Abstract
Background
Rice is one of the most important crops for global food security but its productivity is adversely affected by salt stress prevalent in about 30 % of the cultivated land. For developing salt-tolerant rice varieties through conventional breeding or biotechnological interventions, there is an urgent need to identify natural allelic variants that may confer salt tolerance. Here, 299 wild rice accessions collected from different agro-climatic regions of India were evaluated during growth under salt stress. Of these 95 representative accessions were sequenced for members of HKT ion transporter family genes by employing Ion Torrent PGM sequencing platform.
Results
Haplotype analysis revealed haplotypes H5 and H1 of HKT1;5 and HKT2;3, respectively associated with high salinity tolerance. This is the first study of allele mining of eight members of HKT gene family from Indian wild rice reporting a salt tolerant allele of HKT2;3. HKT1;5 also showed a salt tolerant allele from wild rice. Phylogenetic analysis based on the nucleotide sequences showed different grouping of the HKT family genes as compared to the prevailing protein sequence based classification.
Conclusions
The salt tolerant alleles of the HKT genes from wild rice may be introgressed into modern high yielding cultivars to widen the existing gene pool and enhance rice production in the salt affected areas.
Electronic supplementary material
The online version of this article (doi:10.1186/s12284-016-0083-8) contains supplementary material, which is available to authorized users.
Keywords: Allele mining, HKT, Association, Salt stress, NaCl, Wild rice
Background
Rice (Oryza sativa L.) is cultivated around the world and consumed by more than 50 % of the global human population (Mohanty 2013). The current status of rice production is 495.63 million tonnes (MT) which increased by only 9 MT in the preceding 4 year block (2011–2014) as compared to 80 MT increase in two such 4 year blocks during 2004–2011 (http://faostat3.fao.org/browse/Q/*/E). In contrast, the global population is projected to increase by 25 % to 9.2 billion by 2050 (Schroeder et al. 2013). Rice production must increase correspondingly by 70 % to fulfil the growing demand (FAOSTAT 2009). Fifteen percent of the land currently used for agricultural practices is at saturation point to maintain environmentally sustainable production, therefore innovations are needed to enhance crop productivity to meet the projected demand (Rockström et al. 2009). However, multiple abiotic stresses specifically salinity changes the soil texture and creates unfavourable conditions for rice production by severe inhibition of plant growth and development (Miller and Donahue 1995). Globally 45 Mha of irrigated and 32 Mha of rain fed agriculture are affected by salinity (http://www.fao.org/soils-portal/en/). Furthermore, irrigation with brackish water, tidal waves and tsunami continue to increase the soil salinity (Schroeder et al. 2013). According to one estimate up to 50 % of the present arable land may be affected by salinity by 2050 (Wang et al. 2003). Therefore, we need to explore natural genetic resources to find novel genes and alleles that can help withstand high salt concentration and maintain crop productivity.
Salinity is characterised by presence of exchangeable sodium ions (Na+) which imposes both ion toxicity and osmotic stress on the plant and alters the physiological status and ionic homeostasis of the cell. Crop plants are vulnerable to salinity and further their level of sensitivity and mechanism of tolerance depends on the growth stage of the plant (Hossain et al. 2015). Rice is most sensitive at the seedling stage and beyond 3 dSm−1 of electrical conductivity, seedlings are considered under stress and yield loss due to salinity is measured at twelve percent reduction per dSm−1 above threshold level (Lutts et al. 1995; Maas and Hoffman 1977). However, in most of the modern high yielding rice varieties, 50 % yield reduction occurs at 6 dSm−1 and they become totally unproductive beyond 12 dSm−1 (Linh et al. 2012). A limited number of large scale screening of germplasm for salinity stress has been conducted (Platten et al. 2013). Most of the genetic studies have focused on Saltol QTL derived from Indian rice landrace Pokkali which provides seedling stage salt tolerance (Thomson et al. 2010). There is very little work on the genetics of reproductive stage salt tolerance in rice (Mohammadi et al. 2013). The present understanding of salt tolerance mechanisms has facilitated exploitation of spatially located membrane transporters (Zhu 2001; Munns and Tester 2008). The known mechanisms include sequestration of ions in vacuole and exclusion of ions from root and leaves (Munns and Tester 2008). Thus, allelic variations in the sequence of ion transporter genes are likely to play a significant role in providing effective tolerance to salt stress.
In the present study we selected eight different transporters of the HKT gene family. These genes are further classified into two subfamilies based on their amino acid sequence similarity and differences in their Na+ and K+ selectivity (Horie et al. 2001), though product of each of these is a transmembrane protein (Horie et al. 2009; Mäser et al. 2001; Platten et al. 2006). Functionally, products of subfamily 1 [HKT1;1, HKT1;2, HKT1;3, HKT1;4 and HKT1;5] are Na + specific transporters and have S-G-G-G signature while products of subfamily 2 [HKT2;1, HKT2;2, HKT2;3 and HKT2;4] are Na+-K+ co-transporters or Na+-K+ uniporters with G-G-G-G signature (Jabnoune et al. 2009; Mäser et al. 2002). Oomen et al. 2012 has reported a hybrid of HKT2;1 and HKT2;2 gene as HKT2;2/1 from Nona Bokra which has strong permeability to Na+ and K+ even at high external Na+ concentrations. Spatial localization and differential expression of these genes further enhances their importance. Horie et al. (2001) reported two isoforms of HKT transporters, a Na + transporter OsHKT1 and a Na+- K+-coupled transporter OsHKT2, which may act harmoniously in the salt tolerant indica rice. Further studies reported that OsHKT1;4 is expressed around xylem in the leaf sheath while OsHKT1;5 is expressed around the root xylem (Cotsaftis et al. 2011). Similarly, OsHKT2;1 and OsHKT2;4 are expressed in the outer part of the root and in the root hairs and may provide an entry point for Na+ into plant roots from the soil (Lan et al. 2010; Schachtman and Schroeder 1994). HKT2;1 expression is significantly upregulated in the root cortex under K+ starvation and high Na+ concentration (Almeida et al. 2013; Horie et al. 2001). Further, it was proven that the Na+ influx into K+-starved rice roots was primarily OsHKT2;1-dependent while at concentration >30 mM NaCl, it was permeable to Na+ only (Jabnoune et al. 2009). Another class 2 transporter gene OsHKT2;4 shows 93 % similarity with OsHKT2;3 and mediates K+ transport independent of Na+ concentration (Horie et al. 2011).
The level of salt tolerance provided by known transporter genes in the modern rice cultivars is insufficient and hence new genes or novel allelic variants of these transporters are needed for enhanced salinity tolerance. Crop domestication has selected only a few agronomically desirable alleles and left behind vast pool of genetic diversity in the wild progenitor species due to domestication bottleneck (Tanksley and McCouch 1997). Potential implication of germplasm and crop wild relatives under extreme environment conditions has been reviewed (Mickelbart et al. 2015). Allelic variant of members of HKT transporters such TmHKT1;5-A (Munns et al. 2012), TaKHT1;5-D (Yang et al. 2014) has been introgressed from wild relatives that led to increase in yield of the plant. Allele mining identifies superior alleles from related genotypes that may have been the effect of mutations in the process of evolution. The superior alleles can be used to develop allele specific markers and use them in marker assisted selection and also in tracing the evolution of alleles. Sequencing based allele mining and association analysis is an effective strategy to unravel the hidden potential of wild rice germplasm. Allele mining has been used across the crop species and novel alleles have been identified for abiotic stress tolerance genes in rice (Latha et al. 2004; Negrão et al. 2013; Platten et al. 2013; Singh et al. 2015a), maize (Yu et al. 2010) and barley (Cseri et al. 2011). However, a collection of untapped germplasm is required to mine novel desirable alleles and identify nucleotide sequence variations associated with these alleles (Kumar et al. 2010). India has a wealth of untapped wild rice germplasm that requires hasty expeditions to collect and exploit this fast depleting genetic resource (Singh et al. 2013). The genes already exploited from Indian wild rice include grassy stunt virus resistance from Oryza nivara (Khush and Ling 1974), Bph 19(t) from Oryza rufipogon (Li et al. 2010), Xa38 from Oryza nivara (Bhasin et al. 2012), salinity tolerance genes, inositol methyl transferase (Sengupta et al. 2008) and L-myo-inositol 1-phosphate synthase from wild rice Porteresia coarctata (Das-Chatterjee et al. 2006). The male sterility (MS) gene from O. rufipogon was introduced into the cultivated rice, leading to development of high yielding hybrid rice (Yuan et al. 1993). The beneficial alleles derived from wild relatives of rice have been transferred into elite genetic backgrounds leading to enhanced trait performance in rice (McCouch et al. 2007; Xiao et al. 1996; Xiao et al. 1998).
In the present study, 299 accessions of rice comprising O. rufipogon and O. nivara were screened for their level of salt tolerance. A candidate gene based allele mining was used to find natural allelic variants of agronomically important HKT family genes. Genes were re-sequenced with their promoters from 103 accessions of wild and cultivated rice varieties and their nucleotide and haplotype diversity was analysed. SNP based association analysis was done to link traits for salt tolerance e.g., SPAD for chlorophyll, Na+ and K+ concentration in the shoot and root of rice. Further, merit of HKT1;5 and HKT2;3 were discussed for their potentiality in salt tolerance of rice.
Results
Phenotypic Screening of the Wild Rice Germplasm for Salt Tolerance
Based on standard evaluation system (SES) of IRRI (Gregorio et al. 1997), 299 wild rice accessions were screened and classified according to their level of salt tolerance (Fig. 1, Additional file 1: Table S1). Two accessions NKSWR173 and NKSWR202 were found highly tolerant with SES score 1 as compared to the tolerant check FL478 with SES score 3 after 15 days of salt stress, 30 accessions were found tolerant at 10 days of salt stress, of which 14 accessions maintained the tolerance comparable to FL478 till fifteenth day. Total 84 accessions were found moderately tolerant, 115 sensitive and 67 highly sensitive at 10 days of salt treatment. However, at fifteenth day only 28 accessions were moderately tolerant, while 69 accessions became sensitive and 186 highly sensitive. One accession, O. nivara 336676 displayed interesting result showing high tolerance up to tenth day and thereafter it became highly sensitive and died within the next 2 days in all three years of the experiment (Additional file 1: Table S1). After this primary screening, 45 selected accessions, including all tolerant lines and representative lines from the other sensitive classes, were further evaluated for salt tolerance parameters such as biomass, chlorophyll content (using SPAD meter) and Na + and K+ concentrations in replicated tests. The Na+/K+ ratio in shoot under stress ranged from 0.8 to 2.1 with the maximum value in O. rufipogon 336679 and the minimum in CSR27. The lowest Na+/K+ ratio (1.3) in the root was observed in a tolerant line NKSWR232 and the highest ratio (3.3) was in NKSWR144. FL478 had highest chlorophyll content while VSR156 and NKS074 had no chlorophyll as the plants died (Additional file 2: Table S2).
Fig. 1.

Response of wild rice accessions after 15 days of 150 mM of salt stress in hydroponics. a Border; b NKSWR149; c NKSWR112; d NKSWR104; e NKSWR092; f NKSWR119; g NKSWR085; h NKSWR115; i NKSWR132; j NKSWR097; k Oryza nivara330646; l NKSWR143; m NKSWR101; n NKSWR079; o FL478; p VSR156; q Border (NKSWR143 is tolerant other accessions are sensitive, FL478- tolerant check, VSR156- sensitive check)
Nucleotide Diversity, Haplotype Diversity and Haplotype Networks of the HKT Genes
Eight HKT transporter family genes along with their promoters were re-sequenced from 103 accessions of wild rice and cultivated rice varieties and their nucleotide and haplotype diversity was analysed (Table 1). Nucleotide diversity (π) was the highest (0.00169) for HKT2;3 and the lowest (0.00012) for HKT1;1 gene. Maximum haplotype diversity (HD) of 0.891 was found in HKT2;3 gene followed by HKT1;4 (0.692) and the lowest in HKT1;1 (0.084) gene (Table 1). Haplotype analysis showed the maximum 23 haplotypes each for the HKT1;5 and HKT2;3 genes with 45 and 28 SNP sites, respectively. The HKT2;4 gene showed the least number of 5 haplotypes. The Tajima’s D test, Fu and Li’s D* test and Fu and Li’s F* statistics were performed to distinguish between random versus non-random mutations. All the test values were analysed for entire gene, as well as separately for the coding and non-coding regions. Except for the coding region of all accessions, which had positive values of Tajima’s D test, all other regions of the genes among each class individually and in group deviated significantly from neutrality and Tajima’s D test values were negative.
Table 1.
Nucleotide and haplotype diversity and tests of neutrality for eight high affinity potassium transporter (HKT) family genes from 95 Indian wild rice accessions and 8 cultivated varieties
| Candidate gene (Locus ID) | Region | S | π | θω | H | HD | D | D* | F* |
|---|---|---|---|---|---|---|---|---|---|
| HKT1;1 | Coding | 8 | 0.00013 | 0.00084 | 10 | 0.217 | −2.10133 | −3.97252 | −3.94393 |
| (LOC_Os04g51820) | Non-coding | 3 | 0.0001 | 0.00059 | 4 | 0.076 | −1.48732 | −2.01292 | −2.16727 |
| All | 11 | 0.00012 | 0.00076 | 13 | 0.084 | −2.22874 | −4.21517 | −4.17222 | |
| HKT1;2 | Coding | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| Non-coding | 20 | 0.00027 | 0.00135 | 12 | 0.387 | −2.30631 | −4.55086 | −4.42942 | |
| Chr4:30548314..30545885 | All | 20 | 0.00026 | 0.00131 | 12 | 0.387 | −2.30631 | −4.55086 | −4.42942 |
| HKT1;3 | Coding | 8 | 0.00019 | 0.00108 | 9 | 0.25 | −1.99519 | −2.80081 | −2.99198 |
| (LOC_Os02g07830) | Non-coding | 6 | 0.00067 | 0.00137 | 4 | 0.164 | −1.14077 | 0.15513 | −0.32717 |
| All | 14 | 0.00037 | 0.00118 | 10 | 0.29 | −1.88141 | −1.8801 | −2.24114 | |
| HKT1;4 | Coding | 2 | 0.00031 | 0.00091 | 3 | 0.128 | −1.01249 | −1.0804 | −1.23948 |
| (LOC_Os04g51830) | Non-coding | 42 | 0.00059 | 0.00191 | 19 | 0.69 | −2.1536 | −3.07965 | −3.2462 |
| All | 44 | 0.00057 | 0.00182 | 20 | 0.692 | −2.15775 | −3.12193 | −3.27713 | |
| HKT1;5 | Coding | 8 | 0.00025 | 0.00183 | 8 | 0.166 | −2.08122 | −3.61227 | −3.65909 |
| (LOC_Os01g20160) | Non-coding | 37 | 0.0012 | 0.00322 | 18 | 0.571 | −1.94371 | −2.70539 | −2.87895 |
| All | 45 | 0.00094 | 0.00284 | 23 | 0.631 | −2.10258 | −3.40294 | −3.44926 | |
| HKT2;1 | Coding | 5 | 0.00015 | 0.00073 | 5 | 0.18 | −1.66589 | −3.23816 | −3.20982 |
| (LOC_Os06g48810) | Non-coding | 46 | 0.00042 | 0.00241 | 17 | 0.333 | −2.60346 | −4.60994 | −4.55342 |
| All | 51 | 0.00035 | 0.00196 | 18 | 0.421 | −2.61003 | −4.91693 | −4.76765 | |
| HKT2;3 | Coding | 24 | 0.00238 | 0.00353 | 23 | 0.891 | −0.96284 | −1.72038 | −1.71118 |
| (LOC_Os01g34850) | Non-coding | 4 | 0.00013 | 0.00132 | 3 | 0.038 | −1.77201 | −3.90552 | −3.78567 |
| All | 28 | 0.00169 | 0.00284 | 23 | 0.891 | −1.22676 | −2.67757 | −2.5308 | |
| HKT2;4 | Coding | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| (LOC_Os06g48800) | Non-coding | 4 | 0.0012 | 0.00448 | 5 | 0.198 | −1.44156 | −1.48409 | −1.72999 |
| All | 4 | 0.0012 | 0.00448 | 5 | 0.198 | −1.44156 | −1.48409 | −1.72999 |
(S SNP/Indel sites, π nucleotide diversity, θω nucleotide diversity with Watterson’s estimator, H number of haplotypes, HD haplotype diversity, D Tajima’s D statistic, D* Fu and Li’s D* statistic, F*: Fu and Li’s F* statistic.)
Haplotype analysis of HKT1;1 gene showed 13 haplotypes with haplotype number H12 having a single highly tolerant accession and a distinguishing guanine residue instead of adenine in the first intron. HKT1;2 gene has diversified to 13 haplotypes, HKT1;3 into 10 haplotypes and HKT1;4 to 20 haplotypes. Network analysis of HKT1;5 gene showed that out of the total 23 haplotypes of this gene two haplotypes, H5 and H22 included all the tolerant accessions. Haplotype H5 had 13 accessions, including tolerant cultivated varieties FL478 and CSR27 along with sensitive variety VSR156 and five sensitive accessions from Gujrat. A unique haplotype H22 was represented by salt tolerant cultivated rice variety CSR11, which also matched with the published sequence of tolerant Indian rice variety Nona Bokra (Ren et al. 2005). The members of HKT sub-family 2 genes, HKT2;1 and HKT2;4 showed 18 and 5 haplotypes, respectively. Further, haplotype analysis of HKT2;3 gene showed that its haplotype H1 included four tolerant wild rice accessions along with the salt tolerant check varieties FL478 and CSR27. However, another salt tolerant variety CSR11 possessing haplotype H10 grouped separately along with tolerant O. nivara accession 330641. A phenotypically unique accession, showing tolerance only up to 10 days of stress followed by extreme sensitivity, had a unique haplotype H5 of the HKT2;3 gene (Fig. 2).
Fig. 2.
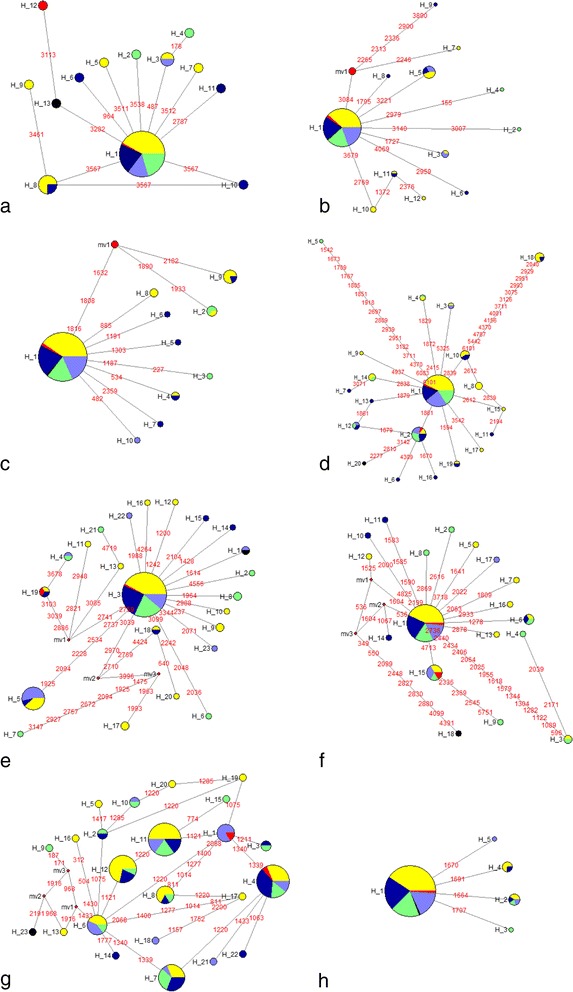
Haplotype networks of eight HKT transporter genes including (a) HKT1;1 (b) HKT1;2 (c) HKT1;3 (d) HKT1;4 (e) HKT1;5 (f) HKT2;1 (g) HKT2;3 (h) HKT2;4. Each circle represents a haplotype and size of each circle is proportional to haplotype (allele) frequency. Colour coding represents phenotype class based on standard evaluation system (SES) score. (Red-highly tolerant, Purple-tolerant, Green moderately tolerant, Blue-Sensitive, Yellow-highly sensitive)
Evolution of the HKT Family Genes
A phylogenetic tree was constructed using nucleotide sequences of the eight HKT family genes using NJ method. Sorghum HKT gene taken from sorghum gene database was included as outgroup. As expected the tree showed total eight clades representing the individual HKT genes that clustered into two major groups. The phylogenetic tree revealed a somewhat different relationship among the HKT transporter genes as compared with their functional classification. The HKT1;3 gene is at the root of divergence as it was most closely related to the sorghum HKT gene. A close relationship was observed between the HKT1;3, HKT1;2 and HKT2;3. The remaining five genes were clustered in group I and showed that HKT1;4 was most primitive followed by HKT1;5 and HKT1;2. The recently evolved genes HKT2;4 and HKT2;1 are located in tandem on chromosome 6 and very closely related to each other diverging from a common branching point (Fig. 3).
Fig. 3.
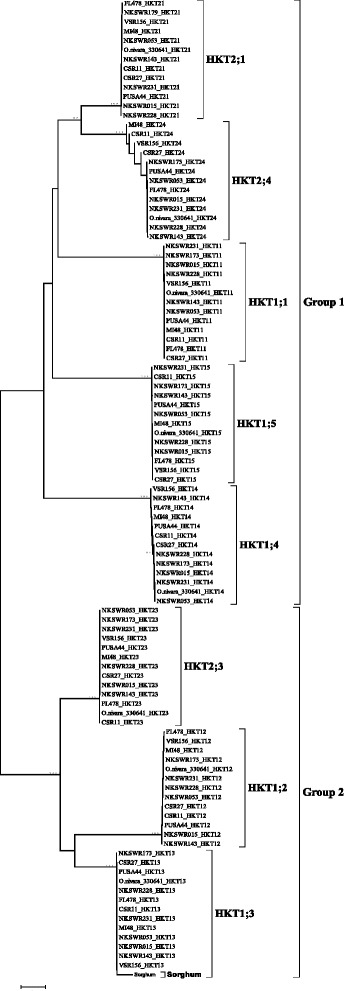
Minimum evolution phylogenetic tree of eight HKT genes re-sequenced from representative wild rice accessions and cultivar checks constructed using MEGA 5.1 software
Association of HKT Gene SNPs with Salt Tolerance
The association study was conducted on 45 accessions including 3 check rice varieties, of which, 18 belonged to tolerant class, 9 moderately tolerant, 7 sensitive and 11 highly sensitive. For association study a Bayesian based analysis of population structure was conducted which used an ad hoc statistics (∆K), the rate of change in log probability of data between successive K values. It showed that the highest log likelihood was at K = 3 suggesting three major sub-population in the analysed Indian wild rice accessions. The individual assignments and Q-matrix in the three different sub-population revealed that sub-population I included 51.4 %, sub-population II 33.8 % and sub-population III 14.8 % of the analysed accessions. The mean Fst values were 0.8981, 0.6154 and 0.7291 for the sub-populations I, II and III, respectively (Fig. 4, Additional file 3: Table S3).
Fig. 4.
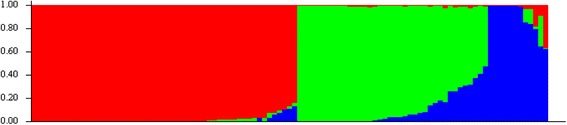
Distribution of 103 wild rice accessions and cultivated rice into three different sub-populations based on 48 genome wide SNP markers. X-axis shows rice accessions and y-axis represents Fst values (Blue: Population I; Green: Population II; Red: Population III)
SNP based association analysis showed significant associations with the salt tolerance traits using MLM_Q + K statistical method. Thus, the results were filtered with association values greater than 5 % and p value less than 0.01. A total of 50 SNPs from all the eight HKT genes showed significant associations with the salt tolerance traits (Table 2, Additional file 4: Figures S1, S2).
Table 2.
Association of SNPs with different salt related traits at 10 days of treatment with HKT genes (with R2 > 5 % and p < 0.01)
| Gene | Trait | SNP | Site | p | R2 (%) | SNP type | MAF | Gene | Trait | SNP | Site | p | R2 (%) | SNP type | MAF |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| HKT 1;1 | N_R | S0231 | 30724332 | 0.003 | 34.43 | Int | 37 | HKT 1;5 | N_S | S0077 | 11459042 | 0.002 | 31.57 | Int | 36 |
| K_S | S1955 | 30726056 | 0.003 | 30.75 | Syn | 5 | N_S | S3893 | 11462858 | 0.005 | 33.00 | P/A140 | 20 | ||
| SPAD | S2094 | 30726195 | 0.003 | 31.90 | S/N258 | 32 | K_S | S0077 | 11459042 | 0.003 | 30.92 | Int | 36 | ||
| SPAD | S2331 | 30726432 | 0.003 | 31.90 | S/F179 | 30 | N_R | S0887 | 11459852 | 0.003 | 34.12 | Int | 13 | ||
| SPAD | S2546 | 30726647 | 0.003 | 31.90 | Syn | 30 | N_R | S0919 | 11459884 | 0.009 | 34.42 | Int | 35 | ||
| HKT 1;2 | K_S | S0221 | 30545988 | 0.003 | 31.83 | 5’ NC | 36 | N_R | S0958 | 11459923 | 0.009 | 34.65 | Int | 35 | |
| K_S | S1430 | 30547197 | 0.009 | 25.16 | Int | 21 | N_R | S0959 | 11459924 | 0.003 | 34.12 | Int | 13 | ||
| K_S | S2282 | 30548049 | 0.003 | 31.54 | Int | 31 | N_R | S0974 | 11459939 | 0.008 | 35.09 | Int | 6 | ||
| HKT 1;3 | N_R | S0337 | 4103487 | 0.005 | 33.93 | C/R16 | 30 | N_R | S1036 | 11460001 | 0.002 | 37.37 | Int | 5 | |
| SPAD | S0091 | 4103241 | 0.009 | 27.83 | 5’ NC | 12 | K_R | S1036 | 11460001 | 0.008 | 26.80 | Int | 5 | ||
| N_R | S0744 | 4103894 | 0.008 | 38.19 | Syn | 22 | N_R | S1379 | 11460344 | 0.003 | 34.44 | K/Q429 | 49 | ||
| SPAD | S0744 | 4103894 | 0.008 | 35.21 | Syn | 22 | N_R | S1913 | 11460878 | 0.003 | 34.62 | Int | 43 | ||
| SPAD | S1682 | 4104832 | 0.009 | 27.83 | Int | 10 | N_R | S2043 | 11461008 | 0.004 | 33.62 | Int | 29 | ||
| SPAD | S1762 | 4104912 | 0.005 | 31.75 | Int | 45 | K_S | S2146 | 11461111 | 0.002 | 34.93 | Int | 45 | ||
| SPAD | S2347 | 4105497 | 0.003 | 34.78 | A/V488 | 23 | K_S | S2347 | 11461312 | 0.006 | 27.43 | Int | 28 | ||
| SPAD | S2374 | 4105524 | 0.004 | 32.49 | R/Q497 | 46 | K_S | S2865 | 11461830 | 0.001 | 36.63 | Int | 14 | ||
| SPAD | S2392 | 4105542 | 0.009 | 27.68 | T/M503 | 13 | K_S | S3893 | 11462858 | 0.009 | 31.17 | Int | 20 | ||
| HKT 1;4 | SPAD | S0886 | 30734275 | 0.003 | 34.72 | 3’NC | 32 | HKT 2;1 | K_S | S3062 | 29541671 | 0.008 | 32.35 | 5’ NC | 7 |
| SPAD | S1267 | 30734656 | 0.003 | 34.72 | 3’NC | 32 | N_R | S3062 | 29541671 | 0.004 | 39.70 | 5’ NC | 7 | ||
| SPAD | S2284 | 30735673 | 0.002 | 37.42 | Int | 32 | SPAD | S4153 | 29542762 | 0.009 | 25.80 | 5’ NC | 38 | ||
| SPAD | S2804 | 30736193 | 0.003 | 34.72 | Int | 32 | SPAD | S4377 | 29542986 | 0.002 | 33.96 | 5’ NC | 36 | ||
| SPAD | S2974 | 30736363 | 0.002 | 37.03 | Int | 33 | K_S | S5815 | 29544424 | 0.002 | 40.52 | 5’ NC | 21 | ||
| SPAD | S3185 | 30736574 | 0.002 | 37.03 | Int | 32 | K_S | S5817 | 29544426 | 0.007 | 32.66 | 5’ NC | 7 | ||
| SPAD | S3228 | 30736617 | 0.007 | 36.36 | Int | 33 | HKT 2;3 | K_S | S0356 | 19241884 | 0.002 | 34.84 | Int | 10 | |
| SPAD | S3307 | 30736696 | 0.003 | 34.87 | Int | 28 | SPAD | S0761 | 19242289 | 0.003 | 31.88 | I/T77 | 34 | ||
| SPAD | S3544 | 30736933 | 0.003 | 34.72 | Int | 32 | K_S | S1001 | 19242529 | 0.009 | 25.08 | I/T157 | 5 | ||
| SPAD | S4678 | 30738067 | 0.003 | 35.09 | Int | 35 | SPAD | S2053 | 19243581 | 0.003 | 31.79 | Syn | 34 | ||
| HKT 2;4 | K_S | S0334 | 29534433 | 0.004 | 37.20 | Int | 37 |
(N_S sodium concentration in shoot, K_S potassium concentration in shoot, N_R sodium concentration in root, K_S potassium concentration in root, Int intron, Syn synonymous, NC non-coding, MAF minor allele frequency in %)
Among the HKT subfamily I members, analysis of associated SNPs of OsHKT1;1 gene revealed two non-synonymous substitutions, (i) S2094 (Ser258 to Asp258) and (ii) S2331 (Ser179 to Phe179) that were significantly associated with SPAD chlorophyll content. These two SNPs are spatially located, one (S2094) on the extracellular side and the other on the cytoplasmic side (S2331). In addition, one SNP (S0231) in the intronic region showed highly significant association with sodium concentration in the root. Further, two synonymous SNPs were associated with potassium concentration in the shoot (S1955) and SPAD chlorophyll content (S2546). In case of HKT1;2 gene three SNPs were associated with potassium concentration in the shoot, two of these (S1430 and S2282) were present in the intronic region while third one (S221) was in the upstream promoter region. In case of OsHKT1;3 a non-synonymous SNP S337 (Cys16 to Arg16) present in the extracellular domain of the protein explained 34 % of phenotypic variance (R2 value) and was associated with sodium concentration in the root while, three non-synonymous SNPs in the cytoplasmic domain, S2374 (Ala488 to Val488), S2374 (Arg479 to Glu479) and S2392 (Thr503 to Met503) showed association with SPAD chlorophyll content. A synonymous SNP (S744) was associated with potassium concentration in the shoot and SPAD chlorophyll content. Interestingly, the SNP in the promoter region (S91) and those in the intronic region (S1682 and S1762) were also associated with the SPAD chlorophyll value. All SNPs identified in the OsHKT1;4 gene were associated with SPAD chlorophyll content, among them, two were in the downstream promoter region while eight were in the intronic region. Of the two SNPs in the downstream promoter region (S886 and S1267), one was in the consensus GT-1 binding site and the other one in the ACGT sequence. In the OsHKT1;5 gene total 14 SNPs were associated with the salt tolerance traits. Of which 12 were in the intronic region while remaining 2, S1379 (Lys429 Glu429) and S3893 (Pro140 Ala140), were non-synonymous. Both the non-synonymous SNPs were associated with sodium concentration: S1379 was located on extracellular side and showed significant association with the concentration of sodium in the root while, the other S3893 was on the cytoplasmic side and showed association with concentration of sodium in shoot, explaining 32 % (R2 value) of the phenotypic variance (Table 2).
From the HKT subfamily II, OsHKT2;1 gene had five SNPs in the promoter region showing association with the salt tolerance traits. Three of these (S3062, S5815, and S5817) were associated with shoot potassium concentration, of which S3062 was also associated with root sodium concentration, while the other two, S4153, and S4377, were associated with SPAD chlorophyll content. In OsHKT2;3 a total of 4 SNPs were associated with the salt tolerance traits. Of the two non-synonymous SNPs, S1001 was associated with potassium concentration in shoot (extracellular side) while S761 was associated with SPAD chlorophyll content (cytoplasmic side), both the SNPs resulted in Isoleucine to Threonine substitution at amino acid positions 157 and 77, respectively. In addition, one synonymous SNP S2053 was associated with SPAD chlorophyll content and another SNP S356 in the intronic region was associated with potassium concentration in shoot. Only one SNP S334 present in the intronic region of OsHKT2;4 gene was associated with potassium concentration in the shoot (Table 2).
Discussion
Wild rice is expected to play important role in rice improvement in the coming years. In this study we explored untapped diversity of Indian wild rice to identify natural alleles of the HKT transporter family genes. Analysis of nucleotide sequence variations for eight HKT family genes in wild rice showed higher nucleotide and haplotypic diversity as compared to the cultivated rice varieties (Table 1, Fig. 2). This supports the notion that wild relatives are genetically much more diverse than their cultivated counterparts (Hoisington et al. 1999). However, nucleotide variations of a gene is also associated with respect to position in the gene (McNally et al. 2006). Platten et al. (2013) observed comparatively lower nucleotide diversity and haplotypic diversity in cultivated rice and identified relationship between different haplotypes and salt tolerance for HKT1;5 gene. Our results indicate that nucleotide diversity was quite different from haplotype diversity as only effective SNPs participated in haplotypic group determination (Goodall-Copestake et al. 2012). Differences between nucleotide and haplotype diversity has been measured across different genes such as Sucrose Synthase 3 in rice (Lestari et al. 2011) and OsDREB1F in wild and cultivated rice (Singh et al. 2015a).
To understand the natural selection process in the evolutions of HKT genes, Tajima’s D test, Fu and Li’s D* test and Fu and Li’s F* statistics were calculated. Negative values obtained for wild rice population shows an excess of rare polymorphisms which still undergoes some population expansion and positive selection pressure and were further validated by Ka/Ks ratio and Jukes and Counter correction values (>1). However, positive value for coding region in cultivated rice revealed that alleles had evolved either by balancing selection or population bottlenecks (Table 1, Akey et al. 2004).
Among the eight genes only two HKT1;5 and HKT2;3 showed tolerant haplotypes. Functionally, HKT1;5 is a Na+ specific transporter that maintains Na+/K+ homeostasis in the leaves under salt stress (Ren et al. 2005; Mickelbart et al. 2015), whereas HKT2;3 regulates K+ transport independent of Na+ concentration (Horie et al. 2011). The genotypes having tolerant allele of HKT1;5 showed lower Na+/K+ concentration in the shoot as compared to root (Thomson et al. 2010). Six wild rice accessions that showed lower shoot Na+ grouped into four separate haplotypic group. In addition, four other accessions that showed low Na+ concentration in shoot comparable to the tolerant checks fell into separate haplotypic groups indicating allelic variation for the HKT1;5 gene among the accessions (Fig. 2, Additional file 2: Table S2). An exceptionally higher Na+ and K+ ratio was observed in the shoots of a tolerant accession O. rufipogon 336679 indicating presence of an alternate mechanism to sustain such a high level of salt concentration in shoot. Mian et al. (2011) identified role of HKT 2;1 gene in barley and reported increased shoot Na + concentrations and improved biomass production under salt stress. Haplotype analysis of HKT2;3 gene showed almost even distribution of accessions into different haplotypic groups, but it also formed a separate haplotype for the reference sequence. Of the total 23 observed haplotypes, one was associated with tolerant and two were associated with moderately tolerant genotypes. A unique accession of O. nivara 336676 showing tolerance only up to tenth day of applying the stress separated in a different haplotype indicating that it is possibly following some different tolerance mechanism which breaks down beyond 10 days of stress. Salt susceptibility of this accessions after 10 days might be due to higher accumulation of Na+ in root (Additional file 2: Table S2). Another plausible explanation for the tolerance in this accession is presence of root specific tonoplast transporters with some accessory factors associated with the genes for providing energy to transport ions into vacuole but with limited accumulation capacity of ions in vacuole. Functionally, HKT2;3 mediates K+ transport independent of Na+ concentration hence, even at high salt concentration the physiological functions that are vital to crop plants relating to K+ e.g., stomatal movement, CO2 uptake and cofactor to make enzymes etc. are maintained inside the cell. It also maintains ionic homeostasis (Horie et al. 2007; Roy et al. 2014). However, haplotype analysis revealed uneven distribution of tolerant genotypes among different haplotypic group for different genes. It shows that an unknown type of mechanism might be present among the Indian wild rice that could help to enhance our understanding towards salt tolerance mechanism and be used for improvement of cultivated rice.
Phylogenetic analysis describes the evolutionary relationship among genes and different clustering patterns indicate the level of their functional divergence (Mäser et al. 2001). Here, two genes of phylogenetic group I (HKT2;1 and HKT 2;4) showed high nucleotide similarity with each other. They most likely have evolved from recent duplication of a single gene as also suggested by their tandem location in the rice chromosome 6. Interestingly, in the phylogenetic tree functionally diverse, HKT1;3 and HKT1;2 genes grouped together along with HKT2;3. This suggests that members of functionally different groups might have evolved from a common ancestor through duplication and subsequent functional divergence.
Population structure analysis revealed three distinct subpopulations in the Indian wild rice accessions with extensive genetic variation between and also within the populations as was also shown recently using a high density 50 K SNP chip (Singh et al. 2015b). Higher average values of Fst indicated that a high level of unshared allelic variation was present in the population (Holsinger and Weir 2009). Here sub-population I comprising more than 50 % of the wild rice accessions was the largest of the three sub-populations.. Linkage disequilibrium (LD) based association analysis is supposed to be the best method for association analysis (Yu and Buckler 2006). Here, LD based association analysis showed strong associations of multiple SNPs in different candidate HKT genes with the salt tolerance traits.
Haplotype based association analysis for identification of allelic variants associated with specific traits is considered better over single-allele studies (Johnson et al. 2001). Overall, only three tolerant haplotypes were identified, one for the HKT1;5 gene and two for the HKT 2;3 gene, but, multiple SNPs were found to be associated with different salt tolerance traits. It may be because of less number of accessions grouping in many haplotypes resulting in low minor allele frequencies (<2 %), and synonymous or noncoding nature of the SNPs. Negrão et al. (2013) identified allelic variants for HKT1;5 gene by setting filter for associated SNPs and individually assessing the results. Here total 14 filtered SNPs were found to be associated with traits for HKT1;5, and among them 12 SNPs were in the intronic regions, one was synonymous and two were non- synonymous, S1379 (Lys429 to Glu429) and S3893 (Pro140 to Ala140) leading to amino acid substitutions. Replacement of proline causes changes in the protein backbone that is responsible to introduce tight turns or kinks into alpha helices (Betts and Russell 2003). Conversion of proline to alanine was also found in Pokkali and Nona Bokra alleles of the HKT1;5 gene which is supposed to increase the substrate specificity (Cotsaftis et al. 2012; Negrão et al. 2013). Cotsaftis et al. (2012) reported change in conformation of protein with change in overall charge.
Two SNPs in the promoter region of OsHKT1;4 gene, S886 (GT-1 binding site region) and S1267 (downstream to the promoter ACGT sequence), were significantly associated with SPAD chlorophyll content value. It has been reported that in many light-regulated genes like PHYA in oat and rice are regulated by GT-1 binding site (Villain et al. 1996). Further, Park et al. (2004) has reported this site to be salt induced. Downstream to the promoter region is the ACGT sequence that is required for etiolation induced expression of erd1 (early responsive to dehydration) gene in Arabidopsis (Simpson et al. 2003), hence it is speculated that OsHKT1;4 gene may regulate dehydration stress imposed by salinity.
Two haplotypes (H1 and H10) of the OsHKT2;3 were associated with salt tolerance SES score. In addition, total 4 SNPs were associated with the analysed salt tolerance traits, one synonymous, one intronic and two non-synonymous SNPs, S761 and S1001, both leading to Iso to Thr substitution at amino acid positions 77 and 157, respectively (Table 2). The isoleucine to threonine substitution is known to impact post translation modifications such as phosphorylation. An increase in the phosphorylation level has been observed with increase in Thr in a protein sequence (Vlad et al. 2008). The non-synonymous mutations outside functional domain of genes may alter structure of the protein and consequently its function (Negrão et al. 2013). Both synonymous and non-synonymous SNPs showed significant association with salt tolerance traits, perhaps affecting the RNA splicing, mRNA stability, and post-translational modification of protein function (Negrão et al. 2013). Some of this could be due to close linkage of a non-functional SNP with the functional SNP. A large number of rare alleles and haplotypes were observed for different HKT genes, whose association with trait could not be studied due to low minor allele frequency. Rare alleles contributing to the gain or enhancement of the trait value may be useful for future adaptability of the rice crop, these may involve novel mechanisms of salt tolerance. Introgression of rare alleles through marker-assisted backcross breeding (MABB) techniques may help develop new genetic resources for breeding of rice for tolerance to extreme salt stress. To find out effective rare alleles, bi-parental mapping populations involving these lines will be needed to validate the function of rare alleles and also to understand their genetic control mechanisms (Semagn et al. 2010).
Conclusions
The wild rice accessions screened in this study have been collected from different ecological habitats including salt-affected areas. An accessions (NKSWR173) from upland and another accession (NKSWR202) from upper gangetic plain region showed high level of tolerance phenotypic patterns and may have different salt tolerance mechanisms. Haplotype analysis indicated a substantial level of natural diversity for the HKT family of genes, especially for the HKT1;5 and HKT2;3 genes among the Indian wild rice accessions. The novel haplotypes showing association with salt tolerance may have great impact on rice salinity breeding. A programme for introgression of the identified haplotypes into high yielding but salt sensitive rice varieties has been initiated. It will help breed rice genotypes with higher level of salt tolerance beyond the existing salt tolerant varieties.
Methods
Plant Material
A total of 299 wild rice accessions including 244 new accessions collected from different geographical regions of India along with their passport data and 58 accessions from NBPGR gene bank, New Delhi, India were analysed. In addition, 6 cultivated O. sativa (salt tolerant FL478, CSR27, CSR11 with different tolerance mechanisms, MI48 as a moderate check while VSR156 and Pusa 44 as sensitive checks) were used. The collection sites and other detailed information on each wild rice accession is available in a database at http://nksingh.nationalprof.in/ (Additional file 1: Table S1).
Phenotyping for Salt Tolerance
Phenotyping for all the accessions was done at National Phytotron Facility, IARI, New Delhi, India for three times. Experiment was planned according to IRRI protocol with minor modifications (Gregorio et al. 1997). Each set of experiment had three replicates with 10 plants of each accession per replicate. The seeds were germinated in petri plates and then transferred into thermocol trays after two days of germination. Each tray had a positive check and a negative check. The seedlings were allowed to grow for 14 days in Hoagland’s solution and on 15th day, they were transferred to Hoagland’s solution with 150 mM NaCl concentrations. Rice seedlings were allowed to grow and scoring was done after 10 days and 15 days of stress according to the 1–9 scale of Standard Evaluation System (SES) developed by Gregorio et al. (1997). Most of the SES were same for all the replicates but in case of variation mode was taken. Based on the above data and geographical location 45 representative accessions from each class were selected to evaluate physiological parameters to reduce the cost and labour. Sodium and Potassium ion concentrations from roots and shoots were estimated using a flame photometer LABTRONICS model LT66 after digestion of 0.1 gm of dried plant sample with 1:3 ratio of perchloric acid and nitric acid. Chlorophyll content of leaves was determined using SPAD meter.
PCR Amplification and re-Sequencing
After phenotypic screening of 299 accessions, 103 were selected based on their response to salt stress. All the salt tolerant lines and representative accessions from moderately tolerant and sensitive classes were taken for re-sequencing of HKT genes. Genomic DNA was extracted from leaf tissue using the CTAB method described by Murray and Thompson (1980). Eight HKT genes were amplified using primer walking method (Additional file 5: Table S4). Nucleotide sequence of the genes were retrieved from NCBI database (http://www.ncbi.nlm.nih.gov/) and primers were designed using Primer3 software (http://bioinfo.ut.ee/primer3-0.4.0/). Specific amplification and validation of the primers was done by NCBI Primer BLAST (http://www.ncbi.nlm.nih.gov/tools/primer-blast/) against Oryza taxon and high stringency conditions during PCR amplification. The PCR amplifications were carried out in 25 μl reaction consisting of 1 Unit SpeedStar™HS DNA Polymerase from TAKARA BIO INC, 1x Fast Buffer, 2 μl of dNTP mixture, 0.5 Pico moles of each primers and 80 ng of template DNA. The PCR reaction was carried out in BIORAD Thermal cycler under following conditions: Initial denaturation at 98 °C for 3 min followed by 38 cycles of denaturation for 10 s at 98 °C, annealing for1 min at 64 °C and extension for 1 min at 68 °C, and for final extension for 10 min at 68 °C. The amplified products were further checked by electrophoresis in 1 % agarose gel in 1X TBE buffer.
PCR products were directly sequenced by Ion Torrent PGM sequencer (Life Technologies) after fragmentation, library preparation, purification and cycle sequencing according to manufacturer’s instructions. Briefly, the amplicons of different genes of the same genotype were pooled in equimolar concentration, sheared to size of ~200 bp and then barcoded to identify individual accessions. Since each gene has unique sequence no barcoding was needed for individual gene. The sheared and barcoded products were now size selected, pooled together in equi-molar ratio and then PCR sequenced. At each step purification was done using Agencourt AMPure XP reagent using Kingfisher Flex. Ion OneTouch2™ was used for emulsion PCR to clone the library on the beads and thereafter for enrichment. The enriched library was loaded on Ion PGM 316 chip and sequencing was performed on Ion PGM™ sequencer.
Sequence Data Analysis
The depth of sequencing obtained was approximately 88x. Coverage analysis and variant caller plugins were run and the sequences were viewed in IGV (Robinson et al. 2011; Thorvaldsdóttir et al. 2012). SamTools mpileup command was used to generate consensus sequences against the reference for each gene and alignment of OsHKT genes was done by ClustalW (Thompson et al. 1994) in BioEdit (Hall 2011). Sequences were deposited to NCBI GenBank database (KT795544-KT796361). Nucleotide polymorphisms were analysed using the DnaSP software version 5.10 (Rozas et al. 2003). Level of silent-site nucleotide diversities per site (π) (Nei 1987) and population mutation parameter (θ) (Watterson 1975) was estimated. Sliding window analysis was performed to examine nucleotide polymorphism across the genes in all accessions using DnaSP software. Statistical tests of neutrality such as Tajima’s D (Tajima 1989), Fu and Li’s D* and F were calculated to examine the selection pressure at this locus. A haplotype network was constructed for comparison of genealogical relationships among the haplotypes using Network software (Bandelt et al. 1999) (http://www.fluxus-engineering.com). The nucleotide sequences were translated into amino acid sequences, and the protein variants were identified as compared with the reference protein. The bootstrap consensus tree inferred from 500 replicates was constructed to represent the evolutionary history of the HKT genes by using the Minimum Evolution Method in MEGA5 (Tamura et al. 2011).
Population Structure and SNP-Trait Association Analysis
Association analysis was performed with the MLM model, considering both kinship (K) and population structure (Q), implemented in TASSEL software. The kinship (K) and population structure (Q) were generated from a genome wide 48-plex Illumina GoldenGate SNP genotyping assay (B. Singh, unpublished) and analysing data with Structure software (Pritchard et al. 2000). To overcome the problems of interpreting the real value of K, a range of ad hoc K values were tested and analysed using Evanno plot (Evanno et al. 2005). The results of estimated likelihood values for a given K (from 2 to 10) in five independent runs were harvested with structure harvester an online program (Earl 2012). For association mapping filtered sites within OsHKT genes were used to determine linkage disequilibrium (LD) by correlation between alleles at two loci in TASSEL 5.0 (Bradbury et al. 2007) software and significance of LD among SNPs was determined by Fisher’s exact test. The mixed model showed least deviation of observed P-values from expected P-values in Q-Q plot when compared with that of Q (population structure) or K (kinship) model only. A probability value of 0.01 was used as the threshold for significance of SNP–trait associations. Functions of associated sites in the promoter region was identified by PLACE (Higo et al. 1999).
Acknowledgements
The financial assistance from Indian Council of Agricultural Research through National Professor B.P. Pal Chair project is gratefully acknowledged. We are thankful to National Bureau of Plant Genetic Resources, New Delhi, India for providing seeds of 58 wild rice accessions.
Additional files
List of wild rice accessions, their place of collection and Standard Evaluation System (SES) score obtained during screening. (CSV 13 kb)
Phenotype (SES) score and evaluated salt related traits among Indian wild rice at 10th day of 150mM salt treatment. (CSV 3 kb)
Set of overlapping primers used for amplification of HKT genes. (CSV 3 kb)
Linkage Disequilibrium plots for HKT genes Figure S2. Q-Q plots of HKT genes obtained after MLM based association of SNP with traits. (PDF 1156 kb)
List of varieties and their classification in different sub-populations using Structure software. (CSV 1 kb)
Footnotes
Competing Interests
The authors declare that they have no competing interests.
Authors’ Contributions
SM: Design and conduct of experiments, Sequencing, Data Analysis and Manuscript preparation; BS: Collection and characterization of wild rice, Conduct of experiments and manuscript drafting; KP: Database maintenance; BPS: Collection of Wild Rice; NS: Design of 48-plex SNP genotyping assay; PM: Manuscript editing; VR: Planning of experiment, Manuscript editing; NKS: Wild rice collection, planning and supervision of the experiments, manuscript editing and finalization. All authors read and approved the final manuscript.
References
- Akey JM, Eberle MA, Rieder MJ, Carlson CS, Shriver MD, Nickerson DA, Kruglyak L. Population history and natural selection shape patterns of genetic variation in 132 genes. PLoS Biol. 2004;2:1591–1599. doi: 10.1371/journal.pbio.0020286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almeida P, Katschnig D, de Boer AH. HKT transporters—state of the art. Int J Mol Sci. 2013;14:20359–20385. doi: 10.3390/ijms141020359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandelt H-J, Forster P, Röhl A. Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol. 1999;16:37–48. doi: 10.1093/oxfordjournals.molbev.a026036. [DOI] [PubMed] [Google Scholar]
- Betts MJ, Russell RB. Amino acid properties and consequences of substitutions. Bioinformatics Genet. 2003;317:289. [Google Scholar]
- Bhasin H, Bhatia D, Raghuvanshi S, Lore JS, Sahi GK, Kaur B, Vikal Y, Singh K. New PCR-based sequence-tagged site marker for bacterial blight resistance gene Xa38 of rice. Mol Breed. 2012;30:607–611. doi: 10.1007/s11032-011-9646-y. [DOI] [Google Scholar]
- Bradbury PJ, Zhang Z, Kroon DE, Casstevens TM, Ramdoss Y, Buckler ES. TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics. 2007;23:2633–2635. doi: 10.1093/bioinformatics/btm308. [DOI] [PubMed] [Google Scholar]
- Cotsaftis O, Plett D, Johnson AAT, Walia H, Wilson C, Ismail AM, Close TJ, Tester M, Baumann U. Root-specific transcript profiling of contrasting rice genotypes in response to salinity stress. Mol Plant. 2011;4:25–41. doi: 10.1093/mp/ssq056. [DOI] [PubMed] [Google Scholar]
- Cotsaftis O, Plett D, Shirley N, Tester M, Hrmova M. A Two-staged model of Na (+) exclusion in rice explained by 3D modeling of HKT transporters and alternative splicing. PLoS One. 2012;7 doi: 10.1371/journal.pone.0039865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cseri A, Cserháti M, Von KM, Nagy B, Horváth GV, Palágyi A, Pauk J, Dudits D, Törjék O. Allele mining and haplotype discovery in barley candidate genes for drought tolerance. Euphytica. 2011;181:341–356. doi: 10.1007/s10681-011-0445-7. [DOI] [Google Scholar]
- Das-Chatterjee A, Goswami L, Maitra S, Dastidar KG, Ray S, Majumder AL. Introgression of a novel salt-tolerant L-myo-inositol 1-phosphate synthase from Porteresia coarctata (Roxb.) Tateoka (PcINO1) confers salt tolerance to evolutionary diverse organisms. FEBS Lett. 2006;580:3980–3988. doi: 10.1016/j.febslet.2006.06.033. [DOI] [PubMed] [Google Scholar]
- Earl DA. Structure harvester: a website and program for visualizing structure output and implementing the Evanno method. Conserv Genet Resour. 2012;4:359–361. doi: 10.1007/s12686-011-9548-7. [DOI] [Google Scholar]
- Evanno G, Regnaut S, Goudet J. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol. 2005;14:2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x. [DOI] [PubMed] [Google Scholar]
- FAOSTAT D. Food and Agriculture Organization of the United Nations Year book. 2009. [Google Scholar]
- Goodall-Copestake W, Tarling G, Murphy E. On the comparison of population-level estimates of haplotype and nucleotide diversity: a case study using the gene cox1 in animals. Heredity. 2012;109:50–56. doi: 10.1038/hdy.2012.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregorio GB, Senadhira D, Mendoza RD. Screening rice for salinity tolerance. Metro Manila: International Rice Research Institute discussion paper series; 1997. [Google Scholar]
- Hall T. BioEdit: an important software for molecular biology. GERF Bull Biosci. 2011;2:60–61. [Google Scholar]
- Higo K, Ugawa Y, Iwamoto M, Korenaga T. Plant cis-acting regulatory DNA elements (PLACE) database. Nucleic Acids Res. 1999;27:297–300. doi: 10.1093/nar/27.1.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoisington D, Khairallah M, Reeves T, Ribaut J-M, Skovmand B, Taba S, Warburton M. Plant genetic resources: what can they contribute toward increased crop productivity? Proc Natl Acad Sci. 1999;96:5937–5943. doi: 10.1073/pnas.96.11.5937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holsinger KE, Weir BS. Genetics in geographically structured populations: defining, estimating and interpreting FST. Nat Rev Genet. 2009;10:639–650. doi: 10.1038/nrg2611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horie T, Yoshida K, Nakayama H, Yamada K, Oiki S, Shinmyo A. Two types of HKT transporters with different properties of Na + and K+ transport in Oryza sativa. Plant J. 2001;27:129–138. doi: 10.1046/j.1365-313x.2001.01077.x. [DOI] [PubMed] [Google Scholar]
- Horie T, Costa A, Kim TH, Han MJ, Horie R, Leung HY, Miyao A, Hirochika H, An G, Schroeder JI. Rice OsHKT2; 1 transporter mediates large Na + influx component into K + −starved roots for growth. EMBO J. 2007;26:3003–3014. doi: 10.1038/sj.emboj.7601732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horie T, Hauser F, Schroeder JI. HKT transporter-mediated salinity resistance mechanisms in Arabidopsis and monocot crop plants. Trends Plant Sci. 2009;14:660–668. doi: 10.1016/j.tplants.2009.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horie T, Brodsky DE, Costa A, Kaneko T, Schiavo FL, Katsuhara M, Schroeder JI. K+ transport by the OsHKT2; 4 transporter from rice with atypical Na + transport properties and competition in permeation of K+ over Mg2+ and Ca2+ ions. Plant Physiol. 2011;156:1493–1507. doi: 10.1104/pp.110.168047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hossain H, Rahman M, Alam M, Singh R. Mapping of quantitative trait loci associated with reproductive‐stage salt tolerance in rice. J Agron Crop Sci. 2015;201:17–31. doi: 10.1111/jac.12086. [DOI] [Google Scholar]
- Jabnoune M, Espeout S, Mieulet D, Fizames C, Verdeil J-L, Conéjéro G, Rodríguez-Navarro A, Sentenac H, Guiderdoni E, Abdelly C. Diversity in expression patterns and functional properties in the rice HKT transporter family. Plant Physiol. 2009;150:1955–1971. doi: 10.1104/pp.109.138008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson GC, Esposito L, Barratt BJ, Smith AN, Heward J, Di Genova G, Ueda H, Cordell HJ, Eaves IA, Dudbridge F. Haplotype tagging for the identification of common disease genes. Nat Genet. 2001;29:233–237. doi: 10.1038/ng1001-233. [DOI] [PubMed] [Google Scholar]
- Khush GS, Ling K. Inheritance of resistance to grassy stunt virus and its vector in rice. J Hered. 1974;65:135–136. [Google Scholar]
- Kumar GR, Sakthivel K, Sundaram RM, Neeraja CN, Balachandran SM, Rani NS, Viraktamath BC, Madhav MS. Allele mining in crops: prospects and potentials. Biotechnol Adv. 2010;28:451–461. doi: 10.1016/j.biotechadv.2010.02.007. [DOI] [PubMed] [Google Scholar]
- Lan W-Z, Wang W, Wang S-M, Li L-G, Buchanan BB, Lin H-X, Gao J-P, Luan S. A rice high-affinity potassium transporter (HKT) conceals a calcium-permeable cation channel. Proc Natl Acad Sci. 2010;107:7089–7094. doi: 10.1073/pnas.1000698107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latha R, Rubia L, Bennett J, Swaminathan M. Allele mining for stress tolerance genes in Oryza species and related germplasm. Mol Biol. 2004;27:101–108. doi: 10.1385/MB:27:2:101. [DOI] [PubMed] [Google Scholar]
- Lestari P, Lee G, Ham TH, Woo MO, Piao R, Jiang W, Chu SH, Lee J, Koh HJ. Single nucleotide polymorphisms and haplotype diversity in rice sucrose synthase 3. J Hered. 2011;102:735–746. doi: 10.1093/jhered/esr094. [DOI] [PubMed] [Google Scholar]
- Li R, Li L, Wei S, Wei Y, Chen Y, Bai D, Yang L, Huang F, Lu W, Zhang X. The evaluation and utilization of new genes for brown plant hopper resistance in common wild rice (Oryza rufipogon Griff.) Mol Entomol. 2010 [Google Scholar]
- Linh LH, Linh TH, Xuan TD, Ham LH, Ismail AM, Khanh TD. Molecular breeding to improve salt tolerance of rice (Oryza sativa L.) in the Red River Delta of Vietnam. Intern J Plant genom. 2012;2012 doi: 10.1155/2012/949038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutts S, Kinet J, Bouharmont J. Changes in plant response to NaCl during development of rice (Oryza sativa L.) varieties differing in salinity resistance. J Exp Bot. 1995;46:1843–1852. doi: 10.1093/jxb/46.12.1843. [DOI] [Google Scholar]
- Maas E, Hoffman G. Crop salt tolerance: evaluation of existing data. J Irrigat Drainage. 1977;15:187. [Google Scholar]
- Mäser P, Thomine S, Schroeder JI, Ward JM, Hirschi K, Sze H, Talke IN, Amtmann A, Maathuis FJM, Sanders D. Phylogenetic relationships within cation transporter families of Arabidopsis. Plant Physiol. 2001;126:1646–1667. doi: 10.1104/pp.126.4.1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mäser P, Eckelman B, Vaidyanathan R, Horie T, Fairbairn DJ, Kubo M, Yamagami M, Yamaguchi K, Nishimura M, Uozumi N. Altered shoot/root Na + distribution and bifurcating salt sensitivity in Arabidopsis by genetic disruption of the Na + transporter AtHKT1. FEBS Lett. 2002;531:157–161. doi: 10.1016/S0014-5793(02)03488-9. [DOI] [PubMed] [Google Scholar]
- McCouch SR, Sweeney M, Li J, Jiang H, Thomson M, Septiningsih E, Edwards J, Moncada P, Xiao J, Garris A. Through the genetic bottleneck: O. rufipogon as a source of trait-enhancing alleles for O. sativa. Euphytica. 2007;154:317–339. doi: 10.1007/s10681-006-9210-8. [DOI] [Google Scholar]
- McNally KL, Bruskiewich R, Mackill D, Buell CR, Leach JE, Leung H. Sequencing multiple and diverse rice varieties. Connecting whole-genome variation with phenotypes. Plant Physiol. 2006;141:26–31. doi: 10.1104/pp.106.077313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mian A, Oomen RJ, Isayenkov S, Sentenac H, Maathuis FJ, Véry AA. Over‐expression of an Na + −and K + −permeable HKT transporter in barley improves salt tolerance. Plant J. 2011;68:468–479. doi: 10.1111/j.1365-313X.2011.04701.x. [DOI] [PubMed] [Google Scholar]
- Mickelbart MV, Hasegawa PM, Bailey-Serres J. Genetic mechanisms of abiotic stress tolerance that translate to crop yield stability. Nat Rev Genet. 2015;16(4):237–251. doi: 10.1038/nrg3901. [DOI] [PubMed] [Google Scholar]
- Miller RW, Donahue RL. Soils in our environment. Upper Saddle River: Prentice hall; 1995. [Google Scholar]
- Mohammadi R, Mendioro MS, Diaz GQ, Gregorio GB, Singh RK. Mapping quantitative trait loci associated with yield and yield components under reproductive stage salinity stress in rice (Oryza sativa L.) J Genet. 2013;92:433–443. doi: 10.1007/s12041-013-0285-4. [DOI] [PubMed] [Google Scholar]
- Mohanty S. Trends in global rice consumption. Rice Today. 2013;12(1):44–45. [Google Scholar]
- Munns R, Tester M. Mechanisms of salinity tolerance. Annu Rev Plant Biol. 2008;59:651–681. doi: 10.1146/annurev.arplant.59.032607.092911. [DOI] [PubMed] [Google Scholar]
- Munns R, James RA, Xu B, Athman A, Conn SJ, Jordans C, Byrt CS, Hare RA, Tyerman SD, Tester M, Plett D. Wheat grain yield on saline soils is improved by an ancestral Na + transporter gene. Nat Biotechnol. 2012;30:360–364. doi: 10.1038/nbt.2120. [DOI] [PubMed] [Google Scholar]
- Murray M, Thompson WF. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 1980;8:4321–4326. doi: 10.1093/nar/8.19.4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negrão S, Cecília AM, Pires IS, Abreu IA, Maroco J, Courtois B, Gregorio GB, McNally KL, Margarida OM. New allelic variants found in key rice salt‐tolerance genes: an association study. Plant Biotechnol J. 2013;11:87–100. doi: 10.1111/pbi.12010. [DOI] [PubMed] [Google Scholar]
- Nei M. Molecular evolutionary genetics. New York: Columbia University Press; 1987. [Google Scholar]
- Oomen RJ, Benito B, Sentenac H, Rodríguez‐Navarro A, Talón M, Véry AA, Domingo C. HKT2; 2/1, a K + −permeable transporter identified in a salt‐tolerant rice cultivar through surveys of natural genetic polymorphism. Plant J. 2012;71:750–762. doi: 10.1111/j.1365-313X.2012.05031.x. [DOI] [PubMed] [Google Scholar]
- Park HC, Kim ML, Kang YH, Jeon JM, Yoo JH, Kim MC, Park CY, Jeong JC, Moon BC, Lee JH. Pathogen-and NaCl-induced expression of the SCaM-4 promoter is mediated in part by a GT-1 box that interacts with a GT-1-like transcription factor. Plant Physiol. 2004;135:2150–2161. doi: 10.1104/pp.104.041442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platten JD, Cotsaftis O, Berthomieu P, Bohnert H, Davenport RJ, Fairbairn DJ, Horie T, Leigh RA, Lin H-X, Luan S. Nomenclature for HKT transporters, key determinants of plant salinity tolerance. Trends Plant Sci. 2006;11:372–374. doi: 10.1016/j.tplants.2006.06.001. [DOI] [PubMed] [Google Scholar]
- Platten JD, Egdane JA, Ismail AM. Salinity tolerance, Na + exclusion and allele mining of HKT1; 5 in Oryza sativa and O. glaberrima: many sources, many genes, one mechanism? BMC Plant Biol. 2013;13:32. doi: 10.1186/1471-2229-13-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren Z-H, Gao J-P, Li L-G, Cai X-L, Huang W, Chao D-Y, Zhu M-Z, Wang Z-Y, Luan S, Lin H-X. A rice quantitative trait locus for salt tolerance encodes a sodium transporter. Nature Genet. 2005;37:1141–1146. doi: 10.1038/ng1643. [DOI] [PubMed] [Google Scholar]
- Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP. Integrative genomics viewer. Nat Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockström J, Falkenmark M, Karlberg L, Hoff H, Rost S, Gerten D. Future water availability for global food production: the potential of green water for increasing resilience to global change. Water Resour Res. 2009;45 [Google Scholar]
- Roy SJ, Negrão S, Tester M. Salt resistant crop plants. Curr Opin Biotechnol. 2014;26:115–124. doi: 10.1016/j.copbio.2013.12.004. [DOI] [PubMed] [Google Scholar]
- Rozas J, Sánchez-DelBarrio JC, Messeguer X, Rozas R. DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics. 2003;19:2496–2497. doi: 10.1093/bioinformatics/btg359. [DOI] [PubMed] [Google Scholar]
- Schachtman DP, Schroeder JI. Structure and transport mechanism of a high-affinity potassium uptake transporter from higher plants. Nature. 1994;370(6491):655–658. doi: 10.1038/370655a0. [DOI] [PubMed] [Google Scholar]
- Schroeder JI, Delhaize E, Frommer WB, Guerinot ML, Harrison MJ, Herrera-Estrella L, Horie T, Kochian LV, Munns R, Nishizawa NK. Using membrane transporters to improve crops for sustainable food production. Nature. 2013;497:60–66. doi: 10.1038/nature11909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semagn K, Bjørnstad Å, Xu Y. The genetic dissection of quantitative traits in crops. Electron J Biotechnol. 2010;13:16–17. doi: 10.2225/vol13-issue5-fulltext-14. [DOI] [Google Scholar]
- Sengupta S, Patra B, Ray S, Majumder AL. Inositol methyl tranferase from a halophytic wild rice, Porteresia coarctata Roxb. (Tateoka): regulation of pinitol synthesis under abiotic stress. Plant Cell Environ. 2008;31:1442–1459. doi: 10.1111/j.1365-3040.2008.01850.x. [DOI] [PubMed] [Google Scholar]
- Simpson SD, Nakashima K, Narusaka Y, Seki M, Shinozaki K, Yamaguchi‐Shinozaki K. Two different novel cis‐acting elements of erd1, a clpA homologous Arabidopsis gene function in induction by dehydration stress and dark‐induced senescence. Plant J. 2003;33:259–270. doi: 10.1046/j.1365-313X.2003.01624.x. [DOI] [PubMed] [Google Scholar]
- Singh A, Singh B, Panda K, Rai VP, Singh AK, Singh SP, Chouhan SK, Rai V, Singh PK, Singh NK. Wild rices of Eastern Indo-Gangetic plains of India constitute two sub-populations harbouring rich genetic diversity. POJ. 2013;6(2):121–127. [Google Scholar]
- Singh BP, Jayaswal PK, Singh B, Singh PK, Kumar V, Mishra S, Singh N, Panda K, Singh NK. Natural allelic diversity in OsDREB1F gene in the Indian wild rice germplasm led to ascertain its association with drought tolerance. Plant Cell Rep. 2015;34:993–1004. doi: 10.1007/s00299-015-1760-6. [DOI] [PubMed] [Google Scholar]
- Singh N, Jayaswal PK, Panda K, Mandal P, Kumar V, Singh B, Mishra S, Singh Y, Singh R, Rai V, Gupta A, Sharma TR, Singh NK. Single-copy gene based 50 K SNP chip for genetic studies and molecular breeding in rice. Sci Rep. 2015;5 doi: 10.1038/srep11600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989;123:585–595. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanksley SD, McCouch SR. Seed banks and molecular maps: unlocking genetic potential from the wild. Science. 1997;277:1063–1066. doi: 10.1126/science.277.5329.1063. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson MJ, de Ocampo M, Egdane J, Rahman MA, Sajise AG, Adorada DL, Tumimbang-Raiz E, Blumwald E, Seraj ZI, Singh RK. Characterizing the Saltol quantitative trait locus for salinity tolerance in rice. Rice. 2010;3:148–160. doi: 10.1007/s12284-010-9053-8. [DOI] [Google Scholar]
- Thorvaldsdóttir H, Robinson JT, Mesirov JP. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform. 2012;14(2):178–192. doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villain P, Mache R, Zhou D-X. The mechanism of GT element-mediated cell type-specific transcriptional control. J Biol Chem. 1996;271:32593–32598. doi: 10.1074/jbc.271.51.32593. [DOI] [PubMed] [Google Scholar]
- Vlad F, Turk BE, Peynot P, Leung J, Merlot S. A versatile strategy to define the phosphorylation preferences of plant protein kinases and screen for putative substrates. Plant J. 2008;55:104–117. doi: 10.1111/j.1365-313X.2008.03488.x. [DOI] [PubMed] [Google Scholar]
- Wang W, Vinocur B, Altman A. Plant responses to drought, salinity and extreme temperatures: towards genetic engineering for stress tolerance. Planta. 2003;218:1–14. doi: 10.1007/s00425-003-1105-5. [DOI] [PubMed] [Google Scholar]
- Watterson G. On the number of segregating sites in genetical models without recombination. Theor Popul Biol. 1975;7:256–276. doi: 10.1016/0040-5809(75)90020-9. [DOI] [PubMed] [Google Scholar]
- Xiao J, Li J, Yuan L, Tanksley S. Identification of QTLs affecting traits of agronomic importance in a recombinant inbred population derived from a subspecific rice cross. Theor Appl Genet. 1996;92:230–244. doi: 10.1007/BF00223380. [DOI] [PubMed] [Google Scholar]
- Xiao J, Li J, Grandillo S, Ahn SN, Yuan L, Tanksley SD, McCouch SR. Identification of trait-improving quantitative trait loci alleles from a wild rice relative, Oryza rufipogon. Genetics. 1998;150:899–909. doi: 10.1093/genetics/150.2.899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C, Zhao L, Zhang H, Yang Z, Wang H, Wen S, Zhang C, Rustgi S, von Wettstein D, Liu B. Evolution of physiological responses to salt stress in hexaploid wheat. Proc Natl Acad Sci. 2014;111:11882–11887. doi: 10.1073/pnas.1412839111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Buckler ES. Genetic association mapping and genome organization of maize. Curr Opin Biotechnol. 2006;17:155–160. doi: 10.1016/j.copbio.2006.02.003. [DOI] [PubMed] [Google Scholar]
- Yu Y-T, Wang R-H, Shi Y-S, Song Y-C, Wang T-Y, Yu L. Haplotypic structure and allelic variation of rab17, an ABA-responsive gene, in a mini core set of Chinese diversified maize inbred lines. Agric Sci China. 2010;9:1726–1738. doi: 10.1016/S1671-2927(09)60271-6. [DOI] [Google Scholar]
- Yuan S-C, Zhang Z-G, He H-H, Zen H-L, Lu K-Y, Lian J-H, Wang B-X. Two photoperiodic-reactions in photoperiod-sensitive genic male-sterile rice. Crop Sci. 1993;33:651–660. doi: 10.2135/cropsci1993.0011183X003300040002x. [DOI] [Google Scholar]
- Zhu J-K. Plant salt tolerance. Trends Plant Sci. 2001;6:66–71. doi: 10.1016/S1360-1385(00)01838-0. [DOI] [PubMed] [Google Scholar]


