Abstract
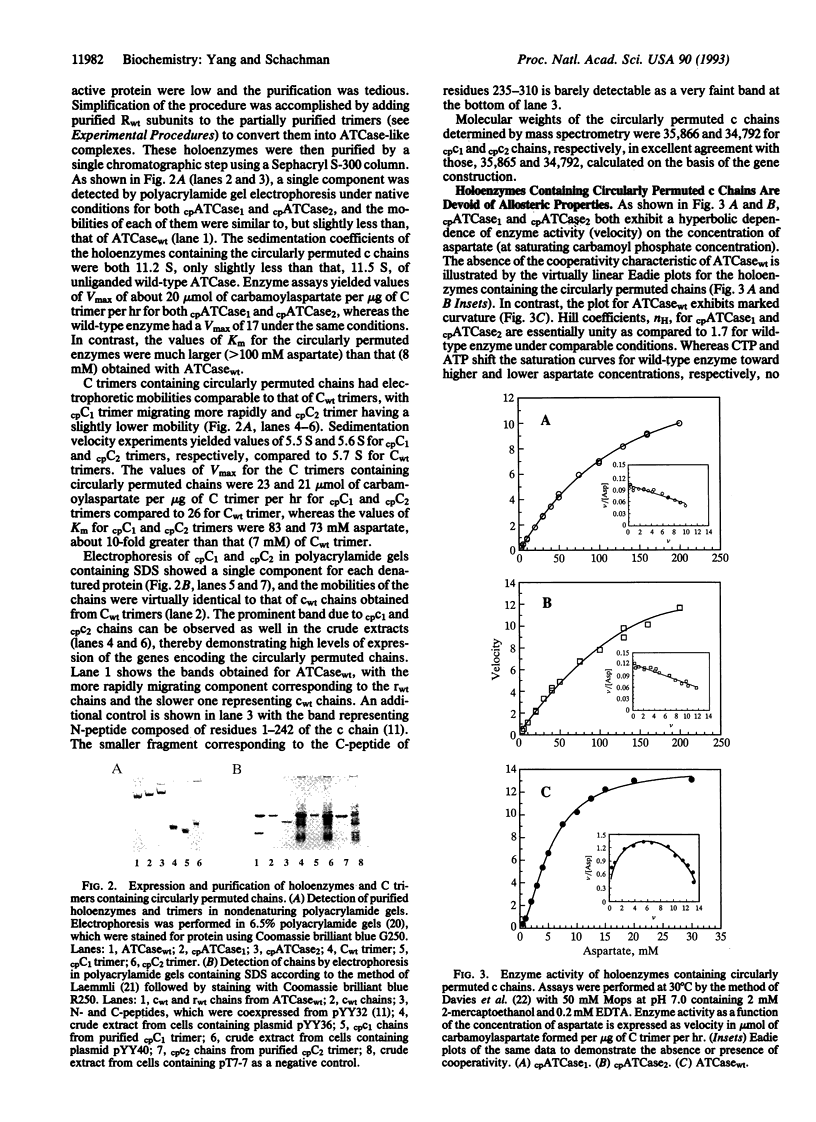
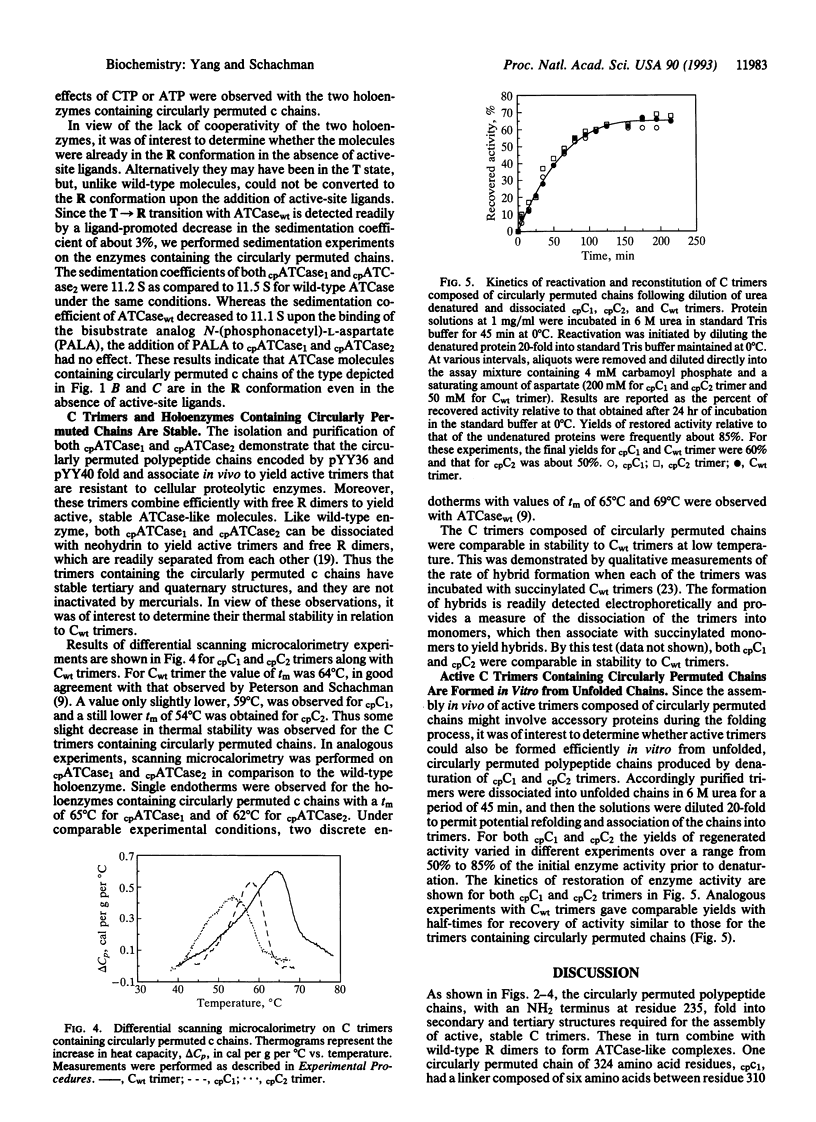
Based on the demonstration that active enzyme is formed in vitro and in vivo from polypeptide fragments of the catalytic chains of aspartate transcarbamoylase (ATCase; EC 2.1.3.2) and the evidence that NH2 and COOH termini of wild-type chains are in close proximity, we constructed altered genes to determine whether circularly permuted catalytic chains could fold and assemble into active catalytic trimers. Two slightly different genetic constructs led to the expression in good yield of circularly permuted catalytic chains, which associated in vivo into active trimers. They, in turn, combined in vitro with wild-type regulatory dimers to form ATCase-like molecules. Both polypeptide chains began at residue 235 in a different domain from the NH2 terminus of wild type and had an overlapping sequence of eight residues at the COOH terminus. One had a six-amino-acid linker, and the other had a deletion of four residues. Enzymes containing rearranged chains were similar to their wild-type counterparts in physical properties. Whereas values of Vmax were close to those of wild-type trimers and ATCase, the Km values were more than 10-fold greater. Also the allosteric properties characteristic of wild-type ATCase were lacking in the enzymes containing permuted chains. Denaturation of trimers by urea was reversible, and recovery of activity in both rate and yield was comparable to that of wild-type trimers. The experiments demonstrate that folding of chains into clearly defined domains and the assembly of active, thermodynamically stable oligomers are not dependent on the positions of NH2 and COOH termini; the folded structures are a consequence of the final sequence and not the order of biosynthetic addition of amino acids.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Buchwalder A., Szadkowski H., Kirschner K. A fully active variant of dihydrofolate reductase with a circularly permuted sequence. Biochemistry. 1992 Feb 18;31(6):1621–1630. doi: 10.1021/bi00121a006. [DOI] [PubMed] [Google Scholar]
- Chung C. T., Miller R. H. A rapid and convenient method for the preparation and storage of competent bacterial cells. Nucleic Acids Res. 1988 Apr 25;16(8):3580–3580. doi: 10.1093/nar/16.8.3580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies G. E., Vanaman T. C., Stark G. R. Aspartate transcarbamylase. Stereospecific restrictions on the binding site for L-aspartate. J Biol Chem. 1970 Mar 10;245(5):1175–1179. [PubMed] [Google Scholar]
- Goldenberg D. P., Creighton T. E. Circular and circularly permuted forms of bovine pancreatic trypsin inhibitor. J Mol Biol. 1983 Apr 5;165(2):407–413. doi: 10.1016/s0022-2836(83)80265-4. [DOI] [PubMed] [Google Scholar]
- Honzatko R. B., Crawford J. L., Monaco H. L., Ladner J. E., Ewards B. F., Evans D. R., Warren S. G., Wiley D. C., Ladner R. C., Lipscomb W. N. Crystal and molecular structures of native and CTP-liganded aspartate carbamoyltransferase from Escherichia coli. J Mol Biol. 1982 Sep 15;160(2):219–263. doi: 10.1016/0022-2836(82)90175-9. [DOI] [PubMed] [Google Scholar]
- Imai Y., Matsushima Y., Sugimura T., Terada M. A simple and rapid method for generating a deletion by PCR. Nucleic Acids Res. 1991 May 25;19(10):2785–2785. doi: 10.1093/nar/19.10.2785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke H. M., Lipscomb W. N., Cho Y. J., Honzatko R. B. Complex of N-phosphonacetyl-L-aspartate with aspartate carbamoyltransferase. X-ray refinement, analysis of conformational changes and catalytic and allosteric mechanisms. J Mol Biol. 1988 Dec 5;204(3):725–747. doi: 10.1016/0022-2836(88)90365-8. [DOI] [PubMed] [Google Scholar]
- Kim K. H., Pan Z. X., Honzatko R. B., Ke H. M., Lipscomb W. N. Structural asymmetry in the CTP-liganded form of aspartate carbamoyltransferase from Escherichia coli. J Mol Biol. 1987 Aug 20;196(4):853–875. doi: 10.1016/0022-2836(87)90410-4. [DOI] [PubMed] [Google Scholar]
- Kunkel T. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc Natl Acad Sci U S A. 1985 Jan;82(2):488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Luger K., Hommel U., Herold M., Hofsteenge J., Kirschner K. Correct folding of circularly permuted variants of a beta alpha barrel enzyme in vivo. Science. 1989 Jan 13;243(4888):206–210. doi: 10.1126/science.2643160. [DOI] [PubMed] [Google Scholar]
- Peterson C. B., Schachman H. K. Role of a carboxyl-terminal helix in the assembly, interchain interactions, and stability of aspartate transcarbamoylase. Proc Natl Acad Sci U S A. 1991 Jan 15;88(2):458–462. doi: 10.1073/pnas.88.2.458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers V. M., Yang Y. R., Fogli M. J., Schachman H. K. Reconstitution of active catalytic trimer of aspartate transcarbamoylase from proteolytically cleaved polypeptide chains. Protein Sci. 1993 Jun;2(6):1001–1012. doi: 10.1002/pro.5560020613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schachman H. K., Pauza C. D., Navre M., Karels M. J., Wu L., Yang Y. R. Location of amino acid alterations in mutants of aspartate transcarbamoylase: Structural aspects of interallelic complementation. Proc Natl Acad Sci U S A. 1984 Jan;81(1):115–119. doi: 10.1073/pnas.81.1.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studier F. W., Rosenberg A. H., Dunn J. J., Dubendorff J. W. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- Thornton J. M., Sibanda B. L. Amino and carboxy-terminal regions in globular proteins. J Mol Biol. 1983 Jun 25;167(2):443–460. doi: 10.1016/s0022-2836(83)80344-1. [DOI] [PubMed] [Google Scholar]
- Yang Y. R., Kirschner M. W., Schachman H. K. Aspartate transcarbamoylase (Escherichia coli): preparation of subunits. Methods Enzymol. 1978;51:35–41. doi: 10.1016/s0076-6879(78)51007-0. [DOI] [PubMed] [Google Scholar]
- Yang Y. R., Schachman H. K. Hybridization as a technique for studying interchain interactions in the catalytic trimers of aspartate transcarbamoylase. Anal Biochem. 1987 May 15;163(1):188–195. doi: 10.1016/0003-2697(87)90111-4. [DOI] [PubMed] [Google Scholar]
- Yang Y. R., Schachman H. K. In vivo formation of active aspartate transcarbamoylase from complementing fragments of the catalytic polypeptide chains. Protein Sci. 1993 Jun;2(6):1013–1023. doi: 10.1002/pro.5560020614. [DOI] [PMC free article] [PubMed] [Google Scholar]



