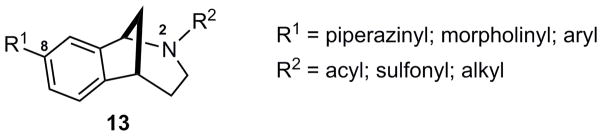Abstract
A novel structural class with high affinity and subtype selectivity for the sigma 2 receptor has been discovered. Preliminary structure affinity relationship data are presented showing that 8-substituted 2,3,4,5-tetrahydro-1,5-methanobenzazepine (norbenzomorphan) derivatives elicit modest to high selectivity for the sigma 2 receptor over the sigma 1 receptor. Indeed, piperazine analog 8-(4-(3-ethoxy-3-oxopropyl)piperazin-1-yl)-1,3,4,5-tetrahydro-1,5-methanobenzazepine-2-carboxylate (SAS-1121) is 574-fold selective for the sigma 2 receptor over the sigma 1 receptor, thereby establishing it as one of the more subtype selective sigma 2 binding ligands reported to date. Emerging evidence has implicated the sigma 2 receptor in multiple health disorders, so the drug-like characteristics of many of the selective sigma 2 receptor ligands disclosed herein, coupled with their structural similarity to frameworks found in known drugs, suggest that norbenzomorphan analogs may be promising candidates for further development into drug leads.
Keywords: Sigma 2 receptor; norbenzomorphan; 1,5-methanobenzazepine; Sig2R/PGRMC1
Graphical Abstract
1. Introduction
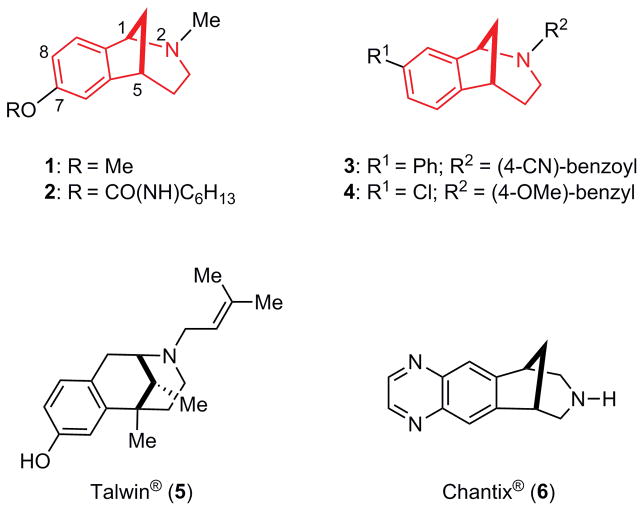
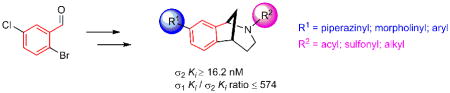
In a series of investigations directed toward discovering biologically active small molecules, we developed a general platform for the rapid synthesis of small collections of functionalized heterocyclic scaffolds that can be further diversified by cross-coupling reactions and refunctionalizations [1,2]. One class of heterocycles that piqued our interest is the substituted norbenzomorphan ring system (Fig. 1, red highlight in 1–4) [2], which has drug-like features, including low molecular weight and conformational rigidity [3,4]. Investigational compounds embodying this framework include 1, which induces antinociception in an animal pain model [5] and 2, which inhibits acetylcholinesterase in vivo [6]. Screening novel norbenzomorphans prepared in our lab led to the discovery of hit compounds with potentially useful medicinal properties. For example, amide 3 is an inhibitor of striatal-enriched protein tyrosine phosphatase (STEP) [2b], an enzyme that is overactive in Alzheimer’s disease (AD) [7], whereas benzylamine 4 is an antagonist of dopamine-3 [8], a target currently being evaluated for treating addiction [9].
Figure 1.
Norbenzomorphans 1-4 and structurally-related psychoactive drugs 5 and 6.
The structural resemblance of the norbenzomorphan framework to the closely related scaffolds in psychoactive drugs, including Talwin® (5) and Chantix® (6), suggests that norbenzomorphan derivatives may possess the favorable pharmacokinetic attributes required of leads for treating neurological disorders. Accordingly, a small set of norbenzomorphans were screened against a comprehensive panel of CNS-based proteins at the Psychoactive Drug Screening Program (PDSP; UNC-Chapel Hill) [10]. Although a number of compounds were identified that exhibit selective binding profiles, we were particularly intrigued by those with high affinity and subtype selectivity for sigma receptors [11], which had not been associated with this class of compounds.
Sigma receptors are a distinct class of non-GPCR receptors that are involved in a variety of critical cellular processes, including regulation of ion concentration, stabilization of cell surface receptors, and induction of apoptosis [12]. Two receptor subtypes, sigma 1 receptor (Sig1R) and sigma 2 receptor (Sig1R), have been identified, and whereas the former has been cloned and sequenced, the latter is not well characterized. Sig2R was reported to reside in the progesterone receptor membrane component 1 (PGRMC1) [13,14], and although this finding has not been universally accepted [15,16], the term Sig2R/PGRMC1 is commonly used to refer to this receptor, thus this convention will be adopted in this report.
This controversy notwithstanding, there is accumulating evidence that Sig2R/PGRMC1is involved in a number of disease states [17,18]. For example, Sig2R/PGRMC1is highly overexpressed in proliferating tumor cells and has been identified as an attractive target for cancer diagnostics. Because agonists of Sig2R/PGRMC1induce cell death in a variety of cancer cell lines, they are also garnering increasing interest as potential chemotherapeutics. Moreover, in several preliminary experiments, we have discovered that ligands that bind to Sig2R/PGRMC1 can exhibit CNS effects and traverse the blood brain barrier (BBB). For example, we discovered that several antagonists of σ2R are neuroprotective in a C. elegans model of neurodegeneration [19,20]. In a preliminary pharmacokinetic assessment, we also found that SAS-0132 (32) achieves a brain concentration of 3.8 μM within 3 h after a single subcutaneous injection (10 mg/kg) [11,21]. Sig2R/PGRMC1 was recently implicated in AD because it appears to mediate amyloid-β induced-synaptotoxicity [22,23]. Toward discovering brain penetrant Sig2R/PGRMC1 binding ligands that may hold promise for treating a spectrum of CNS disorders, we now describe a set of substituted norbenzomorphans that exhibit high selectivity for Sig2R/PGRMC1over Sig1R.
2. Chemistry
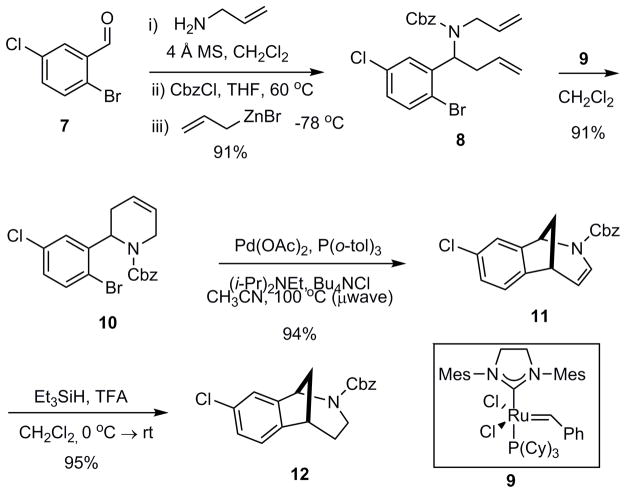
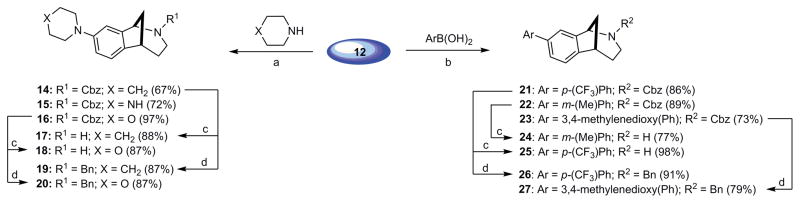
As a point of embarkation, we prepared the key intermediate 12 via a Mannich-type multicomponent assembly process (MCAP) followed by sequential ring closing metathesis, Heck cyclization, and olefin reduction as previously described (Scheme 1) [2b]. Scaffold 12 proved to be well-suited for generating a variety of analogs, including those represented by general structure 13 (Fig. 2). The aryl chloride functional handle present in 12 enabled derivatization via palladium catalyzed cross-coupling reactions to deliver analogs having a range of electrostatic properties and varying degrees of lipophilicity, such as the anilines 14–16 and the biaryls 21–23 (Scheme 2). The cyclic secondary amines piperidine, piperazine, and morpholine were selected as coupling partners in Buchwald-Hartwig reactions to provide analogs having amino groups at C(8) (Fig. 2) of the norbenzomorphan nucleus. Six-membered, cyclic amines were used in these studies to minimize conformational variables amongst the different aryl amino analogs, which vary primarily in the Lewis basic nature of the C(8) substituent. Boronic acid coupling partners for the Suzuki reactions were chosen to provide both electron-rich and electron-deficient biaryl products (Scheme 2). The Cbz group on N(2) (Fig. 2) of 14, 16 and 21–23 was then removed using iodotrimethylsilane (TMSI) followed by either acidic or basic workup as previously described [2] to give the corresponding tertiary N-benzyl compounds 19, 20, 26, and 27 and the secondary amines 17, 18, 24, and 25.
Scheme 1.
Synthesis of chloro norbenzomorphan 12.
Figure 2.
A variety of norbenzomorphan analogs prepared via cross-coupling and N-derivatization reactions.
Scheme 2.
Cross-coupling reactions of aryl chloride 12 to generate aniline and biaryl analogs.a
aReagents and Conditions: a) Pd(OAc)2, JohnPhos®, NaOt-Bu, toluene, 100 °C. b) Pd[P(t-Bu)3]2, Cs2CO3, 1,4-dioxane, 100 °C. c) TMSI, CH2Cl2, 0o C → rt, then HCl. d) TMSI, CH2Cl2, 0o C → rt, then NaHCO3 (aq).
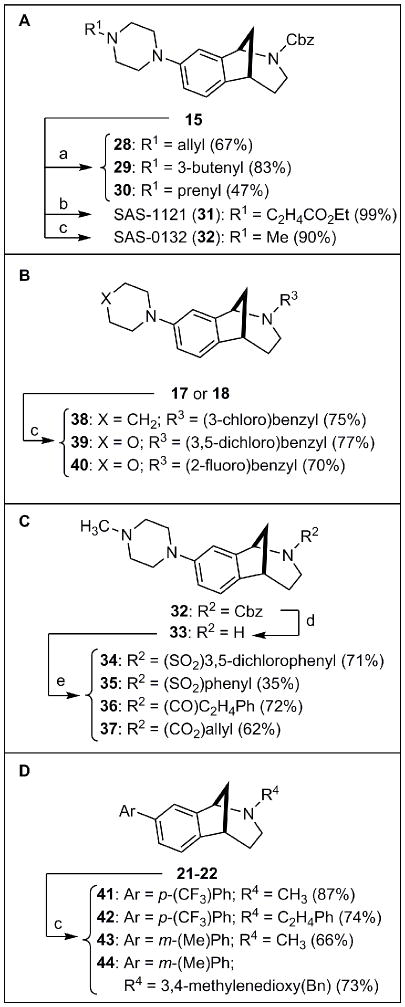
The substituents on the two nitrogen atoms of 15 were then diversified. In the event, the secondary amino group on the piperazine ring of 15 was first N-alkylated using standard procedures to give tertiary amines 28–30, SAS-1121 (31), and SAS-0132 (32) (Fig. 3A). Substitution at the carbamoyl nitrogen atom of 32 was then varied by removing the Cbz group using TMSI followed by an acidic workup to give 33 (Fig. 3C). Subsequent N-sulfonylation or N-acylation of 33 under standard conditions delivered 34–37. Reductive amination of 17 and 18 using several aryl aldehydes and sodium triacetoxyborohydride [Na(OAc)3BH] provided 38–40 (Fig. 3B). Similarly, reductive aminations of 21 and 22 led to analogs 41–44 (Fig. 3D).
Figure 3.
N-Functionalization Reactions.
Nitrogen functionalization reactions. Reagents and Conditions: (a) alkyl bromide, CH3CN, K2CO3; (b) ethyl acrylate, EtOH; (c) aldehyde, Na(OAc)3BH, CH3COOH, CH2Cl2; (d) TMSI, CH2Cl2, 0 °C → rt, then HCl; (e) acyl or sulfonyl chloride, CH2Cl2, Et3N. A. Alkylation and reductive amination of piperazine 15. B. Reductive amination of 2° amines 17 or 18. C. Acylation and sulfonylation of 2° amine 33. D. Reductive amination of biaryls 21 or 22.
3. Pharmacology and Drug-like Attributes
These novel norbenzomorphans were then screened against a comprehensive panel of CNS-based proteins at the PDSP (Table 1) [10]. A cursory examination of the results in Table 1 reveals a number of compounds that exhibit high affinity and subtype selectivity for Sig2R/PGRMC1versus Sig1R. Indeed, Sig2R/PGRMC1 affinity and subtype selectivity is maintained over a considerable range of substituents on the aromatic ring and the nitrogen atom of the norbenzomorphan nucleus (vide infra). Although prevailing models of the putative pharmacophore for Sig2R/PGRMC1 binding are generally restricted to a specific ligand class [24], the features that are common to structurally diverse Sig2R/PGRMC1 ligands (e.g., an ionizable nitrogen atom, a hydrogen bond acceptor, and hydrophobic aromatic residues) are present in the vast majority of these norbenzomorphans [25].
Table 1.
Sigma receptor affinity of norbenzomorphan analogs.
| |||||
|---|---|---|---|---|---|
| Cmpd | R1 | R2 | Sig1R Ki(nM)a pKi ± SEMb |
Sig2R Ki(nM) pKi ± SEM |
Ki(Sig1R)/Ki(Sig2R) |
| 12 | Cl |
|
* c | * | - |
| 16 |
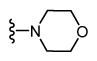
|
|
* | * | - |
| 20 |
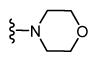
|
|
3,777 5.42 ± 0.09 |
1,034 5.99 ± 0.07 |
4 |
| 39 |
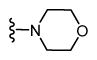
|
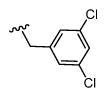
|
2,975 5.53 ± 0.09 |
92 7 ± 0.1 |
32 |
| 40 |
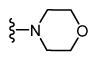
|
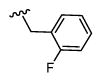
|
4,544 5.34 ± 0.09 |
1,258 5.9 ± 0.1 |
4 |
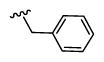
| 19 |
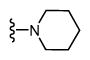
|
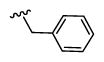
|
2,429 5.61 ± 0.06 |
318 6.5 ± 0.1 |
8 |
| 38 |
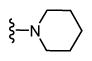
|
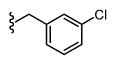
|
3,519 5.5 ± 0.1 |
746 6.1 ± 0.1 |
5 |
| 27 |
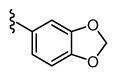
|
|
3,039 5.5 ± 0.1 |
723 6.14 ± 0.09 |
4 |
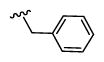
| 41 |
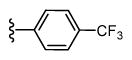
|
CH3 | 156 6.81 ± 0.09 |
43 7.36 ± 0.08 |
4 |
| 42 |
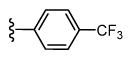
|
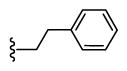
|
1,058 6.0 ± 0.1 |
166 6.8 ± 0.1 |
6 |
| 43 |
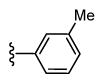
|
CH3 | 391 6.41 ± 0.08 |
133 6.88 ± 0.07 |
3 |
| 44 |
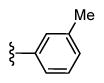
|
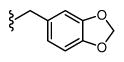
|
4,612 5.34 ± 0.08 |
828 6.08 ± 0.08 |
6 |
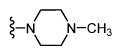
| SAS-0132 (32) |
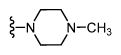
|
|
497 6.31 ± 0.06 |
71 7.1 ± 0.1 |
7 |
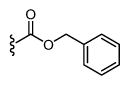
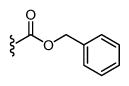
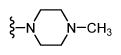
| 33 |
|
H | * | * | - |
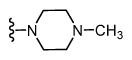
| 34 |
|
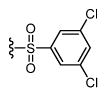
|
442 6.36 ± 0.06 |
27 7.58 ± 0.08 |
16 |
| 35 |
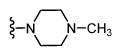
|
|
* | 764 6.12 ± 0.07 |
- |
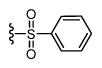
| 36 |
|
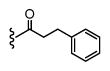
|
294 6.53 ± 0.08 |
357 6.45 ± 0.06 |
1 |
| 37 |
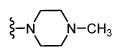
|
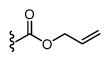
|
2105 5.68 ± 0.07 |
442 6.37 ± 0.07 |
5 |
| 28 |
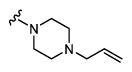
|
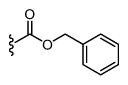
|
589 6.23 ± 0.06 |
34 7.47 ± 0.08 |
17 |
| 29 |
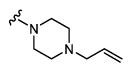
|
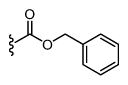
|
172 6.76 ± 0.05 |
16 7.79 ± 0.05 |
11 |
| 30 |
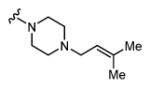
|
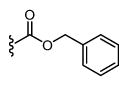
|
62 7.21 ± 0.05 |
23 7.64 ± 0.07 |
3 |
| SAS-1121 (31) |
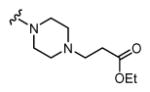
|
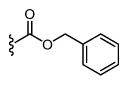
|
9294d 5.1 ± 0.1 |
16.2e 7.8 ± 0.16 |
574 |
Ki values obtained from non-linear regression of radioligand competition binding isotherms;
SEM calculated for pKi;
Less than 50% inhibition of radioligand binding with 10 μM test ligand;
average of two IC50 determinations;
average of three IC50 determinations.
A brief summary of the results presented in Table 1 is instructive. Within the morpholine series comprising compounds 16, 20, 39, and 40, there appears to be a requirement for a second basic nitrogen atom in the molecule. This tentative assessment is based upon the observation that 16 does not bind to the receptor, whereas the two N-benzyl derivatives 20 and 40 exhibit modest Sig2R/PGRMC1 affinity (1,034 nM and 1,258 nM) and a 4-fold preference for σ2R relative to σ1R. The 3,5-dichlorobenzyl analog 39 benefits from a large increase in Sig2R/PGRMC1 binding affinity (92 nM) coupled with a 32-fold increase in selectivity for Sig2R/PGRMC1 over Sig1R. The aryl piperidine derivative 19 displays moderate σ2R affinity (318 nM) and about 8-fold subtype selectivity favoring Sig2R/PGRMC1, and replacing the benzyl group of 19 with a 3-chlorobenzyl substituent (e.g., 38) affects a marginal decrease in both Sig2R/PGRMC1 affinity and selectivity. For the series of biaryl compounds 27 and 41–44, SigR affinity appears to be markedly dependent upon the size of the alkyl group at N(2). For example, the N-methyl derivatives 41 and 43 exhibit significantly enhanced binding affinity at both Sig1R and Sig2R/PGRMC1 relative to 42 and 44.
When the C(8) position of the norbenzomorphan ring is substituted with a piperazino moiety, Sig2R/PGRMC1 binding affinity is typically high, and the subtype selectivity can be tuned by altering either R2 or the alkyl group on the aliphatic nitrogen atom of the piperazine ring. Changing the nature of the alkyl group leads to only modest variations in Sig2R/PGRMC1 affinities, whereas the effects on Sig1R affinity are more pronounced. This is dramatically illustrated by comparing SAS-1121 (31), which exhibits 574-fold selectivity for Sig2R/PGRMC1, with other members of this series. To our knowledge, this represents one of the most selective Sig2R/PGRMC1 ligands reported to date [26]. Whereas replacing the Cbz group of SAS-0132 (32) with a hydrocinnamoyl (e.g., 36) or an allyloxycarbonyl (e.g., 37) moiety leads to a loss in σ2R binding affinity and selectivity, substituting a 3,5-dichlorobenzenesulfonamide group for Cbz affords a 2- to 3-fold increase in Sig2R/PGRMC1 binding affinity and selectivity. Notably, the corresponding nor-chloro analog 35 displays much lower affinity for both SigR subtypes, and the presence of a secondary amino group (e.g., 33) completely abolished binding at Sig1R and Sig2R/PGRMC1.
The binding affinities for the compounds in Table 1 reveal that an array of different substituents at R1 and R2 are tolerated in ligands having modest to excellent preference for Sig2R/PGRMC1 over Sig1R, and a number of compounds exhibit < 50 nM affinity for Sig2R/PGRMC1. Notably, many compounds display low affinity relative to Sig2R/PGRMC1 at a broad range of other CNS proteins, including serotonin, adrenergic, dopamine, opioid, and neurotransmitter transporters (See Table S1, Supplementary Information). Some compounds, however, display significant affinity for non-sigma binding sites. For example, 34 exhibits high affinity for 5HT1D (Ki = 195 nM), 5HT6 (Ki = 82 nM), and α2a (Ki = 162 nM), and 41 binds with sub-micromolar affinity to α2c (Ki = 755 nM) and NET (Ki = 524 nM). As might be expected, the more promiscuous ligands are typically highly lipophilic (i.e., ClogP > 5) [27].
A majority of the sigma ligands described herein have properties suggestive of moderate solubility and good membrane- and BBB permeability [28]. For example, the ClogD (pH 7.4) values range from 2.9–4.5 for the bulk of the compound set, so a high degree of CNS exposure may be anticipated (See Table S2, Supplementary Information). This prediction is supported by the observation that SAS-0132 (32) is highly brain penetrant. Moreover, ligands in this collection lack a highly basic amine function (e.g., pKa > 9), which can be predictive of low hERG channel inhibition [29]. These data collectively suggest that R1 and R2 on the norbenzomorphan scaffold can be tuned for Sig2R/PGRMC1 binding to generate promising leads for drug discovery.
4. Conclusion
In summary, a modular synthetic platform was used to rapidly access a variety of substituted norbenzomorphans that exhibit high potency and selectivity for Sig2R/PGRMC1 relative to Sig1R. Notably, it appears to be possible to modulate SigR subtype selectivity by varying the groups at C(8) and N(2) of the norbenzomorphan scaffold. SAS-1121 displays a 574-fold preference for Sig2R/PGRMC1 over Sig1R, suggesting that exceptional Sig2R/PGRMC1 subtype selectivity can be achieved. A representative ligand, SAS-0132, is highly brain penetrant [11] and, like many of the Sig2R/PGRMC1 subtype selective ligands disclosed herein, it has molecular attributes likely to impart desirable absorption, distribution, metabolism, and excretion (ADME) properties [3,4]. Although the important question of whether Sig2R/PGRMC1 can be safely targeted in man remains unanswered, it is significant that the EU approved anxiolytic/antidepressant opipramol has appreciable affinity for Sig2R/PGRMC1 and has been safely used for decades [30]. Previous clinical trials with investigational compounds that bind to Sig2R/PGRMC1, such as siramesine and rimcazole, suggest that pharmacological modulation of Sig2R/PGRMC1 may be safe in man [31]. Ongoing clinical trials with drug candidates that bind to Sig2R/PGRMC1 will further determine if the receptor is drug-gable. For instance, Minerva Neuroscience® has advanced MIN-101 to phase IIb trials for schizophrenia [32], and Cognition Therapeutics® recently launched a phase I study with their AD drug lead, Cog1812 [33]. The discovery of novel Sig2R/PGRMC1 ligands with drug-like features is thus an important component to validating the clinical significance of this receptor.
Supplementary Material
Acknowledgments
We thank the National Institutes of Health (GM 86192 and GM 24539) and the Robert A. Welch Foundation (F-0652) for their generous support of this work. We are also grateful to Dr. Richard Pederson (Materia, Inc.) for catalyst support and to Dr. Mehrdad Shamloo (Stanford Behavioral and Functional Neuroscience Lab) for conducting pharmacokinetic studies of 32. Determination of receptor binding profiles was generously provided by the National Institute of Mental Health’s Psychoactive Drug Screening Program, Contract # HHSN-271-2013-00017-C (NIMH PDSP). The NIMH PDSP is Directed by Bryan L. Roth MD, PhD at the University of North Carolina at Chapel Hill and Project Officer Jamie Driscoll at NIMH, Bethesda MD, USA.
Footnotes
Associated Content
General experimental procedures and full characterization is provided for all new, representative compounds. 1H NMR and MS data are provided for all other new compounds.
References
- 1.(a) Sunderhaus JD, Dockendorff C, Martin SF. Applications of multicomponent reactions for the synthesis of diverse heterocyclic scaffolds. Org Lett. 2007;9:4223–4226. doi: 10.1021/ol7018357. [DOI] [PubMed] [Google Scholar]; (b) Sunderhaus JD, Dockendorff C, Martin SF. Synthesis of diverse heterocyclic scaffolds via tandem additions to imine derivatives and ring-forming reactions. Tetrahedron. 2009;65:6454–6469. doi: 10.1016/j.tet.2009.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Donald JR, Martin SF. Synthesis and diversification of 1,2,3-triazole-fused 1,4-benzodiazepine scaffolds. Org Lett. 2011;13:852–855. doi: 10.1021/ol1028404. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Donald JR, Wood RR, Martin SF. Application of a sequential multicomponent assembly process/Huisgen cycloaddition strategy to the preparation of libraries of 1,2,3-triazole-fused 1,4-benzodiazepines. ACS Comb Sci. 2012;14:135–143. doi: 10.1021/co2002087. [DOI] [PMC free article] [PubMed] [Google Scholar]; (e) Hardy S, Martin SF. Multicomponent assembly and diversification of novel heterocyclic scaffolds derived from 2-arylpiperidines. Org Lett. 2011;13:3102–3105. doi: 10.1021/ol201010s. [DOI] [PMC free article] [PubMed] [Google Scholar]; (f) Granger BA, Kaneda K, Martin SF. Multi-component assembly strategies for the synthesis of diverse tetrahydroisoquinoline scaffolds. Org Lett. 2011;13:4542–4545. doi: 10.1021/ol201739u. [DOI] [PMC free article] [PubMed] [Google Scholar]; (g) Granger BA, Wang Z, Kaneda K, Fang Z, Martin SF. Multicomponent assembly process for the synthesis of diverse yohimbine and corynanthe alkaloid analogs. ACS Comb Sci. 2013;15:379–386. doi: 10.1021/co400055b. [DOI] [PMC free article] [PubMed] [Google Scholar]; (h) Granger BA, Kaneda K, Martin SF. Libraries of 2,3,4,6,7,11b-hexahydro-1H-pyrido[2,1-a]isoquinolin-2-amine derivatives via a multicomponent assembly process/1,3-dipolar cycloaddition strategy. ACS Comb Sci. 2012;14:75–79. doi: 10.1021/co2001636. [DOI] [PMC free article] [PubMed] [Google Scholar]; (i) Sahn JJ, Granger BA, Martin SF. Evolution of a strategy for preparing bioactive small molecules by sequential multicomponent assembly processes, cyclizations, and diversification. Org Biomol Chem. 2014;12:7659–7672. doi: 10.1039/c4ob00835a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.(a) Sahn JJ, Su J, Martin SF. Facile and unified approach to skeletally diverse, privileged scaffolds. Org Lett. 2011;13:2590–2593. doi: 10.1021/ol200709h. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Sahn JJ, Martin SF. Expedient synthesis of norbenzomorphan library via multicomponent assembly process coupled with ring-closing reactions. ACS Combi Sci. 2012;14:496–502. doi: 10.1021/co300068a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lipinski CA, Lombardo F, Dominy BW, Feeney PJ. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv Drug Delivery Rev. 2001;46:3–26. doi: 10.1016/s0169-409x(00)00129-0. [DOI] [PubMed] [Google Scholar]
- 4.Veber DF, Johnson SR, Cheng HY, Smith BR, Ward KW, Kopple KD. Molecular properties that influence the oral bioavailability of drug candidates. J Med Chem. 2002;45:2615–2623. doi: 10.1021/jm020017n. [DOI] [PubMed] [Google Scholar]
- 5.Jacobson AE, Mokotoff M. Azabicyclochemistry I “Synthesis of 1,5-methano-7-methoxy-2,3,4,5-tetrahydro-1H-2-benzazepines. B-norbenzomorphans. J Med Chem. 1970;13:7–9. doi: 10.1021/jm00295a002. [DOI] [PubMed] [Google Scholar]
- 6.Chen YL, Liston D, Nielsen J, Chapin D, Dunaiskis A, Hedberg K, Ives J, Johnson J, Jr, Jones S. Syntheses and anticholinesterase activity of tetrahydrobenzazepine carbamates. J Med Chem. 1994;37:1996–2000. doi: 10.1021/jm00039a013. [DOI] [PubMed] [Google Scholar]
- 7.Xu J, Chatterjee M, Baguley TD, Brouillette J, Kurup P, Ghosh D, Kanyo J, Zhang Y, Seyb K, Ononenyi C, Foscue E, Anderson GM, Gresack J, Cuny GD, Glicksman MA, Greengard P, Lam TT, Tautz L, Nairn AC, Ellman JA, Lombroso PJ. Inhibitor of the tyrosine phosphatase STEP reverses cognitive deficits in a mouse model of Alzheimer’s disease. PLoS biology. 2014;12:e1001923. doi: 10.1371/journal.pbio.1001923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.See:http://pubchem.ncbi.nlm.nih.gov/assay/asis.cgi?reqid=1071805557102354222&q=r&version=1.1
- 9.Furman CA, Roof RA, Moritz AE, Miller BN, Doyle TB, Free RB, Banala AK, Paul NM, Kumar V, Sibley CD, Newman AH, Sibley DR. Investigation of the binding and functional properties of extended length D3 dopamine receptor-selective antagonists. Eur Neuropsychopharmacol. 2015;25:1448–1461. doi: 10.1016/j.euroneuro.2014.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Binding affinities determined through competition binding assay in the presence of radioligand. See Supplementary Information for summary. Detailed protocols can be found within the PDSP Assay Protocol Book (version II), see: http://pdsp.med.unc.edu/pdspw/binding.php.
- 11.Martin SF, Sahn JJ, Scott L, Pierce-Shimomura JT. WO2015009742A2. Preparation of norbenzomorphan compounds as sigma receptor inhibitor and methods for treating cancer, neurological disorders, ethanol withdrawal, anxiety, depression, and neuropathic pain. 2015
- 12.Matsumoto RR, Bowen WD, Su T-P, editors. Sigma receptors: Chemistry, cell biology, and clinical implications. Springer; New York: 2007. [Google Scholar]
- 13.Xu J, Zeng C, Chu W, Pan F, Rothfuss JM, Zhang F, Tu Z, Zhou D, Zeng D, Vangveravong S, Johnston F, Spitzer D, Chang KC, Hotchkiss RS, Hawkins WG, Wheeler KT, Mach RH. Identification of the PGRMC1 protein complex as the putative sigma-2 receptor binding site. Nat Commun. 2011;2:380. doi: 10.1038/ncomms1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cahill MA. Progesterone receptor membrane component 1: an integrative review. J Steroid Biochem Mol Biol. 2007;105:16–36. doi: 10.1016/j.jsbmb.2007.02.002. [DOI] [PubMed] [Google Scholar]
- 15.Abate C, Niso M, Infantino V, Menga A, Berardi F. Elements in Support of the ‘Non-Identity’ of the PGRMC1 Protein with the σ2 Receptor. Eur J Pharmacol. doi: 10.1016/j.ejphar.2015.03.067. In Press. [DOI] [PubMed] [Google Scholar]
- 16.Chu UB, Ramachandran S, Hajipour AR, Ruoho AE. Photoaffinity Labeling of the Sigma-1 Receptor with N-[3-(4-Nitrophenyl)propyl]-N-dodecylamine: Evidence of Receptor Dimers. Biochemistry. 2013;52:859–868. doi: 10.1021/bi301517u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guo L, Zhen X. Sigma-2 receptor ligands: neurobiological effects. Cur Med Chem. 2015;22:989–1003. doi: 10.2174/0929867322666150114163607. [DOI] [PubMed] [Google Scholar]
- 18.Mach RH, Zeng C, Hawkins WG. The sigma2 receptor: A Novel Protein for the Imaging and Treatment of Cancer. J Med Chem. 2013;56:7137–7160. doi: 10.1021/jm301545c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chan JZ, Pierce-Shimomura J, Martin SF, Scott LL, Sahn JJ. Investigating sigma 2 receptor ligands for the targeted therapeutics of Alzheimer’s disease by utilizing a novel C. elegans model of AD. American Chemical Society; 2014. p. MEDI-97. [Google Scholar]
- 20.Sahn JJ, Scott LL, Zuniga G, Satarasinghe T, Wong TA, Ardestani PM, Pierce-Shimomura J, Shamloo M, Martin SF. Sigma Receptors: Emerging Roles in Health and Disease. Chicago, IL: Society for Neuroscience; 2015. Development of Novel Therapeutics Targeting Sig2R/PGRMC1 for Alzheimer’s Disease. Program no. 555.05. Neuroscience 2015 Abstracts. Online. [Google Scholar]
- 21.The pharmacokinetic study of 32 was performed at the Stanford Behavioral and Functional Neuroscience Laboratory; Director Mehrdad Shamloo, Ph.D.
- 22.Izzo NJ, Staniszewski A, To L, Fa M, Teich AF, Saeed F, Wostein H, Walko T, 3rd, Vaswani A, Wardius M, Syed Z, Ravenscroft J, Mozzoni K, Silky C, Rehak C, Yurko R, Finn P, Look G, Rishton G, Safferstein H, Miller M, Johanson C, Stopa E, Windisch M, Hutter-Paier B, Shamloo M, Arancio O, LeVine H, 3rd, Catalano SM. Alzheimer’s therapeutics targeting amyloid beta 1-42 oligomers I: Abeta 42 oligomer binding to specific neuronal receptors is displaced by drug candidates that improve cognitive deficits. PloS one. 2014;9:e111898. doi: 10.1371/journal.pone.0111898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Izzo NJ, Xu J, Zeng C, Kirk MJ, Mozzoni K, Silky C, Rehak C, Yurko R, Look G, Rishton G, Safferstein H, Cruchaga C, Goate A, Cahill MA, Arancio O, Mach RH, Craven R, Head E, LeVine H, 3rd, Spires-Jones TL, Catalano SM. Alzheimer’s therapeutics targeting amyloid beta 1-42 oligomers II: Sigma-2/PGRMC1 receptors mediate Abeta 42 oligomer binding and synaptotoxicity. PloS one. 2014;9:e111899. doi: 10.1371/journal.pone.0111899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Laurini E, Zampieri D, Mamolo MG, Vio L, Zanette C, Florio C, Posocco P, Fermeglia M, Pricl S. A 3D-pharmacophore model for sigma2 receptors based on a series of substituted benzo[d]oxazol-2(3H)-one derivatives. Bioorg Med Chem Lett. 2010;20:2954–2957. doi: 10.1016/j.bmcl.2010.03.009. [DOI] [PubMed] [Google Scholar]
- 25.Rhoades DJ, Kinder DH, Mahfouz TM. A comprehensive ligand based mapping of the σ2 receptor binding pocket. Med Chem. 2014;10:98–121. doi: 10.2174/1573406409999131119103621. [DOI] [PubMed] [Google Scholar]
- 26.For a detailed accounting of selective Sig2R/PGRMC1 ligands, see: Huang YS, Lu HL, Zhang LJ, Wu Z. Sigma-2 receptor ligands and their perspectives in cancer diagnosis and therapy. Med Res Rev. 2014;34:532–66. doi: 10.1002/med.21297.Narayanan S, Bhat R, Mesangeau C, Poupaert JH, McCurdy CR. Early development of sigma-receptor ligands. Future Med Chem. 2011;3:79–94. doi: 10.4155/fmc.10.279.
- 27.ClogP, ClogD, and pKa calculations were performed with ACD I-Lab software: http://www.acdlabs.com/resources/ilab/
- 28.Kerns EH, Di L. Drug-like Properties: Concepts, Structure Design and Methods: from ADME to Toxicity Optimization. Academic Press; Boston: 2008. [Google Scholar]
- 29.Wager TT, Chandrasekaran RY, Hou X, Troutman MD, Verhoest PR, Villalobos A, Will Y. Defining desirable central nervous system drug space through the alignment of molecular properties, in vitro ADME, and safety attributes. ACS Chem Neurosci. 2010;1:420–434. doi: 10.1021/cn100007x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Möller HJ, Volz HP, Reimann IW, Stoll KD. Opipramol for the Treatment of Generalized Anxiety Disorder. A Placebo-Controlled Trial Including an Alprazolam Treated Group. J Clinical Phramacol. 2001;21:59–65. doi: 10.1097/00004714-200102000-00011. [DOI] [PubMed] [Google Scholar]
- 31.(a) Volz HP, Stoll KD. Clinical trials with sigma ligands. Pharmacopsychiatry. 2004;37(Suppl 3):S214–20. doi: 10.1055/s-2004-832680. [DOI] [PubMed] [Google Scholar]; (b) Gilmore DL, Liu Y, Matsumoto RR. ”Review of the pharmacological and clinical profile of rimcazole. CNS Drug Rev. 2004;10:1–22. doi: 10.1111/j.1527-3458.2004.tb00001.x. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Luthringer RH, Pellegrini L, Karabelas AN. WO2012012543A1. Methods of using cyclic amide derivatives to treat sigma receptor-mediated disorders. 2012
- 32.See: http://ir.minervaneurosciences.com/releasedetail.cfm?ReleaseID=947593
- 33.See: http://www.cogrx.com/docs/US_biotech_Cognition_Therapeutics_tests...r%E2%80%99s_drug_in_Australia_The_Australian.pdf
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.