Significance
Molecular chaperones play key roles in maintaining protein homeostasis within cells. Membrane protein chaperones face particular challenges, as they not only protect highly aggregation-prone membrane protein substrates, but also need to achieve tight spatiotemporal coordination of their chaperone cycle. In this work, biochemical and NMR analyses address these questions and for the first time, to our knowledge, define the complete chaperone cycle for cpSRP43, an ATP-independent chaperone dedicated to integral membrane proteins. The study reveals that cpSRP43’s substrate binding domain samples at least three distinct conformations. This property enables it to be readily switched on by positive regulators in the soluble phase to ensure tight substrate binding and be switched off by the translocase at the membrane to ensure facile and productive substrate release.
Keywords: membrane protein biogenesis, molecular chaperone, signal recognition particle, protein dynamics, NMR spectroscopy
Abstract
Membrane protein biogenesis poses enormous challenges to cellular protein homeostasis and requires effective molecular chaperones. Compared with chaperones that promote soluble protein folding, membrane protein chaperones require tight spatiotemporal coordination of their substrate binding and release cycles. Here we define the chaperone cycle for cpSRP43, which protects the largest family of membrane proteins, the light harvesting chlorophyll a/b-binding proteins (LHCPs), during their delivery. Biochemical and NMR analyses demonstrate that cpSRP43 samples three distinct conformations. The stromal factor cpSRP54 drives cpSRP43 to the active state, allowing it to tightly bind substrate in the aqueous compartment. Bidentate interactions with the Alb3 translocase drive cpSRP43 to a partially inactive state, triggering selective release of LHCP’s transmembrane domains in a productive unloading complex at the membrane. Our work demonstrates how the intrinsic conformational dynamics of a chaperone enables spatially coordinated substrate capture and release, which may be general to other ATP-independent chaperone systems.
Protein homeostasis is essential for all cells and requires proper control of the folding, localization, and interactions of proteins. The biogenesis of membrane proteins poses a particular challenge to protein homeostasis. Before arrival at the membrane, newly synthesized membrane proteins need to traverse aqueous cellular compartments where they are highly prone to aggregation. Thus, the posttranslational targeting of membrane proteins relies critically on effective molecular chaperones that maintain nascent membrane proteins in translocation competent states. Many examples illustrate the intimate link between chaperone function and membrane protein biogenesis: SecB, Skp, and SurA protect bacterial outer membrane proteins (1–5), and Hsp70 homologs assist the import of mitochondrial or chloroplast proteins (6).
Our understanding of membrane protein chaperones lags far behind that for soluble proteins, such as DnaK and GroEL. All chaperones need to switch between “open” and “closed” conformations to allow substrate release and binding, respectively. For many chaperones that promote the folding of soluble proteins, these switches can be driven either by ATPase cycles, such as Hsp70 (7) and GroEL (8), or by changes in environmental conditions, such as the acid-induced HdeA (9, 10) and oxidation-induced Hsp33 (11). In contrast, membrane protein chaperones must regulate their action spatially: they must effectively capture substrate proteins in the aqueous phase, and then facilely and productively release them at the target membrane. With few exceptions (1, 2), how membrane protein chaperones achieve spatiotemporal coordination of their chaperone cycle is not well understood.
The light harvesting chlorophyll a/b-binding proteins (LHCPs) provide an excellent model system to address these questions. Like >95% of organellar proteins, LHCPs are initially synthesized in the cytosol and imported across the chloroplast envelope in a largely unfolded state with the assistance of the LHCP translocation defect protein (12). In the stroma, LHCPs are protected in a soluble “transit complex” by the chloroplast signal recognition particle (cpSRP), comprised of cpSRP43 and cpSRP54 (13). Via interactions between the GTPase domains of cpSRP54 and its receptor cpFtsY, LHCPs are delivered to the Alb3 translocase and inserted into the thylakoid membrane (13–17). LHCPs comprise more than 50% of the proteins in the thylakoid membrane and are the most abundant membrane protein family on earth. Their sheer abundance, high aggregation propensity, and crucial roles in energy generation of green plants demand highly effective chaperone(s) during their biogenesis, making this a robust system to understand the function and mechanism of membrane protein chaperones.
Previous work showed that the cpSRP43 subunit in cpSRP binds tightly to and quantitatively prevents the aggregation of multiple members of the LHCP family, demonstrating that cpSRP43 is responsible for chaperone function (18, 19). cpSRP43 is comprised of multiple protein-interaction domains: three chromodomains (CDs) and an ankyrin repeat domain (A1–A4) between CD1 and CD2 (Fig. 1A) (14). Biochemical and crystallographic analyses showed that a conserved Tyr204 in the third ankyrin repeat recognizes a FDPLGL motif in L18, a conserved 18-amino acid sequence between TM2 and TM3 of LHCP (20–22). In addition, aromatic cages in CD2 provide binding sites for a conserved RRKR motif in the C terminus of cpSRP54 (23). A recent study found that cpSRP54 can induce compaction of cpSRP43 and enhance L18 peptide binding threefold, suggesting that cpSRP54 could positively regulate cpSRP43 (24). Finally, cpSRP43 also interacts directly with the C-terminal stromal domain of the Alb3 translocase (termed Alb3CT) that mediates the membrane insertion of LHCP (25–29), suggesting a potential mechanism to couple substrate release to the correct localization of LHCP and its imminent membrane insertion (30). The ability of cpSRP43 to directly bind Alb3 may also explain findings in earlier genetic studies that, when both cpSRP54 and cpFtsY are deleted, cpSRP43 by itself can mediate the targeting and insertion of some LHCP family members, albeit less efficiently (30).
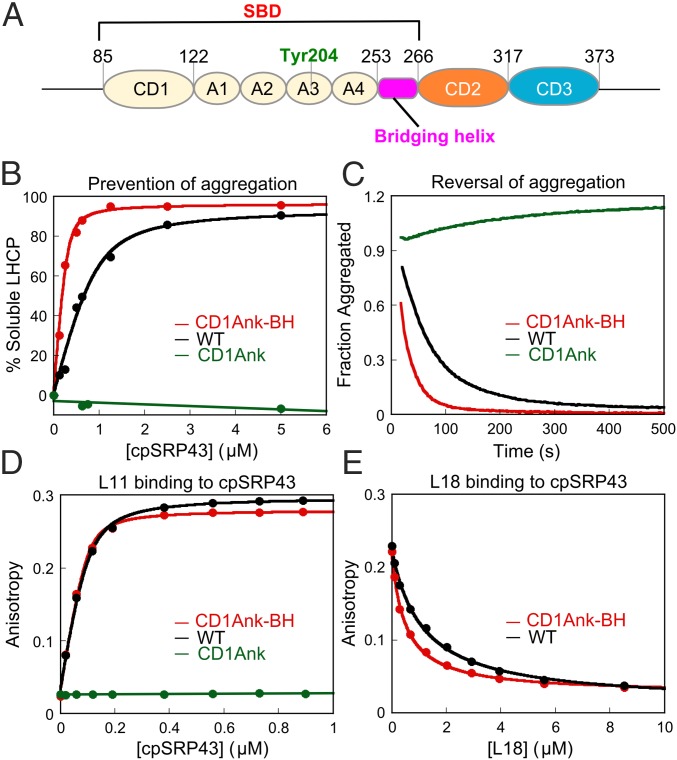
Fig. 1.
The CD1Ank-BH fragment is necessary and sufficient for the chaperone activity of cpSRP43. (A) Schematic of cpSRP43. CD, chromodomain; A1–A4, ankyrin repeats 1–4; BH, bridging helix; SBD, substrate binding domain. (B) Binding of LHCP to wildtype cpSRP43 (black) and to the CD1Ank (green) and CD1Ank-BH (red) fragments of cpSRP43 were measured by the ability of cpSRP43 to prevent LHCP aggregation (Materials and Methods). The data were fit to Eq. 1 and gave apparent Kd values of 170 and 32 nM for LHCP binding to cpSRP43 and to the CD1Ank-BH fragment, respectively. (C) CD1Ank-BH (red) can reverse LHCP aggregation more efficiently than WT cpSRP43 (black), but the CD1Ank (green) fragment cannot. (D) Binding of HiLyte-Fluor 488–labeled L11 peptide to WT and mutant cpSRP43, detected by fluorescence anisotropy. The data were fit to Eq. 2 and gave Kd values of 25 and 15 nM for the binding of dye-labeled L11 peptide to cpSRP43 (black) and to the CD1Ank-BH (red) fragment, respectively. (E) Binding of the L18 peptide to WT and mutant cpSRP43 was measured using L18 as a competitor of dye-labeled L11. The data were fit to Eq. 3 and gave values of 1.1 and 0.5 µM for cpSRP43 (black) and the CD1Ank-BH (red) fragment, respectively. Errors of Kd and values were estimated to be ±10% (SD) based on at least two measurements (technical replicates).
Nevertheless, the molecular mechanism of cpSRP43’s chaperone function remains elusive. Where is the substrate binding domain of this chaperone located? How does it interact with the targeting (cpSRP54) and translocation (Alb3) machineries to achieve accurate spatiotemporal regulation of its activity? More fundamentally, in the absence of an ATPase module, what propels the substrate binding and release cycle for this chaperone? In this work, a combination of biochemical and solution NMR studies addresses these questions and for the first time, to our knowledge, defines the complete chaperone cycle for a chaperone dedicated to integral membrane proteins. Our results show that cpSRP43 inherently exchanges between three distinct conformations; this allows it to be readily turned “on” by cpSRP54 in the aqueous stroma to enable tight substrate binding and to be readily switched to less active conformations by Alb3 at the membrane to enable facile substrate unloading. Furthermore, we show that Alb3 specifically induces the release of substrate TMDs, but not the L18 motif, from cpSRP43, suggesting a highly productive, stepwise mechanism for handover of the membrane protein substrates to the translocation machinery.
Results
Defining the Substrate-Binding Domain for cpSRP43.
In previous work, a fragment comprising CD1 and the ankyrin repeat domain of cpSRP43 (CD1Ank) could not chaperone LHCP (19). Nevertheless, crystallographic analysis (20) revealed an additional 20-amino acid-long helix, not present in other ankyrin repeat proteins, that bridges the ankyrin repeat domain and CD2 (here termed the bridging helix or BH; Fig. 1A). Intriguingly, addition of this BH to CD1Ank was necessary and sufficient to restore chaperone activity. Using a light scattering assay (19), which directly monitors the formation of large protein aggregates, we found that a protein fragment containing CD1, the ankyrin repeat domain, and BH (CD1Ank-BH) prevented the aggregation of LHCP fivefold more efficiently than full-length cpSRP43 (Fig. 1B). In a more stringent disaggregation assay (19), which monitors the ability of cpSRP43 to reverse preformed large LHCP aggregates, CD1Ank-BH was also more efficient than full-length cpSRP43 (Fig. 1C). Consistent with previous results (19), removal of BH (i.e., CD1Ank) abolished chaperone activity in both assays (Fig. 1 B and C).
A key substrate recognition motif of cpSRP43 is the L18 sequence between TM2 and TM3 of LHCP (21, 31). To test whether the BH is important for this recognition, we measured cpSRP43 binding with L18 based on the increase in fluorescence anisotropy of a HiLyte-Fluor488–labeled L11 peptide, which comprises the minimal binding motif in L18 (20, 22). The CD1Ank-BH fragment binds tightly to the dye-labeled L11 peptide, with a Kd value (15 nM) twofold lower than that of WT cpSRP43 (25 nM) (Fig. 1D). Consistent with the results of the light scattering assay, removal of the BH abolished this binding (Fig. 1D). To rule out potential artifacts introduced by dye labeling, we also measured the binding of the WT L18 sequence to cpSRP43 by using it as a competitor of dye-labeled L11 in the anisotropy assay. These experiments showed that L18 effectively bound to both WT cpSRP43 and CD1Ank-BH and competed with dye-labeled L11 (Fig. 1E).
Together with previous mutational analyses (19), these results establish that the CD1Ank-BH fragment comprises the minimal substrate binding domain (SBD) of cpSRP43 and that the bridging helix connecting the ankyrin repeat domain to CD2 is crucial for substrate binding.
Chromodomain 2 Regulates Chaperone Activity.
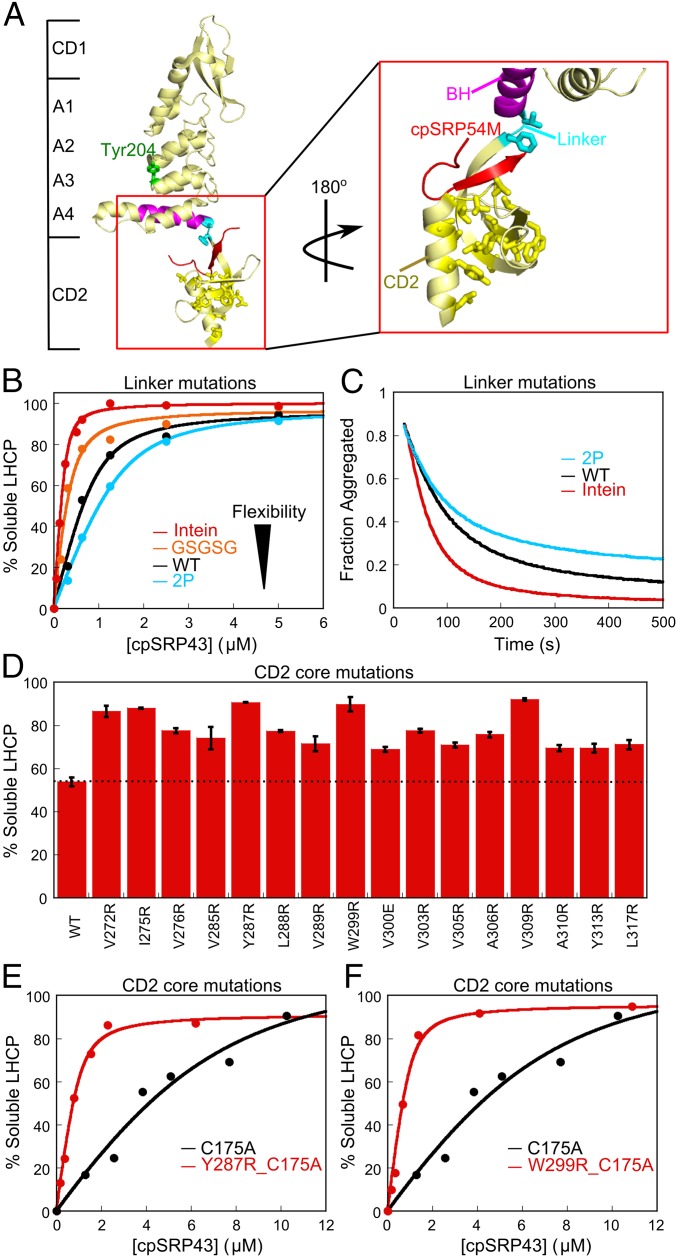
Intriguingly, we isolated many mutations in the neighboring CD2 that affect chaperone activity of the SBD. One class of these mutants resides in the linker connecting BH to CD2 (Fig. 2A, cyan). The chaperone activity of cpSRP43 is strongly correlated with the flexibility of the linker sequence, with longer and more flexible linkers [GSCFNGT (the Intein construct) or GSGSG insertion], leading to higher activities of cpSRP43 in preventing (Fig. 2B) and reversing (Fig. 2C) LHCP aggregation, whereas a conformationally more restricted linker 2P (two Pro replacing the natural QV) leads to lower activity (Fig. 2 B and C).
Fig. 2.
Mutations in CD2 hyperactivate the chaperone. (A) Crystal structure of cpSRP43 CD1Ank-BH-CD2 fragment bound to 54M peptide [Protein Data Bank (PDB) ID code 3UI2]. Cyan shows the linker (V266, F267) between BH and CD2, yellow shows residues mutated in the hydrophobic core of CD2, magenta highlights the BH, and red shows the 54M peptide. (B and C) Chaperone activity of cpSRP43 linker mutants, measured by prevention (B) and reversal (C) of LHCP aggregation. The data in B were fitted to Eq. 1 and gave apparent Kd values of 33, 100, 177, and 256 nM for LHCP binding to intein-cpSRP43 (red), GSGSG-cpSRP43 (orange), WT cpSRP43 (black), and 2P-cpSRP43 (cyan), respectively. (D) Many point mutations in the conserved hydrophobic core of CD2 activate chaperone activity. Chaperone activities were measured by prevention of LHCP aggregation using 1 µM LHCP and 4 µM WT or mutant cpSRP43. Error bars represent SD, with n = 2. To rule out potential involvements of CD3, all measurements were made with the ΔCD3 construct. A soft mutation, C175A, was present in all constructs to increase the sensitivity of the mutational screen under these conditions. (E and F) Mutants Y287R (E) and W299R (F) enhance LHCP binding to cpSRP43 >10-fold. Chaperone activities were measured and analyzed as in B and gave apparent Kd values of 1.9 µM, 168 nM, and 141 nM for cpSRP43(C175A), cpSRP43(Y287R_C175A), and cpSRP43(W299_C175A), respectively. Errors of Kd values were estimated to be ±10% (SD) based on at least two measurements (technical replicates).
A second class of mutants resides in the conserved hydrophobic core of CD2 (Fig. 2A, yellow, and Fig. S1). Virtually every single mutation introduced into this core hyperactivates the chaperone (Fig. 2D). Quantitative analysis of some of the mutants further showed that cpSRP43’s substrate binding is enhanced 10- to 20-fold in each mutant (Fig. 2 E and F). Thus, although CD2 does not directly bind the substrate protein, it regulates the conformation and activity of the SBD.
Fig. S1.
Sequence alignment of cpSRP43 CD1, CD2, CD3, Polycomb (Pc), and heterochromatin protein 1 (HP1). Green highlights conserved hydrophobic residues in CD2 that were mutated in this study. Red asterisks denote aromatic cage residues in canonical chromodomains. Black asterisks denote aromatic cage residues involved in 54M binding (23). Secondary structure information is labeled above the sequence highlighting β-sheet (β1, β2, β3) and α-helix (α1).
The cpSRP54 M Domain Drives cpSRP43 to the Active Conformation.
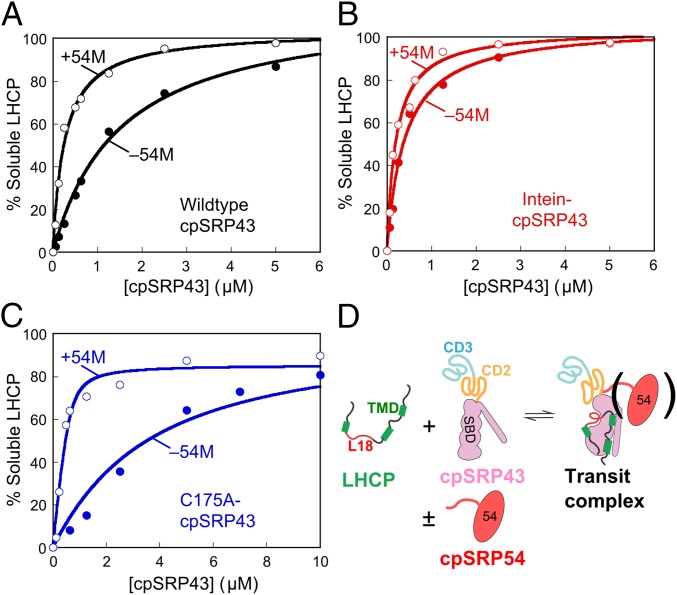
In the stroma, cpSRP43 is bound to cpSRP54, the other subunit of cpSRP. The C terminus of the cpSRP54 M-domain (termed 54M) binds at CD2 in the vicinity of the BH (Fig. 2A) (23, 32), placing it in an optimal position for regulating interdomain interactions of cpSRP43. In support of this notion, 54M enhances the binding affinity of LHCP to WT cpSRP43 sixfold under stringent low salt conditions (100 mM NaCl; Fig. 3A). This result is consistent with a recent study, which also reported that cpSRP43 becomes more compact and binds the L18 peptide threefold more strongly in the presence of cpSRP54 (24). Importantly, the superactive mutant, intein-cpSRP43, is not further stimulated by 54M (Fig. 3B). In contrast, a soft mutation C175A reduced cpSRP43’s chaperone activity ∼18-fold; this partially crippled mutant is much more strongly activated by 54M (Fig. 3C). Together, these results strongly suggest that 54M and superactive mutations in CD2 drive the same activating conformational change in cpSRP43 and that interdomain dynamics control cpSRP43’s chaperone activity (Fig. 3D).
Fig. 3.
The cpSPR54 M-domain activates cpSRP43 for substrate binding. The abilities of chaperone to prevent LHCP aggregation were measured for WT cpSRP43 (A), superactive intein-cpSRP43 (B), and partially defective mutant cpSRP43(C175A) (C) in the absence (●) and presence (○) of 54M. The data were fit to a Michaelis–Menton equation and gave apparent Kd values of 0.26 and 1.5 µM for WT cpSRP43 with and without 54M (A), 0.20 and 0.41 µM for intein-cpSRP43 with and without 54M (B), and 0.08 and 3.0 µM for cpSRP43(C175A) with and without 54M (C). In A and B, activities were measured under 100 mM NaCl, a stringent condition under which cpSRP43 exhibits slightly reduced activity, to overcome the saturation effects with highly active chaperone constructs and better reveal the stimulatory effects of 54M. (D) Scheme depicting the conformational change of cpSRP43 on substrate binding. TMD, transmembrane domain.
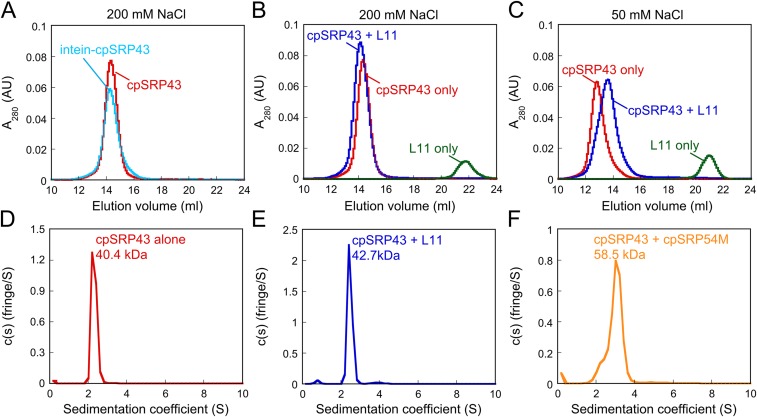
CpSRP43 has been bound to shift between monomer and higher oligomeric forms depending on solution ionic strength (19). To test whether the activating conformational change of cpSRP43 is related to changes in its oligmeric state, we carried out gel filtration chromatography and analytical ultracentrifugation analyses (Fig. S2). The results showed that, under high salt conditions (≥200 mM NaCl), WT or superactive intein-cpSRP43 is predominantly a monomer (Fig. S2 A and D). cpSRP43 also binds the L11 peptide and 54M in monomeric form (Fig. S2 B, E, and F). Under low salt conditions (50 mM NaCl), cpSRP43 predominantly runs on Superdex 200 as a dimer (Fig. S2C) as previously observed (19). Importantly, binding of L11 shifts dimeric cpSRP43 toward the monomeric form (Fig. S2C). These results, together with the previous observation that the shift of cpSRP43 to dimeric complexes at lower ion strength is correlated with loss of chaperone activity (19), strongly suggest that monomeric cpSRP43 is chaperone active. Thus, the substrate- and 54M-induced rearrangement of cpSRP43 reflects an intramolecular conformational change, rather than changes in its oligomeric state.
Fig. S2.
CpSRP43 is active as a monomer. (A) WT (red) and superactive intein-cpSRP43 (cyan) runs as a monomer on Superdex 200 column with buffer containing 200 mM NaCl. (B) The complex of WT cpSRP43 and HiLyte-Fluor488–labeled L11 peptide (blue) was eluted as a 1:1 complex. (C) At lower ionic strength (50 mM NaCl), cpSRP43 exhibits oligomeric forms (red), but L11 binding shifts cpSRP43 to a lower molecular weight complex (blue). (D–F) Sedimentation coefficient distributions calculated from a velocity sedimentation experiment of cpSRP43 alone (D), cpSRP43 with HiLyte-Fluor488–labeled L11 peptide (E), and cpSRP43 with cpSRP54M (F) using buffer containing 200 mM NaCl. The experimental molecular mass is close to the predicted values of cpSRP43 and cpSRP54M (36 and 22 kDa, respectively), suggesting that the active form of cpSRP43 is monomer.
NMR Spectroscopy Directly Reveals Chaperone Conformational Dynamics.
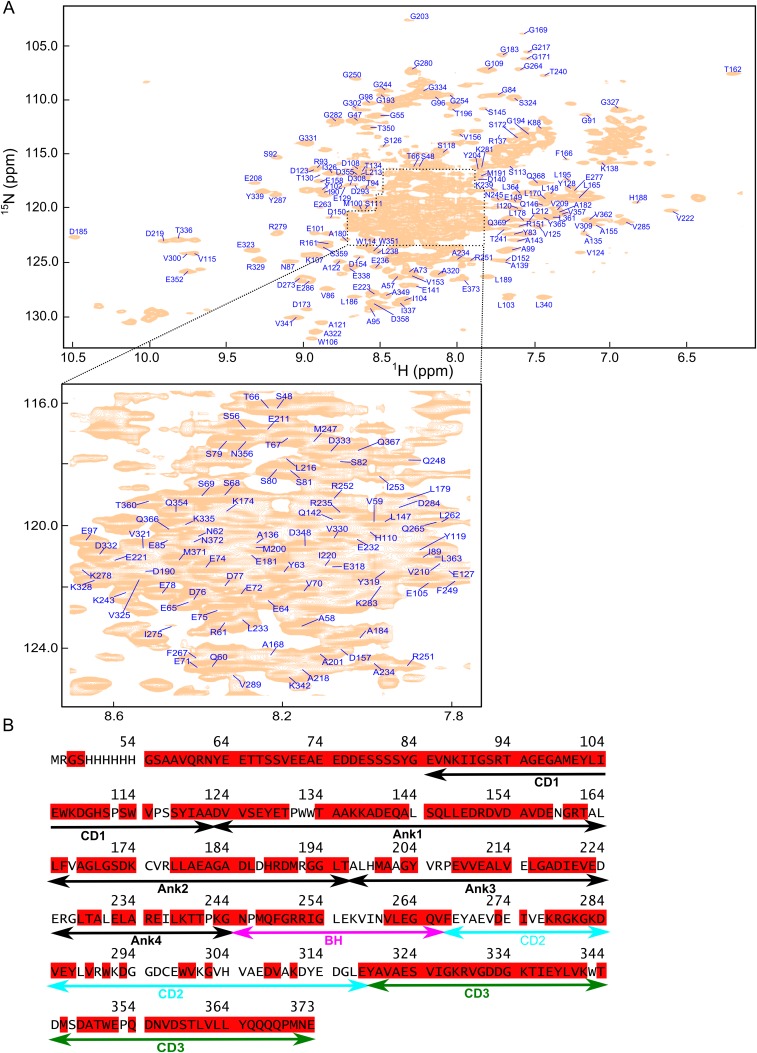
To directly probe for conformational dynamics in cpSRP43, we used transverse relaxation optimized (TROSY) NMR spectroscopy. We assigned the cpSRP43 backbone resonances using 3D triple resonance TROSY spectra of 2H,13C,15N-labeled cpSRP43 and the CD1Ank-BH fragment; assignments for several CD2 cross-peaks in full-length cpSRP43 were obtained by transferring published assignments of the isolated CD2CD3 fragment (33) (SI Materials and Methods). We were able to assign the amide 15N, 1HN, 13Cα, and 13Cβ resonances for 80% of residues (Fig. S3).
Fig. S3.
Assignments of the cpSRP43 spectra. (A) 800 MHz 1H-15N TROSY spectrum of full-length 15N, 13C, 2H-labeled cpSRP43 is shown with the assigned residues indicated. The central area of the spectrum is enlarged for clarity. (B) Assigned residues are highlighted in red in the cpSRP43 sequence.
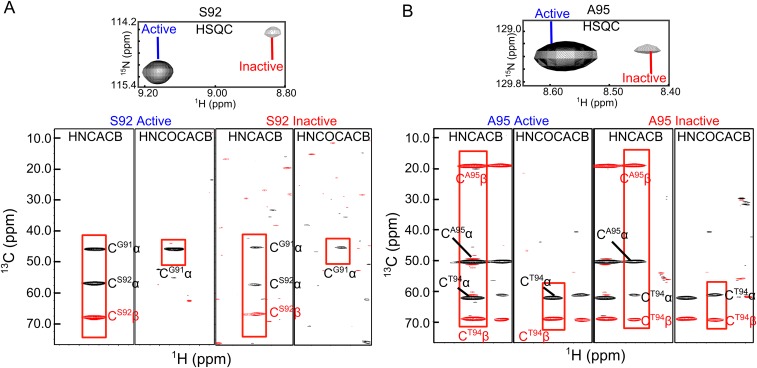
Intriguingly, the number of NMR cross-peaks in the 1H-15N TROSY spectrum of cpSRP43 far exceeded the number of residues in the protein. This puzzle was resolved during NMR assignments: we found that at least 12 residues in CD1 (highlighted in Fig. 4G in blue) give rise to two 1H-15N cross-peaks in the TROSY spectrum that have identical 13Cα and 13Cβ chemical shifts (examples for S92 and A95 are shown in Fig. S4 A and B). This observation indicates that the component cross-peaks in the TROSY spectra arise from the same amino acids and represent two distinct conformations of cpSRP43 in slow exchange on the chemical shift timescale.
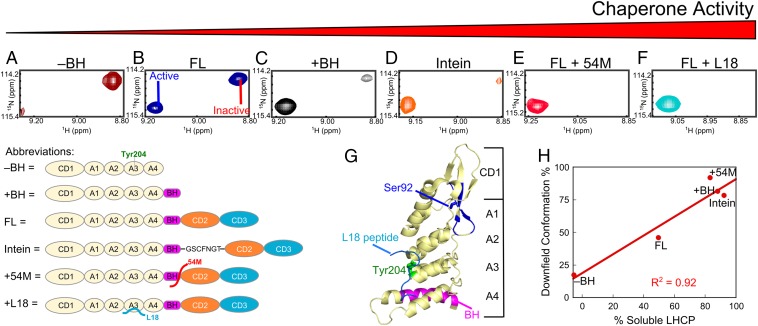
Fig. 4.
Conformational dynamics of cpSRP43 correlates with its chaperone activity. (A–F) Component cross-peaks for Ser92 in the TROSY spectrum of 2H,15N-labeled CD1Ank fragment (A), full-length cpSRP43 (B), CD1Ank-BH fragment (C), intein-cpSRP43 (D), cpSRP43 bound to 54M peptide (E), and cpSRP43 bound to the L18 peptide (F). (G) Crystal structure of CD1Ank-BH fragment (PDB ID code 3DEP) highlighting the residues in CD1 (blue) for which component cross peaks reflecting the conformational dynamics of cpSRP43 have been unambiguously assigned. Green shows Tyr204 in Ank3, which binds L18 (cyan). Magenta highlights the bridging helix. (H) Correlation between the chaperone activity (% soluble LHCP observed with 0.625 µM chaperone) of each construct or complex and the relative intensity of the downfield component cross-peak.
Fig. S4.
Pairs of component cross-peaks in CD1 arise from the same amino acids. (Upper) Component cross-peaks of residues Ser92 (A) and Ala95 (B) in the TROSY spectrum of 2H, 15N, 13C-labeled CD1Ank-BH. (Lower) Strips from the HNCACB and HN(CO)CACB spectra of 15N, 13C, 2H-labeled CD1Ank-BH for the two component peaks of S92 and A95. For each residue, the two component peaks have connectivities to identical Cα and Cβ chemical shifts for residues i and i-1, indicating that they arise from the same amino acid. The red rectangles highlight the connectivities for the active state (Left) and inactive state (Right).
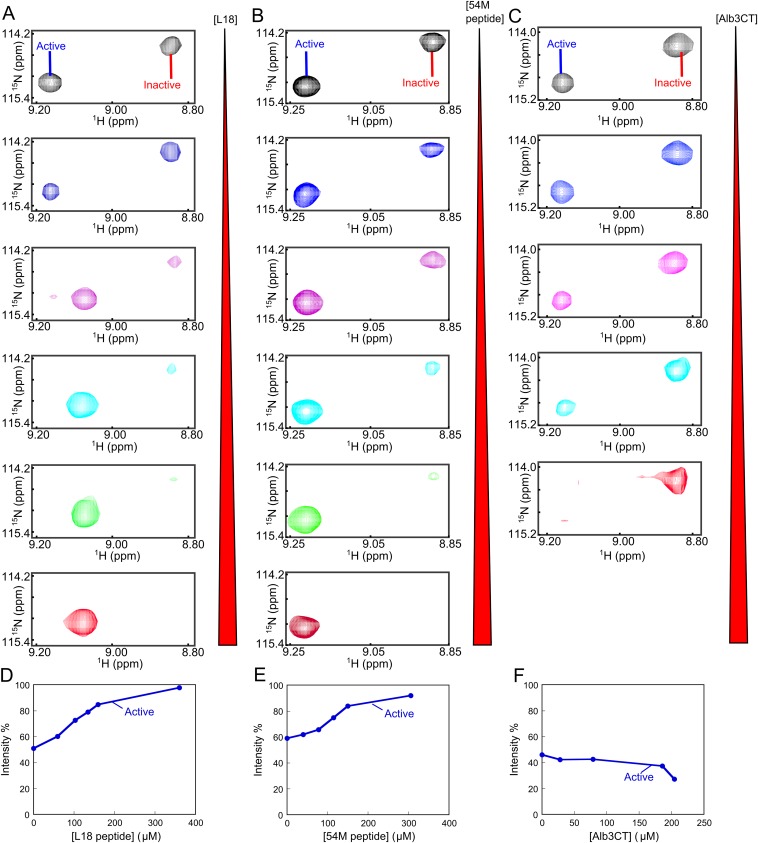
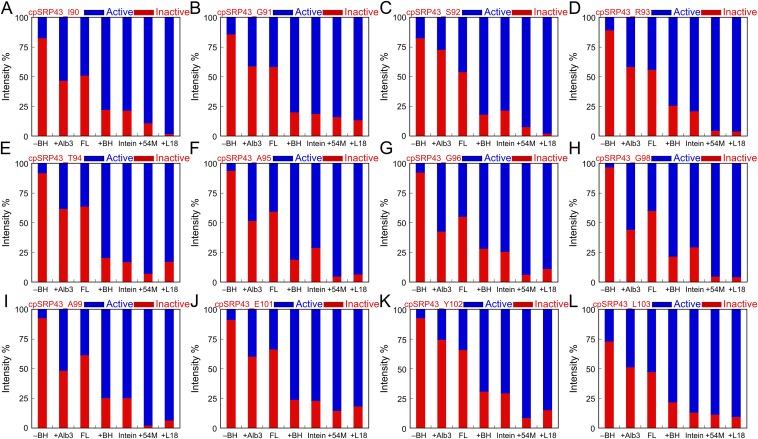
Importantly, the relative intensities of the component cross-peaks change in different constructs and in response to different ligands. An example for S92 is shown in Fig. 4, additional examples are shown in Fig. S5, and a summary for all 12 residues is shown in Fig. S6. In full-length cpSRP43, the intensities of the two component cross-peaks are comparable (Fig. 4B and Fig. S5). In the inactive CD1Ank construct, the upfield peak is dominant (Fig. 4A). In contrast, in the CD1Ank-BH construct or the superactive mutant intein-cpSRP43, which binds LHCP two- to fivefold more tightly than full-length cpSRP43 (Figs. 1B and 2B), the downfield peak becomes more intense (Fig. 4 C and D). When cpSRP43 is bound to the L18 peptide or to the activator, 54M, or the C-terminal peptide of 54M that binds cpSRP43 (54M peptide), the downfield peak is dominant (Fig. 4 E and F and Fig. S5 A and B). Additional titration experiments with L18 and 54M peptide (Fig. S5 A and B) corroborated that the downfield peak can be assigned to the active conformation of cpSRP43 conducive to substrate binding. Further, the relative intensity of this peak is strongly correlated with chaperone activity (Fig. 4H and Fig. S6).
Fig. S5.
The conformation of cpSRP43 is regulated by binding partners. (A–C) Titration of the L18 peptide (A), 54M peptide (B), and Alb3CT (C) into the 2H, 15N-labeled cpSRP43 during the TROSY experiment. Component cross-peaks are shown for Ser92. (D–F) Quantification of the relative intensity of the component cross-peaks from the data in A–C.
Fig. S6.
(A–L) Quantification of the relative intensities of component cross peaks for 12 residues in various chaperone constructs and with various ligands bound. –BH, CD1Ank fragment; +Alb3, full-length cpSRP43 + Alb3CT; FL, full-length cpSRP43; +BH, CD1Ank-BH fragment; Intein, Intein-cpSRP43; +54M, full-length cpSRP43 + 54M peptide; +L18, full-length cpSRP43 + L18 peptide.
Collectively, the biochemical and NMR data demonstrate that the SBD of cpSRP43 exchanges between active and inactive conformations that are regulated by CD2, by the substrate, and by interaction with the cpSRP54 M-domain.
Bidentate Interactions of Alb3 with cpSRP43 Drive Substrate Release.
Recent data showed that cpSRP43 directly interacts with the C-terminal stromal domain of Alb3 (Alb3CT). However, which site(s) in cpSRP43 interact with Alb3CT is extensively debated (25–27, 29). To resolve this question, we titrated Alb3CT into cpSRP43 and monitored changes in the TROSY spectra. We found that Alb3CT substantially (>90%) broadened a specific set of cross-peaks in CD3 (Fig. S7 A and C), in contrast to the absence of any substrate- or 54M-induced perturbations in this domain. Many perturbed cross-peaks map to an extensive surface displaying conserved acidic residues and two aromatic residues (Fig. 5A), suggesting this to be a binding site for Alb3CT. This observation is consistent with a recent study in which a peptide corresponding to motif IV in Alb3CT is found to bind at an acidic surface in CD3 (29).
Fig. S7.
Summary of the effect of Alb3CT on cross-peak intensities in the 1H-15N TROSY spectra of full-length cpSRP43 (A) and the CD1Ank-BH fragment (B). The intensities of each cross-peak in the presence and absence of Alb3CT [I(+Alb3CT) and I(–Alb3CT), respectively] were quantified and normalized to those of residues 1–63, a highly unstructured region whose intensities were unaffected by any binding partners, and their ratios were plotted. The gray bars denote unassigned residues (including all of the prolines), and the dashed lines show the cutoff where the peak is broadened ≥70%. (C) Structural model for full-length cpSRP43 (19). Dark red highlights residues whose cross-peaks are broadened >70% (from A) on binding to Alb3CT. Green highlights Tyr204 that binds L18.
Fig. 5.
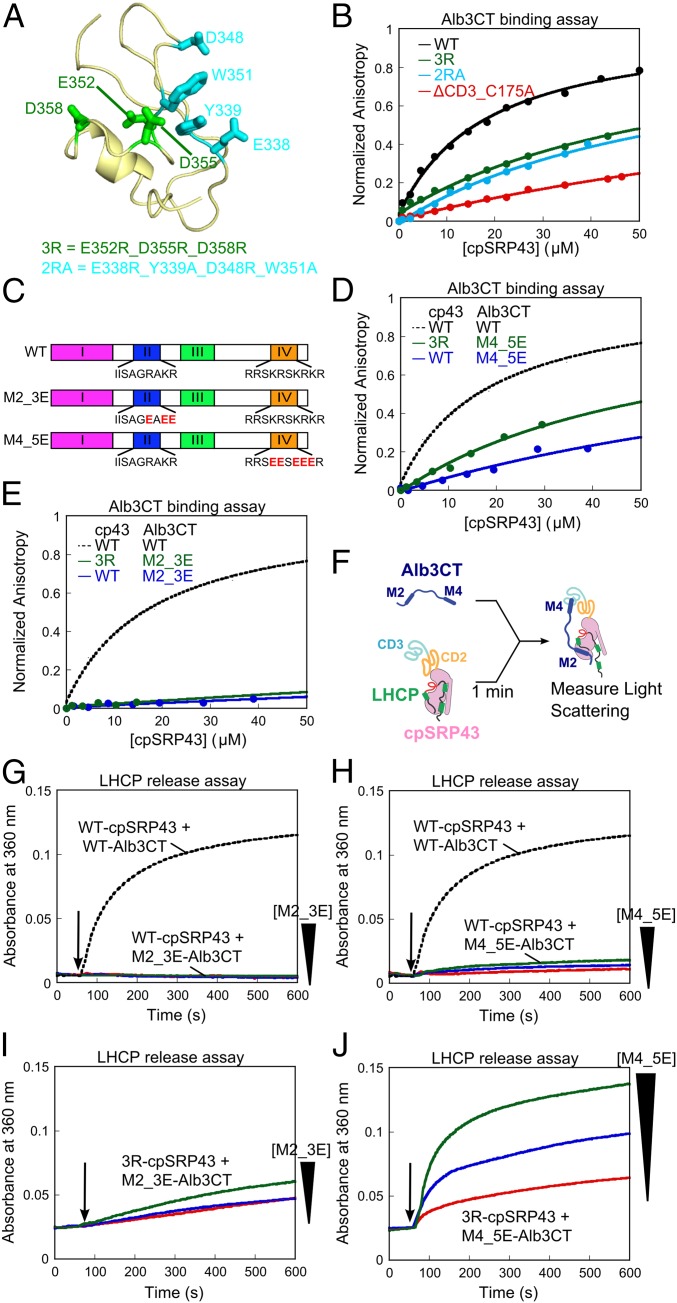
Chromodomain 3 of cpSRP43 binds motif IV of Alb3CT. (A) A structural model of CD3 (PDB ID code 1X32) highlighting the mutated residues. (B) Binding of fluorescein-labeled Alb3CT(S371C) to WT and mutant cpSRP43 were measured by changes in fluorescence anisotropy. The data were fit to Eq. 2 and gave Kd values of 18, >60, >63, and >160 µM for WT cpSRP43 and mutants 2RA, 3R, and ΔCD3, respectively. The anisotropy value of mutants was normalized to the same end point as WT cpSRP43. (C) Schematics of WT and charge reversal mutants of Alb3CT. (D and E) Binding of mutant Alb3CT(M4_5E) (D) and Alb3CT(M2_3E) (E) to WT cpSRP43 and mutant cpSRP43(3R), measured and analyzed as in B. The dashed lines indicate binding between WT Alb3CT and cpSRP43 from B and are shown for comparison. (F) Scheme for the LHCP TMD release assay. M2 and M4 denote Alb3CT motifs II and IV, respectively. (G–J) Alb3CT-induced TMD release from cpSRP43 for the reaction of WT cpSRP43 with mutant Alb3CT(M2_3E) (G), WT cpSRP43 with Alb3CT(M4_5E) (H), mutant cpSRP43(3R) with mutant Alb3CT(M2_3E) (I), and mutant cpSRP43(3R) with mutant Alb3CT(M4_5E) (J). The arrows indicate time of Alb3CT addition. Red, blue, and green indicate addition of 5, 10, and 20 µM Alb3CT, respectively. The dashed lines denote data of WT cpSRP43 with 5 µM Alb3CT and are shown for comparison.
To biochemically test this model, we generated two charge reversal mutants in CD3, E352R/D355R/D358R (3R; Fig. 5A, green), and E338R/Y339A/D348R/W351A (2RA; Fig. 5A, cyan). We measured the binding of Alb3CT to WT and mutant cpSRP43 based on the anisotropy change of fluorescein labeled at Alb3CT(S371C). Consistent with previous measurements (25, 26), WT cpSRP43 bound Alb3CT with an equilibrium dissociation constant (Kd) of 18 µM in this assay. Binding was weakened ninefold on deletion of CD3 (Fig. 5B, red), consistent with an important role of CD3 in cpSRP43-Alb3 binding (25). Both charge reversal mutants in CD3, cpSRP43(3R) and cpSRP43(2RA), weakened the binding of Alb3CT fourfold (Fig. 5B).
Alb3CT contains two conserved motifs, II and IV, that could interact with cpSRP43 (Fig. 5C) (25). Both motifs, especially motif IV, are enriched in basic residues, providing strong candidates for interacting with the acidic patch in CD3. We therefore introduced charge reversal mutations into motif II (M2_3E) or motif IV (M4_5E) of Alb3CT (Fig. 5C). Both mutations significantly weakened the binding of Alb3CT to cpSRP43 (Fig. 5 D and E, blue). Importantly, when the charge reversal mutants cpSRP43(3R) and Alb3CT(M4_5E) were combined to restore electrostatic complementarity, binding was partially rescued (Fig. 5D, green). In contrast, rescue was not observed when cpSRP43(3R) was combined with mutant Alb3CT(M2_3E) (Fig. 5E, green). These results suggest a specific electrostatic interaction between motif IV in Alb3CT and the acidic surface in CD3 of cpSRP43.
To provide additional evidence for this model and to probe the function of Alb3CT, we developed an independent assay in which a preformed, soluble cpSRP43•LHCP complex is challenged with Alb3CT. If interaction with Alb3CT releases substrate from cpSRP43, this would lead to aggregation of LHCP that can be monitored in real time using light scattering (Fig. 5F). Indeed, addition of Alb3CT led to the reappearance of large LHCP aggregates in a dose-dependent manner (Fig. S8A). This result indicates that the hydrophobic TMDs of LHCP are no longer protected by cpSRP43 on Alb3CT binding and provides a robust assay to analyze the interaction and activity of Alb3CT. Both charge reversal mutants of Alb3CT, M2_3E and M4_5E, display severely compromised activities in this release assay (Fig. 5 G and H), supporting the importance of both motifs in Alb3CT activity. Importantly, combining the charge reversal mutant cpSRP43(3R) with Alb3CT(M4_5E) restored release activity (Fig. 5J), whereas the combination with Alb3CT(M2_3E) did not (Fig. 5I). Thus, in agreement with the recent structural analysis (29), cpSRP43 CD3 provides a platform to specifically bind motif IV in Alb3CT. Our results further show that this interaction is electrostatic in nature and important for the ability of Alb3CT to induce substrate release from cpSRP43.
Fig. S8.
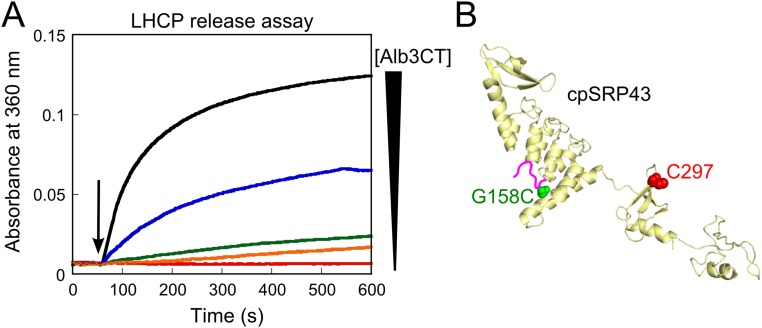
(A) Alb3CT induces dose-dependent release of LHCP TMDs from cpSRP43, performed as outlined in Fig. 5F and described in Materials and Methods. Red, orange, green, blue, and black denote data obtained with 0, 0.625, 1.25, 2.5, and 5.0 µM Alb3CT, respectively. Errors were estimated to be ±10% (SD) based on three technical replicates. (B) Structural model of cpSRP43 highlighting the positions of the FRET probes. Residue G158 in the L18 motif of LHCP (green) was mutated to cysteine and labeled with Atto488 (donor), and the native C297 in cpSRP43 (red) was labeled with TMR.
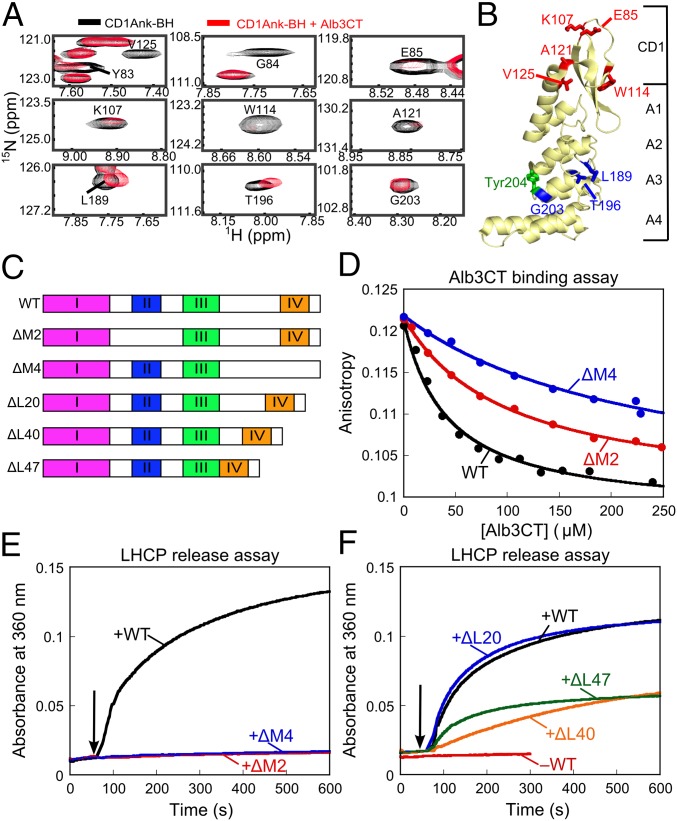
In addition to CD3, Alb3CT also induced substantial broadening of 17 cross-peaks in the TROSY spectrum of cpSRP43 that map to the SBD (Fig. S7 A and C). This observation was corroborated when the TROSY experiment was repeated with the CD1Ank-BH construct: even in the absence of CD2 and CD3, addition of Alb3CT-induced broadening or shifts of a specific set of cross-peaks in the TROSY spectrum of CD1Ank-BH (Fig. 6A and Fig. S7B). Seven of the Alb3CT-perturbed cross-peaks map to CD1; another three map to Ank3 (Fig. 6B). Thus, Alb3CT also contacts the SBD.
Fig. 6.
Bidentate interaction of Alb3CT with cpSRP43. (A) Alb3CT-induced perturbation of cross-peaks in the TROSY spectra of 2H,15N-labeled CD1Ank-BH fragment. Spectra in the absence and presence of Alb3CT are shown in black and red, respectively. (B) Structure of CD1Ank-BH (PDB ID code 3DEO) highlighting residues for which NMR cross-peaks are perturbed by Alb3CT. Red denotes residues for which cross-peaks are broadened >70%; blue denotes residues whose cross-peaks are shifted by Alb3CT. Green shows Tyr204 in Ank3 where L18 binds. (C) Schematic of the mutant Alb3CT constructs used in this study. ΔM2 and ΔM4 denote deletion of motifs II and IV, respectively, ΔL denotes deletion of linker sequence. (D) Binding of WT Alb3CT (black) and mutant ΔM2 (red) and ΔM4 (blue) to cpSRP43, measured by their ability to compete with fluorescein-labeled Alb3CT(S371C) for binding to cpSRP43 and detected by fluorescence anisotropy. The data were fit to Eq. 3 and gave values of 41, 99, and 237 µM for WT Alb3CT and mutants ΔM2 and ΔM4, respectively. (E and F) LHCP release assays were carried out as outlined in Fig. 5F for WT Alb3CT (black), mutants ΔM2 (red) and ΔM4 (blue) (E), and mutants ΔL20 (blue), ΔL40 (orange) and ΔL47 (green) (F).
The simplest model to explain these results is that Alb3CT makes bidentate interactions with cpSRP43 in both the SBD and CD3. To test this model, we deleted motif II or IV of Alb3CT (Fig. 6C, ΔM2 and ΔM4, respectively). We measured the binding of WT and mutant Alb3CT to cpSRP43 by using them as competitors of fluorescein-labeled Alb3CT in the anisotropy assay (Fig. 6D). ΔM2 and ΔM4 weakened the binding of Alb3CT to cpSRP43 two- and sixfold, respectively (Fig. 6D). Both mutations also abolished the ability of Alb3CT to trigger LHCP release from cpSRP43 (Fig. 6E). The same effects were observed with charge reversal mutations in Alb3CT motifs II and IV (Fig. 5 D, E, G, and H). Together, these data provide strong evidence that Alb3CT uses motifs II and IV to make bidentate interactions with cpSRP43. The much larger effects of each mutant in the LHCP release assay than on cpSRP43-Alb3 binding (Fig. 6 D vs. E) further suggests that, although each motif could bind cpSRP43 at sufficiently high concentrations, the ability of Alb3CT to trigger substrate release requires the interactions mediated by both motifs.
If a bidentate interaction with Alb3CT was required to release LHCP from cpSRP43, then the spacing between motifs II and IV would also be important for this activity. In WT Alb3, these two motifs are bridged by a ∼72-amino acid unstructured linker sequence, rendering it plausible that the two motifs can span the distance between the SBD and CD3 of cpSRP43. To test the importance of this spacing, we shortened the linker sequence (Fig. 6C, mutants ΔL20, ΔL40, and ΔL47). Although mutant ΔL20 did not exhibit a significant defect, further deletion of the linker sequence in mutants ΔL40 and ΔL47 compromised the ability of Alb3CT to induce LHCP release from cpSRP43 (Fig. 6F). Collectively, these data provide strong evidence for a bidentate interaction between Alb3CT and cpSRP43 that are essential to release LHCP from cpSRP43 at the membrane translocase.
Alb3CT Drives cpSRP43 to a Less Active Conformation.
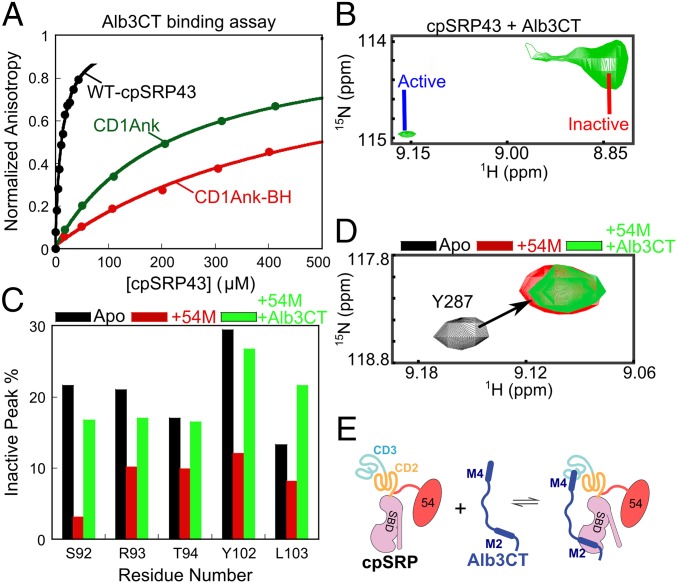
How does Alb3CT trigger substrate release from cpSRP43? Given that cpSRP43 samples between active and inactive conformations (Figs. 2–4), an attractive hypothesis is that Alb3CT drives cpSRP43 to a less active form. To test this model, we used two independent approaches. First, we took advantage of cpSRP43 variants, CD1Ank-BH and CD1Ank, which reside predominantly in the active and inactive conformations, respectively, and tested their binding with Alb3CT using the fluorescence anisotropy assay. We found that Alb3CT exhibits significant interactions with the CD1Ank fragment, although the interaction is 18-fold weaker than with full-length cpSRP43, consistent with the model that the SBD provides another binding site for Alb3CT (Fig. 7A). Importantly, Alb3CT bound to the CD1Ank fragment two- to threefold more strongly than to the CD1Ank-BH fragment, suggesting that it preferentially interacts with a cpSRP43 in a less active conformation. The stronger interaction of CD1Ank with Alb3CT than CD1Ank-BH also ruled out the possibility that irreversible misfolding of CD1Ank is responsible for its lack of chaperone activity.
Fig. 7.
Alb3CT preferentially binds and induces a less active conformation of cpSRP43. (A) Binding of Alb3CT to full-length cpSRP43 (black) and to the CD1Ank (green) and CD1Ank-BH (red) fragments, measured as in Fig. 5B. The data were fit to Eq. 2 and gave Kd values of 12, 216, and >530 µM for WT cpSRP43, CD1Ank, and CD1ANk-BH, respectively. Errors for Kd were estimated to be ±10% (SD) based on at least two technical replicates. (B) Region of the TROSY spectrum showing the component cross-peaks for Ser92 in 2H, 15N-labeled cpSRP43 in complex with Alb3CT. (C) The relative intensities of the cross-peaks corresponding to the inactive chaperone conformation are reduced by 54M (red bars) and enhanced by subsequent addition of Alb3CT (green bars). Data are shown for indicated residues in 2H, 15N-labeled intein-cpSRP43. Relative intensities were determined from cross-peak heights. (D) 54M and Alb3CT bind to intein-cpSRP43 simultaneously. The cross-peaks for Tyr287 in the TROSY spectra of 2H, 15N-labeled intein-cpSRP43 are shown. The peak was shifted on 54M binding (red), and this perturbation persists on subsequent addition of Alb3CT (green). (E) Scheme depicting the cobinding of 54M and Alb3CT to cpSRP43. M2, Alb3CT motif II; M4, Alb3CT motif IV.
In the second approach, we took advantage of the TROSY spectra of cpSRP43, in which the relative intensity of component cross-peaks from individual amide groups directly detects cpSRP43 subpopulations in active and inactive conformations. Of the 12 residues in cpSRP43 for which component cross-peaks can be unambiguously assigned, residues S92, R93, Y102, L103 exhibited an Alb3CT-induced increase in the relative intensity of the peak that arise from the inactive conformation (Fig. 7B and Figs. S5C and S6). These results provide independent support for the model that Alb3CT biases cpSRP43 to a less active conformation.
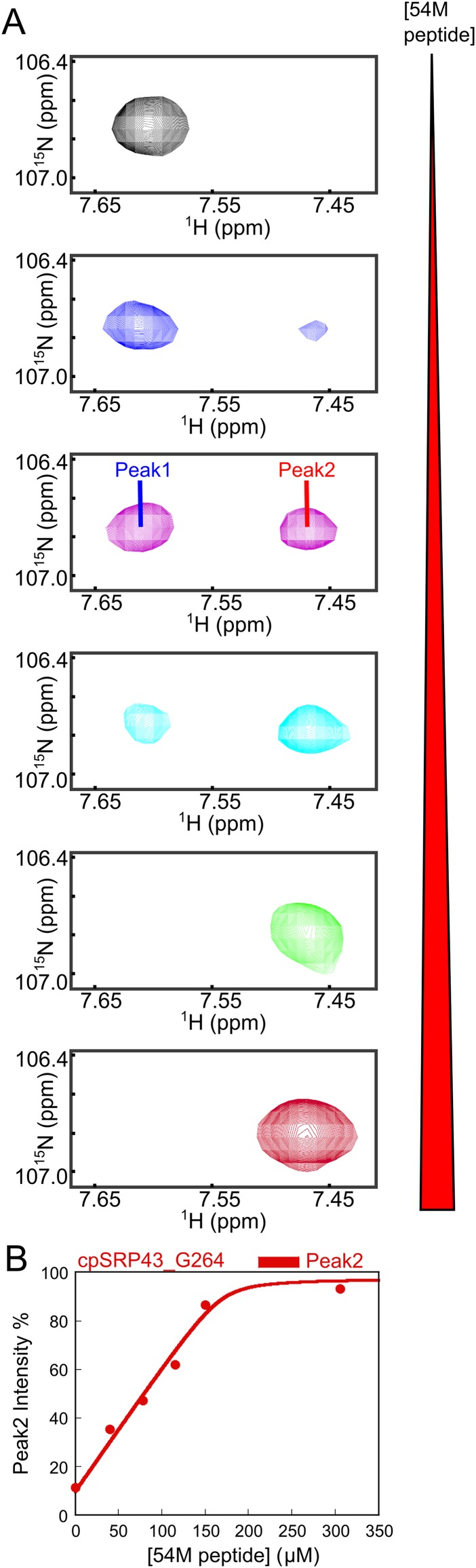
In the targeting pathway, Alb3CT must interact with and release substrates from the LHCP•cpSRP43•cpSRP54 complex. To test whether this is possible, we sequentially added the 54M peptide, followed by Alb3CT, during 2D-TROSY experiments with the superactive intein-cpSRP43 (to mimic the conformation of substrate-bound cpSRP43). For five residues (S92, R93, T94, Y102, and L103), the intensity of the component cross-peaks representing the inactive conformation was significantly reduced on the addition of 54M, but these inactive peaks regained significant intensity on subsequent addition of Alb3CT (Fig. 7C). The effect of Alb3CT did not arise from dissociation of 54M from cpSRP43, as the binding of 54M to cpSRP43 could be detected by shifts of specific cross-peaks corresponding to residues in and near CD2 (Fig. 7D and Fig. S9), and the 54M-induced perturbation of these cross-peaks remained even after the addition of Alb3CT (Fig. 7D).
Fig. S9.
Effects of 54M binding on the intensity of G264 (in BH). (A) Region of the TROSY spectrum of 2H, 15N-labeled cpSRP43 showing the effects of increasing concentrations of the 54M peptide on the G264 cross-peaks corresponding to unbound and 54M-bound cpSRP43 (labeled as peak 1 and peak 2, respectively). (B) Quantification of the relative intensities for the 54M-bound peak (peak 2) for G264. The line is a fit of the data to Eq. 1.
Collectively, the biochemical and NMR data strongly suggest that Alb3CT induces cpSRP43 to a less active conformation (Fig. 7E). The fact that this transition was observed with only a subset of residues further suggests that Alb3CT does not drive cpSRP43 into the completely inactive form but rather induces structural transitions in part of the cpSRP43 molecule. This notion is further supported by results in the following section.
Alb3CT Specifically Releases Substrate TMDs from cpSRP43.
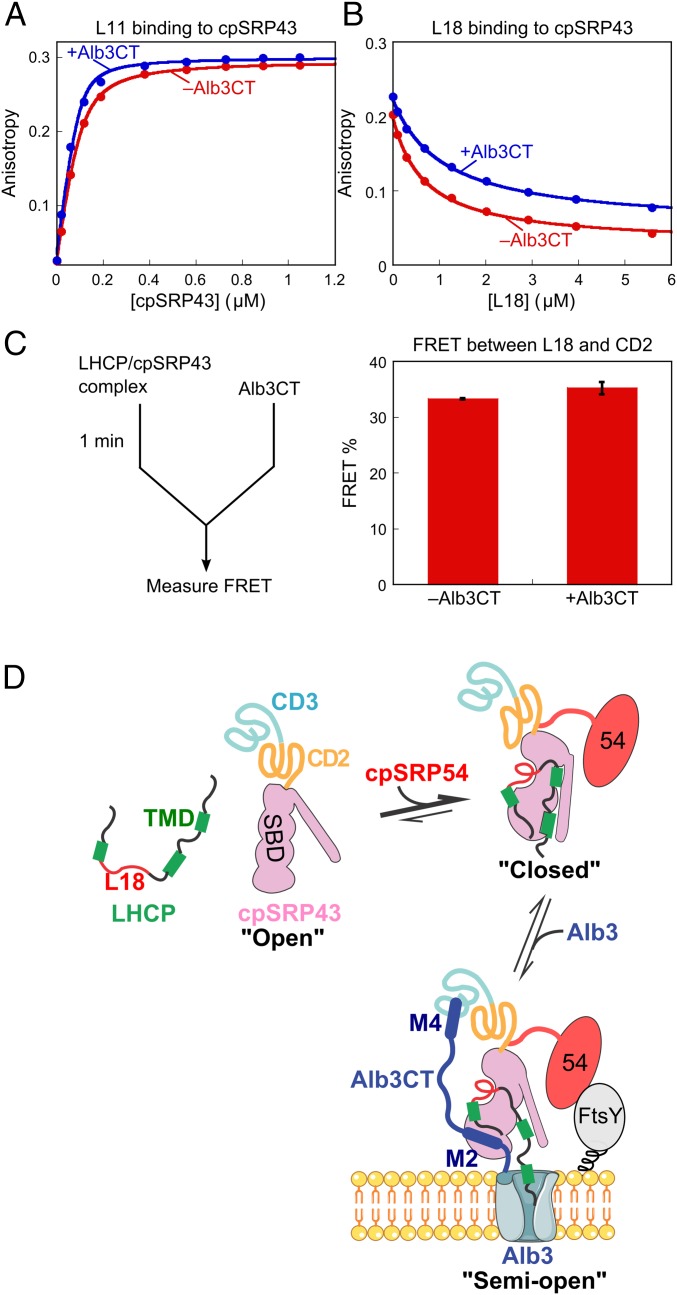
Our previous work showed that cpSRP43 binds LHCP via two sets of interactions: recognition of the L18 motif in loop 2 and promiscuous hydrophobic interactions with the TMDs in LHCP (22). When the cpSRP43•LHCP complex was challenged by Alb3CT in the release assay, the light scattering data indicated that Alb3CT antagonizes the interaction of substrate TMDs with cpSRP43 such that they are no longer protected from aggregation (Fig. 5F and Fig. S8A). To test whether Alb3CT also antagonizes the interaction of cpSRP43 with the L18 motif, we measured the binding of HiLyte-Fluor488–labeled L11 peptide to cpSRP43 based on fluorescence anisotropy (Fig. 8A). Intriguingly, the binding of L11 to cpSRP43 was unaffected, if not slightly stronger, in the presence of Alb3CT (Fig. 8A). To rule out artifacts from dye labeling, we also measured the binding of unlabeled L18 peptide to cpSRP43 by using it as a competitor of dye-labeled L11 (Fig. 8B). This experiment yielded the same result: L18 binds to cpSRP43 with comparable affinity with or without Alb3CT present. These data indicate that Alb3CT specifically antagonizes the interaction of cpSRP43 with the substrate TMDs, but does not affect the interaction of this chaperone with the L18 motif.
Fig. 8.
Alb3CT uncouples the interaction of cpSRP43 with the L18 motif and TMD of LHCP. (A) Binding of HiLyte-Fluor488–labeled L11 peptide to cpSRP43 with (blue) and without (red) Alb3CT present, measured using fluorescence anisotropy. The data were fit to Eq. 2 and gave Kd values of 22 and 11 nM in the absence and presence of 20 µM Alb3CT, respectively. (B) Binding of the L18 peptide to cpSRP43 in the absence (red) and presence (blue) of Alb3CT, measured by using L18 as a competitor of HiLyte-Fluor488–labeled L11 peptide in binding cpSRP43. The data were fit to Eq. 3 and gave values of 1.1 and 0.68 µM in the presence and absence of Alb3CT, respectively. (C) (Left) Scheme of the FRET-based LHCP release assay (Materials and Methods). (Right) FRET efficiency between Atto488-labeled LHCP (at L18) and TMR labeled cpSRP43 before and after challenge by Alb3CT. Values reported are mean ± SD, with n = 2. (D) Model for the chaperone cycle of cpSRP43 during LHCP targeting and insertion, as described in the text. TMD, transmembrane domain; SBD, substrate binding domain; M2, Alb3CT motif II; M4, Alb3CT motif IV.
To test this model in the context of full-length LHCP, we reperformed the LHCP release assay except that, instead of monitoring the release reaction by light scattering, we monitored FRET between a donor dye (Atto488) labeled in the L18 motif of LHCP (G158C) and an acceptor dye [tetramethylrhodamine (TMR)] labeled at the native Cys297 of cpSRP43 (Fig. S8B). In contrast to the release of TMDs detected by light scattering, the FRET assay showed no changes in, if not slightly higher, FRET efficiency between LHCP-L18 and cpSRP43-CD2 when the cpSRP43•LHCP complex was challenged with Alb3CT (Fig. 8C), suggesting that Alb3CT did not induce the release of L18 from cpSRP43. Together, these results support a model in which Alb3CT induces cpSRP43 into a distinct conformation in which the substrate TMDs are released, whereas the L18 motif of LHCP remains bound to the chaperone.
SI Materials and Methods
Protein Production and Purification for NMR.
Isotope-labeled full-length cpSRP43 and CD1Ank-BH were overexpressed in BL21(DE3) cells at 37 °C in M9 D2O media supplemented with 15NH4SO4 and 12C or 13C-glucose. Protein expression was induced at OD600 = 0.7 by addition of 1 mM Isopropyl beta-D-1-thiogalactopyranoside (IPTG) for 15 h. Isotope-labeled cpSRP43 and CD1Ank-BH were purified using the same protocol as unlabeled cpSRP43. Proteins were exchanged into NMR buffer (50 mM phosphate, 150 mM NaCl, pH 6.5) using a PD MidiTrap G-25 column (GE Healthcare).
Assignment of cpSRP43 Backbone.
Triple labeled (2H, 15N, 13C) cpSRP43 and CD1Ank-BH (∼0.8 mM) were prepared in NMR buffer containing 10% (vol/vol) D2O. Backbone resonance assignments were made using 3D TROSY-HNCA, TROSY-HN(CO)CA, TROSY-HNCACB, and TROSY-HN(CO)CACB spectra (44, 45). A total of 252 of 323 nonproline backbone residues were assigned using RunAbout in NMRview java. Many of the cross-peaks associated with CD2 are weak, and only a subset (residues 278–287) could be assigned from the triple resonance spectra. Assignments for additional CD2 residues (F267, D273, I275, E277, L288, V289, W291, D293, G294, W299, V300, G302, D308, V309, and K311) were made by transferring published assignments for the isolated CD2 domain (33).
Characterization of the Oligomeric State of cpSRP43.
The oligomeric state of cpSRP43 was assessed by two methods. First, cpSRP43 in the presence or absence of equal molar HiLyte-Fluor488–labeled L11 peptide were analyzed by size exclusion chromatography using Superdex 200 column (GE Healthcare). The column was equilibrated in 50 mM KHepes, pH 7.5, with either 200 mM or 50 mM NaCl. The protein elution profile was confirmed by SDS/PAGE, and the molecular mass on the column was further calibrated using the LMW kit (GE Healthcare). Second, velocity sedimentation-type analytical ultracentrifugation was performed using Optima XL-I (Beckman Coulter) with an absorbance optical detection system (280 nm). The sample was spun at 201,600 × g at 20 °C with 7 μM of cpSRP43 alone and in complex with equal molar of HiLyte-Fluor488–labeled L11 peptide or cpSRP54M. Buffer viscosity, protein partial specific volumes, and density were calculated using the SEDNTERP (46). The observed sedimentation data were fitted to a single component system by using the SEDFIT software (47), and the sedimentation coefficient distribution was extracted from the fitting.
Discussion
Membrane proteins pose special challenges to protein homeostasis during their posttranslational targeting and require highly effective chaperones. Compared with chaperones that facilitate the folding of soluble proteins, membrane protein chaperones not only handle much more aggregation-prone client proteins, but must also regulate their substrate binding and release in response to spatial cues. Here, we define the complete chaperone cycle for cpSRP43, an ATP-independent chaperone dedicated to integral membrane proteins. Our results reveal a remarkably modular nature of this chaperone, wherein substrate binding and spatial regulations are mediated by distinct domains. Most importantly, the SBD of cpSRP43 intrinsically samples at least three distinct conformations. This conformational sampling enables this chaperone to be readily switched on by activators in the stroma and switched off by a negative regulator at the target membrane, driving highly coordinated substrate capture and release despite the lack of ATPase cycles.
Substrate Binding Domain Samples Multiple Conformations.
Remarkably, a 25-kDa fragment in cpSRP43 comprised of CD1, the ankyrin repeats, and the bridging helix was sufficient for stoichiometric binding and chaperoning of LHCP by cpSRP43. Combined with previous mutational work that tested the effects of deleting CD1 and individual ankyrin repeats (19), this defines the CD1Ank-BH fragment as the minimal substrate binding domain for cpSRP43. How this small chaperone domain protects client proteins, which match its own size and contain three TMDs, will be the next challenging question.
Importantly, the SBD of cpSRP43 intrinsically samples multiple conformations. This property is most directly visualized in the 1H-15N TROSY spectra of cpSRP43, in which at least two distinct conformations in slow exchange gave rise to pairs of component cross-peaks for the same backbone NH. Although component cross-peaks are unambiguously assigned for 12 residues reported here, the number of cross-peaks in the TROSY spectrum is ∼50% greater than the number of residues in cpSRP43, indicating that many more residues undergo analogous conformational sampling. A recent single molecule study also revealed a high degree of conformational fluctuation in cpSRP43 (24). Our ability to isolate a large set of superactive and defective mutations that lock this chaperone into distinct conformations provides further evidence for this model. Importantly, the relative intensity of the component cross-peaks in the TROSY spectra strongly correlate with chaperone activity in different cpSRP43 variants and in the presence of different regulators (more discussions below). Based on these observations, we propose that the SBD samples between an open conformation unable to bind substrates and a closed conformation conducive to tight substrate binding (Fig. 8D, Upper).
SBD Is Activated by cpSRP54 in the Soluble Phase.
Although CD2 does not directly bind substrate proteins, molecular events in this domain regulate substrate binding in the neighboring SBD. Intriguingly, CD2 by itself reduces substrate binding in the SBD and biases cpSRP43 toward the less active open conformation. Combined with the ability of a large number of point mutations in CD2 to hyperactivate cpSRP43 and drive the SBD to the closed conformation, these observations suggest the presence of strong evolutionary pressure to maintain a substantial population of apo-cpSRP43 in the inactive conformation, i.e., to keep this chaperone at the tipping point of conformational transitions.
This conformational property enables cpSRP43 to be readily turned on by cpSRP54 in the stroma. The C-terminal tail of the cpSRP54 M-domain intercalates between the bridging helix and CD2, placing it at an optimal position for regulating interdomain interactions (23). In support of this model, we found here that 54M or the 54M peptide stimulates the chaperone activity of SBD. A recent study also found that cpSRP54 enhanced binding of L18 to cpSRP43 threefold (24). Our observation that the stimulatory effect of 54M is largely bypassed in superactive chaperone mutants and becomes more pronounced in defective chaperone mutants further indicates that 54M and superactive mutations in CD2 drive the same activating conformational change in the SBD. This 54M-driven rearrangement allows cpSRP43 to efficiently capture and tightly bind its substrates in the stroma, effectively protecting LHCPs from aggregation in the aqueous environment (Fig. 8D, Upper Right).
The structural basis of the communication between CD2, 54M, and SBD remains a challenging question for future studies. Nevertheless, the available data provide intriguing clues. As mutations in the linker bridging the SBD and CD2 can lead to gain or loss of function, communication likely involves reorientation of CD2 relative to the SBD. This model is consistent with previous small angle X-ray scattering (SAXS) analysis (23) and with the observation of enhanced FRET efficiency between dye pairs in Ank3 and CD2 on cpSRP54 binding (24). The BH, which connects the ankyrin repeat domain to CD2 and whose deletion drives cpSRP43 into the inactive state, provides a prime candidate for mediating interdomain communication. Nevertheless, multiple residues in CD1 can sense the binding of L18 and 54M, which binds >50 Å away, indicating that substrate and 54M induces long-range communications that propagates through the entire length of the SBD.
Bidentate Interactions of Alb3CT Drive Coordinated Substrate Release.
At the target membrane, cpSRP43 must facilely release its substrate to the Alb3 translocase. The finding that cpSRP43 directly interacts with the stromal domain of Alb3 (25, 27, 30) provides an attractive mechanism to couple the release of substrate to its correct localization and imminent insertion into membrane. Nevertheless, where and how cpSRP43 interacts with Alb3 has been highly controversial, with the ankyrin repeat domain or the chromodomains alternatively proposed as Alb3 binding sites (25–27, 29). Here, high-resolution NMR combined with biochemical analyses resolved this issue and showed that Alb3CT uses motifs II and IV to make bidentate interactions with both the SBD and CD3 of cpSRP43, respectively. As these sites are distinct from the 54M binding site (located in CD2), this rationalizes the observation that cpSRP43 can form a ternary complex with both cpSRP54 and Alb3CT in the NMR studies here and in previous pull-down experiments (25). The fact that Alb3CT and cpSRP54 biases cpSRP43 to different conformations also explains the anti-cooperative binding between these two factors (24, 25). Given the predominantly electrostatic nature of the interaction between CD3 and Alb3 motif IV, it is likely that the acidic patch on CD3 provides the site for initial recruitment of Alb3CT (29), which then enables Alb3 motif II to further contact the SBD to induce substrate release.
Intriguingly, Alb3CT specifically induces the release of substrate TMDs from cpSRP43 without disrupting the interaction of the L18 motif with the chaperone. This observation has several important implications. First, Alb3CT does not simply reverse the effect of 54M and drive cpSRP43 to the completely inactive open conformation. Instead, Alb3CT induces a third, semiopen state of cpSRP43 in which its interactions with the L18 and TMDs on substrate protein are uncoupled (Fig. 8D, Lower). Second, this suggests a highly coordinated mechanism of substrate release at the membrane, in which the TMDs are first released and could initiate their membrane insertion via Alb3, whereas the L18 loop of LHCP remains bound to cpSRP43. Formation of such a LHCP•cpSRP•Alb3 release complex at the membrane is consistent with the previous observation that cpSRP43 was trapped into high molecular weight species with LHCP on addition of Alb3CT (27). In contrast to the current mechanisms in which LHCP is completely released from cpSRP43, such a stepwise mechanism would minimize irreversible aggregation of LHCP and abortive targeting reactions, providing a more productive route for unloading the membrane protein substrate onto the translocase.
Summary.
Our work here defines a rigorous framework for the chaperone cycle of cpSRP43 (Fig. 8D). The SBD of cpSRP43 samples at least three conformational states: an open state (Upper Left), a closed state (Upper Right), and semiopen state that binds tightly to L18 but not the TMDs of LHCP (Lower). CD2 biases the SBD to the open state, whereas interaction with cpSRP54 drives cpSRP43 to the closed state that binds LHCP tightly. When the transit complex is targeted to the thylakoid membrane via the interaction of cpSRP54 with cpFtsY, Alb3CT uses motifs II and IV (M2 and M4) to make bidentate interactions with SBD and CD3 of cpSRP43, respectively. These interactions induce cpSRP43 to a semiopen conformation, triggering the release of the substrate TMDs from cpSRP43 and initiating their membrane insertion, whereas the L18 motif remains bound to cpSRP43. Ultimate release of L18 from cpSRP43 might be driven by folding of LHCP in the membrane and binding of photosynthetic pigments.
Although cpSRP43 is dedicated to the LHCP family of proteins, it embodies multiple new concepts that have emerged from the recent discovery of a wide array of chaperones: (i) the use of protein interaction energy instead of ATPase cycles to regulate substrate binding/release, which is found in cyclophilins (34), small heat-shock proteins (35–38), and all of the chaperones in the bacterial periplasmic or eukaryotic extracellular space; and (ii) the use of conformational flexibility for activation, which was also found in HdeA, Hsp33, and Hsp26 (39, 40), and may be a general feature of ATP-independent chaperones. The mechanism we describe here for cpSRP43 could facilitate understanding of these conceptually analogous chaperone systems, as well as methods for their investigation.
Materials and Methods
Protein Expression and Purification.
cpSRP43 and Alb3CT mutants were constructed using the QuikChange procedure (Stratagene). CD1Ank constructs were deleted residues from 253 to 266 compared with CD1Ank-BH. In the linker mutants Intein (GSCFNGT) and GSGSG, the indicated sequences were inserted between V266 and F267. In mutant 2P, two prolines replaced the original linker residues (Q265 and V266). WT and mutant cpSRP43 and LHCP were overexpressed and purified as previous described (41). Alb3CT was overexpressed in BL21(DE3) cells and purified as previously described (25). 54M peptide (QKAPPGTARRKRKAC) was from Eton Bioscience (99% purity). Peptides L11 (GSFDPLGLADD), L18 (VDPLYPGGSFDPLGLADD), and L11 labeled with HiLyte-Fluor488 were purchased from AnaSpec (>95% purity). Single cysteine mutants of cpSRP43 and LHCP were labeled with fluorescent dyes via maleimide chemistry with 80–90% efficiency. Labeled proteins were purified through Sephadex G25 to remove free dye.
Chaperone Activity of cpSRP43.
Two types of experiments were used. First, we measured the ability of cpSRP43 to prevent LHCP aggregation, as described previously (19, 22). Briefly, urea denatured LHCP was diluted into buffer containing varying concentrations of cpSRP43, and light scattering at 360 nm was measured over time until equilibrium was reached. The percentage of soluble LHCP (% soluble) at equilibrium was plotted as a function of cpSRP43 concentration. The data were fit to
| [1] |
in which [pro] is cpSRP43 concentration, ΔA is the soluble% at saturating cpSRP43 concentrations, and Kd is the apparent dissociation constant of LHCP interaction with cpSRP43.
Second, we measured the ability of cpSRP43 to reverse preformed LHCP aggregates. LHCP (1 µM) was aggregated in buffer for 1 min, followed by addition of 4–5 µM WT or mutant cpSRP43, and the clearance of large LHCP aggregates was followed in real time by light scattering at 360 nm.
Measurement of L18 Binding.
The interaction of L18 with cpSRP43 was assessed by two methods. First, binding of cpSRP43 to HiLyte-Fluor488–labeled L11 peptide, which contains the minimal interaction sequence in L18, was detected by changes in fluorescence anisotropy of the dye. Anisotropy measurements were conducted in buffer D (50 mM KHepes, pH 7.5, 200 mM NaCl) on Fluorolog 3–22 (Yobin Yvon), using 100 nM HiLyte-Fluor488–labeled L11 and varying concentrations of cpSRP43. The samples were excited at 500 nm, and the fluorescence anisotropy was recorded at 527 nm. The data were fitted to
| [2] |
in which Aobsd is the observed anisotropy value, A0 is the anisotropy value without cpSRP43, ΔA is the change in anisotropy at saturating cpSRP43 concentrations, and Kd is the equilibrium dissociation constant for cpSRP43 interaction with L11-HiLyte-Fluor488.
To independently measure the L18-cpSRP43 interaction without perturbations from the dye, unlabeled L18 peptide was used as a competitor for the binding of L11-HiLyte-Fluor488 to cpSRP43; 100 nM L11-HiLyte-Fluor488 was preincubated with 120 nM cpSRP43 for 5 min, and the complex was challenged with an increasing concentration of L18 peptide. Anisotropy values were recorded at equilibrium and plotted as a function of [L18]. The data were fit to
| [3] |
in which Aobsd is the observed anisotropy value, A0 is the anisotropy value without L18 present, ΔA is the change in anisotropy at saturating L18 concentrations, and is the apparent inhibition constant.
Alb3CT Binding to cpSRP43.
Binding was detected by two methods. First, Alb3CT(S371C) was labeled with fluorescein-5′-maleimide (Invitrogen), and labeled protein was purified through Sephadex G25 (Sigma) to remove free dye. Binding of fluorescein-labeled Alb3CT(S371C) to cpSRP43 was detected by changes in fluorescence anisotropy, measured as described for L11 binding except that 200 nM Alb3CT(S371C)-fluorescein was used instead of L11. Samples were excited at 495 nm, and the fluorescence anisotropy was recorded at 512 nm. The data were fit to Eq. 2, with the exception that [Alb3CT] replaces [L11].
Second, unlabeled Alb3CT was used as competitors in the interaction of cpSRP43 with Alb3CT(S371C)-fluorescein; 200 nM Alb3CT-S371C-fluorescein was preincubated with 10 µM cpSRP43 for 5 min, and the complex was chased with increasing concentrations of WT or mutant Alb3CT. Fluorescence anisotropy values were recorded at equilibrium and plotted as a function of competitor concentration. The data were fit to Eq. 3, with the exception that [Alb3CT] replaces [L18].
Alb3-Induced Substrate Release.
LHCP release was evaluated by two methods. First, a soluble LHCP•cpSRP43 complex was performed in buffer D using 1 µM LHCP and 5 µM cpSRP43. After 1-min incubation, the complex was chased with Alb3CT, and the reaction was monitored in real time by light scattering at 360 nm. Second, a single cysteine LHCP mutant (C80A,G158C) was labeled with Atto488 maleimide in the middle of L18. A single cysteine mutant of cpSRP43 (C175A-C297) was labeled with TMR-5-maleimide (Invitrogen). Labeled LHCP (50 nM) and cpSRP43 (2 µM) were preincubated for 5 min in buffer D with 0.1 mg/mL BSA, followed by addition of 5 µM Alb3CT. Fluorescence was monitored on a Fluorolog 3–22 spectrofluorometer (Jobin Yvon) using an excitation wavelength of 505 nm and emission wavelength of 525 nm. A control release reaction was performed using unlabeled cpSRP43 to obtain the intensity for the donor sample (ID). FRET efficiency was calculated as
| [4] |
in which IDA is the fluorescence intensity of donor in the presence of acceptor-labeled cpSRP43.
NMR Spectroscopy.
Multidimensional NMR spectra were recorded on Bruker Avance spectrometers operating at 800 and 900 MHz. All NMR spectra were acquired at 17 °C using 2H, 15N-labeled or 2H, 15N, 13C-labeled protein (∼0.2 mM) in NMR buffer containing 10% (vol/vol) D2O at pH 6.5. Titration experiments were performed by serial addition of unlabeled ligands into the NMR sample containing 2H, 15N-labeled cpSRP43. NMR data were processed with NMRPipe (42) and analyzed with NMRView Java (43). Details on NMR sample preparation and assignments are described in SI Materials and Methods.
Acknowledgments
We thank R. Dalbey and P. Jaru-Ampornpan for plasmid encoding Alb3CT, V. Q. Lam and M. Yamout for initial optimization of the isotope labeling condition, P. Aoto for advice on NMR assignments, S. Chandrasekar and S. Lieblich (S. Mayo’s laboratory) for help with analytical ultracentrifugation, and members of the S.-o.S. laboratory for comments on the manuscript. This work was supported by fellowships from the Gordon and Betty Moore Foundation and American Federation for Aging Research (to S.-o.S.) and the Skaggs Institute of Chemical Biology (to P.E.W.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1524777113/-/DCSupplemental.
References
- 1.Hardy SJ, Randall LL. A kinetic partitioning model of selective binding of nonnative proteins by the bacterial chaperone SecB. Science. 1991;251(4992):439–443. doi: 10.1126/science.1989077. [DOI] [PubMed] [Google Scholar]
- 2.Randall LL, Hardy SJ. SecB, one small chaperone in the complex milieu of the cell. Cell Mol Life Sci. 2002;59(10):1617–1623. doi: 10.1007/PL00012488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Walton TA, Sandoval CM, Fowler CA, Pardi A, Sousa MC. The cavity-chaperone Skp protects its substrate from aggregation but allows independent folding of substrate domains. Proc Natl Acad Sci USA. 2009;106(6):1772–1777. doi: 10.1073/pnas.0809275106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hennecke G, Nolte J, Volkmer-Engert R, Schneider-Mergener J, Behrens S. The periplasmic chaperone SurA exploits two features characteristic of integral outer membrane proteins for selective substrate recognition. J Biol Chem. 2005;280(25):23540–23548. doi: 10.1074/jbc.M413742200. [DOI] [PubMed] [Google Scholar]
- 5.Thoma J, Burmann BM, Hiller S, Müller DJ. Impact of holdase chaperones Skp and SurA on the folding of β-barrel outer-membrane proteins. Nat Struct Mol Biol. 2015;22(10):795–802. doi: 10.1038/nsmb.3087. [DOI] [PubMed] [Google Scholar]
- 6.Mihara K, Omura T. Cytoplasmic chaperones in precursor targeting to mitochondria: The role of MSF and hsp 70. Trends Cell Biol. 1996;6(3):104–108. doi: 10.1016/0962-8924(96)81000-2. [DOI] [PubMed] [Google Scholar]
- 7.Zhuravleva A, Clerico EM, Gierasch LM. An interdomain energetic tug-of-war creates the allosterically active state in Hsp70 molecular chaperones. Cell. 2012;151(6):1296–1307. doi: 10.1016/j.cell.2012.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fenton WA, Horwich AL. GroEL-mediated protein folding. Protein Sci. 1997;6(4):743–760. doi: 10.1002/pro.5560060401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tapley TL, et al. Structural plasticity of an acid-activated chaperone allows promiscuous substrate binding. Proc Natl Acad Sci USA. 2009;106(14):5557–5562. doi: 10.1073/pnas.0811811106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tapley TL, Franzmann TM, Chakraborty S, Jakob U, Bardwell JC. Protein refolding by pH-triggered chaperone binding and release. Proc Natl Acad Sci USA. 2010;107(3):1071–1076. doi: 10.1073/pnas.0911610107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reichmann D, et al. Order out of disorder: Working cycle of an intrinsically unfolded chaperone. Cell. 2012;148(5):947–957. doi: 10.1016/j.cell.2012.01.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ouyang M, et al. LTD is a protein required for sorting light-harvesting chlorophyll-binding proteins to the chloroplast SRP pathway. Nat Commun. 2011;2:277. doi: 10.1038/ncomms1278. [DOI] [PubMed] [Google Scholar]
- 13.Schuenemann D, et al. A novel signal recognition particle targets light-harvesting proteins to the thylakoid membranes. Proc Natl Acad Sci USA. 1998;95(17):10312–10316. doi: 10.1073/pnas.95.17.10312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schünemann D. Structure and function of the chloroplast signal recognition particle. Curr Genet. 2004;44(6):295–304. doi: 10.1007/s00294-003-0450-z. [DOI] [PubMed] [Google Scholar]
- 15.Franklin AE, Hoffman NE. Characterization of a chloroplast homologue of the 54-kDa subunit of the signal recognition particle. J Biol Chem. 1993;268(29):22175–22180. [PubMed] [Google Scholar]
- 16.Li X, Henry R, Yuan J, Cline K, Hoffman NE. A chloroplast homologue of the signal recognition particle subunit SRP54 is involved in the posttranslational integration of a protein into thylakoid membranes. Proc Natl Acad Sci USA. 1995;92(9):3789–3793. doi: 10.1073/pnas.92.9.3789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tu CJ, Schuenemann D, Hoffman NE. Chloroplast FtsY, chloroplast signal recognition particle, and GTP are required to reconstitute the soluble phase of light-harvesting chlorophyll protein transport into thylakoid membranes. J Biol Chem. 1999;274(38):27219–27224. doi: 10.1074/jbc.274.38.27219. [DOI] [PubMed] [Google Scholar]
- 18.Falk S, Sinning I. cpSRP43 is a novel chaperone specific for light-harvesting chlorophyll a,b-binding proteins. J Biol Chem. 2010;285(28):21655–21661. doi: 10.1074/jbc.C110.132746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jaru-Ampornpan P, et al. ATP-independent reversal of a membrane protein aggregate by a chloroplast SRP subunit. Nat Struct Mol Biol. 2010;17(6):696–702. doi: 10.1038/nsmb.1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stengel KF, et al. Structural basis for specific substrate recognition by the chloroplast signal recognition particle protein cpSRP43. Science. 2008;321(5886):253–256. doi: 10.1126/science.1158640. [DOI] [PubMed] [Google Scholar]
- 21.DeLille J, et al. A novel precursor recognition element facilitates posttranslational binding to the signal recognition particle in chloroplasts. Proc Natl Acad Sci USA. 2000;97(4):1926–1931. doi: 10.1073/pnas.030395197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jaru-Ampornpan P, et al. Mechanism of an ATP-independent protein disaggregase: II. distinct molecular interactions drive multiple steps during aggregate disassembly. J Biol Chem. 2013;288(19):13431–13445. doi: 10.1074/jbc.M113.462861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Holdermann I, et al. Chromodomains read the arginine code of post-translational targeting. Nat Struct Mol Biol. 2012;19(2):260–263. doi: 10.1038/nsmb.2196. [DOI] [PubMed] [Google Scholar]
- 24.Gao F, et al. Regulation of structural dynamics within a signal recognition particle promotes binding of protein targeting substrates. J Biol Chem. 2015;290(25):15462–15474. doi: 10.1074/jbc.M114.624346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Falk S, Ravaud S, Koch J, Sinning I. The C terminus of the Alb3 membrane insertase recruits cpSRP43 to the thylakoid membrane. J Biol Chem. 2010;285(8):5954–5962. doi: 10.1074/jbc.M109.084996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Falk S, Sinning I. The C terminus of Alb3 interacts with the chromodomains 2 and 3 of cpSRP43. J Biol Chem. 2010;285(53):le25–le26. doi: 10.1074/jbc.L110.160093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lewis NE, et al. A dynamic cpSRP43-Albino3 interaction mediates translocase regulation of chloroplast signal recognition particle (cpSRP)-targeting components. J Biol Chem. 2010;285(44):34220–34230. doi: 10.1074/jbc.M110.160093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dünschede B, Bals T, Funke S, Schünemann D. Interaction studies between the chloroplast signal recognition particle subunit cpSRP43 and the full-length translocase Alb3 reveal a membrane-embedded binding region in Alb3 protein. J Biol Chem. 2011;286(40):35187–35195. doi: 10.1074/jbc.M111.250746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Horn A, et al. Structural basis for cpSRP43 chromodomain selectivity and dynamics in Alb3 insertase interaction. Nat Commun. 2015;6:8875. doi: 10.1038/ncomms9875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tzvetkova-Chevolleau T, et al. Canonical signal recognition particle components can be bypassed for posttranslational protein targeting in chloroplasts. Plant Cell. 2007;19(5):1635–1648. doi: 10.1105/tpc.106.048959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tu CJ, Peterson EC, Henry R, Hoffman NE. The L18 domain of light-harvesting chlorophyll proteins binds to chloroplast signal recognition particle 43. J Biol Chem. 2000;275(18):13187–13190. doi: 10.1074/jbc.c000108200. [DOI] [PubMed] [Google Scholar]
- 32.Funke S, Knechten T, Ollesch J, Schünemann D. A unique sequence motif in the 54-kDa subunit of the chloroplast signal recognition particle mediates binding to the 43-kDa subunit. J Biol Chem. 2005;280(10):8912–8917. doi: 10.1074/jbc.M409992200. [DOI] [PubMed] [Google Scholar]
- 33.Sivaraja V, et al. Three-dimensional solution structures of the chromodomains of cpSRP43. J Biol Chem. 2005;280(50):41465–41471. doi: 10.1074/jbc.M507077200. [DOI] [PubMed] [Google Scholar]
- 34.Chakraborty A, et al. A single-domain cyclophilin from Leishmania donovani reactivates soluble aggregates of adenosine kinase by isomerase-independent chaperone function. J Biol Chem. 2002;277(49):47451–47460. doi: 10.1074/jbc.M204827200. [DOI] [PubMed] [Google Scholar]
- 35.Kargul J, Laurent GJ. Small heat shock proteins: Molecular protectors against the disease. Int J Biochem Cell Biol. 2012;44(10):1587. doi: 10.1016/j.biocel.2012.06.022. [DOI] [PubMed] [Google Scholar]
- 36.Laskowska E, Matuszewska E, Kuczyńska-Wiśnik D. Small heat shock proteins and protein-misfolding diseases. Curr Pharm Biotechnol. 2010;11(2):146–157. doi: 10.2174/138920110790909669. [DOI] [PubMed] [Google Scholar]
- 37.Mogk A, Deuerling E, Vorderwülbecke S, Vierling E, Bukau B. Small heat shock proteins, ClpB and the DnaK system form a functional triade in reversing protein aggregation. Mol Microbiol. 2003;50(2):585–595. doi: 10.1046/j.1365-2958.2003.03710.x. [DOI] [PubMed] [Google Scholar]
- 38.Veinger L, Diamant S, Buchner J, Goloubinoff P. The small heat-shock protein IbpB from Escherichia coli stabilizes stress-denatured proteins for subsequent refolding by a multichaperone network. J Biol Chem. 1998;273(18):11032–11037. doi: 10.1074/jbc.273.18.11032. [DOI] [PubMed] [Google Scholar]
- 39.Bardwell JC, Jakob U. Conditional disorder in chaperone action. Trends Biochem Sci. 2012;37(12):517–525. doi: 10.1016/j.tibs.2012.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Foit L, George JS, Zhang BW, Brooks CL, 3rd, Bardwell JC. Chaperone activation by unfolding. Proc Natl Acad Sci USA. 2013;110(14):E1254–E1262. doi: 10.1073/pnas.1222458110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jaru-Ampornpan P, Chandrasekar S, Shan SO. Efficient interaction between two GTPases allows the chloroplast SRP pathway to bypass the requirement for an SRP RNA. Mol Biol Cell. 2007;18(7):2636–2645. doi: 10.1091/mbc.E07-01-0037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Delaglio F, et al. NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6(3):277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 43.Johnson BA, Blevins RA. NMR View: A computer program for the visualization and analysis of NMR data. J Biomol NMR. 1994;4(5):603–614. doi: 10.1007/BF00404272. [DOI] [PubMed] [Google Scholar]
- 44.Salzmann M, Pervushin K, Wider G, Senn H, Wüthrich K. TROSY in triple-resonance experiments: new perspectives for sequential NMR assignment of large proteins. Proc Natl Acad Sci USA. 1998;95(23):13585–13590. doi: 10.1073/pnas.95.23.13585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Salzmann M, Wider G, Pervushin K, Senn H, Wuthrich K. TROSY-type triple-resonance experiments for sequential NMR assignments of large proteins. J Am Chem Soc. 1999;121(4):844–848. [Google Scholar]
- 46.Lebowitz J, Lewis MS, Schuck P. Modern analytical ultracentrifugation in protein science: A tutorial review. Protein Sci. 2002;11(9):2067–2079. doi: 10.1110/ps.0207702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schuck P, Perugini MA, Gonzales NR, Howlett GJ, Schubert D. Size-distribution analysis of proteins by analytical ultracentrifugation: Strategies and application to model systems. Biophys J. 2002;82(2):1096–1111. doi: 10.1016/S0006-3495(02)75469-6. [DOI] [PMC free article] [PubMed] [Google Scholar]