Significance
Nasopharyngeal carcinoma (NPC) is a valuable cancer model to study the interaction of host genetics, viral infection, and environment in tumorigenesis. Little is known about the genetic basis for the remarkably distinct geographical distribution of NPC. We used a whole-exome sequencing approach to identify the genetic alterations associated with NPC susceptibility and revealed a strong link between macrophage-stimulating 1 receptor (MST1R) and NPC early-age onset (age of ≤20 y). MST1R is critical for innate immunity and plays an important role for host defense against viral infection. We further discovered that an interaction network involved in the MST1R/14-3-3 complex was frequently deregulated by genetic alterations in NPC. Our findings provide new insights in the pathogenesis of NPC by highlighting the involvement of the MST1R-mediated signaling pathways.
Keywords: nasopharyngeal carcinoma, whole-exome sequencing, early-age onset, MST1R, cancer susceptibility genes
Abstract
Multiple factors, including host genetics, environmental factors, and Epstein–Barr virus (EBV) infection, contribute to nasopharyngeal carcinoma (NPC) development. To identify genetic susceptibility genes for NPC, a whole-exome sequencing (WES) study was performed in 161 NPC cases and 895 controls of Southern Chinese descent. The gene-based burden test discovered an association between macrophage-stimulating 1 receptor (MST1R) and NPC. We identified 13 independent cases carrying the MST1R pathogenic heterozygous germ-line variants, and 53.8% of these cases were diagnosed with NPC aged at or even younger than 20 y, indicating that MST1R germ-line variants are relevant to disease early-age onset (EAO) (age of ≤20 y). In total, five MST1R missense variants were found in EAO cases but were rare in controls (EAO vs. control, 17.9% vs. 1.2%, P = 7.94 × 10−12). The validation study, including 2,160 cases and 2,433 controls, showed that the MST1R variant c.G917A:p.R306H is highly associated with NPC (odds ratio of 9.0). MST1R is predominantly expressed in the tissue-resident macrophages and is critical for innate immunity that protects organs from tissue damage and inflammation. Importantly, MST1R expression is detected in the ciliated epithelial cells in normal nasopharyngeal mucosa and plays a role in the cilia motility important for host defense. Although no somatic mutation of MST1R was identified in the sporadic NPC tumors, copy number alterations and promoter hypermethylation at MST1R were often observed. Our findings provide new insights into the pathogenesis of NPC by highlighting the involvement of the MST1R-mediated signaling pathways.
Nasopharyngeal carcinoma (NPC) is a malignant tumor that emerges from the epithelium of the nasopharynx. It has remarkable ethnic and geographical distribution. Southern China is among one of the highest incidence regions for NPC. Familial clustering of NPC is often observed, and the age of diagnosis of the familial cases is generally younger than for sporadic cases (1). The risk of the individuals with first-degree relatives of NPC is dramatically increased (4- to 10-fold) compared with those individuals without a family history (2), indicating the important role of genetics in NPC development. Moreover, some patients are the early-age onset (EAO) cases who have been diagnosed with NPC at a much younger age (≤20 y) than the upper 40s, as in most NPC patients. According to the statistics from the Hong Kong Cancer Registry, 24 cases younger than 20 y of age were reported from 2003 to 2012. Due to the rarity of the EAO cases in Southern Chinese populations, little is known about the etiology and epidemiology of these patients. Multiple factors, including host genetics, environmental factors, and Epstein–Barr virus (EBV) infection, contribute to the development of NPC. The association of particular human leukocyte antigen (HLA) subtypes with NPC susceptibility has been extensively studied in the high-risk regions of Southern China (3). HLA polymorphisms reported so far contributed to the modest risk of NPC in sporadic cases for patients generally in their upper 40s at diagnosis. We hypothesize that other important genes contributing to NPC genetic susceptibility may be uncovered in the EAO cases and the family history-positive (FH+) cases because the onset of NPC in these two cohorts occurs earlier than in the sporadic cases. Importantly, losses of chromosomes 3p and 9p were frequently observed in the histologically normal nasopharyngeal epithelium with multiple genetically aberrant lesions and are thus generally considered as involved in crucial early events in NPC tumorigenesis (4, 5). Previous studies by others and us discovered the deletion and/or methylation of a number of tumor suppressors, including Ras association domain family member 1 (RASSF1) (6), ADAM metallopeptidase with thrombospondin type 1 motif 9 (ADAMTS9) (7), protein tyrosine phosphatase, receptor type G (PTPRG) (8), zinc finger MYND-type containing 10 (ZMYND10) (BLU) (9, 10), and fibulin 2 (FBLN2) (11) on chromosome 3p, and p16 on chromosome 9p (12, 13). These findings suggest that the potential susceptibility loci associated with NPC may reside on these chromosome regions. To identify the NPC genetic susceptibility genes, we performed whole-exome sequencing (WES) in 161 NPC cases compared with 895 noncancer controls. A strong link between macrophage-stimulating 1 receptor (MST1R), mapped to the chromosome 3p21.3 region, and NPC genetic susceptibility was discovered in this study. MST1R encodes a cell surface receptor for macrophage-stimulating 1 (MST1, also known as MSP) with tyrosine kinase activity. Previous studies show that MST1R plays a critical function in host defense: (i) It is predominantly expressed in the tissue-resident macrophages, which promote tissue repair and inhibit inflammation by repressing the production of proinflammatory molecules induced by pathogens (14–17) and (ii) MST1R expression is detected in the ciliated epithelial cells in the normal nasal mucosa, and activation of MST1R signaling can increase the ciliary motility to prevent chronic infection (18). Previous studies and our WES data strongly support the role of MST1R as an NPC susceptibility gene.
Results
Gene-Level Analysis Identified the Association of MST1R Deleterious Variants with NPC.
We sequenced the blood samples from 161 NPC cases, including 39 EAO cases, 63 FH+ cases from 52 independent families, and 59 sporadic cases by WES and achieved an average coverage of 49-fold on target (range of 32- to 76-fold) (SI Appendix, Table S1). The accuracy of the variant calling was evaluated by Sanger sequencing or targeted resequencing in 936 SNPs and 63 INDELs. The verification rate was 100% for SNPs and 98.2% for INDELs. The WES data of the noncancer control cohort included 895 individuals of Southern Chinese descent (SI Appendix, Table S2). The homogenous population structure of the EAO, FH+, and sporadic NPC patients, together with the control individuals, is revealed by the multidimensional scaling (MDS) and principal component analysis (PCA) using the linkage disequilibrium (LD)-pruned common variants with minor allele frequency (MAF) of >0.05 and genotyping rate of >95% (SI Appendix, Fig. S1). The variants with an MAF of >1% in the 1000 Genomes project and National Heart, Lung, and Blood Institute (NHLBI) Grand Opportunity Exome Sequencing Project (ESP6500) were excluded. We screened for the presence of deleterious variants (damaging missense, nonframeshift INDELs, and loss-of-function variants). In the EAO cases, we identified 157 genes with rare deleterious variants in at least 10% of the cases (SI Appendix, Fig. S2). These genes are enriched in the multiple gene sets that are potentially relevant to cancer development, including ABC transporters and DNA repair (enrichment test, P < 0.01 and q ≤ 0.1) (SI Appendix, Table S3). Interestingly, out of these 157 genes, five genes (COL7A1, MST1R, PTPRG, SCN5A, and UQCRC1) are from the chromosome 3p21-14 region [Human Genome Organization (HUGO) Gene Nomenclature Committee (HGNC) cytogenetic band, enrichment test, P = 0.01] whereas no other region/chromosome reached the significance level of P < 0.05 in the enrichment test. We then examined the FH+ cases and identified 62 genes with deleterious variants in at least 10% of the cases (SI Appendix, Fig. S2); no enrichment of DNA repair and ABC transporter was found among these genes. In total, we identified 200 genes with deleterious variants in at least 10% of the cases in either EAO or FH+ groups. These genes were searched against the GeneRIF database for their relevance to NPC; three genes have previously been reported to have relevance to NPC genetic susceptibility (ERCC2) or tumor suppression (ADAMTS9 and PTPRG) (SI Appendix, Table S4).
To further identify the genetic susceptibility genes in NPC, we subsequently performed gene-based burden tests in 895 noncancer controls and 161 NPC cases using all of the rare deleterious variants identified in the study (MAF of ≤0.01). The association between the genes and NPC was evaluated by the variant test (VT), SNP-set kernel association test (SKAT), and combined and multivariate collapsing test (CMC). We excluded the genes not expressed in both nasopharyngeal nontumor and tumor tissues (19) or that were reported to be false positives in other WES studies (20). Fifteen genes showed a suggestive association with NPC (P value of <0.001 in at least two out of three tests) (SI Appendix, Table S5). The genes were ranked according to the number of NPC patients carrying the rare deleterious variants. The MST1R gene from the chromosome 3p21.3 region was the top hit in the analysis, with multiple deleterious variants identified in 13 (8.7%) independent NPC cases (VT, P = 0.0009; SKAT, P = 0.0205; CMC, P = 0.0003). In total, 11 MST1R deleterious variants, including one frameshift deletion and 10 damaging missense variants, were observed in NPC (Fig. 1 A and B). The patients carrying the MST1R deleterious variants are listed in Table 1. Out of 13 NPC cases carrying the MST1R deleterious variants, 7 are EAO cases, indicating that MST1R germ-line deleterious variants were often associated with disease early-age onset (Fisher's exact test for enrichment, P = 0.041). In total, five heterozygous missense variants at MST1R were observed in 17.9% of the EAO cases and only 1.2% of the controls (P = 7.94 × 10−12). These variants are c.G2917A:p.A973T, c.G2113A:p.E705K, c.T2009G:p.V670G, c.G979A:p.A327T, and c.G917A:p.R306H (GenBank accession no. NM_002447) (Table 1 and Fig. 1A); all were confirmed by Sanger sequencing (SI Appendix, Fig. S3). They are located at the evolutionally conserved residues (SI Appendix, Fig. S4) and are predicted to be pathogenic by KGGSEq (21).
Fig. 1.
Multiple rare deleterious variants at MST1R identified in NPC cases. (A) The schematic plot of the MST1R deleterious variants at the protein level in the EAO, FH+, and sporadic NPC patients. *, The EAO NPC patient with family history of NPC. (B) The percentage of the cases carrying the MST1R deleterious variants in EAO (17.9%), FH+ (7.1%), sporadic (5.1%) cases, and controls (3.4%). (C) The schematic plot of five MST1R variants at the transcript level identified from the EAO cases. The graph is adapted from the University of California, Santa Cruz (UCSC) Genome Browser (43). The MST1R transcripts were plotted according to the UCSC known gene database (hg19). MST1R promoter has two CGIs (CGI 1 and CGI 2) defined by the UCSC database.
Table 1.
MST1R germ-line rare deleterious variants identified in NPC patients
| Family | Subject | Age at diagnosis | Gender | Group | Variant | Protein alteration | Exon | MAF in controls (n = 895) | MAF in 1000 Genomes (Asian) | MAF in ESP6500 |
| 1 | HK_FH_49 | 55 | M | FH+ | 834_835del | T278fs | 1 | 0.0006 | ND | ND |
| 2 | HK_12 | 17 | F | EAO | G917A | R306H | 1 | 0.0006 | ND | ND |
| 3 | HK_6 | 19 | F | EAO/FH+ | G979A | A327T | 1 | 0.0017 | 0.005 | ND |
| 4 | GX_2 | 19 | M | EAO | G979A | A327T | 1 | 0.0017 | 0.005 | ND |
| 5 | NPCHKE48_N | 58 | F | Sporadic | G1067A | G356D | 1 | 0.0012 | ND | ND |
| 6 | HK_FH_35 | 34 | M | FH+ | C1510T | R504C | 3 | ND | ND | ND |
| 7 | NPCHKE39_N | 38 | F | Sporadic | G1769A | C590Y | 5 | ND | ND | ND |
| 8 | ST_1 | 15 | F | EAO | T2009G | V670G | 6 | 0.0034 | 0.004 | ND |
| 9 | HK_10 | 19 | F | EAO | T2009G | V670G | 6 | 0.0034 | 0.004 | ND |
| 10 | HK_17 | 20 | M | EAO | G2113A | E705K | 7 | ND | ND | ND |
| 11 | FJ_5 | 12 | M | EAO | G2917A | A973T | 12 | 0.0006 | ND | ND |
| 12 | NPCHKE29_N | 58 | M | Sporadic | A3718C | S1240R | 18 | 0.0006 | ND | ND |
| 13* | HK5_FH_1 | 39 | F | FH+ | C4192T | R1398W | 20 | ND | 0.001 | 7.70E−05 |
| 13* | HK5_FH_2 | 39 | M | FH+ | C4192T | R1398W | 20 | ND | 0.001 | 7.70E−05 |
ND, not detected.
Two cases are siblings from an NPC FH+ family.
MST1R Germ-Line Variant c.G917A:p.R306H Mapping to a CpG Site Important for Regulating MST1R Expression Is Highly Associated with NPC in the Validation Cohort.
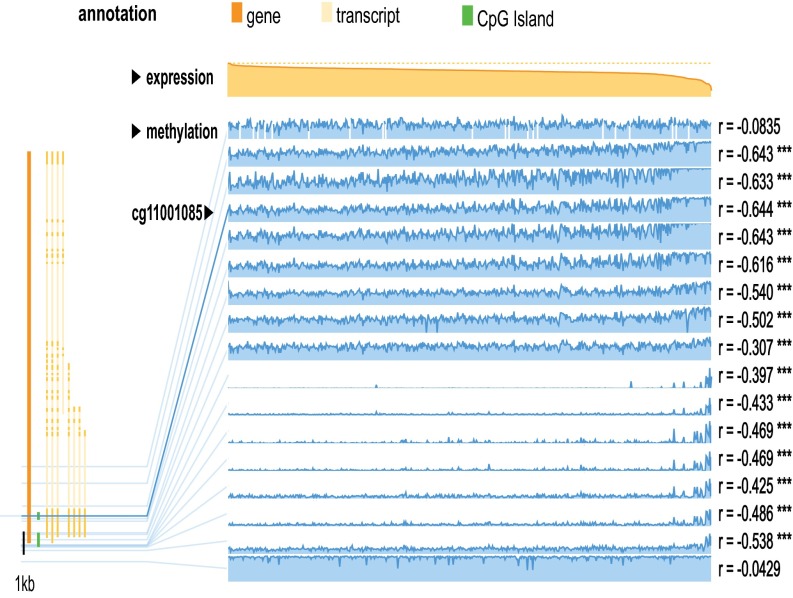
Out of five variants identified in the EAO cases, two variants (c.G979A:p.A327T and c.G917A:p.R306H) are only 62 bp apart. Both are located at the Sema domain, important for ligand binding and receptor activation, and are predicted to be pathogenic by KGGSeq, SIFT, and PolyPhen algorithms. The p.A327T variant can destabilize MST1R, as predicted by mutation Cutoff Scanning Matrix (mCSM) (ΔΔG, −1.34 Kcal/mol), Site Directed Mutator (SDM) (ΔΔG, −1.46 Kcal/mol), and an integrated computational approach (DUET) (ΔΔG -1.33 Kcal/mol). The protein stability with the p.R306H alteration cannot be predicted because this region is absent in the PDB file for the Sema domain. Interestingly, we found this variant mapped to exon 1 within a CpG site in a CpG island (CGI 2) (Fig. 1C). Besides the germ-line genetic change (c.G917A:p.R306H), we also found a reduced methylation level at this CpG site in the nasopharyngeal tissues compared with the peripheral blood from healthy individuals and other morphologically normal tissues obtained from breast, kidney, liver, and lung cancer patients in The Cancer Genome Atlas (TCGA) studies (SI Appendix, Fig. S5). Similar reduction of methylation was also observed in the normal surrounding tissues from the head and neck and bladder cancer patients. We examined the methylome and RNASeq data in TCGA studies. The data showed that increased methylation of this CpG was correlated with decreased MST1R expression in both cancers (head and neck cancer, n = 542; bladder cancer, n = 425) (Fig. 2 and SI Appendix, Fig. S6). The results suggested the importance of this locus for regulating the MST1R expression.
Fig. 2.
Correlation between MST1R methylation and expression in head and neck squamous cell carcinoma in a TCGA study (n = 542). The CpG site (cg11001085) is highlighted in the solid blue line. Increased methylation of this CpG site was significantly correlated with decreased expression of MST1R (r = −0.644, ***P < 0.001). The two CGIs at promoter are shown in green lines.
We further examined the variant p.R306H in an independent NPC patient cohort, including 2,160 cases and 2,433 controls and found an additional 8 (0.4%) unrelated cases, but only one control who carried this heterozygous variant (association, P = 0.0079, Fisher's exact test, one tailed, odds ratio = 9.0). This variant has not been reported in the 1000 Genomes Project and ESP6500. In the ExAC exome database, the MAF of this variant is 0.0002 in the East Asian population and only 0.00002 in the European population. These results indicate that this heterozygous variant is highly associated with NPC.
An Interaction Network Associated with MST1R/14-3-3 Complex in NPC Genetic Susceptibility.
To further explore the NPC genetic susceptibility genes that directly or indirectly associate with MST1R, we examined all 303 genes with P < 0.01 in the gene-based burden test by Disease Association Protein–Protein Link Evaluator (DAPPLE) analysis (22) and found a protein interaction network that is linked to MST1R (Fig. 3). Interestingly, we identified another important gene YWHAB, which potentially contributed to NPC susceptibility (VT, P = 0.0062; SKAT, P = 0.0047; CMC P = 0.0035). The gene YWHAB encodes the 14-3-3 protein α and β. The MST1R C-terminal amino acid sequence fits with the canonical 14-3-3 consensus binding sequences (RXRXXpSXP) to allow formation of a protein complex (23). We identified two rare missense variants (c.A229C:p.K77Q and c.C620T:p.T207M) at YWHAB (GenBank accession no. NM_003404), accounting for 5.4% of the FH+ cases and 3.4% of the sporadic cases. Together with MST1R, YWHAB, and another eight genes most closely linked to MST1R or YWHAB in the DAPPLE analysis, deregulation of the MST1R/14-3-3 signaling by rare deleterious variants was found in 35.4% of the cases, but only in 14.3% of the noncancer controls [overall odds ratio = 3.3, 95%; confidence interval (CI), 2.3–4.8; P = 4.3 × 10−10; permutation, P = 0.002] (Fig. 3 and SI Appendix, Fig. S7).
Fig. 3.
The MST1R/14-3-3 interaction network associated with NPC genetic susceptibility (P value of <0.01 in the burden test). Only the genes most closely linked to MST1R and YWHAB are shown on the graph. The percentage of the cases (E, EAO; F, FH+; S, sporadic) carrying the rare deleterious variants is listed on the graph. The P values shown in color keys indicate the probability that the seed protein would be connected to the other seed proteins by chance.
Somatic Changes of MST1R in Sporadic NPC Tumors.
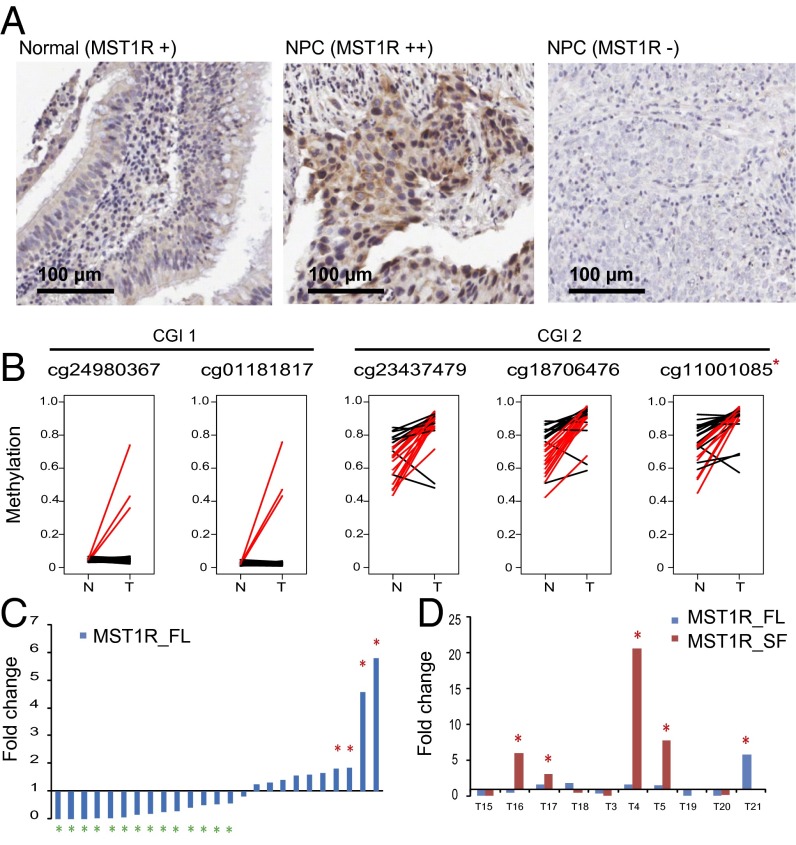
In normal nasopharyngeal mucosa, we detected a weak expression of MST1R in the ciliated epithelial cells by immunohistochemical (IHC) staining (Fig. 4A). We further examined the MST1R somatic mutations using the WES data in the sporadic tumors. However, no somatic mutation at MST1R was identified in either the Hong Kong cohort (n = 59) or the Singapore cohort reported previously (n = 56) (24). MST1R maps to the chromosome 3p21.3 region; loss of heterozygosity (LOH) was frequently reported in this region as an early event in tumorigenesis (4, 9). We detected the allelic imbalance of copy numbers at the MST1R locus by WES data in 64% of the tumors compared with blood samples (SI Appendix, Fig. S8). Consistently, the allelic imbalance of MST1R expression was detected by RNASeq data in the tumors (SI Appendix, Fig. S9). Moreover, two CpG islands (CGI 1 and CGI 2) are located at the proximal promoter of MST1R. Significant increase of methylation at CGI 2 was detected in 44–56% of the tumors (n = 25) (Fig. 4B) (25). These results suggested that MST1R expression may be deregulated by copy number and/or methylation changes in NPC. In line with these findings, the full-length MST1R canonical transcript was down-regulated in 56% of the tumors and up-regulated in 16% (n = 25) (Fig. 4C). At the protein level, we detected weak to moderate expression of MST1R in 12% of the cases by IHC (n = 101) (Fig. 4A and SI Appendix, Fig. S10). Noticeably, at least a threefold increase of the MST1R short isoform expression was found in 40% of the tumors analyzed by RNASeq (Fig. 4D and SI Appendix, Fig. S11), and a twofold increase was observed in the NPC cell line C666 by real-time quantitative reverse transcription polymerase chain reaction (QPCR) (SI Appendix, Fig. S12).
Fig. 4.
Somatic changes of MST1R in sporadic NPC tumors. (A) MST1R expression in the normal nasopharyngeal mucosa and NPC tumors detected by IHC staining. The MST1R expression level is measured as weak (+), moderate (++), strong (+++), and no expression (−). (Left) There is a weak expression of MST1R in the ciliated epithelial cells in normal nasopharyngeal mucosa. (Middle) NPC tumor with MST1R moderate expression. Weak to moderate expression of MST1R was detected in 12% of the tumors (n = 101). (Right) NPC tumor without detectable MST1R expression. (B) Methylation level of CGI 1 and CGI 2 at the proximal promoter of MST1R. The methylation level was measured by Illumina HumanMethylation450 BeadChip. The CGI 1 became methylated in 12% of the NPC patients whereas the CGI 2 was hypermethylated in 44–56% of the Hong Kong NPC patients (n = 25). *, The CpG site (cg11001085) was at the same genomic location as MST1R variant c.G917A:p.R306H. The red line indicates the patient with more than 20% increase of methylation in tumor (T) compared with nontumor (N) sample. (C) Expression of MST1R full-length transcript (MST1R_FL) measured by QPCR in 25 pairs of tumor/normal surrounding samples from NPC patients. The experiments were done in triplicate. MST1R full-length transcript is up-regulated (fold change of >1.8) in 4/25 (16%) of the cases and down-regulated (fold change of <0.56) in 14/25 (56%) of the cases. (D) Expression of MST1R full-length transcript (MST1R_FL, uc003cxy.4) and short-isoform transcript (MST1R_SF, uc031rzv.1) were measured by RNASeq as fold change in 10 pairs of tumor/normal surrounding samples from NPC patients.
Discussion
We examined 161 NPC cases and 895 noncancer controls using a WES approach and gene-based burden test to reveal the association between MST1R, which maps to the chromosome 3p21.3 region, and NPC genetic susceptibility. Similarly, family-based association analysis in 18 NPC families from Southern China identified the 3p21.31–3p22.1 region as a genetic susceptibility locus for NPC (26). In our study, the patients carrying MST1R germ-line variants are enriched in the EAO cases; the age of these patients is younger than the remaining cases in the study cohort (median age ± SD, 27 ± 17 y vs. 40 ± 17 y), indicating that MST1R abnormalities are strongly associated with early-disease onset.
In the sporadic cases, MST1R germ-line deleterious variants were found in 5.1% of the blood samples whereas no somatic mutation at MST1R was detected in the tumor samples. Copy number alterations and proximal promoter hypermethylation at MST1R are often observed in the tumors, which may contribute to the frequent down-regulation of the full-length MST1R in NPC. In addition, a noticeable up-regulation of the short isoform was found in tumors. This isoform encodes a truncated protein (sf-RON), lacking most of the receptor extracellular domain, but retaining the whole transmembrane and intracellular domains. sf-RON is highly and constitutively activated by autophosphorylation at the protein kinase domain. It can interact with the oncoprotein from Friend leukemia virus, which has emerged as a mechanism for MST1R-mediated tumorigenesis in animals (27). In humans, this isoform is often observed in the clinical cancer samples and has been linked to an aggressive phenotype (28).
In healthy individuals, MST1R plays an important role in host defense, and MST1R-depleted mice show a compromised cell-mediated immunity, increased inflammation, and expression of IL-6, TNF-α, MCP-1, and MIP-2 after lung injury (17, 29). MST1R is predominantly expressed in the tissue-resident macrophages (17). Binding with the ligand MSP, MST1R enables activation of a number of signal transduction pathways important for regulating the balance of the macrophage state (30). Accumulating evidence shows the direct link between macrophages and tumor initiation (31–33). It is believed that the macrophages with persistent inflammatory phenotypes release cytotoxic molecules, causing extensive tissue damage. The DNA damage in the surrounding epithelial cells may predispose these cells to premalignant transformation and tumor initiation (33). So it is possible that the MST1R deleterious variants impair the MST1R function by protein destabilization in the tissue-resident macrophages, leading to the loss of macrophage homeostasis and thus promoting tumorigenesis. Moreover, in this current study and from previous findings of others (18), MST1R is detected in the ciliated epithelial cells in the nasal cavity. Activation of MST1R by MSP leads to a significant increase in ciliary beat frequency (18). Cilia have an important role as a part of the host defense mechanisms. The deleterious variants may impair the MST1R host defense function by reducing the nasal cilia motility. In addition, the short-isoform MST1R is up-regulated in a proportion of NPC tumors, which can potentially promote invasive tumor growth in the established tumor. We speculate that these multiple factors may function collectively to accelerate NPC tumor development in the EAO cases.
We also discovered deleterious variants in ABC transporters and DNA repair genes in the EAO cases, suggesting the possibility of multigenic effects in NPC genetic susceptibility. Besides MST1R, four additional genes including PTPRG map to the 3p21-14 region. Our early studies on PTPRG identified it as a tumor suppressor gene that functionally suppresses NPC tumorigenesis (34), indicating that it is a potential candidate gene relevant to NPC genetic susceptibility. However, unlike MST1R, none of the other genes reached the significance level in our gene-based burden tests. The relatively small sample size for EAO cases, genetic heterogeneity, and moderate effect of these genes may contribute to the nonsignificant results. Investigation of the deleterious variants in these genes in a larger NPC cohort is warranted.
To our knowledge, this is the first study to fully characterize the germ-line variants in NPC patients using the WES approach and highlights the importance of MST1R signaling in the pathogenesis of NPC. The study sheds light on the identification of the rare and disease-associated variants in NPC. Future challenges will be further functional characterization of MST1R deleterious variants and elucidation of the mechanisms for the role of MST1R-mediated signaling in NPC development.
Materials and Methods
Study Subjects.
We studied 161 NPC cases, including 39 NPC cases diagnosed at 20 y of age or younger, 63 cases from 52 history-positive (FH+) NPC families, and 59 sporadic cases, as well as a control group including 895 noncancer individuals of Southern Chinese descent (SI Appendix, Table S2) by WES. Four EAO cases also had a family history of NPC, making a total of 56 independent FH+ families. An additional 2,160 NPC cases and 2,433 healthy controls from Hong Kong were further examined for the selected candidate variants. The FH+ cases were defined as the patients from a family with at least two family members diagnosed with NPC. Eighty-eight percent of the FH+ cases in this WES study came from families with at least two generations of NPC cases. The study was approved by the Institutional Review Board (IRB) of the University of Hong Kong. All study subjects provided a signed consent form for participation in the study.
Whole-Exome Sequencing.
The germ-line DNA was isolated from the peripheral blood lymphocytes using a QIAamp DNA blood Mini Kit (Qiagen). The quantity and quality of the genomic DNA were measured by Nanodrop 1000 (Thermo Scientific) and Qubit (Life Technologies). The library preparation, capture, and sequencing were performed by the Centre for Genomic Sciences (CGS) at the University of Hong Kong (HKU). In brief, 250–1,000 ng of genomic DNA were fragmented by an ultrasonicator (Covaris). The consistent results of 250 ng and 1,000 ng of DNA input were confirmed using duplicates (SI Appendix, Table S6). These fragments were amplified using a NEBNext Ultra DNA Library Prep Kit (NEB) and then hybridized to the Illumina TruSeq capture kit for enrichment. In each capture, we included six blood samples. The magnitude of the enrichment was estimated using real-time PCR. Paired end, 100-bp read-length sequencing was performed by a HiSEq 1500 sequencer (Illumina). One captured library was sequenced in one lane to ensure that each sample met the desired average-fold coverage.
WES Data Analysis.
The WES data of both cases and noncancer controls were processed according to GATK Best Practices recommendations (35). The sequence reads for the exome sequence of each individual were aligned to the reference human genome (hg19) using Burrows–Wheeler Aligner (36). Picards were applied to sort output bamfiles and mark duplicates. GATK was applied for INDEL realignment, base quality recalibration, variant discovery, and variant quality score recalibration (VQSR) in both NPC cases and noncancer controls. Quality of variants was checked by GATK and PLINK (37, 38) to remove abnormal samples or contamination. A total of five annotations were used for VQSR, and the model was evaluated for discordant rate between a pair of monozygotic twins, Mendelian error, ratio of known-to-novel variants, and transition-to-transversion (ti/tv) ratio. Sensitivity thresholds of 99.5% and 97% were chosen for SNPs and INDELs, respectively. Genotypes with read depth (DP) < 8 or genotype quality (GQ) < 20 were considered as ambiguous calls and therefore were considered as missing values. Variants missing in more than 20% of the samples in either case or control groups were excluded. Multiallelic variants were not considered for downstream analysis due to uncertainty in calling quality. The multidimensional scaling (MDS) and principal component analysis (PCA) were performed by PLINK using the LD-pruned variants with a MAF of >0.05 and genotyping rate of more than 95%. The qualified variants were annotated by ANNOVAR (39). The rare variants were defined as the variants with a MAF of ≤0.01 in any of the databases in the 1000 Genomes Project (all population, Asian population, European population, and African population) and ESP6500. We defined the deleterious variants as the loss-of-function (stopgain, frameshift insertion, and frameshift deletion) variants, nonframeshift INDELs, or missense variants that are predicted to be damaging by KGGSEq (21). KGGSeq combines the prediction scores from five algorithms (SIFT, Polyphen-2, LRT, MutationTaster, and PhyloP) by the logistic regression method to estimate a probability of a variant being pathogenic.
Sanger Sequencing.
All deleterious MST1R variants identified by WES were confirmed by Sanger sequencing. The primers and PCR conditions are listed in SI Appendix, Table S7.
LightSNiP Assay.
The LightSNiP assay targeting the variant c.G917A:p.R306H was designed by TIB MOLBIOL and performed using the Roche LightCycler 480 instrument and LightCycler FastStart DNA Master HybProbe on the LightCycler 480 Multiwell plate 384. The melting curve analysis is illustrated in SI Appendix, Fig. S13. We repeated 2% of the samples (n = 96) in the validation study using this assay. The result was highly consistent between duplicates. The accuracy of the assay was further evaluated in 7% of the samples (n = 326) by Sanger sequencing. The sensitivity and specificity of the assay are 100%.
RNA Sequencing.
Total RNA was extracted from fresh-frozen tissues (10 NPC tumors and matched nontumor biopsies) using an AllPrep DNA/RNA Micro Kit (Qiagen). Sequencing data were mapped to hg19 using RSEM (40). The MST1R expression was measured by transcripts per million (TPM) values.
Gene-Based Burden Test.
The genes with at least five rare mutant alleles (damaging missense or loss-of-function variants) in the combined cohort, including 161 NPC cases and 895 noncancer controls, were considered in the burden test using the Efficient and Parallelizable Association Container Toolbox (EPACTS) software (v.3.2.6). Three tests, including the variant test, SNP-set kernel association test (SKAT), and combined and multivariate collapsing test, were used to evaluate the association between genes and NPC risk.
Protein Stability Prediction.
The model of the Sema domain was based on the X-ray crystal structure of the Sema domain from the Protein Data Bank (PDB ID code 4FWW). The effects of the germ-line variants on the protein stability were analyzed by DUET (41).
DAPPLE Analysis and Permutation Test.
All of the genes with a P value of <0.01 in the gene-based burden test were imported in the DAPPLE analysis with default parameters. The indirect network was simplified in the plot. We used the Monte Carlo method to examine the probability of the association of the deleterious variants in the MST1R-relevant genes with NPC by chance. We randomly selected 10 genes and calculated the odds ratio of the patients against controls carrying the deleterious variants at these genes for 10,000 times. The permutation p was estimated as the times-with-odds ratio above a certain level divided by 10,000.
TCGA Data Analysis.
The TCGA methylome and RNASeq data from head and neck and bladder cancers were analyzed by MEXPRESS (42).
Supplementary Material
Acknowledgments
We acknowledge the Research Grants Council Area of Excellence (AoE) Hong Kong NPC Research Tissue Bank for providing us with NPC blood samples and the patient demographics, as well as the NPC tissue microarrays. All the Hong Kong NPC patient specimens were collected by the AoE Hong Kong NPC Tissue Bank from Queen Mary, Queen Elizabeth, Pamela Youde Nethersole Eastern, Princess Margaret, and Tuen Mun hospitals. We thank the Fujian Provincial Cancer Hospital, Cancer Hospital of Shantou University Medical College, Guangxi Medical University, and Cancer Research Institute of Zhongshan City, People’s Republic of China for collecting samples from the EAO and FH+ patients. We acknowledge our collaborators J. Wong, P. Sham, and S. Cherny (Department of Psychiatry), M. Garcia-Barcelo and P. Tam (Department of Surgery), and K. Cheah (Department of Biochemistry, University of Hong Kong), L. Baum (University of Hong Kong), and P. Kwan (Department of Medicine, The University of Melbourne) for sharing the TruSeq whole-exome sequencing (WES) data for the noncancer controls. The RNASeq and methylome data for head and neck squamous cell carcinoma and bladder urothelial carcinoma were obtained from The Cancer Genome Atlas (TCGA) studies. This study was funded by Hong Kong Research Grants Council Grant AoE/M-06/08, Hong Kong Health and Medical Research Fund (HMRF) Grant 01121496, and the Hong Kong Cancer Fund (to M.L.L.). Noncancer controls from the epilepsy study are funded by Hong Kong Research Grants Council/General Research Fund (GRF) Grant HKU 7630/12M (to S. Cherny). Noncancer controls from the studies of congenital diseases are funded by Hong Kong Research Grants Council/General Research Fund (GRF) Grant HKU 766913M (to M. Garcia-Barcelo) and Grant HKU 778213M (to P. Tam), and Hong Kong Health and Medical Research Fund (HMRF) Grant 01121576 (to P. Tam).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: Whole-exome sequencing data for the early-age onset cases have been deposited in the Sequence Read Archive (SRA) database (accession ID SRA291701).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1523436113/-/DCSupplemental.
References
- 1.Bei JX, Jia WH, Zeng YX. Familial and large-scale case-control studies identify genes associated with nasopharyngeal carcinoma. Semin Cancer Biol. 2012;22(2):96–106. doi: 10.1016/j.semcancer.2012.01.012. [DOI] [PubMed] [Google Scholar]
- 2.Chang ET, Adami HO. The enigmatic epidemiology of nasopharyngeal carcinoma. Cancer Epidemiol Biomarkers Prev. 2006;15(10):1765–1777. doi: 10.1158/1055-9965.EPI-06-0353. [DOI] [PubMed] [Google Scholar]
- 3.Bei JX, et al. A genome-wide association study of nasopharyngeal carcinoma identifies three new susceptibility loci. Nat Genet. 2010;42(7):599–603. doi: 10.1038/ng.601. [DOI] [PubMed] [Google Scholar]
- 4.Chan AS, et al. High frequency of chromosome 3p deletion in histologically normal nasopharyngeal epithelia from southern Chinese. Cancer Res. 2000;60(19):5365–5370. [PubMed] [Google Scholar]
- 5.Chan AS, et al. Frequent chromosome 9p losses in histologically normal nasopharyngeal epithelia from southern Chinese. Int J Cancer. 2002;102(3):300–303. doi: 10.1002/ijc.10689. [DOI] [PubMed] [Google Scholar]
- 6.Chow LS, et al. RASSF1A is a target tumor suppressor from 3p21.3 in nasopharyngeal carcinoma. Int J Cancer. 2004;109(6):839–847. doi: 10.1002/ijc.20079. [DOI] [PubMed] [Google Scholar]
- 7.Lo PH, et al. Identification of a tumor suppressive critical region mapping to 3p14.2 in esophageal squamous cell carcinoma and studies of a candidate tumor suppressor gene, ADAMTS9. Oncogene. 2007;26(1):148–157. doi: 10.1038/sj.onc.1209767. [DOI] [PubMed] [Google Scholar]
- 8.Cheung AK, et al. PTPRG suppresses tumor growth and invasion via inhibition of Akt signaling in nasopharyngeal carcinoma. Oncotarget. 2015;6(15):13434–13447. doi: 10.18632/oncotarget.3876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheng Y, et al. Functional evidence for a nasopharyngeal carcinoma tumor suppressor gene that maps at chromosome 3p21.3. Proc Natl Acad Sci USA. 1998;95(6):3042–3047. doi: 10.1073/pnas.95.6.3042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cheng Y, et al. Anti-angiogenic pathway associations of the 3p21.3 mapped BLU gene in nasopharyngeal carcinoma. Oncogene. 2015;34(32):4219–4228. doi: 10.1038/onc.2014.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Law EW, et al. Anti-angiogenic and tumor-suppressive roles of candidate tumor-suppressor gene, Fibulin-2, in nasopharyngeal carcinoma. Oncogene. 2012;31(6):728–738. doi: 10.1038/onc.2011.272. [DOI] [PubMed] [Google Scholar]
- 12.Lo KW, Huang DP, Lau KM. p16 gene alterations in nasopharyngeal carcinoma. Cancer Res. 1995;55(10):2039–2043. [PubMed] [Google Scholar]
- 13.Cheng Y, et al. A functional investigation of tumor suppressor gene activities in a nasopharyngeal carcinoma cell line HONE1 using a monochromosome transfer approach. Genes Chromosomes Cancer. 2000;28(1):82–91. doi: 10.1002/(sici)1098-2264(200005)28:1<82::aid-gcc10>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- 14.Morrison AC, Wilson CB, Ray M, Correll PH. Macrophage-stimulating protein, the ligand for the stem cell-derived tyrosine kinase/RON receptor tyrosine kinase, inhibits IL-12 production by primary peritoneal macrophages stimulated with IFN-gamma and lipopolysaccharide. J Immunol. 2004;172(3):1825–1832. doi: 10.4049/jimmunol.172.3.1825. [DOI] [PubMed] [Google Scholar]
- 15.Wilson CB, et al. The RON receptor tyrosine kinase regulates IFN-gamma production and responses in innate immunity. J Immunol. 2008;181(4):2303–2310. doi: 10.4049/jimmunol.181.4.2303. [DOI] [PubMed] [Google Scholar]
- 16.Ray M, et al. Inhibition of TLR4-induced IκB kinase activity by the RON receptor tyrosine kinase and its ligand, macrophage-stimulating protein. J Immunol. 2010;185(12):7309–7316. doi: 10.4049/jimmunol.1000095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang MH, Zhou YQ, Chen YQ. Macrophage-stimulating protein and RON receptor tyrosine kinase: Potential regulators of macrophage inflammatory activities. Scand J Immunol. 2002;56(6):545–553. doi: 10.1046/j.1365-3083.2002.01177.x. [DOI] [PubMed] [Google Scholar]
- 18.Sakamoto O, et al. Role of macrophage-stimulating protein and its receptor, RON tyrosine kinase, in ciliary motility. J Clin Invest. 1997;99(4):701–709. doi: 10.1172/JCI119214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sengupta S, et al. Genome-wide expression profiling reveals EBV-associated inhibition of MHC class I expression in nasopharyngeal carcinoma. Cancer Res. 2006;66(16):7999–8006. doi: 10.1158/0008-5472.CAN-05-4399. [DOI] [PubMed] [Google Scholar]
- 20.Fuentes Fajardo KV, et al. NISC Comparative Sequencing Program Detecting false-positive signals in exome sequencing. Hum Mutat. 2012;33(4):609–613. doi: 10.1002/humu.22033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li MX, Gui HS, Kwan JS, Bao SY, Sham PC. A comprehensive framework for prioritizing variants in exome sequencing studies of Mendelian diseases. Nucleic Acids Res. 2012;40(7):e53. doi: 10.1093/nar/gkr1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rossin EJ, et al. International Inflammatory Bowel Disease Genetics Constortium Proteins encoded in genomic regions associated with immune-mediated disease physically interact and suggest underlying biology. PLoS Genet. 2011;7(1):e1001273. doi: 10.1371/journal.pgen.1001273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Santoro MM, Gaudino G, Marchisio PC. The MSP receptor regulates alpha6beta4 and alpha3beta1 integrins via 14-3-3 proteins in keratinocyte migration. Dev Cell. 2003;5(2):257–271. doi: 10.1016/s1534-5807(03)00201-6. [DOI] [PubMed] [Google Scholar]
- 24.Lin DC, et al. The genomic landscape of nasopharyngeal carcinoma. Nat Genet. 2014;46(8):866–871. doi: 10.1038/ng.3006. [DOI] [PubMed] [Google Scholar]
- 25.Dai W, et al. Comparative methylome analysis in solid tumors reveals aberrant methylation at chromosome 6p in nasopharyngeal carcinoma. Cancer Med. 2015;4(7):1079–1090. doi: 10.1002/cam4.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zeng Z, et al. Family-based association analysis validates chromosome 3p21 as a putative nasopharyngeal carcinoma susceptibility locus. Genet Med. 2006;8(3):156–160. doi: 10.1097/01.gim.0000196821.87655.d0. [DOI] [PubMed] [Google Scholar]
- 27.Nishigaki K, Thompson D, Hanson C, Yugawa T, Ruscetti S. The envelope glycoprotein of friend spleen focus-forming virus covalently interacts with and constitutively activates a truncated form of the receptor tyrosine kinase Stk. J Virol. 2001;75(17):7893–7903. doi: 10.1128/JVI.75.17.7893-7903.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bardella C, et al. Truncated RON tyrosine kinase drives tumor cell progression and abrogates cell-cell adhesion through E-cadherin transcriptional repression. Cancer Res. 2004;64(15):5154–5161. doi: 10.1158/0008-5472.CAN-04-0600. [DOI] [PubMed] [Google Scholar]
- 29.Cary DC, Clements JE, Henderson AJ. RON receptor tyrosine kinase, a negative regulator of inflammation, is decreased during simian immunodeficiency virus-associated central nervous system disease. J Immunol. 2013;191(8):4280–4287. doi: 10.4049/jimmunol.1300797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chaudhuri A. Regulation of macrophage polarization by RON receptor tyrosine kinase signaling. Front Immunol. 2014;5:546. doi: 10.3389/fimmu.2014.00546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Murray PJ, Wynn TA. Protective and pathogenic functions of macrophage subsets. Nat Rev Immunol. 2011;11(11):723–737. doi: 10.1038/nri3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nowarski R, Gagliani N, Huber S, Flavell RA. Innate immune cells in inflammation and cancer. Cancer Immunol Res. 2013;1(2):77–84. doi: 10.1158/2326-6066.CIR-13-0081. [DOI] [PubMed] [Google Scholar]
- 33.Biswas SK, Sica A, Lewis CE. Plasticity of macrophage function during tumor progression: Regulation by distinct molecular mechanisms. J Immunol. 2008;180(4):2011–2017. doi: 10.4049/jimmunol.180.4.2011. [DOI] [PubMed] [Google Scholar]
- 34.Cheung AK, et al. Functional analysis of a cell cycle-associated, tumor-suppressive gene, protein tyrosine phosphatase receptor type G, in nasopharyngeal carcinoma. Cancer Res. 2008;68(19):8137–8145. doi: 10.1158/0008-5472.CAN-08-0904. [DOI] [PubMed] [Google Scholar]
- 35.DePristo MA, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43(5):491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Purcell S, et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3):559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chang CC, et al. Second-generation PLINK: Rising to the challenge of larger and richer datasets. Gigascience. 2015;4:7. doi: 10.1186/s13742-015-0047-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang K, Li M, Hakonarson H. ANNOVAR: Functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38(16):e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li B, Dewey CN. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pires DE, Ascher DB, Blundell TL. DUET: A server for predicting effects of mutations on protein stability using an integrated computational approach. Nucleic Acids Res. 2014;42(Web Server issue):W314–319. doi: 10.1093/nar/gku411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Koch A, De Meyer T, Jeschke J, Van Criekinge W. MEXPRESS: Visualizing expression, DNA methylation and clinical TCGA data. BMC Genomics. 2015;16:636. doi: 10.1186/s12864-015-1847-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Speir ML, et al. The UCSC Genome Browser database: 2016 update. Nucleic Acids Res. 2016;44(D1):D717–725. doi: 10.1093/nar/gkv1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.