Abstract
Background
The thylakoid membrane of higher plant chloroplasts is made of membrane lipids synthesized in the chloroplast envelope. As the inner envelope membrane and the thylakoid are separated by the aqueous stroma, a system for transporting newly synthesized lipids from the inner envelope membrane to the thylakoid is required. Ultrastructural as well as biochemical studies have indicated that lipid transport inside the chloroplast could be mediated by a system similar in characteristics to vesicular trafficking in the cytosol. If indeed the chloroplast system is related to cytosolic vesicular trafficking systems, a certain degree of sequence conservation between components of the chloroplast and the cytosolic systems could be expected. We used the Arabidopsis thaliana genome and web-based subcellular localization prediction tools to search for chloroplast-localized homologues of cytosolic vesicular trafficking components.
Results
Out of the 28952 hypothetical proteins in the A. thaliana genome sequence, 1947 were predicted to be chloroplast-localized by two different subcellular localization predictors. In this chloroplast protein dataset, strong homologues for the main coat proteins of COPII coated cytosolic vesicles were found. Homologues of the small GTPases ARF1 and Sar1 were also found in the chloroplast protein dataset.
Conclusion
Our database search approach gives further support to that a system similar to cytosolic vesicular trafficking is operational inside the chloroplast. However, solid biochemical data is needed to support the chloroplast localization of the identified proteins as well as their involvment in intra-chloroplast lipid trafficking.
Background
The thylakoid membrane of higher plant chloroplasts contains a high proportion of galactolipids, which are synthesized in the chloroplast envelope membranes. Newly synthesized lipids are rapidly transported from the chloroplast envelope to the thylakoid membrane [1-4]. In theory, lipid transport across the aqeous stroma could be mediated by lipid transfer at sites of physical contact between the envelope and the thylakoid, by monomer diffusion facilitated by lipid transfer proteins or by a vesicular mechanism. Ultrastructural studies have failed to demonstrate any apparent physical contacts between the inner envelope and the thylakoid membrane in mature chloroplasts, and chloroplast-localized lipid transport proteins have not been demonstrated. Regarding a vesicular transfer mode, however, support comes from both ultrastructural and biochemical studies. When leaf tissue was incubated at low temperatures, vesicle-like structures accumulated in situ inside chloroplasts, in the stroma between the chloroplast envelope and thylakoid[5], similarily to the accumulation at low temperature of transitory vesicles between the endoplasmic reticulum (ER) and the cis-Golgi compartment in animal cells [6]. When the temperature was increased, from 12 to 21°C, the vesicles dissapeared. The low temperature-dependent accumulation of vesicles is considered to reflect that fusion of vesicles with the target membrane (in the examples above thylakoid and cis-Golgi, respectively) is blocked at a higher temperature than vesicle fission from the donor membrane (chloroplast envelope and ER, respectively) [5]. It was subsequently shown that the transfer of lipids from envelope to thylakoid in organello was strongly inhibited at the temperatures where vesicles accumulated in the stroma [4]. A cell-free reconstitution of lipid transport from envelope to thylakoid demonstrated a requirement for stromal proteins and ATP [7] and the release of lipids from isolated envelope required stromal proteins, ATP and GTP and was stimulated by acyl-CoA [8]. Vesicular structures were observed also in isolated chloroplasts and their abundance was affected by inhibitors of vesicular trafficking in the secretory pathway [9]. Vesicular trafficking in the secretory pathway is mediated by COPI, COPII and clathrin coated vesicles [10,11]. Coat assembly and vesicle formation in the secretory pathway is regulated by small GTP binding proteins, such as ARF and SAR, whereas correct targeting and fusion of cargo vesicles in the secretory pathway is mediated by syntaxins and small GTPases [12]. Although not studied in the same degree of detail, plant cytosolic vesicular trafficking seem to require essentially the same proteinaseous components as mammalian and yeast cytosolic vesicular trafficking [13-15]. The putative vesicular transport system in the chloroplast stroma thus appears to resemble, as inferred by the evidence at hand, the transport system between the ER and the Golgi apparatus. Given the biochemical characteristics, the molecular machinery behind intra-chloroplast vesicular transport could be evolutionary related to the machinery that drives vesicle trafficking in the secretory pathway. In addition to membrane lipids, the vesicles could also be expected to transport other hydrophobic substances, such as quinones and carotenoids, synthesized in the envelope membrane to the thylakoid [16]. Several studies on the unicellular algae Chlamydomonas reinhardtii also underline the importance of the inner envelope as biogenic structure for the thylakoid membrane, including photosytem assembly [17-19] and synthesis of chlorophyll b [20]. We aimed to find chloroplast-localized A. thaliana homologues to known components of cytosolic vesicular trafficking. The web based chloroplast localization prediction tools TargetP [21] and Predotar (version 0.5; http://www.inra.fr/predotar/) were used to extract putative chloroplast localized proteins from the dataset of full non-redundant A. thaliana predicted proteins. The resulting set of sequences was searched for putative vesicle trafficking components.
Results and discussion
Prediction of chloroplast-localized protein
Of the 28952 protein sequences in the non-redundant A. thaliana dataset, 4780 and 4582 were predicted to be chloroplast localized by TargetP and Predotar respectively (not regarding the different reliability classes). Of these, 1947 sequences were predicted to be chloroplast localized by both predictors. Combining the output from more than one predictor is likely to significantly reduce the number of false positives, but is also very likely to produce a significant number of false negatives. That this is in fact the case has been experimentally shown for the mitochondrial proteome [22]. Our dataset contained 202 of 362 experimentally verified envelope proteins [23] and 128 of 213 experimentally identified thylakoid-localized proteins [24]. Subsequently, we added the "missing" sequences to our dataset. Finally, we also added the sequences for all the 88 predicted chloroplast encoded proteins. We tested our chloroplast protein sequence dataset for the presence of some well established chloroplast proteins and found that e.g. the rubisco small subunit, light harvesting complex proteins and protochlorophyllide oxidoreductase were represented in the dataset.
Vesicle budding components
Vesicle budding in the secretory pathway is mediated by the assembly of three different kinds of protein coats, COPII, COPI or clathrin. Formation of COPII-coated vesicles from isolated ER or chemically defined liposomes has been shown to require three soluble cytosolic components, the Sec13-Sec31 complex, the Sec24-Sec23 complex and the small GTPase Sar1 [25]. The formation of COPI-coated vesicles requires the presence of two soluble complexes consisting of a total five different subunits [11,26] and is regulated by the small GTPase ARF. Assembly of clathrin coats is similarly controlled by small GTPases and requires the presence of clathrin monomers and adaptins that link the clathrin coat to activated cargo receptors in the vesicle bud [11]. A simple BLAST search against the full A. thaliana peptide dataset retrieved highly conserved homologues of the sequences for the key components of the three different vesicle coats in yeast (Saccharomyces cerevisae; Table 1). The identified sequences agree well with previously published studies on protein components of higher plant cytosolic vesicular trafficking [14,15,27]. Having established the conservation of the cytosolic vesicle coats between yeast and A. thaliana, our next step was to search the putative chloroplast protein sequence dataset for vesicle coat components.
Table 1.
Putative A. thaliana homologues of yeast vesicular trafficking components
| S. cerevisaea vesicular trafficking component (predicted size in kD) | A. thaliana homologue | BLAST-score (%identity / %positives) | E-value |
| Sec13 | At3g01340 | 265 (47/63) | 2e-071 |
| At2g30050 | 258 (46/61) | 3e-069 | |
| Sec31 | At3g63460 | 236 (23/41) | 2e-061 |
| At1g18830 | 228 (23/43) | 2e-059 | |
| Sec23 | At4g14160 | 577 (39/58) | 1e-165 |
| At1g05520 | 577 (40/57) | 1e-165 | |
| At3g23660 | 574 (39/58) | 1e-164 | |
| At5g43670 | 545 (37/55) | 1e-155 | |
| At2g21630 | 530 (38/56) | 1e-150 | |
| Sec24 | At3g07100 | 355 (32/52) | 5e-098 |
| Sar1 | At1g56330 | 237 (63/75) | 3e-063 |
| At4g02080 | 232 (63/74) | 1e-061 | |
| At3g62560 | 229 (62/75) | 5e-061 | |
| At1g09180 | 220 (60/72) | 4e-058 | |
| At1g02620 | 113 (55/70) | 7e-026 | |
| Ret1 | At1g62020 | 936 (41/59) | 0.0 |
| At2g21390 | 932 (42/59) | 0.0 | |
| Sec26 | At4g31490 | 650 (39/59) | 0.0 |
| At4g31480 | 650 (38/58) | 0.0 | |
| Sec27 | At1g79990 | 751 (47/66) | 0.0 |
| At1g52360 | 749 (46/67) | 0.0 | |
| At3g15980 | 742 (46/66) | 0.0 | |
| Sec21 | At4g34450 | 488 (35/54) | e-138 |
| Ret2 | At5g05010 | 131 (41/71) | 9e-031 |
| Sec28 | At2g34780 | 35 (24/41) | 0.067 |
| Ret3 | At1g60970 | 95 (34/59) | 3e-020 |
| At3g09800 | 90 (33/54) | 9e-019 | |
| At4g08520 | 89 (32/57) | 2e-018 | |
| Arf1 | At3g62290 | 296 (78/89) | 4e-081 |
| Clathrin | At3g08530 | 1419 (42/66) | 0.0 |
| heavy chain | At3g11130 | 1408 (42/65) | 0.0 |
| Clathrin light chain | At3g28770 | 41 (19/38) | 7e-004 |
We found strong homologues for all the COPII coat subunits in our chloroplast protein sequence dataset (Table 2). The similarities were in general extensive throughout the length of the proteins. Thus the low E-values were not only results of short stretches of nearly exact matches. In particular, yeast Sec23 and Sec24 had extremely good homologues in our chloroplast dataset. Alignment of the chloroplast localized Sec13, 23 and 24 with their yeast homologues as well as the best match from the full A. thaliana peptide dataset, revealed N-terminal extensions in the chloroplast-localized peptides (Fig. 1 and 2 and not shown) that may well correspond to the N-terminal targeting sequences required for post-translational import of the protein into the chloroplast. The putative chloroplast-localized Sec31 homologues, however, were both shorter than the yeast query sequence and the overall homology was much lower than for the other putative chloroplast COPII subunits. The chloroplast-localized COPII subunit homologues are all probably highly expressed, judging by the number of ESTs present in the TAIR database (Table 2).
Table 2.
Putative chloroplast-localized A. thaliana homologues of yeast COPII coat proteins.
| S. cerevisaea COPII component (predicted size in kD) | A. thaliana chloroplast homologue | BLAST-score (%identity / %positives) | E-value | Proposed name | Predicted size (kD) | Number of ESTs | TargetP reliability class (transit peptide length) |
| Sec13 (33) | At3g49660.1 | 65 (26/40) | 5e-012 | cpSec13a | 49.6 | 7 | 3 (41) |
| Sec31 (139) | At5g38560.1 | 54 (27/37) | 5e-008 | cpSec31a | 72.3 | 12 | 2 (13) |
| At2g45000.1 | 51 (22/36) | 4e-007 | cpSec31b | 73.5 | 1 | 1 (27) | |
| Sec23 (85) | At4g01810.1 | 100 (20/36) | 2e-022 | cpSec23 | 95.9 | 26 | 3 (8) |
| Sec24 (104) | At3g44340.1 | 279 (26/47) | 4e-076 | cpSec24a | 117.7 | 22 | 2 (27) |
| At4g32640.1 | 276 (28/48) | 2e-075 | cpSec24b | 114.7 | 8 | 3 (47) |
Figure 1.
Multiple alignment of the yeast Sec23, the best match in the whole A. thaliana proteome At4g14160.2 and the best match in the predicted chloroplast proteome At4g01810.1. Identical residues are shown in black and conserved residues are shown in gray.
Figure 2.
Multiple alignment of the yeast Sec24, the best match in the whole A. thaliana proteome At3g07100.1 and the two best matches in the predicted chloroplast proteome At3g44340.1 and At4g32640.1. Identical residues are shown in black and conserved residues are shown in gray.
BLAST searches retrieved no significant hits for five out of seven yeast COPI coat subunits, yeast light and heavy clathrin subunits nor any yeast adaptin. The two yeast COPI subunits that did retrieve significant hits (Ret1 and Sec27), retrieved the same polypeptides as did yeast Sec13 and Sec31. To rule out cyanobacterial origin of the chloroplast-localized COPII coat components, the genome of the cyanobacteria Synecocystis sp. PCC 6803 was searched for homologues of the yeast COPII subunits. This search yielded only very short stretches of matching sequence; nothing like full length conserved proteins emerged (not shown). These data fit with a recent suggestion, that lack of observable vesicular structures in photosynthetic organisms beside the embryophytes point to that the vesicular transport system was acquired by the chloroplast from the host eucaryotic cell rather than having evolved from a cyanobacterial mechanism [28].
Membrane fission
The polymerization of the COPI and COPII coats provides force enough to induce deformation and eventually the fission of membrane vesicles from isolated membranes or liposomes [11,25,26]. The formation of clathrin-coated vesicles, however, requires the presence of the GTPase dynamin [10]. It is believed that dynamin polymerizes to form a ring, which through GTP hydrolysis pinches off the transport vesicle [29]. Dynamin homologue(s) have been experimentally shown to be chloroplast-localized in A. thaliana [30]. The Arabidopsis Dynamin Like Protein 2 (ADL2) was predicted to be chloroplast-localized by TargetP but not by Predotar. A BLAST search with various dynamin sequences from several organisms did not retrieve any new dynamin homologues from our chloroplast protein sequence dataset.
Vesicle targeting and fusion
Vesicle targeting and fusion in the secretory pathway is thought to be mediated by SNARE proteins and regulated by RAB GTPases [11]. A few putative chloroplast-localized SNAREs were identified by a BLAST search of the sequences for chloroplast-targeted proteins against the sequences of several yeast and human SNAREs (not shown). Cytoplasmic SNAREs are generally membrane-spanning proteins (with the exception of one; SNAP-25). The web based membrane spanning helix prediction service TopPred [31] predicts one specific membrane spanning helix in all the different yeast and human SNAREs used for the BLAST search. However, of the putative chloroplast-localized SNAREs, only two contained putative trans-membrane helices and these were not in the same regions as in the query SNAREs. When the putative chloroplast-localized SNAREs were used as query sequences for BLAST searching GeneBank, no SNAREs were retrieved. All of the found proteins were much larger than any of the tested authentic SNAREs. Apparently, no authentic SNARE sequences were present in the chloroplast protein sequence dataset. This could be due to that no SNARES actually are present in the chloroplast or to that the SNAREs simply were missed in the prediction of subcellular localization.
Small GTPases
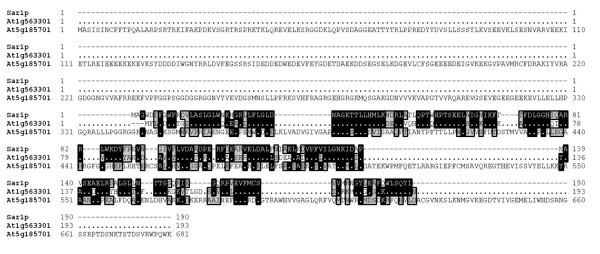
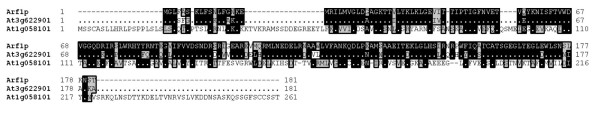
Both vesicle budding and fusion in the secretory pathway are regulated by small GTPases [10,11]. Formation of COPII coated vesicles is regulated by the GTPase SAR, whereas formation of COPI and clathrin coated vesicles is regulated by ARF GTPases. We could retrieve one homologue of yeast Sar1 from the A. thaliana chloroplast protein sequence dataset (Table 3). The putative A. thaliana chloroplast Sar1 homologue sequence was predicted to encode a protein substantially larger than yeast Sar1. Alignment of yeast Sar1, A. thaliana cytoplasmic Sar1 and the chloroplast Sar1 homologue reveals that the Sar1 homology is situated in the C-terminus of the chloroplast Sar1 sequence (Fig. 3). The chloroplast protein sequence dataset contained one protein sequence with significant homology to ARF1 (Table 3). The predicted size of this protein was quite similar in size to the yeast sequence. Alignment of the yeast ARF1, the A. thaliana cytoplasmic homologue and the chloroplast homologue revealed an N-terminal extension in the chloroplast homologue (Fig. 4), again pointing to an N-terminal extension that targets the protein to the chloroplast. Overall similarity was much higher between ARF1 and its chloroplast homologue than between Sar1 and its putative chloroplast homologue. The chloroplast Sar1 homologue is probably a GTPase, but whether it performs the same functions as Sar1 appears less certain.
Table 3.
Putative chloroplast-localized A. thaliana homologues of yeast small GTPases.
| S. cerevisaea small GTPase (predicted size in kD) | A. thaliana chloroplast homologue | BLAST-score (%identity / %positives) | E-value | Proposed name | Predicted size (kD) | Number of ESTs | TargetP reliability class (transit peptide length) |
| Sar1 (21) | At5g18570.1 | 37 (25/42) | 8e-004 | cpSar1a | 75.6 | 5 | 1 (32) |
| ARF1 (21) | At1g05810.1 | 47 (27/48) | 5e-007 | cpARF1 | 24.2 | 3 | 3 (42) |
Figure 3.
Multiple alignment of the yeast Sar1, the best match in the whole A. thaliana proteome At1g56330.1 and the best match in the predicted chloroplast proteome At5g18570.1. Identical residues are shown in black and conserved residues are shown in gray.
Figure 4.
Multiple alignment of the yeast Arf1, the best match in the whole A. thaliana proteome At3g62290.1 and the best match in the predicted chloroplast proteome At1g05810.1. Identical residues are shown in black and conserved residues are shown in gray.
Conclusions
Morphological [5,9,28], and biochemical evidence [4,7,8] suggest that lipid transport from the chloroplast envelope to the thylakoid membrane is mediated by a vesicular transport mechanism. The bio-informatics data presented herein suggest that homologues of the components required for formation of COPII coated vesicles are present in the chloroplasts of higher plants. Biochemical studies of the identified components and A. thaliana T-DNA insertion lines will establish whether the proteins identified in the present study are in fact involved in intra-chloroplast lipid trafficking. The one component of the minimum vesicular transport system still missing is a SNARE. On the other hand fusion may be mediated by other mechanisms beside SNARE-SNARE interaction [12]. SNAREs in the secretory pathway are activated by NSF (N-ethylmaleimide sensitive factor) [12]. A NSF homologue has been cloned from chromoplasts isolated from red pepper and was found to be required for fusion of chromoplast inner membrane vesicles [32]. This indicates that a system similar to the SNARE system in cytosolic vesicular trafficking nevertheless mediates fusion events inside the chloroplast.
Methods
The non-redundant A. thaliana protein dataset (file: ATH1.pep) was retrieved from the TIGR homepage http://www.tigr.org. The full dataset comprising 28 952 sequences was divided into sets of 1400 sequences each. The sets were submitted to TargetP http://www.cbs.dtu.dk/services/TargetP/ and Predotar http://www.inra.fr/predotar/. Sequences predicted to be chloroplast-localized by both predictors were compiled and used for local BLAST searches using the BLAST software package [33] downloaded from the NCBI web page ftp://ftp.ncbi.nih.gov/blast/. S. cerevisae and Homo sapiens sequences for components of cytoplasmic vesicle transport were downloaded from Gene Bank. Sequence alignment was performed with ClustalW [34,35] at the web service offered by PBIL http://npsa-pbil.ibcp.fr/. Sequences were compiled and edited with the BioEdit software package. The datasets used in this study will be made available upon request.
Authors' contributions
MA conceived the study and performed the bioinformatics work. MA and ASS contributed equally to the interpretation of the results and the preparation of the manuscript.
Contributor Information
Mats X Andersson, Email: mats.andersson@botany.gu.se.
Anna Stina Sandelius, Email: annastina.sandelius@botany.gu.se.
References
- Siebertz HP, Heinz E, Joyard J, Douce R. Labeling in vivo and in vitro of molecular species of lipids from chloroplast envelopes and thylakoids. European Journal of Biochemistry. 1980;108:177–186. doi: 10.1111/j.1432-1033.1980.tb04710.x. [DOI] [PubMed] [Google Scholar]
- Rawyler A, Meylan M, Siegenthaler PA. Galactolipid export from envelope to thylakoid membranes in intact chloroplasts 1. Characterization and involvement in thylakoid lipid asymmetry. Biochimica Et Biophysica Acta. 1992;1104:331–341. doi: 10.1016/0005-2736(92)90048-Q. [DOI] [PubMed] [Google Scholar]
- Rawyler A, Meylanbettex M, Siegenthaler PA. (Galacto)Lipid export from envelope to thylakoid membranes in intact chloroplasts 2. A general process with a key role for the envelope in the establishment of lipid asymmetry in thylakoid membranes. Biochimica Et Biophysica Acta. 1995;1233:123–133. doi: 10.1016/0005-2736(94)00248-N. [DOI] [PubMed] [Google Scholar]
- Andersson MX, Kjellberg JM, Sandelius AS. Chloroplast biogenesis. Regulation of lipid transport to the thylakoid in chloroplasts isolated from expanding and fully expanded leaves of pea. Plant Physiology. 2001;127:184–193. doi: 10.1104/pp.127.1.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morré DJ, Selldén G, Sundqvist C, Sandelius AS. Stromal low-temperature compartment derived from the inner membrane of the chloroplast envelope. Plant Physiology. 1991;97:1558–1564. doi: 10.1104/pp.97.4.1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreau P, Juguelin H, Cassagne C, Morré DJ. Molecular basis for low-temperature compartment formation by transitional endoplasmic reticulum of rat liver. Febs Letters. 1992;310:223–228. doi: 10.1016/0014-5793(92)81337-L. [DOI] [PubMed] [Google Scholar]
- Morré DJ, Morré JT, Morré SR, Sundqvist C, Sandelius AS. Chloroplast biogenesis – Cell-free transfer of envelope monogalactosylglycerides to thylakoids. Biochimica Et Biophysica Acta. 1991;1070:437–445. doi: 10.1016/0005-2736(91)90084-l. [DOI] [PubMed] [Google Scholar]
- Räntfors M, Evertsson I, Kjellberg JM, Sandelius AS. Intraplastidial lipid trafficking: Regulation of galactolipid release from isolated chloroplast envelope. Physiologia Plantarum. 2000;110:262–270. doi: 10.1034/j.1399-3054.2000.110217.x. [DOI] [Google Scholar]
- Westphal S, Soll J, Vothknecht UC. A vesicle transport system inside chloroplasts. Febs Letters. 2001;506:257–261. doi: 10.1016/S0014-5793(01)02931-3. [DOI] [PubMed] [Google Scholar]
- Kirchhausen T. Three ways to make a vesicle. Nature Reviews Molecular Cell Biology. 2001;1:187–198. doi: 10.1038/35043117. [DOI] [PubMed] [Google Scholar]
- Bonifacino JS, Glick BS. The mechanisms of vesicle budding and fusion. Cell. 2004;116:153–166. doi: 10.1016/S0092-8674(03)01079-1. [DOI] [PubMed] [Google Scholar]
- Mayer A. Membrane fusion in eukaryotic cells. Annual Review of Cell and Developmental Biology. 2002;18:289–314. doi: 10.1146/annurev.cellbio.18.032202.114809. [DOI] [PubMed] [Google Scholar]
- Nebenfuhr A. Vesicle traffic in the endomembrane system: a tale of COPs, Rabs and SNAREs. Current Opinion in Plant Biology. 2002;5:507–512. doi: 10.1016/S1369-5266(02)00303-5. [DOI] [PubMed] [Google Scholar]
- Pimpl P, Movafeghi A, Coughlan S, Denecke J, Hillmer S, Robinson DG. In situ localization and in vitro induction of plant COPI-coated vesicles. Plant Cell. 2000;12:2219–2235. doi: 10.1105/tpc.12.11.2219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Movafeghi A, Happel N, Pimpl P, Tai GH, Robinson DG. Arabidopsis Sec21p and Sec23p homologs. Probable coat proteins of plant COP-coated vesicles. Plant Physiology. 1999;119:1437–1445. doi: 10.1104/pp.119.4.1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joyard J, Teyssier E, Miege C, Berny-Seigneurin D, Maréchal E, Block MA, Dorne AJ, Rolland N, Ajlani G, Douce R. The biochemical machinery of plastid envelope membranes. Plant Physiology. 1998;118:715–723. doi: 10.1104/pp.118.3.715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoober JK, Eggink LL. Assembly of light-harvesting complex II and biogenesis of thylakoid membranes in chloroplasts. Photosynthesis Research. 1999;61:197–215. doi: 10.1023/A:1006313703640. [DOI] [Google Scholar]
- Hoober JK, Boyd CO, Paavola LG. Origin of thylakoid membranes in Chlamydomonas-reinhardtii Y-1 at 38°C. Plant Physiology. 1991;96:1321–1328. doi: 10.1104/pp.96.4.1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoober JK, White RA, Marks DB, Gabriel JL. Biogenesis of thylakoid membranes with emphasis on the process in Chlamydomonas. Photosynthesis Research. 1994;39:15–31. doi: 10.1007/BF00027139. [DOI] [PubMed] [Google Scholar]
- Eggink L, LoBrutto R, Brune D, Brusslan J, Yamasato A, Tanaka A, Hoober JK. Synthesis of chlorophyll b: Localization of chlorophyllide a oxygenase and discovery of a stable radical in the catalytic subunit. BMC Plant Biology. 2004;4:5. doi: 10.1186/1471-2229-4-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuelsson O, Nielsen H, Brunak S, von Heijne G. Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. Journal of Molecular Biology. 2000;300:1005–1016. doi: 10.1006/jmbi.2000.3903. [DOI] [PubMed] [Google Scholar]
- Heazlewood JL, Tonti-Filippini JS, Gout AM, Day DA, Whelan J, Millar AH. Experimental analysis of the Arabidopsis mitochondrial proteome highlights signaling and regulatory components, provides assessment of targeting prediction programs, and indicates plant-specific mitochondrial proteins. Plant Cell. 2004;16:241–256. doi: 10.1105/tpc.016055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Froehlich JE, Wilkerson CG, Ray WK, McAndrew RS, Osteryoung KW, Gage DA, Phinney BS. Proteomic study of the Arabidopsis thaliana chloroplastic envelope membrane utilizing alternatives to traditional two-dimensional electrophoresis. Journal of Proteome Research. 2003;2:413–425. doi: 10.1021/pr034025j. [DOI] [PubMed] [Google Scholar]
- Friso G, Giacomelli L, Ytterberg AJ, Peltier J-B, Rudella A, Sun Q, Wijk KJv. In-depth analysis of the thylakoid membrane proteome of Arabidopsis thaliana chloroplasts: New proteins, new functions, and a plastid proteome database. Plant Cell. 2004;16:478–499. doi: 10.1105/tpc.017814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuoka K, Orci L, Amherdt M, Bednarek SY, Hamamoto S, Schekman R, Yeung T. COPII-coated vesicle formation reconstituted with purified coat proteins and chemically defined liposomes. Cell. 1998;93:263–275. doi: 10.1016/S0092-8674(00)81577-9. [DOI] [PubMed] [Google Scholar]
- Spang A, Matsuoka K, Hamamoto S, Schekman R, Orci L. Coatomer, Arf1p, and nucleotide are required to bud coat protein complex I-coated vesicles from large synthetic liposomes. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:11199–11204. doi: 10.1073/pnas.95.19.11199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreeva AV, Kutuzov MA, Evans DE, Hawes CR. Proteins involved in membrane transport between the ER and the Golgi apparatus: 21 putative plant homologues revealed by dbEST searching. Cell Biology International. 1998;22:145–160. doi: 10.1006/cbir.1998.0235. [DOI] [PubMed] [Google Scholar]
- Westphal S, Soll J, Vothknecht UC. Evolution of chloroplast vesicle transport. Plant and Cell Physiology. 2003;44:217–222. doi: 10.1093/pcp/pcg023. [DOI] [PubMed] [Google Scholar]
- Praefcke GJK, McMahon HT. The dynamin superfamily: Universal membrane tubulation and fission molecules? Nature Reviews Molecular Cell Biology. 2004;5:133–147. doi: 10.1038/nrm1313. [DOI] [PubMed] [Google Scholar]
- Park JM, Cho JH, Kang SG, Jang HJ, Pih KT, Piao HL, Cho MJ, Hwang I. A dynamin-like protein in Arabidopsis thaliana is involved in biogenesis of thylakoid membranes. EMBO European Molecular Biology Organization Journal. 1998;17:859–867. doi: 10.1093/emboj/17.4.859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claros MG, VonHeijne G. Toppred-II – an improved software for membrane-protein structure predictions. Computer Applications in the Biosciences. 1994;10:685–686. doi: 10.1093/bioinformatics/10.6.685. [DOI] [PubMed] [Google Scholar]
- Hugueney P, Bouvier F, Badillo A, Dharlingue A, Kuntz M, Camara B. Identification of a plastid protein involved in vesicle fusion and/or membrane-protein translocation. Proceedings of the National Academy of Sciences of the United States of America. 1995;92:5630–5634. doi: 10.1073/pnas.92.12.5630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. Journal of Molecular Biology. 1990;215:403–410. doi: 10.1006/jmbi.1990.9999. [DOI] [PubMed] [Google Scholar]
- Higgins DG, Thompson JD, Gibson TJ. Using CLUSTAL for multiple sequence alignments. Computer Methods for Macromolecular Sequence Analysis. 1996;266:383–402. doi: 10.1016/S0076-6879(96)66024-8. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. Clustal-W – Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Research. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]