ABSTRACT
Steroids are ubiquitous in natural environments and are a significant growth substrate for microorganisms. Microbial steroid metabolism is also important for some pathogens and for biotechnical applications. This study delineated the distribution of aerobic steroid catabolism pathways among over 8,000 microorganisms whose genomes are available in the NCBI RefSeq database. Combined analysis of bacterial, archaeal, and fungal genomes with both hidden Markov models and reciprocal BLAST identified 265 putative steroid degraders within only Actinobacteria and Proteobacteria, which mainly originated from soil, eukaryotic host, and aquatic environments. These bacteria include members of 17 genera not previously known to contain steroid degraders. A pathway for cholesterol degradation was conserved in many actinobacterial genera, particularly in members of the Corynebacterineae, and a pathway for cholate degradation was conserved in members of the genus Rhodococcus. A pathway for testosterone and, sometimes, cholate degradation had a patchy distribution among Proteobacteria. The steroid degradation genes tended to occur within large gene clusters. Growth experiments confirmed bioinformatic predictions of steroid metabolism capacity in nine bacterial strains. The results indicate there was a single ancestral 9,10-seco-steroid degradation pathway. Gene duplication, likely in a progenitor of Rhodococcus, later gave rise to a cholate degradation pathway. Proteobacteria and additional Actinobacteria subsequently obtained a cholate degradation pathway via horizontal gene transfer, in some cases facilitated by plasmids. Catabolism of steroids appears to be an important component of the ecological niches of broad groups of Actinobacteria and individual species of Proteobacteria.
IMPORTANCE
Steroids are ubiquitous growth substrates for environmental and pathogenic bacteria, and bacterial steroid metabolism has important pharmaceutical and health applications. To date, the genetics and biochemistry of microbial steroid degradation have mainly been studied in a few model bacteria, and the diversity of this metabolism remains largely unexplored. Here, we provide a bioinformatically derived perspective of the taxonomic distribution of aerobic microbial steroid catabolism pathways. We identified several novel steroid-degrading bacterial groups, including ones from marine environments. In several cases, we confirmed bioinformatic predictions of metabolism in cultures. We found that cholesterol and cholate catabolism pathways are highly conserved among certain actinobacterial taxa. We found evidence for horizontal transfer of a pathway to several proteobacterial genera, conferring testosterone and, sometimes, cholate catabolism. The results of this study greatly expand our ecological and evolutionary understanding of microbial steroid metabolism and provide a basis for better exploiting this metabolism for biotechnology.
INTRODUCTION
Microbial steroid degradation is an important process in several ways. Steroids constitute a highly abundant class of organic molecules in natural environments. Sterols are a major constituent of the membranes of all eukaryotic cells (1). Other steroids such as bile salts and steroid hormones are excreted into the environment by vertebrates. Thus, during decomposition of biomass and excreta, these steroids are available to microorganisms as significant potential growth substrates. Bacterial steroid degradation is also medically relevant, as a functioning cholesterol degradation pathway is essential for the survival of phagocytized Mycobacterium tuberculosis, the pathogen behind the tuberculosis epidemic (2). And, in the pharmaceutical industry, bacterial steroid biotransformation has been explored as a source of biocatalysts for the production of steroid-based drugs (3).
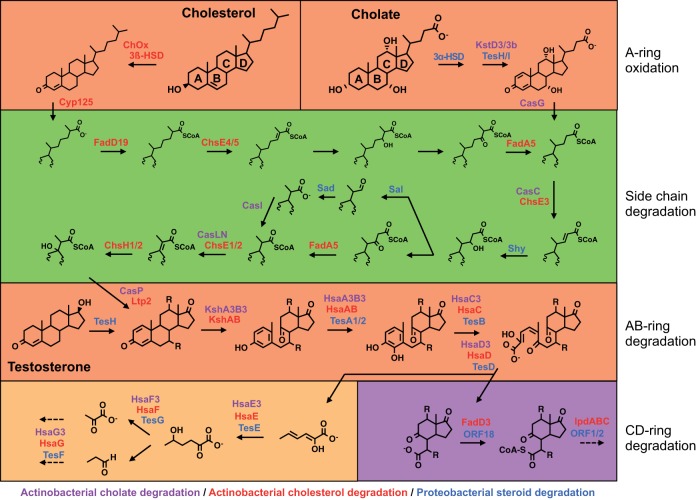
Knowledge of steroid catabolism pathways is based mainly on the investigation of a few Actinobacteria and Proteobacteria species, which have been shown to grow on and, in several cases, mineralize steroids (4–10). Testosterone degradation and bile salt degradation have been best studied in Comamonas testosteroni strains CNB-2 and TA441 (4) and Pseudomonas sp. strain Chol1 (11). Cholesterol degradation has been best studied in Rhodococcus jostii RHA1 (9) and Mycobacterium tuberculosis H37Rv (12–14). Recently, bile salt degradation has also been studied in strain RHA1 (8). In all of these cases, degradation of the steroid nucleus follows very similar progressions, using the 9,10-seco pathway (Fig. 1). Where present, side chains are degraded by a beta-oxidation process. In the case of cholesterol, the alkyl side chain is initially activated by a monooxygenase (15–17). Bacterial steroid uptake is poorly understood. Actinobacteria spp. appear to take up cholesterol with a complex ABC transporter comprised of many proteins (18). The genes encoding steroid catabolism that have been identified tend to occur in large clusters encoding major components of the degradation pathways (see Fig. S1 in the supplemental material). In the cholesterol pathway, the genes encoding C and D ring degradation are in a distinct regulon (19–21). It is not known how well these characterized pathways represent steroid catabolism in other microorganisms.
FIG 1 .
Aerobic 9,10-seco degradation pathways for cholesterol, cholate, and testosterone. The steroid ring structure is degraded by oxygen-dependent opening and subsequent hydrolytic cleavage of rings A and B. Subsequent degradation of the C and D rings occurs by a mechanism not yet described. In Actinobacteria, side chain degradation and ring opening can occur simultaneously. Characterized or annotated enzymes involved in the degradation of cholesterol by Actinobacteria are red, those involved in the degradation of cholate by Actinobacteria are lilac, and those involved in the degradation of testosterone or cholate by Proteobacteria are blue. Protein nomenclature is based on that of Rhodococcus jostii RHA1, Mycobacterium tuberculosis H37Rv, Comamonas testosteroni TA441, and Pseudomonas sp. strain Chol1, and not all proteins are named.
Several denitrifying Proteobacteria spp. have been described to degrade cholesterol and testosterone under anaerobic conditions (22–24) using dioxygen-independent reactions to degrade the steroidal core (25, 26), in contrast to the aerobic 9,10-seco pathway. Unfortunately, the genetic and biochemical background of the anaerobic steroid degradation pathway is largely unknown, and genome sequences are not available for these bacteria.
Given the ubiquity of steroids in the natural environment, it is conceivable that diverse microbial taxa possess steroid degradation capabilities and occupy a range of ecological niches. One expedient, culture-independent method of discovering new steroid-degrading taxa is mining genomic databases for steroid degradation gene clusters homologous to those found in known steroid degraders. Although biased toward medically or economically important organisms, genome databases now represent diverse microbial taxa. Thus, analysis of genomes can potentially determine the occurrence of biochemical pathways among taxa and may lend insight into the evolution of these pathways. In this study, we explored the distribution of pathways homologous to known steroid degradation pathways among genomes in the curated RefSeq database hosted by the National Center for Biotechnology Information. All fungal, archaeal, and bacterial genomes from RefSeq were searched using hidden Markov models (HMMs), and a subset was searched using reciprocal BLAST. The results were used to infer the distribution of the pathways among taxa and to deduce aspects of the evolution of the pathways. Several newly identified steroid degraders were tested in vitro to validate predictions of their steroid degradation capacities.
RESULTS
Identification of steroid-degrading organisms.
Hidden Markov models (HMMs) were used to search sequenced microbial genomes in order to identify putative steroid-degrading organisms. A total of twenty-five HMMs were used, representing variants of eight key enzymes involved in steroid nucleus degradation (Table 1), to search 8,277 complete and draft bacterial, archaeal, and fungal genomes downloaded from RefSeq. A total of 265 putative steroid-degrading organisms were identified, their genomes encoding at least six of the eight key enzymes, including KshA/CtCNB1_1306, the oxygenase subunit of the 3-ketosteroid-9α-hydroxylase, and HsaC/TesB, an extradiol dioxygenase required for A/B ring degradation. The organisms identified were Actinobacteria spp. as well as alpha-, beta-, and gammaproteobacteria (see Table S1 in the supplemental material). The putative steroid-degrading bacteria were mainly from soil, eukaryotic host, and marine environments, with the majority of host-associated ones being pathogens (see Fig. S2). No putative steroid-degrading fungi or Archaea were detected.
TABLE 1 .
Table listing eight orthologous groups of proteins from Rhodococcus jostii RHA1, Mycobacterium tuberculosis H37Rv, and Comamonas testosteroni CNB-2 used as references for HMM generationa
| RHA1/H37Rv name | CNB-2 name | CNB-2 testosterone/cholate cluster | RHA1cholate cluster | RHA1cholesterol cluster | H37Rvcholesterol cluster | Annotation |
|---|---|---|---|---|---|---|
| KstD | TesH | CtCNB1_1357 | RHA1_ro05798 | RHA1_ro04532 | Rv3537 | 3-Ketosteroid-dehydrogenase |
| (CTCNB1_RS06925) | (RHA1_RS28305) | (RHA1_RS22090) | ||||
| RHA1_ro05813 | ||||||
| (RHA1_RS28380) | ||||||
| KshA | No name | CtCNB1_1306 | RHA1_ro05811 | RHA1_ro04538 | Rv3526 | 3-Ketosteroid-9-alpha hydroxylase (oxygenase) |
| (CTCNB1_RS06665) | (RHA1_RS28370) | (RHA1_RS22120) | ||||
| HsaA | TesA2 | CtCNB1_1356 | RHA1_ro05802 | RHA1_ro04539 | Rv3570c | 3-Hydroxy-9,10-seconandrost-1,3,5(10)-triene-9,17-dione-4-hydroxylase (oxygenase) |
| (CTCNB1_RS06920) | (RHA1_RS28325) | (RHA1_RS22125) | ||||
| HsaC | TesB | CtCNB1_1275 | RHA1_ro05803 | RHA1_ro04541 | Rv3568c | 3,4-Dihydroxy-9,10-secoandrosta-1,3,5(10)-triene-9,17-dione-4,5-dioxygenase |
| (CTCNB1_RS06510) | (RHA1_RS28330) | (RHA1_RS22135) | ||||
| HsaD | TesD | CtCNB1_1354 | RHA1_ro05797 | RHA1_ro04540 | Rv3569c | 4,5-9,10-Diseco-3-hydroxy-5,9,17-trioxoandrosta-1(10),2-diene-4-oate hydrolase |
| (CTCNB1_RS06910) | (RHA1_RS28300) | (RHA1_RS22130) | ||||
| HsaE | TesE | CtCNB1_1353 | RHA1_ro05799 | RHA1_ro04533 | Rv3536c | 2-Hydroxyhexa-2,4-dienoate hydratase |
| (CTCNB1_RS06905) | (RHA1_RS28310) | (RHA1_RS22095) | ||||
| HsaF | TesG | CtCNB1_1351 | RHA1_ro05801 | RHA1_ro04535 | Rv3534c | 4-Hydroxy-2-oxohexanoate aldolase |
| (CTCNB1_RS06905) | (RHA1_RS28320) | (RHA1_RS22105) | ||||
| HsaG | TesF | CtCNB1_1352 | RHA1_ro05800 | RHA1_ro04534 | Rv3535c | Propanol dehydrogenase |
| (CTCNB1_RS06900) | (RHA1_RS28315) | (RHA1_RS22100) |
Locus tags of the respective proteins are listed as used in this study and after subsequent GenBank reannotation (the latter are indicated in parentheses).
A total of 212 putative steroid-degrading Actinobacteria spp. were identified, representing 16 genera. These included most genera in the suborder Corynebacterineae (Amycolicicoccus, Dietzia, Gordonia, Mycobacterium, Nocardia, Rhodococcus, and Tsukamurella) as well as the genera Actinoplanes, Aeromicrobium, Amycolatopsis, Arthrobacter, Nocardioides, Saccharomonospora, Salinispora, Streptomyces, and Thermomonospora (see Table S1 in the supplemental material). With few exceptions, all available genomes from these genera appear to encode at least one steroid catabolism pathway. The exceptions lacking such pathways were Rhodococcus sp. strain AW25M09 (affiliated with R. fascians), all draft and complete genomes of Mycobacterium leprae, 5 of 6 draft and complete genomes of Saccharomonospora spp., 3 of 4 complete genomes of Actinoplanes spp., and 55 of 57 draft and complete genomes of Streptomyces spp.
A total of 53 putative steroid-degrading Proteobacteria spp. were identified. These were individual species within the genera Burkholderia, Comamonas, Cupriavidus, Glaciecola, Hydrocarboniphaga, Marinobacterium, Novosphingobium, Pseudoalteromonas, Pseudomonas, Shewanella, and Sphingomonas (see Table S1 in the supplemental material). They also included unclassified species of the alphaproteobacteria and gammaproteobacteria as well members of the SAR86 clade of gammaproteobacteria and the OM60 clade of the oligotrophic marine Gammaproteobacteria (OMG) group. Notably, genomes of Glaciecola, Marinobacterium, Pseudoalteromonas, and Shewanella and of OM60 and SAR86 all represent organisms from marine environments. In contrast to the actinobacterial genera, only one or a few genomes from each proteobacterial genus appear to encode steroid catabolism. The only exception is the genus Comamonas, in which steroid catabolism genes were found in four of four genomes.
Identification of steroid catabolism genes.
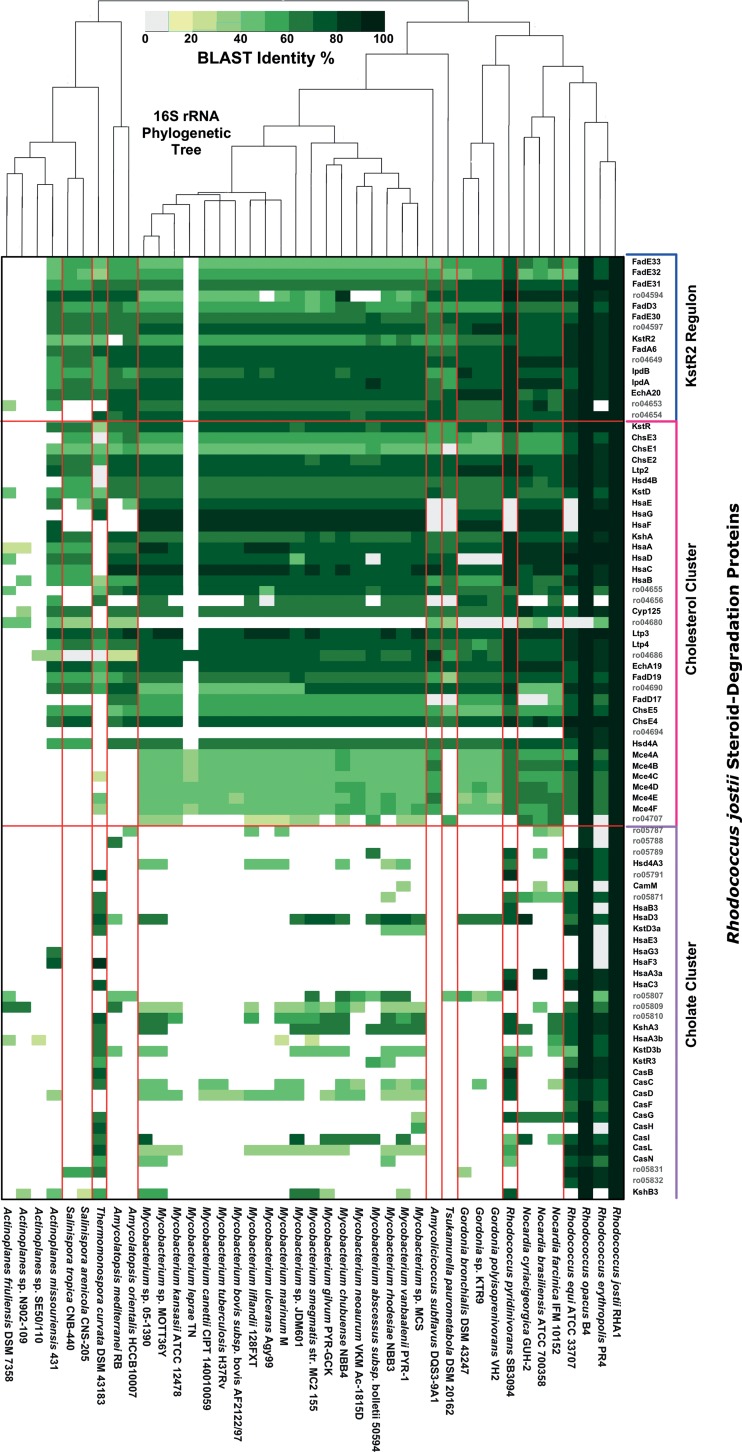
Genomes of putative steroid-degrading organisms were subsequently searched by best reciprocal BLASTp analysis to more comprehensively identify steroid catabolism genes and match them to their orthologs among the reference genes. A total of 124 complete genomes were analyzed by BLASTp (see Table S2 in the supplemental material). These included the genomes of all species within each genus identified by HMM analysis. Where species were represented by multiple genomes, a single representative strain was analyzed by BLASTp. The only exception was Pseudomonas putida, for which genomes of all strains were analyzed by BLASTp, because only a subset of strains were identified by the HMM analysis. In addition, 75 draft genomes were analyzed by BLASTp (Table S2). These included the genomes of a single strain from each species identified by HMM analysis. Draft Mycobacterium genomes were not analyzed, as this genus was very well represented by complete genomes. We additionally conducted BLASTp analysis of 24 Rhodococcus fascians genomes. Actinobacterial genomes were queried using 114 protein sequences deduced from the R. jostii cholate and cholesterol degradation gene clusters (see Fig. S1), while proteobacterial genomes were queried using 93 protein sequences deduced from the C. testosteroni CNB-2 cholate and testosterone degradation gene cluster. The phylogeny of the bacteria was assessed using 16S rRNA gene sequences. Nearly all complete genomes and draft genomes identified by the previous HMM analysis had reciprocal hits to a large majority of query sequences from at least one gene cluster. The only two exceptions among complete genomes were those of Novosphingobium pentaromativorans and Arthrobacter gangotriensis (not shown). None of the 24 R. fascians draft genomes were found to encode a steroid catabolism pathway (not shown).
In all Actinobacteria confirmed to have steroid catabolism genes via BLASTp analysis, the hits included orthologs of cholesterol catabolism genes from R. jostii (Fig. 2; see also Fig. S3 in the supplemental material). The only exception to this was Thermomonospora curvata. The distribution of the actinobacterial cholate pathway was much more restricted than that of the cholesterol pathway, as it was identified only in genomes of Rhodococcus spp., T. curvata, Gordonia rubripertincta, and Saccharomonospora paurometabolica.
FIG 2 .
Heat map showing BLAST identity for best reciprocal BLASTp hits to Rhodococcus jostii RHA1 steroid degradation proteins in 41 actinobacterial complete genomes.
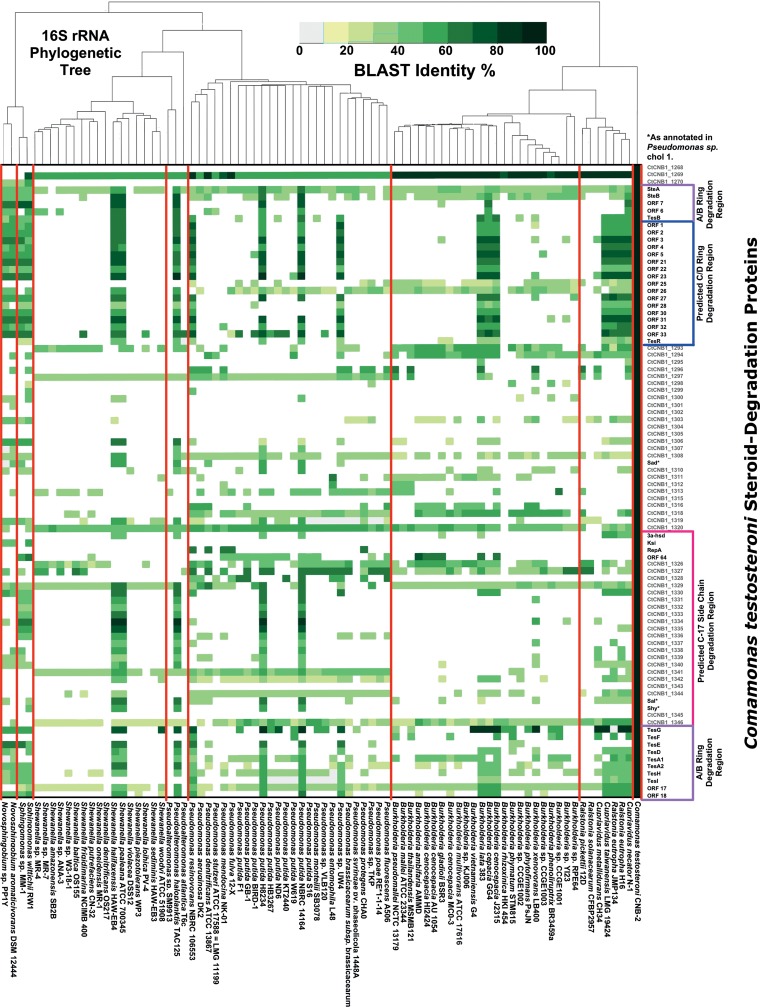
In all Proteobacteria spp. confirmed to have steroid catabolism genes via BLASTp analysis, the hits included orthologs of testosterone/cholate catabolism genes from strain CNB-2 (Fig. 3; see also Fig. S3 in the supplemental material). In contrast to actinobacterial genera, proteobacterial genera represented by multiple genomes had a minority of members with putative steroid catabolism pathways. And, within the species Pseudomonas putida, some strains were predicted to have a steroid catabolism pathway, whereas others were not. Of the Proteobacteria with steroid catabolism pathways, all members of Burkholderia, Ralstonia, Cupriavidus, and Novosphingobium as well as two of four members of Pseudomonas lacked most of the genes associated with degradation of the cholate side chain. Further, the two Sphingomonas strains had only a subset of the side chain degradation genes.
FIG 3 .
Heat map showing BLAST identity for best reciprocal BLASTp hits to Comamonas testosteroni CNB-2 steroid degradation proteins in 82 proteobacterial complete genomes.
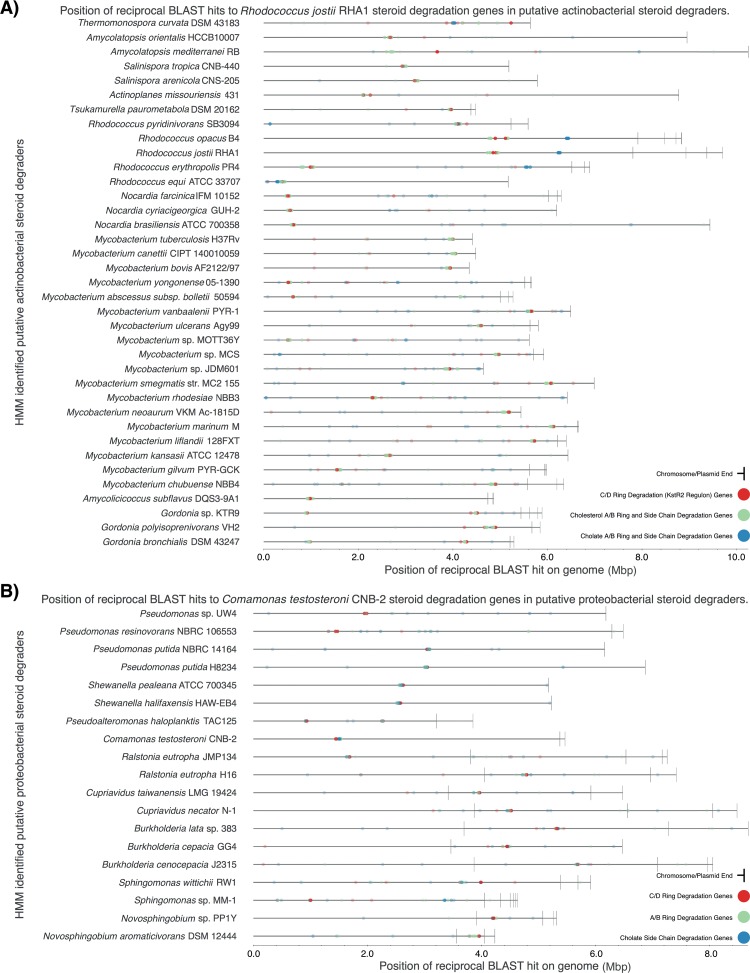
Gene localization.
The positions in each genome of the BLASTp hits described above were mapped, and there was a strong tendency, as in the reference genomes, for genes to reside in one or a few clusters. Most of the actinobacterial genomes had a single cluster containing most of the cholesterol catabolism genes (Fig. 4A), including genes encoding C/D ring degradation, which are organized in a distinct regulon regulated by KstR2, a Tet-like repressor, in R. jostii and M. tuberculosis H37Rv. Rhodococcus spp. additionally had the cholate catabolism genes in a separate cluster, remote from the cholesterol catabolism cluster and lacking C/D ring degradation genes. R. equi was an exception, having a single gene cluster with predicted cholate and cholesterol catabolism genes. There were several other actinobacterial genomes with distinct clustering patterns. Thermomonospora curvata had a cluster with cholate degradation genes and a separate one with C/D ring degradation genes. Mycobacterium abscessus subsp. bolletii had two gene clusters, both with cholesterol catabolism genes. Finally, in Amycolatopsis mediterranei, the gene cluster with C/D ring degradation genes was distant from the one encoding the remainder of the cholesterol pathway. Most of the proteobacterial genomes had a single cluster with all of predicted testosterone/cholate catabolism genes (Fig. 4B). Exceptions were Sphingomonas wittichii and Pseudoalteromonas haloplanktis, in which A/B ring and C/D ring degradation genes are located in separate clusters.
FIG 4 .
Localization of steroid catabolism genes in genomes. The genes and their functional groupings are as described for Fig. 2 and 3. Each gene is mapped by a translucent dot, so areas of intense color indicate clusters with many genes. (A) Rhodococcus jostii RHA1. (B) Comamonas testosteroni CNB-2.
In the vast majority of cases, steroid degradation genes were chromosomally located. One exception is a cluster of genes putatively encoding only A/B ring degradation located on a large linear plasmid, pRHL1, of R. jostii (27). These genes are most similar to those encoding the cholate pathway, but their function, if any, is unknown (8). Another exception is a gene cluster putatively encoding the testosterone/cholate pathway in two Novosphingobium spp. located on plasmids. Steroid degradation genes were not found on the chromosomes of these two strains.
Phylogeny of steroid degradation genes.
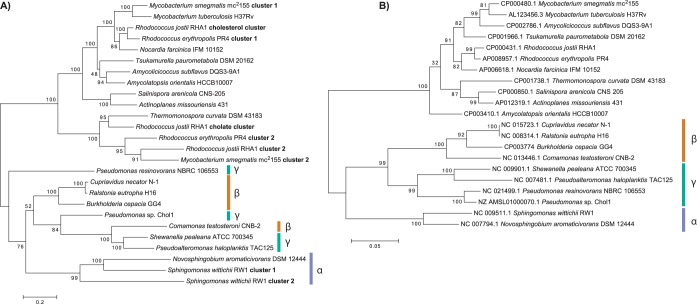
The phylogeny of a subset of four key steroid degradation enzymes, KshA/CtCNB1_1306, HsaA/TesA1, HsaC/TesB, and HsaD/TesD, plus a set of their orthologs was investigated by multilocus sequence analysis. The phylogeny of the proteins reveals two distinct clades for Actinobacteria and Proteobacteria (Fig. 5). The actinobacterial proteins form subclades corresponding to the steroid substrate (cholesterol or cholate). A third subclade includes proteins of unknown function encoded in gene cluster 2 of R. jostii plus proteins from R. erythropolis and M. smegmatis. The phylogeny of the actinobacterial proteins is congruent with that of the corresponding 16S rRNA genes. In contrast, the phylogeny of the proteobacterial proteins is not congruent with that of the corresponding 16S rRNA genes. Further, the proteins form subclades that include proteins from both beta- and gammaproteobacteria. Only the proteins from alphaproteobacteria form a coherent subclade, which includes proteins encoded in two separate gene clusters found in Sphingomonas wittichii.
FIG 5 .
Phylogeny of key steroid degradation enzymes and the corresponding organisms. Bootstrap values are given as percentages of 2,500 repetitions. (A) Phylogeny of orthologs of KshA/CtCNB1_1306, HsaA/TesA1, HsaC/TesB, and HsaD/TesD. The dendrogram is based on concatenated sequences of the four proteins from each steroid catabolism gene cluster found in each bacterium. The scale corresponds to 0.2 substitutions per amino acid. (B) Phylogeny of the 16S rRNA genes of each organism. The scale corresponds to 0.05 substitutions per nucleotide.
Growth on steroids.
To verify our predictions of steroid degradation capacity, we tested nine putative steroid-degrading bacteria, newly identified by the analyses described above, for their ability to grow on or otherwise metabolize cholesterol, cholate, and testosterone. These phylogenetically diverse strains were isolated from a range of environments. Growth was determined as an increase in protein levels attributable to the steroid substrate, and metabolism was confirmed by removal of steroids from the medium or transformation to metabolites. In some cases additional organic substrates were required in the medium, and in some cases tyloxapol or methyl-β-cyclodextrin was required to make steroids bioavailable.
As predicted, all nine strains were able to grow on, or metabolize, at least one of the steroid substrates (Table 2; see also Fig. S4 in the supplemental material). The three nonmarine proteobacterial strains all grew on testosterone, while the marine proteobacterium Shewanella pealeana, with the cluster of genes predicted to encode cholate side chain degradation (Fig. 3), metabolized both testosterone and cholate. None of the Proteobacteria spp. metabolized cholesterol under any conditions tested.
TABLE 2 .
Growth on or metabolism of three steroids by nine predicted steroid degradersa
| Phylum | Strain | Steroid substrate |
||
|---|---|---|---|---|
| Cholesterol | Cholate | Testosterone | ||
| Proteobacteria | Pseudomonas resinovorans NRBC106553 | – | – | G, R |
| Cupriavidus necator ATCC 17699 | – | – | G, R | |
| Sphingomonas wittichii RW1 | – | – | G, R | |
| Shewanella pealeana ATCC 700345 | – | R, a | T, a | |
| Actinobacteria | Thermomonospora curvata ATCC 19995 | – | G, R | T |
| Actinoplanes missouriensis 431 | G, R, t | – | T | |
| Salinispora arenicola CNS-205 | R, a, c/t | – | T | |
| Amycolicicoccus subflavus DQS3-9A1T | R, a, c | – | – | |
| Amycolatopsis sp. strain ATCC 39116 | G, R | R, a | G, R, c | |
Symbols: −, no growth or metabolism; G, growth on steroid as sole organic substrate; T, transformation of steroid to metabolites; R, complete removal of steroid with no detectable metabolite accumulation; t, tyloxapol required; c, methyl-β-cyclodextrin required; c/t, either tyloxapol or cyclodextrin required; a, metabolism in the presence of an additional organic substrate.
Substrate use by the five tested actinobacterial strains was more multifarious, but all strains grew on, or metabolized, either cholesterol or cholate (Table 2; see also Fig. S4 in the supplemental material). As predicted, Actinoplanes missouriensis, Salinispora arenicola, Amycolicicoccus subflavus, and Amycolatopsis sp. all grew on or otherwise metabolized cholesterol. Amycolatopsis sp. additionally grew on testosterone and metabolized cholate. As expected, Thermomonospora curvata grew on cholate but not on cholesterol or testosterone.
DISCUSSION
Novel steroid degraders.
This study characterized the occurrence of aerobic steroid degradation pathways among more than 8,000 microbes with high-quality genome sequences. We found such pathways only in members of the Actinobacteria and Proteobacteria, while they do not appear to exist in Archaea, fungi, or other bacterial phyla. This taxonomic distribution is consistent with previous studies reporting the enrichment and isolation of microbial steroid degraders (5, 6) and the phylogeny of the known steroid-degrading bacteria. However, within these two phyla, the results of this study substantially expand the range of taxa known to be capable of, or predicted to be capable of, steroid degradation. We provide the first evidence for this capacity in members of the genera Actinoplanes, Aeromicrobium, Amycolatopsis, Amycolicicoccus, Burkholderia, Cupriavidus, Glaciecola, Hydrocarboniphaga, Marinobacterium, Nocardia, Nocardioides, Ralstonia, Saccharomonospora, Salinispora, Shewanella, Streptomyces, and Thermomonospora. Our growth experiments confirmed predictions of steroid metabolism by Actinoplanes, Cupriavidus, Salinispora, Shewanella, Thermomonospora, and Amycolicicoccus (Table 2; see also Fig. S4 in the supplemental material). As discussed below, our growth experiments also generally confirmed predictions of substrate specificities of the pathways.
A caveat of this analysis is that it cannot identify steroid degradation pathways that are nonhomologous to, or extremely divergent from, the reference pathways. Furthermore, microbial taxa are not equally represented by RefSeq genome sequences, so the probability of identifying steroid degradation pathways in poorly represented taxa was lower. Since genome sequences were not available for the denitrifying Proteobacteria spp. mentioned above that anaerobically degrade cholesterol and testosterone, we could not include them in our analysis. Thus, additional pathways may remain to be discovered.
Distribution of pathways among taxa.
The cholesterol pathway genes are part of a core genome shared by members of most genera within the suborder Corynebacterineae (Fig. 2; see also Fig. S3 in the supplemental material). Accordingly, some members of Dietzia, Mycobacterium, Gordonia, Rhodococcus, and Tsukamurella were previously shown to degrade cholesterol and use it as a growth substrate (5, 6, 9, 28). The cholesterol pathway was found in additional suborders within the Actinobacteria, but its distribution there is generally unclear due to the limited number of genome sequences representing most of these taxa. The cholesterol pathway occurs but is not conserved in some actinobacterial genera, as it was found in only 1 in 4 Actinoplanes spp., 1 in 6 Saccharomonospora spp., and 1 in over 50 Streptomyces spp.
Additionally, the cholate pathway genes are part of the core genome of the genus Rhodococcus. Interestingly, although Rhodococcus spp. do not appear to be monophyletic (29), the cholate pathway is conserved among Rhodococcus spp. and not among members of closely related genera, such as Nocardia and Gordonia (Fig. 2). Rhodococcus fascians is an exception, as it lacks both the cholesterol and cholate pathways. R. fascians comprises plant pathogens and other plant-associated strains (30), which presumably would not benefit from the ability to catabolize the two animal steroids and therefore have lost the corresponding genes. The cholate pathway genes may help to resolve the complex taxonomy of Rhodococcus and related genera.
In contrast to the distribution of steroid degradation pathways in Actinobacteria, the distribution of the testosterone/cholate pathway among proteobacterial taxa is generally patchy. Thus, in proteobacterial genera represented by multiple genome sequences, we found the testosterone/cholate pathway genes in only one or a few of those genomes (Fig. 3; see also Fig. S3 in the supplemental material). And, among strains of Pseudomonas putida, we found those genes in only a few of many strains. However, exceptions to this trend are the genus Comamonas, in which all four species with sequenced genomes have the pathway, and the SAR86 cluster, in which all three strains with sequenced draft genomes have the pathway. Recently, genotypic analysis of 14 Comamonas testosteroni strains revealed that the testosterone/cholate degradation pathway is part of the core genome of this species (31). Unfortunately, most of these genomes were not available at the time of our analysis.
Specificity of pathways.
Culture-based experiments were largely consistent with the bioinformatic predictions of the abilities of strains to metabolize particular steroids. Thus, the three nonmarine proteobacterial strains tested consistently grew on testosterone, while Shewanella pealeana, with genes encoding cholate side chain degradation, metabolized both testosterone and cholate. The inability of these Proteobacteria spp. to metabolize cholesterol is in agreement with our prediction and previous reports of steroid-degrading Proteobacteria unable to degrade sterols, such as cholesterol, with alkyl side chains (4, 5). In Proteobacteria spp., the inability to degrade cholesterol is consistent with the absence of orthologs of the P450 monooxygenases Cyp125 and Cyp142. These enzymes are used by Actinobacteria to oxidize steroid alkyl side chains to initiate their degradation (15–17, 32, 33). In the Proteobacteria spp. that have been examined, degradation of the steroid side chain is a prerequisite for subsequent steroid nucleus degradation (34, 35). The lack of a transporter, such as the Mce4 system, may also contribute to the inability of Proteobacteria to metabolize cholesterol. Overall, there is currently no evidence suggesting that members of the Proteobacteria can catabolize cholesterol via the 9,10-seco pathway.
As predicted, four actinobacterial strains with cholesterol degradation gene clusters all metabolized cholesterol. Amycolatopsis sp. additionally grew with testosterone, suggesting that the pathway for cholesterol catabolism can also support catabolism of testosterone in some bacteria, an ability that was not previously recognized. Salinispora arenicola and Amycolicicoccus subflavus completely removed cholesterol from their medium but failed to grow. This unexpected result may be related to the fact that they are very slow growing, even on rich LB medium. The ability of Amycolatopsis sp. to metabolize cholate despite its lacking an actinobacterial cholate degradation gene cluster indicates that the actinobacterial cholesterol degradation cluster can also support degradation of cholate for some organisms.
As predicted, Thermomonospora curvata grew on cholate but not on cholesterol. It also metabolized testosterone, indicating that the actinobacterial cholate degradation pathway also has this capacity. This strain offers a rare opportunity to examine the actinobacterial cholate degradation pathway in isolation, verifying that it is sufficient for catabolism of cholate. Most actinobacterial genomes encoding the cholate pathway also encode the cholesterol pathway, and some Rhodococcus spp. have further clusters of steroid degradation genes of unknown function (8, 9). Overall, our culture-based experiments add credibility to the bioinformatic predictions of steroid catabolism by Proteobacteria and Actinobacteria. Further supporting our predictions, Gordonia sp. strain KTR9, predicted to have a cholesterol degradation cluster, was previously shown to grow with cholesterol but not with cholate or testosterone (36).
Evolution of pathways.
The distributions of steroid degradation pathways among taxa suggest a possible scenario for evolution and dissemination of the pathways. This scenario involves evolution of a single ancestral pathway, since the known aerobic pathways are all homologous. The most parsimonious interpretation of our results is that the pathway originated in an ancestor of the Corynebacterineae and gave rise to the cholesterol pathway. This ancestry is consistent with our findings, including the nearly ubiquitous occurrence of the pathway in most genera within this suborder. However, this ancestry is speculative, and even the possibility of an origin in the Proteobacteria cannot be excluded. A more comprehensive phylogenetic analysis, and perhaps more genome sequences, would be required to better establish ancestry.
The distribution of the actinobacterial cholate pathway suggests that it originated via a duplication of the cholesterol pathway genes in an ancestor of Rhodococcus. The presence of more than two clusters of homologous steroid degradation genes in several Rhodococcus spp. suggests multiple duplications of these genes. Genes encoding steroid A/B ring degradation are found in gene clusters for both the cholesterol and cholate pathways, while, notably, the cluster of genes encoding steroid C/D ring degradation did not duplicate and is found linked only to the cholesterol pathway genes. The occurrence of the cholesterol or cholate pathway in Actinobacteria beyond the Corynebacterineae could be due to either vertical or horizontal transmission, as the limited availability of genome sequences representing these taxa does not strongly support either possibility. The patchy distribution of the monophyletic testosterone/cholate pathway among Proteobacteria spp. suggests a single horizontal transfer of an actinobacterial steroid pathway to a proteobacterium, followed by horizontal distribution among Proteobacteria. The concept of horizontal transfer among Proteobacteria is further supported by the phylogeny of four key proteins in the three reference pathways, which is not congruent with the phylogeny of the corresponding 16S rRNA genes (Fig. 5). The observed clustering of steroid degradation genes within genomes (Fig. 4) and the location of some clusters on plasmids both likely facilitated horizontal transfer of entire pathways or major components of pathways. In particular, a gene cluster with greatest similarity to that of the actinobacterial cholate pathway genes located on plasmid pRHL1 in R. jostii may have facilitated horizontal transfer of that pathway, although the function of those genes in R. jostii is unclear. It is noteworthy that such large, linear, single-copy plasmids may exist unrecognized in draft or incompletely assembled genomes. The location of testosterone/cholate pathway genes on plasmids in Novosphingobium spp. is consistent with horizontal transfer of the pathway to this genus. While the evolutionary scenario described above is most parsimonious with respect to the available evidence, it remains speculative.
Ecology.
The majority of putatively steroid-degrading bacteria that we identified were isolated from soil, host, and aquatic environments (see Fig. S2 in the supplemental material). Our analysis particularly extends knowledge of steroid-degrading marine bacteria, including members of Glaciecola, Marinobacterium, Pseudoalteromonas, Shewanella, OM60, and SAR86, which originated from marine environments (37–42). Although some Vibrio species within the gammaproteobacteria, without sequenced genomes, have been characterized as marine steroid degraders (43, 44), we did not find steroid degradation genes in 186 genomes of marine Vibrio spp. that we analyzed, which is consistent with the patchy distribution of steroid degradation pathways among proteobacterial taxa.
In soil and aquatic environments, steroids constitute a significant resource for heterotrophic bacteria, and steroid degraders function in decomposition of eukaryotic biomass and excreta from vertebrates. Accordingly, many taxa identified in this study are associated with biomass decomposition. Notably, the R. jostii type strain was isolated from the sarcophagus of a medieval knight (45), so it is tempting to speculate that R. jostii participated in degrading the corpse and then survived centuries of dormancy. The strong conservation of the genes encoding steroid degradation pathways in core genomes of actinobacterial taxa indicates that this catabolism is fundamental to their life history. Thus, cholesterol catabolism appears important to niches of members of several actinobacterial genera, including most in the Corynebacterineae, while cholate catabolism additionally appears to be important to the niches of most members of Rhodococcus. In contrast, the patchy distribution of the testosterone/cholate pathway among members of Proteobacteria indicates that individual, distantly related proteobacterial species or strains have adopted niches involving steroid catabolism.
In addition to free-living species, a substantial proportion of the cholesterol-degrading Actinobacteria that we identified, including members of Mycobacterium, Rhodococcus, and Nocardia, are pathogens of mammals (46–48). These pathogenic species tend to have reduced genomes, but within these genera, only the obligately intracellular pathogen Mycobacterium leprae has lost the steroid degradation genes. Recent studies showed that M. leprae has indeed lost the ability to metabolize cholesterol as a carbon and energy source (49) but still requires host cholesterol for intracellular survival (50). Conservation of the pathway in all other pathogenic species in those genera suggests that cholesterol catabolism is important to their niches. Indeed, several lines of evidence indicate that catabolism of host cholesterol is essential for M. tuberculosis survival in vivo (9, 13, 51) and for pathogenesis of R. equi, which infects foals (52).
Further delineation of steroid degradation genes.
Our comparisons of a large number of steroid catabolism gene clusters shed new light on the involvement of particular genes in the respective pathways. Uncharacterized genes ro04680 and ro04694 from the R. jostii cholesterol pathway gene cluster are not conserved in other putative cholesterol degraders and so are unlikely to be associated with the pathway. The mce4 genes, encoding a cholesterol/sitosterol uptake system (18), are conserved only in mycolic acid bacteria within the Corynebacterineae. The only complete genome from a mycolic acid bacterium with the cholesterol pathway but lacking the mce4 genes is that of Tsukamurella paurometabola. The Mce4 uptake system may be specifically adapted for transport across the distinct cell envelope of mycolic acid bacteria, which includes a true outer membrane (53). Steroid uptake systems in other taxa or systems for other steroid substrates have not been identified.
The restricted distribution of the actinobacterial cholate pathway limits the comparison of genomes to verify genes associated with the pathway, but some conclusions are possible. The lack of conservation of ro05807 and ro05809 supports the previous conclusion, based on transcriptomic analysis, that these genes are not associated with the pathway (8). Similarly, camM is not conserved, in accordance with the previous study showing that CamM is a transporter functioning in reassimilation of a cholate metabolite by R. jostii which is not essential for growth on cholate (54). It also appears that the casH gene may not be essential to the pathway.
A single large cluster of genes in the genomes of C. testosteroni strains TA441 and CNB-2 encodes the proteobacterial testosterone/cholate pathway (see Fig. S1 in the supplemental material). Our analysis suggests that a subset of 26 genes in this cluster (C. testosteroni CNB-2 CtCNB1_1293 through CtCNB1_1320) is not associated with the pathway. An exception within that subset is CtCNB1_1306, which is conserved in most genomes encoding the pathway but not in genomes of related organisms that do not encode the pathway. The product of this gene is associated via BLASTp clustering with actinobacterial KshA, the large subunit of 3-ketosteroid-9α-hydroxylase (KSH), which cleaves steroid B ring (55). The putative role of this gene in C. testosteroni and other Proteobacteria was not previously recognized. However, the ORF17 gene in strain CNB-2 was predicted to encode KshB, the small subunit of KSH (4), a conclusion which is further supported by our analysis.
We further predict that genes CtCNB1_1308, CtCNB1_1309, CtCNB1_1310, ORF6, ORF7, CtCNB1_1347, and CtCNB1_1348 in strain CNB-2 are involved in cholate degradation but not in testosterone degradation, as they are conserved in Proteobacteria predicted to degrade both substrates but not in those predicted to degrade only testosterone. Accordingly, CtCNB1_1309, CtCNB1_1347, and CtCNB1_1348 are homologs of the cholate side chain degradation genes sad, sal, and shy, respectively, which were identified in Pseudomonas sp. strain Chol1 (11). Similarly, ORF6 and ORF7 are homologs of the genes sor1 and hsh1, respectively, which were identified in strain Chol1 and are required for the degradation of steroids with a hydroxyl group at C12 (56). Our analysis suggests that these genes are not associated with testosterone degradation but only with cholate degradation. Nevertheless, Horinouchi et al. have shown that transcription of ORF6 is induced during growth with both substrates (35). In addition, our analysis supports the previous prediction that CtCNB1_1330 to CtCNB1_1340 encode the remaining steps of cholate side chain degradation (4, 57), because these genes are conserved only in organisms predicted to degrade cholate. Furthermore, CtCNB1_1308 encodes a putative MFS transporter, which could be involved in uptake of cholate or one of its degradation intermediates. Among the cluster of genes in strain CNB-2 predicted to encode C/D ring degradation (4), our analysis supports the previously published notion (31) that the ORF25 and ORF26 genes are not associated with the testosterone/cholate pathway.
Finally, homologs of genes hsaE and tesE, genes hsaF and tesG, and genes hsaG and tesF are not conserved in many of the steroid catabolism gene clusters. These genes encode a series of reactions that are common to many catabolic pathways that involve meta-cleavage of aromatic rings (58, 59). Thus, it appears that many steroid degraders have genes elsewhere in their genomes encoding enzymes catalyzing one or more of these three reactions. Accordingly, hsaE, hsaF, and hsaG from the cholesterol degradation gene cluster of M. tuberculosis strain H37Rv have been shown to be nonessential for growth on cholesterol (60).
MATERIALS AND METHODS
Bioinformatic software environments and packages as well as growth experiments used in this study are described in the supplemental Methods (see Text S1 in the supplemental material).
Steroid catabolism reference proteins.
Predicted proteins encoded by previously characterized steroid catabolism genes from three bacterial strains served as initial reference proteins. These strains were Rhodococcus jostii RHA1 (RefSeq NC_008268.1), Mycobacterium tuberculosis H37Rv (RefSeq NC_000962.3), and Comamonas testosteroni CNB-2 (RefSeq NC_013446.2). Strain CNB-2 was the only C. testosteroni strain with a complete genome in NCBI’s GenBank at the time of this investigation, and it was selected due to the fact that the sequenced and characterized steroid degradation genes of C. testosteroni TA441 were mapped to the strain CNB-2 genome in a recent publication (4).
The initial reference proteins were binned into homologous groups using BLASTp (61) (v2.2.29; http://blast.ncbi.nlm.nih.gov/Blast.cgi?PAGE=Proteins), applying a minimum identity filter of 30% and a maximum E value filter of 10−30. The latter two parameters were selected empirically in order to yield clear clusters within the network, which shared common GenBank annotations. Additional reference genes were obtained from the genomes of organisms related to those three strains. On 6 February 2014, 105 genomes (see Table S3 in the supplemental material) were downloaded from NCBI’s GenBank, comprising all draft and complete genomes from Rhodococcus, Mycobacterium, and Comamonas spp. plus a subset from Streptomyces spp. A custom program, BackBLAST (v1.0, Lee Bergstrand; https://github.com/LeeBergstrand/BackBLAST_Reciprocal_BLAST), was used to search the predicted proteins of these genomes for putative orthologs (reciprocal BLASTp hits) of the initial reference proteins. A maximum E value of 10−30 and a minimum identity of 25% were used to maximize stringency while not removing annotated orthologs of the reference proteins, and all other BLASTp settings were default values.
Generation of HMMs.
Eight proteins were selected for HMM development (Table 1), because (i) they unambiguously clustered in orthologous groups, (ii) they occur in all three reference pathways, and (iii) they have known functions in steroid nucleus degradation. Each of these proteins from R. jostii, M. tuberculosis, and C. testosteroni plus their reciprocal BLASTp hits were subclustered with CD-hit (62) (v4.6.1; https://github.com/weizhongli/cdhit), using a minimum sequence identity of 50%, a word size value of 3, and all other parameters left at default, yielding between 1 and 5 smaller subclusters per input protein. Typically, these subclusters represented proteins of similar taxonomic origins and/or substrate specificities. As an additional filtering step, potentially nonorthologous proteins that did not cluster with the initial eight proteins were removed. Sequences from each of the resulting 25 subclusters were aligned and manually trimmed using Mega (63) (v5.2.2; http://www.megasoftware.net/). The sets of aligned protein sequences were used to generate 25 hidden Markov model (HMMs) using HMMER (64) (v3.1b1; http://hmmer.janelia.org). The HMMs developed are available online (https://github.com/MohnLab/Mohn_Lab_Steroid_Degradation_HMM_Analysis_2015).
HMM searches.
On 15 May 2014, all complete bacterial and archaeal genomes, plus all complete and draft fungal genomes, were downloaded from NCBI’s curated RefSeq database (2,788 genomes). On 30 July 2014, all incomplete bacterial and archaeal genomes were downloaded from RefSeq (5,489 genomes). Annotated proteins from these genomes were searched with a custom program, HMMER-DB (v1.0, Lee Bergstrand; https://github.com/LeeBergstrand/HMMER-DB), which stores HMM hits generated by HMMER’s hmmsearch in a searchable database. A maximum HMMER E value was empirically optimized to 10−25, which identified previously known steroid catabolism genes while providing maximum stringency against false positives. All proteins of organisms identified as potential steroid degraders by HMM searches were subsequently searched for best reciprocal BLAST hits to initial reference proteins via BackBLAST, filtering for a minimum identity of 25% and maximum E value of 10−25 in accordance with previous BLAST and HMM criteria. Additionally, 24 Rhodococcus fascians genomes were downloaded from GenBank on 12 August 2015 and searched for best reciprocal BLAST hits.
Phylogenetic analysis.
The protein sequences for KshA/CtCNB1_1306, HsaA/TesA1, HsaC/TesB, and HsaD/TesD from the reference strains and sequences of their orthologs from 18 additional strains, identified by HMM analysis, were used for phylogenetic analysis. Homologous sequences were aligned using the Muscle algorithm (65) from Mega v6.06 and manually trimmed. The resulting four sequences inferred from each gene cluster were concatenated. Phylogenetic reconstruction was performed with the concatenated protein sequences as well as the 16S rRNA gene sequences of the corresponding organisms, using the maximum likelihood model with default parameters and 2,500 bootstrap replications in Mega v6.06.
SUPPLEMENTAL MATERIAL
Steroid catabolism gene clusters of Rhodococcus jostii RHA1, Mycobacterium tuberculosis H37Rv, Comamonas testosteroni CNB-2, and Pseudomonas sp. strain Chol1. Filled arrows indicate characterized genes and proteins, open arrows indicate annotated genes, and gray arrows indicate genes and proteins probably not involved in steroid degradation. Gene names correspond to protein names in Fig. 1 or gene locus tags, which have been abbreviated to the last digits for strains CNB-2 and Chol1. Download
Sources of isolates whose genomes were found to have steroid catabolism genes via HMM analysis. Download
Heat map showing BLAST identity for best reciprocal BLASTp hits to Rhodococcus jostii RHA1 and Comamonas testosteroni CNB-2 steroid degradation proteins in 75 bacterial draft genomes. Download
Growth on or metabolism of three steroid substrates by nine predicted steroid degraders. (A) Growth of six strains, measured as protein production, on cholate or cholesterol and removal of the steroid substrates. For A. missouriensis 431, differential protein yield was calculated by subtracting the protein yield in control cultures without a steroid substrate. (B) Removal of cholesterol or cholate in the presence of additional organic substrates by four strains. The results from two representative replicate cultures (A and B) and a noninoculated control (ni) are shown. Download
Table listing 265 putatively steroid-degrading bacteria identified by HMM analysis.
Table listing 124 complete and 75 draft genomes of putatively steroid-degrading bacteria that were analyzed by reciprocal BLASTp analysis.
Table listing 105 genomes from GenBank of Rhodococcus, Mycobacterium, Comamonas, and Streptomyces spp. that were sources of reference genes.
Materials and methods used for bioinformatic analysis and growth experiments. Download
ACKNOWLEDGMENTS
We thank Gordon R. Stewart for technical assistance in culturing bacteria and chemical analyses and Paul Jensen from the Scripps Institution of Oceanography (University of California, San Diego, San Diego, CA) for providing Salinispora arenicola CNS-205. L.H.B. carried out most of the bioinformatic analyses and codrafted the manuscript. E.C. provided direction and advice in bioinformatic analyses. J.H. performed the growth experiments and analysis of gene functions and codrafted the manuscript. J.D.V. edited the manuscript. W.W.M. conceived of the study, supervised the work, and codrafted the manuscript. We all read and approved the final manuscript.
This study was funded by an NSERC Discovery Grant and Accelerator Supplement to W.W.M. E.C. was supported by a fellowship from the Tula Foundation. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Footnotes
Citation Bergstrand LH, Cardenas E, Holert J, Van Hamme JD, Mohn WW. 2016. Delineation of steroid-degrading microorganisms through comparative genomic analysis. mBio 7(2):e00166-16. doi:10.1128/mBio.00166-16.
REFERENCES
- 1.Dufourc EJ. 2008. Sterols and membrane dynamics. J Chem Biol 1:63–77. doi: 10.1007/s12154-008-0010-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nesbitt NM, Yang X, Fontán P, Kolesnikova I, Smith I, Sampson NS, Dubnau E. 2010. A thiolase of Mycobacterium tuberculosis is required for virulence and production of androstenedione and androstadienedione from cholesterol. Infect Immun 78:275–282. doi: 10.1128/IAI.00893-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Donova MV, Egorova OV. 2012. Microbial steroid transformations: current state and prospects. Appl Microbiol Biotechnol 94:1423–1447. doi: 10.1007/s00253-012-4078-0. [DOI] [PubMed] [Google Scholar]
- 4.Horinouchi M, Hayashi T, Kudo T. 2012. Steroid degradation in Comamonas testosteroni. J Steroid Biochem Mol Biol 129:4–14. doi: 10.1016/j.jsbmb.2010.10.008. [DOI] [PubMed] [Google Scholar]
- 5.Merino E, Barrientos A, Rodríguez J, Naharro G, Luengo JM, Olivera ER. 2013. Isolation of cholesterol- and deoxycholate-degrading bacteria from soil samples: evidence of a common pathway. Appl Microbiol Biotechnol 97:891–904. doi: 10.1007/s00253-012-3966-7. [DOI] [PubMed] [Google Scholar]
- 6.Holert J, Yücel O, Suvekbala V, Kulić Z, Möller H, Philipp B. 2014. Evidence of distinct pathways for bacterial degradation of the steroid compound cholate suggests the potential for metabolic interactions by interspecies cross-feeding. Environ Microbiol 16:1424–1440. doi: 10.1111/1462-2920.12407. [DOI] [PubMed] [Google Scholar]
- 7.Fernández de las Heras L, García Fernández E, María Navarro Llorens J, Perera J, Drzyzga O. 2009. Morphological, physiological, and molecular characterization of a newly isolated steroid-degrading actinomycete, identified as Rhodococcus ruber strain Chol-4. Curr Microbiol 59:548–553. doi: 10.1007/s00284-009-9474-z. [DOI] [PubMed] [Google Scholar]
- 8.Mohn WW, Wilbrink MH, Casabon I, Stewart GR, Liu J, van der Geize R, Eltis LD. 2012. Gene cluster encoding cholate catabolism in Rhodococcus spp. J Bacteriol 194:6712–6719. doi: 10.1128/JB.01169-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Van der Geize R, Yam K, Heuser T, Wilbrink MH, Hara H, Anderton MC, Sim E, Dijkhuizen L, Davies JE, Mohn WW, Eltis LD. 2007. A gene cluster encoding cholesterol catabolism in a soil actinomycete provides insight into Mycobacterium tuberculosis survival in macrophages. Proc Natl Acad Sci U S A 104:1947–1952. doi: 10.1073/pnas.0605728104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li W, Ge F, Zhang Q, Ren Y, Yuan J, He J, Li W, Chen G, Zhang G, Zhuang Y, Xu L. 2014. Identification of gene expression profiles in the actinomycete Gordonia neofelifaecis grown with different steroids. Genome 57:345–353. doi: 10.1139/gen-2014-0030. [DOI] [PubMed] [Google Scholar]
- 11.Holert J, Jagmann N, Philipp B. 2013. The essential function of genes for a hydratase and an aldehyde dehydrogenase for growth of Pseudomonas sp. strain Chol1 with the steroid compound cholate indicates an aldolytic reaction step for deacetylation of the side chain. J Bacteriol 195:3371–3380. doi: 10.1128/JB.00410-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Griffin JE, Gawronski JD, Dejesus MA, Ioerger TR, Akerley BJ, Sassetti CM. 2011. High-resolution phenotypic profiling defines genes essential for mycobacterial growth and cholesterol catabolism. PLoS Pathog 7:e1002251. doi: 10.1371/journal.ppat.1002251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wipperman MF, Sampson NS, Thomas ST. 2014. Pathogen roid rage: cholesterol utilization by Mycobacterium tuberculosis. Crit Rev Biochem Mol Biol 49:269–293. doi: 10.3109/10409238.2014.895700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thomas ST, VanderVen BC, Sherman DR, Russell DG, Sampson NS. 2011. Pathway profiling in Mycobacterium tuberculosis: elucidation of cholesterol-derived catabolite and enzymes that catalyze its metabolism. J Biol Chem 286:43668–43678. doi: 10.1074/jbc.M111.313643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Capyk JK, Kalscheuer R, Stewart GR, Liu J, Kwon H, Zhao R, Okamoto S, Jacobs WR, Eltis LD, Mohn WW. 2009. Mycobacterial cytochrome P450 125 (Cyp125) catalyzes the terminal hydroxylation of C27 steroids. J Biol Chem 284:35534–35542. doi: 10.1074/jbc.M109.072132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rosłoniec KZ, Wilbrink MH, Capyk JK, Mohn WW, Ostendorf M, van der Geize R, Dijkhuizen L, Eltis LD. 2009. Cytochrome P450 125 (CYP125) catalyses C26-hydroxylation to initiate sterol side-chain degradation in Rhodococcus jostii RHA1. Mol Microbiol 74:1031–1043. doi: 10.1111/j.1365-2958.2009.06915.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.García Fernández E, Frank DJ, Galán B, Kells PM, Podust LM, García JL, Ortiz de Montellano PR. 2013. A highly conserved mycobacterial cholesterol catabolic pathway. Environ Microbiol 15:2342–2359. doi: 10.1111/1462-2920.12108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mohn WW, van der Geize R, Stewart GR, Okamoto S, Liu J, Dijkhuizen L, Eltis LD. 2008. The actinobacterial Mce4 locus encodes a steroid transporter. J Biol Chem 283:35368–35374. doi: 10.1074/jbc.M805496200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.García Fernández EN, Galán B, Medrano FJ, García JL. 2015. Characterization of the KstR2 regulator responsible of the lower cholesterol degradative pathway in Mycobacterium smegmatis. Environ Microbiol Rep 7:155–163. doi: 10.1111/1758-2229.12255. [DOI] [PubMed] [Google Scholar]
- 20.Casabon I, Zhu S-H, Otani H, Liu J, Mohn WW, Eltis LD. 2013. Regulation of the KstR2 regulon of Mycobacterium tuberculosis by a cholesterol catabolite. Mol Microbiol 89:1201–1212. doi: 10.1111/mmi.12340. [DOI] [PubMed] [Google Scholar]
- 21.Kendall SL, Burgess P, Balhana R, Withers M, Bokum Ten A, Lott JS, Gao C, Uhia-Castro I, Stoker NG. 2010. Cholesterol utilization in mycobacteria is controlled by two TetR-type transcriptional regulators: kstR and kstR2. Microbiology 156:1362–1371. doi: 10.1099/mic.0.034538-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harder J, Probian C. 1997. Anaerobic mineralization of cholesterol by a novel type of denitrifying bacterium. Arch Microbiol 167:269–274. doi: 10.1007/s002030050442. [DOI] [PubMed] [Google Scholar]
- 23.Tarlera S, Denner EB. 2003. Sterolibacterium denitrificans gen. nov., sp. nov., a novel cholesterol-oxidizing, denitrifying member of the beta-proteobacteria. Int J Syst Evol Microbiol 53:1085–1091. doi: 10.1099/ijs.0.02039-0. [DOI] [PubMed] [Google Scholar]
- 24.Fahrbach M, Kuever J, Remesch M, Huber BE, Kämpfer P, Dott W, Hollender J. 2008. Steroidobacter denitrificans gen. nov., sp. nov., a steroidal hormone-degrading gammaproteobacterium. Int J Syst Evol Microbiol 58:2215–2223. doi: 10.1099/ijs.0.65342-0. [DOI] [PubMed] [Google Scholar]
- 25.Wang P-H, Leu Y-L, Ismail W, Tang S-L, Tsai C-Y, Chen H-J, Kao A-T, Chiang Y-R. 2013. Anaerobic and aerobic cleavage of the steroid core ring structure by Steroidobacter denitrificans. J Lipid Res 54:1493–1504. doi: 10.1194/jlr.M034223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang P-H, Yu C-P, Lee T-H, Lin C-W, Ismail W, Wey S-P, Kuo A-T, Chiang Y-R. 2014. Anoxic androgen degradation by the denitrifying bacterium Sterolibacterium denitrificans via the 2,3-seco pathway. Appl Environ Microbiol 80:3442–3452. doi: 10.1128/AEM.03880-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McLeod MP, Warren RL, Hsiao WW, Araki N, Myhre M, Fernandes C, Miyazawa D, Wong W, Lillquist AL, Wang D, Dosanjh M, Hara H, Petrescu A, Morin RD, Yang G, Stott JM, Schein JE, Shin H, Smailus D, Siddiqui AS, Marra MA, Jones SJM, Holt R, Brinkman FSL, Miyauchi K, Fukuda M, Davies JE, Mohn WW, Eltis LD. 2006. The complete genome of Rhodococcus sp. RHA1 provides insights into a catabolic powerhouse. Proc Natl Acad Sci U S A 103:15582–15587. doi: 10.1073/pnas.0607048103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Drzyzga O, Fernández de las Heras L, Morales V, Navarro Llorens JM, Perera J. 2011. Cholesterol degradation by Gordonia cholesterolivorans. Appl Environ Microbiol 77:4802–4810. doi: 10.1128/AEM.05149-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gürtler V, Seviour RJ. 2010. Systematics of members of the genus Rhodococcus (Zopf 1891) emend. Goodfellow et al. 1998. In Alvarez HM (ed), Biology of Rhodococcus. Springer, New York, NY. doi: 10.1007/978-3-642-12937-7. [DOI] [Google Scholar]
- 30.Creason AL, Davis EW, Putnam ML, Vandeputte OM, Chang JH. 2014. Use of whole genome sequences to develop a molecular phylogenetic framework for Rhodococcus fascians and the Rhodococcus genus. Front Plant Sci 5:406. doi: 10.3389/fpls.2014.00406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu L, Zhu W, Cao Z, Xu B, Wang G, Luo M. 2015. High correlation between genotypes and phenotypes of environmental bacteria Comamonas testosteroni strains. BMC Genomics 16:110. doi: 10.1186/s12864-015-1314-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Johnston JB, Ouellet H, Ortiz de Montellano PR. 2010. Functional redundancy of steroid C26-monooxygenase activity in Mycobacterium tuberculosis revealed by biochemical and genetic analyses. J Biol Chem 285:36352–36360. doi: 10.1074/jbc.M110.161117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McLean KJ, Lafite P, Levy C, Cheesman MR, Mast N, Pikuleva IA, Leys D, Munro AW. 2009. The structure of Mycobacterium tuberculosis CYP125: molecular basis for cholesterol binding in a P450 needed for host infection. J Biol Chem 284:35524–35533. doi: 10.1074/jbc.M109.032706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Holert J, Kulić Ž, Yücel O, Suvekbala V, Suter MJ, Möller HM, Philipp B. 2013. Degradation of the acyl side chain of the steroid compound cholate in Pseudomonas sp. strain Chol1 proceeds via an aldehyde intermediate. J Bacteriol 195:585–595. doi: 10.1128/JB.01961-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Horinouchi M, Hayashi T, Koshino H, Malon M, Yamamoto T, Kudo T. 2008. Identification of genes involved in inversion of stereochemistry of a C-12 hydroxyl group in the catabolism of cholic acid by Comamonas testosteroni TA441. J Bacteriol 190:5545–5554. doi: 10.1128/JB.01080-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen H-P, Zhu S-H, Casabon I, Hallam SJ, Crocker FH, Mohn WW, Indest KJ, Eltis LD. 2012. Genomic and transcriptomic studies of an RDX (hexahydro-1,3,5-trinitro-1,3,5-triazine)-degrading actinobacterium. Appl Environ Microbiol 78:7798–7800. doi: 10.1128/AEM.02120-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Qin Q-L, Xie B-B, Yu Y, Shu Y-L, Rong J-C, Zhang Y-J, Zhao D-L, Chen X-L, Zhang X-Y, Chen B, Zhou B-C, Zhang Y-Z. 2014. Comparative genomics of the marine bacterial genus Glaciecola reveals the high degree of genomic diversity and genomic characteristic for cold adaptation. Environ Microbiol 16:1642–1653. doi: 10.1111/1462-2920.12318. [DOI] [PubMed] [Google Scholar]
- 38.Choi Y-U, Kwon Y-K, Ye B-R, Hyun J-H, Heo S-J, Affan A, Yoon K-T, Park H-S, Oh C, Kang D-H. 2012. Draft genome sequence of Marinobacterium stanieri S30, a strain isolated from a coastal lagoon in Chuuk state in Micronesia. J Bacteriol 194:1260. doi: 10.1128/JB.06703-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Médigue C, Krin E, Pascal G, Barbe V, Bernsel A, Bertin PN, Cheung F, Cruveiller S, D’Amico S, Duilio A, Fang G, Feller G, Ho C, Mangenot S, Marino G, Nilsson J, Parrilli E, Rocha EP, Rouy Z, Sekowska A, Tutino ML, Vallenet D, Heijne von G, Danchin A. 2005. Coping with cold: the genome of the versatile marine Antarctica bacterium Pseudoalteromonas haloplanktis TAC125. Genome Res 15:1325–1335. doi: 10.1101/gr.4126905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fredrickson JK, Romine MF, Beliaev AS, Auchtung JM, Driscoll ME, Gardner TS, Nealson KH, Osterman AL, Pinchuk G, Reed JL, Rodionov DA, Rodrigues JL, Saffarini DA, Serres MH, Spormann AM, Zhulin IB, Tiedje JM. 2008. Towards environmental systems biology of Shewanella. Nat Rev Microbiol 6:592–603. doi: 10.1038/nrmicro1947. [DOI] [PubMed] [Google Scholar]
- 41.Yan S, Fuchs BM, Lenk S, Harder J, Wulf J, Jiao N-Z, Amann R. 2009. Biogeography and phylogeny of the NOR5/OM60 clade of Gammaproteobacteria. Syst Appl Microbiol 32:124–139. doi: 10.1016/j.syapm.2008.12.001. [DOI] [PubMed] [Google Scholar]
- 42.Dupont CL, Rusch DB, Yooseph S, Lombardo M-J, Richter RA, Valas R, Novotny M, Yee-Greenbaum J, Selengut JD, Haft DH, Halpern AL, Lasken RS, Nealson K, Friedman R, Venter JC. 2012. Genomic insights to SAR86, an abundant and uncultivated marine bacterial lineage. ISME J 6:1186–1199. doi: 10.1038/ismej.2011.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sang Y, Xiong G, Maser E. 2012. Identification of a new steroid degrading bacterial strain H5 from the Baltic Sea and isolation of two estradiol inducible genes. J Steroid Biochem Mol Biol 129:22–30. doi: 10.1016/j.jsbmb.2011.01.018. [DOI] [PubMed] [Google Scholar]
- 44.Zhang T, Xiong G, Maser E. 2011. Characterization of the steroid degrading bacterium S19-1 from the Baltic Sea at Kiel, Germany. Chem Biol Interact 191:83–88. doi: 10.1016/j.cbi.2010.12.021. [DOI] [PubMed] [Google Scholar]
- 45.Takeuchi M, Hatano K, Sedlácek I, Pácová Z. 2002. Rhodococcus jostii sp. nov., isolated from a medieval grave. Int J Syst Evol Microbiol 52:409–413. doi: 10.1099/00207713-52-2-409. [DOI] [PubMed] [Google Scholar]
- 46.Rue-Albrecht K, Magee DA, Killick KE, Nalpas NC, Gordon SV, MacHugh DE. 2014. Comparative functional genomics and the bovine macrophage response to strains of the Mycobacterium genus. Front Immunol 5:536. doi: 10.3389/fimmu.2014.00536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Howard ST. 2013. Recent progress towards understanding genetic variation in the Mycobacterium abscessus complex. Tuberculosis 93(Suppl):S15–S20. doi: 10.1016/S1472-9792(13)70005-2. [DOI] [PubMed] [Google Scholar]
- 48.Weinstock DM, Brown AE. 2002. Rhodococcus equi: an emerging pathogen. Clin Infect Dis 34:1379–1385. doi: 10.1086/340259. [DOI] [PubMed] [Google Scholar]
- 49.Marques MA, Berrêdo-Pinho M, Rosa TL, Pujari V, Lemes RM, Lery LM, Silva CA, Guimarães AC, Atella GC, Wheat WH, Brennan PJ, Crick DC, Belisle JT, Pessolani MC. 2015. The essential role of cholesterol metabolism in the intracellular survival of Mycobacterium leprae is not coupled to central carbon metabolism and energy production. J Bacteriol 197:3698–3707. doi: 10.1128/JB.00625-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mattos KA, Oliveira VC, Berrêdo-Pinho M, Amaral JJ, Antunes LC, Melo RC, Acosta CC, Moura DF, Olmo R, Han J, Rosa PS, Almeida PE, Finlay BB, Borchers CH, Sarno EN, Bozza PT, Atella GC, Pessolani MC. 2014. Mycobacterium leprae intracellular survival relies on cholesterol accumulation in infected macrophages: a potential target for new drugs for leprosy treatment. Cell Microbiol 16:797–815. doi: 10.1111/cmi.12279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.VanderVen BC, Fahey RJ, Lee W, Liu Y, Abramovitch RB, Memmott C, Crowe AM, Eltis LD, Perola E, Deininger DD, Wang T, Locher CP, Russell DG. 2015. Novel inhibitors of cholesterol degradation in Mycobacterium tuberculosis reveal how the bacterium’s metabolism is constrained by the intracellular environment. PLoS Pathog 11:e1004679. doi: 10.1371/journal.ppat.1004679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Van der Geize R, Grommen AW, Hessels GI, Jacobs AA, Dijkhuizen L. 2011. The steroid catabolic pathway of the intracellular pathogen Rhodococcus equi is important for pathogenesis and a target for vaccine development. PLoS Pathog 7:e1002181. doi: 10.1371/journal.ppat.1002181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Barry CE, Lee RE, Mdluli K, Sampson AE, Schroeder BG, Slayden RA, Yuan Y. 1998. Mycolic acids: structure, biosynthesis and physiological functions. Prog Lipid Res 37:143–179. doi: 10.1016/S0163-7827(98)00008-3. [DOI] [PubMed] [Google Scholar]
- 54.Swain K, Casabon I, Eltis LD, Mohn WW. 2012. Two transporters essential for reassimilation of novel cholate metabolites by Rhodococcus jostii RHA1. J Bacteriol 194:6720–6727. doi: 10.1128/JB.01167-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Petrusma M, van der Geize R, Dijkhuizen L. 2014. 3-Ketosteroid 9α-hydroxylase enzymes: Rieske non-heme monooxygenases essential for bacterial steroid degradation. Antonie Van Leeuwenhoek 106:157–172. doi: 10.1007/s10482-014-0188-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Holert J, Yücel O, Jagmann N, Prestel A, Möller HM, Philipp B. 2015. Identification of bypass reactions leading to the formation of one central steroid degradation intermediate in metabolism of different bile salts in Pseudomonas sp. strain Chol1. Environ Microbiol. doi: 10.1111/1462-2920.13192. [DOI] [PubMed] [Google Scholar]
- 57.Horinouchi M, Hayashi T, Koshino H, Malon M, Hirota H, Kudo T. 2014. Identification of 9α-hydroxy-17-oxo-1,2,3,4,10,19-hexanorandrostan-5-oic acid in steroid degradation by Comamonas testosteroni TA441 and its conversion to the corresponding 6-en-5-oyl coenzyme A (CoA) involving open reading frame 28 (ORF28)- and ORF30-encoded acyl-CoA dehydrogenases. J Bacteriol 196:3598–3608. doi: 10.1128/JB.01878-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fuchs G, Boll M, Heider J. 2011. Microbial degradation of aromatic compounds—from one strategy to four. Nat Rev Microbiol 9:803–816. doi: 10.1038/nrmicro2652. [DOI] [PubMed] [Google Scholar]
- 59.Díaz E, Jiménez JI, Nogales J. 2013. Aerobic degradation of aromatic compounds. Curr Opin Biotechnol 24:431–442. doi: 10.1016/j.copbio.2012.10.010. [DOI] [PubMed] [Google Scholar]
- 60.Griffin JE, Pandey AK, Gilmore SA, Mizrahi V, McKinney JD, Bertozzi CR, Sassetti CM. 2012. Cholesterol catabolism by Mycobacterium tuberculosis requires transcriptional and metabolic adaptations. Chem Biol 19:218–227. doi: 10.1016/j.chembiol.2011.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL. 2009. BLAST+: architecture and applications. BMC Bioinformatics 10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fu L, Niu B, Zhu Z, Wu S, Li W. 2012. CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics 28:3150–3152. doi: 10.1093/bioinformatics/bts565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Finn RD, Clements J, Eddy SR. 2011. HMMER web server: interactive sequence similarity searching. Nucleic Acids Res 39:W29–W37. doi: 10.1093/nar/gkr367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Steroid catabolism gene clusters of Rhodococcus jostii RHA1, Mycobacterium tuberculosis H37Rv, Comamonas testosteroni CNB-2, and Pseudomonas sp. strain Chol1. Filled arrows indicate characterized genes and proteins, open arrows indicate annotated genes, and gray arrows indicate genes and proteins probably not involved in steroid degradation. Gene names correspond to protein names in Fig. 1 or gene locus tags, which have been abbreviated to the last digits for strains CNB-2 and Chol1. Download
Sources of isolates whose genomes were found to have steroid catabolism genes via HMM analysis. Download
Heat map showing BLAST identity for best reciprocal BLASTp hits to Rhodococcus jostii RHA1 and Comamonas testosteroni CNB-2 steroid degradation proteins in 75 bacterial draft genomes. Download
Growth on or metabolism of three steroid substrates by nine predicted steroid degraders. (A) Growth of six strains, measured as protein production, on cholate or cholesterol and removal of the steroid substrates. For A. missouriensis 431, differential protein yield was calculated by subtracting the protein yield in control cultures without a steroid substrate. (B) Removal of cholesterol or cholate in the presence of additional organic substrates by four strains. The results from two representative replicate cultures (A and B) and a noninoculated control (ni) are shown. Download
Table listing 265 putatively steroid-degrading bacteria identified by HMM analysis.
Table listing 124 complete and 75 draft genomes of putatively steroid-degrading bacteria that were analyzed by reciprocal BLASTp analysis.
Table listing 105 genomes from GenBank of Rhodococcus, Mycobacterium, Comamonas, and Streptomyces spp. that were sources of reference genes.
Materials and methods used for bioinformatic analysis and growth experiments. Download