Abstract
Liver regeneration is a highly orchestrated process which can be regulated by microRNAs (miRNAs, miRs), though the mechanisms are largely unclear. This study was aimed to identify miRNAs responsible for hepatocyte proliferation during liver regeneration. Here we detected a marked elevation of miR-382 in the mouse liver at 48 hrs after partial hepatectomy (PH-48h) using microarray analysis and qRT-PCRs. miR-382 overexpression accelerated the proliferation and the G1 to S phase transition of the cell cycle both in mouse NCTC1469 and human HL7702 normal liver cells, while miR-382 downregulation had inverse effects. Moreover, miR-382 negatively regulated PTEN expression and increased Akt phosphorylation both in vitro and in vivo. Using PTEN siRNA and Akt activator/inhibitor, we further found that PTEN inhibition and Akt phosphorylation were essential for mediating the promotive effect of miR-382 in the proliferation and cell growth of hepatocytes. Collectively, our findings identify miR-382 as a promoter for hepatocyte proliferation and cell growth via targeting PTEN-Akt axis which might be a novel therapeutic target to enhance liver regeneration capability.
Keywords: microRNA, liver regeneration, proliferation, PTEN, Akt
INTRODUCTION
Liver possesses a tremendous regenerative capability after injury or surgical resection [1, 2]. In rodent model of 70% partial hepatectomy (PH), adult hepatocytes immediately enter G1 phase and transverse to S phase of the cell cycle with a peak of DNA synthesis at 24-40 hrs after PH, and ultimately grow to its pre-resection mass in 7-10 days [3, 4]. However, liver regenerative capability can usually be impaired under certain circumstances such as liver cirrhosis and liver failure [5, 6], highlighting the necessity to identify novel approaches to enhance liver regenerative capacity.
MicroRNAs (miRNAs, miRs), a group of small non-coding RNAs, are negative regulators of their target genes at posttranscriptional level, principally through combining to the 3′-UTR of target messenger RNA (mRNA) and reducing mRNA stability and/or translation [7-10]. Recently, miRNAs have increasingly been reported to control the process of liver regeneration [11, 12]. Among the aberrantly expressed miRNAs, miR-21 [11, 13-15], −23b [16], −122 [17], −203 [18] and −221 [19] have been shown as promoters for hepatocyte proliferation, while miR-26a [20], −33 [21], −34a [22], −127 [23], −150 [24] and −378 [11] have inverse effect. However, the mechanisms mediating the regulatory effect of miRNAs in liver regeneration are still largely unclear.
In this study, we found a significant elevation of miR-382 in the mouse liver at 48 hrs after 70% PH using microarrays and quantitative reverse transcription-polymerase chain reactions (qRT-PCRs). Our data showed that miR-382 could promote hepatocyte proliferation and cell growth in vitro. Additionally, miR-382 overexpression negatively correlated with PTEN expression at post transcriptional level both in vivo and in vitro. Using PTEN siRNA and Akt activator/inhibitor, we further confirmed that PTEN inhibition and Akt phosphorylation were essential for mediating the promotive effect of miR-382 in hepatocyte proliferation and cell growth. Collectively, these data indicate that miR-382 is a promoter for hepatocyte proliferation and liver regeneration via targeting PTEN-Akt axis.
RESULTS
miR-382 is upregulated in mouse regenerating liver
Our group has previously demonstrated that hepatocyte proliferation peaked at 48 hrs after PH in the mouse liver as represented by a peak of PCNA protein level and EdU positive cells [4]. In the current study, the microarray analysis was used to evaluate miRNA profiles in the mouse liver during the proliferative phase of liver regeneration (PH-48h) in comparison to those in the control liver (PH-0h). As shown in the heat map (Figure 1A), miR-1946a, -296-3p, -504-3p, -128-3p, -674-3p, -421-3p, -3473a, -1907, and -382-5p were found to be upregulated, while miR-3068-3p, -664-5p, -342-5p, -5100, and -720 downregulated in the mouse liver at 48 hrs post PH (for data see Table 1). Next, qRT-PCRs were further conducted, verifying that the expression of miR-382-5p was significantly increased, while miR-3068-3p, -664-5p, and -5100 decreased in the mouse liver at PH-48h (Figure 1B). As miR-504-3p was found to be downregulated in the mouse liver at PH-48h using qRT-PCR which was contrary to the result of microarrays (Figure 1A and 1B), miR-504-3p was then excluded for subsequent functional assays. Among the aberrantly expressed miRNAs, miR-3068-3p, -664-5p, and -5100 did not impact the proliferation of mouse NCTC1469 liver cells in vitro as measured by EdU incorporation assay (data not shown). Noteworthy, miR-382-5p (previous IDs: miR-382) overexpression or suppression was found to be effective to modify hepatocyte proliferation (for detail see following results). Thus, miR-382 has been screened out for further investigation regarding its role in hepatocyte proliferation and liver regeneration in the present study.
Figure 1. The miRNA profiles in the mouse liver at 48 hrs after PH (PH-48h) compared to those at PH-0h.

A. Heat map demonstrated the aberrantly expressed microRNAs in the mouse liver at PH-48h vs. those in the control liver at PH-0h (n=4). B. qRT-PCRs showed that miR-382-5p was upregulated, while miR-504-3p, -3068-3p, -664-5p, and -5100 were downregulated in the mouse liver at PH-48h (n=5). *, P<0.05; **, P<0.01.
Table 1. Microarray analysis showing dysregulated miRNAs in the mouse liver at 48 hrs after PH (PH-48h) versus those in the control liver at PH-0h (n=4).
| Systematic Name | P value | Fold-change | Regulation |
|---|---|---|---|
| mmu-miR-1946a | 0.014 | 5.238 | up |
| mmu-miR-3068-3p | 0.013 | 4.103 | down |
| mmu-miR-296-3p | 0.042 | 3.958 | up |
| mmu-miR-504-3p | 0.025 | 3.299 | up |
| mmu-miR-128-3p | 0.036 | 2.881 | up |
| mmu-miR-664-5p | 0.037 | 2.656 | down |
| mmu-miR-674-3p | 0.022 | 2.361 | up |
| mmu-miR-342-5p | 0.031 | 2.358 | down |
| mmu-miR-421-3p | 0.003 | 2.355 | up |
| mmu-miR-3473a | 0.013 | 2.279 | up |
| mmu-miR-5100 | 0.044 | 2.240 | down |
| mmu-miR-720 | 0.036 | 2.185 | down |
| mmu-miR-1907 | 0.023 | 2.071 | up |
| mmu-miR-382-5p | 0.030 | 2.001 | up |
miR-382 promotes hepatocyte proliferation and cell growth in vitro
To investigate the effects of miR-382 on the cell growth and proliferation of hepatocytes, miR-382 mimics, inhibitor, or their negative controls were transfected to mouse NCTC1469 and human HL7702 normal liver cells, respectively. MiR-382 mimics increased, while miR-382 inhibitor reduced miR-382 level in NCTC1469 and HL7702 liver cells, confirming that these mimics and inhibitor took effects (Figures 2A and 3A). CCK-8 cell counting, EdU cell proliferation assay, and Ki67 immunostaining showed that miR-382 mimics promoted, while miR-382 inhibitor reduced the proliferation of NCTC1469 liver cells (Figure 2B-2D). We further found that miR-382 overexpression was associated with a reduced cell population in G1 phase and an increased cell population in S phase using flow cytometry, indicating that miR-382 promoted the transition of NCTC1469 cell population from G1 phase to S phase of the cell cycle, while miR-382 downregulation had inverse effect (Figure 2E). Meanwhile, the promotive effect of miR-382 in the proliferation and cell growth of hepatocytes were also found in human HL7702 liver cells (Figure 3B-3E). Thus, miR-382 is validated as a promoter for cell growth and proliferation of hepatocytes in vitro.
Figure 2. miR-382 promotes the proliferation and cell growth of mouse NCTC1469 liver cells.

A. qRT-PCR analysis for miR-382 level in NCTC1469 cells transfected with miR-382 mimics (miR-382-m), miR-382 inhibitor (miR-382-i), or their respective negative controls (nc-m or nc-i) (n=5). CCK-8 cell counting (n=10) B., EdU (green) cell proliferation assay (n=5) C., and Ki67 (red) immunostaining (n=5) D. demonstrated that miR-382 mimics promoted while miR-382 inhibitor reduced NCTC1469 cell proliferation. Nuclei were counterstained with Hoechst (blue). Original magnification 100x, scale bar = 50 μm. E. Flow cytometry showed that miR-382 mimics induced a G1 to S phase transition of the cell cycle of NCTC1469 cells, while miR-382 inhibitor had inverse effect (n=5). *, P<0.05; **, P<0.01.
Figure 3. miR-382 promotes the proliferation and cell growth of human HL7702 liver cells.
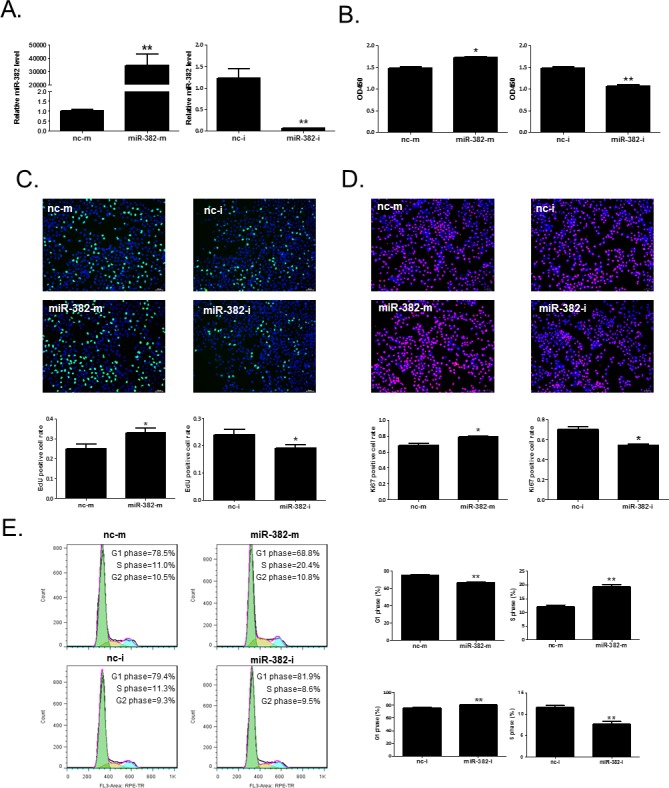
A. qRT-PCR analysis for miR-382 level in HL7702 cells transfected with miR-382 mimics (miR-382-m), miR-382 inhibitor (miR-382-i), or their respective negative controls (nc-m or nc-i) (n=5). CCK-8 cell counting (n=10) B., EdU (green) cell proliferation assay (n=5) C., and Ki67 (red) immunostaining (n=5) D. demonstrated that miR-382 mimics promoted while miR-382 inhibitor reduced HL7702 cell proliferation. Nuclei were counterstained with Hoechst (blue). Original magnification 100x, scale bar = 50 μm. E. Flow cytometry showed that miR-382 mimics induced a G1 to S phase transition of the cell cycle of HL7702 cells, while miR-382 inhibitor had inverse effect (n=5). *, P<0.05; **, P<0.01.
miR-382 overexpression negatively correlates with PTEN protein level and parallels with increased Akt phosphorylation both in vitro and in vivo
PTEN has previously been confirmed as a direct target of miR-382 in HIF-1α-stimulated vascular endothelial cells [25]. Here we demonstrated that miR-382 mimics reduced, while miR-382 inhibitor increased PTEN expression at protein level in NCTC1469 liver cells (Figure 4A and 4B). It is well known that Akt phosphorylation is negatively regulated by PTEN, which contributes to the cell growth and proliferation [26, 27]. We further found that miR-382 overexpression increased Akt phosphorylation in NCTC1469 cells, while miR-382 downregulation showed inverse effect (Figure 4A and 4B). The total Akt protein level was found unchanged in NCTC1469 cells either with miR-382 mimics or inhibitor transfection (Figure 4A and 4B). Intriguingly, PTEN expression was also found to be reduced, while Akt phosphorylation was enhanced in the mouse liver at PH-48h compared to control mouse liver (PH-0h), though total Akt was not altered (Figure 4C). These data indicate a potential relationship between miR-382 overexpression and the PTEN/Akt signaling pathway during the proliferative phase of liver regeneration that might contribute to the cell growth and proliferation of hepatocytes.
Figure 4. miR-382 negatively correlates with PTEN expression at protein level both in vitro and in vivo.
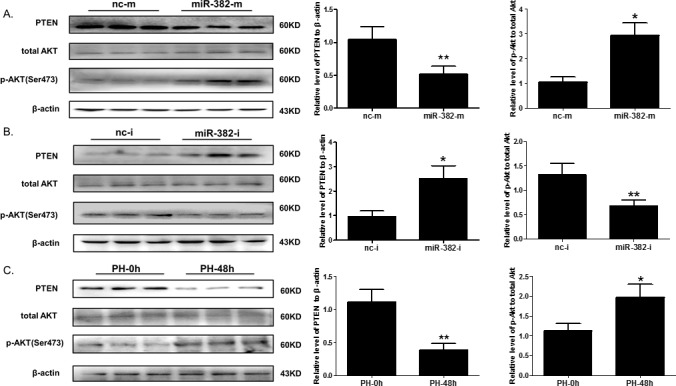
A. Western blot analysis for PTEN, p-AKT, and total AKT in NCTC1469 cells transfected with miR-382 mimics (miR-382-m) or negative control (nc-m) (n=3). B. Western blot analysis for PTEN, p-AKT, and total AKT in NCTC1469 cells transfected with miR-382 inhibitor (miR-382-i) or negative control (nc-i) (n=3). C. Western blot analysis for PTEN, p-AKT, and total AKT in the mouse liver at 48 hrs after PH (PH-48h) compared with control mouse liver (PH-0h) (n=3). β-actin was used as loading control. *, P<0.05; **, P<0.01.
miR-382 promotes hepatocyte proliferation and cell growth via targeting PTEN
Using siRNA-PTEN (sequence-01 or -02), the PTEN mRNA and protein levels were significantly reduced in NCTC1469 liver cells (Figure 5A and 5B). CCK-8 cell counting, EdU cell proliferation assay, and Ki67 immunostaining showed that NCTC1469 cell proliferation was reduced by miR-382 inhibitor, while increased by siRNA-PTEN-01 or -02 (Figure 5C-5E). Meanwhile, miR-382 inhibitor caused a G1 phase arrest in NCTC1469 cells, while siRNA-PTEN-01 or -02 induced a transition of the cell population from G1 to S phase (Figure 5F). More importantly, co-transfection with miR-382 inhibitor and siRNA-PTEN (sequence-01 or -02) abolished the inhibitory effect of miR-382 inhibitor on hepatocyte proliferation and cell growth (Figure 5C-5F).
Figure 5. siRNA-PTEN reverses miR-382 inhibition-induced NCTC1469 cell growth arrest.
siRNA-PTEN (sequence-01 or -02) reduced PTEN mRNA A. and protein B. levels in NCTC1469 cells as measured by qRT-PCRs (n=5) and Western blot analysis (n=3). CCK-8 cell counting (n=10) C., EdU (green) cell proliferation assay (n=5) D., and Ki67 (red) immunostaining (n=5) E. demonstrated that siRNA-PTEN (sequence-01 or -02) reversed the suppressive effect of miR-382 inhibitor (miR-382-i) on the proliferation of NCTC1469 cells. Nuclei were counterstained with Hoechst (blue). Original magnification 100x, scale bar = 50 μm. F. Flow cytometry showed that miR-382 inhibitor-induced G1 phase arrest of NCTC1469 cells was significantly abolished by siRNA-PTEN (sequence-01 or -02) (n=5). *, P<0.05 vs. control; **, P<0.01 vs. control; #, P<0.05 vs. miR-382-i; ##, P<0.01 vs. miR-382-i.
To further examine to which extent PTEN modulation mediates the role of miR-382 on hepatocyte proliferation, miR-382 mimics and siRNA-PTEN were co-transfected to NCTC1469 liver cells. Using CCK-8 cell counting, EdU cell proliferation assay, and Ki67 immunostaining, we found that either miR-382 mimics or siRNA-PTEN (sequence-01 or -02) promoted hepatocyte proliferation (Figure 6A-6C). Transfection with miR-382 mimics or siRNA-PTEN (sequence-01 or -02) alone also induced a transition of cell population from G1 to S phase in NCTC1469 cells (Figure 6D). However, co-transfection with miR-382 mimics and siRNA-PTEN did not further enhance the proliferation or cell growth of hepatocytes (Figure 6A-6D). These data suggest that the promotive effect of miR-382 on hepatocyte proliferation and cell growth is closely related to PTEN inhibition.
Figure 6. siRNA-PTEN does not further enhance the promotive effect of miR-382 on the proliferation and cell growth of NCTC1469 cells.
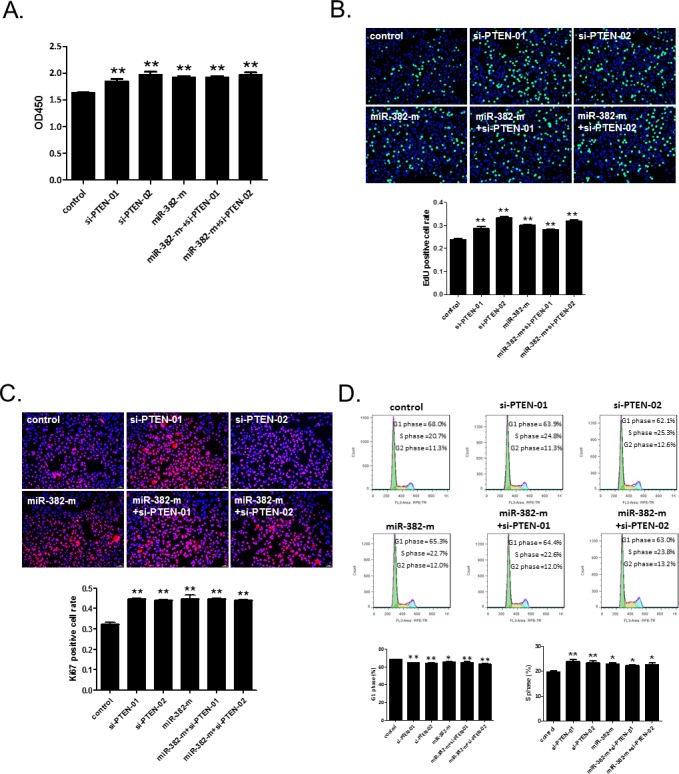
CCK-8 cell counting (n=10) A., EdU (green) cell proliferation assay (n=5) B., and Ki67 (red) immunostaining (n=5) C. demonstrated that co-transfection with miR-382 mimics (miR-382-m) and siRNA-PTEN (sequence-01 or -02) did not show an additional effect on the proliferation of NCTC1469 cells as compared to those transfected with miR-382-mimics or siRNA-PTEN alone. Nuclei were counterstained with Hoechst (blue). Original magnification 100x, scale bar = 50 μm. D. Flow cytometry showed that siRNA-PTEN (sequence-01 or -02) did not further enhance the miR-382 mimics-induced G1 to S phase transition of the cell cycle of NCTC1469 cells (n=5). *, P<0.05 vs. control; **, P<0.01 vs. control.
Akt activation is required for miR-382 overexpression-induced hepatocyte proliferation
As PTEN is an inhibitor for Akt phosphorylation, we further investigated whether Akt contributes to the promotive effect of miR-382 on hepatocyte proliferation and cell growth. Here we showed that miR-382 mimics or Akt activator enhanced, while miR-382 inhibitor or Akt inhibitor reduced the proliferation of NCTC1469 liver cells (Figure 7A-7C). Meanwhile, miR-382 mimics or Akt activator induced a G1 to S phase transition of NCTC1469 cells, while miR-382 inhibitor or Akt inhibitor caused a G1 phase arrest (Figure 7D). Noteworthy, co-treatment with miR-382 mimics and Akt inhibitor significantly abolished miR-382 overexpression-induced hepatocyte proliferation and cell growth (Figure 7A-7D). Also, Akt activator could reverse miR-382 inhibition-induced NCTC1469 cell growth arrest (Figure 7A-7D). These data, together with the results of PTEN, fully support that the PTEN/Akt signaling pathway is a downstream mechanism mediating the role of miR-382 in hepatocyte proliferation and cell growth.
Figure 7. Akt activation is involved in miR-382 overexpression-induced hepatocyte proliferation.

CCK-8 cell counting (n=10) A., EdU (green) cell proliferation assay (n=5) B., Ki67 (red) immunostaining (n=5) C., and flow cytometry (n=5) D. demonstrated that Akt activator (Akt-a) reversed miR-382 inhibitor (miR-382-i)-induced suppression of NCTC1469 cell proliferation and cell cycle progression, while Akt inhibitor (Akt-i) abolished miR-382 mimics (miR-382-m)-induced cell proliferation and cell growth. Nuclei were counterstained with Hoechst (blue). Original magnification 100x, scale bar = 50 μm. *, P<0.05 vs. control; **, P<0.01 vs. control; #, P<0.05 vs. miR-382-m or miR-382-i; ##, P<0.01 vs. miR-382-m or miR-382-i.
DISCUSSION
Liver regeneration after PH is principally mediated by the proliferation of hepatocytes which can be impacted by dysregulated miRNAs, though the underlying molecular mechanisms are still largely unclear. In the current study, we found a marked induction of miR-382 in the mouse liver at 48 hrs after 70% PH. miR-382 overexpression promotes hepatocyte proliferation and G1/S phase transition of the cell cycle in vitro. Moreover, miR-382 negatively regulates PTEN expression and increases Akt phosphorylation both in vitro and in vivo. We further confirm that PTEN inhibition and Akt phosphorylation are required for miR-382 overexpression-induced hepatocyte proliferation and cell cycle progression. Thus, the present study identifies miR-382 as a promoter for hepatocyte proliferation and cell growth via targeting PTEN-Akt axis.
We previously established the mouse model of PH demonstrating that hepatocyte proliferation peaked at 48 hrs after PH as represented by a peak of PCNA protein level and EdU-positive cells [4]. In the current study, we particularly focused on the roles of miRNAs in the proliferative phase of liver regeneration. Thus, miRNA array and qRT-PCRs were performed to identify dysregulated miRNAs in liver tissues at 48 hrs post hepatectomy. The upregulation of miR-382 during the proliferative phase of liver regeneration is intriguing. miR-382, one of the miRNA located in chromosome 14q32 locus, has been reported to be involved in angiogenesis as well as in cancer growth and invasion [25, 28-31]. Previously, miR-382 induced by HIF-1α has been identified as an angiogenic miRNA in vascular endothelial cells [25]. Besides that, increased serum miR-382 has been suggested to be a potential biomarker for breast cancer [32]. On the other hand, miR-382 has been identified as metastasis-suppressive miRNA in melanoma [33]. Also, miR-382 inhibits tumor growth and metastasis in osteosarcoma [34, 35]. Thus, the biological functions of miR-382 are tissue and cell dependent. In the present study, we found a marked elevation of miR-382 in mouse liver at PH-48h, which prompted us to further investigate the role of miR-382 in liver regeneration.
Our data show that miR-382 overexpression increases cell proliferation and induces a G1 to S phase transition of the cell cycle in both mouse NCTC1469 and human HL7702 liver cells, while miR-382 inhibition exhibits inverse effects, indicating miR-382 as a promoter for hepatocyte proliferation and cell cycle progression that might contribute to liver regeneration at proliferative stage. Previous studies have reported that miRNAs could impact hepatocyte proliferation and cell cycle progression via regulating cyclins and cyclin-dependent kinases (CDK). For example, miR-21 and miR-221 are responsible for cyclin D1 induction, leading to a rapid G1 to S phase transition of the cell cycle of hepatocytes during liver regeneration [14, 19]. On the contrary, miR-33 could inhibit cyclin D1 and CDK6 at mRNA level, thus causing reduced cell proliferation and cell cycle progression during liver regeneration [21]. As certain cyclins such as cyclin D1 and cyclin E are key regulators for G1/S phase transition, further study is needed to examine the molecular mechanisms by which miR-382 impacts the proliferation and cell cycle progression of hepatocytes [36-39].
PTEN-regulated Akt activation is a key signaling pathway which controls cell growth, survival, and proliferation [40-42]. PTEN, has previously been identified as a direct target gene of miR-382 contributing to hypoxia-induced angiogenesis [25]. Also, PTEN has been shown to be inversely regulated by miR-21, and the latter promotes hepatocyte proliferation in vitro [13]. However, whether the PTEN/Akt axis is regulated by miR-382 in hepatocytes is unknown. Our data show that PTEN protein level is downregulated, while Akt phosphorylation is enhanced in the mouse liver at 48 hrs after PH. Furthermore, we demonstrate that miR-382 negatively regulates PTEN expression and increases Akt phosphorylation in cultivated hepatocytes. Using PTEN siRNA and Akt activator/inhibitor, our data further provide key evidence indicating that Akt phosphorylation, at least in part associated with PTEN inhibition, is essential for miR-382 overexpression-induced hepatocyte proliferation and cell growth.
Several limitations of this study need to be highlighted. First, as multiple secreted and soluble factors, such as tumor necrosis factor (TNF), interleukin-6 (IL-6), hepatocyte growth factor (HGF), epidermal growth factor (EGF) and transforming growth factor-α (TGF-α), are responsible for initiating and promoting the liver regeneration process [43], it will be of interest to examine whether miR-382 upregulation during liver regeneration is related to these factors. Second, as we know, in vitro normal hepatocytes are already primed for mitosis which is sensitive for growth factors like HGF, however hepatocytes in vivo has a low sensitivity to these factors unless they are primed or activated by TNF and IL-6 [44, 45]. Indeed, it will be highly needed to further examine the in vivo effect of miR-382 in liver regeneration in the future. Finally, non-parenchymal cells as well as oval/progenitor cells also contribute to liver regeneration [46, 47]. Whether miR-382 regulates newborn hepatocytes generated from liver stem cells remains a topic for further investigation.
In conclusion, the present study shows an induction of miR-382 in the mouse liver during the proliferative phase of liver regeneration, and further demonstrates that miR-382 overexpression promotes hepatocyte proliferation and cell growth via targeting PTEN-Akt axis. The overexpression of miR-382 may be considered as a prospective novel therapeutic target to improve liver regeneration.
MATERIALS AND METHODS
Mouse model of partial hepatectomy (PH)
Eight-week-old pathogen-free male C57BL/6 mice were purchased from Shanghai Laboratory Animal Center (SLAC). 70% PH was conducted as previously described [4]. Briefly, mice were anaesthetized with intraperitoneal injection of 1% pentobarbital sodium (50 mg/kg), followed by abdominal median incision and hepatectomy of the median and left lobes of the liver. After the liver was resected, the abdominal incision was closed and mice were maintained in 37°C environment for anesthesia recovery. At 48 hrs after PH (PH-48h), mice were sacrificed and the livers were harvested and immediately kept into liquid azote. The liver tissues were then conserved at −80°C until RNA or protein extraction. The control mice received the same 70% PH but sacrificed at 0 hr after PH (PH-0h). This study was approved by the local ethical committees and all animal experiments were conducted under the guidelines on humane use and care of laboratory animals for biomedical research published by National Institutes of Health (No. 85-23, revised 1996).
miRNA microarray analysis
Total RNAs were isolated from liver tissues and quantified by the NanoDrop ND-2100 (Thermo Scientific). After the control of RNA integrity using Agilent 2100 (Agilent Technologies), total RNAs were tailed with Poly A, labeled with Biotin, and then hybridized for 16 hrs at 48°C on Affemetrix miRNA 3.0 Array. GeneChips were washed and stained in the Affymetrix Fluidics Station 450. The arrays were scanned by the Affymetrix Scanner 3000 (Affymetrix) and the array images were analyzed using Affymetrix GeneChip Command Console 4.0 software (Affymetrix) to get raw data and then provide RMA normalization. Using Genespring 12.5 software (Agilent Technologies), the probes that at least 75% of samples in any 1 condition out of 2 conditions have flags in “P” were chosen for further data analysis. The differentially expressed miRNAs, with a fold change>= 2.0 and a P value < 0.05 between the groups PH-0h and PH-48h, were chosen for further validation using qRT-PCRs. The MIAME-compliant data have been submitted to Gene Expression Omnibus (GEO, platform ID: GSE68451).
Cell culture and treatment
Mouse NCTC1469 normal liver cells were maintained in Dulbecco's Modified Eagle's Medium (Hyclone, USA) supplemented with 10% fetal bovine serum (Hyclone, USA) and 1% penicillin-streptomycin (Keygen, China) at 37°C in 5% CO2 environment. Human HL7702 normal liver cells were maintained in RPMI-1640 medium (Hyclone, USA) supplemented with 20% fetal bovine serum (Hyclone, USA) and 1% penicillin-streptomycin (Keygen, China) at 37°C in 5% CO2 environment.
For overexpression or suppression of miR-382, cells were transfected with miR-382 mimics (50 nM, RiboBio, China), miR-382 inhibitor (100 nM, Ribobio, China), or their negative controls for 48 hrs, respectively. To further investigate the role of PTEN and Akt in miR-382-associated hepatocyte proliferation and cell growth, cells were transfected with siRNA-PTEN (sequence-01 or 02, 75 nM, Ribobio, China) or siRNA-negative control for 48 hrs, or treated with Akt activator SC 79 (4 μg/mL, No. 4635, Tocris, UK) or Akt inhibitor LY 294002 (25 μM, No. 1130, Tocris, UK) for 24 hrs, respectively.
Cell counting kit (CCK-8) for cell proliferation analysis
A proliferation assay was performed using CCK-8 (Dojindo, Japan) according to the manufacturer's instructions. Briefly, NCTC 1469 or HL7702 cells were seeded in 96-well plates at a density of 2×105/ml. After 48 hrs of treatment, CCK-8 solution was added to each well and incubated for 30 min at 37°C. The absorbance was measured at 450 nm using a spectrophotometer.
EdU incorporation assay
To detect the DNA synthesis of hepatocytes, NCTC 1469 or HL7702 cells were seeded in 96-well plates at a density of 1×105/ml. 50 μM EdU was added into the medium and incubated for 2 hrs before the end of 48 hrs of cell treatment. After cells were washed 3 times with PBS and fixed with 4% paraformaldehyde (PFA) for 30 min, EdU staining was conducted using Cell-Light™ EdU Apollo®488 In Vivo Imaging Kit (RiboBio, China). Nuclei were counterstained with Hoechst (Sigma, USA). Digital images were acquired under fluorescence microscopy (Leica, Germany) with original magnification of 100x. The results were presented as EdU positive cell rate which was analyzed with ImageJ software.
Immunostaining for Ki67
To determine the proliferation of hepatocytes, NCTC 1469 or HL7702 cells were seeded in 96-well plates at a density of 1×105/ml. After 48 h of treatment, cells were washed 3 times with PBS and fixed with 4% PFA for 30 min. Subsequently, cells were washed again with PBS for 3 times and permeabilized with 0.5% Triton X-100 for 20 min. Next, cells were pretreated with 5% BSA in PBS for 1 hr and incubated with rabbit anti-Ki67 antibody (1:100, ab15580, Abcam, USA) overnight at 4°C. Then cells were incubated with Rhodamine-conjugated goat anti-rabbit secondary antibody (1:200, Keygen, China) for 1 hr. Finally, nuclei were counterstained with Hoechst (Sigma, USA). Digital images were acquired under fluorescence microscopy (Leica, Germany) with original magnification of 100x. The results were presented as Ki67 positive cell rate which was analyzed with ImageJ software.
Flow cytometry for cell cycle analysis
NCTC 1469 or HL7702 cells were seeded in 12-well plates at a density of 2×105/ml. After 48 hrs of treatment, cells were detached with trypsin, washed 3 times with ice cold PBS, and fixed in 70% ethanol at −20°C overnight. Cells were then suspended in a solution containing 0.5 mg/L propidium iodide (PI) and ribonuclease A. Cellular DNA content was analyzed using MoFlo XDP Cell Sorter (Beckman Coulter). The results were presented as the percentage of cell population in each phase of the cell cycle which was determined using FlowJo 7.6 software (Treestar Inc., USA).
RNA extraction and qRT-PCRs
Total RNAs were isolated using the RNAiso Reagent extraction kit (Takara, Japan). For mRNA detection, a total of 400 ng RNA was subjected to reverse transcription-PCR reaction using iScript™ cDNA Synthesis Kit (Bio-Rad, USA). qRT-PCR was performed in triplication using SYBR Green PCR Master Mix (BioRad, USA) and CFX96 Touch™ Real-Time PCR Detection System (BioRad, USA) with the following cycle parameters: 95°C 3 min, (95°C 15 s, 60°C 30 s, 72°C 30s) for 40 cycles. The primer sequences (forward and reverse) are as follows: PTEN: TGGATTCGACTTAGACTTGACCT, GCGGTGTCATAATGTCTCTCAG; β-actin: GGCTGTATTCCCCTCCATCG, CCAGTTGGTAACAATGCCATGT. For miRNA detection, a total of 400 ng RNA was subjected to reverse transcription-PCR with Bulge-Loop™ miRNA qRT-PCR Primer Set (RiboBio, China) and CFX96 Touch™ Real-Time PCR Detection System (BioRad, USA). The following cycle parameters were used: 95°C 20 s, (95°C 10 s, 60°C 20 s, 72°C 10s) for 40 cycles. β-actin or U6 was used as internal control to equal cDNA content. The fold change of each gene expression was calculated using the 2−ΔΔCT method.
Western blot analysis
The total protein extracts of liver cells or liver tissues were prepared for Western blot analysis. Briefly, cells or liver tissues were lysed in RIPA lysis buffer (Beyotime, China) supplemented with 1 mM PMSF (Beyotime, China). A total of 30 μg of protein was subjected to electrophorese on 12% SDS-PAGE gels, transferred to PVDF membranes, and incubated with primary antibodies against Akt (1:1000, AP0059, Bioworld, USA), phospho-Akt Ser473 (1:1000, BS4007, Bioworld, USA) and PTEN (1:1000, ab32199, Abcam, USA) at 4°C overnight. Blots were then incubated with corresponding HRP-conjugated secondary antibodies for 1 hr. The protein levels were detected using enhanced chemiluminescence (ECL) system (Pierce Biotechnology Inc., USA) and analyzed with Image Lab software (Bio-Rad). β-actin (1:1000, AP0060, Bioworld, USA) was used as loading control.
Statistical analysis
Data are presented as the mean ± standard error of mean (SEM). Statistical analysis was performed using independent student T-test or ANOVA with post hoc tests. P value of <0.05 was accepted as statistically significant. Statistical analysis was carried out with SPSS 20.0 software.
Acknowledgments
This work was supported by the grants from National Natural Science Foundation of China (81400647 to Y. Bei; 81200169 to J. Xiao; 81370926 to Y. Xiang; 81070343 and 81370559 to C. Yang; 81400635 to F. Wang), Innovation Program of Shanghai Municipal Education Commission (13YZ014 to J. Xiao), Foundation for University Young Teachers by Shanghai Municipal Education Commission (year 2012, to J. Xiao), Innovation fund from Shanghai University (sdcx2012038 to J. Xiao), Program for the integration of production, teaching and research for University Teachers supported by Shanghai Municipal Education Commission (year 2014, to J. Xiao), China Postdoctoral Science Foundation (2014M561456 to Y. Bei), Jonit Projects in Major Diseases funding from Shanghai Municipal Commission of Health and Family Planning (2014ZYJB0201 to C. Yang), Jonit Projects for Novel Frontier Technology in Shanghai Municipal Hospital from Shanghai Municipal Commission of Health and Family Planning (SHDC1204122 to C. Yang), Shanghai Medical Guide Project from Shanghai Science and Technology Committee (14411971500 to F. Wang), grants from Chinese Foundation for Hepatitis Prevention and Control (TQGB20140141 to F. Wang), and funds from Shanghai Innovation Program (12431901002 to C. Yang).
Footnotes
CONFLICTS OF INTEREST
The authors declare that they have no competing interests.
REFERENCES
- 1.Mao SA, Glorioso JM, Nyberg SL. Liver regeneration. Transl Res. 2014;163:352–362. doi: 10.1016/j.trsl.2014.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fausto N, Campbell JS, Riehle KJ. Liver regeneration. J Hepatol. 2012;57:692–694. doi: 10.1016/j.jhep.2012.04.016. [DOI] [PubMed] [Google Scholar]
- 3.Bucher NL, Swaffield MN. The rate of incorporation of labeled thymidine into the deoxyribonucleic acid of regenerating rat liver in relation to the amount of liver excised. Cancer Res. 1964;24:1611–1625. [PubMed] [Google Scholar]
- 4.Wang F, Song Y, Bei Y, Zhao Y, Xiao J, Yang C. Telocytes in liver regeneration: possible roles. J Cell Mol Med. 2014;18:1720–1726. doi: 10.1111/jcmm.12355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang DW, Yin YM, Yao YM. Advances in the management of acute liver failure. World J Gastroenterol. 2013;19:7069–7077. doi: 10.3748/wjg.v19.i41.7069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Duncan AW, Soto-Gutierrez A. Liver repopulation and regeneration: new approaches to old questions. Curr Opin Organ Transplant. 2013;18:197–202. doi: 10.1097/MOT.0b013e32835f07e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Van Rooij E. The art of microRNA research. Circ Res. 2011;108:219–234. doi: 10.1161/CIRCRESAHA.110.227496. [DOI] [PubMed] [Google Scholar]
- 8.Farh KK, Grimson A, Jan C, Lewis BP, Johnston WK, Lim LP, Burge CB, Bartel DP. The widespread impact of mammalian MicroRNAs on mRNA repression and evolution. Science. 2005;310:1817–1821. doi: 10.1126/science.1121158. [DOI] [PubMed] [Google Scholar]
- 9.Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 10.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 11.Song G, Sharma AD, Roll GR, Ng R, Lee AY, Blelloch RH, Frandsen NM, Willenbring H. MicroRNAs control hepatocyte proliferation during liver regeneration. Hepatology. 2010;51:1735–1743. doi: 10.1002/hep.23547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lakner AM, Bonkovsky HL, Schrum LW. microRNAs: fad or future of liver disease. World J Gastroenterol. 2011;17:2536–2542. doi: 10.3748/wjg.v17.i20.2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yan-nan B, Zhao-yan Y, Li-xi L, jiang Y, Qing-jie X, Yong Z. MicroRNA-21 accelerates hepatocyte proliferation in vitro via PI3K/Akt signaling by targeting PTEN. Biochem Biophys Res Commun. 2014;443:802–807. doi: 10.1016/j.bbrc.2013.12.047. [DOI] [PubMed] [Google Scholar]
- 14.Ng R, Song G, Roll GR, Frandsen NM, Willenbring H. A microRNA-21 surge facilitates rapid cyclin D1 translation and cell cycle progression in mouse liver regeneration. J Clin Invest. 2012;122:1097–1108. doi: 10.1172/JCI46039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Castro RE, Ferreira DMS, Zhang X, Borralho PM, Sarver AL, Zeng Y, Steer CJ, Kren BT, Rodrigues CM. Identification of microRNAs during rat liver regeneration after partial hepatectomy and modulation by ursodeoxycholic acid. Am J Physiol Gastrointest Liver Physiol. 2010;299:G887–897. doi: 10.1152/ajpgi.00216.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yuan B, Dong R, Shi D, Zhou Y, Zhao Y, Miao M, Jiao B. Down-regulation of miR-23b may contribute to activation of the TGF-β1/Smad3 signalling pathway during the termination stage of liver regeneration. FEBS Lett. 2011;585:927–934. doi: 10.1016/j.febslet.2011.02.031. [DOI] [PubMed] [Google Scholar]
- 17.John K, Hadem J, Krech T, Wahl K, Manns MP, Dooley S, Batkai S, Thum T, Schulze-Osthoff K, Bantel H. MicroRNAs play a role in spontaneous recovery from acute liver failure. Hepatology. 2014;60:1346–1355. doi: 10.1002/hep.27250. [DOI] [PubMed] [Google Scholar]
- 18.Da Silva CG, Studer P, Skroch M, Mahiou J, Minussi DC, Peterson CR, Wilson SW, Patel VI, Ma A, Csizmadia E, Ferran C. A20 promotes liver regeneration by decreasing SOCS3 expression to enhance IL-6/STAT3 proliferative signals. Hepatology. 2013;57:2014–2025. doi: 10.1002/hep.26197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yuan Q, Loya K, Rani B, Möbus S, Balakrishnan A, Lamle J, Cathomen T, Vogel A, Manns MP, Ott M, Cantz T, Sharma AD. MicroRNA-221 overexpression accelerates hepatocyte proliferation during liver regeneration. Hepatology. 2013;57:299–310. doi: 10.1002/hep.25984. [DOI] [PubMed] [Google Scholar]
- 20.Zhou J, Ju W, Wang D, Wu L, Zhu X, Guo Z, He X. Down-regulation of microRNA-26a promotes mouse hepatocyte proliferation during liver regeneration. PloS One. 2012;7:e33577. doi: 10.1371/journal.pone.0033577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Iwakiri Y. A role of miR-33 for cell cycle progression and cell proliferation. Cell Cycle. 2012;11:1057–1058. doi: 10.4161/cc.11.6.19744. [DOI] [PubMed] [Google Scholar]
- 22.Chen H, Sun Y, Dong R, Yang S, Pan C, Xiang D, Miao M, Jiao B. Mir-34a is upregulated during liver regeneration in rats and is associated with the suppression of hepatocyte proliferation. PloS One. 2011;6:e20238. doi: 10.1371/journal.pone.0020238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pan C, Chen H, Wang L, Yang S, Fu H, Zheng Y, Miao M, Jiao B. Down-regulation of MiR-127 facilitates hepatocyte proliferation during rat liver regeneration. PloS One. 2012;7:e39151. doi: 10.1371/journal.pone.0039151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yu ZY, Bai YN, Luo LX, Wu H, Zeng Y. Expression of microRNA-150 targeting vascular endothelial growth factor-A is downregulated under hypoxia during liver regeneration. Mol Med Rep. 2013;8:287–293. doi: 10.3892/mmr.2013.1493. [DOI] [PubMed] [Google Scholar]
- 25.Seok JK, Lee SH, Kim MJ, Lee YM. MicroRNA-382 induced by HIF-1α is an angiogenic miR targeting the tumor suppressor phosphatase and tensin homolog. Nucleic Acids Res. 2014;42:8062–8072. doi: 10.1093/nar/gku515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ohta K, Hoshino H, Wang J, Ono S, Iida Y, Hata K, Huang SK, Colquhoun S, Hoon DS. MicroRNA-93 activates c-Met/PI3K/Akt pathway activity in hepatocellular carcinoma by directly inhibiting PTEN and CDKN1A. Oncotarget. 2015;6:3211–3224. doi: 10.18632/oncotarget.3085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Qadir XV, Han C, Lu D, Zhang J, Wu T. miR-185 inhibits hepatocellular carcinoma growth by targeting the DNMT1/PTEN/Akt pathway. Am J Pathol. 2014;184:2355–2364. doi: 10.1016/j.ajpath.2014.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kriegel AJ, Fang Y, Liu Y, Tian Z, Mladinov D, Matus IR, Ding X, Greene AS, Liang M. MicroRNA-target pairs in human renal epithelial cells treated with transforming growth factor beta 1: a novel role of miR-382. Nucleic Acids Res. 2010;38:8338–8347. doi: 10.1093/nar/gkq718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thayanithy V, Sarver AL, Kartha RV, Li L, Angstadt AY, Breen M, Steer CJ, Modiano JF, Subramanian S. Perturbation of 14q32 miRNAs-cMYC gene network in osteosarcoma. Bone. 2012;50:171–181. doi: 10.1016/j.bone.2011.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sarver AL, Thayanithy V, Scott MC, Cleton-Jansen AM, Hogendoorn PC, Modiano JF, Subramanian S. MicroRNAs at the human 14q32 locus have prognostic significance in osteosarcoma. Orphanet J Rare Dis. 2013;8:7. doi: 10.1186/1750-1172-8-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li Z, Lu J, Sun M, Mi S, Zhang H, Luo RT, Chen P, Wang Y, Yan M, Qian Z, Neilly MB, Jin J, Zhang Y, et al. Distinct microRNA expression profiles in acute myeloid leukemia with common translocations. Proc Natl Acad Sci U S A. 2008;105:15535–15540. doi: 10.1073/pnas.0808266105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mar-Aguilar F, Mendoza-Ramírez JA, Malagón-Santiago I, Espino-Silva PK, Santuario-Facio SK, Ruiz-Flores P, Rodríguez-Padilla C, Reséndez-Pérez D. Serum circulating microRNA profiling for identification of potential breast cancer biomarkers. Dis Markers. 2013;34:163–169. doi: 10.3233/DMA-120957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hanniford D, Segura MF, Zhong J, Philips E, Jirau-Serrano X, Darvishian F, Berman RS, Shapiro RL, Pavlick AC, Brown B, Osman I, Hernando E. Identification of metastasis-suppressive microRNAs in primary melanoma. J. Natl. Cancer Inst. 2015 doi: 10.1093/jnci/dju494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xu M, Jin H, Xu CX, Sun B, Mao Z, Bi WZ, Wang Y. miR-382 inhibits tumor growth and enhance chemosensitivity in osteosarcoma. Oncotarget. 2014;5:9472–9483. doi: 10.18632/oncotarget.2418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xu M, Jin H, Xu CX, Sun B, Song ZG, Bi WZ, Wang Y. miR-382 inhibits osteosarcoma metastasis and relapse by targeting Y box-binding protein 1. Mol Ther. 2015;23:89–98. doi: 10.1038/mt.2014.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Albrecht JH, Hansen LK. Cyclin D1 promotes mitogen-independent cell cycle progression in hepatocytes. Cell Growth Differ. 1999;10:397–404. [PubMed] [Google Scholar]
- 37.Taub R. Liver regeneration 4: transcriptional control of liver regeneration. FASEB J. 1996;10:413–427. [PubMed] [Google Scholar]
- 38.Wang W, Du Z, Yan J, Ma D, Shi M, Zhang M, Peng C, Li H. Mesenchymal stem cells promote liver regeneration and prolong survival in small-for-size liver grafts: involvement of C-Jun N-terminal kinase, cyclin D1, and NF-κB. PloS One. 2014;9:e112532. doi: 10.1371/journal.pone.0112532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hanse EA, Nelsen CJ, Goggin MM, Anttila CK, Mullany LK, Berthet C, Kaldis P, Crary GS, Kuriyama R, Albrecht JH. Cdk2 plays a critical role in hepatocyte cell cycle progression and survival in the setting of cyclin D1 expression in vivo. Cell Cycle. 2009;8:2802–809. doi: 10.4161/cc.8.17.9465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fine B, Hodakoski C, Koujak S, Su T, Saal LH, Maurer M, Hopkins B, Keniry M, Sulis ML, Mense S, Hibshoosh H, Parsons R. Activation of the PI3K pathway in cancer through inhibition of PTEN by exchange factor P-REX2a. Science. 2009;325:1261–1265. doi: 10.1126/science.1173569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tan W, Gu Z, Shen B, Jiang J, Meng Y, Da Z, Liu H, Tao T, Cheng C. PTEN/Akt-p27(kip1) signaling promote the BM-MSCs senescence and apoptosis in SLE patients. J Cell Biochem. 2015;116:1583–1594. doi: 10.1002/jcb.25112. [DOI] [PubMed] [Google Scholar]
- 42.Zhang W, Neo SP, Gunaratne J, Poulsen A, Boping L, Ong EH, Sangthongpitag K, Pendharkar V, Hill J, Cohen SM. Feedback regulation on PTEN/AKT pathway by the ER stress kinase PERK mediated by interaction with the Vault complex. Cell Signal. 2015;27:436–442. doi: 10.1016/j.cellsig.2014.12.010. [DOI] [PubMed] [Google Scholar]
- 43.Koniaris LG, McKillop IH, Schwartz SI, Zimmers TA. Liver regeneration. J Am Coll Surg. 2003;197:634–659. doi: 10.1016/S1072-7515(03)00374-0. [DOI] [PubMed] [Google Scholar]
- 44.Roos F, Ryan AM, Chamow SM, Bennett GL, Schwall RH. Induction of liver growth in normal mice by infusion of hepatocyte growth factor/scatter factor. Am J Physiol. 1995;268:G380–386. doi: 10.1152/ajpgi.1995.268.2.G380. [DOI] [PubMed] [Google Scholar]
- 45.Webber EM, Godowski PJ, Fausto N. In vivo response of hepatocytes to growth factors requires an initial priming stimulus. Hepatology. 1994;19:489–497. [PubMed] [Google Scholar]
- 46.Miyajima A, Tanaka M, Itoh T. Stem/progenitor cells in liver development, homeostasis, regeneration, and reprogramming. Cell Stem Cell. 2014;14:561–574. doi: 10.1016/j.stem.2014.04.010. [DOI] [PubMed] [Google Scholar]
- 47.Miyaoka Y, Miyajima A. To divide or not to divide: revisiting liver regeneration. Cell Div. 2013;8:8. doi: 10.1186/1747-1028-8-8. [DOI] [PMC free article] [PubMed] [Google Scholar]



