Abstract
The rate of single nucleotide polymorphism varies substantially across the human genome and fundamentally influences evolution and incidence of genetic disease. Previous studies have only considered the immediate flanking nucleotides around a polymorphic site –the site’s trinucleotide sequence context– to study polymorph levels across the genome. Moreover, the impact of larger sequence contexts has not been fully clarified, even though context substantially influences rates of polymorphism. Using a new statistical framework and data from the 1000 Genomes Project, we demonstrate that a heptanucleotide context explains >81% of variability in substitution probabilities, revealing new mutation-promoting motifs at ApT dinucleotide, CAAT, and TACG sequences. Our approach also identifies previously undocumented variability in C-to-T substitutions at CpG sites, which is not immediately explained by differential methylation intensity. Using our model, we present informative substitution intolerance scores for genes and a new intolerance score for amino acids, and we demonstrate clinical use of the model in neuropsychiatric diseases.
INTRODUCTION
Measured at the level of the chromosome down to the individual base, rates of single nucleotide substitution vary substantially by position across mammalian genomes, including the humans1. An exquisite example of the role for sequence context in contributing variability in substitution rate is provided by CpG dinucleotides, where spontaneous deamination of 5-methylcytosine results in ~14 fold higher C-to-T substitution rates compared to the genome-wide average1,2,3. Modeling the variability in nucleotide substitution rates will inform our understanding of evolutionary processes, help identify functional noncoding regions4 and mutation promoting motifs, suggest mechanisms behind spontaneous mutation, and aid in prediction of the clinical impact of polymorphisms discovered through resequencing5. Such models will need to determine not only the optimal window of local sequence context but should also integrate knowledge of functional constraint on the genome due to pressure from purifying selection.
Studies of complex human disease have incorporated a simple trinucleotide sequence context6,7 into models to quantify the probability of de novo mutational events8–10, to clarify the distribution of somatic mutational events segregating in different cancers11, and to model the purifying selective pressure on gene sequences12. As their focus was clinical, these reports did not determine if this context model best captured the extent to which flanking nucleotides influence the variability in genome-wide nucleotide substitution rates. Here, we report a statistical framework that compares the extent to which different local sequence lengths influence the probability of nucleotide substitution, tested using data from the 1000 Genomes (1KG) Project13, apply our models to the coding genome, and demonstrate utility to interpret de novo mutations identified in studies of neuropsychiatric disorders. We define the probability of nucleotide substitution as the chance that a nucleotide in the human genome reference is polymorphic, that is, the nucleotide position segregates alternative nucleotides within the population. This probability depends upon population history, selection, sample ascertainment, and local context features that influence the rate of mutation.
RESULTS
Sequence context modeling of substitution probabilities
We hypothesized that local sequence context –the nucleotides that flank a polymorphic site– could explain the observed variability in nucleotide substitution probabilities. To test this hypothesis, we defined a statistical model (Supplementary Fig. 1, Methods) whereby the probability that a nucleotide substitution occurs at a genomic site varies according to (i) the identities of the nucleotides that flank the site and (ii) the size of the 5′-to-3′ local sequence context window. To minimize the impact of natural selection, we focused on intergenic noncoding regions of the genome (Methods). As the estimated nucleotide substitution probabilities were robust (Supplementary Table 1a), we developed a likelihood-ratio testing procedure to evaluate competing local sequence context models (Methods).
First, we calculated the likelihood of the observed data assuming a “1-mer” model, which allowed different substitution classes (e.g., A-to-G, C-to-T, etc.) to occur at different rates but ignored effects of sequence context on substitution probabilities. We compared the 1-mer model to the trinucleotide (“3-mer”) sequence context model where single 5′ and 3′ nucleotides flanking the polymorphic middle position impact the rate of substitution. As expected, the 3-mer model significantly improved fit to the data (log likelihood ratio, LLR = 6,070,948, P ≪ 10−100, Supplementary Table 1a). Next, we evaluated if additional local nucleotides could further improve fit to the observed data. We demonstrate that, when compared to the 3-mer model or the pentanucleotide (‘5-mer’) model (with two flanking nucleotides on each side), the larger, heptanucleotide (‘7-mer’) model (with three flanking nucleotides on each side) fit the data better (both LLR > 494,212, P ≪ 10−100, Supplementary Table 1b). To further validate the models, we estimated substitution probabilities using 1,659,929 HapMap14 variants found in our noncoding regions (Methods), and observed that 7-mer context probabilities strongly correlated with probabilities estimated from 1KG data (Supplementary Fig. 2, Supplementary Table 2), and provided the best fit to the observed polymorphisms (Supplementary Table 3). Our model recapitulates expected shifts in probabilities consistent with population histories15 (Supplementary Fig. 3) and the downward shift in the average substitution probability for the X chromosome16 relative to autosomes (Supplementary Table 4) due to the smaller effective population size at the X chromosome. Taken collectively, our analyses demonstrate for the first time, to our knowledge, that a 7-mer sequence context model explains the observed distribution of polymorphisms found in human populations.
To incorporate prior information, we developed a Bayesian formulation using objective conjugate priors for analysis of the noncoding genome (Methods). Consistent with our previous analysis, the 7-mer context model proved superior compared to all other models (Approximate Bayes Factor (ABF) ≫ 1,000, Supplementary Table 1c). In subsequent analyses, we use these posteriors for the nucleotide substitutions probabilities.
7-mer context predicts noncoding substitution rates
To quantify the variance in the posterior probabilities that a 7-mer sequence context model could explain, we considered each substitution class separately, as well as CpG site contexts (nine classes total). We employed forward regression (Methods) to select features from a 7-mer context window to predict substitution probabilities and considered up to four-way interactions at positions within the window. When compared to single-base and position models without interactions, incorporating higher-order interactions substantially improved the fit to data (Supplementary Table 5). Specifically, we found that our selected models in a separately held test data set explained a median of 81% of the variability (as compared to 30% explained by the 3-mer context) in probabilities across all substitution classes, covering 84% of all mutational events and fitting well the probability of C-to-T substitution at CpGs (Table 1, Fig. 1A). Although we identified a common set of interactions across classes (Supplementary Table 6), many common features did not always influence substitution probabilities in the same way, and others had class-specific effects. These observations indicate that core and class-specific features based on sequence context are predictive of the potential for nucleotide substitution.
Table 1.
Summary and performance of forward regression model for feature selection using the 7-mer context in the intergenic noncoding genome. % Substitutions represents the percentage of substitutions for that class observed in the genome. # Parameters represents the number of features selected in the best 7-mer model. Model R2 (7-mer) reflects prediction accuracy in the test dataset alone (not used for model training) with the best model using heptanucleotide sequence context features. Model R2 (3-mer) denotes the prediction accuracy with only trinucleotide sequence context features.
| Substitution Class | # Contexts | % Substitutions | # Parameters | Model R2 (7-mer) | Model R2 (3-mer) |
|---|---|---|---|---|---|
| Outside CpG Dinucleotide Context | |||||
| A-to-C | 4,096 | 7.3 | 266 | 56.5 | 11.2 |
| A-to-G | 4,096 | 28.2 | 366 | 91.5 | 40.9 |
| A-to-T | 4,096 | 7.1 | 197 | 58.7 | 37.4 |
| C-to-A | 3,072 | 8.5 | 282 | 83.5 | 30.0 |
| C-to-G | 3,072 | 7.5 | 268 | 81.0 | 17.1 |
| C-to-T | 3,072 | 24.4 | 254 | 86.8 | 37.6 |
| Within CpG Dinucleotide Context | |||||
| C to A | 1,024 | 1.0 | 26 | 58.3 | 19.0 |
| C to G | 1,024 | 0.8 | 95 | 48.7 | 9.5 |
| C to T | 1,024 | 15.2 | 96 | 93.1 | 44.4 |
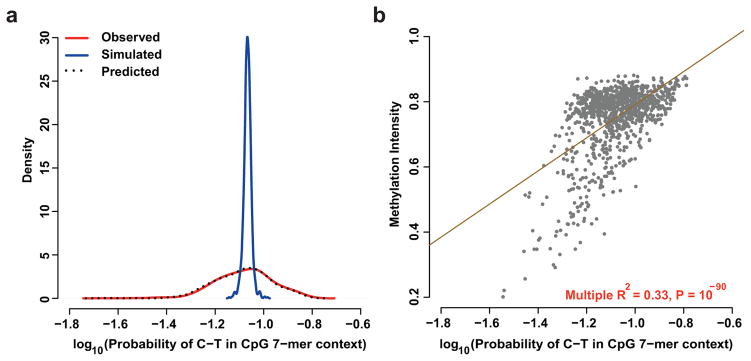
Figure 1.
C-to-T substitution probabilities and methylation patterns within 7-mer CpG sequence contexts. (a) Simulations based on a fixed C-to-T substitution rate (blue) at CpG contexts do not capture the observed distribution of substitution probabilities (red) within the 7-mer sequence context. Rates predicted from our regression model (black) closely match the substitution probabilities observed under the 7-mer sequence context (R2 = 0.93). (b) Correlation between average methylation intensity versus probability of C-to-T substitution in CpG 7-mer context.
Methylation cannot fully explain patterns at CpG sites
The spontaneous deamination of 5-methylcytosine at CpG sites results in ~14-fold higher rates of C-to-T substitutions generally3,17. Although a previous report indicated that divergence at CpG sites varies as a function of local context, the focus was on introns, and did not consider population-level polymorphisms in humans18. Thus, we hypothesized that the surrounding sequence context further influences the probability of nucleotide substitution at CpGs, and examined the C-to-T substitution class within the subset contexts that contain CpG at position 4 and 5 in the 7-mer. Simulations using a model that ignored additional genomic context, or considered the 3-mer context (Supplementary Fig. 4), using a fixed CpG substitution probability generated significantly less variability in 7-mer CpG substitution probabilities than was empirically observed (empirical P ≪ 10−10, Fig. 1A). These data indicate that (i) not all CpG sites accrue substitutions at the same rate and (ii) that the sequence context surrounding CpG sites correlate with biological features or mechanisms that influence this rate.
To explore the possibility that the excess variability depends upon variation in methylation intensity across sequence contexts, we reanalyzed whole-genome bisulfite sequencing data obtained from germline and other tissues of healthy individuals19,20. Comparing the CpG sites that are consistently methylated versus consistently unmethylated across subjects, we observed as expected that methylation correlates with an increase in the probability of C-to-T substitution (P ≪ 10−100, Supplementary Fig. 5). Unexpectedly, when we compared the methylation intensity in sperm at 7-mer CpG contexts with the probability of substitutions, we found a positive but imperfect correlation (R2 = 0.33, P < 10−90, Fig. 1B), with similar results in other tissues (Supplementary Fig. 6), noting instances of methylation status decoupled from substitution probabilities. For example, nearly every genomic instance of the sequence contexts GTACGCA and GATCGCA showed consistent methylation signals (both methylated in >94% of occurrences in sperm), the probability of C-to-T transition was more than two-fold different for these two contexts (0.148 vs. 0.07, respectively). These data are consistent with the hypothesis that local context features beyond DNA methylation influence probabilities of C-to-T transitions at CpG sites, though we cannot exclude the possibility that sub-tissue methylation differences could explain these patterns.
Identification of novel mutation-promoting motifs
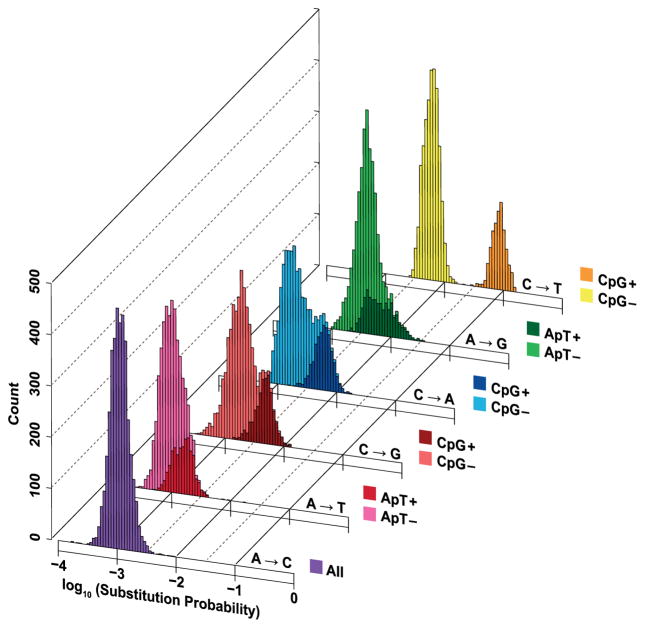
We next investigated the substitution probabilities for 7-mer contexts partitioned by substitution class (Fig. 2, Supplementary Table 7). First, we noted that several classes, C-to-A, and C-to-G in addition to C-to-T, appeared to segregate as mixtures of two distributions, explainable by CpG effects. These observations are consistent with studies demonstrating elevated substitutions at CpGs in humans21, though this early work was not powered to measure context dependencies surrounding CpG sites as we are here. As the methylation transition state intermediate 5-formylcytosine can induce spontaneous C-to-A or C-toG substitutions22, one possibility is that methylation also elevates these rates in this context. We next determined if local sequence context motifs –analogous to but beyond CpG dinucleotides– correlate with variable substitution probabilities across classes (Methods). We noted that poly-CG sequences in the lower tail of C-to-T substitutions for the CpG context were enriched (P < 10−16, Table 2). This observation is consistent with previous reports23 as this context is found proximal to genes (Supplementary Fig. 7) and is associated with lower methylation intensities (Supplementary Fig. 8). In the upper tail of the A-to-T substitution class, we observed a poly(T) + poly(A) motif in the outlier sequences (P < 10−5, Table 2). We also observed a similar quad-A motif in the lower tail of the A-to-G class (P < 10−10). One possible mechanism that may contribute is the ‘slippage’ of protein machinery during DNA replication24. Our analysis also revealed motifs without an obvious contributing mechanism. First, in the upper tail of CpG rates, we observed enrichment of a TACG motif (P < 10−10, Table 2) that was strongly methylated (Supplementary Fig. 8), but curiously, a similar motif shifted by one position was enriched in the lower tail of the A-to-C class (P < 10−4). Second, the ApT dinucleotide was found to elevate the substitution probabilities (Fig. 2) for the A-to-G (P < 10−25) and A-to-T classes (P < 10−17), though not statistically significantly so for A-to-C. Finally, we observed a CAAT motif also enriched in the upper tail of the A to G substitution class (P < 10−53), reported in an earlier study of dbSNP variants25. These latter cases indicate potentially new mechanisms contributing to elevated nucleotide substitutability, not documented by the commonly utilized trinucleotide context model. As a final robustness analysis, keeping in mind limitations due to variant ascertainment, we estimated the substitution probabilities using HapMap variants and found similar mutation promoting motifs across substitution classes (Supplementary Table 8).
Figure 2.
Posterior probabilities of all classes of nucleotide substitution in the intergenic noncoding genome, estimated using the 7-mer context model. Sequences contexts are further stratified by color to indicate either the presence of a CpG (C at the polymorphic 4th position and G at the 5th position, for C-to-A, C-to-G and C-to-T substitution classes = CpG+; else CpG−) or the ApT state (A at the polymorphic 4th position and T at the 5th position, for A-to-G and A-to-T substitution classes = ApT+; else ApT−). For A-to-C, the ApT state did not significantly contribute to variability in the estimated probability distribution.
Table 2.
Enrichment of motifs identified in posterior nucleotide substitution probabilities for the 7-mer sequence context models inferred from intergenic noncoding genome. CpG+ indicates the distribution of sequence contexts which include a CpG site (4th position polymorphic site is C, 5th position fixed as G). Enrichment P-value is based on the enrichment of the motif in the 1% tail of the given substitution class: “Higher” implies enrichment in the upper 1% tail of the sequence context probability distribution, “Lower” implies enrichment in the lower 1% tail. Odds ratio and [95% CI] denotes the odds ratio (and 95% confidence interval) of enrichment of motif in the upper or lower 1% tail of the sequence context probability distribution. Fold change in substitution rate denotes the fold increase or decrease in substitution rates for the motif relative to its substitution class.
| Motif | Substitution Class | Effect on Substitution Probability | Enrichment P-value | Odds ratio and [95% CI] | Fold change in substitution rate |
|---|---|---|---|---|---|
| NNNCGNN | C-to-T | Higher | 2 × 10−26 | 134.4 [18.4–977.4] | 13.9 |
| C-to-G | Higher | 1 × 10−13 | 12.8 [5.9–27.7] | 2.4 | |
| C-to-A | Higher | 9 × 10−22 | 60.8 [14.6–252.1] | 2.7 | |
| N[A/C/G][C/G/T]CGCG | C-to-T (CpG+) | Lower | 7 × 10−16 | 366.3 [45.6–2,939.5] | 1.5 |
| Poly T and Poly A combination (AAAATTT, TTTAAAA) | A-to-T | Higher | 9 × 10−5 | 304.2 [31.0–2,987.6] | 12.7 |
| Quad A (AAAANNN, NAAAANN, NNAAAAN, NNNAAAA) | A-to-G | Lower | 5 × 10−10 | 10.2 [7.3–14.1] | 1.9 |
| NTACG[C/G][A/C/G] | C-to-T (CpG+) | Higher | 1 × 10−10 | 102.5 [27.4–383.2] | 1.7 |
| NNTACGN | A-to-C | Lower | 3 × 10−4 | 9.4 [3.6–24.8] | 1.5 |
| NNNATNN | A-to-T | Higher | 2 × 10−17 | 22.3 [8.7–57.1] | 1.6 |
| A-to-G | Higher | 1 × 10−25 | 131.2 [18.0–954.2] | 2.0 | |
| [C/T]CAAT[C/G/T]N | A-to-G | Higher | 8 × 10−53 | 5966 [2,091–17,021] | 5.1 |
Experiments to validate the noncoding rate model
If the estimated noncoding substitution probabilities reflect properties of mutation, one would expect that these rates should (a) not influenced by rates of recombination (b) strongly correlate with rates of species divergence26, (c) be consistent for both rare and common genetic variants, and (d) also be reflected in de novo mutational events. We explored each of these predictions in turn. First, we estimated the 7-mer substitution rates from all intergenic noncoding variants separately for high and low recombination rate regions, and found a strong correlation between the two (R2 = 0.97, P ≪ 10−100, Supplementary Fig. 9, Methods), indicating that substitution probabilities estimated from the noncoding genome are correlated across high and low rates of recombination. Next, using human-chimpanzee and human-macaque alignments over intergenic noncoding sequences, we found a strong correlation between divergence and substitution probabilities for our 7-mer contexts (both R2 = 0.96, P ≪ 10−100, Supplementary Fig. 10, Supplementary Table 9, Methods). We then estimated 7-mer probabilities from all intergenic noncoding rare variants (singletons and doubletons) separately from low and high frequency variants (>1%), and found a strong correlation (R2 = 0.98, P ≪ 10−100, Supplementary Fig. 11, Methods), as well as a superior 7-mer context fit to data across variant frequencies (Supplementary Table 10). Finally, we obtained 4,748 de novo mutational events from a high quality pedigree sequencing dataset on 78 parent-offspring trios27. We tested for the presence of motifs we identified in Table 2 around de novo events, and observed a significant enrichment (Supplementary Table 11, Methods). Taken collectively, these findings provide additional validation for the hypothesis that our substitution probabilities capture features of germline mutation.
7-mer context also predicts exonic substitution rates
Assuming that the processes that generate spontaneous mutations apply uniformly across the genome, we hypothesized that sequence context could explain variability in substitution probabilities in the coding genome. We therefore extended our initial framework (Supplementary Fig. 1, Methods) to the coding genome by (i) using information obtained from our model on the noncoding genome as prior and (ii) allowing for context dependence of codons and local sequence context in our estimates of substitution probabilities to accommodate purifying selective pressure28. Our new model substantially improved the fit to the data compared to either 3-mer sequence context models with or without codon context (ABF ≫ 1,000, Supplementary Table 12). To further validate, we tested our model on a different large scale exome-sequencing dataset from ~4,300 individuals29, and noted that our 7-mer model fit patterns of exonic polymorphisms better than competing models (ABF ≫ 1,000, Supplementary Table 12, Methods). These results demonstrate for the first time, that a broader sequence context –beyond simple codon or trinucleotide context– captures the forces that shape variability in nucleotide substitutions in the coding genome.
We then examined the posterior distribution of substitution probabilities for all contexts stratified by the type of amino acid substitution (Supplementary Fig. 12, Supplementary Table 13), and found excess variability in each class than expected under simulation (Supplementary Table 14, Methods). Next, we enumerated the substitution probability profiles for each amino acid change, and found certain nonsense and missense substitution probabilities to be higher than synonymous levels (Supplementary Fig. 13), partially explained by CpG contexts. These observations caution against the practice –invoked in rare-variant association tests– of ignoring codon and sequence context when testing for the burden of functional substitutions. Our results here demonstrate that functional substitutions may not be equally likely or tolerated with respect to purifying selection.
7-mer context improves power to detect pathogenic variants
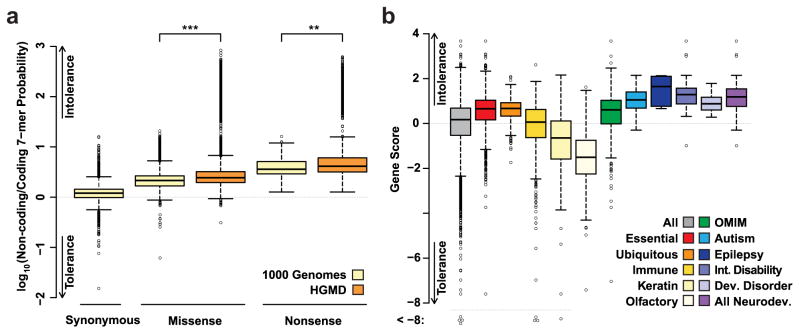
We now turn to applications of our model to improve the interpretation of variation discovered by clinical re-sequencing. Efforts to prioritize variants from such studies often rely on classifying variants that are deleterious with respect to population genetic fitness, hypothesizing that such variants are more likely pathogenic30. As our coding substitution probabilities are influenced both by forces of mutation (estimated from the noncoding genome) and selection, we hypothesized that the ratio of these probabilities quantifies the action of selective pressure, and could be used to prioritize pathogenic variants. To test this hypothesis, we calculated the log ratio of intergenic noncoding and coding substitution probabilities, defined as sequence constraint score, for missense (n = 48,450) and nonsense (n = 12,054) variants present in the Human Gene Mutation Database (HGMD, Methods)31. We observed that the distribution of sequence constraint scores for HGMD variants was shifted towards larger values (intolerance) compared to 1KG variants (P ≪ 10−100, Fig. 3A), compatible with the “intolerant variant, pathogenic variant” hypothesis. Moreover, the distribution of scores based on our 7-mer model was further shifted towards intolerance with a thicker tail, compared to a 3-mer model (P ≪ 10−100, Supplementary Fig. 14). These data demonstrate that a coding model that includes codon and a 7-mer context improves identification of variants that are potentially pathogenic.
Figure 3.
Prioritizing pathogenic variants and causal genes using constraint scores. (a) log10 ratios of substitution probabilities from the 7-mer sequence context model using coding sequences matched to the intergenic noncoding sequences, for each type of substitution (synonymous, missense and nonsense) for all variants in the 1KG project or Human Gene Mutation Database (HGMD). Larger values indicate fewer substitutions in the coding genome than expected from matched noncoding sequences, consistent with the action of selective constraint. *** represents P ≪ 10−100 and ** represents P < 10−29. (b) Box and whisker plot of gene scores from the model, stratified into statistically significant gene classes. Positive gene scores indicate intolerance to substitutions that change an amino acid. For the boxplot, the center line in each box denotes the median. The inter-quartile range (25th and 75th) is indicated by the ends of each box. The whiskers extend 1.5x the inter-quartile range, and data points beyond this range are plotted as open circles.
Describing genic intolerance to mutation via 7-mer context
Several groups have argued that the power to identify causal disease genes from clinical resequencing data could be enhanced by incorporating estimates of selective constraint on genes12,32,33. The underlying hypothesis behind this concept is that genes that are under selective constraint are more likely to have functional consequences and are therefore most likely to be pathogenic and have fewer functional variants (“intolerant gene, pathogenic gene”). The community has successfully applied this concept to neurodevelopmental and psychiatric disorders34, however the existing approaches have not incorporated the 7-mer sequence or codon context in their models.
Therefore, we applied our 7-mer coding substitution probabilities to develop an intolerance score (Supplementary Table 15, Methods) quantifying the difference between the expected and observed number of functional variants at a gene, with higher scores consistent with functional constraint. To further validate, we found gene scores on a separate, larger exome sequencing data set and observed a strong correlation between the two (Supplementary Fig. 15). We found that genes belonging to putatively essential or ubiquitously expressed categories, scored strongly for genic intolerance (P ≪ 10−100, Fig. 3B). In contrast, gene sets representing Keratin and Olfactory categories were found to be highly tolerant of functional changes (Fig. 3B). Next, we applied this to OMIM genes or known genes behind several neuropsychiatric disorders like Autism35, Epilepsy36, Developmental disorder37 and Intellectual disability38–40, and found them to have significantly higher intolerance scores (P ≪ 10−100, Fig. 3B). We then compared our gene scores to previously reported scores (Supplementary Fig. 16, Methods), and found that our approach improved classification or performed comparably to other approaches32 for genes in each set, including the disease categories (Supplementary Table 16). These results demonstrate that the most accurate scoring of genic tolerance to functional substitution can be achieved by modeling 7-mer sequence and coding context.
An amino acid score for pathogenic variant prioritization
Beyond the average rate of amino acid replacement that a gene might tolerate, genes could be further intolerant to specific types of amino-acid substitutions, signifying added localized selective constraint or importance for gene functionality. Therefore, we developed a score measuring the intolerance at amino acid replacement level in a gene (Supplementary Table 17, Methods), after quantifying the difference between the expected and observed number of functional variants for a specific amino acid at a gene. Across all genes represented in HGMD with a large number of putatively pathogenic amino acid changes for a specific substitution, we found they segregate larger intolerance scores for that amino acid (empirical P < 10−10). Moreover, a gene might score “tolerant” for functional substitution, but intolerant for specific amino acid changes. For example, Von Willebrand Factor (VWF), a blood glycoprotein involved in hemostasis, is tolerant to substitution overall (within top 8% of gene tolerance) but intolerant to cysteine substitution (within top 3.5% of cysteine intolerance). This data is consistent with a causal mechanism for von Willebrand disease; protein misfolding when cysteine residues are substituted41. We note that 5,652 genes segregate a profile similar to VWF: average genic tolerance, but amino acid intolerance.
Interpretation of de novo mutations discovered in Autism
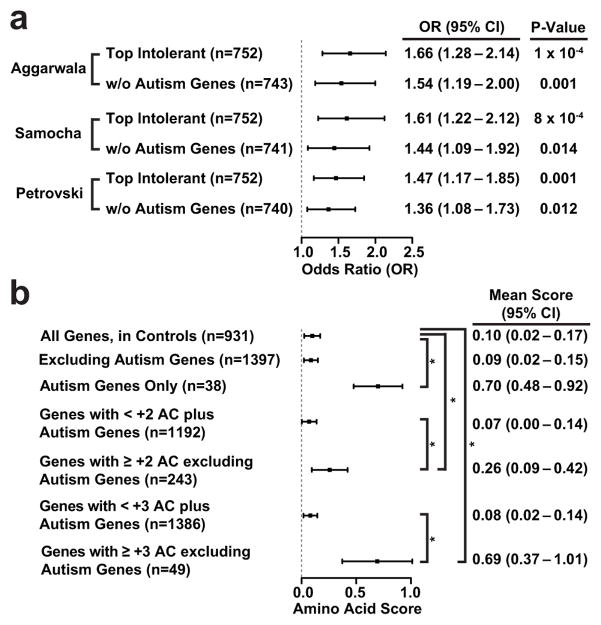
Autism spectrum disorder is a disease with complex etiology, and recent efforts have aimed to identify de novo mutational events that may contribute to disease. To highlight the utility of gene12,32 and amino-acid scores, we applied them to interpret de novo mutations collected from 2,508 Autism spectrum disorder42 cases and 1,911 control family trios. First, we found that the most intolerant genes based on our gene score segregated a significant burden of de novo mutations in cases as opposed to controls (OR = 1.66, P < 0.0001, Fig. 4A, Methods), even after removing known autism genes35 (OR = 1.54, P < 0.001), and similar, though slightly attenuated burden using other scores (Fig. 4A). Next, we found that the average amino acid scores for de novo mutations at Autism genes in cases was higher (more intolerant) than that found in controls, or at other genes in cases (P = 0.002, Fig. 4B, Methods). We further observed higher (intolerant) average amino acid scores for variants in genes with a positive variant burden in cases, relative to controls (+2 or +3 allele count excess in cases, both P < 0.01, Fig. 4B). Finally, several genes from the excess allele count set stood out with amino-acid specific intolerance (all within top 4 percentile of intolerance): MYO9B, WDFY3, NAV2, STIL, and SCUBE2. Aside from WDFY3, these genes are generally ‘tolerant’, based on their gene-score, indicating utility of sub-gene wise measurement of functional intolerance. While MYO9B has been implicated in autism35 and WDFY3 deletions in a murine model has been shown to cause Autism like symptoms43, our analysis points to the remaining candidates for future follow-up.
Figure 4.
Applications of gene and amino acid intolerance scores on de novo ASD mutational data. (a) Forest plot of the odds ratios (ORs), 95% confidence intervals (CIs), and p-values when comparing the de novo mutational burden in cases versus controls, on intolerant genes using different gene scoring methods. Scores are calculated including and excluding known Autism genes, as indicated. “Aggarwala” indicates gene scores from this report, while “Samocha” and “Petrovski” refers to the intolerant gene list from those works12,32. (b) Forest plots of the mean amino acid scores (with 95% CIs) found from de novo mutations in various gene collections. Average scores were based on variants ascertained in cases, except where noted (i.e., the first row: all genes in controls). W/o: without. +AC: excess count of missense or nonsense changes in cases relative to controls. For example, +3 indicates that a gene has 3 more missense or nonsense changes in cases relative to controls. *: P < 0.01.
DISCUSSION
We report a sequence context model that explains patterns of nucleotide substitution observed in the human genome. Our motivation was based on the need to statistically evaluate competing models for sequence context. We demonstrate that the commonly used context that includes one nucleotide flanking a polymorphic site does not fully capture the complete spectrum of where, what type, and how frequently nucleotides are expected to change. Furthermore, by using population level data, rather than de novo or somatic events, we were able to improve the resolution of substitution models and identify novel mutation promoting motifs. Our approach also characterized average selective pressures operating in the coding genome at a finer level of detail. Our model indicates substantial variability across all amino acid replacement classes, and, in some cases, synonymous substitutions that were less prone to change than missense or even nonsense substitutions. We suggest that inference of the presence and strength of selection on genes might further benefit by incorporating information at this resolution.
One question in the field has been how much sequence context can explain patterns of nucleotide substitution in genomes44. Our results suggest that a substantial fraction can be robustly predicted by sequence context alone, although specific substitution classes may require more than sequence context for their prediction. In evolutionary genetics studies, the set of substitutions that occur at nearly constant rates proportional to the lineage (i.e., most “clock-like”) is important for accurate dating divergence events45. While we did not apply our model to other species, the strong correlation with divergence suggests our features are potentially conserved across primates.
We acknowledge that a number of features remain to be formally evaluated in the genome46, for example, recombination in the coding genome47 or replication timing48. Our framework has the flexibility to model the complexity found in any sequences that contain features hypothesized to be important. We also acknowledge that context models beyond three flanking nucleotides were not considered. The regression approach we presented does suggest that the 7-mer models could be refined, perhaps allowing broader context to be considered.
With an appropriate background model for nucleotide substitution, novel statistics for clinical re-sequencing studies can be envisioned, based on the occurrence of discovered variation. Such approaches may complement statistics that assay allele frequency differences between cases and controls at one or more polymorphic sites. Moreover, comparative genomics applications to identify non-neutrally evolving regions, genome alignments, or tree reconstruction49, would benefit from accurate models of nucleotide substitution. While the underlying mechanisms that determine how nucleotide sequences change over time remain to be addressed, we posit that features identified from our model provide important clues in elucidating these fundamental principles.
ONLINE METHODS
Sourcing population samples
Samples were obtained from phase 1 of the 1KG Project. We considered only the variants from African, European, and East Asian ancestries.
Selection of intergenic noncoding sequences
Intergenic sequences were defined as the full set of genomic sequences that are not annotated in ENSEMBL Biomart (version 75) and RefSeq Genes. We then removed centromeric, telomeric, repetitive regions and sequences not present in the accessibility mask (version 20120824) filter of the 1KG project. Within these intergenic regions, we identified variants for the three populations for use in downstream analysis. More details in Supplementary Note.
Statistical framework to model substitution probabilities for intergenic noncoding regions
We initially describe a simple model that does not take into account local sequence context, and then build upon this by incorporating additional local sequence contexts.
Suppose that we observe nC occurrences of nucleotide C in the reference genome. A subset of these nC sites will be polymorphic within the population of individuals. Let nCA represent the number of sites where a nucleotide change C-to-A has occurred. Similarly, nCG is the number of sites where a change C-to-G has occurred and nCT is the number of sites where a change C-to-T has occurred. Then the probability of nucleotide substitution or polymorphism within the population can be described using a multinomial distribution:
| (1) |
where the probabilities of observing a substitution from C-to-A, C-to-G, and C-to-T are expressed as αCA, αCG, and αCT, respectively. After iterating over all possible substitutions (i.e., A-to-C, A-to-G, A-to-T, C-to-A, C-to-G, C-to-T, T-to-A, T-to-G, T-to-C, G-to-A, G-to-C, G-to-T), we merged the reverse-complementary pairs (e.g., A-to-C was merged with T-to-G, etc.) to yield 6 “substitution classes” as parameters for the simple model, which we refer to as the “1-mer” model. We then use maximum-likelihood estimation (MLE) to find the substitution probability estimates for all possible substitutions.
This model can be naturally extended to consider the effects of local sequence context by replacing the count of nx occurrences of nucleotide X with the count of occurrences of a particular nucleotide sequence context. For example, if we want to consider the local sequence context ACA, then we count the number of times the 3-mer sequence ACA occurs (nACA) in the reference genome. A subset of nACA will be polymorphic at the middle position C within a given population. Thus, let nACA→AAA represent the number of sites where a nucleotide change C-to-A has occurred at the middle position, nACA→AGA for changes from C-to-G and nACA→ATA for changes from C-to-T at the middle position. After iterating over all possible nucleotides combinations at the two ends (4 possibilities at either side for a total of 16) and substitutions at the middle position (3 possible changes per nucleotides for a total of 12), we merged the reverse complementary pairs yielding 96 substitution classes as parameters for the “3-mer” model.
Analogously, we extended the size of the sequence context window to evaluate the “5-mer” model and the “7-mer” model by considering additional fixed nucleotides (2 and 3, respectively) on either side of the polymorphic site, thereby estimating a total of 1,536 parameters for the 5-mer model and 24,576 parameters for the 7-mer model. More details in Supplementary Note.
Log-likelihood ratio testing for model comparison
We initially find the likelihood of the observed distribution of polymorphic sites using the substitution rate parameters for a sequence context model. We then calculate the likelihood ratio test statistic as:
| (2) |
where S1 and S2 represent parameters estimated from two competing sequence context models. The test is chi-squared distributed, with degrees of freedom equal to the difference in the number of parameters between the two models (e.g., comparing the 3-mer model versus the 1-mer model requires 90 degrees of freedom; comparing the 7-mer model versus the 3-mer model requires 24,480 degrees of freedom).
Selection of HapMap variants
Single nucleotide polymorphic variants were obtained from phase 3 release of the HapMap project. We considered only the variants from African ancestry present in our intergenic noncoding sequences. More details in Supplementary Note.
Incorporating prior information into the statistical framework
Since the likelihood of our framework is based on a multinomial distribution, we utilize its conjugate prior, i.e., the dirichlet distribution, for different sequence context models. For inference in the intergenic, noncoding genome, we selected the objective version of the prior for our analysis, with all concentration parameters of the dirichlet prior as 1. We then use MAP to find the substitution probability estimates for all possible substitutions. More details in Supplementary Note.
Bayes Factor analysis for model comparison
We calculated the approximate posterior likelihood, using the Chib’s method, on the overall data using the maximum a posteriori (MAP) estimates of the substitution probabilities for a specific sequence context model found before. We then calculate the approximate Bayes factor as:
| (3) |
where S1 and S2 represent parameters estimate from two competing sequence context models. We use the Jefferey’s scale for interpreting the approximate Bayes Factors, where the ratio if greater than 100 is considered to be decisive evidence against the Model1. More details in Supplementary Note.
Regression modeling and feature selection
We considered each substitution class separately and created an additional substitution class for each of the three possible changes within a CpG context, resulting in nine substitution classes. For each substitution class, we considered the initial regression model:
| (4) |
where the probability that a nucleotide changes from X1 to X2 is modeled using a position-base variable p, a set of bases (e.g., {C, G, or T} where A is the reference base) denoted by the superscript for p, each position (= 1, 2, 3, 5, 6, or 7) denoted by the subscript for p within sequence context S, intercept α, and error term ε. We assigned A as the reference nucleotide at each position and encoded the single nucleotide present at each position as the combination of three thermometer variables (e.g., 0,0,0 = A; 0,0,1 = C; 0,1,0 = G; 1,0,0 = T). Next, we examined non-additivity (i.e., interactions) between nucleotides at sequence context positions. Rather than including all possible interaction terms, we employed feature selection (i.e., model training and testing to select the most informative features) and incorporated these terms into the final model. We considered 2-way, 3-way, and 4-way interactions across positions within the 7-mer as:
| (5) |
where the probability that a nucleotide changes from X1 to X2 is modeled as described in Equation 4, and a set of additional terms related to interactions is also incorporated.. The effect of the interaction is represented by terms βa for 2-way interactions, βb for 3-way interactions, and βc for 4-way interactions. We then divided the genome into two distinct sets for feature selection, using all even-numbered chromosomes for training and all odd-numbered chromosomes for model testing. During training, we performed stepwise forward regression for each level of interaction in order of increasing complexity (i.e., first 2-way, then 3-way, and finally 4-way). For each level of interaction, we further trained the model by sequentially incorporating interaction terms, one at a time, and evaluating whether each term improved the model using the ANOVA F-test. The most informative interaction term was added to the model at each step. For higher-order (3-way and 4-way) interactions, we ensured that a proposed feature maintained the hierarchy constraint (i.e., a selected 4-way term must bring with it all of its associated 3-way and 2-way terms), thereby adding degrees of freedom to our F-test assessment. We repeated this process until no additional features further improved the model (i.e., all proposed features were P > 0.001 by the F-test). As our final model, we selected the trained model with the lowest mean-squared error, calculated via cross-validation within each substitution class. The 3-mer calculations considered all 2-way interactions plus single (i.e., position 3 and 5 only) features. More details in Supplementary Note.
Sourcing CpG methylation data
We obtained CpG methylation data for our intergenic regions of interest from whole genome bisulphite sequencing studies performed on germline19 (sperm, oocyte), blastocyst, blood and brain20 tissues. We performed our analysis on the 7,059,740 intergenic CpG sites that were methylated and the 651,479 intergenic CpG sites that were unmethylated in all 3 samples in the sperm tissue. We summarized the methylation signal across all samples for a tissue by calculating the mean intensity.
Sequence motif Identification
We examined the top and bottom 10 sequences for each substitution class, and manually identified a total of 6 motifs that we tested in each substitution class, stratified by CpG context. This results in a total of (9 substitution classes) * (2 tails, high and low) * (6 motifs) = 108 total tests. Note that we required a nominal P = 4.6 × 10−4 (Bonferroni correction for multiple testing). Testing was performed via Fisher’s exact test. More details in Supplementary Note.
Recombination and substitution rates
We obtained recombination rate map of the YRI population from the phase 1 release of the 1KG project, and segregated our intergenic noncoding regions of interest into high (rate >3 cM/Mb) and low recombination rate (rate < 0.05 cM/Mb) regions. More details in Supplementary Note.
Human and primate divergence
We obtained human-chimpanzee and human-macaque chain and netted alignments from the golden path directories in the UCSC genome browser and found divergence between the human-primate pair by calculating fixed differences between the aligned intergenic noncoding sequences at each 7-mer sequence context. More details in Supplementary Note.
Variants across the frequency spectrum
We defined the rare variants as those occurring fewer than two times in the population, and low or high frequency variants as those with MAF >1%. We only considered the intergenic noncoding variants present in 1KG project belonging to the African ancestry, and found 2,789,383 rare and 8,019,893 low/high frequency variants. More details in Supplementary Note.
De novo mutations
We only considered the de novo mutations occurring in the accessible regions of the 1KG project. For each motif class, we found the expected number of mutations under a normalized 1-mer sequence context model. More details in Supplementary Note.
Extension of the substitution probability framework in the coding region
To model substitution probabilities for the coding genome, we utilized the statistical model developed for intergenic regions with the following modifications: First, we accounted for codon position-effects (i.e., a given sequence context around a polymorphic site may occur at three different positions on a codon), which can lead to amino acid changes that may be subject to different levels of selective constraint. Second, we utilized probabilities learned from the intergenic noncoding region model as our Bayesian prior for the coding model. The parameters for this dirichlet distribution prior include the weighted baseline probabilities from the intergenic noncoding region as shape parameters. More details in Supplementary Note.
Selection of coding sequences
We selected exonic coordinates of the longest transcript for each gene annotated in ENSEMBL Biomart (version 75). We only considered those transcripts where (i) total exonic region length was a multiple of 3 and (ii) 90% or larger of it was present in the combined accessibility mask (version 20120824) filter of the 1KG project. This yielded 16,386 autosomal transcripts and 679 transcripts from the X chromosome.
To test our model in a different data set, SNP sites for ~4300 individuals of European ancestry were obtained from the Exome Variant Server (EVS, downloaded on August 26th 2013). For EVS data, to obtain a representative spectrum of allele frequencies (and impact of background selection) observed from the smaller set of individuals found in the 1KG data, we only considered variants with frequency greater than 0.03%. More details in Supplementary Note.
Annotation of SNP variants in the autosomal coding genome
For both 1KG and EVS data, we manually annotated the type of codon change caused by each variant specific to the transcript.
Scaling the substitution probability estimates for a larger sample
To calibrate our model (built using the 1KG dataset) for use with the larger EVS dataset, we rescaled the substitution probabilities estimated using 1KG data to make them proportional to the EVS dataset. We used a constant scaling factor defined as:
| (7) |
on all substitution probabilities in the new dataset.
Simulating variability in substitution probabilities within amino acid replacement classes
We start by randomly distributing the observed substitutions within the amino acid replacement class, using a fixed rate model. We then calculate the respective 7-mer probabilities from the randomized data set using our multinomial distribution model for randomization, and then find the variance in the new substitution probability estimates for that class. We use 106 simulations to generate the distribution of substitution probabilities.
Measuring the effects of selection on polymorphisms in the coding region
To minimize the effects of selection on initial estimates of substitution probabilities, we selected intergenic noncoding intervals for model development. Assuming that the mechanisms that introduce new mutations into coding regions are similar to those at work in the noncoding genome, we inferred that the relative ratio of coding-to-noncoding substitution probabilities could indicate natural selection occurring in the coding genome. To quantify the effect of selection on substitution probabilities, we measured the log10 ratio of coding-to-noncoding substitution probabilities using all coding variants observed in the 1KG African group. More details in Supplementary Note.
Calculating tolerance scores for genes
We find the expected distribution of polymorphism levels for each gene by performing simulations from the standard multinomial distribution using our coding substitution probability estimates. We then normalize the difference between the observed levels of polymorphism and those generated from simulations, to obtain gene tolerance score defined as:
| (8) |
where μNS and σNS represent the mean and standard deviation of nonsynonymous polymorphisms generated from simulations based on our model, and nNS is the empirical number of nonsynonymous polymorphism observed in the data. A positive gene score indicates that the number of observed substitutions is fewer than expected, and identifies genes experiencing stronger than average purifying selection.
Categorizing genes based on tolerance scores
We subdivided genes into various categories – i.e., essential genes (where the mouse homolog knock-out is lethal), ubiquitously expressed genes, genes with known phenotypes described in OMIM, immune-related genes, keratin genes, and olfactory genes. The dataset from33 was used to find the first two categories, while32 was used to classify OMIM genes. OMIM sub-categorizes genes according to mutational models, including de novo, dominant, haploinsufficient, or recessive. In our analysis, we merged OMIM’s de novo, dominant, and haploinsufficient categories, treating them as a single category. We used the DAVID ontology database (version 6.7) to classify immune-related, keratin, and olfactory genes. We considered the gene list published in the latest de novo sequencing analysis papers of Autism35, Epilepsy36, Intellectual disability38–40 and Developmental disorder37, as the gene set belonging to these diseases. We merged the gene lists of the aforementioned diseases, treating them as single category belonging to “All Neuropsychiatric disease”.
AUC comparison between competing gene scores on different gene sets
We used the receiver operating characteristic (ROC) curve to compare the performance of our gene scores against previously annotated scores for classifying genes into the gene sets we described above. We fitted a linear classifier using the three different gene scores, on each gene set and found the area under the curve (AUC) for each. More details in Supplementary Note.
Calculating tolerance scores for amino acids
We find the expected distribution of polymorphism levels for a specific amino acid within a gene by performing simulations from the standard multinomial distribution using our coding substitution probability estimates. Within a given gene, we then normalized the difference between the observed numbers of changes at a specific amino acid versus the number of changes expected from simulation using the equation:
| (9) |
where μAA and σAA represent the mean and standard deviation of the specific amino acid replacement polymorphisms generated from simulations based on our model, and nAA is the empirical number of amino acid replacement polymorphisms observed in the data. We consider the normalized value in Equation 9 as the final tolerance score for that amino acid within the given gene. We interpret a positive amino acid (AA) tolerance score to indicate that the observed number of changes for that specific amino acid within the given gene was even fewer than expected. Thus, the AA tolerance score serves to identify amino acids experiencing stronger than average purifying selection.
Sourcing information about pathogenic variants
We used the Human Gene Mutation Database (HGMD professional 2014.4) to identify pathogenic variants for our autosomal genes of interest, which supplied 60,504 variants distributed over 3,647 genes for 5,359 putative human disorders.
Application of gene and amino acid score on Autism spectrum de novo sequencing data
We used the de novo sequencing data for Autism spectrum disorder42, to test the efficacy of our gene and amino acid score approach in identifying and prioritizing novel genes and variants associated with Autism. We found the de novo mutations belonging to cases and controls separately for each of our genic sequences of interest and considered a total of 2,171 mutations in 2,508 cases and 1,421 mutations in 1,911 controls. For a uniform comparison of gene scores across different approaches12,32, we only considered the top 752 intolerant genes identified from each approach. We choose 752 genes because this was the number of intolerant genes identified in12, which mapped to our autosomal genic sequences of interest (i.e., which pass the stringent criteria of sequencing quality in the 1KG project). We used the Odds ratio to find the burden of de novo mutations in cases as opposed to controls, in the set of intolerant genes. Fisher’s exact test was used to compare the significance of burden. For amino acid score, all statistical comparisons were performed using the Wilcoxon sum ranked test. More details in Supplementary Note.
Code Availability
The computational pipelines used for probability estimation for the noncoding and coding genomes, and for forward regression and feature selection are available on request.
Supplementary Material
Acknowledgments
The authors would like to thank Casey Brown, Maja Bucan, Paul Babb, Katie Siewert, Kelsey Johnson, Stacie Bumgarner, and two anonymous reviewers for helpful comments on the manuscript. B.F.V. is grateful for support of the work from the Alfred P. Sloan Foundation (BR2012-087), the American Heart Association (13SDG14330006), the W.W. Smith Charitable Trust (H1201) and the NIH/NIDDK (R01DK101478).
Footnotes
URLs:
Exome Variant Server (EVS), http://evs.gs.washington.edu/EVS/
AUTHOR CONTRIBUTIONS:
V.A. and B.F.V. conceived and designed the experiments, developed the model, performed the statistical analysis, developed and contributed analysis tools, and wrote the paper. B.F.V. supervised the research.
COMPETING FINANCIAL INTERESTS:
The authors declare no conflict of interest.
References
- 1.Hodgkinson A, Eyre-Walker A. Variation in the mutation rate across mammalian genomes. Nat Rev Genet. 2011;12:756–66. doi: 10.1038/nrg3098. [DOI] [PubMed] [Google Scholar]
- 2.Ehrlich M, Wang RY. 5-Methylcytosine in eukaryotic DNA. Science. 1981;212:1350–7. doi: 10.1126/science.6262918. [DOI] [PubMed] [Google Scholar]
- 3.Rideout WM, Coetzee GA, Olumi AF, Jones PA. 5-Methylcytosine as an endogenous mutagen in the human LDL receptor and p53 genes. Science. 1990;249:1288–90. doi: 10.1126/science.1697983. [DOI] [PubMed] [Google Scholar]
- 4.Arbiza L, et al. Genome-wide inference of natural selection on human transcription factor binding sites. Nat Genet. 2013;45:723–9. doi: 10.1038/ng.2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang Y, et al. Clinical whole-exome sequencing for the diagnosis of mendelian disorders. N Engl J Med. 2013;369:1502–11. doi: 10.1056/NEJMoa1306555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hwang DG, Green P. Bayesian Markov chain Monte Carlo sequence analysis reveals varying neutral substitution patterns in mammalian evolution. Proc Natl Acad Sci U S A. 2004;101:13994–4001. doi: 10.1073/pnas.0404142101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blake RD, Hess ST, Nicholson-Tuell J. The influence of nearest neighbors on the rate and pattern of spontaneous point mutations. J Mol Evol. 1992;34:189–200. doi: 10.1007/BF00162968. [DOI] [PubMed] [Google Scholar]
- 8.Neale BM, et al. Patterns and rates of exonic de novo mutations in autism spectrum disorders. Nature. 2012;485:242–5. doi: 10.1038/nature11011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Michaelson JJ, et al. Whole-genome sequencing in autism identifies hot spots for de novo germline mutation. Cell. 2012;151:1431–42. doi: 10.1016/j.cell.2012.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fromer M, et al. De novo mutations in schizophrenia implicate synaptic networks. Nature. 2014;506:179–84. doi: 10.1038/nature12929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lawrence MS, et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature. 2013;499:214–8. doi: 10.1038/nature12213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Samocha KE, et al. A framework for the interpretation of de novo mutation in human disease. Nat Genet. 2014;46:944–950. doi: 10.1038/ng.3050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Abecasis GR, et al. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.The International HapMap Consortium. A haplotype map of the human genome. Nature. 2005;437:1299–320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Campbell MC, Tishkoff SA. African genetic diversity: implications for human demographic history, modern human origins, and complex disease mapping. Annu Rev Genomics Hum Genet. 2008;9:403–33. doi: 10.1146/annurev.genom.9.081307.164258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schaffner SF. The X chromosome in population genetics. Nat Rev Genet. 2004;5:43–51. doi: 10.1038/nrg1247. [DOI] [PubMed] [Google Scholar]
- 17.Nachman MW, Crowell SL. Estimate of the Mutation Rate per Nucleotide in Humans. Genetics. 2000;156:297–304. doi: 10.1093/genetics/156.1.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mugal CF, Ellegren H. Substitution rate variation at human CpG sites correlates with non-CpG divergence, methylation level and GC content. Genome Biol. 2011;12:R58. doi: 10.1186/gb-2011-12-6-r58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Okae H, et al. Genome-wide analysis of DNA methylation dynamics during early human development. PLoS Genet. 2014;10:e1004868. doi: 10.1371/journal.pgen.1004868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hovestadt V, et al. Decoding the regulatory landscape of medulloblastoma using DNA methylation sequencing. Nature. 2014;510:537–41. doi: 10.1038/nature13268. [DOI] [PubMed] [Google Scholar]
- 21.Walser JC, Furano AV. The mutational spectrum of non-CpG DNA varies with CpG content. Genome Res. 2010;20:875–82. doi: 10.1101/gr.103283.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kamiya H, et al. Mutagenicity of 5-formylcytosine, an oxidation product of 5-methylcytosine, in DNA in mammalian cells. J Biochem. 2002;132:551–5. doi: 10.1093/oxfordjournals.jbchem.a003256. [DOI] [PubMed] [Google Scholar]
- 23.Deaton AM, Bird A. CpG islands and the regulation of transcription. Genes Dev. 2011;25:1010–22. doi: 10.1101/gad.2037511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Levinson G, Gutman GA. Slipped-strand mispairing: a major mechanism for DNA sequence evolution. Mol Biol Evol. 1987;4:203–21. doi: 10.1093/oxfordjournals.molbev.a040442. [DOI] [PubMed] [Google Scholar]
- 25.Panchin AY, Mitrofanov SI, Alexeevski AV, Spirin SA, Panchin YV. New words in human mutagenesis. BMC Bioinformatics. 2011;12:268. doi: 10.1186/1471-2105-12-268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lanfear R, Welch JJ, Bromham L. Watching the clock: studying variation in rates of molecular evolution between species. Trends Ecol Evol. 2010;25:495–503. doi: 10.1016/j.tree.2010.06.007. [DOI] [PubMed] [Google Scholar]
- 27.Kong A, et al. Rate of de novo mutations and the importance of father’s age to disease risk. Nature. 2012;488:471–5. doi: 10.1038/nature11396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bustamante CD, et al. Natural selection on protein-coding genes in the human genome. Nature. 2005;437:1153–7. doi: 10.1038/nature04240. [DOI] [PubMed] [Google Scholar]
- 29.Fu W, et al. Analysis of 6,515 exomes reveals the recent origin of most human protein-coding variants. Nature. 2013;493:216–20. doi: 10.1038/nature11690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cooper GM, Shendure J. Needles in stacks of needles: finding disease-causal variants in a wealth of genomic data. Nat Rev Genet. 2011;12:628–40. doi: 10.1038/nrg3046. [DOI] [PubMed] [Google Scholar]
- 31.Stenson PD, et al. The Human Gene Mutation Database: building a comprehensive mutation repository for clinical and molecular genetics, diagnostic testing and personalized genomic medicine. Hum Genet. 2014;133:1–9. doi: 10.1007/s00439-013-1358-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Petrovski S, Wang Q, Heinzen EL, Allen AS, Goldstein DB. Genic intolerance to functional variation and the interpretation of personal genomes. PLoS Genet. 2013;9:e1003709. doi: 10.1371/journal.pgen.1003709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Georgi B, Voight BF, Bućan M. From mouse to human: evolutionary genomics analysis of human orthologs of essential genes. PLoS Genet. 2013;9:e1003484. doi: 10.1371/journal.pgen.1003484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Uddin M, et al. Brain-expressed exons under purifying selection are enriched for de novo mutations in autism spectrum disorder. Nat Genet. 2014;46:742–7. doi: 10.1038/ng.2980. [DOI] [PubMed] [Google Scholar]
- 35.De Rubeis S, et al. Synaptic, transcriptional and chromatin genes disrupted in autism. Nature. 2014;515:209–215. doi: 10.1038/nature13772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Allen AS, et al. De novo mutations in epileptic encephalopathies. Nature. 2013;501:217–21. doi: 10.1038/nature12439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Large-scale discovery of novel genetic causes of developmental disorders. Nature. 2015;519:223–8. doi: 10.1038/nature14135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hamdan FF, et al. De Novo Mutations in Moderate or Severe Intellectual Disability. PLoS Genet. 2014;10:e1004772. doi: 10.1371/journal.pgen.1004772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rauch A, et al. Range of genetic mutations associated with severe non-syndromic sporadic intellectual disability: an exome sequencing study. Lancet. 2012;380:1674–82. doi: 10.1016/S0140-6736(12)61480-9. [DOI] [PubMed] [Google Scholar]
- 40.De Ligt J, et al. Diagnostic Exome Sequencing in Persons with Severe Intellectual Disability. N Engl J Med. 2012;367:1921–1929. doi: 10.1056/NEJMoa1206524. [DOI] [PubMed] [Google Scholar]
- 41.Ginsburg D, Bowie EJ. Molecular genetics of von Willebrand disease. Blood. 1992;79:2507–19. [PubMed] [Google Scholar]
- 42.Iossifov I, et al. The contribution of de novo coding mutations to autism spectrum disorder. Nature. 2014;515:216–221. doi: 10.1038/nature13908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Orosco LA, et al. Loss of Wdfy3 in mice alters cerebral cortical neurogenesis reflecting aspects of the autism pathology. Nat Commun. 2014;5:4692. doi: 10.1038/ncomms5692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Eyre-Walker A, Eyre-Walker YC. How much of the variation in the mutation rate along the human genome can be explained? G3 (Bethesda) 2014;4:1667–70. doi: 10.1534/g3.114.012849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kimura M, Ohta T. On some principles governing molecular evolution. Proc Natl Acad Sci U S A. 1974;71:2848–52. doi: 10.1073/pnas.71.7.2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ségurel L, Wyman MJ, Przeworski M. Determinants of mutation rate variation in the human germline. Annu Rev Genomics Hum Genet. 2014;15:47–70. doi: 10.1146/annurev-genom-031714-125740. [DOI] [PubMed] [Google Scholar]
- 47.Hussin JG, et al. Recombination affects accumulation of damaging and disease-associated mutations in human populations. Nat Genet. 2015;47:400–404. doi: 10.1038/ng.3216. [DOI] [PubMed] [Google Scholar]
- 48.Koren A, et al. Genetic Variation in Human DNA Replication Timing. Cell. 2014;159:1015–1026. doi: 10.1016/j.cell.2014.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Siepel A, Haussler D. Phylogenetic estimation of context-dependent substitution rates by maximum likelihood. Mol Biol Evol. 2004;21:468–88. doi: 10.1093/molbev/msh039. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.