Abstract
Glycans of prostate-specific antigen (PSA) in prostate cancer were found to be different from that in benign disease. It is difficult to analyze heterogeneous PSA glycoforms in each individual specimen because of low protein abundance and the limitation of detection sensitivity. We developed a method for prostate cancer diagnosis based on PSA glycoforms. Specific glycoforms were screened in each clinical sample based on liquid chromatography-tandem mass spectrometry with ion accumulation. To look for potential biomarkers, normalized abundance of each glycoform in benign prostate hyperplasia (BPH) and in prostate cancer was evaluated. The PSA glycoform, Hex5HexNAc4NeuAc1dHex1, and monosialylated, sialylated, and unfucosylated glycoforms differed significantly between the prostate cancer and BPH samples. The detection sensitivity (87.5%) and specificity (60%) for prostate cancer identification are higher than those of the serum PSA marker. As low as 100 amol PSA could be detected with the ion accumulation method which has not been reported before. The improved detection specificity can help reduce unnecessary examinations.
1. Introduction
Prostate cancer is very common among men worldwide. Prostate-specific antigen (PSA) is a glycoprotein with an N-linked glycosylation site [1], and its level in serum is an FDA-approved prostate cancer marker [2–4]. However, the serum PSA concentration has a low diagnostic specificity for prostate cancer, which leads to many unnecessary patient biopsies [5]. Therefore, there is an urgent need for markers with better specificity. The alterations of protein glycoforms is one of the candidates.
To analyze PSA glycoforms, mass spectrometry (MS) [6–10] has been adopted. Glycan composition, protein binding sites, and peptide backbone sequences can be detected by tandem MS. However, glycoforms identification is hampered by glycan heterogeneity. Liquid chromatography (LC), which is often coupled with mass spectrometry, can be used to separate compounds from a mixture to improve the detection sensitivity. In addition, when the MS is set to detect a specific ion with a known mass-to-charge ratio (m/z), more ions can be accumulated in a trap for MS analysis. In a typical LC-MS analysis, only a small portion of the sample under the specific LC peak is used for mass spectrometry. A typical LC peak lasts from seconds to minutes, but a typical round of ion trapping lasts from only a few milliseconds to a few hundred milliseconds. Therefore, most of the samples eluted from the LC are not used for MS analysis. Because the quantity of each specific PSA glycoform can be very low, ion accumulation can improve the detection sensitivity by orders of magnitude. Using this ion accumulation method, the detection sensitivity can be further improved.
Glycopeptide quantification is a challenge because of the lack of synthetic standards to establish calibration curves. Label-free approaches have been developed for peptide glycoform analysis [11–13]. In LC-MS, the area under an LC peak represents the abundance of the selected molecules. This method has been used to evaluate the specific posttranslational modifications of a protein [14, 15]. To avoid interference from sample processing, the glycopeptide abundance can be evaluated by normalizing to an internal reference peptide from the same protein. The reduced interference from sample processing means that this approach can be used to investigate the relative abundance of each PSA glycoform in an individual clinical sample. Using this approach, the relative abundance of each specific glycoform of haptoglobin and of immunoglobulin G has been accurately measured [11–13]. Nevertheless, no studies have been reported for each PSA glycoform by screening clinical samples using LC-MS.
Recently, PSA glycan profiles have been studied as potential prostate cancer markers [6–8, 16–21]. However, detection of multiple PSA glycoforms in each individual sample was seldom pursued because of the low PSA levels in the samples and because of the heterogeneity of the PSA glycoforms, making the detection of each specific glycoform much more difficult. Urine contains more PSA than serum [22], and it is easier to obtain patients' permissions to collect it. Therefore, each individual sample can be analyzed without the need for pooling. As in semen, urinary PSA exists in its free form [22–24]. It is beneficial to pursue analysis of glycan by measuring glycopeptide. In addition, urinary PSA is directly secreted from the prostate to the urine; unlike serum PSA, it is not renally filtered [24]. Therefore, the PSA composition of the urine should be closer to that of the prostate gland. Urine could be a better specimen than serum for PSA glycosylation analysis in prostate cancer diagnosis.
In this study, we evaluated the relative abundance of each urinary PSA glycoform in clinical samples. Specific glycopeptides and a specific PSA peptide that serves as internal reference were detected using LC-MS. The degree of PSA glycosylation was evaluated by calculating the ratio of the abundance of each glycopeptide to that of the PSA internal reference peptide. Specific PSA glycoforms in clinical samples were identified by ion accumulation method. The analysis of the clinical urine samples showed that the relative abundance of several glycoforms differed between BPH and prostate cancer. Therefore, these glycoforms could be potential biomarkers to distinguish between BPH and prostate cancer.
2. Materials and Methods
To quantify the site-specific PSA glycopeptides in clinical samples, the following processes were performed: human seminal PSA was used to investigate the glycopeptide backbone sequence and to select the internal reference peptide. The peptide backbone sequence of the PSA glycopeptides was identified, and the MS2 profile of each glycoform was obtained. Next, the PSA glycopeptide distributions in pooled BPH and PCa urinary samples were identified. Then, for each individual clinical sample, the ratios of these specific PSA glycopeptides to the reference peptide were determined. Potential glycoform markers that are differentially expressed between BPH and PCa could be discovered.
2.1. Materials and Reagents
Human seminal PSA (P3338, ≥95% purity in SDS-PAGE) and iodoacetamide (IAA, I6125) were purchased from Sigma-Aldrich (St. Louis, MO, USA). Human PSA monoclonal antibodies were purchased from R&D Systems, Inc. (MAB1344, Minneapolis, MN, USA). Dynabeads® Protein G was purchased from Life Technologies (10003, Waltham, MA, USA). Centrifugal filter tubes were purchased from Millipore (Amicon® Ultra-15 centrifugal filter units, 10 K NMWL, Billerica, MA, USA). Chymotrypsin was purchased from Promega (V1062, Madison, WI, USA). The deglycosylation enzyme PNGase F was purchased from NEB (P0704, New England Biolabs, ON, Canada). DTT (1,4-dithiothreitol, 111474) was purchased from Merck (Darmstadt, Germany). The C18 nanospray column (75 µm × 20 cm, 2.5 µm particle size) was packed in-house. The C18 particles were purchased from Dr. Maisch HPLC GmbH (ReproSil-Pur Basic®, 2.5 µm, Ammerbuch-Entringen, Germany).
2.2. Clinical Samples
Urine samples were collected from the National Cheng Kung University Hospital (Tainan, Taiwan). In total, 61 BPH samples and 38 prostate cancer samples were tested in this study. Clinical information and urine specimens were collected with the approval of the Institutional Review Board (IRB) of Academia Sinica (Taipei, Taiwan) and the National Cheng Kung University Hospital. Clinical information such as serum PSA level and ages of patients is shown in Table 1. There are no significant differences of serum PSA level between the patients of BPH and prostate cancer in this study.
Table 1.
Patients information collected in this study.
| BPH | PCa | p value | |
|---|---|---|---|
| Samples | 61 | 38 | |
| Age | 48~84 | 56~82 | |
| Mean ± SD | 66.5 | 71.6 | |
| Median | 67 | 73 | |
| Serum PSA (ng/mL) | 0.73~78.0 | 6.19~27354 | 0.247# |
| Mean ± SD | 13.44 ± 14.42 | 857.93 ± 4430.24 | |
| Median | 9.39 | 26.67 | |
| Gleason score | 6~10 |
#No significant differences.
Urine (50–100 mL per patient) was collected then stored at −20°C before processing. Before being used in experiments, the urine sample was thawed at room temperature and then centrifuged (1500 g, 10 min) to remove cell debris and precipitates. The total protein in the supernatant of 50 mL of urine was concentrated using Amicon Ultra-15 centrifugal filter units (5000 g, 45 min/run at 4°C) and was then ready to be immunoprecipitated. Ten microliters of each concentrated urine sample was pooled to generate the BPH and PCa sample pools.
2.3. Urinary PSA Immunoprecipitation and In-Gel Digestion
The anti-PSA monoclonal antibody (10 µg) was dissolved in 200 µL PBST (phosphate buffered saline plus 0.05% Tween-20) and incubated with protein G magnetic beads (1.5 mg) at room temperature for 30 min. After three washes with PBST, the antibody-coupled beads were incubated with concentrated urine for 1 hour at room temperature to capture the PSA. The protein on the beads was denatured in 20 µL SDS sample buffer by boiling at 95°C for 5 min and then separated by SDS-PAGE (12% separating gel). The gel was then stained with Coomassie Brilliant Blue. Seminal PSA (2 µg) was captured using anti-PSA beads and then denatured by boiling in SDS sample buffer (20 µL) at 95°C for 5 min. The denatured protein was subjected to SDS-PAGE separation followed by in-gel digestion.
In-gel protein digestion was carried out as described previously [25]. Briefly, the stained protein band at 28–30 kDa was removed and cut into small pieces (roughly 1 mm2 squares). These pieces were then destained in 40% acetonitrile (ACN) for 10 min, reduced in 10 mM DTT at 56°C for 60 min, alkylated in 55 mM IAA at room temperature for 45 min in the dark, and then dehydrated in 100% ACN. Then, 100 ng of chymotrypsin dissolved in 40 µL of 50 mM ammonium bicarbonate was added to the dehydrated gel pieces and incubated at 37°C for 18 hours. The digested peptides were eluted twice from the gel pieces using 50 µL of 60% ACN/1% trifluoroacetic acid. The collected products were vacuum dried and then reconstituted in 20 µL of deionized water for MS analysis.
2.4. Mass Spectrometry
LC-MS was used to study the seminal and pooled urinary PSA samples. The peptides and N-glycopeptides generated from the chymotrypsin digestion were analyzed with an LTQ-Orbitrap XL Mass Spectrometer (Thermo Scientific, San Jose, CA, USA) equipped with a nanoelectrospray ion source, an Agilent 1100 Series binary high-performance liquid chromatography pump (Agilent Technologies, Palo Alto, CA, USA), and a FAMOS autosampler (LC Packing, San Francisco, CA, USA). A total of 5 µL of samples were injected into a manually packed precolumn (150 µm ID × 30 mm, 5 µm, 200 Å) at a 10 µL/min flow rate. Chromatographic separation was performed over 90 min on a manually packed reversed phase C18 nanocolumn (75 µm ID × 200 mm, 3 µm, 200 Å) using 0.1% formic acid in water as mobile phase A, 0.1% formic acid in 80% acetonitrile as mobile phase B, and a split flow rate of 300 nL/min. The full-scan mass rage was set from m/z 320 to 2000 with 60,000 resolution at m/z = 400. The top ten most intense ions were sequentially isolated for MS2 by LTQ. The electrospray voltage was maintained at 1.8 kV, and the capillary temperature was set to 200°C. The glycopeptides were detected based on the mass and were confirmed using the MS2 spectra of oxonium ions and of peptide ions with a core N-acetylhexosamine (HexNAc) (Y1 ions) [26, 27]. To confirm the peptide backbone sequences, Y1 ions were captured and fragmented with a Velos Pro to produce MS3 spectra. Full MS/MS scans (m/z 100–2000) of high energy C-trap dissociation (HCD) were pursued. The minimum required signal intensity was set at 10,000 count/sec, and the isolation width of the precursor ion was set at 10 Da. The default activation time was 2 ms, and the normalized collision energy (NCE) for fragmentation was 28%. MS3 fragmentation was achieved in CID mode with a minimum signal intensity of 100 counts/sec, the isolation width of the precursor ion was set to 20 Da, the NCE for fragmentation was set to 35%, and the activation time was set to 10 ms. Y1 ion peptide sequence was obtained using the Mascot search engine for mass spectra and was used to identify the glycopeptide backbone sequence.
To analyze the selected ions in the individual sample, a 7-Tesla LTQ-FT Ultra Mass Spectrometer (linear quadrupole ion trap Fourier transform ion cyclotron resonance, Thermo Scientific, San Jose, CA, USA) equipped with nanoelectrospray ionization was used, and the total LC running time was 30 min. The full-scan MS spectra (m/z 320–1600) were acquired using the FTICR with a mass resolution of 200,000 at m/z = 400. Ions of the specific glycopeptides and the internal reference peptide were trapped with ion accumulation time of 250 ms, the NCE for fragmentation was set at 35%, and the isolation width of the precursor ions was set to 3 Da.
For samples that were difficult to detect due to low quantities of PSA, the internal reference peptide ([M + 2H]2+ = 485.78) was selected and analyzed via the ion accumulation method using a Velos Pro with CID fragmentation and elution from a C18 nanoelectrospray column. The maximum ion accumulation time was set to 3000 ms, and the isolation width of the precursor ions was set to 2 Da.
2.5. Database Search of MS/MS Spectra
The MS/MS data were processed and searched using the Mascot search engine (Matrix Science, Boston, MA, USA). The peak-list files were obtained from the MS/MS data using Extract_msm 5.0 software (Thermo Scientific) and they included the mass values, the intensity, and the charge of the precursor ions (parent ions with +2 or +3 charges in this study). The search parameters used in this study were IPI_Human v. 3.74 as the database, chymotrypsin as the enzyme, up to 2 missed cleavages, a peptide ion mass matching tolerance of 10 ppm, and a fragment ion mass tolerance of 0.8 Da. Oxidation (M) and N-terminal protein acetylation were set as variable modifications, and carbamidomethyl (C) was set as a fixed modification.
2.6. Identification of PSA Glycopeptides
Glycopeptide identification was carried out using software developed in-house. Putative glycopeptides were listed as the molecular weight of the deconvoluted LTQ-Orbitrap XL mass spectra and were matched against an established glycopeptide database. The listed glycopeptides were confirmed by MS2 of the oxonium ions and Y1 ions.
2.7. Quantitation of PSA Glycosylation
Each PSA glycopeptide and the internal reference peptide were quantified with a label-free method that used the peak area of MS1 under the LC curve, and the peak area was calculated using Xcalibur Software (Thermo Scientific). The selected PSA glycopeptides were eluted with a retention time of ±1.5 min within a 30 min LC run. The MS1 peaks of each glycopeptide and of the reference peptide were obtained with error tolerance settings of 500 ppm and 10 ppm, respectively. All selected glycopeptides were validated via MS2 spectra. The normalized abundance of each PSA glycoform was calculated according to the following equation:
| (1) |
2.8. Statistical Analysis
To evaluate the differences in normalized glycopeptide abundance in the clinical samples, two-tailed t-tests were used. Receiver operating characteristic (ROC) analysis and the area under the ROC curve (AUC) were used to evaluate sensitivity, specificity, and performance under optimal conditions. The graph was created by plotting the sensitivity and the false positive rate at various thresholds. AUC is a tool that has been used to describe the discriminatory ability of a test under various thresholds. Student's t-tests, ROC, and AUC were calculated using GraphPad Prism 5 software. The cut-off value was determined by calculating the Youden Index (J), which defines the maximum potential effectiveness of a biomarker. The Youden Index was calculated as the maximum value of “Sensitivity + Specificity − 1.”
3. Results
We developed a method for the quantitation of urinary PSA and its glycoforms based on relative abundance. Purified seminal PSA was used as a standard for studying the correlation of PSA quantity and normalized glycoforms abundance. The method was then applied to pool BPH and PCa urine samples from each individual. Because some glycopeptides were low abundance in some samples, specific molecules were ion accumulated for MS2 to increase detection sensitivity. Using this strategy, the abundance of PSA and the degree of PSA glycosylation was monitored simultaneously.
3.1. Quantitation of PSA Protein and Glycopeptides
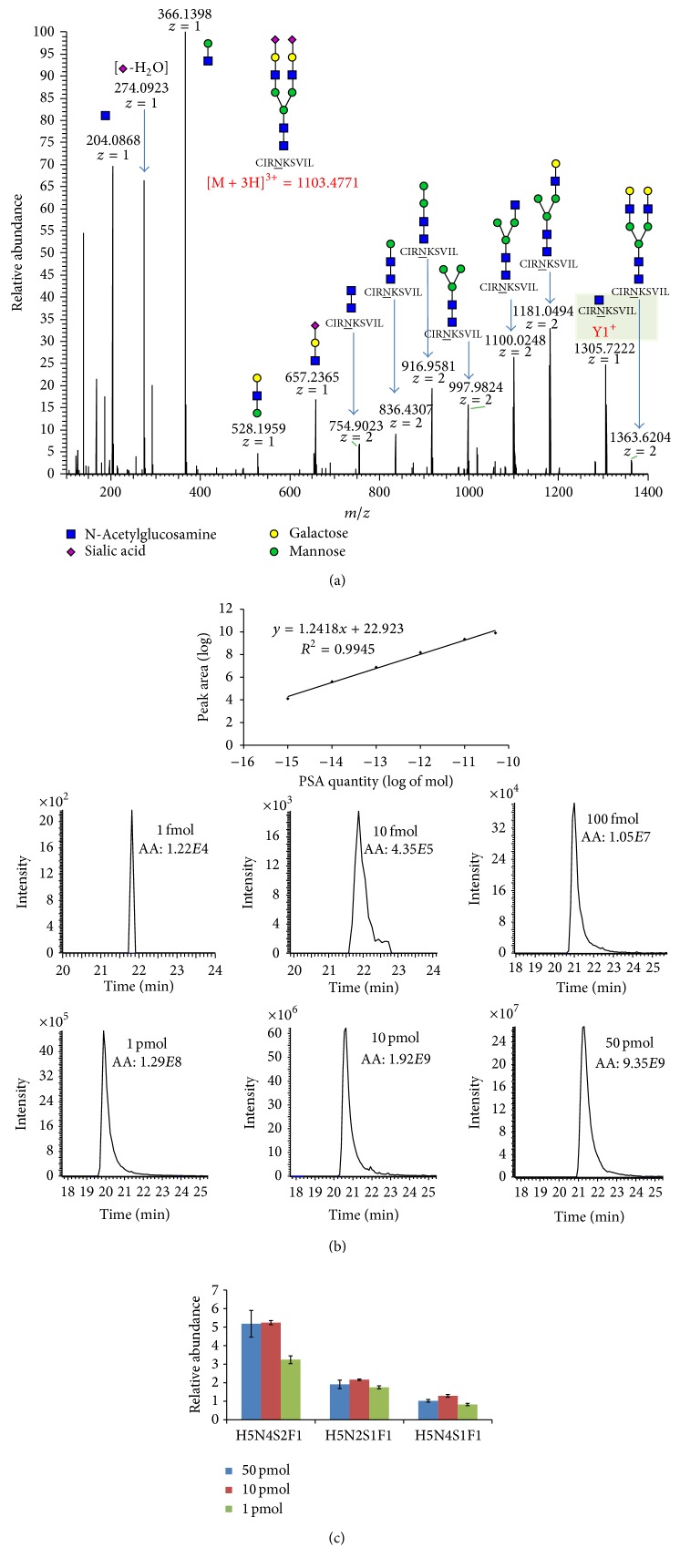
Human seminal PSA was used as a standard to establish the method. The peptides generated by chymotrypsin digestion were analyzed with an LC-LTQ-Orbitrap XL and were searched using Mascot. Glycopeptides were detected based on molecular weight (Table 1) and validated by MS2 fragments with oxonium ions and glycosidic peptide ions (Figure 1(a)). Peptide backbone sequence was validated by deglycosylation and MS3 of Y1 ion.
Figure 1.
Identification and quantitation of PSA glycopeptide via tandem MS. (a) The MS2 of the CIRNKSVIL-Hex5HexNAc4NeuAc2 glycopeptide was identified. Oxonium ions, glycopeptide fragment ions with oligosaccharide fragments, and Y1 ion were all identified. The monosaccharide symbols are N-acetylglucosamine (blue square); mannose (green circle); galactose (yellow circle); and sialic acid (purpule diamond). (b) The dynamic range of the selected PSA internal reference peptide. Each spot is the average of triplicate. The inlet is MS1 peak of the reference peptide detected in varied PSA quantity. Peak intensity and the area under the peak (AA) are shown. (c) The normalized abundance of specific PSA glycopeptides in varying protein quantities. The error bars represent the standard deviations of triplicate results. The glycoform abbreviations are as follows: hexose (H), N-acetyl hexose (N), N-acetylneuraminic acid (sialic acid, S), and fucose (deoxyhexose, F).
To evaluate the relative abundance of the different PSA glycopeptides, an internal reference peptide was selected for normalization based on several criteria. First, the peptide needed to be unique to the PSA protein. The selected peptide had to be consistently detectable, and its abundance should be proportional to the PSA level. The dynamic range of the peptide needed to be large enough to evaluate the protein level. Of the chymotrypsin-digested PSA peptides, a peptide that fits the above criteria, IKDTIVANP ([M + 2H]2+ = 485.7823), was identified. This peptide is located at the C-terminus of PSA and has a sequence that is unique to PSA; its sequence does not overlap with those of other peptides. The peptide's signal was also consistently higher than that of other PSA peptides. The abundance of the peptide (the peak MS1 area under LC curve) was proportional to the quantity of PSA protein (Figure 1(b)). The dynamic range was 1 fmol to 10 pmol (corresponding to 0.3 pg and 300 ng of PSA protein). This peptide was selected as the internal reference peptide for the normalization of glycopeptide abundance.
The signal of the glycopeptides obtained from mass spectrometry is generally lower than that of the peptides without linked glycans [12, 28] which is consistent with our observations. The detection limit for the abundant glycopeptides was approximately 100 fmol. When the PSA protein quantity was lower than 100 fmol, each specific glycopeptide was difficult to detect. An analysis of the abundance of specific glycopeptides relative to the internal reference peptide showed that their abundance was proportional to the overall quantity of PSA (Figure 1(c)). Therefore, specific PSA glycoforms can be evaluated using their normalized abundance.
A total of 23 glycoforms were identified from the seminal PSA (Table 2), most of which were biantennary glycans. No tri- or tetra-antennary glycans were identified. High-mannose glycans were found with low abundance. The most abundant glycoform was Hex5HexNAc4NeuAc2dHex1 (H5N4S2F1), which made up more than 25% of all identified glycoforms. These observed seminal PSA glycoforms were consistent with previous studies [9, 10, 29].
Table 2.
Identified human seminal PSA glycopeptides.
| [M + 3H]3+ (a) | Glycan composition(b) | Abundance | |
|---|---|---|---|
| Peak area | Normalized(c) | ||
| 665.6575 | H3N2 | 3.06E + 06 | 0.51 |
| 720.0094 | H4N2 | 9.44E + 06 | 1.57 |
| 773.6926 | H5N2 | 2.20E + 07 | 3.67 |
| 836.0542 | H4N3F1 | 5.91E + 06 | 0.98 |
| 884.4000 | H4N3S1 | 1.10E + 07 | 1.84 |
| 895.4034 | H6N3 | 5.01E + 06 | 0.83 |
| 898.0755 | H3N4S1 | 4.10E + 06 | 0.68 |
| 933.0860 | H4N3S1F1 | 2.26E + 07 | 3.77 |
| 946.7615 | H3N4S1F1 | 5.84E + 06 | 0.97 |
| 952.0931 | H4N4S1 | 5.41E + 06 | 0.90 |
| 957.7649 | H5N4F1 | 5.80E + 06 | 0.96 |
| 992.4352 | H6N3S1 | 4.24E + 07 | 7.06 |
| 1000.7791 | H4N4S1F1 | 1.10E + 07 | 1.83 |
| 1006.1107 | H5N4S1 | 1.12E + 07 | 1.87 |
| 1014.4546 | H3N5S1F1 | 7.59E + 06 | 1.26 |
| 1019.7863 | H4N5S1 | 4.20E + 06 | 0.70 |
| 1054.7967 | H5N4S1F1 | 1.92E + 07 | 3.20 |
| 1068.4722 | H4N5S1F1 | 7.81E + 06 | 1.30 |
| 1103.1425 | H5N4S2 | 2.80E + 07 | 4.65 |
| 1116.8181 | H4N5S2 | 1.20E + 07 | 1.99 |
| 1151.8285 | H5N4S2F1 | 6.94E + 07 | 11.54 |
| 1157.1601 | H6N4S2 | 9.35E + 06 | 1.56 |
| 1165.5040 | H4N5S2F1 | 2.01E + 07 | 3.34 |
(a) Monoisotopic masses are given throughout.
(b) The abbreviations of glycans are shown: H: hexose (Hex); N: N-acetylhexosamine (HexNAc); S: sialic acid (NeuAc); F: fucose (dHex)
(c) Normalized means the normalized abundance, which is the level of PSA glycosylation.
3.2. Detection of Urinary PSA Glycopeptides in Pooled Clinical Samples
Urinary PSA glycopeptides from pooled BPH and PCa samples were analyzed with an LTQ-Orbitrap XL, which provided a high-accuracy m/z. The glycopeptides and the internal reference peptide were eluted at different times using LC with a run time of 90 minutes. Based on their molecular weights and elution times, eleven glycoforms were identified from both sample types. Eight PSA glycoforms were confirmed with MS2 spectra (Table 3). High-mannose and complex glycoforms were detected, and H5N4S2F1 was the most abundant PSA glycoform. The normalized abundance of each glycoform varied between the pooled BPH and PCa samples, and Hex4HexNAc2 (H4N2) was only detected in the BPH sample, not the PCa sample. However, the differences in the pooled samples cannot reflect the diversity of the individual samples. Therefore, the glycoforms were also investigated in each individual sample.
Table 3.
The urinary PSA glycoforms were identified in pooled samples.
| Glycan composition | Normalized abundance∗ | |
|---|---|---|
| BPH | PCa | |
| H4N2 | 0.52 | ND∗∗ |
| H5N2 | 0.42 | 0.59 |
| H6N3 | 0.18 | 0.08 |
| H6N3S1 | 0.80 | 1.60 |
| H5N4S1F1 | 1.02 | 0.77 |
| H4N5S1F1 | 0.23 | 0.16 |
| H5N4S2 | 0.23 | 0.43 |
| H5N4S2F1 | 1.45 | 2.51 |
∗The results were the average of two measurements.
∗∗ND: not detected.
3.3. Glycoform Validation in Individual Clinical Samples
To investigate glycoforms in each clinical sample, each specific glycopeptide and the reference peptide were selected for MS analysis (Table 4). We focused on these selected ions and screened their distribution in all clinical samples using an LTQ-FT Ultra MS. In total, 61 BPH and 38 prostate cancer samples were analyzed for the six selected glycopeptides and the internal reference peptide using LC with a run time of 30 min.
Table 4.
The normalized abundance of each selected and grouped glycoform detected in each individual sample.
| Selected glycoforms | Grouped glycoforms | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| H4N2 | H5N4S2F1 | H5N4S2 | H5N4S1F1 | H6N3S1 | H5N2 | Unsialylated | Monosialylated | Disialylated | Sialylated | Fucosylated | Unfucosylated | Total | |
| RT (min) | 11.5–13.3 | 13.0–15.5 | 13.5–15.2 | 12.8–13.8 | 12.0–13.0 | 11.1–11.6 | H4N2 + H5N2 | H5N3S1F1 + H5N3S1 | H5N4S2F1 + H5N4S2 | Monosialylated + Disialylated | H5N4S2F1 + H5N4S1F1 | H4N2 + H5N4S2 + H6N3S1 + H5N2 | |
| [M + 3H]3+ | 720.01 | 1152.16 | 1103.47 | 1055.13 | 992.77 | 773.69 | |||||||
| BPH003 | 4.27 | 1.62 | 2.29 | 3.91 | 4.27 | 8.18 | 5.89 | 2.29 | 8.18 | ||||
| BPH004 | 0.70 | 2.53 | 3.58 | 6.12 | 0.70 | 6.81 | 3.23 | 3.58 | 6.81 | ||||
| BPH005 | 0.13 | 0.60 | 0.38 | 0.50 | 0.88 | 0.73 | 1.61 | 0.51 | 1.10 | 1.61 | |||
| BPH006 | 0.71 | 0.21 | 0.94 | 5.95 | 6.89 | 0.93 | 7.81 | 1.65 | 6.16 | 7.81 | |||
| BPH009 | 0.21 | 0.12 | 0.34 | 0.01 | 0.35 | 0.33 | 0.68 | 0.55 | 0.13 | 0.68 | |||
| BPH012 | 0.14 | 0.63 | 0.18 | 0.89 | 0.74 | 0.35 | 0.49 | 1.63 | 0.82 | 2.45 | 1.53 | 1.41 | 2.94 |
| BPH015 | 0.07 | 0.62 | 0.17 | 0.50 | 0.31 | 0.05 | 0.12 | 0.81 | 0.79 | 1.59 | 1.12 | 0.59 | 1.71 |
| BPH016 | 0.38 | 0.02 | 0.61 | 0.17 | 0.78 | 0.40 | 1.18 | 0.99 | 0.19 | 1.18 | |||
| BPH017 | 0.59 | 0.32 | 0.25 | 1.29 | 1.15 | 0.62 | 1.21 | 2.44 | 0.57 | 3.01 | 1.61 | 2.61 | 4.22 |
| BPH019 | 0.68 | 0.28 | 0.25 | 0.10 | 0.81 | 0.68 | 0.91 | 0.53 | 1.44 | 0.38 | 1.74 | 2.12 | |
| BPH020 | 0.23 | 0.55 | 0.78 | 0.78 | 0.23 | 0.55 | 0.78 | ||||||
| BPH021 | 0.95 | 0.31 | 1.12 | 0.30 | 0.29 | 1.24 | 1.42 | 0.31 | 1.73 | 1.43 | 1.54 | 2.97 | |
| BPH022 | 0.16 | 0.65 | 0.42 | 0.51 | 0.45 | 0.15 | 0.32 | 0.96 | 1.07 | 2.03 | 1.16 | 1.19 | 2.35 |
| BPH025 | 0.34 | 0.69 | 0.60 | 1.03 | 0.56 | 0.34 | 1.59 | 1.29 | 2.89 | 1.72 | 1.50 | 3.23 | |
| BPH029 | 0.53 | 0.30 | 0.07 | 0.48 | 0.68 | 0.52 | 1.05 | 1.15 | 0.38 | 1.53 | 0.78 | 1.79 | 2.58 |
| BPH030 | 0.50 | 0.32 | 0.11 | 0.43 | 0.50 | 0.93 | 0.81 | 0.11 | 0.93 | ||||
| BPH031 | 0.34 | 0.24 | 0.12 | 0.65 | 0.44 | 0.58 | 0.91 | 1.10 | 0.36 | 1.46 | 0.89 | 1.48 | 2.37 |
| BPH032 | 0.13 | 0.40 | 0.24 | 0.49 | 0.30 | 0.12 | 0.25 | 0.80 | 0.64 | 1.43 | 0.89 | 0.79 | 1.68 |
| BPH033 | 0.04 | 0.06 | 0.04 | 0.07 | 0.07 | 0.11 | 0.04 | 0.14 | 0.10 | 0.11 | 0.21 | ||
| BPH034 | 0.09 | 0.55 | 0.36 | 0.50 | 0.65 | 0.10 | 0.18 | 1.15 | 0.91 | 2.06 | 1.05 | 1.19 | 2.25 |
| BPH035 | 0.24 | 0.32 | 0.11 | 0.49 | 0.37 | 0.28 | 0.52 | 0.86 | 0.43 | 1.30 | 0.81 | 1.00 | 1.82 |
| BPH036 | 0.06 | 0.17 | 0.25 | 0.25 | 0.17 | 0.06 | 0.23 | 0.23 | 0.25 | 0.48 | |||
| BPH037 | 0.35 | 0.33 | 0.08 | 0.34 | 0.37 | 0.29 | 0.63 | 0.71 | 0.41 | 1.12 | 0.66 | 1.09 | 1.75 |
| BPH038 | 0.75 | 1.16 | 0.89 | 0.44 | 1.30 | 0.75 | 1.74 | 2.05 | 3.80 | 1.60 | 2.94 | 4.55 | |
| BPH039 | 0.13 | 1.37 | 0.02 | 0.32 | 0.32 | 1.39 | 0.13 | 1.52 | 1.50 | 0.34 | 1.84 | ||
| BPH040 | 0.78 | 0.24 | 0.06 | 0.14 | 1.04 | 0.73 | 1.51 | 1.18 | 0.30 | 1.48 | 0.38 | 2.61 | 2.98 |
| BPH042 | 0.52 | 0.14 | 0.66 | 0.66 | 0.52 | 0.14 | 0.66 | ||||||
| BPH044 | 0.24 | 0.53 | 0.25 | 0.49 | 0.68 | 0.34 | 0.58 | 1.16 | 0.78 | 1.95 | 1.02 | 1.51 | 2.53 |
| BPH045 | 0.79 | 6.22 | 2.33 | 2.33 | 7.01 | 9.34 | 0.79 | 8.55 | 9.34 | ||||
| BPH046 | 0.15 | 0.13 | 0.17 | 0.17 | 0.29 | 0.29 | 0.15 | 0.30 | 0.45 | ||||
| BPH047 | 0.10 | 0.41 | 0.35 | 0.49 | 0.58 | 0.10 | 0.21 | 1.07 | 0.76 | 1.83 | 0.90 | 1.13 | 2.03 |
| BPH048 | 0.29 | 0.49 | 0.14 | 0.51 | 0.49 | 0.23 | 0.52 | 1.00 | 0.64 | 1.64 | 1.00 | 1.16 | 2.16 |
| BPH049 | 0.78 | 0.16 | 0.58 | 0.15 | 0.73 | 0.94 | 1.67 | 1.36 | 0.31 | 1.67 | |||
| BPH050 | 0.80 | 0.80 | 0.80 | 0.80 | 0.80 | ||||||||
| BPH051 | 1.91 | 0.96 | 2.09 | 3.06 | 1.91 | 4.96 | 2.87 | 2.09 | 4.96 | ||||
| BPH054 | 0.95 | 0.07 | 0.06 | 0.65 | 0.88 | 1.17 | 2.11 | 1.53 | 0.13 | 1.66 | 0.73 | 3.05 | 3.77 |
| BPH055 | 0.17 | 0.16 | 0.29 | 0.20 | 0.49 | 0.32 | 0.81 | 0.45 | 0.36 | 0.81 | |||
| BPH057 | 0.24 | 0.92 | 0.09 | 0.21 | 0.27 | 0.64 | 0.88 | 0.48 | 1.00 | 1.48 | 1.13 | 1.23 | 2.36 |
| BPH058 | 1.31 | 0.65 | 0.07 | 0.09 | 1.23 | 1.18 | 2.49 | 1.32 | 0.72 | 2.04 | 0.74 | 3.79 | 4.53 |
| BPH060 | 0.10 | 0.59 | 0.19 | 0.39 | 0.29 | 0.06 | 0.16 | 0.67 | 0.78 | 1.45 | 0.97 | 0.64 | 1.61 |
| BPH061 | 0.12 | 0.15 | 0.33 | 0.48 | 0.14 | 0.14 | 0.81 | 0.27 | 1.08 | 0.44 | 0.77 | 1.22 | |
| BPH063 | 0.57 | 2.54 | 2.85 | 5.39 | 0.57 | 5.96 | 3.11 | 2.85 | 5.96 | ||||
| BPH068 | 0.44 | 0.11 | 0.61 | 0.17 | 0.08 | 0.08 | 0.78 | 0.54 | 1.32 | 1.05 | 0.35 | 1.40 | |
| PCa131 | 1.57 | 1.57 | 1.57 | 1.57 | 1.57 | ||||||||
| PCa133 | 0.14 | 0.06 | 0.36 | 0.13 | 0.49 | 0.20 | 0.69 | 0.50 | 0.19 | 0.69 | |||
| PCa135 | 0.85 | 0.09 | 0.16 | 0.24 | 0.34 | 0.85 | 0.58 | 0.25 | 0.83 | 0.33 | 1.35 | 1.68 | |
| PCa137 | 0.40 | 1.04 | 1.04 | 0.40 | 1.44 | 0.40 | 1.04 | 1.44 | |||||
| PCa139 | 0.26 | 0.12 | 0.27 | 0.19 | 0.46 | 0.38 | 0.84 | 0.53 | 0.31 | 0.84 | |||
| PCa143 | 0.62 | 0.62 | 0.62 | 0.62 | 0.62 | ||||||||
| PCa145 | 0.49 | 0.33 | 0.49 | 0.30 | 0.79 | 0.82 | 1.61 | 0.98 | 0.63 | 1.61 | |||
| PCa152 | 0.65 | 0.09 | 0.46 | 0.59 | 1.05 | 0.74 | 1.79 | 1.10 | 0.68 | 1.79 | |||
| PCa157 | 0.13 | 0.07 | 0.37 | 0.17 | 0.55 | 0.20 | 0.75 | 0.51 | 0.24 | 0.75 | |||
| PCa161 | 0.25 | 0.28 | 0.11 | 0.29 | 0.55 | 0.34 | 0.59 | 0.83 | 0.39 | 1.22 | 0.57 | 1.24 | 1.81 |
| PCa162 | 0.17 | 0.42 | 0.11 | 0.28 | 0.24 | 0.19 | 0.36 | 0.52 | 0.52 | 1.04 | 0.69 | 0.71 | 1.40 |
| PCa165 | 0.47 | 0.42 | 0.42 | 0.47 | 0.47 | 0.89 | 0.89 | ||||||
| PCa166 | 0.98 | 0.14 | 0.41 | 0.28 | 1.26 | 0.41 | 0.14 | 0.55 | 0.14 | 1.68 | 1.82 | ||
| PCa171 | 0.28 | 0.26 | 0.02 | 0.34 | 0.26 | 0.41 | 0.69 | 0.60 | 0.28 | 0.87 | 0.60 | 0.97 | 1.57 |
| PCa003 | 0.22 | 0.27 | 0.71 | 0.97 | 0.22 | 1.20 | 0.49 | 0.71 | 1.20 | ||||
| PCa012 | 1.99 | 0.09 | 0.38 | 0.47 | 1.99 | 2.45 | 2.07 | 0.38 | 2.45 | ||||
| PCa013 | 1.01 | 0.09 | 0.21 | 0.79 | 1.00 | 1.11 | 2.11 | 1.22 | 0.89 | 2.11 | |||
| PCa017 | 0.22 | 0.44 | 0.06 | 0.44 | 0.28 | 0.21 | 0.43 | 0.71 | 0.49 | 1.21 | 0.87 | 0.76 | 1.63 |
| PCa019 | 0.51 | 0.21 | 1.01 | 1.21 | 0.51 | 1.72 | 0.71 | 1.01 | 1.72 | ||||
| PCa025 | 0.11 | 0.18 | 0.64 | 0.82 | 0.11 | 0.93 | 0.29 | 0.64 | 0.93 | ||||
| p-value | 0.478 | 0.137 | 0.137 | <0.001∗∗∗ | 0.043∗ | 0.637 | 0.513 | 0.004∗∗ | 0.122 | 0.006∗∗ | 0.054 | 0.003∗∗ | <0.001∗∗∗ |
(1) The retention time (RT) of the selected reference peptide (KIDTIVANP, [M + 2H]2+ = 485.78) was 16–18 min in 30 min LC running time.
(2) Grouped glycoforms are a combination of the level of indicated glycopeptides in each individual sample. “Total” includes all six selected glycopeptides.
(3) Each sample was analyzed and duplicated and the average is shown.
(4) ∗ indicates p < 0.05, ∗∗ indicates p < 0.01, and ∗∗∗ indicates p < 0.001.
The PSA reference peptide could be detected in 59 BPH and 34 PCa samples. Among these samples, at least one selected glycopeptide in 43 BPH and 20 PCa samples was detected (Table 4). For samples that contained at least 100 fmol of PSA one or more of the selected glycopeptides were expected to be detected. No significant differences of serum PSA level between BPH and prostate cancer. Reference peptide but not selected glycopeptides was detected in 14 BPH and 10 PCa samples. Immunoprecipitated urinary PSA was extremely low and difficult to be observed on the gel. However, PSA in these samples was validated by western blot. The reason of glycopeptides undetectable was due to sample variations instead of experimental problems. According to the results, none of the selected glycopeptides were BPH- or PCa-specific in any sample. Among the 6 glycoforms, H5N4S1F1 was the most commonly observed in BPH samples (42/59, 71%), and H6N3S1 was the second most common (18/33, 69%). In the PCa samples, H5N4S2F1 and H6N3S1 were the major isoforms observed (55%). The relative abundance of each glycoform in each sample was estimated (Table 4). Sialylation and fucosylation of PSA glycans were correlated to prostate cancer [19]. These glycan categories were evaluated as grouped glycoforms to sum the level of specific selected glycopeptides. Unsialylated, monosialylated, desialyalted, sialylated, fucosylated, unfucosylated, and all selected glycoforms combined (total) in each individual sample were evaluated, respectively. Among the six selected glycoforms, the normalized relative abundance of H5N4S1F1 and H6N3S1 showed significant differences between BPH and PCa. For grouped glycoforms, monosialylated, unfucosylated, and total showed different expression levels (p < 0.05) (Table 4). However, there are no significant differences between BPH and prostate cancer samples in serum PSA level.
The ROC curve and the AUC of the H5N4S1F1 glycoforms showed significant differences and moderate discrimination power (Table 5). Below the cut-off value, the prostate cancer detection sensitivity is 93%, and the specificity is 59%. The sensitivity is defined as the number of true positives divided by the number of all verified positives. The specificity is defined as the number of true negatives divided by the number of all verified negatives. The AUC of the grouped glycoforms was analyzed based on the samples which was able to detect the grouped glycoforms. For example, grouped monosialylated glycoform was detected in 43 BPH and 19 PCa. The AUC of grouped monosialylated glycoform was analyzed by these 62 samples. By the same way, the AUC of grouped total was analyzed by the results of 43 BPH and 20 PCa. Because the sample number varied between different grouped glycoforms, different groups were considered, respectively. The sialylated, monosialylated, and unfucosylated glycoforms showed lower discrimination power (AUC < 0.7) than that of all groups combined (0.7 < AUC < 0.8) (Table 5). The monosialylated group showed high sensitivity (100%) and weak specificity (47.5%) for prostate cancer detection. The prostate cancer detection sensitivity of all glycoforms combined (total) was 87.5% and the specificity was 60%. Both H5N4S1F1 and the “total” group differed significantly between BPH and PCa.
Table 5.
Areas under the ROC curves of the target PSA glycopeptides.
| Glycan composition | AUC∗ | p value | Cancer threshold | Sensitivity | Specificity |
|---|---|---|---|---|---|
| H5N4S1F1 | 0.7381 ± 0.0661 | 0.009 | <0.470 | 92.9% | 59.0% |
| Monosialylated | 0.7102 ± 0.0681 | 0.015 | <1.060 | 100% | 47.5% |
| Sialylated | 0.6938 ± 0.0763 | 0.025 | <1.260 | 68.8% | 75% |
| Unfucosylated | 0.6758 ± 0.0699 | 0.041 | <1.065 | 87.5% | 60% |
| Total | 0.7242 ± 0.0662 | 0.009 | <1.815 | 87.5% | 60% |
∗AUC was calculated based on the samples which could be detected for the indicated glycoform or grouped glycoforms.
3.4. Further Exploration of Samples without Enough PSA for Routine Analysis
The internal reference peptide for PSA could not be detected in six samples with a routine analysis. To confirm the presence of PSA, the specific peptide was detected with the ion accumulation method using a Velos Pro, which traps a specific m/z and consequently increases sensitivity. The maximum ion accumulation time can be extended from 250 ms to 3000 ms. In principle, the sensitivity could be improved more than 10-fold. Therefore, we focused on analyzing the PSA internal reference peptide using the Velos Pro.
PSA was not able to be detected in 2 BPH samples (BPH066 and BPH069) and 4 prostate cancer samples (PCa010, PCa018, PCa029, and PCa030) by ELISA. However, the internal reference peptide was detected in all 6 samples via validation of its characteristic fragment ions (Table 6). Using the ion accumulation method, the detection limit of the protein peptide fragments was less than 100 amol, which is far lower than the ELISA detection limit of 1 ng/mL.
Table 6.
Velos Pro validation of samples with low PSA levels.
| Sample | ELISA (ng/mL) | Characteristic fragment ions of PSA internal reference peptide (m/z) | |||
|---|---|---|---|---|---|
| 856.48 | 741.45 | 570.41 | 571.35 | ||
| BPH066 | <1 | + | + | + | + |
| BPH069 | <1 | + | + | + | + |
| PCa010 | <1 | + | + | + | + |
| PCa018 | <1 | + | + | + | + |
| PCa029 | <1 | + | + | + | + |
| PCa030 | <1 | + | + | + | ND∗ |
∗ND: not detected.
4. Discussion
An interlaboratory study of human seminal PSA glycosylation was published in 2013 [9], and two of the participating laboratories showed detailed results using bottom-up and top-down approaches [10, 29]. In these studies, major and intermediate glycoforms were detected, and these glycoforms represented more than 80% of the total glycoform content. These abundant glycoforms were also identified in our report on seminal PSA. However, their presence was not validated in clinical serum or urinary PSA samples. In addition, the abundance of each glycoform was estimated based on the relative intensity of each glycan compared with all discovered glycoforms. Depending on the number of discovered PSA glycans, this value varies between experiments. In this work, we not only identify and evaluate these abundant glycoforms in seminal PSA but also analyze each glycoform in clinical urine samples. Highly purified PSA is not necessary for the assay. Furthermore, the normalized abundance of each glycopeptide can provide a standard method to evaluate the abundance of each glycopeptide across different samples. This strategy can also be applied to the analysis of glycoform abundance for other glycoproteins.
The reference peptide but not the selected glycopeptides in some clinical samples was detected. It was due to biological variance or the limitation of sample collection. First, the glycopeptides we screened here were too low to be detected. Some other glycoforms may exist but too low to be detected. To extend the screen list of glycoforms may be helpful. Second, urinary PSA levels in these samples were extremely low. Glycopeptides were difficult to detect in the samples with PSA protein lower than 100 fmole. Collecting higher amounts of urinary PSA, such as first void urine in the morning, would improve the results. As long as the selected glycoforms could be detected, the possibility of prostate cancer could be evaluated.
It is easier to collect large volumes of urine than collect large volumes of serum. However, the quantity of PSA in urine samples varies widely depending on the collection time [30, 31]. The first 5–10 mL of urine in the first morning void contains more PSA [30], whereas randomly collected urine samples and samples collected from midstream contain less PSA. Although more urinary PSA can be collected after prostate massage, this process is inconvenient and uncomfortable for patients. The urine specimens used in this study were collected randomly, potentially leading to low PSA levels. Therefore, the absolute quantity of each glycopeptide in the sample may not provide truly accurate information for disease diagnosis. However, the glycopeptide abundance relative to that of the internal reference peptide can reduce the effects of sample collection and sample processing [12]. In this study, we could evaluate each PSA glycopeptide in samples containing as low as 100 fmol of PSA. To the best of our knowledge, this level of sensitivity has not been reported before.
Although the detection of PSA glycoforms has been published in the past, no reports have measured specific glycoforms in individual samples. Investigating the glycoform distribution in each specimen is extremely difficult. Some glycoforms may be detected in certain samples, but the same glycoforms might not be detected in a pooled sample due to their dilution. For example, glycoform H4N2 was not detectable in the pooled PCa sample but could be detected in six PCa samples. This observation indicates that some low-abundance glycopeptides cannot be efficiently detected by pooling samples. Therefore, the method used here is an improvement because it can screen for the distribution of multiple glycoforms in clinical specimens.
According to the report from the American Cancer Society, the use of serum PSA as a prostate cancer biomarker has a sensitivity and specificity of 63% and 35%, respectively [4]. This low specificity could lead to an incorrect diagnosis and unnecessary treatment for certain patients [5]. In 65% to 75% of patients with elevated PSA, biopsies showed no cancer [32–34]. In these cases, the elevated serum PSA may instead be due to bacterial prostatitis and acute urinary retention [35]. There is a need to develop a highly specific diagnostic approach for prostate cancer. Ideally, the use of PSA testing as a marker would be individually tailored [36]. However, current reports on PSA glycoform analysis are not suitable for this purpose. In this study, multiple glycoforms were simultaneously screened in each sample. In addition, no significant differences of serum PSA level between BPH and PCa patients are analyzed in this study. However, these patients could be differentiated by evaluating the level of PSA glycoforms (H4N5S1F1) or groups of glycoforms with good specificity (60%). This greater specificity may improve prostate cancer diagnosis and reduce unnecessary biopsies. Because no specific glycoform could be detected in every sample, it is reasonable to combine several glycoforms to evaluate the glycan distribution among the samples. Although a large-scale study is needed to reach a definite conclusion, the urinary PSA glycoforms reported here could be a potential choice for prostate cancer diagnosis.
The ion accumulation approach can provide a more sensitive method to detect low levels of PSA (<1 fmol). A similar approach could be applied to study other rare species of known molecular weight in a sample. This approach should be valuable in glycomic and glycoproteomic analyses to find low-abundance biomarkers.
5. Conclusion
We analyzed the urinary PSA glycoforms in individual clinical samples and found significant differences in individual glycoforms and in several groups of glycoforms. Label-free quantitation relative to an internal reference peptide simplifies the search for potential markers. This is the first report to screen for specific PSA glycoforms in each individual clinical sample. The method could be used for large-scale studies that investigate glycoform markers. Compared with other specimens, urine provides a noninvasive choice for prostate cancer diagnosis. In addition, a highly sensitive ion accumulation approach using specific ions can be used to detect low-abundance glycoproteins in clinical samples.
Acknowledgments
This work was supported by the Ministry of Science and Technology (MOST) (MOST 102-2113-M-001-002-MY5), the National Health Research Center (NHRI-EX103-10301EI), and the Genomics Research Center, Academia Sinica.
Abbreviations
- PSA:
Prostate-specific antigen
- PCa:
Prostate cancer
- BPH:
Benign prostate hyperplasia.
Conflict of Interests
The authors declare that they have no conflict of interests.
References
- 1.Zhang H., Li X.-J., Martin D. B., Aebersold R. Identification and quantification of N-linked glycoproteins using hydrazide chemistry, stable isotope labeling and mass spectrometry. Nature Biotechnology. 2003;21(6):660–666. doi: 10.1038/nbt827. [DOI] [PubMed] [Google Scholar]
- 2.Gittes R. F. Prostate-specific antigen. The New England Journal of Medicine. 1987;317(15):954–955. doi: 10.1056/nejm198710083171508. [DOI] [PubMed] [Google Scholar]
- 3.Stamey T. A., Yang N., Hay A. R., McNeal J. E., Freiha F. S., Redwine E. Prostate-specific antigen as a serum marker for adenocarcinoma of the prostate. The New England Journal of Medicine. 1987;317(15):909–916. doi: 10.1056/nejm198710083171501. [DOI] [PubMed] [Google Scholar]
- 4.Wolf A. M. D., Wender R. C., Etzioni R. B., et al. American Cancer Society guideline for the early detection of prostate cancer: update 2010. CA: A Cancer Journal for Clinicians. 2010;60(2):70–98. doi: 10.3322/caac.20066. [DOI] [PubMed] [Google Scholar]
- 5.Wilt T. J., Scardino P. T., Carlsson S. V., Basch E. Prostate-specific antigen screening in prostate cancer: perspectives on the evidence. Journal of the National Cancer Institute. 2014;106(3) doi: 10.1093/jnci/dju010.dju010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.White K. Y., Rodemich L., Nyalwidhe J. O., et al. Glycomic characterization of prostate-specific antigen and prostatic acid phosphatase in prostate cancer and benign disease seminal plasma fluids. Journal of Proteome Research. 2009;8(2):620–630. doi: 10.1021/pr8007545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tajiri M., Ohyama C., Wada Y. Oligosaccharide profiles of the prostate specific antigen in free and complexed forms from the prostate cancer patient serum and in seminal plasma: a glycopeptide approach. Glycobiology. 2008;18(1):2–8. doi: 10.1093/glycob/cwm117. [DOI] [PubMed] [Google Scholar]
- 8.Li Y., Tian Y., Rezai T., et al. Simultaneous analysis of glycosylated and sialylated prostate-specific antigen revealing differential distribution of glycosylated prostate-specific antigen isoforms in prostate cancer tissues. Analytical Chemistry. 2011;83(1):240–245. doi: 10.1021/ac102319g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Leymarie N., Griffin P. J., Jonscher K., et al. Interlaboratory study on differential analysis of protein glycosylation by mass spectrometry: the ABRF glycoprotein research multi-institutional study 2012. Molecular and Cellular Proteomics. 2013;12(10):2935–2951. doi: 10.1074/mcp.m113.030643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Song E., Mayampurath A., Yu C.-Y., Tang H., Mechref Y. Glycoproteomics: identifying the glycosylation of prostate specific antigen at normal and high isoelectric points by LC-MS/MS. Journal of Proteome Research. 2014;13(12):5570–5580. doi: 10.1021/pr500575r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sanda M., Pompach P., Brnakova Z., Wu J., Makambi K., Goldman R. Quantitative liquid chromatography-mass spectrometry-multiple reaction monitoring (LC-MS-MRM) analysis of site-specific glycoforms of haptoglobin in liver disease. Molecular and Cellular Proteomics. 2013;12(5):1294–1305. doi: 10.1074/mcp.m112.023325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hong Q., Lebrilla C. B., Miyamoto S., Ruhaak L. R. Absolute quantitation of immunoglobulin G and its glycoforms using multiple reaction monitoring. Analytical Chemistry. 2013;85(18):8585–8593. doi: 10.1021/ac4009995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee H.-J., Cha H.-J., Lim J.-S., et al. Abundance-ratio-based semiquantitative analysis of site-specific N-linked glycopeptides present in the plasma of hepatocellular carcinoma patients. Journal of Proteome Research. 2014;13(5):2328–2338. doi: 10.1021/pr4011519. [DOI] [PubMed] [Google Scholar]
- 14.Sherrod S. D., Myers M. V., Li M., et al. Label-free quantitation of protein modifications by pseudo selected reaction monitoring with internal reference peptides. Journal of Proteome Research. 2012;11(6):3467–3479. doi: 10.1021/pr201240a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schilling B., Rardin M. J., MacLean B. X., et al. Platform-independent and label-free quantitation of proteomic data using MS1 extracted ion chromatograms in skyline: application to protein acetylation and phosphorylation. Molecular and Cellular Proteomics. 2012;11(5):202–214. doi: 10.1074/mcp.m112.017707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ohyama C., Hosono M., Nitta K., et al. Carbohydrate structure and differential binding of prostate specific antigen to Maackia amurensis lectin between prostate cancer and benign prostate hypertrophy. Glycobiology. 2004;14(8):671–679. doi: 10.1093/glycob/cwh071. [DOI] [PubMed] [Google Scholar]
- 17.Dwek M. V., Jenks A., Leathem A. J. C. A sensitive assay to measure biomarker glycosylation demonstrates increased fucosylation of prostate specific antigen (PSA) in patients with prostate cancer compared with benign prostatic hyperplasia. Clinica Chimica Acta. 2010;411(23-24):1935–1939. doi: 10.1016/j.cca.2010.08.009. [DOI] [PubMed] [Google Scholar]
- 18.Kosanović M. M., Janković M. M. Sialylation and fucosylation of cancer-associated prostate specific antigen. Journal of B.U.ON. 2005;10(2):247–250. [PubMed] [Google Scholar]
- 19.Janković M. M., Kosanović M. M. Glycosylation of urinary prostate-specific antigen in benign hyperplasia and cancer: assessment by lectin-binding patterns. Clinical Biochemistry. 2005;38(1):58–65. doi: 10.1016/j.clinbiochem.2004.09.022. [DOI] [PubMed] [Google Scholar]
- 20.Tabarés G., Radcliffe C. M., Barrabés S., et al. Different glycan structures in prostate-specific antigen from prostate cancer sera in relation to seminal plasma PSA. Glycobiology. 2006;16(2):132–145. doi: 10.1093/glycob/cwj042. [DOI] [PubMed] [Google Scholar]
- 21.Peracaula R., Tabarés G., Royle L., et al. Altered glycosylation pattern allows the distinction between prostate-specific antigen (PSA) from normal and tumor origins. Glycobiology. 2003;13(6):457–470. doi: 10.1093/glycob/cwg041. [DOI] [PubMed] [Google Scholar]
- 22.Bolduc S., Lacombe L., Naud A., Grégoire M., Fradet Y., Tremblay R. R. Urinary PSA: a potential useful marker when serum PSA is between 2.5 ng/ml and 10 ng/ml. Canadian Urological Association Journal. 2007;1(4):377–381. doi: 10.5489/cuaj.444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stephan C., Jung K., Diamandis E. P., Rittenhouse H. G., Lein M., Loening S. A. Prostate-specific antigen, its molecular forms, and other kallikrein markers for detection of prostate cancer. Urology. 2002;59(1):2–8. doi: 10.1016/S0090-4295(01)01449-2. [DOI] [PubMed] [Google Scholar]
- 24.Pannek J., Rittenhouse H. G., Evans C. L., et al. Molecular forms of prostate-specific antigen and human kallikrein 2 (hK2) in urine are not clinically useful for early detection and staging of prostate cancer. Urology. 1997;50(5):715–721. doi: 10.1016/S0090-4295(97)00324-5. [DOI] [PubMed] [Google Scholar]
- 25.Morelle W., Michalski J.-C. Analysis of protein glycosylation by mass spectrometry. Nature Protocols. 2007;2(7):1585–1602. doi: 10.1038/nprot.2007.227. [DOI] [PubMed] [Google Scholar]
- 26.Segu Z. M., Mechref Y. Characterizing protein glycosylation sites through higher-energy C-trap dissociation. Rapid Communications in Mass Spectrometry. 2010;24(9):1217–1225. doi: 10.1002/rcm.4485. [DOI] [PubMed] [Google Scholar]
- 27.Hart-Smith G., Raftery M. J. Detection and characterization of low abundance glycopeptides via higher-energy C-trap dissociation and orbitrap mass analysis. Journal of the American Society for Mass Spectrometry. 2012;23(1):124–140. doi: 10.1007/s13361-011-0273-y. [DOI] [PubMed] [Google Scholar]
- 28.Stavenhagen K., Hinneburg H., Thaysen-Andersen M., et al. Quantitative mapping of glycoprotein micro-heterogeneity and macro-heterogeneity: an evaluation of mass spectrometry signal strengths using synthetic peptides and glycopeptides. Journal of Mass Spectrometry. 2013;48(6):627–639. doi: 10.1002/jms.3210. [DOI] [PubMed] [Google Scholar]
- 29.Behnken H. N., Ruthenbeck A., Schulz J.-M., Meyer B. Glycan analysis of Prostate Specific Antigen (PSA) directly from the intact glycoprotein by HR-ESI/TOF-MS. Journal of Proteome Research. 2014;13(2):997–1001. doi: 10.1021/pr400999y. [DOI] [PubMed] [Google Scholar]
- 30.Iwakiri J., Grandbois K., Wehner N., Graves H. C. B., Stamey T. An analysis of urinary prostate specific antigen before and after radical prostatectomy: evidence for secretion of prostate specific antigen by the periurethral glands. The Journal of Urology. 1993;149(4):783–786. doi: 10.1016/s0022-5347(17)36207-9. [DOI] [PubMed] [Google Scholar]
- 31.Sağlam H. S., Köse O., Özdemir F., Adsan Ö. Do the values of prostate specific antigen obtained from fresh and dried urine reflect the serum measurements? Urology Annals. 2013;5(2):99–102. doi: 10.4103/0974-7796.110006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schröder F. H., Hugosson J., Roobol M. J., et al. Screening and prostate-cancer mortality in a randomized European study. The New England Journal of Medicine. 2009;360(13):1320–1328. doi: 10.1056/nejmoa0810084. [DOI] [PubMed] [Google Scholar]
- 33.Shariat S. F., Scardino P. T., Lilja H. Screening for prostate cancer: an update. The Canadian Journal of Urology. 2008;15(6):4363–4374. [PMC free article] [PubMed] [Google Scholar]
- 34.Grubb R. L., III, Pinsky P. F., Greenlee R. T., et al. Prostate cancer screening in the Prostate, Lung, Colorectal and Ovarian cancer screening trial: update on findings from the initial four rounds of screening in a randomized trial. BJU International. 2008;102(11):1524–1530. doi: 10.1111/j.1464-410x.2008.08214.x. [DOI] [PubMed] [Google Scholar]
- 35.Oesterling J. E. Prostate specific antigen: a critical assessment of the most useful tumor marker for adenocarcinoma of the prostate. The Journal of Urology. 1991;145(5):907–923. doi: 10.1016/s0022-5347(17)38491-4. [DOI] [PubMed] [Google Scholar]
- 36.Andriole G. L., Jr. Screening for prostate cancer. British Medical Journal. 2010;341 doi: 10.1136/bmj.c4538.c4538 [DOI] [PubMed] [Google Scholar]