ABSTRACT
The existence of various highly divergent HIV-1 lineages and of recombination-derived sequence tracts of indeterminate origin within established circulating recombinant forms (CRFs) strongly suggests that HIV-1 group M (HIV-1M) diversity is not fully represented under the current classification system. Here we used a fully exploratory screen for recombination on a set of 480 near-full-length genomes representing the full known diversity of HIV-1M. We decomposed recombinant sequences into their constituent parts and then used maximum-likelihood phylogenetic analyses of this mostly recombination-free data set to identify rare divergent sequence lineages that fall outside the major named HIV-1M taxonomic groupings. We found that many of the sequence fragments occurring within CRFs (including CRF04_cpx, CRF06_cpx, CRF11_cpx, CRF18_cpx, CRF25_cpx, CRF27_cpx, and CRF49_cpx) are in fact likely derived from divergent unclassified parental lineages that may predate the current subtypes, even though they are presently identified as derived from currently defined HIV-1M subtypes. Our evidence suggests that some of these CRFs are descended predominantly from what were or are major previously unidentified HIV-1M lineages that were likely epidemiologically relevant during the early stages of the HIV-1M epidemic. The restriction of these divergent lineages to the Congo basin suggests that they were less infectious and/or simply not present at the time and place of the initial migratory wave that triggered the global epidemic.
IMPORTANCE HIV-1 group M (HIV-1M) likely spread to the rest of the world from the Congo basin in the mid-1900s (N. R. Faria et al., Science 346:56–61, 2014, http://dx.doi.org/10.1126/science.1256739) and is today the principal cause of the AIDS pandemic. Here, we show that large sequence fragments from several HIV-1M circulating recombinant forms (CRFs) are derived from divergent parental lineages that cannot reasonably be classified within the nine established HIV-1M subtypes. These lineages are likely to have been epidemiologically relevant in the Congo basin at the onset of the epidemic. Nonetheless, they appear not to have undergone the same explosive global spread as other HIV-1M subtypes, perhaps because they were less transmissible. Concerted efforts to characterize more of these divergent lineages could allow the accurate inference and chemical synthesis of epidemiologically key ancestral HIV-1M variants so as to directly test competing hypotheses relating to the viral genetic factors that enabled the present pandemic.
INTRODUCTION
All HIV-1 group M (HIV-1M) viruses that infect humans cluster phylogenetically within a clade of SIVcpz sequences sampled in southern Cameroon, leading to the conclusion that it is likely that Cameroon was the site of the cross-species transmission event that gave rise to HIV-1M (1, 2). Consistent with the hypothesis that the Congo basin region was the epicenter of the epidemic, the greatest genetic diversity of HIV-1M in terms of both numbers of subtypes and degree of genetic diversity within subtypes has been observed in this region (3–7). The different subtypes that today account for the vast majority of HIV-1M infections worldwide likely moved out from this region, each to populate different parts of the world, during the 1950s and 1960s (8, 9).
The HIV-1M classification system that we presently employ is based largely on the order in which these various pandemic HIV-1M lineages were discovered. Viruses discovered early on tended to be classified as belonging to “pure” subtypes, and there has been an understandable tendency for more recently discovered viruses with inconsistent degrees of similarity across their genomes to these pure subtypes to be classified as “recombinant forms” (10). While some of these recombinant forms have only ever been sampled from a single individual (in which case they are called unique recombinant forms [URFs]), others have been sampled from multiple unlinked individuals and are thus called circulating recombinant forms (CRFs). This classification system has been both consistent with what is known epidemiologically about the global spread of HIV-1M and a functionally useful means of characterizing subepidemics in parts of the world where only one or a few HIV-1M lineages are circulating; an illustrative example is Cuba, where distinctly Cuban subtype B and subtype G lineages are cocirculating and have yielded a variety of distinctly Cuban CRFs, including CRF20, CRF23, and CRF24 (11).
A potential shortcoming of the present HIV-1M classification system, however, is its capacity to accurately capture the genetic complexity of the HIV-1M epidemics in regions of Africa where large numbers of different subtypes and recombinant forms have been cocirculating for 60 years or more. Many of the recombinant forms that have been found infecting people in such regions have parental viruses belonging to three or more different subtypes. These viruses, termed “complex” CRFs (CRF_cpx) often also contain genome segments that do not phylogenetically cluster with homologous sequences derived from any of the classified HIV-1M subtypes. Crucially, although many of the CRFs (and possibly also many of the URFs) that have been sampled in the Congo basin are circulating at low frequencies, some, such as CRF02_AG in Cameroon, are among the most common HIV-1M variants that are found circulating within particular regions (5, 12).
Another potential shortcoming of the existing HIV-1M classification system is that it has likely been at least partially impacted by sampling biases that have arisen due to both the orders in which lineages were discovered and the undersampling of lineages in the Congo basin HIV-1M diversity hot spot. Further compounding this problem is the fact that it is often very difficult to accurately identify the breakpoint locations and parental subtypes of recombinant sequences (13). Illustrative examples of potential misclassifications having arisen through such issues include subtype G, CRF02_AG and CRF01_AE. It has been proposed that subtype G might be the complex recombinant offspring of subtype A, subtype J, and CRF02_AG parental viruses (14), a possibility that, in the absence of additional lines of evidence, has remained unresolved (15, 16). It has been proposed that CRF01_AE is the recombinant offspring of a subtype A parental virus and an unsampled (and possibly extinct) “subtype E” parent (17) or, alternatively, is a unique nonrecombinant subtype (18). Furthermore, CRF04_cpx, CRF13_cpx, CRF18_cpx, and several URFs contain fragments of sequence that, while closely related among these recombinants, are seemingly not derived from any of the known HIV-1M subtypes, suggesting that all these viruses contain segments that are likely derived from a (formerly) common unsampled (and possibly extinct) parental lineage (19).
The probable existence of multiple epidemiologically relevant but undescribed HIV-1M lineages has previously been suggested (20). In rural Cameroon, where rates of HIV-1M infection are relatively low, Carr and coworkers found a high prevalence of URFs containing genome fragments that had apparently been derived from subtype F, J, H, and K parental viruses (20). Without invoking the existence of numerous undiscovered divergent HIV-1M lineages, it is difficult to explain either how viruses belonging to these apparently low-prevalence parental subtypes have contributed sequences to so many of the HIV-1M recombinant forms that are found in the Congo basin or how intersubtype recombination rates could apparently be so high in rural areas that have such low HIV-1M prevalences. Such mosaic lineages and the presence of sequences of indeterminate origin within some established CRFs provide strong evidence that HIV-1M diversity might not be adequately represented under the current classification system. Concerted efforts to discover and characterize these divergent lineages could allow accurate inference of epidemiologically key ancestral HIV-1M variants so as to directly test competing hypotheses relating to the viral genetic factors that enabled the present pandemic.
In an effort to better characterize HIV-1M diversity within the Congo basin, we reanalyzed the phylogenetic and recombinational relationships of all available near-full-length genome sequences from this region, together with a set of viruses carefully selected to represent as fully as possible the known diversity of HIV-1M found in the rest of the world. From this analysis, we conclude that some of the CRFs from the Congo basin region show strong evidence that they are at least partially derived from major previously unidentified HIV-1M lineages that were likely important components of the early HIV-1M epidemic and which may, even today, be making an epidemiologically relevant contribution to the ongoing diversification of HIV-1M.
MATERIALS AND METHODS
Selection of sequences.
We selected a set of 480 near-full-length genome sequences representative of the known diversity of HIV-1M. These included (i) 423 sequences from 76 taxonomically recognized subtypes/CRFs (21) obtained from the Los Alamos National Laboratory (LANL) database, (ii) 12 sequences classified as “U” (May 2014) and also from the LANL database, and (iii) 45 genetically divergent sequences from GenBank (May 2014). The 423 sequences representative of the known subtype and CRF lineages were specifically selected to include the broadest diversity of sequences previously identified as belonging to these subtypes/CRFs (22). In brief, this was achieved by constructing maximum-likelihood (ML) trees from all available near-full-length sequences for each subtype and CRF using FastTree 2 (23), as implemented in RDP4 (24), and selecting one sequence from each of the up to 20 most basal lineages from the root nodes of these subtype/CRF clades. Along every one of the 10 most basal lineages, we explicitly chose sequences from the most populated branches of these lineages. For subtypes and CRFs with fewer than 16 available near-full-length sequences, all available sequences were included. For the selection of divergent sequences, we retrieved from GenBank all sequences from Angola, Cameroon, the Central African Republic, the Republic of Congo, Zaire, the Democratic Republic of Congo (DRC), Equatorial Guinea, and Gabon. We then constructed an initial ML tree (with FastTree2) with these sequences together with the 435 sequences retrieved from LANL and selected 45 sequences from GenBank that both branched basal to the known subtype/CRF clades (i.e., did not cluster within the sequences of these clades) and were not identical to any of the sequences retrieved from the LANL database.
Recombination analyses.
Sequences between the first 5′ codon of gag and the last 3′ codon of nef were extracted from the 480 near-full-length genomes and aligned using MUSCLE (25). This alignment was manually edited using IMPALE (http://www.cbio.uct.ac.za/∼arjun/). A blinded fully exploratory screen for recombination using RDP4 (24) was performed using this data set. RDP4 uses multiple approaches both to identify recombination signals (the RDP, BOOTSCAN, GENECONV, MAXCHI, CHIMAERA, SISCAN, and 3SEQ methods), and to differentiate between recombinant and parental sequences (the VidRD, PHYLPRO, and EEEP methods); all methods are described in detail in reference 26. In addition, RDP4 employs a recursive fully exploratory recombination screening approach that effectively identifies and erases all evidence of individual detectable recombination events within an input nucleotide sequence alignment. The program was used to sequentially test every sequence in our alignment for evidence of recombination irrespective of whether these had formerly been identified as pure subtypes, CRFs, or URFs. This recombination screen was carried out with default RDP4 settings, and recombination events detected by at least two different methods were taken as credible evidence of possible recombination. Recombination signals arising due to probable misalignment artifacts were identified as outlined previously (26, 27). Briefly this involved realigning pairs of recombinant and parental sequences and then using 2 × 2 χ2 tests (with a χ2 cutoff of 1.96) to detect significant differences in the matched/mismatched status of aligned nucleotide pairs between the individual pairwise alignments and the multiple-sequence alignment.
All the recombinationally derived genome sequence fragments thus identified were then removed to leave just the fragments of sequence within each individual recombinant that were derived from its predominant parental virus (defined here as the parent that contributed most to the genetic material found within a recombinant) (see Fig. S1A in the supplemental material). An ML phylogenetic tree was constructed from these sequences with 1,000 full ML bootstrap replicates using RAxML version 8 (28) as implemented in CIPRES (29). Although RAxML is limited to the use of general time-reversible (GTR)-based nucleotide substitution models (GTRGAMMA in our case), it has been specifically designed to accurately infer phylogenies from alignments containing large amounts of missing data, a factor ideally suited to the analysis of our recombination-free alignment (28, 30). The tree was not rooted in order to best separate the known subtypes from one another while ensuring that the branch distances from the branch tips to the root are kept as low as possible for the majority of sequences in the tree (so as to minimize the amount of white space in Fig. 1).
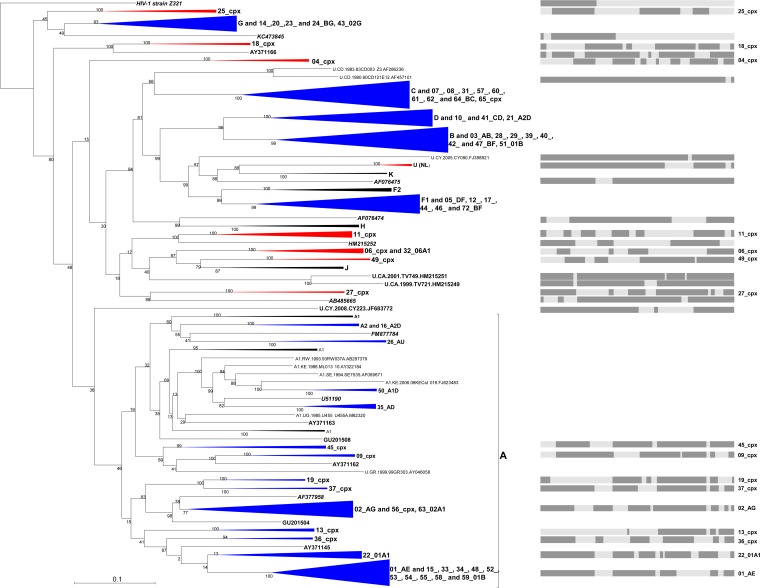
FIG 1.
Maximum-likelihood tree indicating the phylogenetic relationships between 480 HIV-1M near-full-length genomes from which all detectable evidence of individual recombination events has been removed. These sequences represent all near-full-length published subtype, CRF, and unclassified sequences that were available in the LANL database (http://www.hiv.lanl.gov/content/sequence/HIV/mainpage.html) in June 2014 together with a set of 45 divergent sequences from GenBank (obtained in May 2014). Recombination analyses and the stripping from recombinant sequences of fragments of recombinationally derived sequence were performed as stated in Materials and Methods. Some clades have been condensed for the sake of clarity. The tree is unrooted and was constructed with 1,000 full maximum-likelihood bootstrap replicates using RAxML (28). Numbers associated with tree branches indicate degrees of bootstrap support for these branches. Clades that do not cluster within the existing “pure” subtype-based taxonomy are colored in red, while those that cluster within are in blue. Graphics represent the “parental” (i.e., nonrecombinant) segments (in dark gray) from the genome length of clades that do not cluster within the “pure” subtypes and some major CRFs embedded in subtype A.
Divergent sequences within the phylogenetic tree produced by RAxML were defined as either (i) those residing on isolated branches outside subtrees containing previously defined HIV-1 subtype or CRF lineages or (ii) those forming basal branches within subtrees containing previously defined HIV-1 subtypes or CRF lineages. The phylogenetic placement of the latter group of divergent sequences might be due to their being descended from a lineage that diverged close to the origin of their associated subtype or CRF lineages.
RESULTS
Identification of new highly divergent HIV-1M lineages within CRFs.
We first set out to create a set of representative reference sequences that were specifically selected to include the broadest diversity of HIV-1M sequences that had previously been identified as belonging to taxonomically recognized HIV-1 group M subtypes and CRFs (as described in reference 22). Briefly, this selection involved the construction of maximum-likelihood (ML) trees from all available full-length sequences for each subtype (A, B, C, D, F, H, J, and K) and CRF (01 through 72, except for CRF30 and CRF66 to CRF71, which were not included in the Los Alamos National Laboratory database by the time of data collection) and selecting one sequence from each of the up to 20 most basal lineages from the root of these clades, or all sequences if fewer than 16 were initially available. Along every one of the 10 most basal lineages we explicitly chose sequences from the most populated branches. This approach to selecting subtype and CRF reference sequences ensured both that the selected sequences represented the broadest diversity of taxonomically classified HIV-1M sequences and that we did not include in our subsequent analyses too many superfluous sequences that contributed little to the overall diversity of the data set. In addition to these reference sequences, all 12 near-full-length HIV-1M sequences classified as “U” in the Los Alamos National Laboratory sequence database (May 2014) and 45 genetically divergent near-full-length HIV-1M sequences from GenBank (May 2014) originating from countries in the Congo basin were also included in the analysis. This approach resulted in the collation of a set of 480 near-full-length genome sequences.
We analyzed this sequence set by performing a fully exploratory screen for recombination using RDP4 (24), which decomposed recombinant viruses into their constituent parts (the approach is depicted in Fig. S1A in the supplemental material and described in more detail in reference 26). For further analyses, we retained the major parental segment representing >50% of genomic nucleotides for 96% (461/480) of the analyzed sequences. The resulting alignment was then used to construct a mostly intersubtype recombination-free ML phylogenetic tree using RAxML (28). Phylogenetic analyses of this mostly recombination-free HIV-1M sequence data set indicated that the major parental sequences of many of the CRFs (including those of CRF04_cpx, CRF06_cpx, CRF11_cpx, CRF18_cpx, CRF25_cpx, CRF27_cpx, and CRF49_cpx) are not classifiable within the currently established HIV-1M subtypes (Fig. 1; see Fig. S2 in the supplemental material).
Specifically, one of the parental sequences of CRF49_cpx is most closely related to but clusters phylogenetically basal to subtype J and might therefore reasonably be considered to be either a very divergent subtype J sequence (perhaps an as-yet-undiscovered J2 lineage) or a virus that either existed or diverged prior to the diversification of subtype J (Fig. 1). It is similarly apparent that the supposedly subtype J-like parent of CRF06_cpx likely diverged prior to the split of the subtype J/CRF49_cpx lineage. Similarly, the parental lineage from which CRF11_cpx and CRF27_cpx derived the largest portion of their genomes apparently diverged before the most recent common ancestor (MRCA) of CRF06_cpx/J/CRF49_cpx, to which they are more closely related than other named subtype/CRF groups (Fig. 1). Importantly, the major parental viruses of CRF04_cpx and CRF18_cpx do not phylogenetically cluster close to any of the named subtype or CRF groupings (Fig. 1; see Fig. S2 in the supplemental material). Finally, the major parent of CRF25_cpx was apparently either a subtype G-like virus (although this is not phylogenetically well supported) or a virus that diverged shortly prior to the diversification of the subtype G lineage (Fig. 1).
As has been shown for a number of Cameroonian URFs (20, 22), our phylogenetic analyses also indicate that the parental viruses of many HIV-1M genetic variants found in individuals either in or originating from the Congo basin (indicated by bold italic text in Fig. 1 and in Fig. S2 in the supplemental material) contain fragments of sequence that do not cluster phylogenetically within any of the taxonomically recognized HIV-1M subtypes. Specifically, these fragments of sequence are divergent and branch basal to the named HIV-1M subtypes rather than being nested within these subtypes. This suggests either that the recombination events that gave rise to these lineages predated the divergence of the subtypes from one another or that they occurred more recently but involved a variety of currently unsampled divergent HIV-1M lineages that are therefore possibly still circulating at low frequencies within the region.
Confirmation of highly divergent sequence fragments within established CRFs.
In an attempt to reconcile and further understand the analytical issues causing differences between the established genome structures of known recombinants and those described during our exploratory recombination screen, we reexamined CRFs that were apparently derived from parental viruses that are unclassifiable within the current HIV-1M taxonomy. ML trees were constructed using FastTree2 (23) implemented in RDP4 (24) and were used to test whether segments of recombinationally derived sequence were from parental viruses falling within the existing subtype classification system or, alternatively, whether the parental viruses were from divergent currently unclassified lineages (the scheme used for this analysis is depicted in Fig. S1B in the supplemental material). Although these trees were constructed using reference sequences from the same subtypes as the reference sequences that were used in the literature to initially characterize these viruses, the precise reference sequences that we used here were, as in our exploratory recombination screen described above, were selected to represent the full breadth of currently known diversity within each subtype. Since it is was not our intention to completely redefine the mosaic structures of any of the known CRFs (for the most part, previously identified breakpoint locations within these sequences seem very plausible), we opted to use the currently accepted CRF breakpoint locations published in the Los Alamos HIV database (21) for all further analyses of the likely origins of recombinationally derived genome fragments.
The genomes of CRF04_cpx (Fig. 2A), CRF06_cpx (Fig. 2B), CRF11_cpx (Fig. 2C), CRF18_cpx (see Fig. S3A in the supplemental material), CRF25_cpx (see Fig. S3B in the supplemental material), CRF27_cpx (Fig. 2D), and CRF49_cpx (see Fig. S3C in the supplemental material) viruses each contain more than 3,000 nucleotides (nt) of sequence that was likely derived from divergent parental viruses that branch phylogenetically outside the taxonomically recognized HIV-1M subtypes. Eight out of 13 of the recombinationally derived genome segments of CRF04_cpx, representing approximately 4,660 nt of sequence, clustered basal to the subtypes (K, G, and H) that they were most closely related to (Fig. 2A), suggesting that the viruses that donated these genome segments were likely members of divergent HIV-1M lineages.
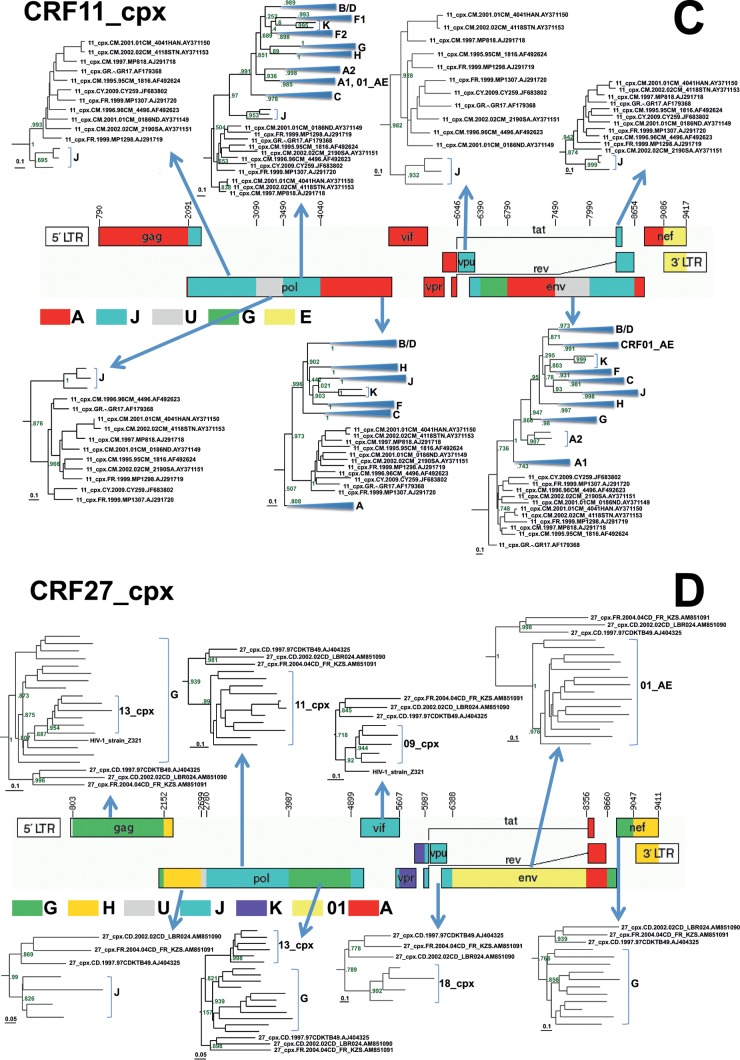
FIG 2.
Reanalysis of the origins of recombinationally derived sequence fragments within some CRFs from the Congo basin that have previously been described in the literature. Maximum-likelihood trees indicating the phylogenetic relationships between segments of recombinationally derived sequence from these CRFs and different HIV-1M subtypes are shown. ML trees were constructed from aligned sequences corresponding to these segments using FastTree 2 (23). Numbers at the tree branches indicate support for these branches using the Shimodaira-Hasegawa-like branch support test. For the sake of clarity, only subtrees that included the divergent, difficult-to-classify sequences were included, except in panel A, where all the subtrees are shown. The graphical genome maps of the CRFs analyzed here were obtained from the Los Alamos HIV database (21) and represent the currently accepted breakpoint and parental subtype annotations for these CRFs.
Similarly, six out of eight of the analyzed genome fragments from the CRF06_cpx viruses, representing ∼5,369 nt of sequence, clustered basal to subtypes G, K, and J (Fig. 2B); the parental sequences that donated these fragments therefore likely originated from a lineage that diverged prior to the diversification of the MRCA of subtypes G, K, and J.
CRF11_cpx (Fig. 2C) was queried against all the standard reference lineages (A to D, F to H, J, K, and 01_AE), as was done during the initial classification of this CRF (31). ML trees from the 12 recombinationally derived genome segments that constitute CRF11 indicated that seven of these segments, representing ∼5,663 nt of sequence, branch basal to the subtypes that they are most closely related to (including one segment previously identified as coming from a subtype A parent; HXB2 nt 4040 to 6046) (Fig. 2B); all of these segments could therefore plausibly have been derived from one or more parents from divergent, currently unclassified HIV-1M lineages.
Among the CRFs analyzed, CRF27_cpx (Fig. 2D) appears to have a particularly interesting ancestry. During the initial analyses of this CRF, it was queried not only against the standard reference subtypes but also against CRF01_AE, CRF02_AG, CRF04_cpx, CRF05_DF, CRF06_cpx, CRF09_cpx, CRF11_cpx, CRF13_cpx, CRF18_cpx, CRF19_cpx, and strains Z321, MAL, and NOGIL3 (32). Eight out of 12 segments, representing 87% of the analyzed genome of CRF27_cpx, branched phylogenetically basal to the MRCAs of the named HIV-1M subtype or CRF groups to which they were most similar (Fig. 2D), clearly suggesting that one or more of its parental viruses were divergent unclassified HIV-1M lineages (21).
Finally, 21 of the analyzed genome fragments comprising CRF18_cpx (see Fig. 3A in the supplemental material), CRF25_cpx (see Fig. S3B in the supplemental material), and CRF49_cpx (see Fig. S3C in the supplemental material), representing ∼5,710, 5,233, and 3,965 nt of sequence, respectively, branched basal to known subtypes, also indicating that the sequences within these segments likely originated from parental viruses that belonged to divergent unclassified HIV-1M lineages.
DISCUSSION
Our analyses imply that there is potentially a far more diverse pool of HIV-1M sequences circulating among humans than the current classified subtypes and CRFs might suggest. It is known that after transmission to humans sometime before 1920 (6), the progenitor of HIV-1M began to diversify extensively into numerous variants and that the absence of geographical barriers likely meant that these “proto-subtype” variants circulated throughout the Congo basin region (33). This is well illustrated by the large genetic distance between ZR59 and DRC60, two sequences identified in the Democratic Republic of Congo (DRC) that predated the global AIDS epidemic, which indicates that there was already an extensive degree of HIV-1M genetic diversity in the Congo basin as early as the late 1950s (6, 33, 34). Following this diversification, but before the global spread of HIV-1M around 1970 (6, 35), it is likely that localized epidemics occurred, and it is plausible that some of these epidemics have remained confined to this region within small groups of infected people. The fact that five of the seven CRF lineages analyzed here, and almost all similarly divergent URF lineages, have been sampled in remote rural regions of the Congo basin is entirely consistent with this hypothesis of multiple localized epidemics. This hypothesis, while not disputing the notion that most of the subsequent global expansion of HIV-1M originated from the region around Kinshasa, does suggest that some HIV-1M lineages were “left behind” in the Congo basin.
In previously described analyses of some CRFs, many divergent segments were found to be difficult to classify due to limited quantities of whole-genome sequence data that were available at the time of their discovery. For example, fragments of CRF11_cpx and CRF13_cpx that were found to be most closely related to subtype J sequences were found to be too different from known subtype J sequences to be considered to belong to subtype J, and it was therefore proposed that they be classified as belonging to a hypothetical subtype J2 progenitor (36). This particular case, however, is exceptional in that most other studies have tended to classify apparently recombinationally derived genome segments as belonging to whichever subtype they were most similar too, irrespective of whether these segments clustered phylogenetically within the sequences of these named subtypes or whether they branched basal to them. We are simply emphasizing here that when a newly discovered sequence fragment is most closely related to those which have been classified as belonging to a particular known subtype, it does not necessarily mean that the new sequence should also be classified as belonging to that subtype. It might therefore be better, as done by Zhang et al. (36), to leave such fragments unclassified. We are also emphasizing that the choice of reference clades for the classification of an unknown sequence (especially from the Congo basin) should not be random; such references should specifically include sequences representing the entire breadth of diversity that exists within a specific clade. In addition, as we have found in our analysis of CRF27_cpx, the reference clades should in some cases also include the broadest possible diversity of HIV-1M CRF sequences.
Accordingly, we hypothesize that CRF04_cpx, CRF06_cpx, CRF11_cpx, CRF18_cpx, CRF25_cpx, CRF27_cpx, and CRF49_cpx are largely (and in some cases predominantly) descended from what were/are major previously unidentified HIV-1M lineages that were likely epidemiologically relevant during the early stages of the HIV-1M epidemic. It remains unclear why these divergent lineages did not undergo the same explosive spread as the known HIV-1M subtypes that are circulating in the Congo basin. One possibility could be that these “missing subtypes” have become extinct. Another possibility is that they are still circulating at low frequencies in the human population but have not undergone the explosive population expansion of the known subtypes either because they were not physically present in order to be part of the initial migratory wave of variants that triggered the global epidemic or because they do not have the same degree of transmissibility as globally circulating HIV lineages. The fact that many CRFs and URFs from the Congo basin apparently have large proportions of their genomes derived from divergent unclassified HIV-1M lineages certainly suggests that large pools of undiscovered HIV-1M genetic diversity likely exist throughout equatorial west Africa.
At a very practical level, a fuller characterization of this diversity is likely to strain the current HIV classification system to the point where it becomes misleading. Our study and some others in recent years (13, 16, 37) indicate that it might be useful to at least consider the establishment of some additional guidelines for the classification of novel complex HIV-1M recombinants and divergent sequences such as those which are continually being discovered in equatorial west Africa (20, 22). Specifically, whenever a novel URF or CRF is discovered, it should be recommended that an effort be made to phylogenetically characterize the portions of these genomes that are apparently derived from divergent parental lineages, using a well-defined standard (and preferably Los Alamos HIV database-approved) set of representative reference sequences. It would also be very illustrative if, in such studies, attempts were made to distinguish (i) recombination events that likely occurred close to (or even before) the time of the progenitors of the major HIV-1M subtypes from (ii) recombination events that likely occurred in recent times between well-characterized circulating parental lineages. Properly representing such viruses would obviously require a slightly more nuanced annotation and naming tool kit than the A-to-K designator-based one that is currently endorsed for HIV classification by the International Committee for Virus Taxonomy and the influential Los Alamos HIV database. For example, a genome segment that has apparently been derived from a virus that branches basal to subtype A might simply be annotated “a” rather than “A,” whereas a parental lineage branching basal to subtypes J and H might be annotated as j|h” rather than “U.” If, for example, this j|h fragment was likely derived during an ancient recombination event, it might additionally be annotated “(j|h)”.
There is much to be gained from properly characterizing the fascinatingly complex HIV-1M variants that are consistently being discovered in the Congo basin. If nothing else, the mere existence of these sequences emphasizes the complexity of devising suitable treatment programs and developing relevant vaccines for this region. Besides being useful in efforts to understand and combat HIV-1M at its geographical origin, viruses presently circulating in equatorial west Africa might also be the source of future global multidrug resistance or vaccine evasion reemergence events. Focused efforts in the Congo basin to discover and characterize more of these highly divergent lineages, whether within the context of genome fragments surviving within CRFs and URFs or within actual novel HIV-1M subtypes, would be invaluable in attempts both to discover the specific viral genetic factors that enabled the initial emergence of HIV-1M and to assess the risks of future reemergence events.
Supplementary Material
ACKNOWLEDGMENTS
Computational analyses were performed using facilities provided by the University of Cape Town's ICTS High Performance Computing Facility (hpc.uct.ac.za). We are also grateful to Wendy Burgers (University of Cape Town) for helpful discussion and critical review of the manuscript and to Sodsai Tovanabutra (U.S. Military HIV Research Program) for kindly providing CRF41_CD sequences.
We declare no conflicts of interest.
This research was supported by the Poliomyelitis Research Foundation (PRF) of South Africa and by the International Centre for Genetic Engineering and Biotechnology (ICGEB). M.T. is an ICGEB Postdoctoral Fellow.
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.02302-15.
REFERENCES
- 1.Keele BF, Van Heuverswyn F, Li Y, Bailes E, Takehisa J, Santiago ML, Bibollet-Ruche F, Chen Y, Wain LV, Liegeois F, Loul S, Ngole EM, Bienvenue Y, Delaporte E, Brookfield JF, Sharp PM, Shaw GM, Peeters M, Hahn BH. 2006. Chimpanzee reservoirs of pandemic and nonpandemic HIV-1. Science 313:523–526. doi: 10.1126/science.1126531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gao F, Bailes E, Robertson DL, Chen Y, Rodenburg CM, Michael SF, Cummins LB, Arthur LO, Peeters M, Shaw GM, Sharp PM, Hahn BH. 1999. Origin of HIV-1 in the chimpanzee Pan troglodytes troglodytes. Nature 397:436–441. doi: 10.1038/17130. [DOI] [PubMed] [Google Scholar]
- 3.Vidal N, Mulanga C, Bazepeo SE, Mwamba JK, Tshimpaka JW, Kashi M, Mama N, Laurent C, Lepira F, Delaporte E, Peeters M. 2005. Distribution of HIV-1 variants in the Democratic Republic of Congo suggests increase of subtype C in Kinshasa between 1997 and 2002. J Acquir Immune Defic Syndr 40:456–462. doi: 10.1097/01.qai.0000159670.18326.94. [DOI] [PubMed] [Google Scholar]
- 4.Kalish ML, Robbins KE, Pieniazek D, Schaefer A, Nzilambi N, Quinn TC, St Louis ME, Youngpairoj AS, Phillips J, Jaffe HW, Folks TM. 2004. Recombinant viruses and early global HIV-1 epidemic. Emerg Infect Dis 10:1227–1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tongo M, Martin DP, Zembe L, Mpoudi-Ngole E, Williamson C, Burgers WA. 2013. Characterization of HIV-1 gag and nef in Cameroon: further evidence of extreme diversity at the origin of the HIV-1 group M epidemic. Virol J 10:29. doi: 10.1186/1743-422X-10-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Faria NR, Rambaut A, Suchard MA, Baele G, Bedford T, Ward MJ, Tatem AJ, Sousa JD, Arinaminpathy N, Pepin J, Posada D, Peeters M, Pybus OG, Lemey P. 2014. HIV epidemiology. The early spread and epidemic ignition of HIV-1 in human populations. Science 346:56–61. doi: 10.1126/science.1256739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vidal N, Peeters M, Mulanga-Kabeya C, Nzilambi N, Robertson D, Ilunga W, Sema H, Tshimanga K, Bongo B, Delaporte E. 2000. Unprecedented degree of human immunodeficiency virus type 1 (HIV-1) group M genetic diversity in the Democratic Republic of Congo suggests that the HIV-1 pandemic originated in Central Africa. J Virol 74:10498–10507. doi: 10.1128/JVI.74.22.10498-10507.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hemelaar J. 2012. The origin and diversity of the HIV-1 pandemic. Trends Mol Med 18:182–192. doi: 10.1016/j.molmed.2011.12.001. [DOI] [PubMed] [Google Scholar]
- 9.Tebit DM, Arts EJ. 2011. Tracking a century of global expansion and evolution of HIV to drive understanding and to combat disease. Lancet Infect Dis 11:45–56. doi: 10.1016/S1473-3099(10)70186-9. [DOI] [PubMed] [Google Scholar]
- 10.Robertson DL, Anderson JP, Bradac JA, Carr JK, Foley B, Funkhouser RK, Gao F, Hahn BH, Kalish ML, Kuiken C, Learn GH, Leitner T, McCutchan F, Osmanov S, Peeters M, Pieniazek D, Salminen M, Sharp PM, Wolinsky S, Korber B. 2000. HIV-1 nomenclature proposal. Science 288:55–56. [DOI] [PubMed] [Google Scholar]
- 11.Sierra M, Thomson MM, Posada D, Perez L, Aragones C, Gonzalez Z, Perez J, Casado G, Najera R. 2007. Identification of 3 phylogenetically related HIV-1 BG intersubtype circulating recombinant forms in Cuba. J Acquir Immune Defic Syndr 45:151–160. doi: 10.1097/QAI.0b013e318046ea47. [DOI] [PubMed] [Google Scholar]
- 12.Hemelaar J, Gouws E, Ghys PD, Osmanov S. 2011. Global trends in molecular epidemiology of HIV-1 during 2000-2007. AIDS 25:679–689. doi: 10.1097/QAD.0b013e328342ff93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kosakovsky Pond SL, Posada D, Stawiski E, Chappey C, Poon AF, Hughes G, Fearnhill E, Gravenor MB, Leigh Brown AJ, Frost SD. 2009. An evolutionary model-based algorithm for accurate phylogenetic breakpoint mapping and subtype prediction in HIV-1. PLoS Comput Biol 5:e1000581. doi: 10.1371/journal.pcbi.1000581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Abecasis AB, Lemey P, Vidal N, de Oliveira T, Peeters M, Camacho R, Shapiro B, Rambaut A, Vandamme AM. 2007. Recombination confounds the early evolutionary history of human immunodeficiency virus type 1: subtype G is a circulating recombinant form. J Virol 81:8543–8551. doi: 10.1128/JVI.00463-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bulla I, Schultz AK, Schreiber F, Zhang M, Leitner T, Korber B, Morgenstern B, Stanke M. 2010. HIV classification using the coalescent theory. Bioinformatics 26:1409–1415. doi: 10.1093/bioinformatics/btq159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang M, Foley B, Schultz AK, Macke JP, Bulla I, Stanke M, Morgenstern B, Korber B, Leitner T. 2010. The role of recombination in the emergence of a complex and dynamic HIV epidemic. Retrovirology 7:25. doi: 10.1186/1742-4690-7-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carr JK, Salminen MO, Koch C, Gotte D, Artenstein AW, Hegerich PA, St Louis D, Burke DS, McCutchan FE. 1996. Full-length sequence and mosaic structure of a human immunodeficiency virus type 1 isolate from Thailand. J Virol 70:5935–5943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Anderson JP, Rodrigo AG, Learn GH, Madan A, Delahunty C, Coon M, Girard M, Osmanov S, Hood L, Mullins JI. 2000. Testing the hypothesis of a recombinant origin of human immunodeficiency virus type 1 subtype E. J Virol 74:10752–10765. doi: 10.1128/JVI.74.22.10752-10765.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thomson MM, Casado G, Posada D, Sierra M, Najera R. 2005. Identification of a novel HIV-1 complex circulating recombinant form (CRF18_cpx) of Central African origin in Cuba. AIDS 19:1155–1163. doi: 10.1097/01.aids.0000176215.95119.1d. [DOI] [PubMed] [Google Scholar]
- 20.Carr JK, Wolfe ND, Torimiro JN, Tamoufe U, Mpoudi-Ngole E, Eyzaguirre L, Birx DL, McCutchan FE, Burke DS. 2010. HIV-1 recombinants with multiple parental strains in low-prevalence, remote regions of Cameroon: evolutionary relics? Retrovirology 7:39. doi: 10.1186/1742-4690-7-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Los Alamos National Laboratory. 2015. HIV circulating recombinant forms (CRFs). http://www.hiv.lanl.gov/content/sequence/HIV/CRFs/CRFs.html Accessed 19 January 2015. [Google Scholar]
- 22.Tongo M, Dorfman JR, Abrahams MR, Mpoudi-Ngole E, Burgers WA, Martin DP. 2015. Near full-length HIV type 1M genomic sequences from Cameroon: evidence of early diverging under-sampled lineages in the country. Evol Med Public Health 2015:254–265. doi: 10.1093/emph/eov022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Price MN, Dehal PS, Arkin AP. 2009. FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol Biol Evol 26:1641–1650. doi: 10.1093/molbev/msp077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martin DP, Murrell B, Golden M, Khoosal A, Muhire B. 26 May 2015. RDP4: detection and analysis of recombination patterns in virus genomes. Virus Evol doi: 10.1093/ve/vev003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Edgar RC. 2004. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5:113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Martin DP. 2015. RDP4 instruction manual. http://web.cbio.uct.ac.za/∼darren/RDP4Manual.pdf Accessed 13 November 2015. [Google Scholar]
- 27.Varsani A, van der Walt E, Heath L, Rybicki EP, Williamson AL, Martin DP. 2006. Evidence of ancient papillomavirus recombination. J Gen Virol 87:2527–2531. doi: 10.1099/vir.0.81917-0. [DOI] [PubMed] [Google Scholar]
- 28.Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miller MA, Pfeiffer W, Schwartz T. 2010. Creating the CIPRES Science Gateway for inference of large phylogenetic trees, p 1–8. In Proceedings of the Gateway Computing Environments Workshop (GCE) IEEE, Piscataway, NJ. [Google Scholar]
- 30.Stamatakis A, Alachiotis N. 2010. Time and memory efficient likelihood-based tree searches on phylogenomic alignments with missing data. Bioinformatics 26:i132–i139. doi: 10.1093/bioinformatics/btq205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Montavon C, Vergne L, Bourgeois A, Mpoudi-Ngole E, Malonga-Mouellet G, Butel C, Toure-Kane C, Delaporte E, Peeters M. 2002. Identification of a new circulating recombinant form of HIV type 1, CRF11-cpx, involving subtypes A, G, J, and CRF01-AE, in Central Africa. AIDS Res Hum Retroviruses 18:231–236. doi: 10.1089/08892220252781301. [DOI] [PubMed] [Google Scholar]
- 32.Vidal N, Frange P, Chaix ML, Mulanga C, Lepira F, Bazepeo SE, Goujard C, Meyer L, Rouzioux C, Delaporte E, Peeters M. 2008. Characterization of an old complex circulating recombinant form, CRF27_cpx, originating from the Democratic Republic of Congo (DRC) and circulating in France. AIDS Res Hum Retroviruses 24:315–321. doi: 10.1089/aid.2007.0241. [DOI] [PubMed] [Google Scholar]
- 33.Worobey M, Gemmel M, Teuwen DE, Haselkorn T, Kunstman K, Bunce M, Muyembe JJ, Kabongo JM, Kalengayi RM, Van Marck E, Gilbert MT, Wolinsky SM. 2008. Direct evidence of extensive diversity of HIV-1 in Kinshasa by 1960. Nature 455:661–664. doi: 10.1038/nature07390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhu T, Korber BT, Nahmias AJ, Hooper E, Sharp PM, Ho DD. 1998. An African HIV-1 sequence from 1959 and implications for the origin of the epidemic. Nature 391:594–597. doi: 10.1038/35400. [DOI] [PubMed] [Google Scholar]
- 35.Gray RR, Tatem AJ, Lamers S, Hou W, Laeyendecker O, Serwadda D, Sewankambo N, Gray RH, Wawer M, Quinn TC, Goodenow MM, Salemi M. 2009. Spatial phylodynamics of HIV-1 epidemic emergence in east Africa. AIDS 23:F9–F17. doi: 10.1097/QAD.0b013e32832faf61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang M, Wilbe K, Wolfe ND, Gaschen B, Carr JK, Leitner T. 2005. HIV type 1 CRF13_cpx revisited: identification of a new sequence from Cameroon and signal for subsubtype J2. AIDS Res Hum Retroviruses 21:955–960. doi: 10.1089/aid.2005.21.955. [DOI] [PubMed] [Google Scholar]
- 37.Jia L, Li L, Li H, Liu S, Wang X, Bao Z, Li T, Zhuang D, Liu Y, Li J. 2014. Recombination pattern reanalysis of some HIV-1 circulating recombination forms suggests the necessity and difficulty of revision. PLoS One 9:e107349. doi: 10.1371/journal.pone.0107349. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.