ABSTRACT
Oropouche virus (OROV) is a midge-borne human pathogen with a geographic distribution in South America. OROV was first isolated in 1955, and since then, it has been known to cause recurring outbreaks of a dengue-like illness in the Amazonian regions of Brazil. OROV, however, remains one of the most poorly understood emerging viral zoonoses. Here we describe the successful recovery of infectious OROV entirely from cDNA copies of its genome and generation of OROV mutant viruses lacking either the NSm or the NSs coding region. Characterization of the recombinant viruses carried out in vitro demonstrated that the NSs protein of OROV is an interferon (IFN) antagonist as in other NSs-encoding bunyaviruses. Additionally, we demonstrate the importance of the nine C-terminal amino acids of OROV NSs in IFN antagonistic activity. OROV was also found to be sensitive to IFN-α when cells were pretreated; however, the virus was still capable of replicating at doses as high as 10,000 U/ml of IFN-α, in contrast to the family prototype BUNV. We found that OROV lacking the NSm protein displayed characteristics similar to those of the wild-type virus, suggesting that the NSm protein is dispensable for virus replication in the mammalian and mosquito cell lines that were tested.
IMPORTANCE Oropouche virus (OROV) is a public health threat in Central and South America, where it causes periodic outbreaks of dengue-like illness. In Brazil, OROV is the second most frequent cause of arboviral febrile illness after dengue virus, and with the current rates of urban expansion, more cases of this emerging viral zoonosis could occur. To better understand the molecular biology of OROV, we have successfully rescued the virus along with mutants. We have established that the C terminus of the NSs protein is important in interferon antagonism and that the NSm protein is dispensable for virus replication in cell culture. The tools described in this paper are important in terms of understanding this important yet neglected human pathogen.
INTRODUCTION
Bunyaviruses form a large group of single-stranded negative-sense RNA viruses consisting of important human and veterinary pathogens, such as the recently emerged severe fever with thrombocytopenia syndrome virus (SFTSV) and Schmallenberg virus (SBV). The family is divided into genera Hantavirus, Nairovirus, Phlebovirus, Tospovirus, and the largest genus Orthobunyavirus, to which Oropouche virus (OROV) belongs (1, 2). OROV causes an acute febrile illness in humans, with signs and symptoms of fever, headache, malaise, myalgia, arthralgia, photophobia, nausea, vomiting, dizziness, and in some cases encephalitis and meningitis. Symptoms last between 2 and 7 days, with some patients reporting a recurrence of these symptoms (3–10). OROV is transmitted among humans via the biting midge Culicoides paraensis and is maintained in the wild by circulating in nonhuman primates, such as the pale-throated three-toed sloth (Bradypus tridactylus) and the black-tufted marmoset (Callithrix penicillata), though the vectors remain largely unknown (3–5, 10–12). Laboratory experiments and epidemiological surveys have reported that mosquitoes Aedes serratus, Aedes scapularis, Aedes albopictus, Culex fatigans, Culex quiquefaciatus, Coquilettidia venezuelensis, and Psorophora ferox are susceptible to OROV infection (13–16). Neutralizing antibodies against OROV have also been detected in both wild and domestic birds (10, 14, 15), leading to speculation that birds could be carriers of the virus (A. Barrett, University of Texas Medical Branch, personal communication).
Oropouche fever (OROF) outbreaks have mainly been reported in Brazil's Amazonian cities. OROV, however, was first recorded in Trinidad in 1955 (13). In Brazil, the virus was isolated in 1960 from a dead sloth found near one of the Belem-Brasilia highway construction sites. The following year (1961), in Belem, 11,000 people were reported ill in what became the first OROF outbreak (17). Between 1961 and 2009, over 30 OROF outbreaks were recorded, with an estimated 500,000 cases (13, 17, 18). Outside of Brazil, OROF was reported for the first time in Panama in 1989 and Peru in 1992. The geographic distribution of OROV today includes Brazil, Panama, Peru, and Argentina. Serological evidence suggests that the virus may also be circulating in Ecuador and Bolivia and in nonhuman primates in Colombia (7, 18–23). However, without a differential surveillance system to distinguish infections with similar clinical symptoms, such as OROV and dengue, chikungunya, and Mayaro fevers, the exact epidemiology of OROV in Central and South America remains unclear. OROV reassortant viruses have also been isolated in Peru and Venezuela and outside the epidemic zone within Brazil (24–26).
The lack of a reverse genetics system has, until now, limited research on OROV at a molecular level. In order to address this issue, we previously reported the establishment of a minigenome and virus-like particle production assay for OROV (27). In the present paper, we report the recovery of infectious OROV entirely from cDNA plasmids. Like all bunyaviruses, OROV contains a tripartite RNA genome with a large (L) segment that encodes the viral RNA-dependent RNA polymerase, a medium (M) segment that encodes the viral glycoproteins Gn and Gc, and a small (S) segment that encodes the nucleocapsid (N) protein. OROV also encodes two nonstructural proteins, NSm, which is a cotranslationally cleaved product formed along with Gn and Gc from the M segment, and NSs, which is encoded from a downstream AUG site on the same mRNA transcript as the N protein. The rescue system described in this paper is based on a T7 RNA polymerase-driven plasmid system (28). Using this, we have successfully recovered wild-type OROV, along with mutant viruses lacking the NSm or NSs protein. We report here the characterization of these recombinant viruses in cultured cells, as a way to contribute to the understanding of this important yet poorly understood emerging viral zoonosis.
MATERIALS AND METHODS
Cells and viruses.
A549 (human alveolar adenocarcinoma epithelial cells), A549/BVDV-NPro (A549 cells that express bovine viral diarrhea virus NPro protein), A549/V (derived from A549 and expressing simian virus 5 V protein), CPT-Tert (sheep choroid plexus cells), DF-1 (chicken embryo fibroblasts), HeLa (human cervical adenocarcinoma epithelial cells), LLC-MK2 (Macaca mulatta kidney epithelial cells), MDCK (canine kidney epithelial cells), MRK101 (gray red-backed vole kidney cells), QT-35 (Japanese quail fibrosarcoma cells), Vero-E6 (African green monkey kidney cells), and 2fTGH (human epithelial fibrosarcoma cells) cells were grown in Dulbecco's modified Eagle's medium (DMEM; Invitrogen) supplemented with 10% fetal bovine serum (FBS). BHK-21 (baby hamster kidney fibroblasts) cells were grown in Glasgow minimal essential medium (GMEM; Invitrogen) supplemented with 10% newborn calf serum (NCS) and 10% tryptose phosphate broth (TPB; Invitrogen). BSR-T7/5 cells, which stably express T7 RNA polymerase (29), were grown in GMEM supplemented with 10% FBS, 10% TPB, and 1 mg/ml of G418 (Geneticin; Invitrogen). All mammalian cell lines were grown at 37°C with 5% CO2.
Mosquito U4.4 cells (Aedes albopictus neonatal larvae cells) were grown in Leibovitz 15 medium (Gibco) supplemented with 10% FBS and 10% TPB, while Aag-2 cells (derived from Aedes aegypti neonatal larvae) were grown in Schneider's Drosophila medium with l-glutamine (Gibco) supplemented with 10% FBS. Both cell lines were maintained at 28°C.
OROV strain BeAn19991 was kindly donated by Luiz Tadeu Moraes Figueiredo (University of São Paulo School of Medicine, Ribeirao Preto, Brazil). OROV isolate BeH759025 was kindly provided by Pedro Vasconcelos (Department of Arboviruses and Hemorrhagic Fevers, Evandro Chagas Institute, Ministry of Health, Ananindeua, Brazil).
All experiments with infectious viruses were conducted under biosafety level 3 (BSL3) conditions.
Plasmids.
Plasmids pTVTOROVL, pTVTOROVM, and pTVTOROVS used for OROV rescue have previously been described (27). pTVTOROVdelNSm was created from pTVTOROVM by excision PCR using 5′-phosphorylated primers to delete nucleotides (nt) 1039 to 1476 of the M segment, hence removing the entire NSm open reading frame (ORF) after the first transmembrane domain up to the third transmembrane domain while maintaining the predicted cleavage site that resides between NSm and Gc. pTVTOROVdelNSs and pTVTOROV246NSs were generated by QuikChange site-directed mutagenesis on pTVTOROVS to insert stop codons at specific regions in order to generate viruses with truncated NSs proteins. These plasmids contain mutations at cDNA nt position 116 for pTVTOROVdelNSs and 313 for pTVTOROV246NSs. The N ORF remains unchanged. pTVTOROV2080S was generated by amplifying the full-length S segment of clinical isolate BeH759025 (24). Primers and cloning strategy were the same as for pTVTOROVS (27). All PCRs were carried out using KOD Hot Start DNA polymerase (Merck). Products were gel purified (Promega) and, where needed, ligated using T4 DNA ligase (Promega), as per the manufacturer's protocol. Plasmids were confirmed by nucleotide sequencing (Source Bioscience). Primers used are listed in Table 1.
TABLE 1.
Oligonucleotides used in this study
| Primer | Sequence (5′→3′) | Plasmid |
|---|---|---|
| delNSmOROVR | TCCTGCAATTGGTGAGATGAATTC | pTVTOROVdelNSm |
| delNSmOROVF | GATGAAGATTGCTTATCTAAAGAT | pTVTOROVdelNSm |
| OROV48NSsF | CAGCATATGTAGCATTTGAAGCTAGATACG | pTVTOROV48delNSs |
| OROV48NSsR | CGTATCTAGCTTCAAATGCTACATATGCTG | pTVTOROV48delNSs |
| OROV246NSsF | CGGACAACGGTCTAACCCTGCACCGTCTGT | pTVTOROV246NSs |
| OROV246NSsR | ACAGACGGTGCAGGGTTAGACCGTTGTCCG | pTVTOROV246NSs |
| OROVdelNSs2F | GAGTTCATTTTCAACGACGTACCACAACGGACTACATCTACATTTGATCCGGAGGCAGCATACGTAGCATTTGAAGC | pTVTOROVdelNSs2 |
| OROVdelNSs2R | GCTTCAAATGCTACGTATGCTGCCTCCGGATCAAATGTAGATGTAGTCCGTTGTGGTACGTCGTTGAAAATGAACTC | pTVTOROVdelNSs2 |
Generation of recombinant OROV from cDNA.
Recombinant OROVs were generated by transfecting BSR-T7/5 cells (1.5 × 105 cells/ml) with 1 μg of pTVTOROVL, pTVTOROVM (or pTVTOROVdelNSm), and pTVTOROVS (or pTVTOROV2080S) using 3 μl of transfection reagent TransIT-LT1 (Mirus Bio LLC) per μg of DNA. By replacing the wild-type (wt) S segment (pTVTOROVS) with 1.5 μg of pTVTOROVdelNSs or pTVTORO246NSs, mutant NSs viruses were generated. Supernatants were harvested 7 days posttransfection (p.t.). Rescue outcome was assessed by plaque assay on BHK-21 cells. All recovered viruses were grown in Vero E6 cells, and genome segments were amplified by reverse transcription-PCR (RT-PCR) for sequence determination (Sanger sequencing).
Plaque assay.
Viruses were titrated on BHK-21 cells seeded at a density of 3 × 105 per ml in a 12-well plate, while virus phenotype assays (with various cells) were carried out in 6-well plates. Cell monolayers were infected with either 150 μl (12-well plate) or 200 μl (6-well plate) of virus diluted in phosphate-buffered saline (PBS)–2% NCS. Cells were then overlaid using 0.6% Avicell (FMC) in 2× minimum essential medium (MEM)–2% NCS. Three days postinfection (p.i.), cells were fixed with 4% formaldehyde and stained using crystal violet to visualize plaques.
Virus propagation.
Working stocks of recombinant and wt viruses were grown in Vero E6 cells at a multiplicity of infection (MOI) of 0.001. Cells were maintained at 37°C and 5% CO2 until visible cytopathic effect (CPE) was observed. Virus-containing supernatant was clarified by low-speed centrifugation, and aliquots were stored at −80°C. Recombinant viruses used were at passage 2. All the introduced mutations were confirmed by amplifying the segment in question by RT-PCR, followed by Sanger sequencing with primers covering the region of interest.
RT-PCR.
Virion RNA of each passaged virus was extracted using the RNeasy minikit (Qiagen) as per the manufacturer's protocol. RT-PCR was then carried out using segment-specific forward or reverse primers (27), with Moloney murine leukemia virus (MMLV) reverse transcriptase (Promega). PCR was carried out using KOD Hot Start DNA polymerase (Merck), and the amplified products were agarose gel purified (Promega), as per the manufacturer's protocol. Specific regions of each segment were then sequenced via Sanger sequencing (Source Bioscience).
Viral infection.
Cells were infected with viruses at the desired MOI for 1 h at 37°C. The cell monolayer was washed three times using PBS, and then complete growth medium was added. At desired time points, virus was harvested or cell lysates were collected. Viral titers were determined by plaque assay on BHK-21 cells for 3 days.
Western blotting.
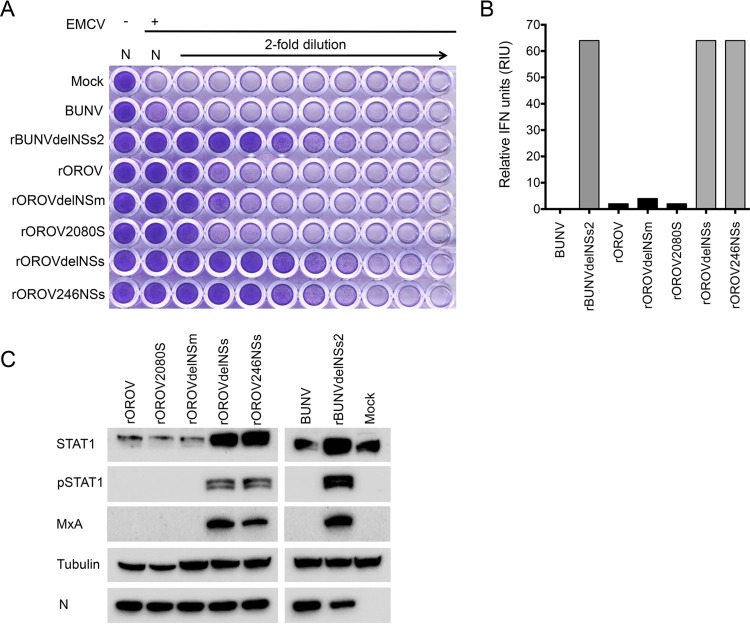
Cell lysates were prepared in lysis buffer (100 mM Tris-HCl [pH 6.8], 4% SDS, 20% glycerol, 200 mM dithiothreitol [DTT], 0.2% bromophenol blue, and 25 U/ml of Benzonase; Novagen), and proteins were then separated on a 4 to 12% gradient NuPAGE bis-Tris gel (Invitrogen) at 180 V for 50 min. Proteins were transferred to a nitrocellulose membrane (Amersham), and a semidry transfer was performed using the Trans-Blot Turbo transfer system (Bio-Rad) at 10 V for 50 min. Membranes were then blocked for 1 h in 5% milk–PBS 0.1% (vol/vol) Tween 20 and incubated in primary antibody overnight at 4°C. Secondary antibody was added for 1 h at 37°C. Proteins were then visualized using SuperSignal WestPico chemiluminescent substrate (Pierce), followed by exposure on a Bio-Rad ChemiDoc imager. Primary antibodies included BUNV anti-N-Rb (1:5,000) (30), anti-MxA (1:500, catalogue no. sc-50509; Santa Cruz Biotech), anti-pSTAT1 (1:750, catalogue no. 9167S; Cell Signaling), anti-STAT1 (1:750, catalogue no. 9172; Cell Signaling), and antitubulin monoclonal antibody (1:5,000, catalogue no. T5168, Sigma). OROV anti-N polyclonal rabbit antibody (1:1,000; GenScript) was a kind gift from Massimo Palmarini (MRC-University of Glasgow Centre for Virus Research). Horseradish peroxidase (HRP)-coupled secondary anti-rabbit (catalogue no. A0545; Sigma) and anti-mouse (catalogue no. A4416; Sigma) antibodies were used at 1:5,000.
Metabolic radiolabeling.
Vero E6 cells were grown in 12-well plates and infected at an MOI for 3 of each virus, and at the desired time points, supernatant was removed and cells were starved in methionine- and cysteine-free DMEM at 37°C for 30 min. Cells were then washed and labeled with 10 μCi per well of EasyTag EXPRESS35S mixed in methionine- and cysteine-free DMEM for 2 h at 37°C. Cell were then lysed in 150 μl of lysis buffer (100 mM Tris-HCl [pH 6.8], 4% SDS, 20% glycerol, 200 mM DTT, 0.2% bromophenol blue, and 25 U/ml of Benzonase; Novagen), and proteins were separated by SDS-PAGE. Gels were fixed and dried, and then labeled proteins were visualized by phosphorimaging (Storm840 phosphorimager; Molecular Dynamics).
Biological IFN production assay.
A549 cells were grown in 12-well plates and infected at an MOI of 1 for each virus. Twenty-four hours postinfection, supernatant was harvested and treated with UV light (8 W, 254 nm, and 2-cm distance) for 4 min with shaking after 2 min to inactivate any virus. Cells were lysed in 150 μl of lysis buffer (100 mM Tris-HCl [pH 6.8], 4% SDS, 20% glycerol, 200 mM DTT, 0.2% bromophenol blue, and 25 U/ml of Benzonase; Novagen) to check viral N and STAT1 protein levels. Phosphorylated STAT1 and MxA production in each sample was also probed. A549/BVDV-NPro cells grown in a 96-well plate were then treated with the UV-inactivated supernatant for 24 h. These cells were then infected with interferon (IFN)-sensitive encephalomyocarditis virus (EMCV; 0.05 PFU/cell) and incubated for 3 days at 37°C. Cells were fixed in 4% formaldehyde and stained with crystal violet to visualize CPE.
IFN-α sensitivity assay.
Vero E6 cells at 1.5 × 105/ml were treated either pre- or post-viral infection with various concentrations of universal type 1 IFN-α (catalogue no. 11200, lot no. 5283; Stratech Scientific). Cells were infected with virus at the desired MOI in PBS–2% FBS for 1 h at 37°C. Cells were washed three times in PBS before medium was replaced with medium with or without IFN-α. Cells were incubated at 37°C for 48 h before harvesting and determination of virus yield by plaque assay on BHK-21 cells. For plaque assays, confluent Vero E6 cells were pretreated with IFN-α for 24 h before the virus was titrated. At 4 days p.i., cells were fixed in 4% formaldehyde and stained with crystal violet. IFN-α was maintained in the medium and Avicell overlay for the duration of infection.
RESULTS
Recovery of wild-type OROV strain BeAn19991.
OROV is a negative-sense virus, and previously (27), we described the cloning of full-length antigenomic sense cDNA copies of its L, M, and S segments into the T7 RNA polymerase-driven plasmid backbone pTVT7R (0, 0) (31). This plasmid contains a single G residue immediately downstream of the T7 promoter sequence to aid efficient transcription. cDNA copies of OROV genome segments were cloned into pTVT7R in the antigenomic sense (Fig. 1A). To recover infectious OROV, BSR-T7/5 cells (29) were transfected with 1 μg of the pTVTOROVL, pTVTOROVM, and pTVTOROVS plasmids. Supernatant was harvested 7 days p.t. once CPE was visible, and success of the rescue attempt was determined by titration of infectious virus by plaque assay. The rescue of OROV was easily reproducible, yielding titers of 2.0 × 107, 4.5 × 106, and 2.3 × 107 PFU/ml in three independent experiments. As a control, BSR-T7/5 cells transfected with only pTVTOROVM and pTVTOROVS did not give rise to infectious virus. To test the authenticity of the recombinant OROV (rOROV), permissive Vero E6 cells were infected with the rescue supernatant and cell extracts were used for Western blotting (data not shown). Furthermore, rOROV-infected Vero E6 cells were fixed and stained at 24 h p.i. using a polyclonal anti-OROV antibody (a kind gift from Luiz Tadeu Moraes Figueiredo, University of São Paulo School of Medicine, Brazil). Substantial amounts of cytoplasmic OROV protein were detectable in the infected cells (data not shown), further confirming the successful recovery of infectious OROV.
FIG 1.
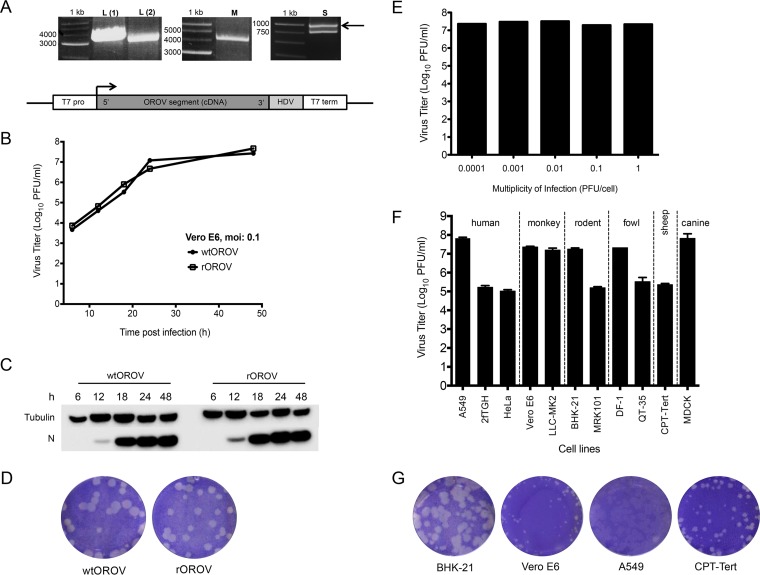
Characterization of recombinant OROV. (A) RT-PCR products derived from the L, M, and S segments of OROV strain BeAN19991. Amplified products were separated on a 1% agarose gel along with a 1-kb marker (Promega). Products were cloned into plasmids containing a T7-RNA polymerase promoter and a hepatitis delta ribozyme as shown in the schematic at the bottom. (B) Growth properties of wild-type (wt) and recombinant (r) OROV in Vero E6 cells. Cells were infected at an MOI of 0.1. At indicated time points samples were harvested and titers determined by plaque assay on BHK-21 cells. The graph shows results of a representative experiment. (C) Western blot showing N protein synthesis from the growth curve (A). Tubulin was probed as a loading control. (D) Comparison of plaque phenotypes of wtOROV and rOROV. A plaque assay was carried out on BHK-21 cells, and at 3 days p.i., cells were fixed and stained with crystal violet. (E) Effects of different MOIs on rOROV yields in Vero E6 cells. Infected cells were harvested 48 h p.i. and titrated on BHK-21 cells. A graph is presented for a representative experiment. (F) Comparison of rOROV growth in various cell lines. Indicated cells were infected at an MOI of 0.001 and at 48 h p.i. harvested and titrated on BHK-21 cells. Bars represent ranges from two experiments. (G) Comparison of rOROV plaque phenotypes on BHK-21, Vero E6, A549, and CPT-Tert cells. Infected cells were fixed and stained with crystal violet at 3 days p.i.
The growth kinetics and plaque phenotype of rOROV were similar to those of the authentic wt virus (Fig. 1B to D). All experiments from this point on were carried out with rOROV.
Growth of recombinant OROV in mammalian cell lines.
The growth properties of rOROV were tested in Vero E6 cells at MOIs ranging from 0.0001 to 1 PFU/cell. Previous work from our group has demonstrated that some viruses show a better fitness in certain cell types and at different MOIs, possibly due to the efficiency at which defective interfering particles are generated (32, 33). rOROV grew to similar titers by 48 h p.i. at all MOIs tested (Fig. 1E) and in a wide range of cell lines derived from several species (MOI, 0.001) (Fig. 1F), similar to other bunyaviruses (32, 33). Lower titers were obtained in human cell lines 2fTGH and HeLa than in A549 cells. Lower virus titers were also obtained from CPT-tert, QT-35, and MRK101 cells; however, due to the specific aims of the current study, these observations were not investigated further. rOROV formed plaques on the rodent, monkey, human, and sheep cell lines that were investigated (Fig. 1G). At 3 days p.i. in BHK-21 cells, rOROV plaques were larger than in the other cell lines, and on A549 cells, the plaques were harder to visualize (Fig. 1G). Based on these results, BHK-21 cells were chosen for virus titration, while Vero E6 and A549 cells were chosen for the purpose of initial characterization of all recombinant viruses in this study. Vero E6 and BHK-21 cells both lack fully functional IFN systems (34–36), while A549 cells are IFN competent (37).
Generation of OROV mutants.
Using our newly established OROV rescue system, we generated OROV mutant viruses as described below.
(i) NSm deletion.
A mutant OROV lacking the entire NSm ORF from the M segment was generated. This was done by deleting the entire NSm coding region immediately after the first NSm transmembrane domain (TMD) and predicted cleavage site up to the third TMD, leaving the predicted cleavage site of the Gc protein intact (Fig. 2A). These sites were predicted using the TMHMM Server v. 2.0 and SignalP 4.1 Server algorithms (http://www.cbs.dtu.dk) based on work done by Xiaohong Shi (MRC-University of Glasgow, Centre for Virus Research) for the characterization of orthobunyavirus M segments (X. Shi and R. M. Elliott, submitted for publication). Primers delNSmOROVF and delNSmOROVR (Table 1) were designed to bind to positions 1475 to 1498 and 1036 to 1013 of the M segment, respectively. This allowed an excision PCR to be performed, thereby deleting the entire NSm region but leaving the first TMD site, so as not to alter the position of the Gc protein in the endoplasmic reticulum and Golgi, during folding. To rescue rOROVdelNSm virus, BSR-T7/5 cells were transfected with 1 μg of pTVTOROVL, pTVTOROVS, and pTVTOROVdelNSm plasmids. At 7 days p.t., infectious virus particles were recovered, titrated, and sequenced (Source Bioscience) to confirm the mutation.
FIG 2.
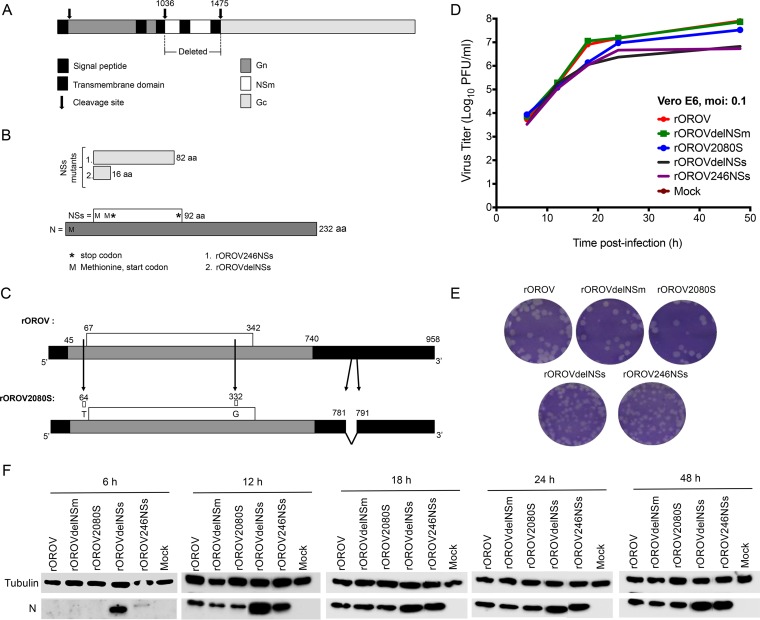
Creation of OROV mutant viruses. (A) Schematic of the M segment showing Gn, NSm, and Gc regions. The arrows depict where cleavage occurs. The patterned box indicates the signal peptide, and the black boxes represent transmembrane domains. Nucleotides 1036 to 1475 were deleted in order to generate delNSm M segment. (B) S segment products N and NSs. NSs is coded from an overlapping reading frame with N. Schematic shows how NSs mutants differ from the wt. rOROV246NSs has a stop codon (asterisk) placed at nucleotide (nt) position 314 of S segment cDNA, changing TTA to TAA and thereby deleting the last 8 aa. rOROVdelNSs has a stop codon at cDNA nt position 116, changing TGG to TAG so that a stop codon is generated immediately after the second start codon (methionine [M]). Numbers are amino acid lengths. (C) rOROV2080S S segment in comparison to wtOROV and rOROV S segment. Numbers are nucleotide positions. Arrows show where changes occur. First two positions generate a variation in the NSs OROF. Black highlights the UTRs. (D) Growth properties of recombinant viruses in Vero E6 cells. Cells were infected at an MOI of 0.1. Samples were harvested at the indicated time points and titrated on BHK-21 cells. The graph shows results of a representative experiment. (E) Plaque phenotype of recombinant viruses in BHK-21 cells. A plaque assay was carried out, and at 3 days p.i., cells were fixed and stained with crystal violet. (F) N production in recombinant viruses. Cell lysates from the growth curve (D) were probed for OROV-N and tubulin.
(ii) NSs mutants.
The following step was the creation of the NSs mutant viruses. As NSs lies in an overlapping reading frame within the N ORF, the positions at which mutations could be introduced were limited. The NSs ORF of OROV has four in-frame methionines; therefore, in an attempt to abrogate NSs transcription, the NSs start codon was left in place and instead a translational stop codon was inserted in frame immediately after the second methionine at position 17 (Fig. 2B, 2. rOROVdelNSs). At the nucleotide level, this is at position 115 and changes TGG (W) to TAG (stop), resulting in a 48-nt NSs ORF. The reason for this approach was that previous work done on BUNV revealed that when the start codon of NSs was removed, the virus was still capable of producing an NSs protein from a downstream methionine (38). The strategy used in this study for OROV is similar to the one used to create an SBV mutant lacking NSs (32). In addition to this, a C-terminally truncated NSs was also engineered. This was generated by introducing a stop codon at nt position 313, changing TTA (L) to TAA (stop). This resulted in a 246-nt NSs ORF and a protein sequence of 82 amino acids (aa), compared to 92 aa for wt NSs (Fig. 2B, 1. rOROV246NSs). Primers used in generating the plasmids are in Table 1. In order to rescue the NSs mutants (named rOROVdelNSs and rOROV246NSs), BSR-T7/5 cells were transfected with 1 μg of pTVTOROVL and pTVTOROVM and 1.5 μg of the mutant S segment (pTVTOROVdelNSs or pTVTORO246NSs). At 7 days p.t., infectious virus particles were recovered and titrated and the entire NSs ORF was sequenced (Source Bioscience) to confirm mutations.
(iii) S-segment mutant.
In a previous work (24), we reported the isolation and sequencing of OROV clinical isolates that differ from the prototype strain (BeAn19991) in the S segment, as they lack 11 nt at position 781 to 791 in the 3′ untranslated region (UTR). The NSs ORFs of these viruses also contain a tandem AUG translation start codon created by a C-U variation at position 332 and a Gln-to-Arg change in the NSs ORF at position 89 (Fig. 2C, rOROV2080S). To test whether these variations altered the in vitro growth properties of the rescued virus, a cDNA plasmid (designated pTVTOROV2080S) containing the S segment of clinical isolate BeH759025 (GenBank accession number KP691614 [24]) was generated using the same cloning strategy as for pTVTOROVS (27). In order to rescue this S-segment mutant (named rOROV2080S), BSR-T7/5 cells were transfected with plasmids pTVTOROVL, pTVTOROVM, and pTVTOROV2080S (1 μg each). At 7 days p.t., infectious virus particles were recovered and titrated and the entire S segment was sequenced (Source Bioscience).
All mutant viruses in this study were passaged three times at a low MOI in Vero E6 cells and sequenced. Introduced mutations were maintained, confirming the stability of these viruses. Subsequent experiments utilized viruses from passage two.
Growth properties of recombinant viruses in mammalian cell lines and their effect on host protein synthesis.
Growth kinetics of rOROV, rOROVdelNSm, rOROVdelNSs, rOROV246NSs, and rOROV2080S were compared in Vero E6 cells at an MOI of 0.1. rOROV, rOROVdelNSm, and rOROV2080S replicate with similar efficiencies; however, mutants rOROVdelNSs and rOROV246NSs appeared attenuated and reached titers that were 1 log lower than that of rOROV (Fig. 2D). Western blotting revealed larger amounts of N protein from rOROVdelNSs at earlier time points, suggesting a possibly increased efficiency of the virus in translating N (Fig. 2F). Plaque morphologies of the recombinant viruses were then compared on BHK-21 cells. rOROV, rOROVdelNSm, and rOROV2080S produced plaques with a round morphology and were clear and similar to each other. The plaques of viruses rOROVdelNSs and rOROV246NSs, on the other hand, were smaller, with corrugated and ill-defined borders (Fig. 2E).
To investigate whether the recombinant viruses caused inhibition of host cell protein synthesis, Vero E6 cells were infected at an MOI of 3, and at 12, 24, and 48 h p.i., cells were radiolabeled with [35S]methionine. Cell extracts were analyzed by SDS-PAGE. rOROV, rOROVdelNSm, and rOROV2080S, as well as the rOROVdelNSs and rOROV246NSs, demonstrated an ability to cause host translation shutoff by 24 h p.i. (Fig. 3). It was also observed that the last two viruses produced noticeably more N protein at this time point than did the other viruses. This result also confirmed that the mutant viruses rOROVdelNSm and rOROVdelNSs do not express NSm and NSs proteins, respectively, and that the rOROV246NSs virus expresses a truncated version of NSs (Fig. 3).
FIG 3.
Host cell protein shutoff. Vero E6 cells were infected with rOROV, rOROVdelNSm, rOROV2080S, rOROVdelNSs, or rOROV246NSs or mock infected. Cells were infected at an MOI of 3 and incubated at 37°C. At the indicated time points, the cells were labeled with [35S]methionine for 2 h. Cells lysates were then separated by SDS-PAGE. Arrows indicate the position of viral proteins Gc, N, NSm, and NSs.
As rOROV2080S behaves similarly to rOROV and rOROV246NSs behaves similarly to rOROVdelNSs in terms of in vitro replication kinetics, only rOROV, rOROVdelNSm, and rOROVdelNSs were focused on for growth comparison in IFN-competent A549 cells. rOROV and rOROVdelNSm grew with similar kinetics and reached comparable titers, whereas rOROVdelNSs growth appeared more restrictive and at 48 h the viral titers were almost 2 logs lower than those of rOROV and rOROVdelNSm (Fig. 4A). Western blot analysis of N expression showed smaller amounts of protein in the rOROVdelNSs-infected cells (Fig. 4B). Next, the growth of rOROV, rOROVdelNSm, and rOROVdelNSs in A549 cells was compared to their growth in IFN-incompetent A549/V cells. These cells express the V protein of parainfluenza type 5 virus, thereby blocking type I IFN signaling via STAT1 degradation (39). Cells were infected at an MOI of 0.001, and titers were measured at 48 h p.i. Cells were also infected with BUNV or a BUNV mutant lacking the NSs protein (rBUNVdelNSs2) for comparison (40, 41). All viruses grew to higher titers in the IFN-incompetent cell line, similar to BUNV. rOROVdelNSs titers were over 1 log higher in A549/V cells than in A549 cells, although this difference was not as high as with rBUNVdelNSs2 (Fig. 4C). Western blotting for N confirmed lower levels of expression in A549 cells infected with rOROVdelNSs and rBUNVdelNSs2, corresponding with the yield assay (Fig. 4D).
FIG 4.
Growth properties of recombinant viruses in A549 cells. (A) Growth kinetics of rOROV, rOROVdelNSm, and rOROVdelNSs in A549 cells at an MOI of 0.1. At the indicated time points, samples were harvested and viral titers determined by plaque assay on BHK-21 cells. The graph presents results for one representative experiment. (B) Western blot for cell lysates from growth curve (A). Lysates were separated by SDS-PAGE and probed for OROV N and tubulin. (C) Comparison of growth properties in A549 and A549/V cells. Cells were infected at an MOI of 0.001 with the indicated viruses. Forty-eight hours p.i., viral titers were determined by plaque assay. BUNV was used for comparison. Bars indicate SDs (n = 3; **, P < 0.01 by Student's t test). (D) Western blot analysis for panel C. Cells lysates were probed for viral N protein. Tubulin was probed as a loading control.
OROV NSs protein inhibits type I IFN production in A549 cells.
We measured IFN production in A549 cells in response to infection with rOROV, rOROVdelNSm, rOROV2080S, rOROVdelNSs, or rOROV246NSs at an MOI of 1. For comparison, we also infected cells with BUNV or rBUNVdelNSs2. At 24 h p.i., the medium from infected monolayers was collected, infectious virus was inactivated by UV treatment, and the amount of IFN present was measured in a biological protection assay as described previously (42). As expected, no IFN was produced from mock- or BUNV-infected cells, and rBUNVdelNSs2-infected cells produced considerable amounts of IFN. While rOROV, rOROVdelNSm, and rOROV2080S induced small amounts of IFN, rOROVdelNSs induced large amounts (Fig. 5A). rOROV246NSs, which lacks only 9 aa at the NSs protein C terminus, induced IFN to the same extent as rOROVdelNSs (Fig. 5A and B). Next, we used Western blotting to probe the A549 cell extracts for STAT1, phosphorylated STAT1 (pSTAT1), and the interferon-stimulated gene (ISG) protein MxA. pSTAT1 and MxA expression was detected in cells infected with rOROVdelNSs or rOROV246NSs but not in cells infected with rOROV, rOROV2080S, or rOROVdelNSm (Fig. 5C), confirming that OROV NSs is an IFN antagonist.
FIG 5.
Biological interferon production assay. A549 cells were infected at an MOI of 1 with BUNV, rBUNdelNSs2, rOROV, rOROVdelNSm, rOROV2080S, rOROVdelNSs, or rOROV246NSs or mock infected. Supernatant was harvested at 24 h p.i., and cell extracts were separated by SDS-PAGE. (A) UV-inactivated supernatant was used to pretreat A549-N pro cells prior to infection with EMCV. At 3 days p.i., cells were fixed and stained with crystal violet. (B) Graph calculated from panel A, presenting relative IFN units expressed as 2N where N is the number of 2-fold dilutions that offered protection. (C) Cells extracts were probed for OROV N, STAT1, pSTAT1, and MxA. Tubulin was probed as a loading control.
OROV is less sensitive to IFN-α treatment than BUNV.
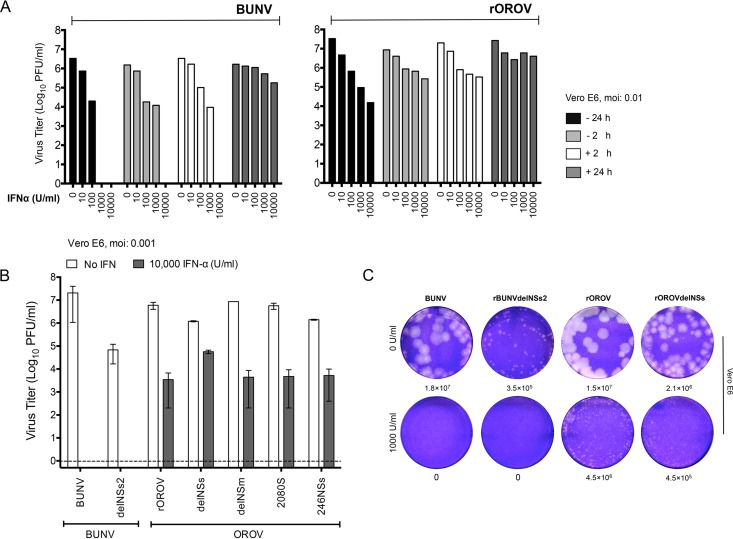
BUNV replication was previously shown to be highly sensitive to IFN-α (43). To test if OROV was equally sensitive, Vero E6 cells (which cannot produce but can respond to IFN [36]) were treated with increasing doses of universal type 1 IFN-α (0, 10, 100, 1,000, and 10,000 U/ml), either preinfection (− 24 or − 2 h) or postinfection (+ 2 or + 24 h). Cells were infected with BUNV or OROV at an MOI of 0.01, and IFN-α was maintained in the medium throughout the infection period. At 48 h p.i., the amount of infectious virus in the culture medium was estimated by plaque assays. While both viruses showed sensitivity to IFN, OROV was clearly less sensitive than BUNV (Fig. 6A). For example, pretreating cells with 10,000 U of IFN-α either 2 or 24 h preinfection completely inhibited BUNV replication, as did treating cells with 10,000 U at 2 h p.i. In contrast, there was only a 1- to 2-log reduction in the titers of OROV in cells pretreated for 2 h with 10,000 U of IFN-α prior to infection and a 3-log reduction in cells pretreated for 24 h. Furthermore, while pretreating cells with 1,000 U of IFN-α for 24 h preinfection completely inhibited BUNV, there was only a 2-log reduction in cells infected with OROV (Fig. 6A). We have repeated the experiment with rOROV using 10,000 U/ml of IFN and at MOIs of 0.001 and 0.01. At 24 or 48 h p.i. at both MOIs, rOROV replication was not completely inhibited, as observed with BUNV, with titers decreased by 2 to 3 logs compared to the values for untreated cells (data not shown). Viruses rOROVdelNSm, rOROV2080S, rOROVdelNSs, and rOROV246NSs demonstrate a sensitivity to IFN-α similar to that of rOROV (Fig. 6B), indicating that the increased resistance of OROV to IFN-α compared to that of BUNV is not due to expression of a functional NSs protein. Next, we investigated the plaque morphology on pretreated Vero E6 cells for rOROV and rOROVdelNSs in comparison to BUNV and rBUNVdelNSs2. A 1,000-U/ml concentration of IFN-α was maintained in the overlay during the infection period. No BUNV or rBUNVdelNSs plaques were observed when the plaque assays were performed in the presence of IFN-α. In contrast, rOROV and rOROVdelNSs plaques were observed in the presence of IFN-α, although they were considerably smaller than those on untreated cells (Fig. 6D). Taken together, these results demonstrate that in the tested cells and with the MOI of virus used, OROV is sensitive to IFN-α in a dose-dependent manner; however, it is significantly more resistant than BUNV. Furthermore, the NSs protein is not responsible for this increased resistance.
FIG 6.
Sensitivity of OROV to IFN-α treatment. (A) IFN-α sensitivity test. Vero E6 cells were treated with increasing concentrations of IFN-α (0, 10, 100, 1,000, and 10,000) either before (−) or after (+) infection. Cells were infected with BUNV or rOROV at an MOI of 0.01. Forty-eight hours p.i., supernatant was harvested and viral titers were determined by plaque assay on BHK-21 cells. Graphs show results of a representative experiment. (B) Vero E6 cells were treated with 10,000 U/ml of IFN-α 24 h prior to infection with indicated viruses at an MOI of 0.001. Samples were harvested at 48 h p.i., and viral titers were determined by plaque assay on BHK-21 cells. Bars represent ranges from two experiments. (C) Vero E6 cells were treated (1,000 U/ml) or not (0 U/ml) with IFN-α 24 h prior to infection. A plaque assay for BUNV, rBUNVdelNSs2, OROV, or rOROVdelNSs was performed. Four days p.i., cells were fixed and stained with crystal violet.
Replication of recombinant viruses in mosquito cell lines.
We have also compared the growth kinetics of rOROV, rOROVdelNSm and rOROVdelNSs in mosquito cell lines U4.4 (Aedes albopictus) and Aag2 (Aedes aegypti). Interestingly, and unlike the situation in mammalian cells, rOROVdelNSs grows to similar levels as rOROV (Fig. 7). In both U4.4 and Aag2, it appears that rOROVdelNSm grows to slightly higher titers than the other viruses. Investigating this further was beyond the scope of the current study.
FIG 7.
Growth kinetics in mosquito cells. Cells were infected with rOROV, rOROVdelNSm, or rOROVdelNSs at an MOI of 0.1. At the indicated time points, samples were harvested and viral titers were determined by plaque assay. Presented graphs are representative experiments. Cell extracts were separated by SDS-PAGE and probed for viral N and tubulin. (A) Replication in U4.4 cells; (B) replication in Aag2 cells; (C) N production in U4.4 cells; (D) N production in Aag2 cells.
DISCUSSION
In this paper, we describe the successful recovery of OROV in cultured cells entirely from cloned cDNAs. OROV is a midge-borne orthobunyavirus that causes a febrile illness in the South American human population. The virus has caused over half a million infections, and though not fatal, its dengue-like symptoms can persist for weeks and in a few cases can progress into more severe symptoms, such as meningitis and encephalitis (18). OROV is closely related to SBV, another Simbu virus also spread by biting midges from the genus Culicoides. SBV causes severe fetal malformations in ruminants in Europe but has not been known to infect humans. Using the previously described reverse genetics system for SBV (32, 44) and our newly established OROV rescue system, we can begin to understand the basis for pathogenicity in humans and study reasons for such a species barrier. In addition to this, we can also begin to study the M-segment variations that are found among Oropouche species. OROV M-segment reassortants Iquitos and Madre de Dios viruses can cause disease in humans (25, 26), while Perdoes virus and the more divergent Jatobal virus have only been isolated from nonhuman primates (Callithrix penicillata) and the South American coati (Nasua nasua), respectively (24, 45). It is interesting that members of the Oropouche species display a broad phylogenetic diversity predominately due to the M segment. The variations observed in the M segments of these viruses could have resulted from either genomic reassortment or extensive adaptation to different hosts and habitats. Using the OROV reverse genetics system established in this study, it would now be possible to study in detail these differences in terms of pathogenesis, virulence outcome, and host range of these viruses. This work would contribute to understanding the evolution of clade A Simbu serogroup viruses within South America.
The recombinant OROV (rOROV) that we have generated replicates similarly to the authentic virus (wild-type OROV [wtOROV]), reaching titers of 107 PFU/ml (Fig. 1B). Using this system, mutant viruses lacking either the NSm or the NSs protein were also generated. Only some bunyaviruses encode these proteins, and the exact role played by the NSm protein in orthobunyavirus infections remains unclear. Work on BUNV NSm demonstrated that the protein can localize to the Golgi efficiently on its own (46) and may play a role in viral assembly (47). In Rift Valley fever virus (RVFV), the NSm protein is important for infection in mosquitoes by allowing the virus to cross the midgut barrier (48, 49). Similarly, in tospoviruses, the NSm protein has been shown to be important for virus cell-to-cell spread (50–52). Results from our work indicate that for OROV, the NSm protein is dispensable for virus replication in cultured cells, as rOROVdelNSm grows and replicates similarly to rOROV (Fig. 2D and E, 4, and 7). We have previously (24) discussed the sequence similarity of the M-segment genes between different OROV reassortants and noted that the NSm region of the M polyprotein of all these viruses is highly conserved compared to the Gn and Gc glycoproteins, which could indicate that this portion of the polyprotein is less prone to mutation due to a common, yet unknown, selective pressure. Future work could include performing mutations on the NSm coding sequence and monitor for effects on virus replication in more relevant primary cell lines and in vivo models, such as insects. Similarly, the rOROV2080S mutant generated in this study would also require in vivo characterization in order to determine if the S-segment difference observed between OROV isolates offers any advantage over the prototype BeAn19991 S segment, as the current study was not sufficient to determine this.
As with other NSs-encoding bunyaviruses, OROV NSs protein is an IFN antagonist, and by deleting the NSs ORF, OROV induces high levels of IFN and thus induces STAT1 phosphorylation and MxA expression (Fig. 5, rOROVdelNSs). Interestingly, the C-terminally truncated NSs mutant was also incapable of inhibiting type I IFN production (Fig. 5, rOROV246NSs). Work on BUNV and RVFV has demonstrated that NSs inhibits IFN-β activation downstream of transcriptional activation through disruption of DNA-dependent RNA polymerase II (RNAPII) activity (53–55). BUNV NSs interacts with subunit MED8 of the RNAPII regulatory module (56), preventing Ser2 phosphorylation and hence preventing elongation and 3′-end processing of the nascent mRNA transcript (57–59). This was initially thought to be due to an interaction of BUNV NSs C terminus (aa 83 to 91) with MED8; however, a BUNV NSs mutant lacking an N terminus of 21 amino acids is also unable to degrade RNAPII, indicating that both the C and the N termini are important for BUNV NSs function (38, 56, 60). The BUNV MED8 binding domain was mapped to a C-terminal amino acid motif LPS, which is conserved in orthobunyavirus NSs proteins (56); interestingly, OROV C-terminal mutant rOROV246NSs also lacks a similar motif, LPC (see Fig. S1A in the supplemental material). This LPC motif is conserved among only the clade A viruses in the Simbu serogroup (see Fig. S1B). Whether the inability of rOROV246NSs to inhibit IFN production is due to its lack of the MED8 binding domain will be investigated in follow-up studies. La Crosse virus (LACV) and SBV NSs function as IFN antagonists by targeting RNAPII for degradation by the proteasome (61–63). Mutations to the C terminus of SBV NSs have also been shown to affect the protein's ability to degrade RNAPII (63). In the phlebovirus RVFV, the NSs protein interacts with subunits of the general transcription factor TFIIH, which also has a role in RNAPI transcription (64). SFTSV NSs forms viral inclusion bodies in the cytoplasm and uses these to capture kinases TBK1 and IKKε and proteins STAT1 and STAT2 (65, 66). Recently, a study comparing 6-week-old C57BL/6 mouse knockout mutants demonstrated that mitochondrial antiviral-signaling protein (MAVS) activation plays a crucial role in type 1 IFN signaling during OROV infection (67); it would be interesting to see how the rOROVdelNSs and rOROV246NSs mutants replicate in such in vivo systems. Interestingly, both rOROVdelNSs and rOROV246NSs are attenuated in Vero E6 (Fig. 2D) and in BHK-21 (Fig. 2E) cells, both of which lack fully functional IFN systems, and from radiolabeling experiments, we know that rOROVdelNSs is capable of causing translational shutoff in Vero E6 cells (Fig. 3). Using the OROV reverse genetics system, we can now begin to study protein-protein interactions and investigate the role of the NSs protein further.
This study also showed that while OROV is sensitive to IFN-α, to see maximal effects, cells have to be treated for 24 h prior to infection (Fig. 6A, rOROV). In contrast, BUNV is highly sensitive to IFN-α (Fig. 6A). These findings are consistent with previously published work demonstrating a resistance of OROV to the antiviral effects of IFN-α both in vivo and in vitro in comparison to other pathogenic orthobunyaviruses (68). The reasons for the differences in relative sensitivity of OROV and BUNV to IFN-α are under investigation, but the differences may, for example, be due to the differential effects of certain ISGs on these viruses or on the ability of OROV to more rapidly switch off host cell gene expression than BUNV. Whatever the reason, the increased resistance of OROV to IFN-α is not due to expression of the NSs protein, as rOROVdelNSs shows a sensitivity to IFN-α similar to that of OROV.
In conclusion, our present work has shown that we are able to generate infectious OROV entirely from cDNA and that similar to the case with other bunyaviruses, OROV NSs is an IFN antagonist. We have also demonstrated that the NSm protein appears to be nonessential for virus replication in the cultured cells that were tested. The work we have presented here will now enable us to study OROV in more detail in order to establish the molecular details involved in viral replication and pathogenesis, and potentially to generate attenuated vaccine strains. The work we present here is an important move forward toward understanding this important yet neglected human pathogen.
Supplementary Material
ACKNOWLEDGMENTS
We thank Xiaohong Shi for advice on creating rOROVdelNSm and Massimo Palmarini for OROV-N antibody and a batch of IFN-α. We thank Stephen Welch for reviewing the manuscript and Mark Tilston for proofreading it.
This paper is dedicated to the memory of Richard M. Elliott, who will forever hold a special place in our hearts as a friend, father figure, and mentor.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.02849-15.
REFERENCES
- 1.Elliott RM. 2014. Orthobunyaviruses: recent genetic and structural insights. Nat Rev Microbiol 12:673–685. doi: 10.1038/nrmicro3332. [DOI] [PubMed] [Google Scholar]
- 2.Elliott RM, Brennan B. 2014. Emerging phleboviruses. Curr Opin Virol 5:50–57. doi: 10.1016/j.coviro.2014.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nunes MR, Martins LC, Rodrigues SG, Chiang JO, Azevedo Rdo S, da Rosa AP, Vasconcelos PF. 2005. Oropouche virus isolation, southeast Brazil. Emerg Infect Dis 11:1610–1613. doi: 10.3201/eid1110.050464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dixon KE, Travassos da Rosa AP, Travassos da Rosa JF, Llewellyn CH. 1981. Oropouche virus. II. Epidemiological observations during an epidemic in Santarem, Para, Brazil in 1975. Am J Trop Med Hyg 30:161–164. [PubMed] [Google Scholar]
- 5.Pinheiro FP, Hoch AL, Gomes ML, Roberts DR. 1981. Oropouche virus. IV. Laboratory transmission by Culicoides paraensis. Am J Trop Med Hyg 30:172–176. [PubMed] [Google Scholar]
- 6.Vasconcelos HB, Azevedo RS, Casseb SM, Nunes-Neto JP, Chiang JO, Cantuaria PC, Segura MN, Martins LC, Monteiro HA, Rodrigues SG, Nunes MR, Vasconcelos PF. 2009. Oropouche fever epidemic in Northern Brazil: epidemiology and molecular characterization of isolates. J Clin Virol 44:129–133. doi: 10.1016/j.jcv.2008.11.006. [DOI] [PubMed] [Google Scholar]
- 7.Baisley KJ, Watts DM, Munstermann LE, Wilson ML. 1998. Epidemiology of endemic Oropouche virus transmission in upper Amazonian Peru. Am J Trop Med Hyg 59:710–716. [DOI] [PubMed] [Google Scholar]
- 8.Mourao MP, Bastos MS, Gimaqu JB, Mota BR, Souza GS, Grimmer GH, Galusso ES, Arruda E, Figueiredo LT. 2009. Oropouche fever outbreak, Manaus, Brazil, 2007-2008. Emerg Infect Dis 15:2063–2064. doi: 10.3201/eid1512.090917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bastos Mde S, Figueiredo LT, Naveca FG, Monte RL, Lessa N, Pinto de Figueiredo RM, Gimaque JB, Pivoto Joao G, Ramasawmy R, Mourao MP. 2012. Identification of Oropouche Orthobunyavirus in the cerebrospinal fluid of three patients in the Amazonas, Brazil. Am J Trop Med Hyg 86:732–735. doi: 10.4269/ajtmh.2012.11-0485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pinheiro FP, Travassos da Rosa AP, Travassos da Rosa JF, Bensabath G. 1976. An outbreak of Oropouche virus disease in the vicinity of Santarem, Para, Brazil. Tropenmed Parasitol 27:213–223. [PubMed] [Google Scholar]
- 11.Pinheiro FP, Travassos da Rosa AP, Gomes ML, LeDuc JW, Hoch AL. 1982. Transmission of Oropouche virus from man to hamster by the midge Culicoides paraensis. Science 215:1251–1253. doi: 10.1126/science.6800036. [DOI] [PubMed] [Google Scholar]
- 12.Roberts DR, Hoch AL, Dixon KE, Llewellyn CH. 1981. Oropouche virus. III. Entomological observations from three epidemics in Para, Brazil, 1975. Am J Trop Med Hyg 30:165–171. [PubMed] [Google Scholar]
- 13.Anderson CR, Spence L, Downs WG, Aitken TH. 1961. Oropouche virus: a new human disease agent from Trinidad, West Indies. Am J Trop Med Hyg 10:574–578. [DOI] [PubMed] [Google Scholar]
- 14.Pinheiro FP, Travassos da Rosa AP, Travassos da Rosa JF, Ishak R, Freitas RB, Gomes ML, LeDuc JW, Oliva OF. 1981. Oropouche virus. I. A review of clinical, epidemiological, and ecological findings. Am J Trop Med Hyg 30:149–160. [PubMed] [Google Scholar]
- 15.Roberts DR, Pinheiro FDPP, Hoch AL, LeDuc JW, Peterson NE, Santos MAV, Western KA. 1977. Vectors and natural reservoirs of Oropouche virus in the Amazon region. US Army Medical Research and Development Command, Washington, DC. [Google Scholar]
- 16.Smith GC, Francy DB. 1991. Laboratory studies of a Brazilian strain of Aedes albopictus as a potential vector of Mayaro and Oropouche viruses. J Am Mosquito Control Assoc 7:89–93. [PubMed] [Google Scholar]
- 17.Pinheiro FP, Pinheiro M, Bensabath G, Causey OR, Shope RE. 1962. Epidemia de vírus Oropouche em Belém. Revista do Serv Especial Saude 12:15–23. [Google Scholar]
- 18.Pinheiro FP, Travassos da Rosa AP, Vasconcelos PF. 2004. Bunyaviridae: other Bunyaviridae. Oropouche fever, p 2418–2423. In Feigin RD, Cherry J, Demmler GJ, Kaplan SL (ed), Textbook of pediatric infectious diseases, 5th ed. Saunders, Philadelphia, PA. [Google Scholar]
- 19.Watts DM, Phillips I, Callahan JD, Griebenow W, Hyams KC, Hayes CG. 1997. Oropouche virus transmission in the Amazon River basin of Peru. Am J Trop Med Hyg 56:148–152. [DOI] [PubMed] [Google Scholar]
- 20.Mercer DR, Castillo-Pizango MJ. 2005. Changes in relative species compositions of biting midges (Diptera: Ceratopogonidae) and an outbreak of Oropouche virus in Iquitos, Peru. J Med Entomol 42:554–558. doi: 10.1093/jmedent/42.4.554. [DOI] [PubMed] [Google Scholar]
- 21.Rosa AP, Rodrigues SG, Nunes MR, Magalhaes MT, Rosa JF, Vasconcelos PF. 1996. Outbreak of oropouche virus fever in Serra Pelada, municipality of Curionopolis, Para, 1994. Rev Soc Bras Med Trop 29:537–541. (In Portuguese.) [DOI] [PubMed] [Google Scholar]
- 22.Pan American Health Organization. 2010. Epidemiological alert. Outbreak of Oropouche fever. Pan American Health Organization, Washington, DC: http://www1.paho.org/hq/dmdocuments/2010/epi_alert_2010_22_June_Oropouche_Fever.pdf. [Google Scholar]
- 23.Forshey BM, Guevara C, Laguna-Torres VA, Cespedes M, Vargas J, Gianella A, Vallejo E, Madrid C, Aguayo N, Gotuzzo E, Suarez V, Morales AM, Beingolea L, Reyes N, Perez J, Negrete M, Rocha C, Morrison AC, Russell KL, Blair JP, Olson JG, Kochel TJ. 2010. Arboviral etiologies of acute febrile illnesses in Western South America, 2000–2007. PLoS Negl Trop Dis 4:e787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tilston-Lunel NL, Hughes J, Acrani GO, da Silva DE, Azevedo RS, Rodrigues SG, Vasconcelos PF, Nunes MR, Elliott RM. 2015. Genetic analysis of members of the species Oropouche virus and identification of a novel M segment sequence. J Gen Virol 96:1636–1650. doi: 10.1099/vir.0.000108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aguilar PV, Barrett AD, Saeed MF, Watts DM, Russell K, Guevara C, Ampuero JS, Suarez L, Cespedes M, Montgomery JM, Halsey ES, Kochel TJ. 2011. Iquitos virus: a novel reassortant orthobunyavirus associated with human illness in Peru. PLoS Negl Trop Dis 5:e1315. doi: 10.1371/journal.pntd.0001315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ladner JT, Savji N, Lofts L, Travassos da Rosa A, Wiley MR, Gestole MC, Rosen GE, Guzman H, Vasconcelos PF, Nunes MR, Lipkin WI, Tesh RB, Palacios G. 2014. Genomic and phylogenetic characterization of viruses included in the Manzanilla and Oropouche species complexes of the genus Orthobunyavirus, family Bunyaviridae. J Gen Virol 95:1055–1066. doi: 10.1099/vir.0.061309-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Acrani GO, Tilston-Lunel NL, Spiegel M, Weidemann M, Dilcher M, da Silva DE, Nunes MR, Elliott RM. 2014. Establishment of a minigenome system for Oropouche virus reveals the S genome segment to be significantly longer than reported previously. J Gen Virol 95:513–523. [DOI] [PubMed] [Google Scholar]
- 28.Bridgen A, Elliott RM. 1996. Rescue of a segmented negative-strand RNA virus entirely from cloned complementary DNAs. Proc Natl Acad Sci U S A 93:15400–15404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Buchholz UJ, Finke S, Conzelmann KK. 1999. Generation of bovine respiratory syncytial virus (BRSV) from cDNA: BRSV NS2 is not essential for virus replication in tissue culture, and the human RSV leader region acts as a functional BRSV genome promoter. J Virol 73:251–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shi X, Elliott RM. 2009. Generation and analysis of recombinant Bunyamwera orthobunyaviruses expressing V5 epitope-tagged L proteins. J Gen Virol 90:297–306. doi: 10.1099/vir.0.007567-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Johnson KN, Zeddam JL, Ball LA. 2000. Characterization and construction of functional cDNA clones of Pariacoto virus, the first Alphanodavirus isolated outside Australasia. J Virol 74:5123–5132. doi: 10.1128/JVI.74.11.5123-5132.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Elliott RM, Blakqori G, van Knippenberg IC, Koudriakova E, Li P, McLees A, Shi X, Szemiel AM. 2013. Establishment of a reverse genetics system for Schmallenberg virus, a newly emerged orthobunyavirus in Europe. J Gen Virol 94:851–859. doi: 10.1099/vir.0.049981-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brennan B, Welch SR, Elliott RM. 2014. The consequences of reconfiguring the ambisense S genome segment of Rift Valley fever virus on viral replication in mammalian and mosquito cells and for genome packaging. PLoS Pathog 10:e1003922. doi: 10.1371/journal.ppat.1003922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Emeny JM, Morgan MJ. 1979. Regulation of the interferon system: evidence that Vero cells have a genetic defect in interferon production. J Gen Virol 43:247–252. [DOI] [PubMed] [Google Scholar]
- 35.Chinsangaram J, Piccone ME, Grubman MJ. 1999. Ability of foot-and-mouth disease virus to form plaques in cell culture is associated with suppression of alpha/beta interferon. J Virol 73:9891–9898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Desmyter J, Melnick JL, Rawls WE. 1968. Defectiveness of interferon production and of rubella virus interference in a line of African green monkey kidney cells (Vero). J Virol 2:955–961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Spann KM, Tran KC, Chi B, Rabin RL, Collins PL. 2004. Suppression of the induction of alpha, beta, and lambda interferons by the NS1 and NS2 proteins of human respiratory syncytial virus in human epithelial cells and macrophages [corrected]. J Virol 78:4363–4369. doi: 10.1128/JVI.78.8.4363-4369.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.van Knippenberg I, Carlton-Smith C, Elliott RM. 2010. The N-terminus of Bunyamwera orthobunyavirus NSs protein is essential for interferon antagonism. J Gen Virol 91:2002–2006. doi: 10.1099/vir.0.021774-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Killip MJ, Young DF, Gatherer D, Ross CS, Short JA, Davison AJ, Goodbourn S, Randall RE. 2013. Deep sequencing analysis of defective genomes of parainfluenza virus 5 and their role in interferon induction. J Virol 87:4798–4807. doi: 10.1128/JVI.03383-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bridgen A, Weber F, Fazakerley JK, Elliott RM. 2001. Bunyamwera bunyavirus nonstructural protein NSs is a nonessential gene product that contributes to viral pathogenesis. Proc Natl Acad Sci U S A 98:664–669. doi: 10.1073/pnas.98.2.664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hart TJ, Kohl A, Elliott RM. 2009. Role of the NSs protein in the zoonotic capacity of Orthobunyaviruses. Zoonoses Public Health 56:285–296. doi: 10.1111/j.1863-2378.2008.01166.x. [DOI] [PubMed] [Google Scholar]
- 42.Hale BG, Knebel A, Botting CH, Galloway CS, Precious BL, Jackson D, Elliott RM, Randall RE. 2009. CDK/ERK-mediated phosphorylation of the human influenza A virus NS1 protein at threonine-215. Virology 383:6–11. doi: 10.1016/j.virol.2008.10.002. [DOI] [PubMed] [Google Scholar]
- 43.Streitenfeld H, Boyd A, Fazakerley JK, Bridgen A, Elliott RM, Weber F. 2003. Activation of PKR by Bunyamwera virus is independent of the viral interferon antagonist NSs. J Virol 77:5507–5511. doi: 10.1128/JVI.77.9.5507-5511.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Varela M, Schnettler E, Caporale M, Murgia C, Barry G, McFarlane M, McGregor E, Piras IM, Shaw A, Lamm C, Janowicz A, Beer M, Glass M, Herder V, Hahn K, Baumgartner W, Kohl A, Palmarini M. 2013. Schmallenberg virus pathogenesis, tropism and interaction with the innate immune system of the host. PLoS Pathog 9:e1003133. doi: 10.1371/journal.ppat.1003133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Figueiredo LT, Da Rosa AP. 1988. Jatobal virus antigenic characterization by ELISA and neutralization test using EIA as indicator, on tissue culture. Mem Inst Oswaldo Cruz 83:161–164. [DOI] [PubMed] [Google Scholar]
- 46.Shi X, Kohl A, Leonard VH, Li P, McLees A, Elliott RM. 2006. Requirement of the N-terminal region of orthobunyavirus nonstructural protein NSm for virus assembly and morphogenesis. J Virol 80:8089–8099. doi: 10.1128/JVI.00579-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shi X, Kohl A, Li P, Elliott RM. 2007. Role of the cytoplasmic tail domains of Bunyamwera orthobunyavirus glycoproteins Gn and Gc in virus assembly and morphogenesis. J Virol 81:10151–10160. doi: 10.1128/JVI.00573-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kading RC, Crabtree MB, Bird BH, Nichol ST, Erickson BR, Horiuchi K, Biggerstaff BJ, Miller BR. 2014. Deletion of the NSm virulence gene of Rift Valley fever virus inhibits virus replication in and dissemination from the midgut of Aedes aegypti mosquitoes. PLoS Negl Trop Dis 8:e2670. doi: 10.1371/journal.pntd.0002670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Crabtree MB, Kent Crockett RJ, Bird BH, Nichol ST, Erickson BR, Biggerstaff BJ, Horiuchi K, Miller BR. 2012. Infection and transmission of Rift Valley fever viruses lacking the NSs and/or NSm genes in mosquitoes: potential role for NSm in mosquito infection. PLoS Negl Trop Dis 6:e1639. doi: 10.1371/journal.pntd.0001639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Soellick T, Uhrig JF, Bucher GL, Kellmann JW, Schreier PH. 2000. The movement protein NSm of tomato spotted wilt tospovirus (TSWV): RNA binding, interaction with the TSWV N protein, and identification of interacting plant proteins. Proc Natl Acad Sci U S A 97:2373–2378. doi: 10.1073/pnas.030548397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kormelink R, Storms M, Van Lent J, Peters D, Goldbach R. 1994. Expression and subcellular location of the NSM protein of tomato spotted wilt virus (TSWV), a putative viral movement protein. Virology 200:56–65. [DOI] [PubMed] [Google Scholar]
- 52.Storms MM, Kormelink R, Peters D, Van Lent JW, Goldbach RW. 1995. The nonstructural NSm protein of tomato spotted wilt virus induces tubular structures in plant and insect cells. Virology 214:485–493. [DOI] [PubMed] [Google Scholar]
- 53.Weber F, Bridgen A, Fazakerley JK, Streitenfeld H, Kessler N, Randall RE, Elliott RM. 2002. Bunyamwera bunyavirus nonstructural protein NSs counteracts the induction of alpha/beta interferon. J Virol 76:7949–7955. doi: 10.1128/JVI.76.16.7949-7955.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kohl A, Clayton RF, Weber F, Bridgen A, Randall RE, Elliott RM. 2003. Bunyamwera virus nonstructural protein NSs counteracts interferon regulatory factor 3-mediated induction of early cell death. J Virol 77:7999–8008. doi: 10.1128/JVI.77.14.7999-8008.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Billecocq A, Spiegel M, Vialat P, Kohl A, Weber F, Bouloy M, Haller O. 2004. NSs protein of Rift Valley fever virus blocks interferon production by inhibiting host gene transcription. J Virol 78:9798–9806. doi: 10.1128/JVI.78.18.9798-9806.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Léonard VH, Kohl A, Hart TJ, Elliott RM. 2006. Interaction of Bunyamwera Orthobunyavirus NSs protein with mediator protein MED8: a mechanism for inhibiting the interferon response. J Virol 80:9667–9675. doi: 10.1128/JVI.00822-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Eick D, Geyer M. 2013. The RNA polymerase II carboxy-terminal domain (CTD) code. Chem Rev 113:8456–8490. doi: 10.1021/cr400071f. [DOI] [PubMed] [Google Scholar]
- 58.Corden JL. 2013. RNA polymerase II C-terminal domain: tethering transcription to transcript and template. Chem Rev 113:8423–8455. doi: 10.1021/cr400158h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Robinson PJJ, Bushnell DA, Trnka MJ, Burlingame AL, Kornberg RD. 2012. Structure of the Mediator Head module bound to the carboxy-terminal domain of RNA polymerase II. Proc Natl Acad Sci U S A 109:17931–17935. doi: 10.1073/pnas.1215241109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Thomas D, Blakqori G, Wagner V, Banholzer M, Kessler N, Elliott RM, Haller O, Weber F. 2004. Inhibition of RNA polymerase II phosphorylation by a viral interferon antagonist. J Biol Chem 279:31471–31477. doi: 10.1074/jbc.M400938200. [DOI] [PubMed] [Google Scholar]
- 61.Verbruggen P, Ruf M, Blakqori G, Overby AK, Heidemann M, Eick D, Weber F. 2011. Interferon antagonist NSs of La Crosse virus triggers a DNA damage response-like degradation of transcribing RNA polymerase II. J Biol Chem 286:3681–3692. doi: 10.1074/jbc.M110.154799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Blakqori G, Delhaye S, Habjan M, Blair CD, Sanchez-Vargas I, Olson KE, Attarzadeh-Yazdi G, Fragkoudis R, Kohl A, Kalinke U, Weiss S, Michiels T, Staeheli P, Weber F. 2007. La Crosse bunyavirus nonstructural protein NSs serves to suppress the type I interferon system of mammalian hosts. J Virol 81:4991–4999. doi: 10.1128/JVI.01933-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Barry G, Varela M, Ratinier M, Blomstrom AL, Caporale M, Seehusen F, Hahn K, Schnettler E, Baumgartner W, Kohl A, Palmarini M. 2014. NSs protein of Schmallenberg virus counteracts the antiviral response of the cell by inhibiting its transcriptional machinery. J Gen Virol 95:1640–1646. doi: 10.1099/vir.0.065425-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Assfalg R, Lebedev A, Gonzalez OG, Schelling A, Koch S, Iben S. 2012. TFIIH is an elongation factor of RNA polymerase I. Nucleic Acids Res 40:650–659. doi: 10.1093/nar/gkr746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ning YJ, Feng K, Min YQ, Cao WC, Wang M, Deng F, Hu Z, Wang H. 2015. Disruption of type I interferon signaling by NSs protein of severe fever with thrombocytopenia syndrome virus via hijacking STAT2 and STAT1 into inclusion bodies. J Virol 89:4227–4236. doi: 10.1128/JVI.00154-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ning YJ, Wang M, Deng M, Shen S, Liu W, Cao WC, Deng F, Wang YY, Hu Z, Wang H. 2014. Viral suppression of innate immunity via spatial isolation of TBK1/IKKepsilon from mitochondrial antiviral platform. J Mol Cell Biol 6:324–337. doi: 10.1093/jmcb/mju015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Proenca-Modena JL, Sesti-Costa R, Pinto AK, Richner JM, Lazear HM, Lucas T, Hyde JL, Diamond MS. 2015. Oropouche virus infection and pathogenesis are restricted by MAVS, IRF-3, IRF-7, and type I interferon signaling pathways in nonmyeloid cells. J Virol 89:4720–4737. doi: 10.1128/JVI.00077-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Livonesi MC, de Sousa RL, Badra SJ, Figueiredo LT. 2007. In vitro and in vivo studies of the interferon-alpha action on distinct Orthobunyavirus. Antiviral Res 75:121–128. doi: 10.1016/j.antiviral.2007.01.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.