ABSTRACT
Rhodopseudomonas palustris is an alphaproteobacterium that has served as a model organism for studies of photophosphorylation, regulation of nitrogen fixation, production of hydrogen as a biofuel, and anaerobic degradation of aromatic compounds. This bacterium is able to transition between anaerobic photoautotrophic growth, anaerobic photoheterotrophic growth, and aerobic heterotrophic growth. As a starting point to explore the genetic basis for the metabolic versatility of R. palustris, we used transposon mutagenesis and Tn-seq to identify 552 genes as essential for viability in cells growing aerobically on semirich medium. Of these, 323 have essential gene homologs in the alphaproteobacterium Caulobacter crescentus, and 187 have essential gene homologs in Escherichia coli. There were 24 R. palustris genes that were essential for viability under aerobic growth conditions that have low sequence identity but are likely to be functionally homologous to essential E. coli genes. As expected, certain functional categories of essential genes were highly conserved among the three organisms, including translation, ribosome structure and biogenesis, secretion, and lipid metabolism. R. palustris cells divide by budding in which a sessile cell gives rise to a motile swarmer cell. Conserved cell cycle genes required for this developmental process were essential in both C. crescentus and R. palustris. Our results suggest that despite vast differences in lifestyles, members of the alphaproteobacteria have a common set of essential genes that is specific to this group and distinct from that of gammaproteobacteria like E. coli.
IMPORTANCE Essential genes in bacteria and other organisms are those absolutely required for viability. Rhodopseudomonas palustris has served as a model organism for studies of anaerobic aromatic compound degradation, hydrogen gas production, nitrogen fixation, and photosynthesis. We used the technique of Tn-seq to determine the essential genes of R. palustris grown under heterotrophic aerobic conditions. The transposon library generated in this study will be useful for future studies to identify R. palustris genes essential for viability under specialized growth conditions and also for survival under conditions of stress.
INTRODUCTION
Essential genes in bacteria and other organisms are those that are absolutely required for viability. Historically, these genes were recognized by the inability of an investigator to generate a knockout mutation in a particular gene. In recent years, rapid and affordable DNA sequencing technologies have stimulated the development of methods to identify virtually all essential genes of a bacterial genome in a single experiment. These methods, referred to as Tn-seq, TraDIS, HITS, or INSeq (1–4), all use saturating transposon (Tn) mutagenesis in a bacterium of interest and identify individual Tn mutants using next-generation sequencing (NGS). Hundreds of thousands of knockout mutations can be identified simultaneously, and those genes that receive few or no mutations are deemed essential. These methods have been used to profile the essential genes in a number of bacterial pathogens, commensals, and model organisms (2–8). However, there have been only a limited number of studies defining the essential genes in metabolically versatile environmental bacteria (9, 10).
Rhodopseudomonas palustris is a facultatively phototrophic alphaproteobacterium (11, 12). It is found in terrestrial soil and water environments and grows phototrophically under anaerobic conditions. Under such conditions, it uses light as a source of energy and either organic compounds or carbon dioxide as carbon sources. R. palustris also grows aerobically as a heterotroph, and some strains grow anaerobically in the dark by nitrate respiration. It has served as a model organism for studies of anaerobic aromatic compound degradation, hydrogen gas production, nitrogen fixation, and photosynthesis (13–21). R. palustris grows by budding in which a motile daughter swarmer cell is pinched off from the pole of a sessile mother cell once chromosome replication is complete (22). Conserved cell cycle genes likely drive this developmental process in a fashion analogous to that studied in Caulobacter crescentus (23).
Here, we used Tn-seq to determine the essential genes of R. palustris grown under heterotrophic aerobic conditions. We generated a library of approximately 175,000 Tn mutants and sequenced the library using NGS to identify the junctions between the Tn insertions and the R. palustris genome. Bioinformatic processing of the sequencing data revealed 552 genes that were disrupted at very low levels, indicating that they are essential for R. palustris viability. Analysis of these genes and comparison to the known essential genes of the alphaproteobacterium C. crescentus and E. coli revealed a common core of essential bacterial genes.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
R. palustris strain CGA009 (11, 16) was used for this study. Cells were grown with illumination from a 60-W incandescent light bulb or in ambient light at 30°C. Photosynthetic medium (PM), a defined mineral medium that contains ammonium sulfate (0.1%) as a nitrogen source and 20 mM sodium acetate as a carbon source (24), was used for growth under anaerobic conditions. Medium was made anoxic by bubbling with argon gas to remove oxygen. It then was aliquoted in 10-ml volumes into 16-ml Hungate tubes in a Coy anaerobic chamber containing N2 gas. For Tn-seq library construction, R. palustris was grown aerobically on CA agar (PM supplemented with Casamino Acids [0.2%] and yeast extract [0.5%]) containing sodium acetate (20 mM). Nutrient agar (Difco) was used to determine viable CFU. Where indicated, R. palustris cultures were supplemented with kanamycin (400 μg/ml) and chloramphenicol (10 μg/ml). E. coli SM10 λpir (25, 26) was used as a donor strain for conjugation with R. palustris. E. coli was grown aerobically at 37°C in Luria-Bertani (LB) medium supplemented with ampicillin (100 μg/ml).
T24 mutant library construction.
R. palustris was grown anaerobically in light to mid-logarithmic phase (A660 of ∼0.2 to 0.7) in four tubes of PM medium (10 ml per tube) supplemented with 20 mM sodium acetate and 0.1% yeast extract. The remaining steps were performed aerobically. The cultures were combined, harvested by centrifugation, and resuspended in 10 ml PM. E. coli SM10 λpir harboring the Tn5-based transposon T24 on a suicide plasmid (pLG107) was grown in 50 ml LB supplemented with ampicillin to mid-logarithmic phase (A600 of ∼0.2 to 0.5), harvested by centrifugation, and washed and resuspended in 10 ml PM. The resuspended R. palustris and E. coli cells were mixed and filtered onto nitrocellulose discs (0.45-μm pore size; Millipore). The discs were incubated mating side up on CA agar for 2 to 3 h at 30°C aerobically in ambient light and subsequently were resuspended in a 1:1 mixture of PM and 50% glycerol prior to storage at −80°C. The mating suspension was thawed and plated on CA agar supplemented with kanamycin (400 μg/ml) and chloramphenicol (10 μg/ml) in 24- by 24-cm QTrays (Molecular Devices). The QTrays were incubated for ∼10 days at 30°C aerobically with light provided by a 60-W bulb. Equal volumes of nine independent matings were pooled to form the final CGA009 Tn mutant library. The library was separated into 500-μl aliquots and stored at −80°C. To obtain DNA for Tn-seq analysis, we thawed two library aliquots and prepared two independent samples of genomic DNA using the PureGene kit (Qiagen). These two technical replicates were treated independently in all preparation and analysis steps.
Tn-seq sample preparation for Illumina sequencing.
R. palustris library DNA was prepared for Illumina sequencing using the Tn-circle method of Gallagher et al. (5), with some modifications. For all steps, DNA was quantified by fluorometry using the broad-range Qubit kit (Life Technologies). DNA was separated into samples of 2 to 3 μg and sheared to an average fragment length of 300 bp using a Covaris E210 (10% duty cycle, intensity of 5, 100 cycles per burst, 500-s duration). Sheared samples were end repaired using the NEBNext end repair module (NEB) and purified using AMPure XP magnetic beads (Agencourt). End-repaired DNA was eluted with 10 mM Tris-Cl (pH 8.5), the beads were retained in the tubes, and eluted samples were adenylated using the NEBNext dA-tailing module (NEB). Bead reactivation buffer (20% polyethylene glycol 8000, 2.5 M NaCl) was added, samples were repurified by the bead purification method and eluted with 10 mM Tris-Cl (pH 8.5), and the beads again were retained in the tubes. Barcoded adaptors (see Table S1 in the supplemental material) were ligated to the adenylated DNA using a Quick Ligation kit (NEB). Ligation reaction mixtures contained a 20-fold molar excess of adaptor relative to DNA; DNA was quantified and the molarity of DNA fragments was estimated using an average fragment length of 300 bp. Beads were reactivated and the reaction mixtures were purified as described above. Samples were eluted in 10 mM Tris-Cl (pH 8.5) and removed from the beads, and like samples were combined.
Adaptor-ligated samples were digested with BamHI (NEB), and the digested reactions were loaded on 6% polyacrylamide Tris-borate-EDTA gels (Invitrogen). Gels were stained with SYBR gold (Invitrogen) and briefly visualized in a UV light box. Fragments between 200 and 450 bp were excised from the gels; the gel fragments then were shredded and incubated in Tris-EDTA buffer for 2 h at 65°C. The gel slurries then were run over Nanosep MF (Pall Corporation) columns to remove gel material. The flowthrough liquid was precipitated with ethanol and resuspended in a small quantity (∼12 μl) of 10 mM Tris-Cl (pH 8.5). The size-selected DNA was quantified and the molarity of DNA fragments was estimated. Two independent ampligase circularization (AC) reactions were prepared for each sample with ∼1-pm size-selected DNA at a concentration of 50 nM, using conditions outlined previously (5). Briefly, total AC reaction volumes were 20 μl and contained 0.5 μl Ampligase enzyme (Epicenter) and a 10-fold molar excess collector probe (T23_PAIR_COLLECT_1; see Table S1 in the supplemental material) relative to size-selected DNA. Following thermocycling, reactions were digested with a combination of three endonucleases as described previously (5). Digested reactions were purified separately using the MinElute PCR purification kit (Qiagen) and eluted in 10.5 μl elution buffer (EB).
For each AC reaction, 3 μl was used as the template for real-time PCR using SsoFast EvaGreen supermix (Bio-Rad) in 50-μl reactions. Amplification primers (T23_SLXA_PAIR_AmpF_3 and SLXA_PAIR_REV_AMP; see Table S1 in the supplemental material) were used at a concentration of 250 nM each. The following real-time PCR cycling procedure was used: 95°C for 2 min 30 s and then 35 cycles of 95°C for 25 s, 59°C for 25 s, and 72°C for 30 s. Test reactions were used to determine the linear range of amplification for each sample. The PCR then was repeated and stopped at an optimal cycle number corresponding to 25 to 50% maximum amplification (usually between 25 and 27 cycles). Control reactions (no exonuclease, no CP, no ampligase) were included. DNA was purified with the MinElute PCR purification kit (Qiagen) and eluted in 12 μl EB. Aliquots (1 μl) of each sample were visualized on 6% polyacrylamide TBE gels (Invitrogen) to verify the expected fragment size range. Samples were sequenced on an Illumina Hi-Seq 2000 with 36-bp single-end reads using sequencing primer T23_SEQ_G (see Table S1).
Analysis of Tn-seq sequencing data.
The standard Illumina preprocessing pipeline was used for initial default filtering (27). Subsequent steps were performed using a custom bioinformatics pipeline, consisting of a series of Perl scripts similar to those described previously (5), with some modifications. The pipeline steps are summarized below. Filtered reads were mapped using Bowtie version 1.0.0 (28) with the following parameters: two mismatches were allowed in the first 28 bp of a read (default -n), a single best alignment according to number of mismatches and Phred quality scores was reported (-best), and the existence of multiple “best” alignments was noted (-M 1) (28 and http://bowtie-bio.sourceforge.net/). For the two replicates, 30 to 50% of the reads mapped to the R. palustris strain CGA009 genome; we attribute this percentage to the proportion of reads that were of sufficiently high quality to be mapped. Analysis after this point was similar to that described previously, with some modifications (5). The total number of mapped reads was normalized to 107 reads per sample, reads per insertion location were counted, insertion locations were annotated, and read counts per gene were generated for the entire open reading frame (ORF) and for the central 5 to 90% of the ORF. In addition, reads mapping to multiple locations were counted for each gene. Finally, a combined list of all insertion locations across the two replicates was generated.
Determination of essential genes and increased or reduced fitness knockouts.
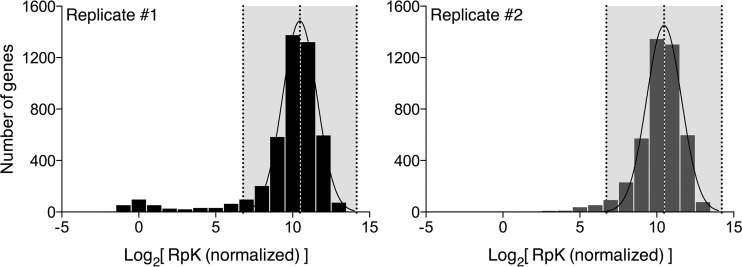
To perform statistical analysis for each Tn-seq sample, reads in the first 5% and last 10% of each gene were discarded and the central 5 to 90% read counts per gene were normalized to gene length (1 kb = 1 gene), resulting in normalized reads per kilobase (RpK) values for each gene. For each replicate, log2 RpK values were analyzed via histogram and fit to a normal distribution using the nonlinear least-squares fit (Prism; GraphPad Software), and the means and standard deviations of the distribution were calculated. Essential genes were those with log2 RpK values falling outside a 99.9% confidence interval (P < 0.001 by two-tailed test) and below the mean in both technical replicates. Genes with reduced insertion fitness (IF) were those with log2 RpK values falling outside a 95% confidence interval (P < 0.05 by two-tailed test) and below the mean in both replicates. Genes with increased IF were those with log2 RpK values falling outside a 95% confidence interval (P < 0.05 by two-tailed test) and above the mean in both replicates.
Bioinformatic assignment of R. palustris CGA009 essential gene homologs in related R. palustris strains and other bacterial species.
We analyzed the genomes of 15 strains of R. palustris for the presence of the 552 essential genes in CGA009 using OrthoMCL (29, 30) in conjunction with custom Perl scripts that we developed. To identify homologs of R. palustris essential genes in E. coli and C. crescentus, we used the 31 and http://www.ncbi.nlm.nih.gov/books/NBK52640/) along with custom Python and Perl scripts. Briefly, the protein sequences of the 552 R. palustris essential genes were individually queried against all protein sequences in E. coli MG1655 and C. crescentus NA1000. Homologs were identified by cutoffs for percent sequence identity (number of identical residues divided by R. palustris query protein length; ≥20%) and expect (E) values (≤0.0005). The highest sequence identity match for each R. palustris query protein was noted for each subject genome and referred to as the best-matching homolog. The essentiality of E. coli homologs was evaluated by consulting the Keio collection construction study (32), while for C. crescentus homologs we referred to a recent Tn-seq study (8).
Accession number.
The primary sequencing data for this study have been deposited in the NCBI BioProject database under accession number PRJNA301497.
RESULTS
Construction of a complex transposon mutant library in R. palustris.
To build a complex Tn mutant library in R. palustris, we introduced a Tn5-based transposon, T24 (ISlacZ-prhaBo/FRT-kan), into R. palustris strain CGA009 by conjugation with an E. coli SM10 λpir strain carrying plasmid pLG107. Transconjugants were plated and incubated aerobically on CA, a mineral medium containing multiple carbon and nitrogen sources (acetate, Casamino Acids, yeast extract, and ammonium sulfate), supplemented with kanamycin to select for the presence of the T24 transposon in the recipient strain and chloramphenicol to select against donor E. coli cells. We found that a high concentration of kanamycin (400 μg/ml) was required to prevent the growth of R. palustris cells that did not acquire a Tn, but this had the effect of slowing the growth of the kanamycin-resistant R. palustris cells to the point where it took weeks for colonies to develop. We found that kanamycin-resistant colonies developed faster when we provided light from a 60-W bulb that was placed approximately 10 cm from the plates. It is likely that this allowed R. palustris cells to carry out some photophosphorylation in the microaerobic environment of the colony interior. R. palustris has been shown to grow faster at reduced oxygen tensions when it is also illuminated (33). Kanamycin-resistant colonies were collected and mixed to produce a mutant pool. We performed nine such matings and combined equal volumes of each pool to produce the final CGA009 T24 mutant library.
To assess the complexity and analyze the makeup of the CGA009 T24 mutant library, two technical replicates were prepared separately and analyzed by Tn-seq. DNA was extracted and treated as described in Materials and Methods to isolate the Tn-genome junctions for Illumina sequencing. The sequencing data were analyzed using our custom bioinformatics pipeline, which consists of a series of custom Perl scripts similar to those previously described by Gallagher et al. (5), with some modifications. The pipeline aligns the sequencing reads to the strain CGA009 genome using the short-read aligner Bowtie (28); each individual read value corresponds to a single insertion mutant present in the population. The pipeline then tallies all insertion locations recognized for each sample. Using this approach, we identified approximately 175,000 individual transposon insertion locations in the CGA009 T24 mutant library (Table 1; also see Table S2 in the supplemental material). This depth of coverage reflects an average of 1 Tn insertion every ∼31 bp over the entire genome. Our analysis is restricted to the chromosome that makes up 99.8% of the R. palustris strain CGA009 genome (11); this strain has one indigenous 8.5 kb plasmid that was not included in our analysis because it is not present in other R. palustris strains and thus is unlikely to be essential. This plasmid (pRPA) carries seven genes, which are predicted to encode a replication protein, a partition protein, a resolvase, a restriction endonuclease, a restriction methylase, and two hypothetical proteins.
TABLE 1.
R. palustris CGA009 transposon library composition
| Parameter | Value for replicate(s): |
||
|---|---|---|---|
| 1 | 2 | 1 and 2 | |
| Reads (no.; filtered) | 46,684,569 | 115,704,516 | |
| Mapped reads (no.) | 13,933,275 | 60,342,240 | |
| Mapping frequency (%) | 29.9 | 52.2 | |
| Unique insertions (no.) | 157,145 | 112,524 | |
| Total unique insertions (no.) | 176,483 | ||
| Genome size (bp) | 5,459,213 | ||
| Insertion frequency (bp/insertion) | 30.9 | ||
Identification and functional characterization of essential R. palustris genes.
To determine the essential genes in R. palustris CGA009, we carried out several computational steps on the Tn-seq data. First, the number of insertions and reads for each gene in the CGA009 genome was counted, and a normalized read value (to a total of 107 reads per sample) was generated for each gene (see Table S3 in the supplemental material) using our bioinformatics pipeline. A comparison of the two technical replicates revealed a high correlation (R = 0.98) between normalized reads per gene values for the two samples, giving us confidence that the methods we used to map the Tn-genome junctions give reproducible results (Fig. 1). A second normalization based on gene length then was performed, resulting in an RpK value for each gene. To identify genes that are required for growth under the conditions of Tn library construction, we examined each technical replicate to find the genes with no or very low RpK values compared to the rest of the genome using a stringent cutoff for essentiality (99.9% CI based on log2 RpK). Genes with RpK values below this cutoff in both replicates were deemed essential (Fig. 2). It is important to note that we used a statistical definition of essentiality; thus, there is a chance that our essential gene set includes both false positives and false negatives, although at a low frequency.
FIG 1.
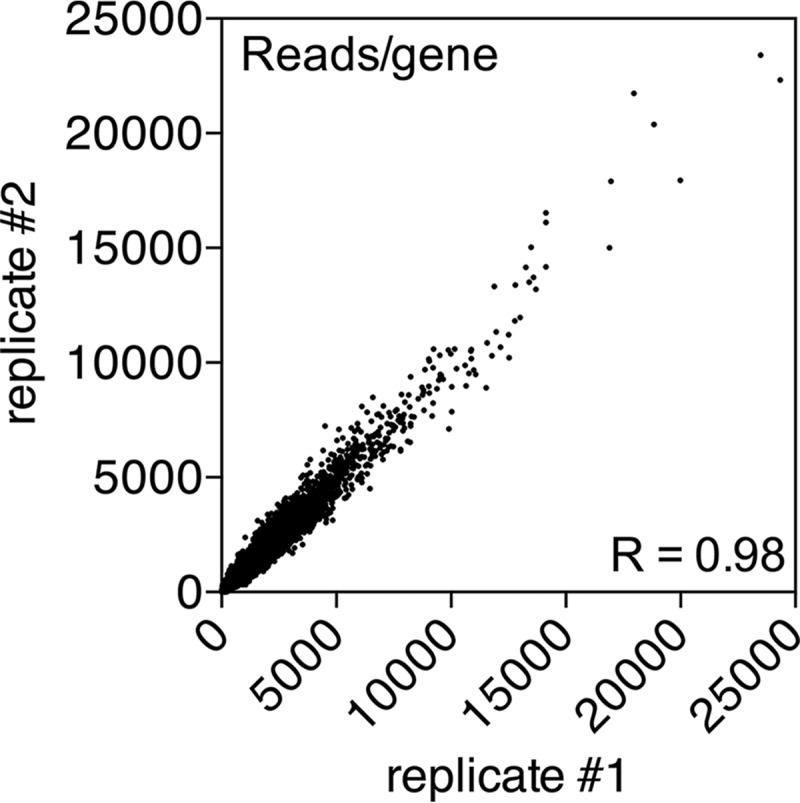
Correlation of Tn-seq sample replicates. Two independent technical replicates of CGA009 T24 library genomic DNA were extracted from cells and prepared for NGS analysis. The read values for each gene were determined for both replicates and plotted to determine the correlation.
FIG 2.
Distribution of RpK values among R. palustris genes. Read counts were normalized for total sample reads and gene length and tallied for the 5 to 90% length window of each gene to give reads-per-kilobase (RpK) values. Log2-transformed RpK values were calculated and histograms were generated. The means and standard deviations of the distribution were calculated and used to determine 99.9% confidence intervals (indicated by shading). Genes with RpK values outside the confidence interval and below the mean are considered essential.
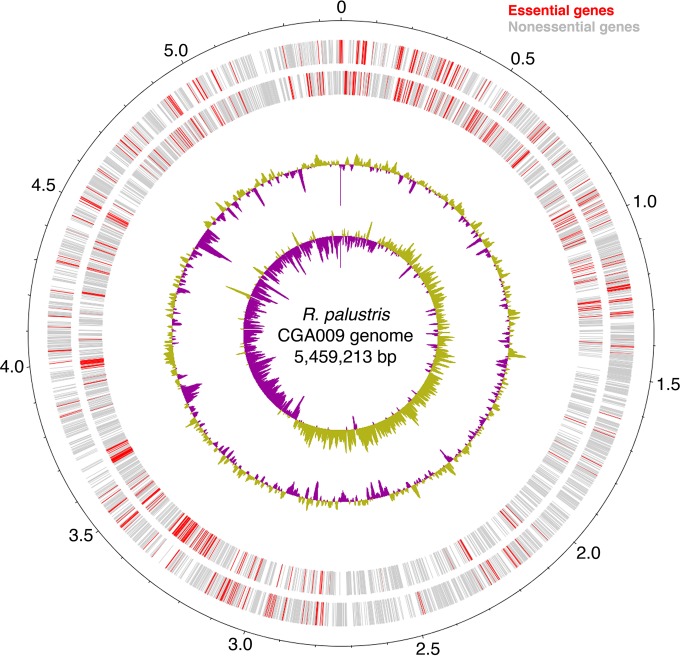
We identified a total of 552 essential genes (see Table S4 in the supplemental material) that are required for growth under the conditions of Tn library construction, that is, aerobic growth in light on medium containing Casamino Acids, yeast extract, and acetate. The physical locations of these essential genes on the R. palustris CGA009 chromosome indicate an organization similar to that of the essential genes in the related organism Caulobacter crescentus (8). The essential genes are concentrated around the origin and the terminus of the bacterial chromosome (Fig. 3).
FIG 3.
Chromosomal arrangement of R. palustris essential genes. The essential R. palustris genes were determined by statistical analysis of the Tn-seq data and are shown in their genomic context. The R. palustris chromosome is depicted (the small plasmid is excluded). From the outside in, the plus (sense) strand, minus (antisense) strand, GC content (%), and GC skew (GC-AT/GC) are shown. Essential genes are depicted on their associated strand in red, while nonessential genes are shown in gray.
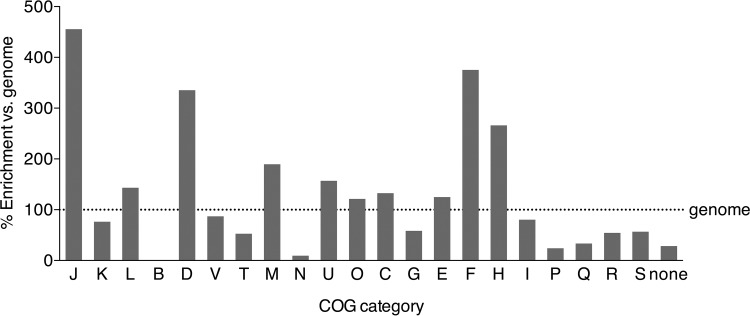
To assess the functional makeup of the R. palustris essential genes, we first referred to the Larimer et al. (11) R. palustris CGA009 genome annotation to obtain Clusters of Orthologous Groups (COG) assignments for the entire genome. To discover enriched or depleted functions in our list of essential genes, we compared the COG category makeup of the essential genes to the entire genome (Fig. 4). We found that several categories were enriched in the essential genes, particularly genes involved in (J) translation, ribosome structure, and biogenesis; (D) cell cycle control, cell division, and chromosome partitioning; (F) nucleotide transport and metabolism; and (H) coenzyme transport and metabolism (Fig. 4). COG categories with less representation in the essential genes compared to the entire genome included (N) cell motility, (G) carbohydrate transport and metabolism, (P) inorganic ion transport and metabolism, and (R) general function predicted only.
FIG 4.
R. palustris essential gene COG category makeup versus the overall CGA009 genome. COG categories for essential genes were determined. The fraction that each COG category contributes to the entire CGA009 genome was set at 100% and compared to the fractions for each of these subsets. The following COG categories were included: J, translation, ribosomal structure, and biogenesis; K, transcription; L, replication, recombination, and repair; B, chromatin structure and dynamics; D, cell cycle control, cell division, and chromosome partitioning; V, defense mechanisms; T, signal transduction mechanisms; M, cell wall/membrane/envelope biogenesis; N, cell motility; U, intracellular trafficking, secretion, and vesicular transport; O, posttranslational modification, protein turnover, and chaperones; C, energy production and conversion; G, carbohydrate transport and metabolism; E, amino acid transport and metabolism; F, nucleotide transport and metabolism; H, coenzyme transport and metabolism; I, lipid transport and metabolism; P, inorganic ion transport and metabolism; Q, secondary metabolite biosynthesis, transport, and catabolism; R, general function prediction only; S, function unknown.
While COG assignments are widely used and allow for comparisons between organisms, COG categories do not exist for genes representing nonuniversal metabolic functions, such as nitrogen fixation and photosynthesis. We manually assigned each gene to an overarching functional group by referring to the online databases EcoCyc (34), InterPro (35), the Kyoto Encyclopedia of Genes and Genomes (KEGG) (36, 37), and Cyanobase (38). The functional category with the greatest representation was metabolism, with 251 genes (Table 2). Essential metabolic genes include those encoding functions in glycolysis, the tricarboxylic acid (TCA) cycle, and the biosynthesis of fatty acids and amino acids. It is important to note that we tested for essentiality under just one growth condition. We expect that some of the biosynthesis genes that were identified as essential here may not be essential for growth in alternative growth media.
TABLE 2.
Functional makeup of R. palustris essential genes
| Functional group | No. of essential genes |
|---|---|
| Catabolite repression | 1 |
| Cell cycle, cell division, and cell envelope | 30 |
| Conserved domain proteins | 9 |
| DNA methylation | 2 |
| DNA replication, recombination, and repair | 21 |
| Hypothetical proteins | 54 |
| Lipoproteins | 6 |
| Metabolism | 251 |
| Outer membrane proteins | 5 |
| Proteases/peptidases | 13 |
| Protein folding/modification | 4 |
| Protein translocation | 2 |
| Secretion | 13 |
| Sensing and signaling | 11 |
| Sigma factors | 3 |
| Stress response | 7 |
| Transcription | 12 |
| Transcriptional regulators | 7 |
| Translation | 88 |
| Transport | 13 |
| Total | 552 |
Even with functional assignment methods at our disposal, a fair number of the R. palustris essential genes have no predicted function. Nearly 10% (54 of 552) of the identified R. palustris essential genes encode hypothetical proteins of unknown function. A protein BLAST search revealed all but one (RPA4735) are conserved in other organisms, mainly other alphaproteobacteria, such as Bradyrhizobium japonicum and Nitrobacter winogradsky. Gene RPA4735 likely has been misannotated in strain CGA009 but correctly called in closely related strains, including strain TIE-1 (Rpal-5217), as a slightly larger gene predicted to encode a cell division protein.
The R. palustris CGA009 genome encodes conserved and unique essential functions.
To determine if R. palustris CGA009 essential genes are conserved and present in other strains, we compared the CGA009 genome to those of 15 other R. palustris strains using the OrthoMCL (29, 30) software package and identified homologs of the 552 essential genes. The 15 strains analyzed are part of our laboratory's collection of R. palustris strains comprising both well-characterized strains (TIE-1) and strains obtained from culture collections and isolated from environmental samples. We found that all but 10 of the 552 essential genes have at least a single homolog in all 15 strains (see Table S5 in the supplemental material). Additionally, 32 of the essential genes have multiple homologs in at least one strain, with one essential gene, RPA3903, having a range of 4 to 10 homologs per R. palustris strain (see Table S5). RPA3903 (mexD) is a resistance-nodulation-division (RND) multidrug efflux transporter, and CGA009 alone has several mexD genes, although only one is essential (see Table S4).
To determine if homologs of R. palustris essential genes are essential in related organisms, we chose two well-studied organisms with identified essential genes: C. crescentus and E. coli. We used BLAST (31 and http://www.ncbi.nlm.nih.gov/books/NBK52640/) to compare the protein sequences of the 552 R. palustris essential genes to all protein sequences in C. crescentus NA1000 and E. coli MG1655 and found all homologs meeting criteria for sequence identity (≥20%) and expect (E) values (≤0.0005). For each R. palustris essential gene, the total number of homologs in each comparison genome was recorded and the best-matching homolog was identified. The average sequence identity for the best-matching homolog in C. crescentus was 52%; for E. coli, the average was 40%. Nearly a quarter (24%) of R. palustris essential genes had no homologs detected by our criteria in E. coli, while 14% had no homologs in C. crescentus. To assess the essentiality of the identified homologs in C. crescentus NA1000, we referred to a Tn-seq study performed under aerobic growth on peptone yeast extract (PYE) (8). For C. crescentus, we identified homologs for 476 essential R. palustris genes; 323 of the identified homologs are essential in C. crescentus, while 30 have high fitness costs when deleted (see Table S4 in the supplemental material). To determine the essentiality of homologs in E. coli MG1655, we referred to the Keio collection study (32). We identified homologs for 422 R. palustris essential genes in E. coli; 187 of the identified homologs are essential in E. coli as well (see Table S4). Overall, our homolog analysis identified 54 more homologs of R. palustris essential genes in C. crescentus than in E. coli. A subset of these, for example, CtrA and CckA, are involved in cell cycle regulation in C. crescentus (8, 39). Essential gene RPA3133 shares 72% identity with essential gene encoding DivK, the response regulator receiver protein involved in cell cycle regulation in C. crescentus. This gene had been misannotated in R. palustris as encoding a CheY chemotaxis protein.
To discover patterns among the essential homologs, we grouped R. palustris genes by COG category and compared their essentiality in C. crescentus and E. coli (see Fig. S1 in the supplemental material). For C. crescentus, the conservation of essential functions was quite high, with an average of 58% shared essentiality across all COG categories. E. coli showed less conservation of essential functions, with an average of 29% shared essentiality across all COGs. However, certain categories were well conserved between all three organisms: the majority of R. palustris essential genes involved in translation, ribosome structure, and biogenesis; intracellular trafficking, secretion, and vesicular transport; and lipid transport and metabolism have essential homologs in both C. crescentus and E. coli. The essentiality of translation functions in particular was highly conserved: of 93 essential translation genes in R. palustris, the majority (84 and 68, respectively) had essential homologs in C. crescentus and E. coli as well (see Table S4).
We wondered if our bioinformatics analysis, which focused on the sequence identity between two proteins, missed functional homologs: proteins with low sequence identity that perform identical or analogous cellular functions. To identify functional homologs of R. palustris essential genes in E. coli, we examined the list of R. palustris genes with no homolog. In E. coli, we were able to identify functional homologs with low sequence identity that serve the same cellular function for 24 R. palustris essential genes by referring to the EcoCyc online database (34). Of these functional homologs, 12 are essential in E. coli in rich medium (LB) (32); these include genes involved in NAD metabolism (nadD), succinate dehydrogenase (sdhD), and ribonucleoside diphosphate reductase (nrd) (Table 3). An additional seven putative functional homologs are essential in E. coli under minimal medium conditions (Table 3) (40, 41).
TABLE 3.
R. palustris essential genes with functional homologs in E. coli
| E. coli essentialitya |
R. palustris CGA009 |
E. coli MG1655 |
Product | ||
|---|---|---|---|---|---|
| Locus | Name | Locus | Name | ||
| Essential in rich medium (LB) | RPA0165 | nadD | b0639 | nadD | NMN adenylyltransferase |
| RPA0279 | b0890 | ftsK | DNA translocase FtsK | ||
| RPA0289 | holA | b0640 | holA | DNA polymerase III δ subunit | |
| RPA0291 | parB1 | b3066 | dnaG | Chromosome partitioning protein | |
| RPA1047 | glyS | b3559 | glyS | Glycyl-tRNA synthetase β subunit | |
| RPA1178 | hisS | b2514 | hisS | Histidyl-tRNA synthetase | |
| RPA1288 | rpoD | b3067 | rpoD | RNA polymerase σ factor | |
| RPA1986 | nadE | b1740 | nadE | NAD synthetase | |
| RPA2006 | psd | b4160 | psd | Phosphatidylserine decarboxylase | |
| RPA2007 | pssA | b2585 | pssA | Phosphatidylserine synthase | |
| RPA2977 | nrd | b2234 | nrdA | Ribonucleotide-diphosphate reductase α subunit | |
| RPA3522 | ftsZ | b0095 | ftsZ | Cell division protein FtsZ | |
| Essential in minimal medium (M9) | RPA0179 | atpH | b3735 | atpH | ATP synthase F0F1 δ subunit |
| RPA0592 | argJ | b2818 | argA | N-Acetylglutamate synthase | |
| b3957 | argE | Acetylornithine deacetylase | |||
| RPA1149 | hisG | b2019 | hisG | ATP phosphoribosyltransferase | |
| RPA2035 | ilvC | b3774 | ilvC | Ketol-acid reductoisomerase | |
| RPA3834 | idh | b1136 | icd | Isocitrate dehydrogenase | |
| RPA4270 | thrB | b0003 | thrB | Homoserine kinase | |
| RPA4309 | serC | b0907 | serC | Phosphoserine aminotransferase | |
| Not essential | RPA0218 | sdhD | b0722 | sdhD | Succinate dehydrogenase cytochrome β subunit |
| RPA0349 | dcm | b1961 | dcm | Site-specific DNA methyltransferase | |
| RPA0822 | gshA | b2688 | gshA | Glutamate-cysteine ligase | |
| RPA0847 | atpI | b3739 | atpI | ATP synthase F0F1 subunit I | |
| RPA1188 | b0143 | pcnB | Poly(A) polymerase | ||
Interestingly, of the 54 essential genes encoding hypothetical proteins in R. palustris CGA009, nearly half (26 of 54) had no homolog in C. crescentus or E. coli, suggesting R. palustris has unique essential functions encoded by these genes. Of 28 hypothetical proteins with a homolog in C. crescentus, 14 were essential, and many have an annotated function in C. crescentus (Table 4). For example, the RPA0595 protein is 52% identical to the BioC biotin biosynthesis protein in C. crescentus (CCNA_00874), while RPA1263 shares 75% identity with SciP (CCNA_00948), a protein that represses the transcription of genes activated by the master cell cycle regulator CtrA. RPA1626 is homologous to ChpT, a histidine phosphotransferase which controls the activity of CtrA (42, 43).
TABLE 4.
R. palustris essential genes encoding hypothetical proteins with essential homologs in C. crescentus
| R. palustris gene | C. crescentus homolog | Sequence identity (%) | Homolog function |
|---|---|---|---|
| RPA0062 | CCNA_00306 | 29 | Lysophospholipid acyltransferase |
| RPA0288 | CCNA_03866 | 22 | LptE superfamily protein |
| RPA0297 | CCNA_03877 | 44 | Conserved hypothetical membrane spannin |
| RPA0423 | CCNA_03837 | 32 | Bacterial SH3 domain protein |
| RPA0595 | CCNA_00874 | 52 | Biotin synthesis protein BioC |
| RPA1157 | CCNA_00309 | 54 | Hypothetical protein |
| RPA1263 | CCNA_00948 | 75 | CtrA inhibitory protein SciP |
| RPA1626 | CCNA_03584 | 33 | Histidine phosphotransferase ChpT |
| RPA2910 | CCNA_01987 | 38 | UDP-2,3-diacylglucosamine pyrophosphatase LpxI |
| RPA3049 | CCNA_01783 | 37 | Chromosome replication regulator protein HdaA |
| RPA4301 | CCNA_02000 | 30 | Small subunit ribosomal protein S2P |
| RPA4344 | CCNA_03865 | 21 | Leucyl-tRNA synthetase |
| RPA4359 | CCNA_00524 | 40 | Conserved hypothetical cytosolic protein |
| RPA4742 | CCNA_01614 | 22 | Hypothetical protein |
Identification of R. palustris genes with reduced or increased insertion fitness.
Our analysis of essential genes used a strict statistical cutoff to characterize the genes required for growth in R. palustris. To more broadly characterize our data set, we extended our analysis to include genes with moderate fitness cost (or benefit) associated with Tn insertion. We identified all genes with normalized read values falling outside a 95% CI (based on log2 RpK values) in both technical replicates. We define genes falling outside this cutoff and below the mean as those with reduced insertion fitness (IF), while those above the mean are increased-IF genes. We identified a total of 193 reduced-IF (see Table S6 in the supplemental material) and 26 increased-IF genes (see Table S7) in R. palustris CGA009 and assessed the functional makeup of the reduced-IF and increased-IF genes using analyses similar to those described above. The identified reduced-IF genes had a functional profile similar to that of the essential genes: a large number are metabolic in nature, and a fair number of them encode hypothetical proteins. One gene, RPA4516, encoding a response regulator receiver-modulated diguanylate phosphodiesterase, is a reduced-IF gene with considerable fitness costs (P < 0.006 in both replicates). To our knowledge, this is the first c-di-GMP phosphodiesterase (PDE) shown to be involved in general bacterial growth. In this context it is worth noting that fluctuations of c-di-GMP recently have been shown to be a key driver of cell cycle progression in C. crescentus (23). For the increased-IF genes, we again identified a number that encode hypothetical proteins. In addition, we identified a phosphate transporter (RPA2281), two cold shock proteins with DNA-binding domains (RPA2525 and RPA3503), and the pucAd light-harvesting complex subunit (RPA3012). We hypothesize that these gene products pose either an energetic or metabolic burden for growing cells that is alleviated by Tn insertion. We also note that the fitness benefit derived from the loss of these genes likely is specific to the conditions tested.
DISCUSSION
Here, we used the Tn circle method of Tn-seq (5) to define the essential genome in the alphaproteobacterium R. palustris. We constructed a transposon mutant library in R. palustris, made up of over 175,000 individual insertion mutants, with a frequency of approximately one insertion per 31 bp throughout the genome. By analyzing this library using Tn-seq and NGS, we identified 552 genes that were essential during aerobic growth on a mineral medium supplemented with acetate, Casamino Acids, and yeast extract.
Identified essential genes reflect Tn-seq library growth conditions.
The growth medium used for the construction of the Tn library dictated that the cells grow heterotrophically using the provided simple and complex carbon and nitrogen sources. Despite the presence of Casamino Acids in the medium, we found that the gene clusters for the synthesis of tryptophan (trpFBA, trpD, and trpG), arginine (argG, argJ, argB, argC, argH, argF, and argD), glutamate (gltBD), and histidine (hisBHAFE and hisGZ) were essential (see Table S4 in the supplemental material). For tryptophan, this result agrees with our knowledge of the composition of Casamino Acids. Casamino Acids are produced by the acid hydrolysis of casein, and tryptophan is destroyed in the hydrolysis process (44). For glutamate, arginine, and histidine, our results suggest that Casamino Acids do not meet the R. palustris requirement for these amino acids despite their survival of acid hydrolysis. In addition, the essential thiOSGE thiamine biosynthesis gene cluster indicates that this vitamin must be synthesized de novo by R. palustris under this growth condition.
R. palustris can grow heterotrophically by aerobic respiration without a contribution from photophosphorylation. However, we found that a light source was necessary to elicit robust growth of the strain CGA009 T24 transconjugants; we recovered a very small number of transconjugants under dark/ambient light conditions. We suspect that the large amount of kanamycin (400 μg/ml) used to select for T24 transconjugants presented a barrier to growth without the addition of energy from light. Supporting this hypothesis, we identified 2 essential genes with possible photosynthetic functions (RPA3742 and RPA3743, both encoding squalene/phytoene synthases that may be involved in carotenoid biosynthesis). In addition, the pufAB light-harvesting complex genes, while not essential by our strict Tn-seq statistical cutoff (see Materials and Methods), exhibit decreased fitness when disrupted by the T24 transposon (see Table S2 in the supplemental material), with the pufA gene meeting our reduced-IF cutoff.
Essential genes in central metabolism reveal redundancies and alternative flux hypotheses.
Recent metabolic flux analysis studies indicate growing R. palustris cells use the glyoxylate shunt to incorporate acetate when it is the only available carbon source (15, 45). Here, we provided R. palustris with complex carbon sources in addition to acetate and found that under these conditions, cells do not require the glyoxylate shunt; isocitrate lyase (aceA) and malate synthase (glcB) are not essential (see Table S4 in the supplemental material). This suggests that the Casamino Acids and yeast extract in CA medium support growth sufficiently such that growing R. palustris cells use the TCA cycle to oxidize acetate fully to CO2. Indeed, all annotated TCA cycle enzyme-encoding genes (gltA, acnA, idh, sucAB, sdhBADC, and mdh) were found to be essential in R. palustris CGA009 except those encoding succinyl-coenzyme A (CoA) synthetase (sucDC) and fumarase (see Table S4). R. palustris CGA009 has single copies of the alpha (sucD) and beta (sucC) subunits of succinyl-CoA synthetase; therefore, redundancy cannot account for the nonessentiality of these genes by Tn-seq. Succinyl-CoA synthetase catalyzes the reversible conversion of succinyl-CoA to succinate and produces 1 GTP or ATP by substrate-level phosphorylation. We hypothesize that sucD::T24 and sucC::T24 cells can compensate for the loss of this activity by producing succinate either from fumarate, via the succinate dehydrogenase complex, or from isocitrate, via isocitrate lyase and the glyoxylate shunt. In addition, succinyl-CoA is the precursor of heme, which is likely required in large quantities by R. palustris for the production of bacteriochlorophyll, providing a metabolic sink for a build-up of intracellular succinyl-CoA in the absence of succinyl-CoA synthetase activity.
Fumarase provides us with another interesting case. There are three predicted fumarate hydratase genes in the R. palustris CGA009 genome (RPA1329, RPA3500, and RPA3876), none of which were found to be essential by Tn-seq. We have two hypotheses for the nonessentiality of fumarase: redundancy and alternative flux. If redundancy is responsible, R. palustris would be expected to survive if one of its fumarase genes were knocked out. Another possibility is that only one of the annotated fumarase genes is required for TCA cycle function, but R. palustris can compensate for its loss by producing malate through the glyoxylate shunt. The maintenance of three copies of fumarase genes is intriguing, and further analysis of their encoded enzymes will be required to verify their predicted functions and to establish how and under what growth conditions each contributes to the physiology of R. palustris. Importantly, these examples underline the misleading notion of labeling a biosynthetic step as nonessential by Tn-seq without also considering the larger genomic context.
Proton-translocating NADH-quinone oxidoreductase, also known as NADH dehydrogenase, plays an important role in transferring electrons from NADH to quinone during aerobic respiration in most living organisms. R. palustris has genes (RPA2937-2952 and RPA4252-4264) that are annotated as encoding paralogous NADH dehydrogenase 1 enzymes. Each enzyme is predicted to be comprised of 13 subunits, consistent with the compositions of other NADH dehydrogenases (46). We were interested to see that RPA2937-2952 are essential genes for R. palustris, whereas RPA4252-4264 are not essential for growth. This implies that the two NADH dehydrogenase complexes have different physiological functions. One possibility is that RPA2937-2952 are required for aerobic respiration, whereas RPA4252-4264 encode an NADH dehydrogenase that operates in the reverse direction to reduce NAD+ to NADH during phototrophic growth. This reaction, known as “reversed electron transport,” is used by other purple nonsulfur bacteria to generate NADH; however, it has not been shown to be accomplished by a dedicated enzyme.
Comparison of essential genes in R. palustris and other organisms.
We compared the essential genes in R. palustris to the well-studied alphaproteobacterium C. crescentus and to E. coli. As expected, there is a great deal of overlap of essential genes among the three microbes. We also found that redundancy plays a role in differences in essential genes between R. palustris and the two comparison organisms. For example, R. palustris encodes an essential phosphoglyceromutase (pgm1; RPA0340). While the E. coli gpmM homolog (b3612) is not essential due to the presence of a second enzyme (gpmA; b0755), an E. coli double gpmAM knockout is nonviable (47), a result which speaks to the similar metabolic requirements of these organisms despite differences in genetic makeup.
In other cases, R. palustris has one protein while other bacteria require two to perform the same function. The R. palustris argJ (RPA0592) gene encodes a bifunctional ornithine acetyltransferase/N-glutamate acetyltransferase which catalyzes the first and fifth steps of arginine biosynthesis. Instead of a bifunctional protein, E. coli encodes two single-function proteins, ArgA (b2818) and ArgE (b3597); both are essential for E. coli strain MG1655 to grow in minimal medium (40, 41). Another interesting case is that of R. palustris hisG, which encodes ATP phosphoribosyltransferase and catalyzes the first step in histidine metabolism. There are two known HisG enzyme clades: in the first, a single enzyme has ATP phosphoribosyltransferase activity, while HisG enzymes in the second clade require a second subunit, HisZ, for enzymatic function (48). E. coli hisG encodes an enzyme from the first clade that is essential in minimal medium (40, 41). However, R. palustris encodes both HisG (RPA1149) and HisZ (RPA1150), and both are essential (see Table S4 in the supplemental material). Interestingly, HisZ-dependent HisG enzymes from Lactococcus lactis, Bacillus subtilis, and Synechocystis are shorter than their counterparts in E. coli and other organisms by approximately 80 amino acids: L. lactis HisG is 208 amino acids, while E. coli HisG is 299 amino acids (48). This difference in length is thought to be the reason behind the requirement for HisZ (48). However, the R. palustris HisG protein is longer (325 amino acids), suggesting that there is a second HisZ-dependent clade of HisG. This warrants further investigation.
We also identified differences in the essentiality of amino acid biosynthesis genes between R. palustris, C. crescentus, and E. coli that likely represent differences in growth conditions that were used to determine gene essentiality. C. crescentus was grown aerobically on peptone-yeast extract (PYE) for Tn-seq experiments (8), while the E. coli Keio collection was built on LB (32). Differences in amino acid composition between LB, peptone, and Casamino Acids may account for the differences in amino acids of essential genes that we observed.
The construction and characterization of an R. palustris Tn library has helped to define core essential genes necessary for the heterotrophic growth of alphaproteobacteria. The construction of the Tn library also sets the stage to study genes that are essential for the growth of R. palustris under more specialized conditions that reflect this organism's metabolic versatility. These include phototrophic growth, growth under nitrogen-fixing conditions, and growth on poorly characterized carbon compounds.
Supplementary Material
ACKNOWLEDGMENTS
We thank Yasuhiro Oda for technical assistance and helpful discussions, Mallory Beightol for technical assistance, the UW Genome Sciences Department for the use of the Covaris instrument, Samuel Miller for the use of the Qubit instrument, and Somsak Phattarasukol for assistance with OrthoMCL. We thank Claudine Baraquet, Jose Mendiola, Amy Schaefer, Kathryn Fixen, and John Scrapper for helpful discussions.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.00771-15.
REFERENCES
- 1.van Opijnen T, Camilli A. 2012. A fine scale phenotype-genotype virulence map of a bacterial pathogen. Genome Res 22:2541–2551. doi: 10.1101/gr.137430.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Langridge GC, Phan MD, Turner DJ, Perkins TT, Parts L, Haase J, Charles I, Maskell DJ, Peters SE, Dougan G, Wain J, Parkhill J, Turner AK. 2009. Simultaneous assay of every Salmonella Typhi gene using one million transposon mutants. Genome Res 19:2308–2316. doi: 10.1101/gr.097097.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gawronski JD, Wong SMS, Giannoukos G, Ward DV, Akerley BJ. 2009. Tracking insertion mutants within libraries by deep sequencing and a genome-wide screen for Haemophilus genes required in the lung. Proc Natl Acad Sci U S A 106:16422–16427. doi: 10.1073/pnas.0906627106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Goodman AL, McNulty NP, Zhao Y, Leip D, Mitra RD, Lozupone CA, Knight R, Gordon JI. 2009. Identifying genetic determinants needed to establish a human gut symbiont in its habitat. Cell Host Microbe 6:279–289. doi: 10.1016/j.chom.2009.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gallagher LA, Shendure J, Manoil C. 2011. Genome-scale identification of resistance functions in Pseudomonas aeruginosa using Tn-seq. mBio 2:e00315-10. doi: 10.1128/mBio.00315-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.van Opijnen T, Bodi KL, Camilli A. 2009. Tn-seq: high-throughput parallel sequencing for fitness and genetic interaction studies in microorganisms. Nat Methods 6:767–772. doi: 10.1038/nmeth.1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kamp HD, Patimalla-Dipali B, Lazinski DW, Wallace-Gadsden F, Camilli A. 2013. Gene fitness landscapes of Vibrio cholerae at important stages of its life cycle. PLoS Pathog 9:e1003800. doi: 10.1371/journal.ppat.1003800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Christen B, Abeliuk E, Collier JM, Kalogeraki VS, Passarelli B, Coller JA, Fero MJ, McAdams HH, Shapiro L. 2011. The essential genome of a bacterium. Mol Syst Biol 7:528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brutinel ED, Gralnick JA. 2012. Anomalies of the anaerobic tricarboxylic acid cycle in Shewanella oneidensis revealed by Tn-seq. Mol Microbiol 86:273–283. doi: 10.1111/j.1365-2958.2012.08196.x. [DOI] [PubMed] [Google Scholar]
- 10.Luo H, Lin Y, Gao F, Zhang C-T, Zhang R. 2014. DEG 10, an update of the database of essential genes that includes both protein-coding genes and noncoding genomic elements. Nucleic Acids Res 42:D574–D580. doi: 10.1093/nar/gkt1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Larimer FW, Chain P, Hauser L, Lamerdin J, Malfatti S, Do L, Land ML, Pelletier DA, Beatty JT, Lang AS, Tabita FR, Gibson JL, Hanson TE, Bobst C, Torres JLTY, Peres C, Harrison FH, Gibson J, Harwood CS. 2004. Complete genome sequence of the metabolically versatile photosynthetic bacterium Rhodopseudomonas palustris. Nat Biotechnol 22:55–61. doi: 10.1038/nbt923. [DOI] [PubMed] [Google Scholar]
- 12.Oda Y, Larimer FW, Chain PSG, Malfatti S, Shin MV, Vergez LM, Hauser L, Land ML, Braatsch S, Beatty JT, Pelletier DA, Schaefer AL, Harwood CS. 2008. Multiple genome sequences reveal adaptations of a phototrophic bacterium to sediment microenvironments. Proc Natl Acad Sci U S A 105:18543–18548. doi: 10.1073/pnas.0809160105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Egland PG, Pelletier DA, Dispensa M, Gibson J, Harwood CS. 1997. A cluster of bacterial genes for anaerobic benzene ring biodegradation. Proc Natl Acad Sci U S A 94:6484–6489. doi: 10.1073/pnas.94.12.6484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fixen KR, Baker AW, Stojkovic EA, Beatty JT, Harwood CS. 2014. Apo-bacteriophytochromes modulate bacterial photosynthesis in response to low light. Proc Natl Acad Sci U S A 111:E237–E244. doi: 10.1073/pnas.1322410111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McKinlay JB, Harwood CS. 2010. Carbon dioxide fixation as a central redox cofactor recycling mechanism in bacteria. Proc Natl Acad Sci U S A 107:11669–11675. doi: 10.1073/pnas.1006175107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rey FE, Oda Y, Harwood CS. 2006. Regulation of uptake hydrogenase and effects of hydrogen utilization on gene expression in Rhodopseudomonas palustris. J Bacteriol 188:6143–6152. doi: 10.1128/JB.00381-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heiniger EK, Oda Y, Samanta SK, Harwood CS. 2012. How posttranslational modification of nitrogenase is circumvented in Rhodopseudomonas palustris strains that produce hydrogen gas constitutively. Appl Environ Microbiol 78:1023–1032. doi: 10.1128/AEM.07254-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rey FE, Heiniger EK, Harwood CS. 2007. Redirection of metabolism for biological hydrogen production. Appl Environ Microbiol 73:1665–1671. doi: 10.1128/AEM.02565-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Scheuring S, Gonçalves RP, Prima V, Sturgis JN. 2006. The photosynthetic apparatus of Rhodopseudomonas palustris: structures and organization. J Mol Biol 358:83–96. doi: 10.1016/j.jmb.2006.01.085. [DOI] [PubMed] [Google Scholar]
- 20.Richter MF, Baier J, Southall J, Cogdell RJ, Oellerich S, Köhler J. 2007. Refinement of the x-ray structure of the RC LH1 core complex from Rhodopseudomonas palustris by single-molecule spectroscopy. Proc Natl Acad Sci U S A 104:20280–20284. doi: 10.1073/pnas.0704599105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moulisová V, Luer L, Hoseinkhani S, Brotosudarmo THP, Collins AM, Lanzani G, Blankenship RE, Cogdell RJ. 2009. Low light adaptation: energy transfer processes in different types of light harvesting complexes from Rhodopseudomonas palustris. Biophys J 97:3019–3028. doi: 10.1016/j.bpj.2009.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brown PJB, de Pedro MA, Kysela DT, Van der Henst C, Kim J, De Bolle X, Fuqua C, Brun YV. 2012. Polar growth in the alphaproteobacterial order Rhizobiales. Proc Natl Acad Sci U S A 109:1697–1701. doi: 10.1073/pnas.1114476109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lori C, Ozaki S, Steiner S, Böhm R, Abel S, Dubey BN, Schirmer T, Hiller S, Jenal U. 2015. Cyclic di-GMP acts as a cell cycle oscillator to drive chromosome replication. Nature 523:236–239. doi: 10.1038/nature14473. [DOI] [PubMed] [Google Scholar]
- 24.Kim MK, Harwood CS. 1991. Regulation of benzoate-CoA ligase in Rhodopseudomonas palustris. FEMS Microbiol Lett 83:199–203. [Google Scholar]
- 25.Miller VL, Mekalanos JJ. 1988. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J Bacteriol 170:2575–2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Simon R, Priefer U, Pühler A. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram negative bacteria. Nat Biotechnol 1:784–791. doi: 10.1038/nbt1183-784. [DOI] [Google Scholar]
- 27.Illumina, Inc. 2011. CASAVA v1.8.2 user guide. Illumina, Inc., San Diego, CA. [Google Scholar]
- 28.Langmead B, Trapnell C, Pop M, Salzberg SL. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen F, Mackey AJ, Stoeckert CJ, Roos DS. 2006. OrthoMCL-DB: querying a comprehensive multi-species collection of ortholog groups. Nucleic Acids Res 34:D363–D368. doi: 10.1093/nar/gkj123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li L, Stoeckert CJ, Roos DS. 2003. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res 13:2178–2189. doi: 10.1101/gr.1224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol 215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 32.Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol 2:2006.0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rey FE, Harwood CS. 2010. FixK, a global regulator of microaerobic growth, controls photosynthesis in Rhodopseudomonas palustris. Mol Microbiol 75:1007–1020. doi: 10.1111/j.1365-2958.2009.07037.x. [DOI] [PubMed] [Google Scholar]
- 34.Keseler IM, Mackie A, Peralta-Gil M, Santos-Zavaleta A, Gama-Castro S, Bonavides-Martinez C, Fulcher C, Huerta AM, Kothari A, Krummenacker M, Latendresse M, Muniz-Rascado L, Ong Q, Paley S, Schroder I, Shearer AG, Subhraveti P, Travers M, Weerasinghe D, Weiss V, Collado-Vides J, Gunsalus RP, Paulsen I, Karp PD. 2013. EcoCyc: fusing model organism databases with systems biology. Nucleic Acids Res 41:D605–D612. doi: 10.1093/nar/gks1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hunter S, Jones P, Mitchell A, Apweiler R, Attwood TK, Bateman A, Bernard T, Binns D, Bork P, Burge S, de Castro E, Coggill P, Corbett M, Das U, Daugherty L, Duquenne L, Finn RD, Fraser M, Gough J, Haft D, Hulo N, Kahn D, Kelly E, Letunic I, Lonsdale D, Lopez R, Madera M, Maslen J, McAnulla C, McDowall J, McMenamin C, Mi H, Mutowo-Muellenet P, Mulder N, Natale D, Orengo C, Pesseat S, Punta M, Quinn AF, Rivoire C, Sangrador-Vegas A, Selengut JD, Sigrist CJA, Scheremetjew M, Tate J, Thimmajanarthanan M, Thomas PD, Wu CH, Yeats C, Yong SY. 2012. InterPro in 2011: new developments in the family and domain prediction database. Nucleic Acids Res 40:D306–D312. doi: 10.1093/nar/gkr948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kanehisa M, Goto S. 2000. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res 28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kanehisa M, Goto S, Sato Y, Kawashima M, Furumichi M, Tanabe M. 2013. Data, information, knowledge and principle: back to metabolism in KEGG. Nucleic Acids Res 42:D199–D205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nakao M, Okamoto S, Kohara M, Fujishiro T, Fujisawa T, Sato S, Tabata S, Kaneko T, Nakamura Y. 2009. CyanoBase: the cyanobacteria genome database update 2010. Nucleic Acids Res 38:D379–D381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brilli M, Fondi M, Fani R, Mengoni A, Ferri L, Bazzicalupo M, Biondi EG. 2010. The diversity and evolution of cell cycle regulation in alpha-proteobacteria: a comparative genomic analysis. BMC Syst Biol 4:52. doi: 10.1186/1752-0509-4-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Patrick WM, Quandt EM, Swartzlander DB, Matsumura I. 2007. Multicopy suppression underpins metabolic evolvability. Mol Biol Evol 24:2716–2722. doi: 10.1093/molbev/msm204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Joyce AR, Reed JL, White A, Edwards R, Osterman A, Baba T, Mori H, Lesely SA, Palsson BO, Agarwalla S. 2006. Experimental and computational assessment of conditionally essential genes in Escherichia coli. J Bacteriol 188:8259–8271. doi: 10.1128/JB.00740-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McAdams HH, Shapiro L. 2011. The architecture and conservation pattern of whole-cell control circuitry. J Mol Biol 409:28–35. doi: 10.1016/j.jmb.2011.02.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Panis G, Murray SR, Viollier PH. 2015. Versatility of global transcriptional regulators in alpha-Proteobacteria: from essential cell cycle control to ancillary functions. FEMS Microbiol Rev 39:120–133. doi: 10.1093/femsre/fuu002. [DOI] [PubMed] [Google Scholar]
- 44.Mueller JH, Johnson ER. 1941. Acid hydrolysates of casein to replace peptone in the preparation of bacteriological media. J Immunol 40:33–38. [Google Scholar]
- 45.McKinlay JB, Oda Y, Rühl M, Posto AL, Sauer U, Harwood CS. 2014. Non-growing Rhodopseudomonas palustris increases the hydrogen gas yield from acetate by shifting from the glyoxylate shunt to the tricarboxylic acid cycle. J Biol Chem 289:1960–1970. doi: 10.1074/jbc.M113.527515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Spero MA, Aylward FO, Currie CR, Donohue TJ. 2015. Phylogenomic analysis and predicted physiological role of the proton-translocating NADH:quinone oxidoreductase (complex I) across bacteria. mBio 6:e00389–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Foster JM, Davis PJ, Raverdy S, Sibley MH, Raleigh EA, Kumar S, Carlow CKS. 2010. Evolution of bacterial phosphoglycerate mutases: nonhomologous isofunctional enzymes undergoing gene losses, gains and lateral transfers. PLoS One 5:e13576. doi: 10.1371/journal.pone.0013576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sissler M, Delorme C, Bond J, Ehrlich SD, Renault P, Francklyn C. 1999. An aminoacyl-tRNA synthetase paralog with a catalytic role in histidine biosynthesis. Proc Natl Acad Sci U S A 96:8985–8990. doi: 10.1073/pnas.96.16.8985. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.