ABSTRACT
Latent infection of B lymphocytes by Epstein-Barr virus (EBV) in vitro results in their immortalization into lymphoblastoid cell lines (LCLs); this latency program is controlled by the EBNA2 viral transcriptional activator, which targets promoters via RBPJ, a DNA binding protein in the Notch signaling pathway. Three other EBNA3 proteins (EBNA3A, EBNA3B, and EBNA3C) interact with RBPJ to regulate cell gene expression. The mechanism by which EBNAs regulate different genes via RBPJ remains unclear. Our chromatin immunoprecipitation with deep sequencing (ChIP-seq) analysis of the EBNA3 proteins analyzed in concert with prior EBNA2 and RBPJ data demonstrated that EBNA3A, EBNA3B, and EBNA3C bind to distinct, partially overlapping genomic locations. Although RBPJ interaction is critical for EBNA3A and EBNA3C growth effects, only 30 to 40% of EBNA3-bound sites colocalize with RBPJ. Using LCLs conditional for EBNA3A or EBNA3C activity, we demonstrate that EBNA2 binding at sites near EBNA3A- or EBNA3C-regulated genes is specifically regulated by the respective EBNA3. To investigate EBNA3 binding specificity, we identified sequences and transcription factors enriched at EBNA3A-, EBNA3B-, and EBNA3C-bound sites. This confirmed the prior observation that IRF4 is enriched at EBNA3A- and EBNA3C-bound sites and revealed IRF4 enrichment at EBNA3B-bound sites. Using IRF4-negative BJAB cells, we demonstrate that IRF4 is essential for EBNA3C, but not EBNA3A or EBNA3B, binding to specific sites. These results support a model in which EBNA2 and EBNA3s compete for distinct subsets of RBPJ sites to regulate cell genes and where EBNA3 subset specificity is determined by interactions with other cell transcription factors.
IMPORTANCE Epstein-Barr virus (EBV) latent gene products cause human cancers and transform B lymphocytes into immortalized lymphoblastoid cell lines in vitro. EBV nuclear antigens (EBNAs) and membrane proteins constitutively activate pathways important for lymphocyte growth and survival. An important unresolved question is how four different EBNAs (EBNA2, -3A, -3B, and -3C) exert unique effects via a single transcription factor, RBPJ. Here, we report that each EBNA binds to distinct but partially overlapping sets of genomic sites. EBNA3A and EBNA3C specifically regulate EBNA2's access to different RBPJ sites, providing a mechanism by which each EBNA can regulate distinct cell genes. We show that IRF4, an essential regulator of B cell differentiation, is critical for EBNA3C binding specificity; EBNA3A and EBNA3B specificities are likely due to interactions with other cell transcription factors. EBNA3 titration of EBNA2 transcriptional function at distinct sites likely limits cell defenses that would be triggered by unchecked EBNA2 prooncogenic activity.
INTRODUCTION
Epstein-Barr virus (EBV) is a herpesvirus that infects over 90% of the population by adulthood. Primary EBV infection usually presents as a nonspecific illness in early childhood but often manifests as infectious mononucleosis in adolescents (1). Thereafter, EBV establishes lifelong latent infection in B lymphocytes and periodically reactivates and is shed in saliva. Rarely, EBV latent infection results in malignancy, including Burkitt and Hodgkin lymphomas, lymphoproliferative disease, nasopharyngeal carcinoma, and gastric cancer. Much of our knowledge of the transforming effects of EBV latent genes derives from the study of EBV latent infection of B lymphocytes in vitro, which results in their growth and transformation into lymphoblastoid cell lines (LCLs). Extensive investigation of the effects of EBV latent genes in LCLs has demonstrated that they constitutively activate growth and survival signals essential for normal B cell development, including the CD40 and B cell receptors (BCR) (reviewed in reference 2).
EBV nuclear antigens (EBNAs) are proteins expressed during latent infection that extensively target the Notch signaling pathway. EBNA2 is a strong transcriptional activator that is targeted to promoters through an interaction with the RBPJ DNA binding protein that normally mediates intracellular Notch (ICN) binding (3, 4). EBNA2 upregulates the other EBV latent gene products, as well as cell oncogenes, such as c-myc, required for LCL growth (5–8). EBNA2 effects are substantially similar, but not identical, to those of ICN (9–12). Unlike that of ICN, EBNA2 activation is constitutive and ligand independent. Remarkably, three other EBV nuclear proteins, EBNA3A, EBNA3B, and EBNA3C, associate with RBPJ in LCLs. The EBNA3 proteins bind RBPJ through their highly homologous N-terminal regions and are thought to have arisen from the triplication of a single ancestral gene (13, 14). Although EBNA3s bind an RBPJ domain that is distinct from the EBNA2/ICN binding site, they nevertheless limit EBNA2 activation by competing for RBPJ binding (15–18). In LCLs and Burkitt lymphoma tumor cells, the EBNA3 proteins have been shown to regulate distinct but extensively overlapping sets of cell genes (19–26). EBNA3 proteins have been implicated in the pathogenesis of Burkitt lymphoma and in attenuating an antiproliferative DNA damage response during EBV transformation of primary B lymphocytes (19, 27–29). Moreover, EBNA3A and EBNA3C repression of the CDKN2A-encoded tumor suppressors p16 and p14 is essential for LCL growth, requires interaction with RBPJ, and is associated with increased H3K27me3 modification at the CDKN2A promoter (22, 23, 30–32).
Despite significant advances in our understanding of the role of EBNA3 proteins in LCL growth, the basis for their different effects via RBPJ remains an area of active investigation. The selectivity in gene regulation suggests that EBNA3 proteins either target different RBPJ-bound sites or exert different effects at the same sites. Chromatin immunoprecipitation (ChIP) experiments have led to a rapid advance in our understanding of this problem. McClellan et al. demonstrated that EBNA3 proteins bound to sites distinct from those of EBNA2 in the Mutu III Burkitt lymphoma cell line (33). However, due to cross-reactivity of the chromatin immunoprecipitation with deep sequencing (ChIP-seq) antibody for multiple EBNA3 proteins, they could only distinguish between EBNA3A, EBNA3B, and EBNA3C at specific EBNA3-bound sites by ChIP-quantitative PCR (qPCR). To overcome this limitation, we derived LCLs that express a single EBNA3 protein with a C-terminal Flag-hemagglutinin (HA) tag (34). Using our EBNA3C-Flag-HA LCLs, Jiang et al. reported that EBNA3C colocalized to BATF/IRF4/Spi1/Runx3 sites and observed that EBNA3C binding signals were stronger at IRF4-cobound sites than at sites without IRF4 binding. Remarkably, EBNA3A-bound sites were also found to be enriched for many of the same factors, although ChIP-re-ChIP experiments supported a critical role for BATF in EBNA3A binding (35). To extend these results, we performed new ChIP-seq experiments from the EBNA3A-Flag-HA, EBNA3B-Flag-HA, and EBNA3C-Flag-HA LCLs; analyzed them concurrently; and now report the first comprehensive analysis of EBNA3 binding in LCLs. Our results demonstrate that EBNA3 proteins bind distinct but partially overlapping genomic sites. In contrast to EBNA2, which exhibits extensive cobinding with RBPJ, only 30 to 40% of EBNA3 sites are also bound by RBPJ. However, we demonstrate that EBNA3A and EBNA3C regulate binding of EBNA2 at distinct sites and do not appear to compete globally with EBNA2 for RBPJ binding. Importantly, we present the first direct evidence that IRF4 is a critical mediator of EBNA3C, but not EBNA3A or EBNA3B, binding to genomic sites. Collectively our results support a model in which EBNA3 proteins regulate partially overlapping but distinct subsets of cell genes via interactions with RBPJ and other cell transcription factors (TFs), such as IRF4, and these non-RBPJ factors are the primary determinants of subset specificity.
MATERIALS AND METHODS
Plasmids.
pCEP-EBNA3A-F-HA, pCEP-EBNA3B-F-HA, and pCEP-EBNA3C-F-HA plasmids were constructed by recloning the EBNA3 C-terminal Flag-HA fusions used to make EBV recombinants into pCEP-EBNA3A, -3B, and -3C, respectively. pVxy-puro-IRF4 (36), a kind gift from Lixin Rui, University of Wisconsin—Madison, expresses the human IRF4 gene, contains a PGK promoter-driven puromycin resistance gene, and was fully sequenced prior to use.
Cell lines.
LCLs expressing EBNA3A-HT (E3A-HT), EBNA3C-HT (E3C-HT), EBNA3A-F-HA, EBNA3B-F-HA, or EBNA3C-F-HA have been previously described (32, 34, 37). BJAB is an EBV-negative BL cell line (38). B cell lines were maintained in RPMI 1640 medium supplemented with 10% fetal bovine serum, streptomycin, and penicillin; for E3A-HT and E3C-HT LCLs, the permissive conditions included addition of 400 nM 4-hydroxytamoxifen (4HT) (Sigma). 293T, a human cell line transformed by adenovirus 5 and simian virus 40 (SV40) large T antigen (39), was cultured in Dulbecco's modified Eagle's medium (Gibco) supplemented with 10% fetal bovine serum, streptomycin, and penicillin. Stable cell lines were generated by first introducing pCEP-EBNA3A-F-HA, pCEP-EBNA3B-F-HA, or pCEP-EBNA3C-F-HA plasmids into BJAB cells, followed by hygromycin (600 μg/ml) selection. Hygromycin-resistant clones were then infected with a retrovirus expressing IRF4 or an empty-vector control, followed by puromycin (0.4 μg/ml) selection. Hygromycin- and puromycin-resistant clones were screened by Western blotting for expression of the appropriate transgenes and maintained in RPMI 1640 medium supplemented with 10% fetal bovine serum, streptomycin, penicillin, 300 μg/ml hygromycin, and 0.2 μg/ml puromycin.
Antibodies.
The following antibodies were used for ChIP-qPCR and Western blotting: anti-HA-tag monoclonal antibody (MAb) magnetic beads (M132-9; MBL), anti-EBNA2 PE2 and anti-EBNA3C A10 mouse monoclonal antibodies (40), anti-EBNA3A antibody (F115P; Exalpha Biologicals), anti-RBPJ rabbit polyclonal sera (3), anti-HA.11 (16B12; Covance), anti-IRF4 antibody (SC-6059), and anti-alpha-tubulin (B-5-1-2; Sigma). The HA probe antibody F-7 (sc-7392; Santa Cruz) was used for ChIP-seq.
Transfection.
BJAB cells (2 × 106) were harvested during log-phase growth, washed with phosphate-buffered saline (PBS), resuspended in 100 μl of buffer V (Lonza), and transferred in a 2-cm cuvette after addition of 2 μg of appropriate plasmid DNA. Program M-013 of Amaxa Nucleofector (Lonza) was used, and the cells were resuspended in RPMI 1640 supplemented with 10% fetal bovine serum, streptomycin, and penicillin after transfection and cultured in a 6-well plate. 293T cells were transfected using Effectene (Qiagen) in a 10-cm culture dish with retroviral packing plasmids, as previously described (41).
Western blot analysis.
Total cell lysates and immunoprecipitated proteins were separated by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis, transferred onto a nitrocellulose membrane, and blotted with appropriate antibodies. After extensive washing, the membrane was probed with horseradish peroxidase-conjugated secondary antibodies (Jackson Immuno Research). The membrane was washed again after 1 h incubation and developed with chemiluminescence reagent (PerkinElmer).
ChIP-qPCR assays.
Cells (2 × 107) were cross-linked in 1% (wt/vol) formaldehyde (Sigma) for 5 min at room temperature, and the cross-linking reaction was stopped by addition of glycine to a final concentration of 0.125 M. The cells were washed twice with cold PBS and lysed in 1 ml of lysis buffer (50 mM Tris-HCl [pH 8.1], 10 mM EDTA, 1% [wt/vol] SDS, 1 mM phenylmethylsulfonyl fluoride [PMSF], 1 μg/ml leupeptin, 20 μg/ml aprotinin) for 30 min on ice before extensive sonication using a Qsonica LLC Q700 sonicator to an average fragment size of 200 to 500 bp. After extract clearing by centrifugation, the supernatants were diluted 1:10 in dilution buffer (16.7 mM Tris-HCl [pH 8.1], 1.2 mM EDTA, 167 mM NaCl, 1.1% [vol/vol] Triton X-100, 0.01% [wt/vol] SDS, 1 mM PMSF, 1 μg/ml leupeptin, 20 μg/ml aprotinin). About 10% of the chromatin lysate was reserved for qPCR and not subjected to any further manipulation. One milliliter of diluted chromatin lysates was incubated with ChIP antibodies with rotation at 4°C overnight; 15 μl protein A/G magnetic beads was added to each 1 ml chromatin lysate and incubated for 1 h at 4°C with rotation. After incubation, protein A/G magnetic beads were pelleted with a magnetic separation rack and then washed once with cold low-salt wash buffer (20 mM Tris-HCl [pH 8.1], 2 mM EDTA, 150 mM NaCl, 1% [vol/vol] Triton X-100, 0.1% [wt/vol] SDS), once with high-salt wash buffer (identical to low-salt wash buffer, except with 500 mM NaCl), once with LiCl wash buffer (10 mM Tris-HCl [pH 8.1], 1 mM EDTA, 0.25 M LiCl, 1% [vol/vol] NP-40, 1% deoxycholic acid), and finally twice with TE buffer (10 mM Tris-HCl [pH 8.1], 1 mM EDTA). Samples were then resuspended in 150 μl of elution buffer (0.1 M NaHCO3, 1% [wt/vol] SDS) and rotated for 20 min at room temperature. Two elutions of protein-DNA complexes were performed and pooled. NaCl and proteinase K were added to each ChIP DNA sample and input to a final concentration of 200 mM and 100 μg/ml, respectively. The ChIP DNA samples were then reverse cross-linked at 65°C for 4 h. DNA was purified using a QIAquick PCR purification kit (28706; Qiagen) and quantified with iTaq universal SYBR green supermix (1725121; Bio-Rad) using a Bio-Rad CFX96 system. Purified DNA inputs were used in real-time PCRs for standardization.
For ChIP-seq, the above-described procedure was scaled up approximately 10-fold as previously described (42). Sequence reads were aligned to the human hg19 genome using Bowtie (http://bowtie-bio.sourceforge.net/index.shtml) to produce SAM files for further analysis.
Peak calling.
Peak calling in the ChIP-seq samples was performed using MOSAiCS (model-based one- and two-sample analysis and inference for ChIP-seq data) (43; http://www.bioconductor.org/packages/release/bioc/html/mosaics.html), controlling the false-discovery rate (FDR) at 0.05. We ran MOSAiCS in two-sample mode, where it estimated the distribution of the background read counts from input data and the distribution of the read counts in peak regions with a two-component negative binomial mixture model.
Peak colocalization.
To perform downstream analysis of the peak lists, we first generated a union peak set by merging peaks that overlapped in the EBNA2, EBNA3A, EBNA3B, EBNA3C, and RBPJ peak lists. Then, we built the incidence matrix by using the union peak set and the peak lists. Each entry of the incidence matrix was set to 1 when a peak in the list was used to construct the region and to 0 otherwise. Additionally, we overlapped the union peak set with 76 GM12878 ChIP-seq peak sets and DNase I-hypersensitive sites (DHS) reported by the ENCODE project for further analysis (the data were from https://www.encodeproject.org).
Histone profile plots.
The average histone modification profiles were calculated using Segvis (https://github.com/keleslab/Segvis) for each 4-kb window centered at the EBNA protein peak summits that overlap DHS. For each coordinate in the window, we calculated the signal as the number of extended reads within a 151-bp window. These signals were then averaged to generate a smooth aggregation profile. Each profile was normalized to 1 million reads, and the profiles for a given peak set were averaged coordinatewise. The histone modification ChIP-seq data sets were downloaded from the ENCODE portal (https://www.encodeproject.org).
Chromatin state classification.
Chromatin states were assigned to EBNA protein peaks by clustering the maximum normalized histone signal values in 250 bp around the peak summit using the partition around medoids algorithm of the cluster package in R (https://cran.r-project.org/web/packages/cluster/index.html).
Motif and gene enrichment analysis.
Motif analysis was performed using the MEME-ChIP (44) tool from the MEME suite (http://meme.sdsc.edu). The input data consisted of 500-bp genomic sequences centered around the peak summits from the top 500 peaks (ranked by the posterior probabilities of binding from MOSAiCS) that overlapped DHS. TFs targeting the discovered motifs were determined by comparing the motifs with annotated motifs in the JASPAR and TRANSFAC databases using the TOMTOM tool of the MEME suite.
Nucleotide sequence accession number.
The ChIP-seq data reported in this paper have been deposited in the GEO database under accession number GSE76166.
RESULTS
Genome-wide binding analysis of EBNA3 proteins in LCLs.
In order to identify EBNA3A-, EBNA3B-, and EBNA3C-bound sites in LCLs, we performed ChIP-seq experiments using three LCLs: EBNA3A-F-HA, EBNA3B-F-HA, and EBNA3C-F-HA, that are transformed with recombinant EBV genomes in which one of the EBNA3 proteins had a Flag-HA epitope fused to its C terminus, as previously described (34). This approach allowed each EBNA3 protein to be precipitated with an anti-HA monoclonal antibody, minimizing the possibility that observed differences were attributable to differences among antibodies used in EBNA3A, EBNA3B, or EBNA3C ChIP.
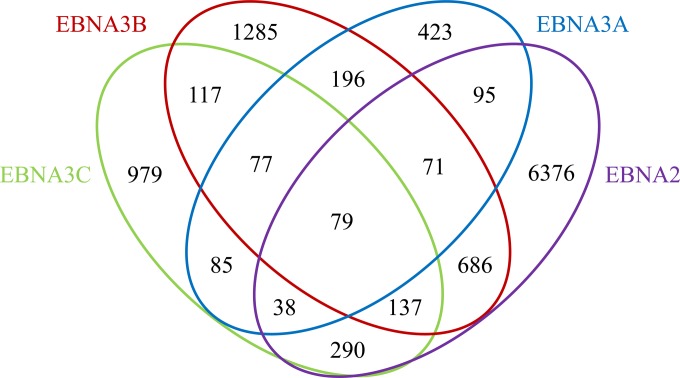
ChIP samples were used to prepare libraries for Illumina sequencing, and the raw reads were aligned to the human hg19 genome using Bowtie (45; http://bowtie-bio.sourceforge.net/index.shtml). Genomic sites bound by each EBNA3 were identified as peaks in the ChIP-seq data set relative to input DNA using MOSAiCS (43). For comparison, we performed an RBPJ ChIP-seq from one of the cell lines (EBNA3C-F-HA LCL) and reanalyzed previously published EBNA2 and RBPJ ChIP-seq data sets (42) using the same bioinformatics pipeline. This identified a total of 1,640 EBNA3A-, 3,033 EBNA3B-, 3,588 EBNA3C-, 8,592 EBNA2-, and 9,938 RBPJ-bound sites. In total, we identified 6,791 distinct genomic sites bound by EBNA3 proteins in LCLs. Although this estimate is comparable to that reported for EBNA3 binding in Mutu III cells (33), only 1,466 (21%) of the sites are in common (see Fig. S1 in the supplemental material). This limited overlap may reflect true differences between EBNA3 binding in LCLs and BL cells or may be due to technical differences, such as undersampling or the use of different antibodies in the ChIP-seq procedure. To facilitate identification of transcription factors that may mediate EBNA3 binding to the human genome, we exploited the observation that the vast majority of cell transcription factor binding sites lie within DHS (46) and defined subsets of EBNA3 binding sites that overlapped DHS in the GM12878 LCL (47) for additional analysis. In total, we identified 1,064 EBNA3A, 2,648 EBNA3B, 1,802 EBNA3C, 7,772 EBNA2, and 8,294 RBPJ binding sites within DHS (Fig. 1; see Tables S1 and S2 in the supplemental material).
FIG 1.
Venn diagram showing colocalization of EBNA3A, EBNA3B, EBNA3C, and EBNA2 binding sites in LCLs. Sites on the human genome bound by EBV EBNA proteins were identified from ChIP-seq data as peaks relative to input using MOSAiCS. The numbers of bound sites and their extents of overlap are indicated for EBNA2, EBNA3A, EBNA3B, and EBNA3C.
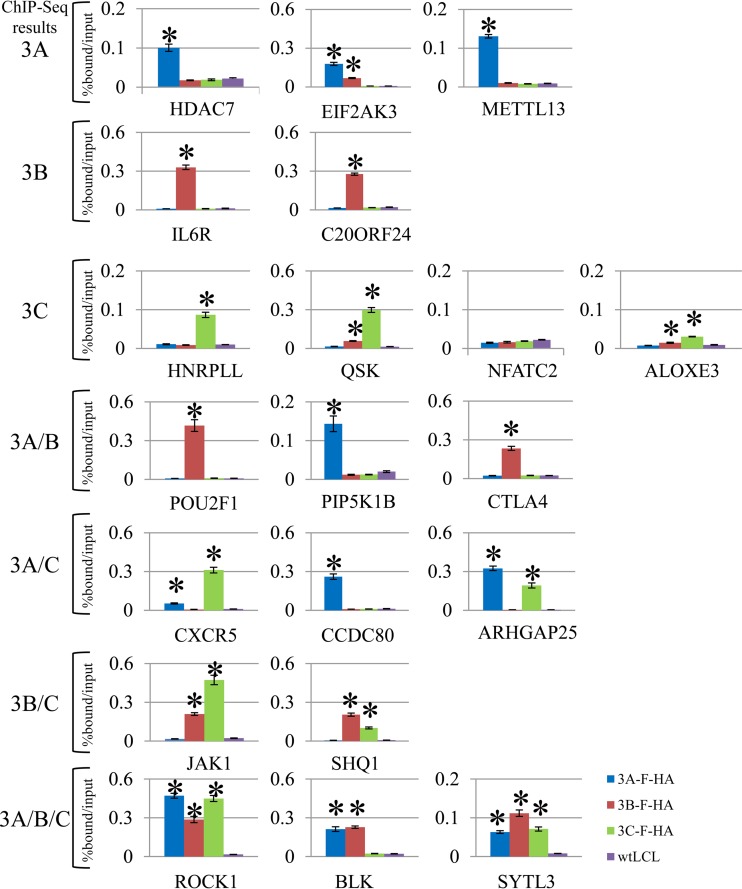
We further validated the specificity of our ChIP-seq peak calling by examining 20 genomic loci bound by one or more EBNA3s. For these experiments, we performed replicate ChIPs using EBNA3-F-HA LCLs and wild-type LCLs and assessed enrichment relative to input by qPCR for each bound site using specific primers (see Table S3 in the supplemental material). EBNA3 binding sites were classified as EBNA3A only (HDAC7, EIF2AK3, and METTL13); EBNA3B only (IL6R and C20ORF24); EBNA3C only (HNRPLL, QSK, ALOXE3, and NFATC2); EBNA3A and -3B (POU2F1, PIP5K1B, and CTLA4); EBNA3B and -3C (JAK1 and SHQ1); EBNA3A and -3C (CXCR5, CCDC80, and ARHGAP25); and EBNA3A, -3B, and -3C (BLK, ROCK1, and SYTL3) based on ChIP-seq results. We considered a peak validated if there was statistically significant enrichment in HA ChIP from the respective EBNA3-F-HA LCL relative to that observed in the wild-type (untagged) LCL by a two-sample t test. Using this approach, we confirmed 10 of 12 EBNA3A-, 9 of 10 EBNA3B-, and 9 of 12 EBNA3C-bound sites identified by ChIP-seq (Fig. 2). In addition, we found evidence of EBNA3B binding at 3 sites (EIF2AK3, QSK, and ALOXE3) by ChIP-qPCR that were not observed by ChIP-seq. Based on these results, we estimated the overall sensitivity and specificity of our EBNA3 ChIP-seq experiments relative to ChIP-qPCR to be 92% and 83%, respectively.
FIG 2.
ChIP-qPCR validation of EBNA3 binding sites in LCLs identified by ChIP-seq. The bar plots show enrichment of genomic DNA from ChIP of EBNA3A, EBNA3B, or EBNA3C relative to input. Each EBNA3 was specifically ChIPed using HA antibody with either the EBNA3A-F-HA, EBNA3B-F-HA, or EBNA3C-F-HA LCL and wild-type (untagged) LCLs as a negative control. Genomic loci were chosen based on the peak patterns observed by ChIP-seq and included EBNA3A-only peaks (HDAC7, EIF2AK3, and METTL13); EBNA3B-only peaks (IL6R and C20ORF24); EBNA3C-only peaks (HNRPLL, QSK, NFATC2, and ALOXE3); EBNA3A- and EBNA3B-cobound peaks (POU2F1, PIP5K1B, and CTLA4); EBNA3A- and EBNA3C-cobound peaks (CXCR5, CCDC80, and ARHGAP25); EBNA3B- and EBNA3C-cobound peaks (JAK1 and SHQ1); and EBNA3A-, EBNA3B-, and EBNA3C-cobound peaks (ROCK1, BLK, and SYTL3). All qPCR signals are reported as percentages of ChIPed DNA relative to input DNA. The results are shown as means ± standard errors of the mean (SEM) of three independent experiments. The asterisks indicate a P value of <0.05 by two-sample t test.
EBNA3-bound sites are overrepresented at promoter and enhancer elements.
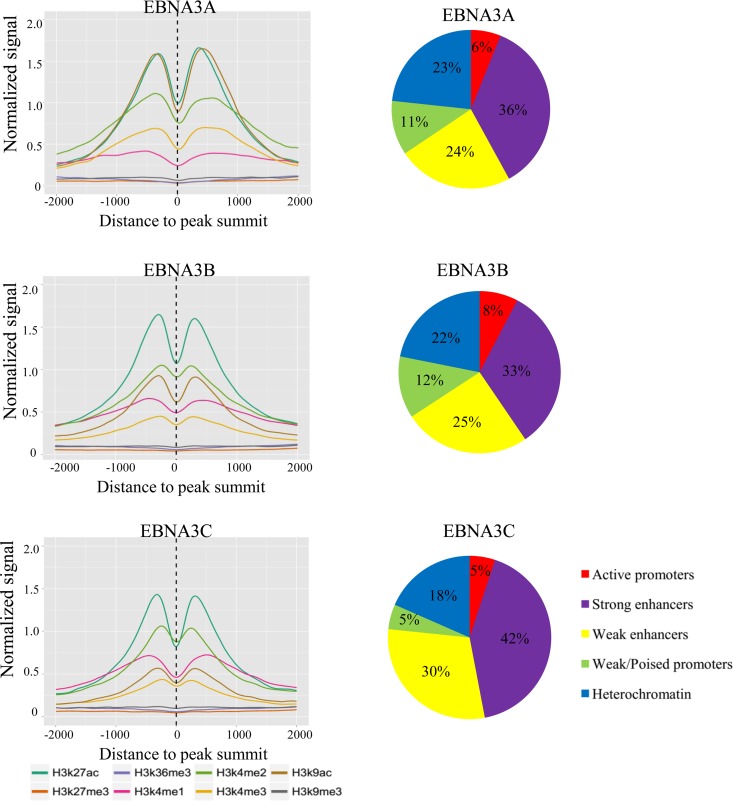
We constructed average histone profile plots for ±2-kb regions centered on EBNA3 peak summits for the activation marks H3K9Ac, H3K27Ac, H3K4me1, H3K4me2, and H3K4me3; the repressive marks H3K27me3 and H3K9me3; and the transcribed-region-associated mark H3K36me3 using ENCODE histone ChIP-seq data sets (48). For each EBNA3 protein, the signals from acetylation marks (H3K9Ac and H3K27Ac) were strong, as were mono-, di-, and trimethylation of H3K4 (Fig. 3A). For all EBNA3s, levels of repressive marks characteristic of facultative (H3K27me3) and constitutive (H3K9me3) heterochromatin were very low, despite the prior observation that H3K27me3 levels are increased at EBNA3A- and EBNA3C-repressed genes, such as CDKN2A and BCL2L11 (BIM). We also annotated EBNA3-bound peaks according to their locations within the epigenetic landscape (49). The results for EBNA3B were typical: 8% of EBNA3B sites reside within active promoters defined by high H3K4me3 and H3K9Ac, 12% within weak and poised promoters characterized by high H3K4me3 and low H3K27Ac or high H3K27me3, 33% within strong enhancers with high H3K4me1 and high H3K27Ac, and 25% within weak enhancers with intermediate H3K4me1 and little H3K27Ac, and 22% were found in heterochromatin regions characterized by the absence of these histone marks (Fig. 3B). Thus, the EBNA3 proteins, despite their roles in the repression of multiple cell genes (19–25), bind predominantly at genomic sites bearing marks of transcriptionally active chromatin.
FIG 3.
Characterization of EBNA3-bound sites. (A) Average histone profile plots for EBNA3A-, EBNA3B-, and EBNA3C-bound sites. The average densities of ChIP-seq reads for the indicated histone modifications are plotted for ±2-kb windows around the summits of the indicated EBNA3-bound sites. The normalized signal strength of each histone modification was derived from ChIP-seq data sets from GM12878 LCLs downloaded from the ENCODE database and is reported in reads per kilobase per million mapped reads (RPKM) for each curve. (B) Pie charts showing the proportions of EBNA3A-, EBNA3B-, or EBNA3C-bound sites located within different functional chromatin domains as defined by Ernst et al. (49).
Overlap among EBNA3A, EBNA3B, EBNA3C, EBNA2, and RBPJ sites in LCLs.
Because the EBNA3 proteins cooperatively regulate many cell genes and because EBNA2, EBNA3A, and EBNA3C must interact with the RBPJ transcription factor to transform B lymphocytes, we wanted to examine the extent of overlap among EBNA3A-, EBNA3B-, EBNA3C-, and EBNA2-bound sites. Significant overlap between EBNA3A and EBNA3C has been noted previously (35). Our experiments estimate that this overlap is about 26% of EBNA3A peaks that are EBNA3C cobound and reveal that the overlap extends to EBNA3B-bound sites. For EBNA3B, 21% are shared with EBNA3A, 22% with EBNA3C, and 37% with EBNA2. McClellan et al. found that 80% of the genes closest to an EBNA3-bound site were also the closest genes to an EBNA2-bound site (33). Using their approach, we found only 36% of EBNA3-bound genes to be EBNA2 cobound, though this proportion rises to 50% for EBNA3B-bound genes (see Table S4 in the supplemental material). Nevertheless, in LCLs, there is substantial overlap among EBNA2-, EBNA3A-, EBNA3B-, and EBNA3C-bound sites, mirroring the overlap observed in the cell genes regulated by these EBNAs.
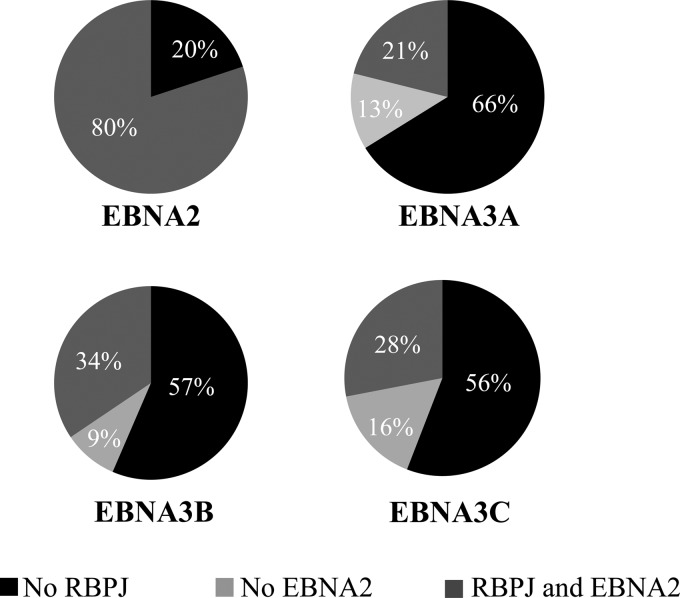
Although EBNA3 proteins are extensively complexed with RBPJ in LCLs (34, 50), we found that 66% of EBNA3A-, 57% of EBNA3B-, and 56% of EBNA3C-bound sites lack RBPJ binding (Fig. 4). RBPJ-independent binding has been noted in prior EBNA3A and EBNA3C ChIP-seq experiments (51, 52). This does not appear to be a technical artifact of the RBPJ ChIP-seq, as we did not observe enrichment for RBPJ cognate binding sequences (e.g., GTGGGAA; see further discussion below) at EBNA3A-, EBNA3B-, or EBNA3C-bound sites such as might occur if the RBPJ ChIP was impaired by EBNA3 cobinding. The EBNA3A-, EBNA3B-, or EBNA3C-bound sites that do colocalize with RBPJ are frequently cobound by EBNA2, as well, suggesting that EBNA3 binding at RBPJ sites may primarily serve to limit EBNA2 binding (Fig. 4, compare 21 to 34% cobound by both with 9 to 16% RBPJ only). We further observed that 27% of EBNA2/RBPJ/EBNA3-bound sites exhibit binding by more than one EBNA3.
FIG 4.
Extent of RBPJ colocalization with EBNA proteins. The pie charts summarize the extents of EBNA2, RBPJ, and EBNA3 cobinding in LCLs. For the EBNA3 charts, the percentages of peaks exhibiting RBPJ and EBNA2 cobinding, as well as RBPJ cobinding without EBNA2, are indicated.
EBNA3A and EBNA3C regulate EBNA2 binding at different RBPJ sites in the genome.
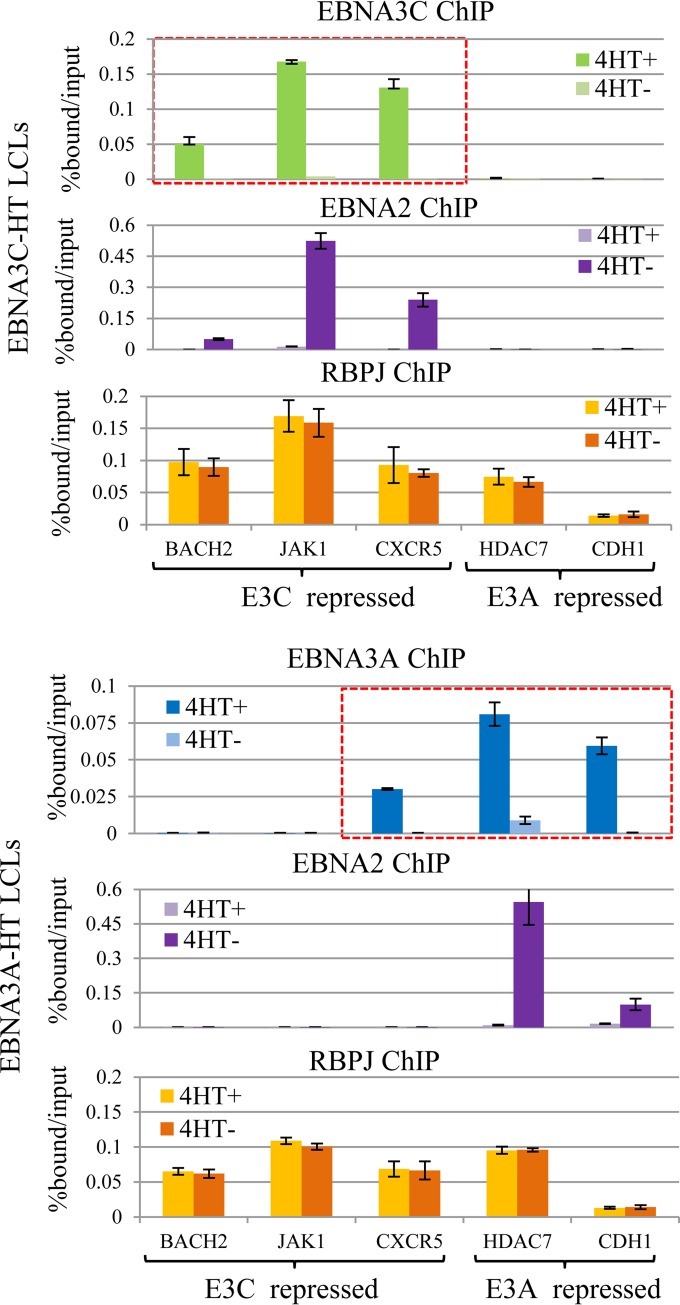
Despite this extensive RBPJ-independent EBNA3 binding, multiple lines of evidence argue that RBPJ interaction is central to EBNA3A- and EBNA3C-mediated gene regulation and LCL growth (30–32, 53). Further, these effects are specific: EBNA3A overexpression cannot compensate for EBNA3C loss, and vice versa (32, 37). The extensive overlap between EBNA2 and RBPJ suggests that each EBNA3 may regulate EBNA2's access to RBPJ at distinct genomic locations. To examine this possibility directly, we determined EBNA2 occupancy at several EBNA3C/RBPJ-cobound sites in the presence or absence of EBNA3C activity using the EBNA3C-HT LCL. EBNA2, EBNA3C, and RBPJ ChIPs were performed in EBNA3C-HT LCLs supplied with 4HT (4HT+) or cultured in medium after withdrawal of 4HT for 2 weeks (4HT−). We used qPCR to examine genomic loci bound by EBNA3C near genes known to be repressed by EBNA3C expression (23, 24), including BACH2, JAK1, and CXCR5 genes (Fig. 5A). As expected, the EBNA3C binding signal was present at each of the three EBNA3C-bound sites (BACH2, JAK1, and CXCR5) in EBNA3C-HT cells grown in the presence of 4HT and markedly declined upon 4HT withdrawal. Although EBNA3C has been observed to reduce RBPJ binding to DNA in gel shift assays, we did not observe any reduction in the RBPJ binding signal due to EBNA3C cobinding under physiological conditions. In contrast, EBNA3C inactivation had a dramatic effect on EBNA2 occupancy at these RBPJ-bound sites. At each location, EBNA2 levels were low in the presence of EBNA3C but increased significantly under the nonpermissive EBNA3C conditions. As controls, we also examined RBPJ and EBNA2 binding at two sites near EBNA3A-regulated genes (HDAC7 and CDH1 genes) that are bound by EBNA3A, but not EBNA3C (21, 24). At these control sites, no significant change in RBPJ occupancy or increase in EBNA2 binding was observed upon EBNA3C inactivation by 4HT withdrawal. These results suggest that EBNA3C does not compete with EBNA2 for global access to RBPJ, but rather, competes in a manner that is highly site specific. It is noteworthy that in the presence of active EBNA3C, EBNA2 binding was not detectible above background at the BACH2, JAK1, and CXCR5 sites and that EBNA2 peaks were not observed at these sites by ChIP-seq (i.e., in cells with EBNA3C expression). Thus, not only is EBNA3C able to reduce EBNA2 binding at specific sites to undetectable levels, but EBNA2/EBNA3C colocalization by static ChIP experiments (Fig. 1 and 4B) likely underestimates the extent of competition for RBPJ-bound sites.
FIG 5.
Regulation of EBNA2 binding by EBNA3A or EBNA3C at specific RBPJ sites. (A) EBNA3C, EBNA2, and RBPJ binding determined by ChIP-qPCR at the indicated loci in EBNA3C-HT LCLs cultured in the presence (4HT+) or absence (4HT−) of 4-hydroxytamoxifen for 2 weeks. The genomic loci included RBPJ binding sites near EBNA3C-repressed genes (BACH2, JAK1, and CXCR5 genes) or EBNA3A-repressed genes (HDAC7 and CDH1 genes), as indicated. The red dashed box indicates sites bound by EBNA3C from ChIP-seq data. All qPCR signals are reported as percentages of ChIPed DNA relative to input DNA. The results are shown as means ± SEM of three independent experiments. (B) Experiment analogous to that shown in panel A using the EBNA3A-HT LCL. EBNA3A, EBNA2, and RBPJ binding were determined by ChIP-qPCR at the indicated sites in the presence or absence of 4-hydroxytamoxifen. The red dashed box indicates sites bound by EBNA3A from ChIP-seq data.
We next examined the specificity of EBNA3A competition with EBNA2 using the EBNA3A-HT LCLs cultured in the presence of 4HT or after 2 weeks of 4HT withdrawal. At EBNA3A-bound and -regulated genes (HDAC7 and CDH1 genes), we observed decreased EBNA3A ChIP signal upon 4HT withdrawal (Fig. 5B, top right). These sites lack EBNA2 binding by ChIP-seq, but EBNA2 binding dramatically increased at these locations upon EBNA3A inactivation. In contrast, EBNA3A exerted no effect on the RBPJ binding signal. As controls, we examined EBNA3A, EBNA2, and RBPJ binding at the EBNA3C-bound and -regulated loci (BACH2, JAK1, and CXCR5). At the BACH2 and JAK1 sites, no EBNA3A binding was observed, and EBNA3A inactivation had no discernible effect on EBNA2 or RBPJ binding (Fig. 5B, left). Notably, CXCR5 is not EBNA3A regulated, despite being EBNA3A bound (Fig. 2 and 5B, top). At this CXCR5 site, we observed a decrease in EBNA3A binding upon 4HT withdrawal, but no corresponding increase in EBNA2 binding. The inability of EBNA3A to affect EBNA2 binding at the CXCR5 site potentially explains why CXCR5 is regulated only by EBNA3C, despite being bound by both EBNA3A and EBNA3C (Fig. 2). Thus, EBNA3A also appears to compete with EBNA2 for occupancy of specific RBPJ sites in the genome, but not for global access to RBPJ. These results are consistent with a prior report that EBNA3A can regulate EBNA2 binding to the CXCL9/10 promoter, but our results further demonstrate that EBNA3A limits EBNA2 binding only to a subset of RBPJ-bound sites where EBNA3A/RBPJ cobinding occurs (53). Further, as with EBNA3C, the absence of EBNA2 at an EBNA3A/RBPJ-cobound site does not exclude the possibility that EBNA3A regulates EBNA2 binding at that locus.
EBNA3 binding sites colocalize with multiple cell transcription factors.
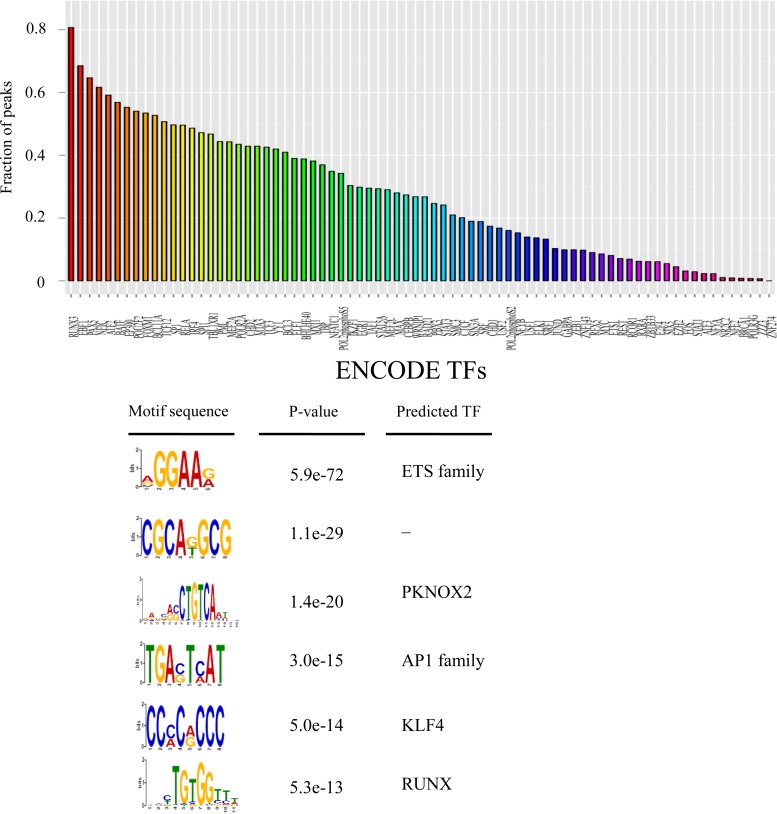
In an effort to identify transcription factors that contribute to EBNA3 binding specificity, we determined cell transcription factor cooccupancy at EBNA3A-, EBNA3B-, and EBNA3C-bound sites using ENCODE transcription factor ChIP-seq data from GM12878 LCLs. RUNX3 (83%), EBF1 (70%), PAX5 (66%), NFIC (63%), ATF2 (60%), BATF (57%), p300 (56%), POU2F2 (54%), FOXM1 (54%), BCL11A (53%), TCF12 (51%), SP1 (50%), and IRF4 (49%) represented the transcription factors with the highest colocalization for EBNA3B (Fig. 6A). Many of the same transcription factors are enriched at EBNA3A and EBNA3C sites (see Fig. S3 in the supplemental material) (35, 51, 52). Because ChIP-seq data are available for only a limited number of transcription factors, we also determined the sequence motifs enriched at EBNA3-bound sites using MEME-ChIP (44). EBNA3B-bound sites were enriched for motifs potentially corresponding to ETS, PKNOX2, AP1, KLF4, and RUNX binding sites (Fig. 6B). Notably, we did not observe enrichment for the RBPJ binding motif at EBNA3B-bound sites. The RBPJ motif is also not enriched at EBNA3A- or EBNA3C-bound sites, even though it can be readily detected at EBNA2-bound sites (12, 42).
FIG 6.
Transcription factor cobinding and motif enrichment at EBNA3B-bound sites. (A) Bar plot showing the fractions of EBNA3B peaks coassociated with the indicated transcription factors (x axis). Colocalization was defined as binding with 200 bp of EBNA3B-bound sites, and cell transcription factor binding locations were derived from ENCODE ChIP-seq data for 76 transcription factors in the GM12878 LCL. (B) EBNA3B-bound sites were analyzed for enriched motifs using MEME-ChIP. The six motifs with the lowest P values are indicated, as well as any transcription factors predicted to recognize them.
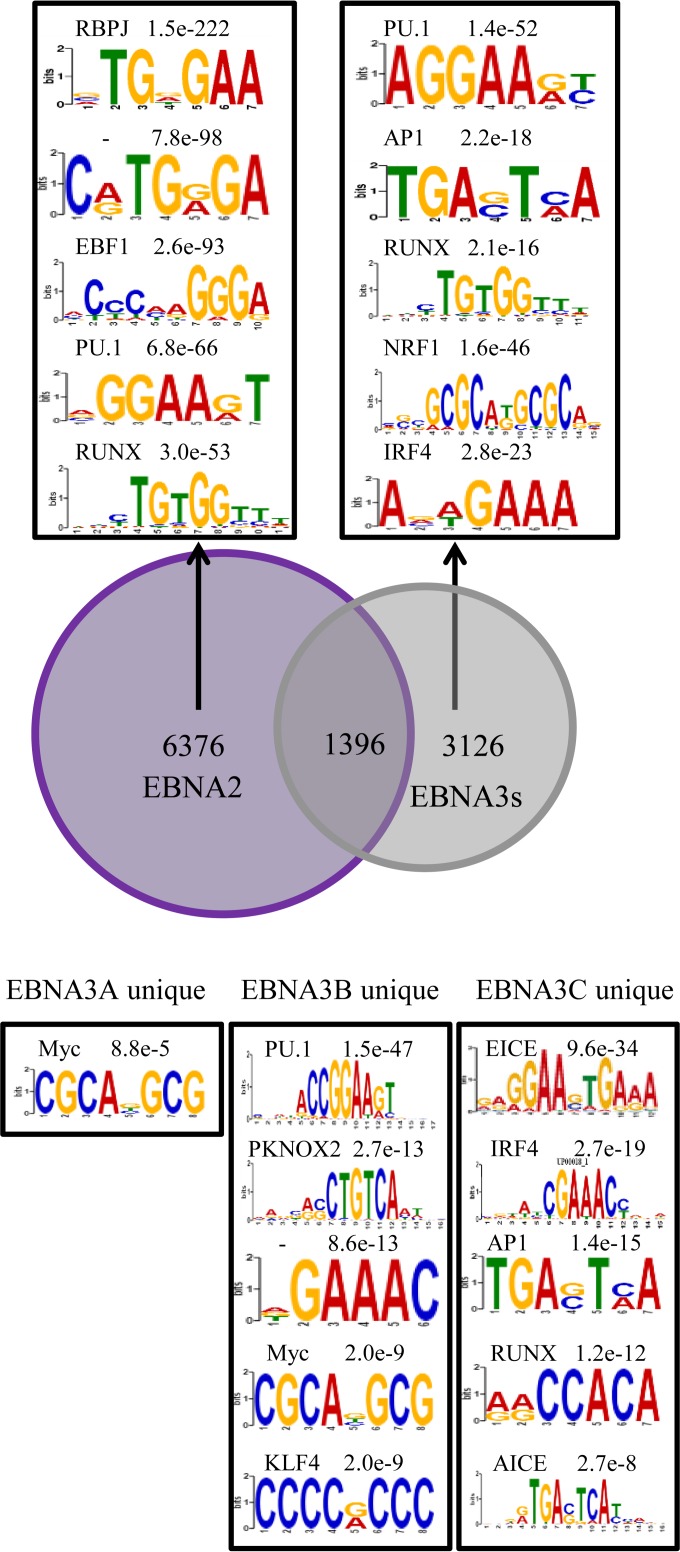
Our observation that transcription factor and motif enrichment at EBNA3B sites is remarkably similar to that present at EBNA3A and EBNA3C sites is consistent with prior results demonstrating similarity between EBNA3A- and EBNA3C-bound sites themselves. These results are perhaps not surprising, given that many of these sites are bound by more than one EBNA3 and hence many of the same cis-acting sites were reanalyzed for each EBNA3. To overcome this, we repeated our motif analyses restricting our attention to sites that did not exhibit cobinding with EBNA2 or the other EBNA3s. We initially sought to determine if any shared binding motifs determined a preference for EBNA3 compared to EBNA2 (Fig. 7A). Uniquely bound EBNA2 and EBNA3 sites still shared many common motifs, including PU.1 and RUNX; however, enrichment for AP1, NRF1, and IRF4 motifs was specifically observed at EBNA3-bound sites that lacked EBNA2 binding. We extended this approach to sites uniquely bound by only EBNA3A, EBNA3B, or EBNA3C (Fig. 7B). The results for IRF4 were particularly striking, as enrichment for IRF4 binding sites and for the IRF4-containing ETS/IRF4 composite element (EICE) and AP1/IRF4 composite element (AICE) was observed only at unique EBNA3C sites. This observation and previous reports that EBNA3C binding signals are stronger at IRF4-cobound sites than at EBNA3C sites without IRF4 binding (35) suggested that IRF4 may be especially important for EBNA3C binding. IRF4 enrichment at other EBNA3-bound sites may be a consequence of overlapping binding among the EBNA3s.
FIG 7.
Enriched motifs at sites bound uniquely by one EBNA protein. (A) Venn diagram summarizing the overlap of EBNA2 binding sites with sites bound by any EBNA3. Enriched motifs at sites bound only by EBNA2 (left box) or bound by any EBNA3, but not by EBNA2 (right box) are indicated, with corresponding P values and the names of any matching transcription factors. (B) Motifs enriched at sites bound uniquely by EBNA3A, EBNA3B, and EBNA3C (but not by EBNA2 or another EBNA3) are indicated, along with corresponding P values and matching transcription factors.
IRF4 is essential for EBNA3C binding specificity.
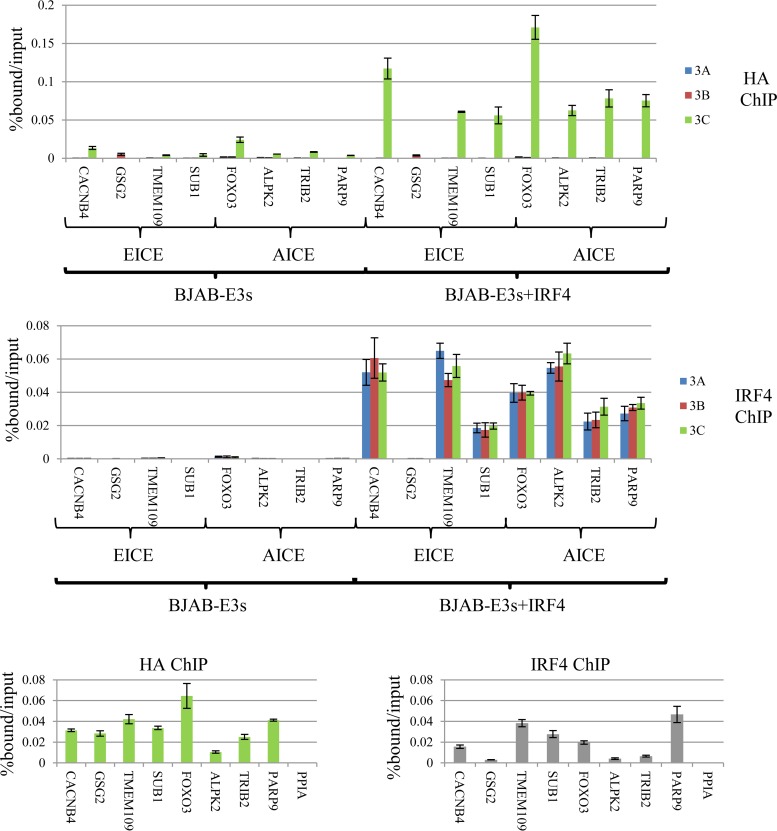
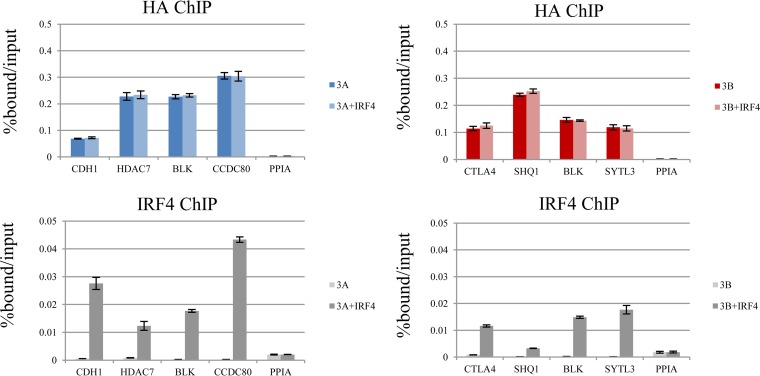
We sought to determine the extent to which IRF4 expression contributes to binding of each of the EBNA3 proteins. Establishing the significance of IRF4/EBNA3C colocalization presented a challenge, since IRF4 knockdown in LCLs promotes apoptosis and decreases proliferation (54). To circumvent this problem, we stably expressed Flag-HA-tagged EBNA3 proteins in the BJAB lymphoma cell line, which lacks IRF4 expression (see Fig. S4 in the supplemental material). The resulting BJAB-E3A-FH, BJAB-E3B-FH, and BJAB-E3C-FH lines expressed epitope-tagged EBNA3A, EBNA3B, and EBNA3C proteins at levels comparable to those of LCLs (see Fig. S5 in the supplemental material). Consistent with LMP1 being the principal inducer of IRF4 during latency III gene expression, none of the EBNA3 proteins resulted in detectable IRF4 expression. Each BJAB-EBNA3-FH line was then converted to stable IRF4 expression by retroviral transduction, which resulted in IRF4 levels comparable to that observed in our LCLs (see Fig. S5 in the supplemental material). Using these stable BJAB cell lines, we examined the dependence of EBNA3A, EBNA3B, and EBNA3C binding on IRF4. We initially examined 8 sites that exhibited EBNA3C/IRF4 cobinding in LCLs, including four EICE (CACNB4, GSG2, TMEM109, and SUB1) and four AICE (FOXO3, ALPK2, TRIB2, and PARP9) sites. In the absence of IRF4 expression, weak EBNA3C binding was observed at some of these locations, particularly FOXO3; however, with IRF4 expression, EBNA3C binding was markedly increased at all AICE and 3 of 4 EICE sites (Fig. 8A). In contrast, IRF4 expression did not result in detectible EBNA3A or EBNA3B binding at any of these sites. Importantly, the one EICE site where EBNA3C binding was absent (GSG2) also lacked IRF4 binding by ChIP-qPCR (Fig. 8A, bottom). Although the reasons for the lack of binding of IRF4 at the GSG2 site are unclear, it probably accounts for the absence of EBNA3C binding at this site and further reinforces the notion that IRF4 plays a critical role in targeting EBNA3C to chromatin.
FIG 8.
IRF4 is essential for EBNA3C binding to specific sites in the human genome. (A) (Top) ChIP-qPCR experiment demonstrating the extent of EBNA3A, EBNA3B, or EBNA3C binding in BJAB cells expressing either Flag-HA-tagged EBNA3A, EBNA3B, or EBNA3C with or without stably coexpressed IRF4. Loci that exhibited EBNA3C and IRF4 cobinding in LCLs and conformed to EICE or AICE, as indicated, were chosen. (Bottom) ChIP-qPCR demonstrating IRF4 binding at the same sites. The error bars indicate SEM. (B) ChIP-qPCR assay demonstrating the extents of EBNA3C (left) and IRF4 (right) binding to the indicated EICE and AICE sites in EBNA3C-F-HA LCLs.
In order to confirm that these AICE- and EICE-containing sites represented authentic EBNA3C binding sites, we performed additional HA ChIPs in the EBNA3C-F-HA LCL. As shown in Fig. 8B, EBNA3C binding was observed at all eight of these sites, including the GSG2 site, but not at a site near the PPIA gene, which has been used in previous EBNA3 ChIP studies as a negative control (52). We additionally confirmed IRF4 binding at each of these sites by ChIP, albeit at low levels at the GSG2, ALPK2, and TRIB2 sites, which were nevertheless significantly above those at the PPIA control site (P < 0.004; two-sample t test).
To more thoroughly evaluate the dependence of EBNA3A and EBNA3B binding on IRF4, we did additional ChIP experiments using the BJAB-E3A-FH and BJAB-E3B-FH cells. We first considered 4 sites that are cobound by EBNA3A and IRF4 in LCLs (CDH1, HDAC7, BLK, and CCDC80). In the absence of IRF4 expression, there was detectible binding above that at the control PPIA site at each of these four locations (Fig. 9A). However, we observed no detectible change in EBNA3A binding with IRF4 coexpression. Additional ChIP-qPCR experiments confirmed IRF4 binding at each site (Fig. 9A, bottom). Using the BJAB-E3B-FH cells, we examined binding to four EBNA3B/IRF4-cobound sites (STLA4, SHQ1, BLK, and SYTL3). Similar to our results for EBNA3A, binding was above that at the PPIA control site at each location, but there was no observed increase in EBNA3B binding with IRF4 coexpression. We confirmed by ChIP-qPCR that IRF4 binding was detectible at each site (Fig. 9B, bottom). Taken together, our data provided direct evidence that IRF4 specifically contributes to EBNA3C binding. In contrast, despite significant overlap between IRF4 and EBNA3A or EBNA3B binding sites, we found no evidence that IRF4 contributes to EBNA3A or EBNA3B binding at any IRF4-cobound site examined in this study.
FIG 9.
IRF4 cobinding does not promote EBNA3A or EBNA3B at specific sites in the human genome. (A) (Top) ChIP-qPCR assay for EBNA3A (HA) in BJAB cells expressing Flag-HA-tagged EBNA3A alone or with IRF4. (Bottom) ChIP-qPCR assay for IRF4 from the same cells. Binding was assessed at multiple sites cobound by EBNA3A and IRF4 in LCLs (CDH1, HDAC7, BLK, and CCDC80). A site near the PPIA that is not bound by EBNAs or IRF4 was included as a negative control. (B) (Top) ChIP-qPCR assay for EBNA3B (HA) in BJAB cells expressing Flag-HA-tagged EBNA3B alone or with IRF4. (Bottom) IRF4 ChIP-qPCR. Representative EBNA3B/IRF4-cobound sites based on LCL data were selected (CTLA4, SHQ1, BLK, and SYTL3), as well as the PPIA negative control. All qPCR signals are reported as percentages of DNA ChIPed relative to the input. The results are shown as means ± SEM of three independent experiments.
DISCUSSION
Here, we report a comprehensive survey of the genome-wide binding of EBNA3 proteins in LCLs. We believe our approach offers several advantages over prior studies, including distinguishing among EBNA3A, EBNA3B, and EBNA3C binding on a genome-wide basis. In addition, using the same epitope tag to ChIP each EBNA3 protein avoids confounding effects of differences in antibody sensitivity and specificity when different sera are used for each EBNA3. Furthermore, as our BJAB/IRF4 experiments highlight, EBNA3 binding is a function of the cell's transcription factor milieu. Thus, the best opportunity to understand the role of EBNA3 proteins in the transformation of resting B lymphocytes into LCLs is afforded by binding data obtained from LCLs. These results have provided important mechanistic insights into the role of EBNA3s in LCL gene regulation. We found that EBNA3A, EBNA3B, and EBNA3C bind to distinct, partially overlapping genome sites, a phenomenon that almost certainly underlies their ability to regulate partially overlapping gene subsets. Second, we demonstrate that EBNA3A and EBNA3C can regulate EBNA2 binding at unique RBPJ-cobound sites. Importantly, we provide direct evidence that IRF4 is a critical determinant of EBNA3C binding but does not affect EBNA3A or EBNA3B binding in BJAB cells. This represents a significant advance in our understanding of the mechanism by which EBNA3C regulates cell gene expression but also supports a more general model where differences in EBNA3 binding specificity are conferred by interactions with cell transcription factors other than RBPJ.
The precise role played by RBPJ in EBNA3 function remains controversial despite extensive study. Since the initial discovery that EBNA3s, like EBNA2, target RBPJ, numerous studies have confirmed that EBNA3s are highly associated with RBPJ in LCLs (14, 16, 50). Each EBNA3 forms a distinct RBPJ complex, though there is some evidence to suggest that they may interact with each other (29, 34). Despite this high degree of association, our ChIP-seq results and those of others reveal that only a minority of EBNA3-bound sites are cobound by RBPJ. This limited overlap and the observation that EBNA3s can disrupt RBPJ binding to DNA by electrophoretic mobility shift assay (EMSA) have led to the interpretation that the RBPJ interaction is not involved in EBNA3 binding to chromatin (18, 35). Several lines of evidence suggest this is not the case. In a detailed study of EBNA3A regulation of the CXCL9/CXCL10 locus, RBPJ expression was demonstrated to be critical for EBNA3A binding to this bidirectional promoter (53). Further, EBNA3A and EBNA3C mutants defective for RBPJ interaction are unable to regulate the CDKN2A gene or support LCL growth. Thus, a model where RBPJ is recruited to DNA by EBNA3s (and not vice versa) seems improbable, since neither EBNA3A nor EBNA3C can compensate for the loss of the other in CDKN2A regulation (30–32). Finally, data presented in Fig. 5 demonstrate that EBNA3A or EBNA3C inactivation produces a dramatic change in EBNA2 binding at multiple genomic sites without significant effect on RBPJ binding. This result, and the specificity with which EBNA3A and EBNA3C regulate RBPJ-bound sites, suggests that EBNA3s are targeted to distinct RBPJ sites via interactions with RBPJ and other cell transcription factors. In this model, the requirement for both EBNA3A and EBNA3C to interact with RBPJ to perform unique roles in CDKN2A coregulation is readily explained if they target two distinct RBPJ sites.
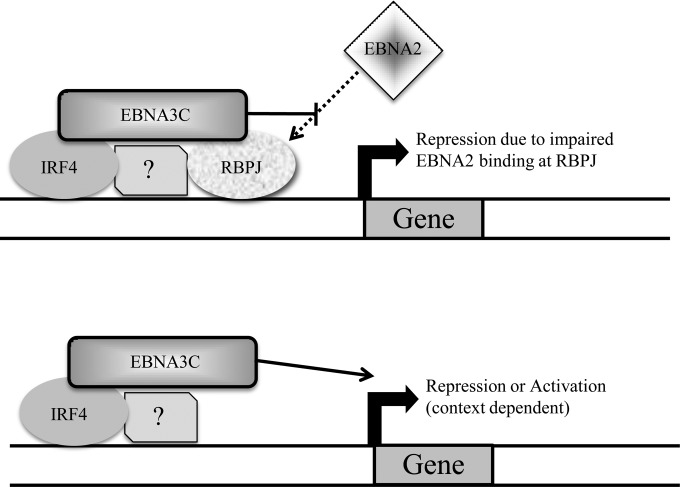
Our observation that EBNA3-EBNA2 competition occurs at RBPJ sites where there is no apparent EBNA2 binding in LCLs has important implications. This finding underscores the ability of EBNA3 proteins to successfully compete for RBPJ-bound sites despite the weaker correlation between EBNA3s and RBPJ binding than with EBNA2. Whether EBNA3 competition for RBPJ affects the ability of Notch to signal in LCLs is an important, unresolved question. In addition, our results imply that the degree to which EBNA3 and EBNA2 compete for genomic sites is underestimated by static ChIP-seq experiments. Thus, gene repression due to binding at EBNA3-only sites may still be attributable to impaired (abrogated) EBNA2 binding. This phenomenon may explain the recent discovery that the increased transforming effects of EBNA2 from type I EBV compared to that from type II EBV are due to increased binding of EBNA2 at ETS/IRF4 sites (12). The “novel” binding sites observed with type I EBNA2 may actually be due to an enhanced ability to compete with EBNA3C for binding at ETS/IRF4 sites. Given the strong transcriptional activation properties of EBNA2, effective competition for RBPJ by EBNA3s should almost always result in repression of target cell genes. At non-RBPJ sites, EBNA3s likely target chromatin via the same factors that determine RBPJ subset specificity. Since competition for EBNA2 binding is infrequent at these sites, EBNA3 binding may cause either activation or repression, depending on the chromatin context (Fig. 10).
FIG 10.
Working model of EBNA3 binding and gene regulation. (Top) At RBPJ sites, EBNA3 binding limits EBNA2 access, resulting in cell gene repression. The specific sites bound by each EBNA3 are determined by interactions with other cell transcription factors, including IRF4 in the case of EBNA3C, as depicted. (Bottom) At non-RBPJ sites, EBNA3 binding would not generally affect EBNA2 binding and produces activation or repression in a context-dependent manner.
The finding that IRF4 expression is a critical mediator of EBNA3C binding to specific genome sites represents an important advance in our understanding of EBNA3 binding specificity. Although IRF4/EBNA3C cobinding has been previously reported, the significance of this correlation remained to be demonstrated. Indeed, other transcription factors, including RUNX3 and ATF2 were more strongly correlated with EBNA3C-bound sites. Moreover, EBNA3A- and EBNA3B-bound sites are also enriched for IRF4 binding (Fig. 6A; see Fig. S2 in the supplemental material) (51). However, some lines of evidence suggested that the IRF4 interaction might be more important for EBNA3C, including the increased EBNA3C binding signals at IRF4 sites compared to non-IRF4 sites and the ability of EBNA3C to bind directly to IRF4 and promote its stabilization (35, 55). When we restricted our analysis to sites uniquely bound by EBNA3A, EBNA3B, or EBNA3C, we observed enrichment for the IRF4 binding motif only in the EBNA3C subset. Using an IRF4-negative BJAB cell line, we provide experimental evidence that IRF4 is determinative of EBNA3C binding. In the absence of IRF4 expression, we observed little to no EBNA3C binding signal at genomic sites normally cobound by EBNA3C and IRF4 in LCLs. Upon IRF4 expression at levels comparable to that seen in LCLs, EBNA3C binding was readily detected. This occurs at both AICEs and EICEs. In contrast, at sites cobound by EBNA3A/IRF4 or EBNA3B/IRF4 in LCLs, we observed no increase in EBNA3A or EBNA3B binding, respectively, upon IRF4 expression. It is important to note that EBNA3C binding was observed by ChIP-seq at only two of the EBNA3B/IRF4 sites (SHQ1 and STYL3) and none of the EBNA3A/IRF4 sites. Thus, IRF4 binding alone does not appear sufficient to confer EBNA3C binding to chromatin. Nevertheless, these results provide strong evidence that IRF4 is a critical mediator of EBNA3C binding specificity. Furthermore, this finding lends implicit support for the model that other cell transcription factors underlie EBNA3A and EBNA3B binding specificity.
The dependence of EBNA3C on IRF4 is further demonstration of EBV targeting of intrinsic B lymphocyte growth and survival pathways. IRF4 suppression of BCL6 and Pax5 is a critical event for B lymphocyte exit from germinal-center reactions (56, 57). Further increases in IRF4 expression upregulate PRDM1 (Blimp1) and XBP1 expression, promoting differentiation into antibody-secreting plasma cells. Although we do not currently know the extent to which EBNA3C effects depend on IRF4, transcriptional-profiling studies suggest that they coregulate many transcription factors responsible for B cell fate, including repression of PAX5, BACH2, NFATC1, SPIB, EBF1, and IL7R, and activation of PRDM1 (Blimp1), AICDA and IL6R (23, 24, 58). This suggests that many key EBNA3C transcriptional effects are dependent on IRF4 and/or EBNA3C and accentuates the IRF4 program. In addition to its role in follicular B cell differentiation, IRF4 has also been found to play a role in marginal-zone B cell development (59). Marginal-zone B cells are unique among B cell subsets in their dependence on Notch signaling and respond rapidly to blood-borne pathogens (60, 61). Interestingly, IRF4-null mice exhibited both increased NOTCH2 expression and enhanced ICN activation in response to CD40 and BCR signaling (59). Although the mechanism of the latter effect is incompletely defined, it suggests that EBNA3C targeting of IRF4 may have been selected for, in part, to limit ICN-like effects due to EBNA2. Given the central role of Notch signaling, and especially RBPJ, in EBNA3 effects, cell transcription factors that modulate Notch signaling in B cells are attractive candidates for transcription factors that, like IRF4, determine EBNA3 specificity.
Supplementary Material
ACKNOWLEDGMENTS
We thank Bill Sugden, Shannon Kenney, Janet Mertz, and Elliott Kieff for helpful discussions. The pVxy-puro-IRF4 plasmid was a kind gift from Lixin Rui.
R.W. received support through a Conacyt graduate studies scholarship. E.J. is supported by R01-DE023939 from the NIDCR. This work was also supported by R01-CA047006.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.02737-15.
REFERENCES
- 1.Johannsen E, Kaye K. 2015. Epstein-Barr virus (infectious mononucleosis, Epstein-Barr virus-associated malignant diseases, and other diseases), p 1754–1771. In Bennett J, Dolin R, Blaser M (ed), Mandell, Douglas, and Bennett's principles and practice of infectious diseases, vol 2 Elsevier Saunders, Philadelphia, PA. [Google Scholar]
- 2.Longnecker R, Kieff E, Cohen J. 2013. Epstein-Barr virus, p 1898–1959. In Knipe D, Howley P, Cohen J, Griffin D, Lamb R, Martin M, Racaniello V, Roizman B (ed), Fields virology, 6th ed, vol 2 Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 3.Grossman SR, Johannsen E, Tong X, Yalamanchili R, Kieff E. 1994. The Epstein-Barr virus nuclear antigen 2 transactivator is directed to response elements by the J kappa recombination signal binding protein. Proc Natl Acad Sci U S A 91:7568–7572. doi: 10.1073/pnas.91.16.7568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Henkel T, Ling PD, Hayward SD, Peterson MG. 1994. Mediation of Epstein-Barr virus EBNA2 transactivation by recombination signal-binding protein J kappa. Science 265:92–95. doi: 10.1126/science.8016657. [DOI] [PubMed] [Google Scholar]
- 5.Kaiser C, Laux G, Eick D, Jochner N, Bornkamm GW, Kempkes B. 1999. The proto-oncogene c-myc is a direct target gene of Epstein-Barr virus nuclear antigen 2. J Virol 73:4481–4484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kempkes B, Spitkovsky D, Jansen-Durr P, Ellwart JW, Kremmer E, Delecluse HJ, Rottenberger C, Bornkamm GW, Hammerschmidt W. 1995. B-cell proliferation and induction of early G1-regulating proteins by Epstein-Barr virus mutants conditional for EBNA2. EMBO J 14:88–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang F, Tsang SF, Kurilla MG, Cohen JI, Kieff E. 1990. Epstein-Barr virus nuclear antigen 2 transactivates latent membrane protein LMP1. J Virol 64:3407–3416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Woisetschlaeger M, Jin XW, Yandava CN, Furmanski LA, Strominger JL, Speck SH. 1991. Role for the Epstein-Barr virus nuclear antigen 2 in viral promoter switching during initial stages of infection. Proc Natl Acad Sci U S A 88:3942–3946. doi: 10.1073/pnas.88.9.3942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rowe M, Raithatha S, Shannon-Lowe C. 2014. Counteracting effects of cellular Notch and Epstein-Barr virus EBNA2: implications for stromal effects on virus-host interactions. J Virol 88:12065–12076. doi: 10.1128/JVI.01431-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Strobl LJ, Hofelmayr H, Marschall G, Brielmeier M, Bornkamm GW, Zimber-Strobl U. 2000. Activated Notch1 modulates gene expression in B cells similarly to Epstein-Barr viral nuclear antigen 2. J Virol 74:1727–1735. doi: 10.1128/JVI.74.4.1727-1735.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hofelmayr H, Strobl LJ, Marschall G, Bornkamm GW, Zimber-Strobl U. 2001. Activated Notch1 can transiently substitute for EBNA2 in the maintenance of proliferation of LMP1-expressing immortalized B cells. J Virol 75:2033–2040. doi: 10.1128/JVI.75.5.2033-2040.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tzellos S, Correia PB, Karstegl CE, Cancian L, Cano-Flanagan J, McClellan MJ, West MJ, Farrell PJ. 2014. A single amino acid in EBNA-2 determines superior B lymphoblastoid cell line growth maintenance by Epstein-Barr virus type 1 EBNA-2. J Virol 88:8743–8753. doi: 10.1128/JVI.01000-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Robertson ES, Lin J, Kieff E. 1996. The amino-terminal domains of Epstein-Barr virus nuclear proteins 3A, 3B, and 3C interact with RBPJ(kappa). J Virol 70:3068–3074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhao B, Marshall DR, Sample CE. 1996. A conserved domain of the Epstein-Barr virus nuclear antigens 3A and 3C binds to a discrete domain of Jkappa. J Virol 70:4228–4236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Calderwood MA, Lee S, Holthaus AM, Blacklow SC, Kieff E, Johannsen E. 2011. Epstein-Barr virus nuclear protein 3C binds to the N-terminal (NTD) and beta trefoil domains (BTD) of RBP/CSL; only the NTD interaction is essential for lymphoblastoid cell growth. Virology 414:19–25. doi: 10.1016/j.virol.2011.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cooper A, Johannsen E, Maruo S, Cahir-McFarland E, Illanes D, Davidson D, Kieff E. 2003. EBNA3A association with RBP-Jkappa down-regulates c-myc and Epstein-Barr virus-transformed lymphoblast growth. J Virol 77:999–1010. doi: 10.1128/JVI.77.2.999-1010.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dalbies-Tran R, Stigger-Rosser E, Dotson T, Sample CE. 2001. Amino acids of Epstein-Barr virus nuclear antigen 3A essential for repression of jkappa-mediated transcription and their evolutionary conservation. J Virol 75:90–99. doi: 10.1128/JVI.75.1.90-99.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Waltzer L, Perricaudet M, Sergeant A, Manet E. 1996. Epstein-Barr virus EBNA3A and EBNA3C proteins both repress RBP-J kappa-EBNA2-activated transcription by inhibiting the binding of RBP-J kappa to DNA. J Virol 70:5909–5915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Anderton E, Yee J, Smith P, Crook T, White RE, Allday MJ. 2008. Two Epstein-Barr virus (EBV) oncoproteins cooperate to repress expression of the proapoptotic tumour-suppressor Bim: clues to the pathogenesis of Burkitt's lymphoma. Oncogene 27:421–433. doi: 10.1038/sj.onc.1210668. [DOI] [PubMed] [Google Scholar]
- 20.Chen A, Zhao B, Kieff E, Aster JC, Wang F. 2006. EBNA-3B- and EBNA-3C-regulated cellular genes in Epstein-Barr virus-immortalized lymphoblastoid cell lines. J Virol 80:10139–10150. doi: 10.1128/JVI.00854-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hertle ML, Popp C, Petermann S, Maier S, Kremmer E, Lang R, Mages J, Kempkes B. 2009. Differential gene expression patterns of EBV infected EBNA-3A positive and negative human B lymphocytes. PLoS Pathog 5:e1000506. doi: 10.1371/journal.ppat.1000506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maruo S, Zhao B, Johannsen E, Kieff E, Zou J, Takada K. 2011. Epstein-Barr virus nuclear antigens 3C and 3A maintain lymphoblastoid cell growth by repressing p16INK4A and p14ARF expression. Proc Natl Acad Sci U S A 108:1919–1924. doi: 10.1073/pnas.1019599108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Skalska L, White RE, Parker GA, Turro E, Sinclair AJ, Paschos K, Allday MJ. 2013. Induction of p16(INK4a) is the major barrier to proliferation when Epstein-Barr virus (EBV) transforms primary B cells into lymphoblastoid cell lines. PLoS Pathog 9:e1003187. doi: 10.1371/journal.ppat.1003187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.White RE, Groves IJ, Turro E, Yee J, Kremmer E, Allday MJ. 2010. Extensive co-operation between the Epstein-Barr virus EBNA3 proteins in the manipulation of host gene expression and epigenetic chromatin modification. PLoS One 5:e13979. doi: 10.1371/journal.pone.0013979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhao B, Mar JC, Maruo S, Lee S, Gewurz BE, Johannsen E, Holton K, Rubio R, Takada K, Quackenbush J, Kieff E. 2011. Epstein-Barr virus nuclear antigen 3C regulated genes in lymphoblastoid cell lines. Proc Natl Acad Sci U S A 108:337–342. doi: 10.1073/pnas.1017419108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bazot Q, Paschos K, Skalska L, Kalchschmidt JS, Parker GA, Allday MJ. 2015. Epstein-Barr virus proteins EBNA3A and EBNA3C together induce expression of the oncogenic microRNA cluster miR-221/miR-222 and ablate expression of its target p57KIP2. PLoS Pathog 11:e1005031. doi: 10.1371/journal.ppat.1005031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tursiella ML, Bowman ER, Wanzeck KC, Throm RE, Liao J, Zhu J, Sample CE. 2014. Epstein-Barr virus nuclear antigen 3A promotes cellular proliferation by repression of the cyclin-dependent kinase inhibitor p21WAF1/CIP1. PLoS Pathog 10:e1004415. doi: 10.1371/journal.ppat.1004415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nikitin PA, Yan CM, Forte E, Bocedi A, Tourigny JP, White RE, Allday MJ, Patel A, Dave SS, Kim W, Hu K, Guo J, Tainter D, Rusyn E, Luftig MA. 2010. An ATM/Chk2-mediated DNA damage-responsive signaling pathway suppresses Epstein-Barr virus transformation of primary human B cells. Cell Host Microbe 8:510–522. doi: 10.1016/j.chom.2010.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Paschos K, Parker GA, Watanatanasup E, White RE, Allday MJ. 2012. BIM promoter directly targeted by EBNA3C in polycomb-mediated repression by EBV. Nucleic Acids Res 40:7233–7246. doi: 10.1093/nar/gks391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee S, Sakakibara S, Maruo S, Zhao B, Calderwood MA, Holthaus AM, Lai CY, Takada K, Kieff E, Johannsen E. 2009. Epstein-Barr virus nuclear protein 3C domains necessary for lymphoblastoid cell growth: interaction with RBP-Jkappa regulates TCL1. J Virol 83:12368–12377. doi: 10.1128/JVI.01403-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maruo S, Johannsen E, Illanes D, Cooper A, Zhao B, Kieff E. 2005. Epstein-Barr virus nuclear protein 3A domains essential for growth of lymphoblasts: transcriptional regulation through RBP-Jkappa/CBF1 is critical. J Virol 79:10171–10179. doi: 10.1128/JVI.79.16.10171-10179.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maruo S, Wu Y, Ito T, Kanda T, Kieff ED, Takada K. 2009. Epstein-Barr virus nuclear protein EBNA3C residues critical for maintaining lymphoblastoid cell growth. Proc Natl Acad Sci U S A 106:4419–4424. doi: 10.1073/pnas.0813134106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McClellan MJ, Wood CD, Ojeniyi O, Cooper TJ, Kanhere A, Arvey A, Webb HM, Palermo RD, Harth-Hertle ML, Kempkes B, Jenner RG, West MJ. 2013. Modulation of enhancer looping and differential gene targeting by Epstein-Barr virus transcription factors directs cellular reprogramming. PLoS Pathog 9:e1003636. doi: 10.1371/journal.ppat.1003636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ohashi M, Holthaus AM, Calderwood MA, Lai CY, Krastins B, Sarracino D, Johannsen E. 2015. The EBNA3 family of Epstein-Barr virus nuclear proteins associates with the USP46/USP12 deubiquitination complexes to regulate lymphoblastoid cell line growth. PLoS Pathog 11:e1004822. doi: 10.1371/journal.ppat.1004822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jiang S, Willox B, Zhou H, Holthaus AM, Wang A, Shi TT, Maruo S, Kharchenko PV, Johannsen EC, Kieff E, Zhao B. 2014. Epstein-Barr virus nuclear antigen 3C binds to BATF/IRF4 or SPI1/IRF4 composite sites and recruits Sin3A to repress CDKN2A. Proc Natl Acad Sci U S A 111:421–426. doi: 10.1073/pnas.1321704111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sciammas R, Shaffer AL, Schatz JH, Zhao H, Staudt LM, Singh H. 2006. Graded expression of interferon regulatory factor-4 coordinates isotype switching with plasma cell differentiation. Immunity 25:225–236. doi: 10.1016/j.immuni.2006.07.009. [DOI] [PubMed] [Google Scholar]
- 37.Maruo S, Johannsen E, Illanes D, Cooper A, Kieff E. 2003. Epstein-Barr Virus nuclear protein EBNA3A is critical for maintaining lymphoblastoid cell line growth. J Virol 77:10437–10447. doi: 10.1128/JVI.77.19.10437-10447.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Menezes J, Leibold W, Klein G, Clements G. 1975. Establishment and characterization of an Epstein-Barr virus (EBC)-negative lymphoblastoid B cell line (BJA-B) from an exceptional, EBV-genome-negative African Burkitt's lymphoma. Biomedicine 22:276–284. [PubMed] [Google Scholar]
- 39.Lebkowski JS, Clancy S, Calos MP. 1985. Simian virus 40 replication in adenovirus-transformed human cells antagonizes gene expression. Nature 317:169–171. doi: 10.1038/317169a0. [DOI] [PubMed] [Google Scholar]
- 40.Maunders MJ, Petti L, Rowe M. 1994. Precipitation of the Epstein-Barr virus protein EBNA 2 by an EBNA 3c-specific monoclonal antibody. J Gen Virol 75:769–778. doi: 10.1099/0022-1317-75-4-769. [DOI] [PubMed] [Google Scholar]
- 41.Rozenblatt-Rosen O, Deo RC, Padi M, Adelmant G, Calderwood MA, Rolland T, Grace M, Dricot A, Askenazi M, Tavares M, Pevzner SJ, Abderazzaq F, Byrdsong D, Carvunis AR, Chen AA, Cheng J, Correll M, Duarte M, Fan C, Feltkamp MC, Ficarro SB, Franchi R, Garg BK, Gulbahce N, Hao T, Holthaus AM, James R, Korkhin A, Litovchick L, Mar JC, Pak TR, Rabello S, Rubio R, Shen Y, Singh S, Spangle JM, Tasan M, Wanamaker S, Webber JT, Roecklein-Canfield J, Johannsen E, Barabasi AL, Beroukhim R, Kieff E, Cusick ME, Hill DE, Munger K, Marto JA, Quackenbush J, Roth FP, DeCaprio JA, Vidal M. 2012. Interpreting cancer genomes using systematic host network perturbations by tumour virus proteins. Nature 487:491–495. doi: 10.1038/nature11288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhao B, Zou J, Wang H, Johannsen E, Peng CW, Quackenbush J, Mar JC, Morton CC, Freedman ML, Blacklow SC, Aster JC, Bernstein BE, Kieff E. 2011. Epstein-Barr virus exploits intrinsic B-lymphocyte transcription programs to achieve immortal cell growth. Proc Natl Acad Sci U S A 108:14902–14907. doi: 10.1073/pnas.1108892108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kuan PF, Chung DJ, Pan GJ, Thomson JA, Stewart R, Keles S. 2011. A statistical framework for the analysis of ChIP-Seq data. J Am Stat Assoc 106:891–903. doi: 10.1198/jasa.2011.ap09706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Machanick P, Bailey TL. 2011. MEME-ChIP: motif analysis of large DNA datasets. Bioinformatics 27:1696–1697. doi: 10.1093/bioinformatics/btr189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Langmead B, Trapnell C, Pop M, Salzberg SL. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thurman RE, Rynes E, Humbert R, Vierstra J, Maurano MT, Haugen E, Sheffield NC, Stergachis AB, Wang H, Vernot B, Garg K, John S, Sandstrom R, Bates D, Boatman L, Canfield TK, Diegel M, Dunn D, Ebersol AK, Frum T, Giste E, Johnson AK, Johnson EM, Kutyavin T, Lajoie B, Lee BK, Lee K, London D, Lotakis D, Neph S, Neri F, Nguyen ED, Qu H, Reynolds AP, Roach V, Safi A, Sanchez ME, Sanyal A, Shafer A, Simon JM, Song L, Vong S, Weaver M, Yan Y, Zhang Z, Lenhard B, Tewari M, Dorschner MO, Hansen RS, Navas PA, Stamatoyannopoulos G, Iyer VR, Lieb JD, Sunyaev SR, Akey JM, Sabo PJ, Kaul R, Furey TS, Dekker J, Crawford GE, Stamatoyannopoulos JA. 2012. The accessible chromatin landscape of the human genome. Nature 489:75–82. doi: 10.1038/nature11232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Natarajan A, Yardimci GG, Sheffield NC, Crawford GE, Ohler U. 2012. Predicting cell-type-specific gene expression from regions of open chromatin. Genome Res 22:1711–1722. doi: 10.1101/gr.135129.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schneider R, Grosschedl R. 2007. Dynamics and interplay of nuclear architecture, genome organization, and gene expression. Genes Dev 21:3027–3043. doi: 10.1101/gad.1604607. [DOI] [PubMed] [Google Scholar]
- 49.Ernst J, Kheradpour P, Mikkelsen TS, Shoresh N, Ward LD, Epstein CB, Zhang X, Wang L, Issner R, Coyne M, Ku M, Durham T, Kellis M, Bernstein BE. 2011. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature 473:43–49. doi: 10.1038/nature09906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Johannsen E, Miller CL, Grossman SR, Kieff E. 1996. EBNA-2 and EBNA-3C extensively and mutually exclusively associate with RBPJkappa in Epstein-Barr virus-transformed B lymphocytes. J Virol 70:4179–4183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schmidt SC, Jiang S, Zhou H, Willox B, Holthaus AM, Kharchenko PV, Johannsen EC, Kieff E, Zhao B. 2015. Epstein-Barr virus nuclear antigen 3A partially coincides with EBNA3C genome-wide and is tethered to DNA through BATF complexes. Proc Natl Acad Sci U S A 112:554–559. doi: 10.1073/pnas.1422580112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.McClellan MJ, Khasnis S, Wood CD, Palermo RD, Schlick SN, Kanhere AS, Jenner RG, West MJ. 2012. Downregulation of integrin receptor-signaling genes by Epstein-Barr virus EBNA 3C via promoter-proximal and -distal binding elements. J Virol 86:5165–5178. doi: 10.1128/JVI.07161-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Harth-Hertle ML, Scholz BA, Erhard F, Glaser LV, Dolken L, Zimmer R, Kempkes B. 2013. Inactivation of intergenic enhancers by EBNA3A initiates and maintains polycomb signatures across a chromatin domain encoding CXCL10 and CXCL9. PLoS Pathog 9:e1003638. doi: 10.1371/journal.ppat.1003638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xu D, Zhao L, Del Valle L, Miklossy J, Zhang L. 2008. Interferon regulatory factor 4 is involved in Epstein-Barr virus-mediated transformation of human B lymphocytes. J Virol 82:6251–6258. doi: 10.1128/JVI.00163-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Banerjee S, Lu J, Cai Q, Saha A, Jha HC, Dzeng RK, Robertson ES. 2013. The EBV latent antigen 3C inhibits apoptosis through targeted regulation of interferon regulatory factors 4 and 8. PLoS Pathog 9:e1003314. doi: 10.1371/journal.ppat.1003314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.De Silva NS, Simonetti G, Heise N, Klein U. 2012. The diverse roles of IRF4 in late germinal center B-cell differentiation. Immunol Rev 247:73–92. doi: 10.1111/j.1600-065X.2012.01113.x. [DOI] [PubMed] [Google Scholar]
- 57.Lenz G, Staudt LM. 2010. Aggressive lymphomas. N Engl J Med 362:1417–1429. doi: 10.1056/NEJMra0807082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang L, Yao ZQ, Moorman JP, Xu Y, Ning S. 2014. Gene expression profiling identifies IRF4-associated molecular signatures in hematological malignancies. PLoS One 9:e106788. doi: 10.1371/journal.pone.0106788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Simonetti G, Carette A, Silva K, Wang H, De Silva NS, Heise N, Siebel CW, Shlomchik MJ, Klein U. 2013. IRF4 controls the positioning of mature B cells in the lymphoid microenvironments by regulating NOTCH2 expression and activity. J Exp Med 210:2887–2902. doi: 10.1084/jem.20131026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Martin F, Kearney JF. 2000. B-cell subsets and the mature preimmune repertoire. Marginal zone and B1 B cells as part of a “natural immune memory.” Immunol Rev 175:70–79. [PubMed] [Google Scholar]
- 61.Tanigaki K, Han H, Yamamoto N, Tashiro K, Ikegawa M, Kuroda K, Suzuki A, Nakano T, Honjo T. 2002. Notch-RBP-J signaling is involved in cell fate determination of marginal zone B cells. Nat Immunol 3:443–450. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.