasser SM. 2014. INO80 and SWR complexes: relating structure to function in chromatin remodeling. Trends Cell Biol 24:619–631. doi: 10.1016/j.tcb.2014.06.004. [DOI] [PubMed] [Google Scholar]
- 60.Euskirchen GM, Auerbach RK, Davidov E, Gianoulis TA, Zhong G, Rozowsky J, Bhardwaj N, Gerstein MB, Snyder M. 2011. Diverse roles and interactions of the SWI/SNF chromatin remodeling complex revealed using global approaches. PLoS Genet 7:e1002008. doi: 10.1371/journal.pgen.1002008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ryme J, Asp P, Böhm S, Cavellán E, Farrants A-KÖ. 2009. Variations in the composition of mammalian SWI/SNF chromatin remodelling complexes. J Cell Biochem 108:565–576. doi: 10.1002/jcb.22288. [DOI] [PubMed] [Google Scholar]
- 62.Morillon A, Karabetsou N, O'Sullivan J, Kent N, Proudfoot N, Mellor J. 2003. Isw1 chromatin remodeling ATPase coordinates transcription elongation and termination by RNA polymerase II. Cell 115:425–435. doi: 10.1016/S0092-8674(03)00880-8. [DOI] [PubMed] [Google Scholar]
- 63.Dastidar RG, Hooda J, Shah A, Cao TM, Henke RM, Zhang L. 2012. The nuclear localization of SWI/SNF proteins is subjected to oxygen regulation. Cell Biosci 2:30. doi: 10.1186/2045-3701-2-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shen X, Xiao H, Ranallo R, Wu W-H, Wu C. 2003. Modulation of ATP-dependent chromatin-remodeling complexes by inositol polyphosphates. Science 299:112–114. doi: 10.1126/science.1078068. [DOI] [PubMed] [Google Scholar]
- 65.Cairns RA, Harris IS, Mak TW. 2011. Regulation of cancer cell metabolism. Nat Rev Cancer 11:85–95. doi: 10.1038/nrc2981. [DOI] [PubMed] [Google Scholar]
- 66.Neigeborn L, Carlson M. 1984. Genes affecting the regulation of SUC2 gene expression by glucose repression in Saccharomyces cerevisiae. Genetics 108:845–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Shain AH, Pollack JR. 2013. The spectrum of SWI/SNF mutations, ubiquitous in human cancers. PLoS One 8:e55119. doi: 10.1371/journal.pone.0055119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, Antipin Y, Reva B, Goldberg AP, Sander C, Schultz N. 2012. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.The Cancer Genome Atlas Research Network. 2012. Comprehensive genomic characterization of squamous cell lung cancers. Nature 489:519–525. doi: 10.1038/nature11404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.The Cancer Genome Atlas Research Network. 2011. Integrated genomic analyses of ovarian carcinoma. Nature 474:609–615. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Eirew P, Steif A, Khattra J, Ha G, Yap D, Farahani H, Gelmon K, Chia S, Mar C, Wan A, Laks E, Biele J, Shumansky K, Rosner J, McPherson A, Nielsen C, Roth AJL, Lefebvre C, Bashashati A, de Souza C, Siu C, Aniba R, Brimhall J, Oloumi A, Osako T, Bruna A, Sandoval JL, Algara T, Greenwood W, Leung K, Cheng H, Xue H, Wang Y, Lin D, Mungall AJ, Moore R, Zhao Y, Lorette J, Nguyen L, Huntsman D, Eaves CJ, Hansen C, Marra MA, Caldas C, Shah SP, Aparicio S. 2014. Dynamics of genomic clones in breast cancer patient xenografts at single-cell resolution. Nature 518:422–426. doi: 10.1038/nature13952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gut P, Verdin E. 2013. The nexus of chromatin regulation and intermediary metabolism. Nature 502:489–498. doi: 10.1038/nature12752. [DOI] [PubMed] [Google Scholar]