Abstract
In mammals, extracellular miRNAs circulate in biofluids as stable entities that are secreted by normal and diseased tissues, and can enter cells and regulate gene expression. Drosophila melanogaster is a proven system for the study of human diseases. They have an open circulatory system in which hemolymph (HL) circulates in direct contact with all internal organs, in a manner analogous to vertebrate blood plasma. Here, we show using deep sequencing that Drosophila HL contains RNase-resistant circulating miRNAs (HL-miRNAs). Limited subsets of body tissue miRNAs (BT-miRNAs) accumulated in HL, suggesting that they may be specifically released from cells or particularly stable in HL. Alternatively, they might arise from specific cells, such as hemocytes, that are in intimate contact with HL. Young and old flies accumulated unique populations of HL-miRNAs, suggesting that their accumulation is responsive to the physiological status of the fly. These HL-miRNAs in flies may function similar to the miRNAs circulating in mammalian biofluids. The discovery of these HL-miRNAs will provide a new venue for health and disease-related research in Drosophila.
Keywords: circulating microRNA, microRNA sequencing, aging, Drosophila, hemolymph
Introduction
Through broad translational repression or mRNA degradation, micro RNAs (miRNAs) control basic cellular processes including cell growth, differentiation, proliferation, and apoptosis1,2 (reviewed by Dhahbi3). In this way, miRNAs modulate physiological processes including the development, cell signaling, immune responses, tumorigenesis, and non-neoplastic disease pathogenesis.4 Recently, miRNAs, as nuclease-resistant entities, were discovered in biofluids including serum, plasma, urine, saliva, and milk.5 Extracellular miRNAs can be contained within exosomes, or they can be complexed with proteins or lipoproteins.6 Because of their contact with plasma, blood cells were at first thought to be the predominant source of extracellular miRNAs. However, the discovery in plasma of miRNAs normally found in the liver, muscle, heart, and brain indicates a multi-tissue origin of the circulating miRNAs.7 Tumors also release miRNAs into the bloodstream.8,9
The presence of miRNAs in extracellular fluids raises the possibility that they may serve as signaling molecules mediating cell-to-cell communication in a paracrine and/or endocrine manner. In support of this view, extracellular miRNAs have been shown to enter cells and alter gene expressions in mammals.10 Vickers et al delivered exogenous miRNAs into hepatocytes, altering the expression of genes involved in lipid metabolism, inflammation, and atherosclerosis.11 Circulating miRNAs are both stable and accessible in serum and plasma. Altered levels of circulating miRNAs are closely associated with multiple types of cancer, suggesting that they may be markers for the diagnosis and prognosis of cancer. Circulating miRNAs are also altered in non-neoplastic diseases, including inflammatory, cardiovascular, and neurological disorders, and in sepsis, rheumatoid arthritis, myocardial infarction, and Alzheimer’s disease.12 Aging is associated with altered circulating miRNA levels, including increased miR-34a in old mice,13 and this miRNA may be involved in age-related neurodegeneration.14
In previous work, we found that multiple miRNAs circulating in mouse serum increase with age, and caloric restriction antagonizes this age-related increase.15 The pathways known to be targeted by these miRNAs include energy metabolism, apoptosis regulation, and Wnt signaling.3 These results raise the possibility that these miRNAs regulate these processes during aging and caloric restriction. Disease database mining suggests that these miRNAs are linked to age-associated pathologies including cancer, as well as neurodegenerative, cardiovascular and inflammatory disorders.15 However, whether the changes in circulating miRNAs are a cause or response to age-associated dysfunction remains to be established, as does their possible role in intercellular signaling.
The genetic amenability of Drosophila melanogaster has made it a useful model system for the study of human diseases and aging.16,17 Drosophila has become an attractive model for studying human microbial, inflammatory, intestinal, and cardiac diseases, as well as neurodegenerative, metabolic, and eating disorders.17,18 Drosophila research has provided invaluable insights into aging. Key mutations and signaling mechanisms that extend lifespan have been identified.19
Flies have an open circulatory system in which hemolymph (HL), the equivalent of blood in vertebrates, circulates in direct contact with internal organs. HL is composed of plasma and a cellular component, namely hemocytes. The plasma component transports hormones, nutrients, metabolic waste, and the proteins that mediate the hemostatic responses to injury and invading microorganisms.20 Hemocytes, which are mostly phagocytes, play a major role in immunity.21 Like clinical blood analysis, HL can be analyzed to evaluate chemical signaling and detect disease markers. HL has been used to characterize the systemic effects of the Drosophila insulin-like peptides and measure glucose tolerance and insulin sensitivity.22 HL protein storage and transportation, enzyme activities, and signaling proteins also have been studied. The effects of HL replacement on longevity have been investigated.23 HL from older Drosophila adversely affects longevity, although HL from younger donors does not extend the recipient’s lifespan.23 However, HL from outbred Drosophila can increase both the fecundity and the adult life span of inbred recipients.24 Although these studies were performed in the 1960s, the factors responsible for the effects on longevity have not been further studied.
Given the importance of Drosophila HL in systemic signaling, and of circulating miRNA in cell-to-cell communication, we investigated the possibly that miRNAs are present in HL and examined their potential regulation by age.
Methods
HL and tissue collection
Stocks of Oregon-R-C D. melanogaster were obtained from Bloomington Drosophila Stock Center (Department of Biology, Indiana University, Bloomington, IN) and maintained as described previously.17 Freshly enclosed flies were sorted under light CO2 anesthesia, and 50 virgin males were placed in each fly bottle; a total of 12 bottles were prepared. After 2 days, four bottles were randomly assigned for the young group used as four replicates to collect HL and tissues. The remaining eight bottles were harvested after 25 days and used as eight replicates for the old group. Flies were individually placed under a Zeiss Stemi 2000 dissection microscope, anesthetized with CO2, punctured in the thorax and head using a fine tungsten needle, and subjected to slight pressure on the abdomen with a fine forceps until a clear droplet of HL was extruded. These drops were collected into Eppendorf tubes containing 20 µL of phosphate buffered saline (PBS), to avoid drying, using a Gilson Minipuls-2 peristaltic pump attached to fine tubing and a needle. Droplets from 20 flies were pooled into a single microcentrifuge tube and used as a biological replicate. Four and eight biological replicates, each from 20 flies, were collected from the young and old groups, respectively. Immediately after the removal of HL, whole flies denuded of HL [termed other body tissue (BT)] were flash-frozen in liquid nitrogen, ground to a fine powder under liquid nitrogen, and stored at −80°C. The volume of HL from each biological replicate was determined, adjusted with PBS to a final volume of 200 µL, centrifuged for 10 minutes at 13,000 g to remove hemocytes, and stored at −80°C.
HL-RNA extraction and small RNA library construction
Total RNA, including miRNA, was extracted from 200 µL of HL prepared as described above, using the Qiagen miRNeasy Serum/Plasma Kit (#217184) according to the manufacturer’s protocol. One-fourth of the RNA from each sample was used to construct small RNA sequencing libraries with the Illumina TruSeq Small RNA Sample Prep Kit (#RS-200-0012) according to the manufacturer’s instructions. Briefly, 3′- and 5′-adapters were sequentially ligated to small RNA and subjected to reverse transcription. The cDNA was amplified by 15 polymerase chain reaction (PCR) cycles using a common primer and a primer containing an index tag, to allow for sample multiplexing. The amplified cDNAs were gel-purified and validated for size, purity, and concentration using the Agilent Bioanalyzer High Sensitivity DNA chip (#5067-4626; Genomics Agilent). The libraries were pooled in equimolar amounts, and 50-base reads were sequenced on an Illumina HiSeq 2000. Read datasets were deposited in the NCBI GEO repository under the accession number GSE77040.
BT-RNA extraction and construction of small RNA and mRNA libraries
Total RNA, including small RNA, was extracted from the frozen, powdered, whole-fly samples using the Qiagen miRNeasy kit (#217004). RNA integrity was confirmed using the Agilent 2100 Bioanalyzer. One-microgram aliquots of total RNA were utilized to construct small RNA sequencing libraries (miRNA-Seq) using the Illumina TruSeq Small RNA Sample Prep Kit (#RS-200-0012). The small RNA libraries were processed and sequenced as described above.
One-microgram aliquots of total RNA were used to construct mRNA sequencing libraries (mRNA-Seq) using the NEBNext Ultra Directional RNA Library Prep Kit for Illumina (New England Biolabs, #E7420S). Briefly, poly(A)+ RNA was purified with oligo-d(T) beads, and fragmented to ~200 nucleotide lengths prior to cDNA synthesis using random primers. Double-stranded cDNA was purified with AMPure XP beads and subjected to end-repair, adapter ligation, and PCR enrichment. The products were purified with AMPure XP Beads, analyzed with the Agilent Bioanalyzer High Sensitivity DNA chip, pooled in equimolar amounts, and used for paired-end sequencing with the Illumina HiSeq 2000 instrument using 50 cycles. Read datasets were deposited in the NCBI GEO repository under the accession number GSE77040.
Real-time quantitative PCR
HL samples, collected and prepared as described above, were spiked with 10 fmoles of synthetic Caenorhabditis elegans cel-miR-39 miScript miRNA (Qiagen #MSY0000010) and incubated for 15 minutes at 37°C with and without addition of 100 ng/mL of RNase A (Thermo Scientific #EN0531) and 4 units of DNase I (Qiagen, #79254). Total RNA, including miRNA, was extracted using the Qiagen miRNeasy Serum/Plasma Kit (#217184) according to the manufacturer’s protocol. Extracted total RNA was reverse-transcribed with the miScript II RT Kit (Qiagen, #218160) using HiFlex Buffer, which enables simultaneous conversion of mRNA and miRNA to cDNA. This makes possible the quantification of miRNA and mRNA from the same sample. The cDNA products were analyzed by quantitative PCR (qPCR) using Qiagen miScript Primer Assays according to the manufacturer’s instructions. We used 10–20 ng of cDNA per qPCR reaction, as suggested by the manufacturer. The levels of miR-14 (Dm_mir-14_1, #MS00017983), miR-8 (Dm_mir-8_1, #MS00018466), miR-184 (Dm_mir-184_1, #MS00017997), and spiked-in C. elegans cel-miR-39 (Ce_mir-39_1, #MS00019789) were measured. QuantiTect Primer Assays were used to measure the mRNA levels of Gapdh1 (Dm_Gapdh1_1_SG, #QT00941465), Actin (Dm_Actn_1_ SG, #QT00496713), and betaTub85D (Dm_betaTub85D _1_ SG, #QT00971572). Real-time qPCR analysis was conducted using a Bio-Rad CFX96 thermocycler. Amplification included an initial activation step at 95°C for 15 minutes, followed by 40 cycles of denaturation at 94°C for 15 seconds, annealing at 55°C for 30 seconds, and extension and data acquisition at 70°C for 30 seconds on the Bio-Rad CFX96 thermocycler.
Analysis of miRNA-Seq
Alignment and annotation of small RNA sequencing reads
Sequencing reads were preprocessed with FASTX-Toolkit (hannonlab.cshl.edu) to trim the adaptor sequences. The reads were mapped to the UCSC Genome Browser dm6 (NCBI genome/47) Drosophila melanogaster genome with Bowtie version 1.0. The mapping was performed using Bowtie “end-to-end k-difference” alignment mode, which allows two mismatches and refrains from reporting any alignments for reads having multiple reportable alignments. Aligned reads of length 18–28 nucleotides were annotated with BEDTools25 using the noncoding RNAs in the “dmel-all-r6.03.gff” file downloaded from ftp://ftp.flybase.net/.
Detection of miRNAs and measurement of their expression levels using miRDeep2
Small RNA sequencing reads were analyzed with miRDeep2,26 a probabilistic algorithm that is based on the miRNA biogenesis model and designed to detect miRNAs from deep sequencing reads. Briefly, the miRD-eep2 analysis involved alignment of the sequencing reads to the dm6 (NCBI genome/47) D. melanogaster genome. Mature miRNA sequences of D. melanogaster and related species of fruit fly were obtained from miRBase v.21 (http://www.mirbase.org) and used as input of known miRNAs. The algorithm aligns reads to potential hairpin structures in a manner consistent with Dicer processing and assigns scores that measure the probability that hairpins are true miRNA precursors. It outputs a scored list of known and novel miRNAs as well as their expression levels.
Differential expression analysis of miRNAs
Expression values of miRNAs generated by miRDeep2 were analyzed with the Bioconductor package edgeR27 to determine the enrichment of miRNAs in HL relative to BT in young and old flies. The algorithm of edgeR uses the negative binomial model to measure differential gene expression. Briefly, expression values were normalized for library sizes with the trimmed mean of M-values (TMM) method. The data were fitted to a generalized linear model (GLM) using a design with two age groups (young and old) and two miRNA locations (HL and BT). Pairwise comparisons between the different experimental groups were used to identify the HL-miRNAs accumulated relative to BT-miRNAs in each of the two age groups. P-values were adjusted for multiple testing using the Benjamini and Hochberg method to control the false discovery rate (FDR).
Retrieval of computationally predicted targets of miRNAs
Predicted miRNA targets were retrieved from the m3RNA database (http://m3rna.cnb.csic.es), which has been developed to improve miRNA–mRNA interactions by combining several different predictive algorithms, as well as integrating experimentally validated interactions.28 The experimentally validated interactions originated from Tarbase, miRTarBase, miRWalk, and miRecords. The predicted miRNA–mRNA interactions were retrieved from nine different algorithms: EIMMo, DIANA-microT, Microcosm, Microrna.org, TargetScan, Mirtarget, PITA, miRWalk-predictive, and TargetSpy. The logistic regression score (LRS) and the weighted scoring by precision (WSP) measure the confidence of predicted miRNA–mRNA interactions. Positive LRS and WSP scores are reliable because they are larger than scores obtained from randomly selected miRNA–mRNA interactions. To increase the statistical confidence and minimize the number of falsely predicted results, we considered only miRNA–mRNA interactions with positive LRS or WSP scores.
Analysis of mRNA-Seq
Paired-end sequencing reads were aligned to the Drosophila reference genome dm6 (NCBI genome/47) with Tophat v229 using the default parameters. The data aligned by Tophat were processed by Cufflinks29 to assemble transcripts and measure their relative abundance in FPKM (fragments per kilobase of exon per million fragments mapped). Transcript expression measurements were used to determine differential gene expression using the Cuffdiff utility in the Cufflinks package with FPKM upper quartile normalization and an FDR threshold of 5%. Cufflinks/Cuffdiff29 compute expression at the gene level by summing the FPKMs of all transcripts within a gene. Genes were considered differentially expressed with age if the fold change is 1.5 or higher and FDR < 5%.
Functional annotation of miRNA targets
Gene lists were imported into DAVID v6.7 (david.abcc.ncifcrf.gov) and analyzed with the Functional Annotation Chart Tool30 to determine which gene ontology (GO) biological processes were potentially involved. This tool measures the similarities among GO terms based on the extent of the associated genes and calculates an enrichment P-value using Fisher’s exact test. The accumulated GO terms were filtered to keep only significant GO biological processes with Benjamini multiple correction test values <0.05. The significant GO biological processes terms and their Benjamini multiple correction test values were processed with REVIGO to remove redundancy.31 REVIGO plots the nonredundant terms in a two-dimensional space using a semantic similarity measure with Uniprot database as background. It generates clusters where the distance between GO terms is a function of their similarity.
Results
RNase-resistant miRNAs circulate in HL
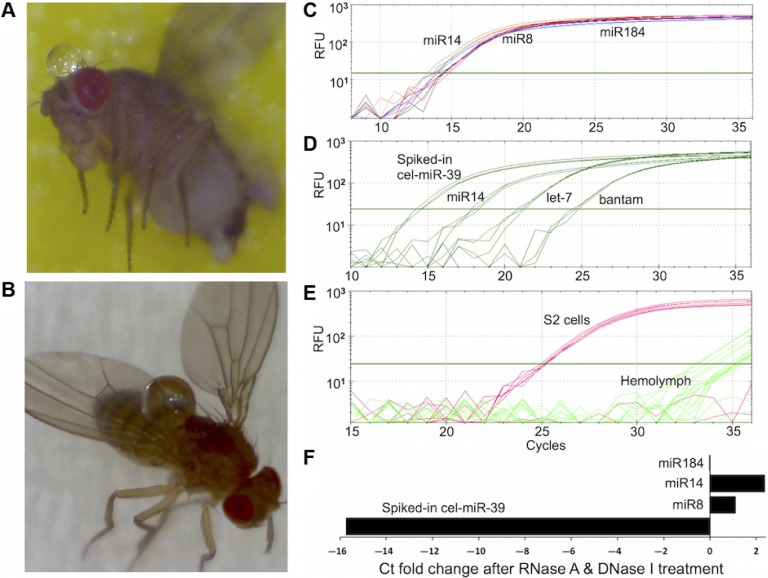
HL droplets were collected after puncture of the head and thorax of anesthetized flies followed by application of slight abdominal pressure (Fig. 1A and B). This technique of HL collection, called bloodletting, minimizes contamination with gut contents which occurs with decapitation techniques.22 Because of its functional similarity to blood plasma, we hypothesized that miRNA might circulate in Drosophila HL. To test this idea, HL was collected and the total RNA, including small RNA, was isolated and analyzed with real-time qPCR. Using readily available primers for Drosophila miRNAs, miR-14, miR-8, miR-184, let-7, and bantam were found at significant levels (Fig. 1C and D). To determine whether these miRNA were carryover from tissue debris or hemocytes, we assayed for the presence of three abundant cellular mRNAs, namely tubulin, actin, and gapdh. These mRNAs were not detected in the HL samples in which miRNAs were observed (Fig. 1E). This indicates that HL miRNAs are not remnants of dead/lysed cells that contain all cellular RNAs including mRNAs; instead, miRNAs accumulated and circulated in HL.
Figure 1.
Presence of stable miRNAs in Drosophila melanogaster hemolymph. (A, B) Clear HL droplets extruded from fly head and thorax. (C–F) Real-time qPCR amplification of selected HL miRNAs and mRNAs. Total RNA including small RNA was extracted from HL samples and analyzed with qPCR to measure the levels of miRNAs and mRNAs. The y-axis represents the relative fluorescence units (RFU) in a semi-log scale. The x-axis represents the cycle at which fluorescence was detected above an automatically determined threshold. (C) Amplification plots for miR-14, miR-8, and miR-184 measured in a representative HL sample. (D) Amplification plots for miR-14, let-7, bantam, and spiked-in synthetic C. elegans cel-miR-39 RNA in representative HL sample. (E) The amplification plots for tubulin, actin, and gapdh mRNAs determined by qPCR using total RNA from HL and S2 cells. Sample from S2 cells are used as a positive control for detecting Drosophila mRNAs by qPCR. The amplification curves of all three mRNAs are superimposed on one another, reflecting the presence of similar amounts of these mRNA in S2 cells. Amplification curves from HL samples show that fluorescent products appear after about 30 cycles, reflecting the significantly lower abundance of these mRNAs in HL relative to S2 cells. (F) The cycle threshold (Ct) fold-change of selected miRNA amplified in the absence or presence of RNase A and DNase I. Total RNA was extracted from HL samples spiked with 10 fmoles of cel-miR-39 RNA. The x-axis represents the ratio of raw Ct values from control samples divided by raw Ct values from samples incubated with RNase A and DNase I. The significantly higher magnitude of the Ct fold change of the spiked-in synthetic miRNA relative to those of the miRNA indicates that the HL-miRNA are present in nuclease-resistant, stable form.
To investigate the stability of these HL-miRNAs, we examined their susceptibility to RNase A and DNase I digestion in isolated HL. Remarkably, levels of each of the three miRNA tested, miR-184, miR-14 and miR-8, were not significantly affected by either nuclease (Fig. 1F). They appear to circulate in HL in vivo in a stable form. In contrast, spiked-in, synthetic cel-miR-39 miRNA was completely degraded (Fig. 1F). Resistance to RNase A digestion suggests the HL-miRNAs are secreted in a protected form, reminiscent of the secretory forms documented in higher animals. Successful amplification of the HL-miRNA after DNase I digestion indicates that their presence is not due to amplification of contaminating cellular DNA derived from cell lysis. Again, these data recall the properties of the stable extracellular miRNAs present in mammalian biofluids.
Sequencing of small RNAs from HL and BT depleted of HL
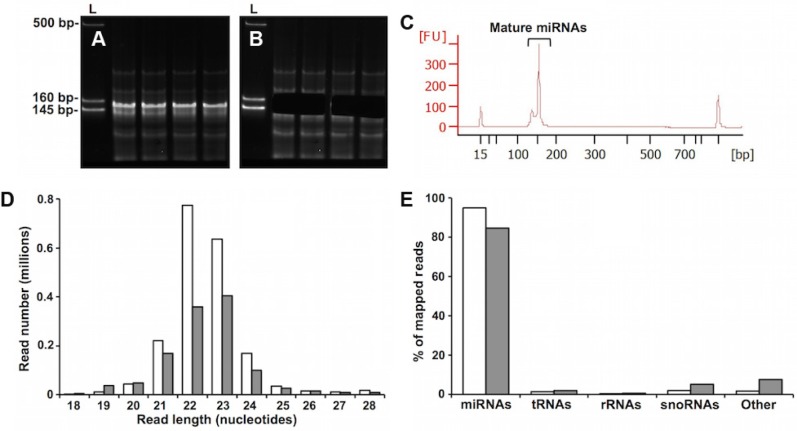
To determine the range of miRNAs circulating in Drosophila HL and to determine their similarity to BT-miRNA, we deep-sequenced small RNAs from both sources. Because aging is a major risk factor for most diseases, we generated cDNA libraries from young and old flies. Aging is used here as a model physiologic state to investigate the possibility that HL-miRNAs are responsive to disease-related risk factors. Reverse transcription and PCR amplifi-cation of HL small RNA yielded cDNA constructs between 145 and 160 bp, which is the length of miRNA cDNA and their 3′ and 5′ adaptors (Fig. 2A). These cDNA were excised from the PAGE gel (Fig. 2B), purified, and analyzed with the Agilent Bioanalyzer High Sensitivity DNA Chip (Fig. 2C). The obtained results revealed high-quality libraries with size, purity, and concentration in accordance with the recommendations for successful cluster generation and sequencing using the Illumina HiSeq platform.
Figure 2.
Small RNA library analysis. (A–C) HL small RNA library purification and validation. (A) Representative samples of amplified cDNA prepared from HL-RNA were chromatographed on a 5% PAGE gel. The first lane in each panel is a marker composed of three dsDNA fragments of 145, 160, and 500 bp. The region between 145 and 160 bps, corresponding to adapter-ligated constructs derived from miRNA, was excised from the gel (B), and purified. The libraries prepared using these purified constructs were validated by analysis with Agilent Bioanalyzer High Sensitivity DNA Chip. (C) A representative electropherogram of a purified library. The peak at ~150 bp indicates the presence of cDNA from HL-miRNA. The peaks other than the ones labeled as miRNAs are the lower and upper markers used by the Bioanalyzer system to determine the size and quantity of the library peak. (D, E) Length distribution and annotation of small RNA sequencing reads from HL and BT. (D) Sequencing reads from all HL or BT miRNA samples were pooled for the purpose of length distribution and annotation analyses. Length distributions by abundance of sequencing reads from BT (clear bars) or HL (filled bars) are shown. Only reads 18–28 nucleotides in size which map uniquely to the UCSC Genome Browser dm6 (NCBI genome/47) Drosophila melanogaster genome are shown. (E) Distribution of RNA biotypes represented as the percentages of reads mapping to the indicated small RNAs. Clear and filled bars represent BT and HL samples, respectively. “Other” represents other Drosophila sncRNAs.
miRNAs are the major fraction of 18-to-28-nucleotide small noncoding RNAs in HL and BT
Sequencing reads from all the samples obtained with the Illumina HiSeq platform were pooled and mapped to the D. melanogaster genome. Pooling was used to examine the general characteristics of the reads, and not to measure the differential expression of miRNAs. Uniquely mapped reads of size 18 to 28 nucleotides were annotated with the noncoding RNA species in the file “dmel-all-r6.03.gff” downloaded from ftp://ftp.flybase.net. The annotated reads had a size distribution peaking at 21–24 nucleotides, consistent with the sizes of miRNAs (Fig. 2D). Annotation analysis showed that more than 80% of the RNAs of 18–28 nucleotides from both HL and BT were miRNAs (Fig. 2E).
Differential accumulation of miRNAs in HL relative to BT of young and old flies
Since the miRNAs in HL must originate from cells, we determined the relative abundance of miRNAs in HL relative to BT in young (2 days) and old (25 days) flies. The level of the small RNAs were quantified with miRDeep2,26 and these data were analyzed statistically with EdgeR.27 The miRNA levels were computed as read counts-per-million (cpm) by the EdgeR cpm() function after normalization to the library sizes. Differential expression analysis with EdgeR identified three types of HL-enriched miRNAs: miRNAs enriched in young flies, those enriched in old flies, and those enriched in both groups. The first category consisted of six miRNAs accumulated in the HL of young flies (Table 1). The second category, consisting of 13 miRNAs, accumulated in the HL of old flies (Table 1). The third consisted of 24 miRNAs, which were enriched in HL irrespective of age (Table 2). Differential and significant age-related accumulation of specific miRNA in HL suggests that these miRNAs are either specifically secreted and/or differentially stable in the HL of flies of different ages. Such differences seem likely to be biologically important, although their role in aging remains to be investigated.
Table 1.
Age-dependent accumulation of specific miRNAs in hemolymph.1
| miRNA2 | PRECURSOR COORDINATES3 | miRDeep2 SCORE4 | CPM5 | FC6 | FDR6 |
|---|---|---|---|---|---|
| miRNAs enriched in hemolymph in young flies | |||||
| dme-miR-14-5p | chr2R:9553764.9553823:+ | 87446.4 | 48201.0 | 2.4 | 0.002 |
| dme-miR-2b-1-5p | chr2L:8258623.8258684:− | 4.4 | 4587.2 | 2.0 | 0.009 |
| dme-miR-2b-2-5p | chr2L:19570189.19570256:− | 5.2 | 4587.2 | 2.0 | 0.009 |
| dme-miR-87-5p | chr2L:9950432.9950504:− | 146.7 | 51.4 | 18.1 | 0.000 |
| dme-miR-982-5p | chrX:4365821.4365883:− | 58.5 | 45.7 | 2.9 | 0.021 |
| dme-miR-9a-5p | chr3L:19565139.19565200:+ | 15636.8 | 6955.3 | 2.8 | 0.002 |
| miRNAs enriched in hemolymph in old flies | |||||
| dme-miR-100-5p | chr2L:18471444.18471505:+ | 1757.8 | 1101.6 | 5.1 | 0.000 |
| dme-miR-1001-5p | chr3R:27642481.27642540:− | 35.4 | 76.7 | 14.6 | 0.000 |
| dme-miR-275-5p | chr2L:7425813.7425875:+ | 16065.1 | 10150.6 | 10.5 | 0.000 |
| dme-miR-317-5p | chr3R:10091138.10091204:+ | 115565.2 | 67884.1 | 1.7 | 0.014 |
| dme-miR-965-5p | chr2L:243054.243118:− | 154.9 | 88.0 | 4.6 | 0.000 |
| dme-miR-966-5p | chr2L:6045640.6045698:− | 26.3 | 33.7 | 10.2 | 0.000 |
| dme-miR-988-5p | chr2R:12158887.12158945:− | 305.8 | 118.3 | 1.8 | 0.012 |
| dme-miR-991-5p | chr2R:20585213.20585275:− | 2.6 | 24.2 | 5.0 | 0.005 |
| chr2R_7074 | chr2R:16887926.16888004:+ | 1.4 | 313.8 | 73.9 | 0.000 |
| chr2R_11226 | chr2R:18180150.18180205:− | 1.4 | 33.2 | 2.3 | 0.037 |
| chr3L_16369 | chr3L:23306664.23306736:− | 1.6 | 36.8 | 5.7 | 0.000 |
| chr3R_18307 | chr3R:1539154.1539245:+ | 2.8 | 18.2 | 5.0 | 0.021 |
| chr3R_20403 | chr3R:22397541.22397596:+ | 1.1 | 1056.8 | 4.7 | 0.000 |
Notes:
miRNAs were considered enriched if FC ≥ 1.5 and FDR < 0.05 between HL and BT.
Names of known miRNAs are from miRBase v.21 (http://www.mirbase.org/). Novel miRNAs are labeled with a unique identification containing the chromosome and an arbitrary number assigned to the hairpin predicted by miRDeep2.
Location of the miRNA precursor in the UCSC Genome Browser dm6 (NCBI genome/47) Drosophila melanogaster genome.
The miRDeep2 score represents the log-odds probability of a sequence being genuine miRNA precursor versus the probability that it is a background hairpin.
Average miRNA read counts-per-million computed over all libraries and taking into account the estimated dispersions and the libraries sizes. It represents a measure of the overall expression level of the miRNA.
Fold change and FDR values for differential expression were computed by EdgeR from pairwise comparisons between HL and BT in the young or old group to identify the miRNAs that are enriched in the hemolymph at young or old age.
Table 2.
Age-independent accumulation of specific miRNAs in hemolymph.1
| miRNA2 | PRECURSOR COORDINATES3 | miRDeep2 SCORE4 | CPM5 | YOUNG | OLD | ||
|---|---|---|---|---|---|---|---|
| FC6 | FDR6 | FC6 | FDR6 | ||||
| dme-let-7-5p | chr2L:18472043.18472103:+ | 8440.8 | 5667.9 | 2.3 | 0.003 | 4.8 | 0.000 |
| dme-miR-11-5p | chr3R:21622502.21622563:− | 31989.8 | 18231.9 | 2.1 | 0.003 | 1.9 | 0.000 |
| dme-miR-125-5p | chr2L:18472342.18472402:+ | 3020.1 | 1751.3 | 2.4 | 0.011 | 3.4 | 0.000 |
| dme-miR-190-5p | chr3L:8571821.8571884:+ | 1852.9 | 1197.1 | 2.4 | 0.001 | 3.1 | 0.000 |
| dme-miR-252-5p | chr3R:13464230.13464296:− | 2716.2 | 1777.4 | 1.9 | 0.011 | 2.4 | 0.000 |
| dme-miR-276a-5p | chr3L:10365243.10365302:+ | 115850.2 | 64277.3 | 2.7 | 0.000 | 1.7 | 0.004 |
| dme-miR-277-5p | chr3R:10100038.10100101:+ | 24458.6 | 8898.9 | 2.3 | 0.003 | 1.6 | 0.011 |
| dme-miR-278-5p | chr2R:15657082.15657142:+ | 5465.5 | 1467.1 | 2.7 | 0.000 | 2.1 | 0.000 |
| dme-miR-279-5p | chr3R:29215605.29215670:+ | 4.8 | 3259.6 | 5.3 | 0.000 | 2.6 | 0.000 |
| dme-miR-282-5p | chr3L:3251037.3251102:+ | 10668.9 | 5833.4 | 2.7 | 0.009 | 11.6 | 0.000 |
| dme-miR-33-5p | chr3L:19797985.19798046:+ | 27140.1 | 18409.2 | 1.8 | 0.030 | 2.2 | 0.000 |
| dme-miR-34-5p | chr3R:10100951.10101025:+ | 20436.6 | 5104.1 | 2.3 | 0.007 | 3.4 | 0.000 |
| dme-miR-8-5p | chr2R:16831446.16831506:+ | 309506.2 | 187622.4 | 2.3 | 0.001 | 2.4 | 0.000 |
| dme-miR-929-5p | chr3R:4295384.4295439:+ | 439.6 | 298.7 | 6.9 | 0.000 | 4.2 | 0.000 |
| dme-miR-970-5p | chrX:12636351.12636416:+ | 1864.8 | 971.3 | 1.7 | 0.027 | 1.8 | 0.000 |
| dme-miR-980-5p | chrX:280188.280254:− | 729 | 412.3 | 17.5 | 0.000 | 21.3 | 0.000 |
| dme-miR-986-5p | chr2R:8444999.8445067:+ | 2392.6 | 1628.6 | 2.2 | 0.001 | 1.8 | 0.000 |
| dme-miR-996-5p | chr3R:29217201.29217265:+ | 4302.4 | 2324.8 | 3.7 | 0.000 | 1.8 | 0.000 |
| chr3L_16646 | chr3L:24539748.24539815:− | 1.1 | 17.3 | 8.9 | 0.023 | 14.6 | 0.000 |
| chr3R_20493 | chr3R:24268348.24268396:+ | 3.4 | 216.0 | 77.8 | 0.000 | 4.0 | 0.002 |
| chrX_28055 | chrX:7302769.7302827:+ | 1.5 | 943.8 | 349.1 | 0.000 | 113.6 | 0.000 |
| chrX_28088 | chrX:8026969.8027019:+ | 1.3 | 4856.6 | 19.5 | 0.000 | 56.6 | 0.000 |
| chrX_32979 | chrX:22373683.22373732:− | 1.3 | 2790.3 | 5.0 | 0.001 | 10.4 | 0.000 |
| chrX_33619 | chrX:23526522.23526569:− | 1557.8 | 2790.3 | 5.0 | 0.001 | 10.4 | 0.000 |
Notes:
miRNAs were considered enriched if FC ≥ 1.5 and FDR < 0.05 between HL and BT.
Names of known miRNAs are from miRBase v.21 (http://www.mirbase.org/). Novel miRNAs are labeled with a unique identification containing the chromosome and an arbitrary number assigned to the hairpin predicted by miRDeep2.
Location of the miRNA precursor in the UCSC Genome Browser dm6 (NCBI genome/47) Drosophila melanogaster genome.
The miRDeep2 score represents the log-odds probability of a sequence being genuine miRNA precursor versus the probability that it is a background hairpin.
Average miRNA read counts-per-million computed over all libraries and taking into account the estimated dispersions and the libraries sizes. It represents a measure of the overall expression level of the miRNA.
Fold change and FDR values for differential expression were computed by EdgeR from pairwise comparisons between HL and BT to identify the miRNAs that are enriched in the hemolymph at young and old ages.
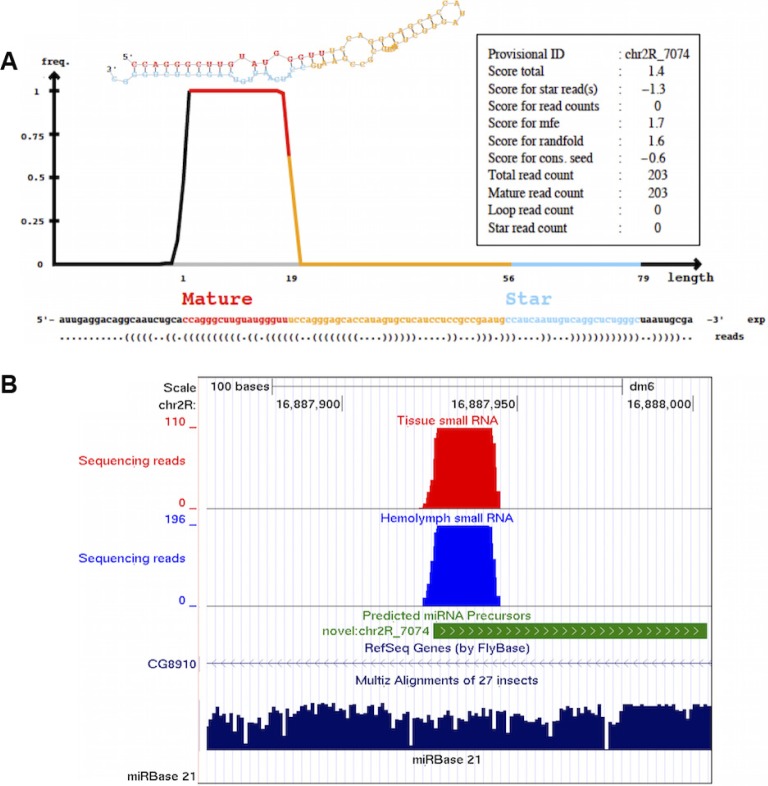
A number of potentially novel miRNAs were predicted by miRDeep2, based on sequence and size.26 Five predicted miRNAs were enriched in the HL of old flies (Table 1) and six were enriched relative to BT independently of age (Table 2). A representative of these novel miRNA, chr2R_7074, which accumulated in the HL of old flies (Table 1), is illustrated in Figure 3. It exhibits a specific read pileup reflecting a mature:star duplex with a two-nucleotide 3′ overhang (Fig. 3A). It is located in a conserved genomic region and is derived from an intron of the CG8910 gene (Fig. 3B). The functions of these novel miRNAs remain to be determined.
Figure 3.
A novel Drosophila melanogaster miRNA predicted by miRDeep2. (A) Graphics of the secondary structure and read distributions for a novel miRNA with the provisional name chr2R_7074 (see Table 1). Both mature and star strands were detected. The information in the text box at the top right represents the miRDeep2 scores and the read counts for each component of the novel miRNA. (B) A UCSC genome browser screenshot showing alignment of reads from HL (blue) and BT (red) to dm6 (NCBI genome/47) Drosophila genome. The precursor of the novel miRNA (green) maps to an intron of the CG8910 gene as annotated in the “RefSeq Genes (by FlyBase)” track. The coverage depth (number of reads, y-axis) shows the “stacks” of sequence reads map to the mature miRNA with no reads mapping to the star region of the precursor. The conservation track at the bottom (deep blue) shows that the novel miRNA maps to a conserved genomic region. A miRBase v.21 custom track was uploaded to the UCSC genome browser to show the absence of known miRNAs in the genomic regions of the predicted novel miRNA.
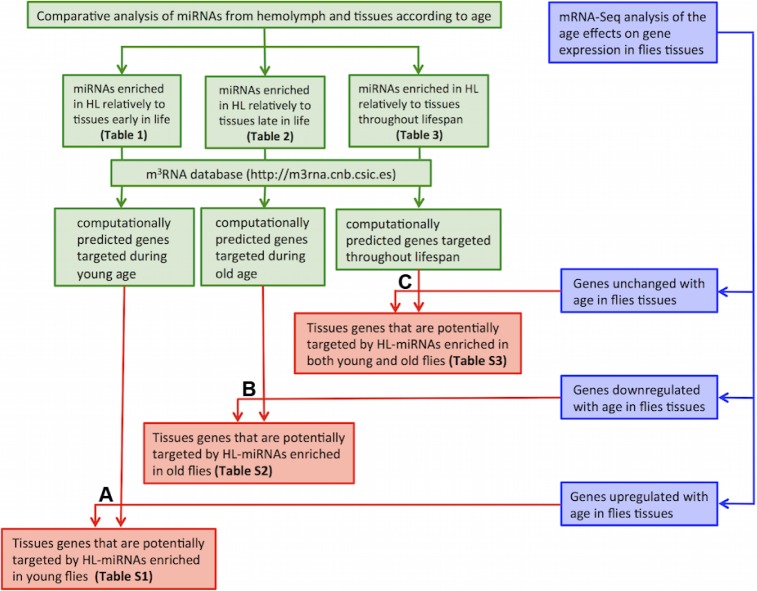
Identification of genes potentially regulated by HL-miRNAs using integration of tissue mRNA sequencing and computational prediction of miRNA target
Because extracellular miRNAs can modulate the expression of specific genes in mammals, we investigated which genes might be regulated by HL-miRNAs. This analysis was performed in three steps (Fig. 4). First, the computationally predicted mRNA targets of the differentially expressed HL-miRNAs were determined. Second, mRNA-Seq was used to determine the age-associated changes in BT mRNA levels. Third, we determined the intersection between the BT-mRNAs that changed with age and the mRNAs predicted to be targets of HL-miRNAs. This analysis identified three groups of genes potentially regulated by HL-miRNA. First were BT-mRNAs possibly downregulated by their cognate HL-miRNA accumulated in young flies (Fig. 4A; Supplementary Table 1). Second were BT-mRNAs potentially downregulated by their cognate HL-miRNAs accumulated in old flies (Fig. 4B; Supplementary Table 2). Third were BT-mRNAs that were unchanged with age and are the predicted targets of HL-miRNA that were also unchanged by age (Fig. 4C; Supplementary Table 3). This final class of genes might be responsive to changes in HL-miRNA induced by homeostatic challenges such as extreme temperatures or infections with bacteria or parasites. The miRNAs and their predicted targets are listed in Supplementary File 1. Further work will be needed to test these predictions.
Figure 4.
Outline of the experimental design and workflow for the integrated analysis of the miRNA-Seq and mRNA-Seq data. Genes that are potentially regulated by the HL-miRNAs were determined by integrating miRNA-Seq and mRNA-Seq data using the following three steps: Step 1 (depicted in green): After identifying the HL-miRNAs enriched relative to BT in either young or old, or both young and old (Tables 1 and 2), we retrieved their computationally predicted target genes from the m3RNA database (http://m3rna.cnb.csic.es). Step 2 (depicted in blue): mRNA-Seq was utilized to identify three groups of genes that were differentially expressed with age, either upregulated, downregulated, or unchanged in BT. Step 3 (depicted in red): The three groups of genes predicted in step1 as targets of HL-miRNAs were intersected with the three groups of genes identified in step 2. In the figure, (A) depicts the derivation of genes that are predicted to be targets of HL-miRNAs enriched in young which are also upregulated with age in BT. These genes (Supplementary Table 1) are likely upregulated in BT of old flies because their expression is no longer repressed by HL-miRNAs that are enriched only in young flies. (B) Depicts the derivation of genes which are the predicted to be targets of HL-miRNAs enriched in old which are also downregulated with age in BT. These genes (Supplementary Table 2) are likely downregulated in BT of old flies because their expression is repressed by HL-miRNAs which are enriched only in old flies. (C) Depicts the derivation of genes which are predicted to be targets of HL-miRNAs enriched in both young and old which also do not change expression with age in BT. These genes (Supplementary Table 3) do not change expression with age because the concentration of their regulatory miRNAs in HL does not change with age.
Discussion
Recently, nuclease-resistant circulating miRNAs have been found in mammalian body fluids, including serum and plasma.5 These miRNA appear to function in cell-to-cell signaling and have potential as disease biomarkers.3,10 By analogy, it is possible the HL-miRNAs identified in this study may function as systemic signaling factors. Like their mammalian counterparts, the Drosophila HL-miRNAs are stable and resistant to RNase A digestion. In mammalian fluids, miRNAs are protected from RNase activity by packaging in exosomes or by binding to protein or lipoprotein factors. The stabilizing factors complexed with extracellular miRNAs include Argonaute-2 and high-density lipoprotein.6 In Drosophila, extracellular exosomes mediate the transmission of Wnt and Evi across the synaptic cleft in neuromuscular junction.32 Two hemocyte-like Drosophila cell lines, Kc167 and S2, have been shown to excrete exosomes containing signaling proteins in a manner similar to that found in higher vertebrate cells.33 However, miRNAs have not yet been described as a part of the exosomal cargo in Drosophila. In mammals, the factors associated with extracellular miRNA not only protect them from nucleases but also mediate their delivery to and uptake by recipient tissues.7 Thus, further investigation of the packaging components and extracellular secretion of Drosophila HL-miRNAs is warranted.
At this time, the relationship between tissue and extracellular miRNAs in mammals remains controversial. Originally, it was assumed that, to be useful as biomarkers, the level of miRNAs in body fluids should reflect their abundance in diseased cells and tissues. However, this is true only in some cases. miRNAs overexpressed in specific cancers have been detected in plasma or serum, and most studies focus on such biomarkers.34 But, other studies find that not all highly abundant cellular miRNAs are represented in the extracellular fluids, suggesting that specific selection mechanisms exist for miRNA.35 Our findings are in accord with this latter hypothesis. Our results are consistent with the idea that specific secretory mechanisms are responsible for the enrichment of specific miRNAs. Further studies of the mechanisms underlying the secretion and stability of miRNAs should aid our understanding of these issues.
If trans-acting, and tissue-specific miRNA do exist in Drosophila as we postulate, their presence would not conflict with the results found by Roignant et al.36 Using a transgene expressing a unique, tissue-specific RNAi, they found that this gene was not inhibited in nontargeted tissues. They concluded that RNAi do not target genes systemically, by which they appear to mean in trans, in other cells. However, a foreign RNAi would be unlikely to be selectively secreted into HL and thereby target other cells.
The functional significance of the changes in HL-miRNA abundance with age remains unclear. However, cells may take up HL-miRNA, in a manner analogous to that found in mammals. To speculate on potentially regulated pathways, we identified gene ontology (GO) biological processes related to the possible gene targets of HL-miRNAs listed in Supplementary Tables 1–3. The potential targets of HL-miRNA in young flies were associated with the GO processes cell cycle regulation, cell division and proliferation, chromatin organization, protein–DNA complex assembly, and several development-associated pathways (Supplementary Fig. 1). This possible involvement of HL-miRNA in the regulation of cell division and proliferation could be important. Although adult flies are largely post-mitotic, dysregulation of intestinal stem cells proliferation can limit the health and lifespan of Drosophila by destroying intestinal integrity.37
The HL-miRNAs accumulated in old age had the putative target GO processes of cell signaling and communication, regulation of G-protein coupled receptor protein signaling pathway and activation of protein kinase activity, and cell adhesion and recognition (Supplementary Fig. 2). Finally, the HL-miRNAs accumulated regardless of age target the GO biological processes metabolism of carbohydrates, lipids, proteins, and nitrogen compounds. Other targeted processes include phosphorus metabolism, protein phosphorylation and modification, posttranscriptional regulation of gene expression, and translational initiation (Supplementary Fig. 3).
Focusing on specific genes, two among the HL-miRNAs abundant in young flies are involved in the control of apoptosis, miR-2b and miR-14 (Table 1). The Drosophila proapoptotic genes reaper and grim are targets of miR-2b,38 and miR-14 is a cell death suppressor in Drosophila. The absence of miR-14 reduces the lifespan of flies.39 Thus, these HL-miRNAs may regulate cell viability and stress resistance in young flies. Other HL-miRNAs accumulated in young include miR-87, which is involved in anti-pathogen and immune responses,40 and miR-9a, which mediates the robust responses to temperature variation by targeting the Drosophila Sens gene.41 The presence of these miRNAs might be indicative of a younger phenotype.
Among HL-miRNAs accumulated in old flies was miR-100 (Table 1), which is induced by ecdysone (a development inducing hormone) and inhibited by juvenile hormone analogs in Drosophila S2 cells.42 These hormones also influence adult longevity,43 suggesting a possible link of miR-100 regulation with longevity. While we did not find any literature describing the genes regulated by miR-100 in Drosophila, in mammals it downregulates the mRNA for mTOR and insulin-like growth factor-I (IGF-I) receptor (IGFR).44 Reduced TOR and IGFR signaling are associated with extended lifespan in worms, flies, mice, and perhaps humans.45 If orthologous genes are similarly regulated by miR-100 in Drosophila, upregulation of the miRNA may be a prolongevity adaptation in old age.
HL-miRNAs that were abundant throughout life are implicated in aging. They are homeostatic regulators that may not be significantly perturbed in adult flies under our culture conditions (Table 2). In Drosophila, Let-7 and miR-125 miRNAs mediate the transition from juvenile to adult flies.46 Let-7 is part of a positive feedback loop that adjusts ecdysteroid signaling strength in response to temperature stress and aging.47 Drosophila miR-125 is the homolog of C. elegans lin-4, which influences C. elegans lifespan, regulates insulin and IGF-I signaling, and regulates cell cycle checkpoints for DNA damage pathways.48 These targets are known regulators of aging.48 miR-34 modulates aging and neurodegeneration in Drosophila,14 miR-277 controls Drosophila lifespan,49 and miR-278 regulates insulin sensitivity and energy homeostasis.50
Conclusion
We demonstrated here that miRNAs circulate in the Drosophila HL in a nuclease-stable form. Specific miRNAs accumulated in HL relative to BT, and the degree of the enrichment of many of these miRNAs changes in an age-dependent manner. Like miRNAs circulating in mammalian biofluids, these HL-miRNAs may be secreted by cells, taken up by others, and thus mediate intercellular communication. Functional analysis suggests that many of the genes that might be targeted in this way are associated with the control of aging and its associated diseases. Since Drosophila is a valuable model system for the investigation of human disease and aging, the discovery of miRNAs in HL provides a new tool in the armamentarium available for disease research.
Supplementary Materials
Supplementary table 1. Genes that are potentially targeted by miRNAs selectively accumulated in HL relative to tissues during young age.
Supplementary table 2. Genes that are potentially targeted by miRNAs selectively accumulated in HL relative to tissues during old age.
Supplementary table 3. Genes that are potentially targeted by miRNAs selectively accumulated in HL relative to tissues independent of age.
Supplementary file 1. miRNAs enriched in hemolymph and their potential targets.
Supplementary figure 1. Gene Ontology (GO) biological processes associated with genes putatively targeted by HL-miRNAs accumulated in young flies. DAVID was initially used to carry out the GO enrichment analysis of the targeted genes, and subsequently REVIGO was used to remove redundancy and plot non-redundant GO terms as a two-dimensional scatter plot. Similar GO terms (depicted by circles) cluster together closer than unrelated ones. GO terms representative of clusters are labeled. Circle size (log size; legend in upper right-hand corner) is proportional to the frequency of a respective GO term in the Uniprot database used as background; circles representing more general terms have larger size. Circle color reflects the significance of GO term enrichment; it indicates the log10 of the P-value derived from DAVID analysis. Ends of the red and blue colors in the upper right-hand corner depict lower and higher log10 P-values, respectively.
Supplementary figure 2. Gene ontology (GO) biological processes associated with genes putatively targeted by HL-miRNAs accumulated in old flies. Description and information about the significant GO Biological Processes are as in Supplementary Figure 1.
Supplementary figure 3. Gene ontology (GO) biological processes associated with genes putatively targeted by HL-miRNAs accumulated independently of age. Description and information about the significant GO Biological Processes are as in Supplementary Figure 1.
Acknowledgments
The authors thank Amber Graham, Karla Mabida, Michelle Gonzales, and Bianca Mabida for feeding and monitoring the flies, and for their technical help.
Footnotes
ACADEMIC EDITOR: Gustavo Caetano-Anollés, Editor in Chief
PEER REVIEW: Four peer reviewers contributed to the peer review report. Reviewers’ reports totaled 1369 words, excluding any confidential comments to the academic editor.
FUNDING: This work was supported by gifts from anonymous donors. The funders had no role in study design, data collection or analysis, decision to publish, or preparation of the manuscript.
COMPETING INTERESTS: Authors disclose no potential conflicts of interest.
Paper subject to independent expert single-blind peer review. All editorial decisions made by independent academic editor. Upon submission manuscript was subject to anti-plagiarism scanning. Prior to publication all authors have given signed confirmation of agreement to article publication and compliance with all applicable ethical and legal requirements, including the accuracy of author and contributor information, disclosure of competing interests and funding sources, compliance with ethical requirements relating to human and animal study participants, and compliance with any copyright requirements of third parties. This journal is a member of the Committee on Publication Ethics (COPE).
Author Contributions
Conceived and designed the experiments: JMD. Analyzed the data: JMD, HA. Wrote the first draft of the manuscript: JMD. Agree with manuscript results and conclusions: JMD, SRS, HA, RL, AY, NG, HTL, PM. Developed the structure and arguments for the paper: JMD. Made critical revisions and approved final version: JMD, SRS. All authors reviewed and approved of the final manuscript.
REFERENCES
- 1.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 2.Garzon R, Marcucci G, Croce CM. Targeting microRNAs in cancer: rationale, strategies and challenges. Nat Rev Drug Discov. 2010;9(10):775–789. doi: 10.1038/nrd3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dhahbi JM. Circulating small noncoding RNAs as biomarkers of aging. Ageing Res Rev. 2014;17:86–98. doi: 10.1016/j.arr.2014.02.005. [DOI] [PubMed] [Google Scholar]
- 4.Koturbash I, Zemp FJ, Pogribny I, Kovalchuk O. Small molecules with big effects: the role of the microRNAome in cancer and carcinogenesis. Mutat Res. 2011;722(2):94–105. doi: 10.1016/j.mrgentox.2010.05.006. [DOI] [PubMed] [Google Scholar]
- 5.Weber JA, Baxter DH, Zhang S, et al. The microRNA spectrum in 12 body fluids. Clin Chem. 2010;56(11):1733–1741. doi: 10.1373/clinchem.2010.147405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arroyo JD, Chevillet JR, Kroh EM, et al. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc Natl Acad Sci U S A. 2011;108(12):5003–5008. doi: 10.1073/pnas.1019055108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Turchinovich A, Weiz L, Burwinkel B. Extracellular miRNAs: the mystery of their origin and function. Trends Biochem Sci. 2012;37(11):460–465. doi: 10.1016/j.tibs.2012.08.003. [DOI] [PubMed] [Google Scholar]
- 8.Healy NA, Heneghan HM, Miller N, Osborne CK, Schiff R, Kerin MJ. Systemic mirnas as potential biomarkers for malignancy. Int J Cancer. 2012;131(10):2215–2222. doi: 10.1002/ijc.27642. [DOI] [PubMed] [Google Scholar]
- 9.Ma R, Jiang T, Kang X. Circulating microRNAs in cancer: origin, function and application. J Exp Clin Cancer Res. 2012;31:38. doi: 10.1186/1756-9966-31-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zernecke A, Bidzhekov K, Noels H, et al. Delivery of microRNA-126 by apoptotic bodies induces CXCL12-dependent vascular protection. Sci Signal. 2009;2(100):ra81. doi: 10.1126/scisignal.2000610. [DOI] [PubMed] [Google Scholar]
- 11.Vickers KC, Palmisano BT, Shoucri BM, Shamburek RD, Remaley AT. MicroRNAs are transported in plasma and delivered to recipient cells by high-density lipoproteins. Nat Cell Biol. 2011;13(4):423–433. doi: 10.1038/ncb2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reid G, Kirschner MB, van Zandwijk N. Circulating microRNAs: association with disease and potential use as biomarkers. Crit Rev Oncol Hematol. 2011;80(2):193–208. doi: 10.1016/j.critrevonc.2010.11.004. [DOI] [PubMed] [Google Scholar]
- 13.Li X, Khanna A, Li N, Wang E. Circulatory miR34a as an RNAbased, noninvasive biomarker for brain aging. Aging. 2011;3(10):985–1002. doi: 10.18632/aging.100371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu N, Landreh M, Cao K, et al. The microRNA miR-34 modulates ageing and neurodegeneration in Drosophila. Nature. 2012;482(7386):519–523. doi: 10.1038/nature10810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dhahbi JM, Spindler SR, Atamna H, et al. Deep sequencing identifies circulating mouse miRNAs that are functionally implicated in manifestations of aging and responsive to calorie restriction. Aging. 2013;5(2):130–141. doi: 10.18632/aging.100540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pandey UB, Nichols CD. Human disease models in Drosophila melanogasterand the role of the fly in therapeutic drug discovery. Pharmacol Rev. 2011;63(2):411–436. doi: 10.1124/pr.110.003293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Spindler SR, Li R, Dhahbi JM, et al. Statin treatment increases lifespan and improves cardiac health in Drosophilaby decreasing specific protein prenylation. PLoS One. 2012;7(6):e39581. doi: 10.1371/journal.pone.0039581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Panayidou S, Apidianakis Y. Regenerative inflammation: lessons from Drosophila intestinal epithelium in health and disease. Pathogens. 2013;2(2):209–231. doi: 10.3390/pathogens2020209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kapahi P, Zid BM, Harper T, Koslover D, Sapin V, Benzer S. Regulation of lifespan in Drosophilaby modulation of genes in the TOR signaling pathway. Curr Biol. 2004;14(10):885–890. doi: 10.1016/j.cub.2004.03.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kounatidis I, Ligoxygakis P. Drosophila as a model system to unravel the layers of innate immunity to infection. Open Biol. 2012;2(5):120075. doi: 10.1098/rsob.120075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ghosh S, Singh A, Mandal S, Mandal L. Active hematopoietic hubs in Drosophila adults generate hemocytes and contribute to immune response. Dev Cell. 2015;33(4):478–488. doi: 10.1016/j.devcel.2015.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haselton AT, Fridell YC. Insulin Injection and Hemolymph Extraction to Measure Insulin Sensitivity in Adult Drosophila melanogaster. J Vis Exp. 2011;(52):e2722. doi: 10.3791/2722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sondhi KC. Studies in aging. II. The effects of injecting hemolymph from younger and older donors on the fecundity and the adult life span of hosts in Drosophila. J Exp Zool. 1966;162(1):89–97. doi: 10.1002/jez.1401620109. [DOI] [PubMed] [Google Scholar]
- 24.Sondhi KC. Studies in aging, 3. The physiological effects of injecting hemolymph from outbred donors into inbred hosts in Drosophila melanogaster. Proc Natl Acad Sci U S A. 1967;57(4):965–971. doi: 10.1073/pnas.57.4.965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26(6):841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Friedlander MR, Mackowiak SD, Li N, Chen W, Rajewsky N. miRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic Acids Res. 2012;40(1):37–52. doi: 10.1093/nar/gkr688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26(1):139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tabas-Madrid D, Muniategui A, Sanchez-Caballero I, et al. Improving miRNA-mRNA interaction predictions. BMC Genomics. 2014;15(suppl 10):S2. doi: 10.1186/1471-2164-15-S10-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Trapnell C, Roberts A, Goff L, et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc. 2012;7(3):562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 31.Supek F, Bosnjak M, Skunca N, Smuc T. REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS One. 2011;1800;6(7):e2. doi: 10.1371/journal.pone.0021800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Korhonen R, Lahti A, Kankaanranta H, Moilanen E. Nitric oxide production and signaling in inflammation. Curr Drug Targets Inflamm Allergy. 2005;4(4):471–479. doi: 10.2174/1568010054526359. [DOI] [PubMed] [Google Scholar]
- 33.Koppen T, Weckmann A, Muller S, et al. Proteomics analyses of microvesicles released by Drosophila Kc167 and S2 cells. Proteomics. 2011;11(22):4397–4410. doi: 10.1002/pmic.201000774. [DOI] [PubMed] [Google Scholar]
- 34.Mitchell PS, Parkin RK, Kroh EM, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci U S A. 2008;105(30):10513–10518. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pigati L, Yaddanapudi SC, Iyengar R, et al. Selective release of microRNA species from normal and malignant mammary epithelial cells. PLoS One. 2010;5(10):e13515. doi: 10.1371/journal.pone.0013515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Roignant JY, Carre C, Mugat B, Szymczak D, Lepesant JA, Antoniewski C. Absence of transitive and systemic pathways allows cell-specific and isoform-specific RNAi in Drosophila. RNA. 2003;9(3):299–308. doi: 10.1261/rna.2154103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang L, Ryoo HD, Qi Y, Jasper H. PERK limits Drosophila lifespan by promoting intestinal stem cell proliferation in response to ER stress. PLoS Genet. 2015;11(5):e1005220. doi: 10.1371/journal.pgen.1005220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stark A, Brennecke J, Russell RB, Cohen SM. Identification of Drosophila MicroRNA targets. PLoS Biol. 2003;1(3):E60. doi: 10.1371/journal.pbio.0000060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xu P, Vernooy SY, Guo M, Hay BA. The Drosophila microRNA Mir-14 suppresses cell death and is required for normal fat metabolism. Curr Biol. 2003;13(9):790–795. doi: 10.1016/s0960-9822(03)00250-1. [DOI] [PubMed] [Google Scholar]
- 40.Liu Y, Zhou Y, Wu J, et al. The expression profile of Aedes albopictus miRNAs is altered by dengue virus serotype-2 infection. Cell Biosci. 2015;5:16. doi: 10.1186/s13578-015-0009-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cassidy JJ, Jha AR, Posadas DM, et al. miR-9a minimizes the phenotypic impact of genomic diversity by buffering a transcription factor. Cell. 2013;155(7):1556–1567. doi: 10.1016/j.cell.2013.10.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sempere LF, Sokol NS, Dubrovsky EB, Berger EM, Ambros V. Temporal regulation of microRNA expression in Drosophila melanogastermediated by hormonal signals and broad-complex gene activity. Dev Biol. 2003;259(1):9–18. doi: 10.1016/s0012-1606(03)00208-2. [DOI] [PubMed] [Google Scholar]
- 43.Simon AF, Shih C, Mack A, Benzer S. Steroid control of longevity in Drosophila melanogaster. Science. 2003;299(5611):1407–1410. doi: 10.1126/science.1080539. [DOI] [PubMed] [Google Scholar]
- 44.Qin C, Huang RY, Wang ZX. Potential role of miR-100 in cancer diagnosis, prognosis, and therapy. Tumour Biol. 2015;36(3):1403–1409. doi: 10.1007/s13277-015-3267-8. [DOI] [PubMed] [Google Scholar]
- 45.Bartke A. Growth hormone, insulin and aging: the benefits of endocrine defects. Exp Gerontol. 2011;46(2–3):108–111. doi: 10.1016/j.exger.2010.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Caygill EE, Johnston LA. Temporal regulation of metamorphic processes in Drosophila by the let-7 and miR-125 heterochronic microRNAs. Curr Biol. 2008;18(13):943–950. doi: 10.1016/j.cub.2008.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Konig A, Shcherbata HR. Soma influences GSC progeny differentiation via the cell adhesion-mediated steroid-let-7-Wingless signaling cascade that regulates chromatin dynamics. Biol Open. 2015;4(3):285–300. doi: 10.1242/bio.201410553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Boehm M, Slack F. A developmental timing microRNA and its target regulate life span in C elegans. Science. 2005;310(5756):1954–1957. doi: 10.1126/science.1115596. [DOI] [PubMed] [Google Scholar]
- 49.Esslinger SM, Schwalb B, Helfer S, et al. Drosophila miR-277 controls branched-chain amino acid catabolism and affects lifespan. RNA Biol. 2013;10(6):1042–1056. doi: 10.4161/rna.24810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Teleman AA, Maitra S, Cohen SM. Drosophila lacking microRNA miR-278 are defective in energy homeostasis. Genes Dev. 2006;20(4):417–422. doi: 10.1101/gad.374406. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary table 1. Genes that are potentially targeted by miRNAs selectively accumulated in HL relative to tissues during young age.
Supplementary table 2. Genes that are potentially targeted by miRNAs selectively accumulated in HL relative to tissues during old age.
Supplementary table 3. Genes that are potentially targeted by miRNAs selectively accumulated in HL relative to tissues independent of age.
Supplementary file 1. miRNAs enriched in hemolymph and their potential targets.
Supplementary figure 1. Gene Ontology (GO) biological processes associated with genes putatively targeted by HL-miRNAs accumulated in young flies. DAVID was initially used to carry out the GO enrichment analysis of the targeted genes, and subsequently REVIGO was used to remove redundancy and plot non-redundant GO terms as a two-dimensional scatter plot. Similar GO terms (depicted by circles) cluster together closer than unrelated ones. GO terms representative of clusters are labeled. Circle size (log size; legend in upper right-hand corner) is proportional to the frequency of a respective GO term in the Uniprot database used as background; circles representing more general terms have larger size. Circle color reflects the significance of GO term enrichment; it indicates the log10 of the P-value derived from DAVID analysis. Ends of the red and blue colors in the upper right-hand corner depict lower and higher log10 P-values, respectively.
Supplementary figure 2. Gene ontology (GO) biological processes associated with genes putatively targeted by HL-miRNAs accumulated in old flies. Description and information about the significant GO Biological Processes are as in Supplementary Figure 1.
Supplementary figure 3. Gene ontology (GO) biological processes associated with genes putatively targeted by HL-miRNAs accumulated independently of age. Description and information about the significant GO Biological Processes are as in Supplementary Figure 1.