Abstract
The evolutionary speed hypothesis (ESH) proposes a causal mechanism for the latitudinal diversity gradient. The central idea of the ESH is that warmer temperatures lead to shorter generation times and increased mutation rates. On an absolute time scale, both should lead to an acceleration of selection and drift. Based on the ESH, we developed predictions regarding the distribution of intraspecific genetic diversity: populations of ectothermic species with more generations per year owing to warmer ambient temperatures should be more differentiated from each other, accumulate more mutations and show evidence for increased mutation rates compared with populations in colder regions. We used the multivoltine insect species Chironomus riparius to test these predictions with cytochrome oxidase I (COI) sequence data and found that populations from warmer regions are indeed significantly more differentiated and have significantly more derived haplotypes than populations from colder regions. We also found a significant correlation of the annual mean temperature with the population mutation parameter θ that serves as a proxy for the per generation mutation rate under certain assumptions. This pattern could be corroborated with two nuclear loci. Overall, our results support the ESH and indicate that the thermal regime experienced may be crucially driving the evolution of ectotherms and may thus ultimately govern their speciation rate.
Keywords: intraspecific diversity, neutral evolution, population differentiation, speciation
1. Introduction
Temperature is probably the most important abiotic factor, because it affects everything that an organism does [1]. This is a direct consequence of the temperature dependence of biochemical processes in general [2], and thus of organismal physiology [3]. However, it is still unclear how temperature dependence of individual physiology influences evolutionary and ecological processes [4,5]. One attempt to link thermal physiology with the latitudinal diversity gradient is the evolutionary speed hypothesis (ESH) proposed by Rensch [6]. He briefly stated that, in his understanding, the temperature-dependent number of generations per year drives the rate of evolution, the effectiveness of selection and thus, eventually speciation [6]. Rohde [7] tried to underpin this rather general statement with a mechanistic basis, and identified three main factors responsible for greater evolutionary speed in the tropics: shorter generation times, increased mutation rates and accelerated selection, leading to faster fixation of favourable mutants.
This assessment combines primary causes (generation time, mutation rate) with an (incomplete) account of the consequences (accelerated selection). In addition to the effectiveness of selection, other evolutionary processes depend on the number of generations per absolute unit of time, such as genetic drift and mutation accumulation. This general dependence suggests that, everything else being equal, population divergence should scale with the number of generations per year, as drift acts in the unit of generations. Even if the mutation rate is constant and independent from thermal conditions, populations with more generations per year in warmer areas should accumulate more mutations relative to their counterparts in colder areas during the same period of time. If, in addition, the spontaneous mutation rate on the molecular level itself depends on ambient temperatures, then the probability and rate of evolutionary processes that involve mutations would further increase with a higher thermal regime.
Strong support for the correlation of molecular evolution with generation time comes from comparative studies on vertebrates, plants and invertebrates [8–12]. However, these studies do not include temperature as a putative environmental driver of this correlation. To assess the temperature/latitude-dependency of genetic or phenotypic diversity, most studies used congeneric species-pair comparisons across a broad range of taxa (reviewed in [13]). The rate of nucleotide substitution in the internal transcribed spacer region of rRNA-encoding DNA was found to be more than twice as high in woody plant species of the tropics compared with woody plant species in temperate latitudes [14]. In flowering plants, temperature and UV radiation were identified as key variables to explain molecular evolutionary rates and species richness [15]. Furthermore, comprehensive studies on vertebrates including mammals, birds, fishes, amphibians and reptiles revealed an increased genetic diversity of mitochondrial marker genes in species from lower latitude regions (reviewed in [16]). In marine foraminifera species, the nucleotide substitution rate was found to be exponentially related to temperature [17].
As mutation rate [18], effective population size [19] and population differentiation [20] are species-specific parameters that may differ substantially among even closely related species, an intraspecific approach to test the ESH may account for these complications. In addition, intraspecific divergence processes are the starting point of speciation [21,22] and may thus present a better model for establishing a functional link between temperature and speciation rate.
Only a few studies have followed the intraspecific approach. Martin & McKay [23] examined geographically dispersed populations of several vertebrate species and found greater intraspecific genetic divergence of populations at lower latitudes. However, this study and others assessing genetic diversity patterns in (endothermic) vertebrates [24,25], found only partial support for the ESH. Eo et al. [26] found greater intraspecific genetic differentiation in plant populations from lower latitudes; however, the authors did not intend to address the causal mechanisms behind this pattern.
We use here intraspecific population genetic data, life cycle experiments and functional species distribution modelling with data from the ectothermic multivoltine insect species Chironomus riparius (Meigen) to test the predictions gained from ESH that:
(1) Populations from warmer sites (i.e. with more generations per year) should be more differentiated from each other, when compared with their counterparts in colder regions.
(2) Haplotypes in populations from warmer sites (i.e. with more generations per year) should, on average, be evolutionarily more derived (i.e. have accumulated more mutations compared with the ancestral haplotype) than haplotypes from populations in colder regions.
(3) Warmer ambient temperatures induce higher mutation rates (μ) per generation. Under the assumption that the effective population size (Ne) is independent of generation time and thermal conditions, this should result in a positive correlation between ambient temperature and the population mutation parameter θ, as a proxy for the per generation mutation rate.
The non-biting midge C. riparius occurs over a large Holarctic range across a pronounced thermal gradient in Europe [27], providing a useful system to examine the ESH at the intraspecific level. We therefore investigated the intraspecific diversity of genetic markers and related these population genetic analyses to a model of the generation time/temperature dependency across Europe.
2. Material and methods
(a). Sample collection
We combined a total of 795 C. riparius larvae sampled from 18 populations from the core range of the species across a climatic gradient in Europe (figure 1a; electronic supplementary material, S2). Specimens were mostly preserved in 96% ethanol with the exception of populations from Bulgaria (SBKP) and Turkey (STC) that were fixed in ethanol–acetic acid (3 : 1). Larvae of the population SI (Italy) were collected alive to create a laboratory culture. Larvae of the first laboratory generation were used for genetic analyses in this population. Specific identity of the cryptic C. riparius was established with COI barcoding [28] (see below).
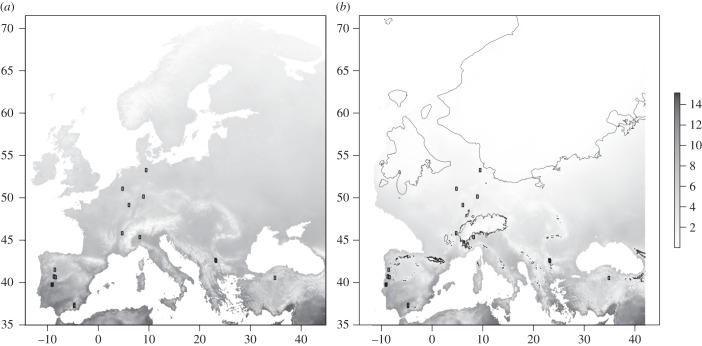
Figure 1.
Potential distribution range of C. riparius along longitudes and latitudes in Europe including the geographical distribution of sampling sites at (a) present time and (b) during the Last Glacial Maximum (LGM) with the borders of glaciation. The grey scale corresponds to the cumulative number of potential generations per year modelled on the basis of monthly mean temperatures and a model of the EmT50-temperature dependency.
(b). Sequencing of genetic markers
Ethanol-acid-preserved larvae from Bulgaria and Turkey were processed with a modified protocol according to Xiong & Kocher [29]. DNA from Portuguese larvae was extracted applying a CTAB protocol (CTAB—cetyl trimethylammonium bromide, modified [30]). DNA from all remaining populations was extracted with the HotSHOT procedure originally developed for zooplanktonic diapausing eggs [31]. DNA was extracted from the head capsules and salivary glands or the last larval abdominal segment to avoid contamination with intestinal bacteria. A fragment of the mitochondrial COI gene was amplified for all populations by polymerase chain reaction (PCR) with the universal primers LCO1490 and HCO2198 [32]. PCR was performed with MOLPol DNA polymerase (Molegene GmbH, Germany) in 10 µl reaction with 5 min incubation at 95°C, 35 amplification cycles (95°C for 30 s, 48°C for 1 min, 72°C for 1 min 30 s) and final elongation for 10 min at 72°C.
Fragments of the nuclear-encoded mitochondrial 39S ribosomal protein L44 (L44) and the plasma membrane calcium-transporting ATPase 3 (PMCA3) were added as nuclear markers for eleven and nine populations. A fragment of L44 was amplified with a species-specific primer-pair (forward: 5′-TGTACGCTTTTGTGCAATC and reverse: 5′-TCTCAACACCGACTGATC). The fragment of PMCA3 covered the fourth and fifth exon, and the intron in between. Amplification was based on a species-specific primer-pair (forward: 5′-ACAAGGCATCCACTCACTTG and reverse: 5′-TGTAGGCATCCAAGTTGACG). PCRs for the two nuclear markers followed the protocol described for COI [28] with modified annealing temperature of 50°C for L44 and 55°C for PMCA3. Sequencing reactions were cleaned according to ABI manufacturer's instructions, PCR products were sequenced on an ABI 3730 Genetic Analyser.
Sequences were trimmed and edited in Geneious 6.1.7. The population-wise and overall alignments were performed with the ClustalW algorithm and IUB cost matrix (Geneious 6.1.7). Sequences of nuclear markers were phased to haplotypes in DnaSP v. 5.10.01 [33].
(c). Estimation of temperature-dependent generation time
The generation time was approximated by the time when 50% of the individuals had emerged (EmT50) during full life cycle experiments at different temperatures. Because the adults reproduce and die shortly after emergence, and the emergence process stretches over several days, this approximation seemed reasonable. A previous common garden study showed that the EmT50 does not differ among populations [34] and experimentally assessed values can be extended to other populations. Full life cycle experiments were performed at 14, 20 and 26°C in separate climate test chambers at a light–dark rhythm of 16 : 8 h. Four different populations were tested with five replicates each in a total of 150 individuals per population and temperature, respectively. Thirty individuals of 1-day-old larvae out of 5–10 egg clutches were reared in one glass vessel filled with 1 l medium (purified water with TropicMarin sea salt adjusted to a conductivity of 520–540 µS cm−1 at pH 8) and washed sand (3 : 1). Emerged imagos were collected from the vessels twice a day. EmT50 values were calculated via nonlinear regression of cumulative emergence against log(time). Additional EmT50 values at 17, 20, 23, 24 and 28°C were taken from the literature [35–37]. These data were fitted to an exponential decay function (electronic supplementary material, S1.1). As water temperatures of small water bodies tend to follow average daily air temperatures [38,39], mean monthly temperatures of current conditions (approx. 1950–2000, WorldClim data [40]) were extracted for every population in 2.5 arc-min resolution. We used the fitted function to calculate the number of possible generations in each month for each site under these temperature conditions. The sum of the monthly values was used as estimate of the average number of potential generations per year for the respective population.
We calculated the number of potential generations per year in Europe in the present by fitting this function to the monthly mean temperatures extrapolated for Europe at a 2.5 arc-min resolution [40]. We then summarized the number of generations projected for each month. For Last Glacial Maximum (LGM) conditions, we calculated monthly mean temperatures from monthly minimum and maximum temperature modelled by three global climate models: MPI-electronic supplementary material-P [41], MIROC-electronic supplementary material [42] and CCSM4 (accessed on 28 September 2015 from http://ccsm.ucar.edu/models/ccsm4.0/). All data for these projections were downloaded from the WorldClim database.
(d). Estimation of genetic parameters
In order to see whether the hypothesized effects of temperature on population genetic parameters via generation time and mutation rate may have a measurable effect on the chosen marker, we initially performed some exploratory simulations (electronic supplementary material, S1.5). We calculated Tajima's D [43] in DnaSP (v. 5.10.01 [33]) for each population and marker separately to test whether the assessed loci were in mutation–migration–drift equilibrium and not unduly influenced by demographic fluctuations, strong unbalanced migration or selection. As an additional line of evidence for possible long-term stability of populations, we calculated the number of potential generations per year under LGM climatic conditions.
Only COI data could be used to estimate the influence of annual mean temperatures on population differentiation. L44 and PMCA3 were not available for a sufficient number of populations to allow a meaningful comparison. Population differentiation was measured as population pairwise ΦST values, to reflect the correlation of haplotypic diversity [44], in Arlequin v. 3.5.1.2 [45]. To account for the effect of geography on population differentiation (isolation-by-distance) expected for this species [34], we regressed the population pairwise ΦSTs against the pairwise geographical distances. Residuals from the ordinary least-squares regression were taken as geographically unbiased population differentiation parameters. Dividing the populations in an evolutionary faster group (greater than 10 generations per year, 10 populations, 45 pairwise comparisons) and a slower group (less than 10 generations, 8 populations, 28 pairwise comparisons) allowed comparison of mean within-group differentiation with a non-parametrical Mann–Whitney U-test. Statistical significance of this statistically non-independent distance data was assessed with Monte Carlo permutation by randomly assigning ΦST residuals 10 000 times to one of the two groups and comparing the resulting null distribution of the U-statistics with the observed value. A threshold of 10 potential generations per year appeared justified, because less than 10 possible generations per year indicate that these populations experience a prolonged developmental pause during cold months (cf. electronic supplementary material, S1.2 for population grouping). However, other thresholds yielded similar results (data not shown).
To estimate mutation accumulation and as an additional measure of intrapopulation drift, we calculated the average population distance from the ancestral haplotype for all populations and all markers. The most likely ancestral haplotype was inferred using the statistical parsimony principles detailed by Pfenninger & Posada [46] using the software TCS [47]. The (uncorrected) p-distance of all other haplotypes to this haplotype was calculated in MEGA 6 [48]. The average of all distances within a population was correlated with their estimated number of potential generations per year.
Temperature dependence of the population-specific per generation mutation rates was estimated by correlating θ (2Neμ, respectively 4Neµ) against average annual mean temperature. θ was calculated from the nucleotide diversity π for each population and marker in DnaSP v. 5.10.01 [33]. Average annual temperature for each site was extracted from the WorldClim database [40]. All statistical analyses were performed with the software package PAST v. 3 [49].
3. Results
(a). Life cycle experiments and species distribution modelling
Full life cycle experiments were successfully performed under 14, 20 and 26°C, and resulted in mean EmT50 values of 33.6, 18.1 and 11.4 days, respectively. Together with additional data from literature (cf. Material and methods), we fitted an exponential decay model (EmT50 [d] = 617.77 × exp(−0.2417 × T[°C]) + 12.382) to the data to model the EmT50-temperature dependency (electronic supplementary material, S1.1). This model captures the effective developmental stall of the larvae below 10°C, because estimated EmT50 becomes practically infinitely large.
The estimated number of potential generations per year ranged between 6.4 (northern Germany, NG) and 15.0 (southern Spain A, SSA—electronic supplementary material, S1.2). Projecting the potential number of generations on the map showed that the species can reproduce at least once per year in most parts of Europe with the exception of the highest altitudes and latitudes (figure 1a).
Projecting the EmT50-temperature dependency model on LGM climatic conditions suggested that the locations investigated could have had at least a few generations per year and thus had the potential to have been permanently occupied for at least the last 100 kyr (figure 1b).
(b). Population genetic analyses
We obtained mitochondrial COI sequences from 795 individuals sampled at 18 sites with an alignment length of 638 bases comprising a set of 60 haplotypes. COI data were successfully sequenced from all populations. Amplification of nuclear markers was successful for a limited number of populations. L44 sequences of 491 individuals from eleven populations were phased to 982 sequences (28 haplotypes) with an alignment length of 551 bases. Phasing of PMCA3 data from 164 individuals sampled at nine sites resulted in 364 haploid sequences of 645 bases.
With the exception of one population (middle France, MF; for COI), the estimates of Tajima's D for all populations and markers suggested that the hypothesis of mutation–migration–drift equilibrium (i.e. neutral evolution could not be rejected; electronic supplementary material, S3). Therefore, the population MF was excluded in subsequent COI analyses.
The population pairwise ΦSTs ranged from zero to 0.71. There was a highly significant isolation-by-distance pattern (r = 0.446, Mantel's test p = 0.0001; electronic supplementary material, S1.3). After accounting for the influence of geographical distance on population divergence, the maternal lineage of populations with more than 10 potential generations per year were significantly more differentiated from each other than slower evolving populations were (Mann–Whitney U-test, z = −6.595, p = 0.0001; figure 2).
Figure 2.
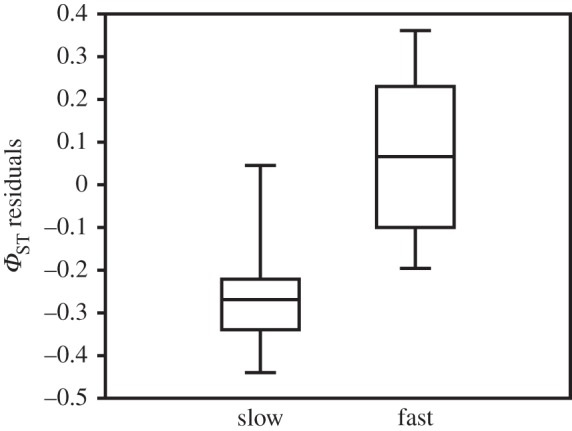
Boxplot of ΦST residuals from the correlation to geographical distance (see electronic supplementary material, S1.3). There was a significant difference between slow-evolving populations with fewer than 10 potential generations per year, and fast-evolving populations with more than 10 potential generations per year (Mann–Whitney U permutation test, p = 0.0001).
We found a significant correlation of average distance from the ancestral COI haplotype to the number of potential generations per year (r = 0.556, p = 0.023; figure 3a). The ancestral haplotype was distributed across all populations; however, additional derived haplotypes with relatively high frequencies were found in populations from warmer sites. In both nuclear markers, there was also a quite strong, albeit marginally non-significant positive relationship between these parameters (L44 r = 0.48, p = 0.12, PMCA3 r = 0.59, p = 0.08; electronic supplementary material, S1.4A).
Figure 3.
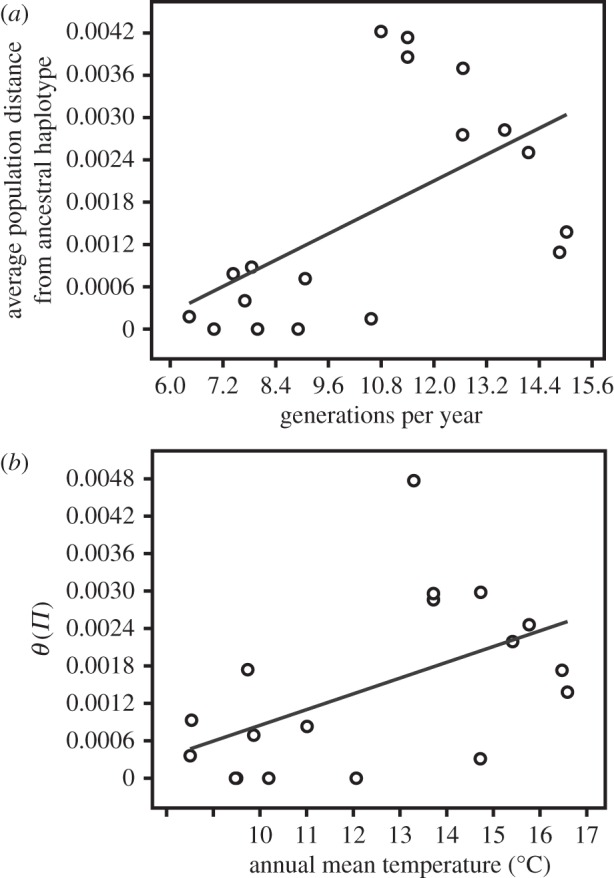
(a) Plot of the average population distance from the ancestral COI haplotype in C. riparius against the number of potential generations per year (r = 0.556, p = 0.023). Average distances are based on a statistical parsimony network and the (uncorrected) p-distance from the ancestral haplotype and numbers of potential generations per year were modelled according to the experimentally assessed EmT50-temperature dependency. (b) Plot of the population mutation parameter θ(π) against the annual mean temperature (r = 0.520, p = 0.028). Climate data were assessed from WorldClim and θ(π) was calculated for the COI data.
θ(π) of the locus COI was significantly positively correlated with the annual mean temperature of the respective sampling site (r = 0.520, p = 0.028; figure 3b). Although the correlations of the two nuclear markers with temperature were positive, the small number of populations assessed yielded not enough statistical power, so they were indistinguishable from zero (L44 r = 0.38, p = 0.23, PMCA3 r = 0.35, p = 0.25; electronic supplementary material, S1.4B).
4. Discussion
Our study is, to our best knowledge, the first to derive and test specific predictions gained from ESH for the distribution of neutral genetic diversity at the intraspecific level. We found support for the hypothesis that population divergence and haplotype diversity scale with the number of generations per year, and thus, with ambient temperature. In addition, under the assumptions that the spontaneous mutation rate is constant between generations and population size is a random factor, there is indirect evidence that the spontaneous mutation rate may also depend on ambient temperature. Our findings were statistically supported for the mitochondrial COI locus. Two additional and independently inherited nuclear loci showed the same trend, so the pattern might not be a locus-specific effect. The lack of statistical support at the nuclear loci was most probably owing to the lower number of populations assayed, which in turn was ultimately caused by DNA preservation conditions for these samples.
(a). Ruling out the influence of population history
The demographic reaction of species to the glacial cycles of the Pleistocene is known to shape genetic diversity across species ranges and may produce similar patterns to those reported here [46]. Therefore, it was of crucial importance to show that the population history of C. riparius was not governed by recent demographic events, like post-glacial range expansions, imbalanced migration or population bottlenecks that would interfere with the expected patterns. All analysed populations were in mutation–migration–drift equilibrium (electronic supplementary material, S3), indicating neutral evolution across Europe for the applied markers. In particular, there was no negative correlation of Tajima's D with latitude, which would be expected in case of a postglacial range expansion (data not shown). Corroborating evidence for a continued long-term persistence of the populations analysed comes from LGM projections of the potential number of generations per year. C. riparius requires small water bodies with organic decomposing matter. During LGM, meltwater from glaciers across Europe (figure 1b) drained into the ice-free regions [50], probably providing the necessary conditions at the current population sites. The estimated origin of C. riparius is about 1.3–1.8 Mya [51]. Given that LGM temperature conditions allowed more than a single generation per year (figure 1b), the populations may have persisted at least since the beginning of the last interglacial and potentially even throughout the Late Pleistocene. Therefore, confounding effects of population history are probably negligible and allow testing the predictions of the ESH. Even though the number of generations per year likely changed according to the climatic cycles, it is reasonable to assume that their relative differential evolutionary speed along the latitudinal gradient persisted and allowed the accumulation of its effect at neutrally evolving loci over time.
(b). Population differentiation in relation to the relative evolutionary age
The ΦST analysis supported the strong overall isolation-by-distance pattern already known for this species [35]. However, subtracting the effect of geographical distance, the analysis of ΦST residuals revealed a significant pattern of increased population differentiation among evolutionary faster populations from warmer sites in comparison with their counterparts from colder sites. Such a pattern could also arise if the southern populations would belong to divergent, geographically limited lineages only one of which colonized the northern part of the range. This is, however, not the case as shown by the absence of diverged lineages and the large geographical spread of all major haplotypes.
Another hypothesis making explicit predictions concerning the expected population structure and distribution of genetic diversity is the ‘abundance centre’ hypothesis [52]. It states that a species's abundance is usually greatest in the centre of its geographical range and uniformly low towards the margins. As a consequence, marginal populations should have higher turnover, smaller Ne and thus harbour less genetic variation and should be stronger differentiated, when compared with central populations [53,54]. The pattern observed here was unlikely to have been produced by this process. First, our sampling covers the central part of this Holarctic distributed species, excluding more northerly (Sweden and Russia [55,56]) and southerly (Morocco [57]) sites. Second, the populations were in mutation–migration–drift equilibrium, which implies that the populations are not subject to frequent extinction/recolonization cycles in association with demographic fluctuations. Third, low genetic diversity was associated with little population differentiation instead of the opposite (see below).
(c). Divergent haplotype age across the species range
The significant correlation of the average distance from the ancestral haplotype with the number of potential generations per year supports the prediction that haplotypes in populations with more generations per year are on average evolutionarily more derived than haplotypes in populations from colder, hence slower evolving sites. Everything else being equal, both the absolute number of annually arising mutations in a population and the change of haplotype frequencies in a population owing to drift depend on the number of generations per year. In a multivoltine species, this relation inevitably links the neutral haplotype turnover with the ambient temperature experienced. The same should be true for the other major generation-dependent process, natural selection [58], though this is empirically much more difficult to quantify and does not apply to the loci examined here.
In principle, all patterns observed could also be due to systematic differences in effective population size Ne across the species range. Smaller Ne means more drift, leading to lower genetic diversity and stronger population differentiation [19]. If Ne would be consistently smaller in colder areas, populations from such sites should therefore be stronger differentiated [59]. However, the opposite is the case (figure 2). Conversely, if Ne would be systematically smaller in warmer areas, genetic variation should be smaller there, which is not what we found (electronic supplementary material, S3). Consequently, the observed pattern contradicts the expectation of a systematic difference in Ne among warmer and colder regions and suggest that variance in Ne can be treated as a random factor. Indeed, C. riparius populations grow to large sizes under favourable conditions [51] and seem to be size limited by habitat availability rather than by climatic conditions, at least in the range investigated here.
(d). Mutation rate
The population mutation parameter θ is the product of Ne and the mutation rate per generation µ [60]. For our study, we estimated θ on the basis of nucleotide diversity π, thus independent of an estimation on Ne and µ. We are not aware of any expectations that Ne depends on temperature [19,61], and variance in Ne is therefore regarded as random factor across generations as well as the species range (also see above). The significant positive correlation of θ (π) from the mitochondrial locus with the annual mean temperature of the respective population site gives an indication that µ, as the remaining variable in the equation (θ = 2Neµ), might thus be primarily responsible for the observed scaling of θ with temperature. With our study, we thus provide additional indirect evidence for this relation, even though the effects of generation time and spontaneous mutation rate cannot be completely disentangled without an experimental approach. The mitochondrial data in our study, supported by a similar pattern from the nuclear data, empirically show that with the increasing number of potential generations per year, evolutionary processes at the molecular level are accelerated in C. riparius. If this process extends to the entire genome, populations from warmer regions thus diverge faster and accumulate more mutations, possibly also owing to an increased mutation rate on an absolute time scale.
Studies on Drosophila melanogaster suggest that indeed temperature-dependent metabolic activities increased the somatic mutation rate as consequence of oxidative stress [62]. However, temperature had no influence on the transposition rate in the same species [63,64]. In Caenorhabditis elegans, higher temperature stress had a significant effect on microsatellite mutation rate [65]. On an interspecific level, the temperature-dependent mass-specific metabolic rate seems to influence the molecular clock rate that depends at least partially on the mutation rate [66].
Although the nuclear data only provided tendential support, our data suggested that the predictions gained from the ESH regarding neutral intraspecific genetic variation apply to C. riparius. This suggests that climate may indeed influence the rate of evolution, at least for ectothermic organisms, i.e. the largest part of biodiversity. Similar patterns were also found in marine prosobranch gastropoda across the Atlantic and Pacific Ocean, where diversity trends are significantly correlated with the mean sea surface temperature [67]. The comparison of 97 recently diverged Centrolenidae species also revealed a positive relationship of mitochondrial and nuclear substitution rates with temperature [68]. Moreover, bacterial diversity is largely generated and maintained by the effects of temperature on the kinetics of metabolism [69]. Taken together, there is increasing empirical support for the ESH from a very diverse group of taxa, implying a common mechanism for the distribution of global biodiversity. The general ESH of Rohde [7] focused on the aspect of selection and neglected the equally important effect of drift on molecular evolution [70]. Showing that neutral evolution in the analysed C. riparius populations depends to some extent on the ambient temperature regime, it is worth extending the ESH with the aspect of drift to a more realistic scenario of evolution. Gillman & Wright [16] proposed an integrated ESH and extended the temperature dependence of the rate of molecular evolution by the influences of water availability, population size and spatial heterogeneity. However, there is already evidence that indeed only temperature might be the crucial and principal factor behind associations with water availability, elevation and UVB radiation [68].
In conclusion, if the temperature dependence of evolutionary speed and mutation rate is found in more comprehensive studies, then this has far-reaching consequences to phylogenetic and phylogeographic reconstructions as well as for population genetic analyses. Even if speciation is not necessarily the inevitable consequence of population divergence, the stronger differentiation among populations from warmer sites nonetheless increases divergence rates [71]. Temperature may thus not only affect everything an organism does [1], but may also crucially influence its evolution. Therefore, further research on the predictions derived from the ESH is needed to finally address the complex interplay between temperature and evolution.
Supplementary Material
Supplementary Material
Supplementary Material
Acknowledgement
We thank Bob O'Hara, Simit Patel and two anonymous reviewers for their helpful comments on the manuscript.
Data accessibility
Sequence data of all markers and all populations are deposited via EMBL-EBI (accession numbers: LN894198–LN896302).
Authors' contributions
A.-M.O. and M.P. conceived the study, performed the statistical analyses and drafted the manuscript. A.-M.O. carried out the life cycle experiments. A.-M.O., J.A.M.P. and J.B.D. performed the molecular work. M.B. carried out the functional species distribution modelling. J.I. and J.L.T.P. provided samples.
Competing interests
We have no competing interests.
Funding
Funding was provided by DFG (PF390/7-1). Furthermore, this work was also partly supported by FEDER through COMPETE- Programa Operacional Factores de Competitividade, and by National funding through FCT-Fundação para a Ciência e Tecnologia, within the research project MIDGE—MIcroevolutionary Dynamics and Genetic Erosion in pollution-affected Chironomus populations (FCOMP-01-0124-FEDER-008954). We also thank FCT and POPH/FSE for the fellowships of J.L.T. Pestana (SFRH/BPD/45342/2008) and J.A.M. Pedrosa (SFRH/BD/75606/2010).
References
- 1.Clarke A. 2003. Costs and consequences of evolutionary temperature adaptation. Trends Ecol. Evol. 18, 573–581. ( 10.1016/j.tree.2003.08.007) [DOI] [Google Scholar]
- 2.Gillooly JF, Brown JH, West GB, Savage VM, Charnov EL. 2001. Effects of size and temperature on metabolic rate. Science 293, 2248–2251. ( 10.1126/science.1061967) [DOI] [PubMed] [Google Scholar]
- 3.Clarke A, Fraser KPP. 2004. Why does metabolism scale with temperature?. Funct. Ecol. 18, 243–251. ( 10.1111/j.0269-8463.2004.00841.x) [DOI] [Google Scholar]
- 4.Gillooly JF, Allen AP. 2007. Linking global patterns in biodiversity to evolutionary dynamics using metabolic theory. Ecology 88, 1890–1894. ( 10.1890/06-1935.1) [DOI] [PubMed] [Google Scholar]
- 5.O'Connor MI, Bruno JF, Gaines SD, Halpern BS, Lester SE, Kinlan BP, Weiss JM. 2007. Temperature control of larval dispersal and the implications for marine ecology, evolution, and conservation. Proc. Natl Acad. Sci. USA 104, 1266–1271. ( 10.1073/pnas.0603422104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rensch B. 1959. Evolution above the species level. London, UK: Methuen & Co Ltd. [Google Scholar]
- 7.Rohde K. 1992. Latitudinal gradients in species-diversity—the search for the primary cause. Oikos 65, 514–527. ( 10.2307/3545569) [DOI] [Google Scholar]
- 8.Nabholz B, Glemin S, Galtier N.. 2008. Strong variations of mitochondrial mutation rate across mammals—the longevity hypothesis. Mol. Biol. Evol. 25, 120–130. ( 10.1093/molbev/msm248) [DOI] [PubMed] [Google Scholar]
- 9.Tsantes C, Steiper ME. 2009. Age at first reproduction explains rate variation in the strepsirrhine molecular clock. Proc. Natl Acad. Sci. USA 106, 18 165–18 170. ( 10.1073/pnas.0906686106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Smith SA, Donoghue MJ. 2008. Rates of molecular evolution are linked to life history in flowering plants. Science 322, 86–89. ( 10.1126/science.1163197) [DOI] [PubMed] [Google Scholar]
- 11.Thomas JA, Welch JJ, Lanfear R, Bromham L. 2010. A Generation time effect on the rate of molecular evolution in invertebrates. Mol. Biol. Evol. 27, 1173–1180. ( 10.1093/molbev/msq009) [DOI] [PubMed] [Google Scholar]
- 12.Nikolaev SI, Montoya-Burgos JI, Popadin K, Parand L, Margulies EH, Antonarakis SE, Program N.. 2007. Life-history traits drive the evolutionary rates of mammalian coding and noncoding genomic elements. Proc. Natl Acad. Sci. USA 104, 20 443–20 448. ( 10.1073/pnas.0705658104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gillman LN, Wright SD. 2014. Species richness and evolutionary speed: the influence of temperature, water and area. J. Biogeogr. 41, 39–51. ( 10.1111/jbi.12173) [DOI] [Google Scholar]
- 14.Wright S, Keeling J, Gillman L.. 2006. The road from Santa Rosalia: a faster tempo of evolution in tropical climates. Proc. Natl Acad. Sci. USA 103, 7718–7722. ( 10.1073/pnas.0510383103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Davies TJ, Savolainen V, Chase MW, Moat J, Barraclough TG. 2004. Environmental energy and evolutionary rates in flowering plants. Proc. R. Soc. Lond. B 271, 2195–2200. ( 10.1098/rspb.2004.2849) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gillman L, Wright S.. 2013. Patterns of evolutionary speed: in search of a causal mechanism. Diversity 5, 811–823. ( 10.3390/d5040811) [DOI] [Google Scholar]
- 17.Allen AP, Gillooly JF, Savage VM, Brown JH. 2006. Kinetic effects of temperature on rates of genetic divergence and speciation. Proc. Natl Acad. Sci. USA 103, 9130–9135. ( 10.1073/pnas.0603587103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bromham L. 2009. Why do species vary in their rate of molecular evolution?. Biol. Lett. 5, 401–404. ( 10.1098/rsbl.2009.0136) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Charlesworth B. 2009. Effective population size and patterns of molecular evolution and variation. Nat. Rev. Genet. 10, 195–205. ( 10.1038/nrg2526) [DOI] [PubMed] [Google Scholar]
- 20.Wright S. 1950. The genetical structure of populations. Ann. Eugenics 15, 323–354. ( 10.1111/j.1469-1809.1949.tb02451.x) [DOI] [PubMed] [Google Scholar]
- 21.Coyne JA, Orr HA. 2004. Speciation. Sunderland, MA: Sinauer Associates. [Google Scholar]
- 22.Avise JC. 2000. Phylogeography: the history and formation of species. Cambridge, MA: Harvard University Press. [Google Scholar]
- 23.Martin PR, McKay JK. 2004. Latitudinal variation in genetic divergence of populations and the potential for future speciation. Evolution 58, 938–945. ( 10.1111/j.0014-3820.2004.tb00428.x) [DOI] [PubMed] [Google Scholar]
- 24.Adams RI, Hadly EA. 2013. Genetic diversity within vertebrate species is greater at lower latitudes. Evol. Ecol. 27, 133–143. ( 10.1007/s10682-012-9587-x) [DOI] [Google Scholar]
- 25.Chek AA, Austin JD, Lougheed SC. 2003. Why is there a tropical-temperate disparity in the genetic diversity and taxonomy of species? Evol. Ecol. Res. 5, 69–77. [Google Scholar]
- 26.Eo SH, Wares JP, Carroll JP. 2008. Population divergence in plant species reflects latitudinal biodiversity gradients. Biol. Lett. 4, 382–384. ( 10.1098/rsbl.2008.0109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pinder LCV. 1986. Biology of freshwater Chironomidae. Annu. Rev. Entomol. 31, 1–23. ( 10.1146/annurev.en.31.010186.000245) [DOI] [Google Scholar]
- 28.Pfenninger M, Nowak C, Kley C, Steinke D, Streit B.. 2007. Utility of DNA taxonomy and barcoding for the inference of larval community structure in morphologically cryptic Chironomus (Diptera) species. Mol. Ecol. 16, 1957–1968. ( 10.1111/j.1365-294X.2006.03136.x) [DOI] [PubMed] [Google Scholar]
- 29.Xiong B, Kocher TD. 1991. Comparison of mitochondrial-DNA sequences of 7 morphospecies of black flies (Diptera, Simuliidae). Genome 34, 306–311. ( 10.1139/g91-050) [DOI] [PubMed] [Google Scholar]
- 30.Jones AS, Walker RT. 1963. Isolation and analysis of the deoxyribonucleic acid of Mycoplasma mycoides var. capri. Nature 198, 588–589. ( 10.1038/198588a0) [DOI] [PubMed] [Google Scholar]
- 31.Montero-Pau J, Gómez A, Munoz J. 2008. Application of an inexpensive and high-throughput genomic DNA extraction method for the molecular ecology of zooplanktonic diapausing eggs. Limnol. Oceanogr.: Methods 6, 218–222. ( 10.4319/lom.2008.6.218) [DOI] [Google Scholar]
- 32.Folmer O, Black M, Hoeh W, Lutz R, Vrijenhoek R. 1994. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol. Mar. Biol. Biotechnol. 3, 294–299. [PubMed] [Google Scholar]
- 33.Librado P, Rozas J.. 2009. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25, 1451–1452. ( 10.1093/bioinformatics/btp187) [DOI] [PubMed] [Google Scholar]
- 34.Nemec S, Hess M, Nowak C, Pfenninger M.. 2012. Experimental evidence for niche segregation in a sister species pair of non-biting midges. Hydrobiologia 691, 203–212. ( 10.1007/s10750-012-1074-4) [DOI] [Google Scholar]
- 35.Nemec S, Patel S, Nowak C, Pfenninger M.. 2013. Evolutionary determinants of population differences in population growth rate × habitat temperature interactions in Chironomus riparius. Oecologia 172, 585–594. ( 10.1007/s00442-012-2517-3) [DOI] [PubMed] [Google Scholar]
- 36.Oetken M, Jagodzinski LS, Vogt C, Jochum A, Oehlmann J.. 2009. Combined effects of chemical and temperature stress on Chironomus riparius populations with differing genetic variability. J. Environ. Sci. Heal A 44, 955–962. ( 10.1080/10934520902996849) [DOI] [PubMed] [Google Scholar]
- 37.Vogt C, Pupp A, Nowak C, Jagodzinski LS, Baumann J, Jost D, Oetken M, Oehlmann J.. 2007. Interaction between genetic diversity and temperature stress on life-cycle parameters and genetic variability in midge Chironomus riparius populations. Clim. Res. 33, 207–214. ( 10.3354/Cr033207) [DOI] [Google Scholar]
- 38.Stefan HG, Preudhomme EB. 1993. Stream temperature estimation from air-temperature. Water Resour. Bull. 29, 27–45. ( 10.1111/j.1752-1688.1993.tb01502.x) [DOI] [Google Scholar]
- 39.Webb BW, Clack PD, Walling DE. 2003. Water–air temperature relationships in a Devon river system and the role of flow. Hydrol. Process 17, 3069–3084. ( 10.1002/hyp.1280) [DOI] [Google Scholar]
- 40.Hijmans RJ, Cameron SE, Parra JL, Jones PG, Jarvis A.. 2005. Very high resolution interpolated climate surfaces for global land areas. Int. J. Climatol. 25, 1965–1978. ( 10.1002/Joc.1276) [DOI] [Google Scholar]
- 41.Giorgetta MA, et al. 2013. Climate and carbon cycle changes from 1850 to 2100 in MPI-ESM simulations for the coupled model intercomparison project phase 5. J. Adv. Model. Earth Syst. 5, 572–597. ( 10.1002/Jame.20038) [DOI] [Google Scholar]
- 42.Watanabe S, et al. 2011. MIROC-ESM 2010: model description and basic results of CMIP5-20c3 m experiments. Geosci. Model Dev. 4, 845–872. ( 10.5194/gmd-4-845-2011) [DOI] [Google Scholar]
- 43.Tajima F. 1989. Statistical-method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123, 585–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Excoffier L, Smouse PE, Quattro JM. 1992. Analysis of molecular variance inferred from metric distances among DNA haplotypes—application to human mitochondrial-DNA restriction data. Genetics 131, 479–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Excoffier L, Laval G, Schneider S. 2005. Arlequin (version 3.0): an integrated software package for population genetics data analysis. Evol. Bioinform. 1, 47–50. [PMC free article] [PubMed] [Google Scholar]
- 46.Pfenninger M, Posada D.. 2002. Phylogeographic history of the land snail Candidula unifasciata (Helicellinae, Stylommatophora): fragmentation, corridor migration, and secondary contact. Evolution 56, 1776–1788. ( 10.1111/j.0014-3820.2002.tb00191.x) [DOI] [PubMed] [Google Scholar]
- 47.Clement M, Posada D, Crandall KA. 2000. TCS: a computer program to estimate gene genealogies. Mol. Ecol. 9, 1657–1659. ( 10.1046/j.1365-294x.2000.01020.x) [DOI] [PubMed] [Google Scholar]
- 48.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S.. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. ( 10.1093/molbev/mst197) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hammer Ø, Harper DAT, Ryan PD. 2001. PAST: paleontological statistics software package for education and data analysis. Palaeontol. Electron. 4, 4. [Google Scholar]
- 50.Küster H. 1999. Geschichte der Landschaft in Mitteleuropa: von der Eiszeit bis zur Gegenwart. Munich, Germany: C.H. Beck. [Google Scholar]
- 51.Schmidt H, Greshake B, Feldmeyer B, Hankeln T, Pfenninger M.. 2013. Genomic basis of ecological niche divergence among cryptic sister species of non-biting midges. BMC Genomics 14, 384 ( 10.1186/1471-2164-14-384) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sagarin RD, Gaines SD. 2002. The ‘abundant centre’ distribution: to what extent is it a biogeographical rule?. Ecol. Lett. 5, 137–147. ( 10.1046/j.1461-0248.2002.00297.x) [DOI] [Google Scholar]
- 53.Eckert CG, Samis KE, Lougheed SC. 2008. Genetic variation across species’ geographical ranges: the central-marginal hypothesis and beyond. Mol. Ecol. 17, 1170–1188. ( 10.1111/j.1365-294X.2007.03659.x) [DOI] [PubMed] [Google Scholar]
- 54.Pfenninger M, Salinger M, Haun T, Feldmeyer B.. 2011. Factors and processes shaping the population structure and distribution of genetic variation across the species range of the freshwater snail Radix balthica (Pulmonata, Basommatophora). BMC Evol. Biol. 11, 135 ( 10.1186/1471-2148-11-135) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gunderina LI, Kiknadze II, Istomina AG, Butler M. 2009. Geographic differentiation of genomic DNA of Chironomus plumosus (Diptera, Chironomidae) in natural holarctic populations. Russ. J. Genet. 45, 54–62. ( 10.1134/S1022795409010086) [DOI] [PubMed] [Google Scholar]
- 56.Jernelov A, Nagell B, Svenson A. 1981. Adaptation to an acid environment in Chironomus riparius (Diptera, Chironomidae) from Smoking Hills, Nwt, Canada. Holarctic Ecol. 4, 116–119. [Google Scholar]
- 57.Kettani K, Langton PH. 2011. New data on the Chironomidae (Diptera) of the Rif (Northern Morocco). Polish J. Entomol. 80, 587–599. ( 10.2478/v10200-011-0046-8) [DOI] [Google Scholar]
- 58.Kimura M. 1980. Average time until fixation of a mutant allele in a finite population under continued mutation pressure—studies by analytical, numerical, and pseudo-sampling methods. Proc. Natl Acad. Sci. USA 77, 522–526. ( 10.1073/pnas.77.1.522) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lanfear R, Kokko H, Eyre-Walker A.. 2014. Population size and the rate of evolution. Trends Ecol. Evol. 29, 33–41. ( 10.1016/j.tree.2013.09.009) [DOI] [PubMed] [Google Scholar]
- 60.Tajima F. 1996. The amount of DNA polymorphism maintained in a finite population when the neutral mutation rate varies among sites. Genetics 143, 1457–1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chesser RK, Rhodes OE, Sugg DW, Schnabel A.. 1993. Effective sizes for subdivided populations. Genetics 135, 1221–1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Garcia AM, Calder RB, Dolle ME, Lundell M, Kapahi P, Vijg J. 2010. Age- and temperature-dependent somatic mutation accumulation in Drosophila melanogaster. PLoS Genet. 6, e1000950 ( 10.1371/journal.pgen.1000950) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Alonso-Gonzalez L, Dominguez A, Albornoz J.. 2006. Direct determination of the influence of extreme temperature on transposition and structural mutation rates of Drosophila melanogaster mobile elements. Genetica 128, 11–19. ( 10.1007/s10709-005-2480-6) [DOI] [PubMed] [Google Scholar]
- 64.Vazquez JF, Albornoz J, Dominguez A.. 2007. Direct determination of the effects of genotype and extreme temperature on the transposition of roo in long-term mutation accumulation lines of Drosophila melanogaster. Mol. Genet. Genomics 278, 653–664. ( 10.1007/s00438-007-0282-5) [DOI] [PubMed] [Google Scholar]
- 65.Matsuba C, Lewis S, Ostrow DG, Salomon MP, Sylvestre L, Tabman B, Ungvari-Martin J, Baer CF. 2012. Invariance (?) of mutational parameters for relative fitness over 400 generations of mutation accumulation in Caenorhabditis elegans. G3-Genes Genom. Genet. 2, 1497–1503. ( 10.1534/g3.112.003947) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gillooly JF, Allen AP, West GB, Brown JH. 2005. The rate of DNA evolution: effects of body size and temperature on the molecular clock. Proc. Natl Acad. Sci. USA 102, 140–145. ( 10.1073/pnas.0407735101) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Roy K, Jablonski D, Valentine JW, Rosenberg G.. 1998. Marine latitudinal diversity gradients: tests of causal hypotheses. Proc. Natl Acad. Sci. USA 95, 3699–3702. ( 10.1073/pnas.95.7.3699) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Dugo-Cota A, Castroviejo-Fisher S, Vila C, Gonzalez-Voyer A.. 2015. A test of the integrated evolutionary speed hypothesis in a Neotropical amphibian radiation. Glob. Ecol. Biogeogr. 24, 804–813. ( 10.1111/geb.12318) [DOI] [Google Scholar]
- 69.Fuhrman JA, Steele JA, Hewson I, Schwalbach MS, Brown MV, Green JL, Brown JH. 2008. A latitudinal diversity gradient in planktonic marine bacteria. Proc. Natl Acad. Sci. USA 105, 7774–7778. ( 10.1073/pnas.0803070105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kimura M. 1985. The neutral theory of molecular evolution. New York, NY: Cambridge University Press. [Google Scholar]
- 71.Danley PD, Kocher TD. 2001. Speciation in rapidly diverging systems: lessons from Lake Malawi. Mol. Ecol. 10, 1075–1086. ( 10.1046/j.1365-294X.2001.01283.x) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequence data of all markers and all populations are deposited via EMBL-EBI (accession numbers: LN894198–LN896302).