SUMMARY
Nearly half of the ribosomes translating a particular bacteriophage T4 mRNA bypass 50 nucleotides in its middle, resuming translation 3’ of this region. How this large-scale, specific hop occurs, and what determines whether a ribosome bypasses, remains unclear. We apply single-molecule fluorescence with zero-mode waveguides to track individual Escherichia coli ribosomes during translation of T4's gene 60 mRNA. Ribosomes that bypass are characterized by a 10- to 20-fold longer pause in a non-canonical rotated state at the take-off codon. During the pause, mRNA secondary structure rearrangements are coupled to ribosome forward movement, facilitated by nascent peptide interactions that disengage the ribosome anticodon-codon interactions for slippage. Close to the landing site, the ribosome then scans the mRNA in search of the optimal base-pairing interactions. Our results provide a mechanistic and conformational framework for bypassing, highlighting a non-canonical ribosomal state to allow for mRNA structure refolding to drive large-scale ribosome movements.
INTRODUCTION
Translation normally occurs sequentially in triplets of nucleotides (codons) with strict maintenance by the ribosome of fidelity and reading frame with error rates of 10−3 to 10−4 per codon (Dunkle and Dunham, 2015; Hansen et al., 2003; Jenner et al., 2010). There are cases when this well-established rule breaks down, where the genetic code can be recoded and altered in an mRNA-specific manner (called programmed). During programmed frameshifting, a portion of translating ribosomes can be stochastically diverted to a different reading frame (Chen et al., 2014b; Marquez et al., 2004; Tinoco et al., 2013). Ribosomes can even be directed to bypass, hopping over a stretch of nucleotides to continue translating a contiguous polypeptide (Herr et al., 2000a). These events increase the richness of information encoded in DNA or RNA, where a coding sequence can specify additional protein products not predicted from the standard readout of the open reading frame, as well as adding a layer of translational control.
The best-documented case of programmed bypassing is the gene 60 mRNA of bacteriophage T4 that codes for a subunit of a viral DNA topoisomerase (Herr et al., 2000a; Huang et al., 1988; Weiss et al., 1990). During translation of the gene 60 mRNA, ribosomes translate the first 45 codons (excluding the initiator fMet tRNA, which we term codon 0) to a Gly GGA codon. Half of the translating ribosomes stop at the subsequent UAG stop codon, while the other half skips the next 50 nucleotides and resumes translation from a downstream Gly codon (Maldonado and Herr, 1998). Instead of stopping at the stop codon, the anticodon of the peptidyl-tRNAGly2 (Gly-2) (Herr et al., 1999) disengages from the mRNA (in a process called “take-off”), the ribosome skips over the 50-nucleotide gap, and the peptidyl-tRNA re-pairs to the mRNA downstream at a GGA codon (called “landing site”). As result, translation resumes at codon 46 to create a single, continuous protein product from a discontinuous open reading frame (Wills, 2010) (Figure 1A).
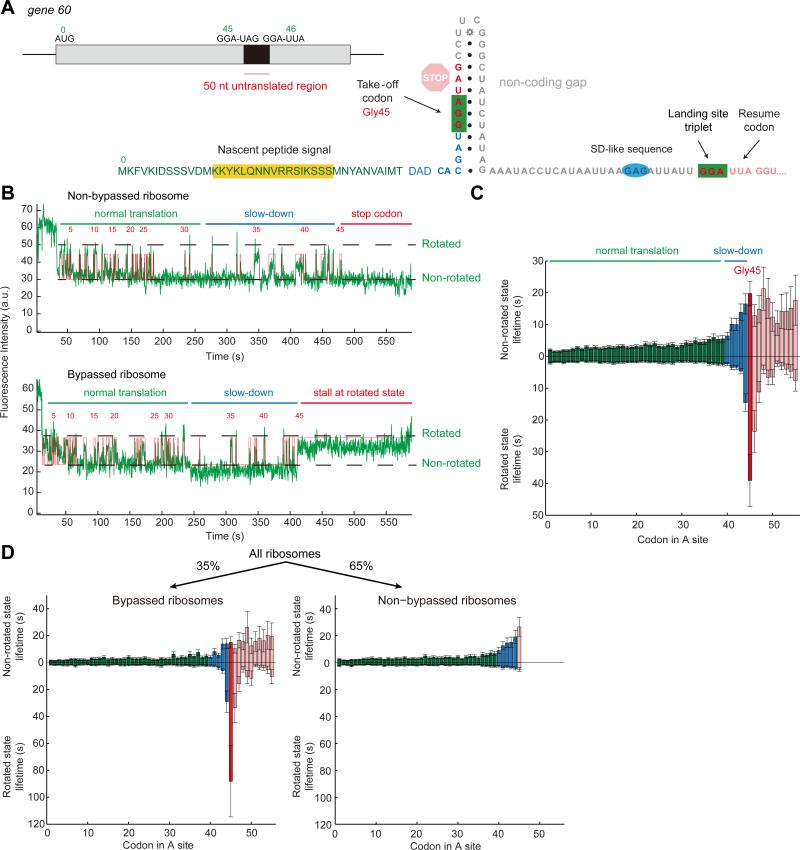
Figure 1. Dynamic pathways of gene60 bypassing.
(A) The elements of gene 60 bypass are labeled: (1) the UAG stop codon immediately 3’ to the take-off GGA site at codon Gly45, (2) the tRNAGly and the matching GGA take-off and landing sites, (3) an upstream nascent peptide signal, (4) a stem-loop consisting of the take-off codon, and possibly (5) a GAG Shine-Dalgarno-like sequence located 6 nucleotides 5’ to the landing site to promote precision of landing. Full sequence of the gene 60 mRNA is shown, where first 42 codons written as their amino acids (with Met being codon 0) and the remaining sequence labeled with nucleotides. The coloring of the codon or nucleotide matches the coloring in part B and C.
(B) Representative traces of ribosomes Cy3B (green) fluorescent intensity for bypassed and non-bypassed ribosomes. For both cases, there is a phase with normal translation (labeled with a green line), a phase of slow down (blue line), and either terminating at a stop codon for non-bypassed ribosomes or go into a rotated state pause at codon Gly45 for bypassed ribosomes. The state assignment is shown in red, with the codon counts above.
(C) The mean state lifetimes. The first 39 codons, when translation occurs normally, are colored in green. Codons 40 to 44, characterized by slow-down due to nascent peptide interaction, are shown in blue. The take-off site at codon 45 is colored in red. At codon 45, there is a long rotated state pause. Codons after bypass are shaded in pink. Number of molecules analyzed, n = 451.
(D) We can parse the subpopulation of ribosomes into bypassed and non-bypassed and separate the lifetimes shown in (C) into the two populations, giving us a bypassing efficiency of 35%. Only the bypassed ribosomes exhibit an increase in rotated state lifetime at codon Gly45. The color scheme is the same as (C). n = 451. See also Figure S1, Figure S2, and Figure S3.
Biochemical, genetic, and mutational analysis relying on detection of protein products both in vitro and in vivo have identified the essential stimulatory elements for programmed bypassing in gene 60: (1) the tRNAGly (Gly-2) and the matching GGA take-off and landing sites bounding the non-coding gap, (2) an upstream nascent peptide sequence, (3) a stem loop consisting of the take-off codon and the adjacent UAG stop codon, and (4) a GAG Shine-Dalgarno-like sequence located 6 nucleotides 5’ to the landing site to promote precision of landing (Figure 1A). With the matched take-off/landing pairs, bypassing is the most efficient for the wild-type GGA codon; other codons are possible, but codons with G or C in the first two positions yield more efficient bypassing (Bucklin et al., 2005). With unmatched take-off/landing pairs, for example GGA/GCA or GCA/GGA, bypassing efficiencies were greatly reduced (Weiss et al., 1990). The take-off codon is located within a potential –UUCG– hairpin stem loop in the 5’ portion of the non-coding gap, which is important for bypassing: mutations that disrupted base pairing reduced bypassing whereas compensatory double mutations restored it. Altering the –UUCG– tetraloop sequence at the top of the stem, extending the length of the stem or increasing loop size also reduced bypassing (Herr et al., 2000b; Weiss et al., 1990; Wills et al., 2008). In addition to the hairpin, a “nascent peptide signal” KKYKLQNNVRRSIKSSS13-29, potentially interacts with the exit-tunnel of the ribosome to stimulate bypassing (Herr et al., 2004; Maldonado and Herr, 1998; Weiss et al., 1990). Lastly, there is an alternative landing site at GGG within the non-coding gap near the top of the stem-loop (positions 9-11 from the take-off codon); however, the bypassing ribosome always lands at the wild-type landing codon (positions 48-50 from the take-off codon). Thus, it has been proposed that the ribosome does not scan the full non-coding gap in search of a potential landing site, but rather hops over the non-coding region (Wills et al., 2008).
How the ribosome traverses the gap remains unclear, with no definitive and testable model proposed for the mechanism of such a large-scale movement. What stimulates the ribosome to initiate bypass and what determines whether a ribosome bypasses or not? What are the roles of the nascent peptide and mRNA secondary structure in inducing bypass? What is the conformational state of the ribosome during bypassing? Prior investigations of frameshifting have underscored the importance of dynamics in translational recoding (Caliskan et al., 2014; Chen et al., 2014b). Here we probe the dynamic and stochastic nature of bypassing using single-molecule fluorescence to track single translating ribosomes in real time, allowing us to define a global mechanism for bypassing.
RESULTS
Real-time observation of ribosome bypassing dynamics
To monitor single Escherichia coli ribosome progression on mRNAs in real-time we used zero-mode waveguide (ZMW) instrumentation (Chen et al., 2014a; Chen et al., 2014b). In this study, we followed conformational changes underlying elongation, involving rotational movements of the small (30S) ribosomal subunit with respect to the large (50S) ribosomal subunit and correlated them with binding and departure of tRNAs and elongation factors. To observe rotational movement, the 30S subunit was labeled with Cy3B on helix 44, and a non-fluorescent quencher, BHQ-2, was placed on helix 101 of the 50S subunit, allowing Förster resonance energy transfer (FRET) between the two dyes (Chen et al., 2013; Chen et al., 2012b) (Figure S1A).
During one elongation cycle, the two subunits start in a non-rotated state (characterized by high FRET, low Cy3B intensity). EF-Tu-GTP-aa-tRNA ternary complex (TC) binds to the vacant A site followed by peptidyl transfer from P-site tRNA to the new A-site aa-tRNA. After peptidyl transfer the ribosomal subunits rapidly rotate relative to each other (rotated state; lower FRET, higher Cy3B intensity). During this stage, the ribosome is “unlocked”, where the ribosome conformation and tRNA spontaneously fluctuates (Blanchard et al., 2004a; Chen et al., 2012a; Cornish et al., 2008), preparing for translocation. mRNA-tRNA interactions and ribosome-tRNA interactions are weaker at this stage (Liu et al., 2011; Valle et al., 2003). Upon translocation catalyzed by EF-G, the two subunits rotate back to their original high-FRET state and the ribosome is “relocked”. Thus, one round of high-low FRET (low-high Cy3B intensity) corresponds to a single ribosome translating one codon, allowing tracking of translation at codon resolution, and providing the timings of individual substeps at each codon (Chen et al., 2013) (Figure S1B). As opposed to previous smFRET studies with probes labeled at ribosomal proteins S6 and L9 showing spontaneous intersubunit rotations after peptidyl-transfer (Cornish et al., 2008), our FRET probe positions possibly monitor a different intersubunit movement that occurs only one cycle per codon. Arrival and departure of the dye-labeled ligands such as tRNAs can be simultaneously observed as a sequence of fluorescent pulses (Chen et al., 2013) (Figure S1C). The correlation of single cycles of FRET to translation of a single codon has been confirmed in multiple studies (Aitken and Puglisi, 2010; Chen et al., 2012a; Marshall et al., 2008).
To follow translating ribosomes, we monitored the intersubunit conformational signal upon delivering total tRNA (tRNAtot) ternary complex (aa-tRNA-EF-Tu-GTP), EF-G, and BHQ-50S to immobilized Cy3B-30S preinitiation complexes on the bottom of the ZMWs, as done previously (Johansson et al., 2014; Tsai et al., 2014). Through statistical analysis of multiple translating single ribosomes, we obtain waiting times of non-rotated and rotated states for each codon. Continuous translation can be observed for more than 50 codons, allowing us to profile the real time dynamics approaching, during, and after bypassing.
Dynamic pathways of bypassing show a rotated state pause for bypassed ribosomes
Translation of the first 40 codons of wild-type gene 60 mRNA proceeds normally, with expected lifetimes of rotated (waiting for translocation) and non-rotated (waiting for peptidyl transfer) states (2~5 s at 3 μM tRNAtot TC and 240 nM EF-G), demonstrating a regular elongation rate at these codons. From codons 40 to 45, i.e. before the take-off site, translation gradually slows with increase in both rotated and non-rotated states lifetimes to roughly 15 s for each state (3-7-fold increase). At the bypass site at codon Gly45, an exceptionally long rotated state pause is observed, with a 20-fold higher mean lifetime of 40 s. For a subset of ribosomes paused at codon Gly45, translation resumes instead of stopping translation at the UAG stop codon after Gly45, indicating that we observe bypassing (Figure 1B, C, Figure S2A, B).
Translating single ribosomes cluster into three major sub-populations: (1) ribosomes that translate 45 codons and stall at the stop codon; these ribosomes do not bypass and do not exhibit the long rotated pause at Gly45, (2) ribosomes that bypass and translate at least codon 46; these ribosomes ubiquitously exhibit a long rotated state at codon 45, or (3) ended the Cy3B signal during the long rotated state due to photobleaching or end of movie. Combining the second and third clusters gives a bypassing efficiency of ~35%, consistent with our in vivo assays (33%), and prior studies (Maldonado and Herr, 1998; Samatova et al., 2014). These results also confirm that bypassing is programmed in the mRNA itself (Samatova et al., 2014); no other auxiliary factors beyond the standard factors added here are required.
Bypassing and non-bypassing ribosomes show distinct dynamics. All ribosomes exhibit the gradual increase in non-rotated state and rotated lifetimes from codons 40 to 45. This increase in lifetime upon approaching the bypass site is reminiscent of the dynamic signatures observed for nascent peptide-ribosome interactions during SecM stalling (Tsai et al., 2014). The long rotated state pause at the bypass site (Gly45) is observed only for ribosomes that undergo bypassing, and is similar to the non-canonical rotated states observed in −1 frameshifting (Chen et al., 2014b). By parsing into two distinct populations of ribosomes, we obtain a more accurate mean lifetime for the rotated state pause (88.2 ± 26.4 s) without the convolution of non-bypassed ribosomes (Figure 1D). Resumption of normal translation post bypassing is not immediate, and the ribosome translates slowly for a few more codons before the rotated state lifetimes return to normal (mean lifetime is 5 s), while the non-rotated state lifetimes remain higher (mean lifetime is 15 s) (Figure 1B, C and see Figure S3).
The role of the nascent peptide signal and its interaction with the ribosome exit tunnel: setting the stage for bypassing
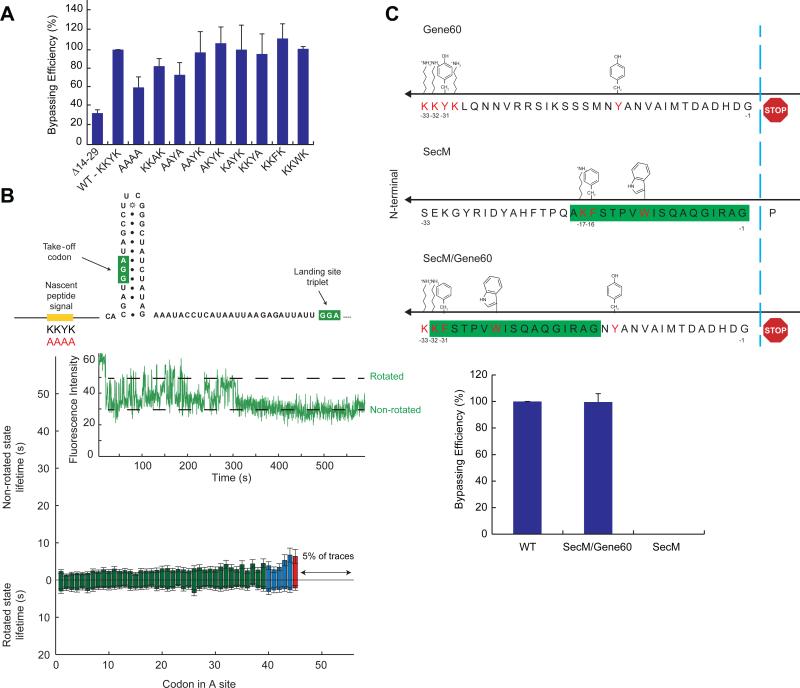
We hypothesized that the general slow-down in translation observed for both non-bypassed and bypassed ribosomes is due to the nascent peptide signal, KYKLQNNVRRSIKSSS14-29 (Weiss et al., 1990; Wills, 2010), interacting with the ribosome exit tunnel. In vivo, deleting from codon 14 to codon 29 causes a 70% decrease in bypassing efficiency (Figure 2A). Mutational analysis of the nascent peptide highlighted the importance of a KKYK13-16 motif (Figure 2A). With our in vitro single-molecule system, deleting the sequence encoding KYKLQNNVRRSIKSSS14-29 eliminates observable bypassing and pausing: translating ribosomes no longer exhibit the increase in rotated and non-rotated state lifetimes and ribosomes now translate 29 codons to the stop codon, with only 1% of the traces showing translation beyond the UAG stop codon (Figure S4). Mutating the critical KKYK motif to AAAA resulted in a similar behavior; the increases in non-rotated and rotated states lifetimes are no longer observed (Figure 2B). Deletion of the non-coding gap while maintaining the nascent peptide signal abrogates bypassing as expected, but increases in rotated and non-rotated state lifetimes approaching codon Gly45 are observed as for the wild-type sequence (Figure S4). These results indicate that the nascent peptide is responsible for the slow-down approaching the bypass site independent of the mRNA sequence and structure at the bypass site and that this slow-down is necessary for the ribosome to undergo the rotated state pause at Gly45 for bypassing. Importantly, these interactions are different from SecM induced stall (Figure 2C).
Figure 2. Mutation of the nascent peptide interaction abolishes the slow-down.
(A) In vivo analysis of bypassing with mutants of the nascent peptide. The absolute value of bypassing in these assays by the WT (2nd from left) is 33% and all other values are of a percentage of it.
(B) Deleting the key interaction of the nascent peptide signal (KKYK) to AAAA do not increase non-rotated and rotated state lifetimes. Most ribosomes terminate at the stop codon after codon Gly45. An example trace is shown. The color scheme is the same as Figure 1. n = 424.
(C) In vivo analysis of bypassing with fusions of gene 60/SecM nascent peptides. The cassette used to generate the result in the middle lane has gene 60 sequence encoding amino acids 32 to 46 in its native location 5’ adjacent to the gene 60 take-off codon. The SecM nascent peptide signal encoding sequence is 5’ adjacent to it. The right lane derives from a cassette with the SecM nascent peptide encoding sequence 5’ adjacent to the gene 60 take-off codon. See also Figure S4.
The role of mRNA hairpin in promoting disengagement of anticodon-codon interactions
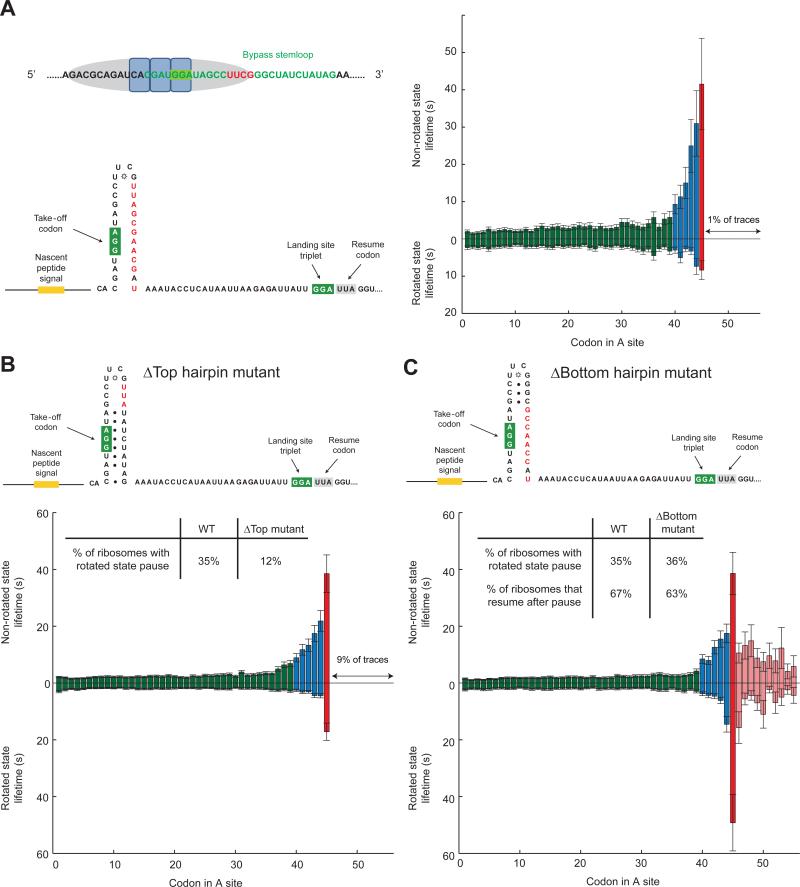
The nascent peptide signal alone is not sufficient to promote bypass; the hairpin at the bypass site is required. Disrupting the potential base pairing in the hairpin stem abolishes the long rotated-state pause at Gly45, but we still observe the slow-down caused by the nascent peptide signal (Figure 3A). This demonstrates that the hairpin stem loop is required for the long pause in the rotated state prior to bypassing, and that the action of the hairpin follows that of the nascent chain.
Figure 3. The –UUCG– hairpin stem loop, especially the top base-pairs, is important for the rotated state pause.
(A) The hairpin is shown in green, and the UUCG tetraloop is marked in red. To investigate the role of the mRNA hairpin, the base pairs were disrupted; the increase in non-rotated state lifetime due to the nascent peptide signal is still observed, but long rotated state pause at Gly45 characteristic of bypassing is no longer detected. n = 244.
(B) Mutation of 3 base pairs below the UUCG tetraloop decreased bypass efficiency to 12%. n = 442.
(C) Mutation of the bottom portion of the hairpin. The bypass efficiency remained the same at 36%. n = 349.
How the mRNA hairpin promotes such a pause is puzzling, since the –UUCG– hairpin stem loop should be fully melted by the ribosome within the mRNA channel at the take-off site, as the ribosome protects 9 nucleotides subsequent to the P-site codon (Qu et al., 2011). We hypothesize here that the unusual stability of a UUCG tetraloop (Ennifar et al., 2000; Todd and Walter, 2013), which has a propensity to form a compact structure, may favor refolding of the apical portions of the hairpin, providing a mechanism for the long rotated pause. If this hypothesis were correct, then the top portion of the hairpin would be sufficient for pausing and bypassing.
To test this hypothesis, we created two mutants, called Δtop hairpin (destabilized the 3 base pairs below the UUCG tetraloop) and Δbottom hairpin (leaving the 3 base pairs below the tetraloop intact but disrupting 7 potential base pairs in the lower part of the stem) (Figure 3B, C). Translation of the Δtop hairpin mutant mRNA resulted in a decrease of ribosomes that go into the rotated state pause (12%). Translation of the Δbottom hairpin mRNA remained similar to the wild-type sequence (36% compared to WT 35%). This highlights the importance of the UUCG tetraloop and the top portions of the hairpin in stimulating bypass, consistent with prior mutagenesis (Weiss et al., 1990; Wills et al., 2008) (~1 ~ 30% of WT). Interestingly, destabilizing the three base pairs located 5 nucleotides from the tetraloop did not significantly reduce bypass efficiency (~60 ~ 90% of WT) (Weiss et al., 1990). Thus, the precise location of the UUCG tetraloop with respect to the ribosome during take-off is critical, in addition to the propensity of the tetraloop to re-fold (see Discussion for speculations on where the hairpin refolds). It is likely that this propensity to re-fold induces a lateral tension on the mRNA-tRNA interaction, which combined with the vertical pull from the nascent peptide interaction, causes the disengagement of the anticodon-codon interaction and “slippage” uncoupling ribosomal motions from tRNA-mRNA movement, causing the ribosome to be trapped in a non-canonical rotated state, reminiscent of the uncoupled translocation in −1 frameshifting (Chen et al., 2014b).
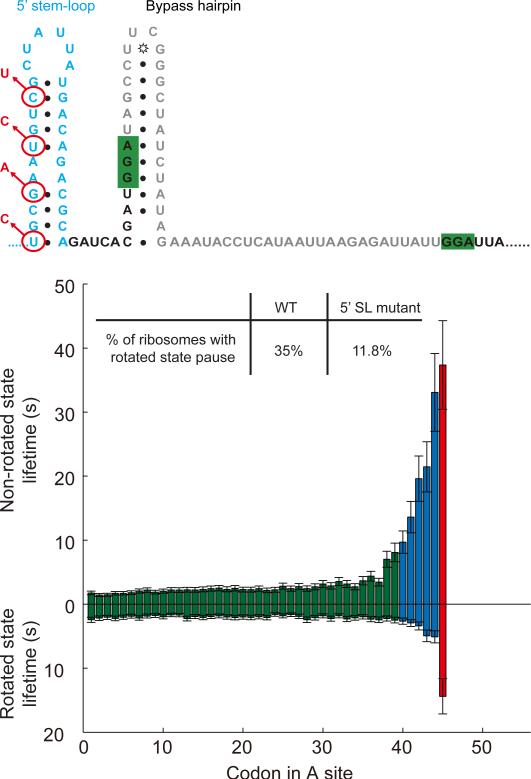
Second hairpin 5’ to the take-off site is required for bypassing
What provides the forward bias for the bypass? To answer this, we focused on a predicted hairpin 5’ to the bypass stem-loop (Figure 4, Figure S5) (Samatova et al., 2014; Todd and Walter, 2013). We introduced synonymous mutations that disrupt this 5’ stem-loop and preserve the amino acid identity, and showed that the percentage of ribosomes that go into the rotated state pause decreases to 11.8%, confirming the importance of this stem-loop. Consistently, Samatova et al. showed that the synonymous mutations that disrupt this 5’ stem-loop reduce bypassing efficiency in vitro (~10% of WT), while compensatory mutations partially restore bypassing (Samatova et al., 2014). When the ribosome is positioned at the take-off Gly45 codon, the 5’ stem-loop is likely partially formed except for the bottom 3~6 base pairs. The refolding of the bottom 3 base pairs may provide a forward bias for the bypass movement. Alternatively, the directionality may be maintained through re-forming of the stem-loop when the ribosome vacates the stem-loop, acting as a block for backward movement.
Figure 4. Effects of the 5’ stem-loop.
Synonymous mutations (shown in red) of the 5’ stem-loop (wild-type sequence shown in blue) to destabilize the secondary structure. The bypass efficiency decreased to 11.8%, with a corresponding decrease in rotated state lifetime at codon Gly45, suggesting that the 5’ stem-loop is important. n = 488. See also Figure S5.
Take-off and landing mechanisms: mRNA refolding causes uncoupled translocation
The concerted effects from the nascent peptide interaction, re-folding of the 5’ hairpin, and the re-folding of the tetraloop induce a long rotated-state pause characteristic of bypassing. Pausing may be caused by translocation that is uncoupled with the ribosome reverse-rotation, similar to what was observed previously for −1 frameshifting in dnaX (Chen et al., 2014b). This leaves the ribosome in a non-canonical rotated state, resulting in the long rotated state pause observed in frameshifting.
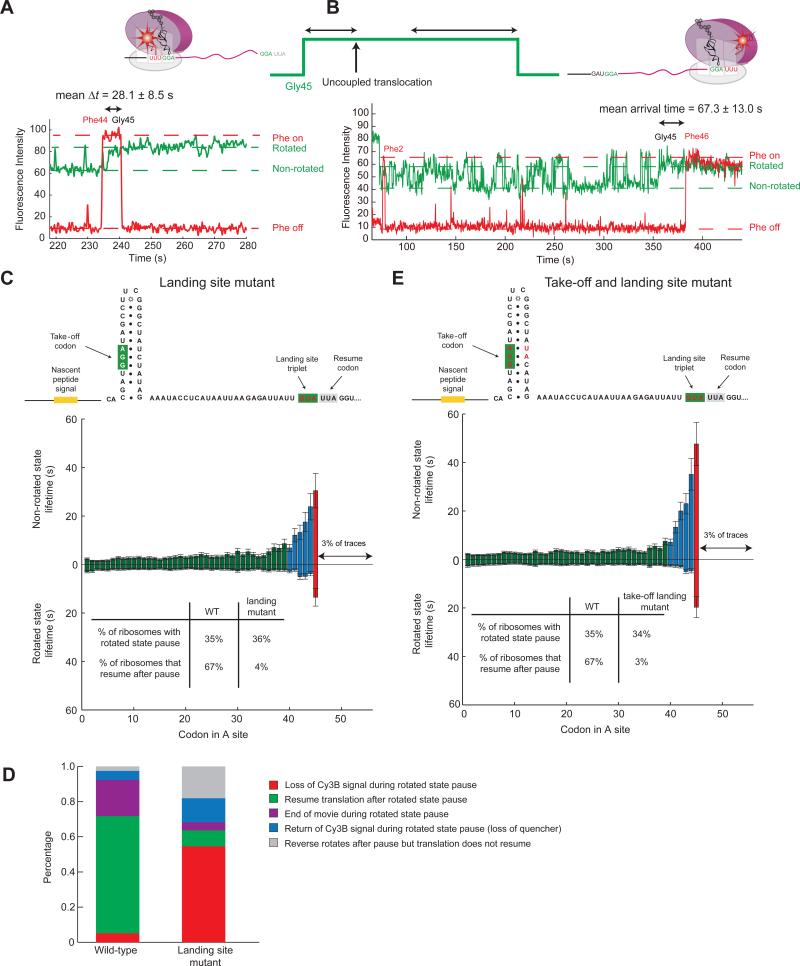
To test whether translocation occurs during the pause, we mutated Asp44 (the codon before Gly45) to Phe; this allows the use of Cy5-labeled tRNAPhe to estimate when translocation occurs during the rotated state pause (through the departure and disappearance of Cy5-tRNAPhe with the Asp44Phe mutant) in correlation with the Cy3B ribosome conformational signal (Figure S1C). Translocation of the P-site tRNA to the E site is typically correlated with ribosome reverse-rotation, hence the rotated state lifetime is equivalent to the time to departure of the P-site tRNA signal and, thus, ribosome reverse-rotation and translocation are usually coupled. Here, we found the Cy5-tRNAPhe departs 28.1 ± 8.5 s after the rotation of the ribosome at codon Gly45, which is much shorter than the lifetime of the rotated state pause (88.2 ± 26.4 s) (Figure 5A), indicating that translocation precedes reverse-rotation and that the two are now uncoupled. Uncoupled translocation results in a ribosome in a non-canonical rotated state with a peptidyl P-site tRNA and an empty A site. During this non-canonical state, recoding events can occur when ribosome-tRNA-mRNA interactions are weaker to allow for the “take-off” to occur and peptidyl-tRNA and mRNA to dissociate.
Figure 5. Timing and mechanism of take-off and landing.
(A) Using the Asp44Phe and Leu46Phe mutant mRNAs introduced in Figure S2, the timing of bypass was probed. Using the Asp45Phe mutant, we can get the timing of when the Cy5-tRNAPhe (red) departs relative to the start of the rotated state pause at Gly45. This gives an upper estimate of when translocation occurs during the pause. The mean departure time is 28.1 ± 8.5 s, which is a lot shorter than the mean lifetime of the pause (90 s), indicating that the translocation is uncoupled with reverse-rotation. This gives an estimate of when the launch occurs.
(B) With Leu46Phe mutant, A-site accessibility could be probed with Cy5-tRNAPhe, giving an estimate on when landing is completed. The mean arrival time is 67.3 ± 13.0 s, which is also during the pause. Thus, bypass and landing is completed during the rotated state pause, making the A site available for tRNA binding.
(C) Landing site was changed from GGA(Gly) to GUA(Val), mRNA sequence is shown. The increase in non-rotated state lifetime due to the nascent peptide signal can be seen. The rotated state pause at Gly45 is shorter than for wild-type. This is due to loss of Cy3B signal during the rotated state pause, when the ribosome fails to find the correct landing codon after launching the bypass and drops-off. Thus, matching take-off and landing codons are required. Consistent with this the percentage of ribosomes undergoing the rotated state pause at Gly45 is the same as wild-type. However, the percentage of ribosomes that resume after the pause is much lower. n = 469.
(D) The end-states of the ribosome after the pause can be parsed to (1) loss of Cy3B signal (due to ribosome drop-off or photobleaching), (2) resume of translation after the pause, (3) end of movie during the pause, (4) return of the Cy3B signal (photobleaching of FRET quencher or dissociation of 50S), and (5), reverse-rotates but translation does not resume.
(E) Both the take-off and landing codons where changed from wild-type GGA(Gly) to GUA(Val). The non-rotated and rotated state lifetimes for the double mutant. Very similar behavior to the landing site mutant can be seen. Thus, the identity of the take-off codon is not critical for initiating bypass. However, for successful landing, the identity of the tRNA is very important. n = 466. See also Figure S6.
Similarly the timing of “landing” was probed through the Leu46Phe mutant, with the codon after the landing codon mutated to Phe. The arrival of Cy5-tRNAPhe after the intersubunit rotation at Gly45 indicates successful landing of the peptidyl-tRNA to the landing Gly45 codon, with an exposed Phe codon in the A site. The arrival time of Cy5-tRNAPhe in this case is 67.3 ± 13.0 s after the rotation at Gly45. These results allow us to determine the timeline of the hop (Figure 5B) – it begins during the long rotated pause and ends within it.
After translational hopping to Gly46, tRNAPhe arrives to the A-site codon 46 with the ribosome in the rotated state unlike during normal translation when tRNAs usually bind to the non-rotated state. The binding of the tRNAPhe is stable, with the lifetime comparable to the remaining lifetime of the rotated state pause. During the remainder of the pause, tRNAs re-pair with the mRNA codon and peptidyl transfer occurs, returning the ribosome to the canonical rotated state with hybrid tRNAs. EF-G can then act on the ribosome and translocate the tRNAs, allowing for normal translation to resume (see Figure S6).
To decipher the mechanism of the bypass during the long rotated state pause, we examined the effects of simultaneous mutations to the take-off and landing codons, as well as mutations of only the landing codon to create a mismatch (Figure 5C). For unmatched take-off and landing codons, the bypassing efficiency decreases to ~5% of wild-type (Weiss et al., 1990). Upon mutation of the landing codon to a GUA (Val) to create a mismatch, 36% of the ribosomes exhibit a pause at Gly45 with the slow-down approaching Gly45 due to the nascent peptide, similar to wild-type mRNA. Thus, the behavior up to the bypass is not affected by the mutated landing codon. However, only 4% of ribosomes in the landing site mutant resume translation after the pause (within observation window) compared to 67% of for wild-type mRNA.
We determined the fate of ribosomes during the pause by examining the ending state of each ribosome at the pause. For wild-type mRNA, the vast majority of the traces (>90%) show ribosomes either resuming translation or the acquisition movie ends during the pause; for the landing site mutant the majority of ribosomes (55%) show a loss of Cy3B signal at Gly45. This loss of signal is not due to photobleaching, since for the wild-type mRNA, under the same experimental conditions, only 5% of the traces showed a loss of Cy3B signal at Gly45. Thus, loss of 30S-Cy3B signal is due to ribosome drop-off on the mutant mRNA, where ribosomes that initiated bypass failed to find the correct landing codon. Since theses ribosomes do not stably form peptidyl-tRNA-mRNA interactions, they dissociate from the mRNA (Herr et al., 2001). Accordingly, the rotated state pause lifetime decreased from 40 s to 15 s (Figure 5C).
Mutation of both the landing and take-off codons from a GGA to GUA (Val) was previously shown to drop the bypass efficiency to 7% of wild-type (Bucklin et al., 2005). Similar results were observed in our experiments if both the take-off and landing codon are changed from GGA to GUA (Val) (Figure 5E). These results suggest that the identity of the take-off tRNA is not critical to start the process of bypassing; the nascent peptide and hairpins induce take-off and the rotated state pause. The identity of the tRNA is important for successful landing; it must match the landing codon, but stable G-C rich pairing is important for successful recognition and re-pairing of the peptidyl-tRNA and mRNA, consistent with earlier data (Bucklin et al., 2005).
Directly monitoring the timing of hop with ribosome-mRNA FRET
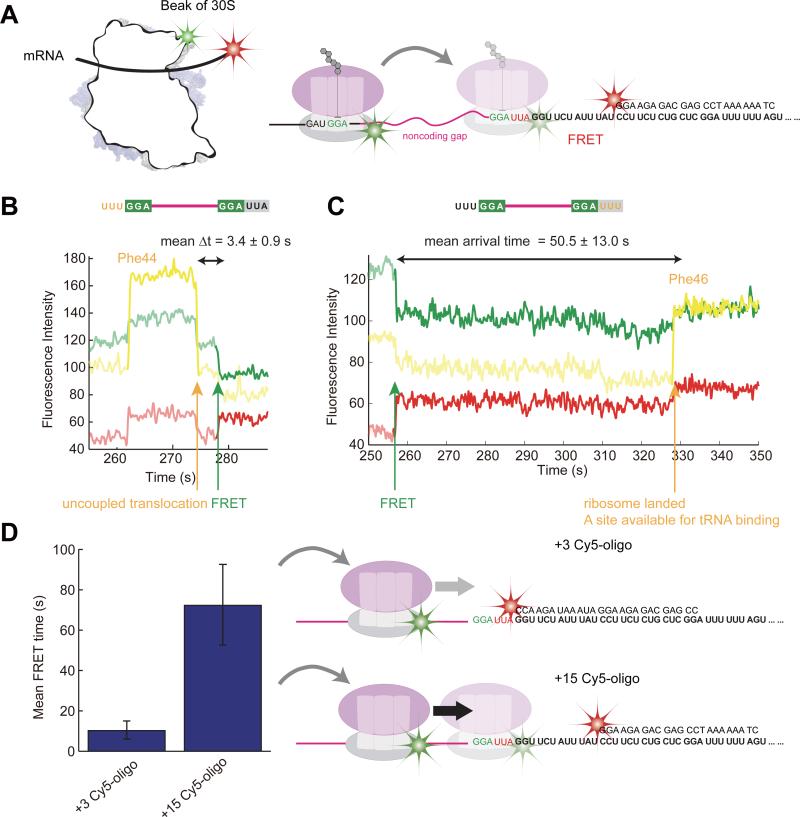
To probe ribosome movement directly during bypassing, we used FRET between the ribosome and mRNA: mRNA was labeled downstream of the landing site by annealing a Cy5-labeled DNA oligonucleotide complimentary to the mRNA (termed +15 Cy5-oligo, 15 nucleotides downstream of the landing GGA codon) and 30S subunits were labeled with Cy3B on helix 33a near the beak domain, which is close to the mRNA entrance channel (Figure 6A, Figure S1). The bypass will bring the Cy3B dye on the ribosome close to the Cy5 dye with the simultaneous appearance of FRET. Translation is followed by stable binding of Cy3.5-labeled Phe-tRNAPhe. Before bypassing, we observed no FRET between translating ribosomes and downstream labels in the mRNA; ribosome-mRNA FRET is thus a hallmark of attempted bypassing.
Figure 6. Timing of ribosomal bypassing and scanning monitored by ribosome-mRNA FRET.
(A) For ribosome-mRNA FRET to monitor the hop, the 30S subunit was labeled with Cy3B on helix 33a, near the beak of 30S subunit, and the mRNA is labeled with Cy5 downstream of the landing site. Landing after bypassing brings the ribosome within FRET distance to the Cy5 dye.
(B) Asp44Phe mutant mRNA allows us to use Cy3.5-labeled (yellow) to track when the tRNA departs at codon 44. This represents the timing of uncoupled translocation during the rotated state pause at Gly45. The ribosome bypasses on average 3.4 ± 0.9 s after uncoupled translocation.
(C) Translation of the Leu46Phe mutant mRNA allows us to use Cy3.5-labeled (yellow) to track when the tRNA arrives to the A site after the bypass. This represents the timing of when successful landing occurs and the A site is available after the bypass (on average 50.5 ± 13.0 s).
(D) With the use of the +15 Cy5-oligo (15 nucleotides downstream of the GGA landing codon, the same used for (B) and (C)), the FRET lifetime is 72.3 ± 20.0 s. By moving the Cy5-oligonucleotide to 3 nucleotides downstream of the take-off GGA codon (called +3 Cy5-oligo) such that the ribosome footprint is blocked upon landing, the FRET average lifetime decreases significantly to 10.2 ± 4.5 s. Since there is still a stable FRET signal, the ribosome must land upstream of the oligonucleotide, then scan to find the landing site, during which the ribosome contact quenches the Cy5 dye.
Using the Asp44Phe mutant and Leu46Phe mutant mRNAs, we can use Cy3.5-labeled tRNAPhe to score for the translation of Phe44 prior to the take-off or Phe46 after successful landing. This allows us to monitor the time between uncoupled translocation (departure of Cy3.5-tRNAPhe from Phe44) and bypassing (appearance of FRET), and also the time between bypassing and successful landing (arrival of Cy3.5-tRNAPhe at Phe46). The hop occurs shortly after uncoupled translocation, on average after 3.4 ± 0.9 s (Figure 6B). The ribosome quickly lands near the landing Gly codon, as demonstrated by the 1~2 frame FRET transition at 100 ms frame-rate. After landing, the resume codon in the A site (Leu in the wild-type and Phe in the Leu46Phe mutant) is not immediately available for binding. Instead, Cy3.5-tRNAPhe binds on average 50.5 ± 13.0 s after the hop (Figure 6C).
Does the ribosome land directly on the landing site, or does the ribosome land upstream and scans to find the optimal landing site? To distinguish these possibilities, we note that the FRET average lifetime for the +15 Cy5-oligo is 72.3 ± 20.0 s. If we move the Cy5-oligonucleotide to 3 nucleotides downstream of the take-off GGA codon (called +3 Cy5-oligo) such that the ribosome footprint is blocked upon landing, the FRET average lifetime decreases significantly to 10.2 ± 4.5 s (Figure 6D). Thus, even when the landing site is blocked, we still see a stable FRET signal, indicating that ribosomes land upstream and then scan before photobleaching or contact quenching the Cy5 dye. Thus bypassing occurs in two steps – a hop in the 3’ direction followed by the scanning, associated with finding the best stable landing site to resume translation.
DISCUSSION
By tracking single ribosomes translating in real time, we delineate here the dynamic events underlying bypassing. All determinants for bypassing are specified by the gene 60 mRNA itself. Translation of the gene 60 sequence results in a branchpoint stimulated by the nascent peptide signal and hairpin. At the take-off codon Gly45, the nascent peptide and the hairpin induces a fraction of the ribosomes (35%) to undergo a long rotated state pause, similar to what was observed for −1 frameshifting (Chen et al., 2014b). During this state, the ribosome-tRNA-mRNA interactions are weaker which allows for unusual and large-scale ribosome reconfiguration events to occur for bypassing. Non-bypassed ribosomes terminate at the stop codon without the pause. In this mechanism both the nascent peptide and the hairpin (especially the UUCG tetraloop with three flanking nucleotides) are critical for bypassing. A recent study by Samatova et al. (Samatova et al., 2014) and our findings confirm the importance of another 5’ stem-loop, which provides directionality for the bypass. Here, we propose a model for bypassing that involves the sequential coupling of the re-folding of the two hairpins to ribosome movement, allowing the ribosome with weakened ribosome-tRNA-mRNA interactions induced by the nascent peptide to bypass the non-coding mRNA region.
The nascent polypeptide of gene 60 causes slowdown in translation as the ribosome approaches the take-off Gly45 codon (from codons 40 to 45), which is a required prelude to bypassing. The interaction causing slowdown begins after a ribosome translates 40 codons when the key KKYK portion of the nascent peptide is ~25 amino acids from the P-site tRNA. The slowdown is defined by increased lifetimes of both the non-rotated and rotated states, indicating increased barriers to tRNA selection/accommodation and translocation, respectively. These barriers increase progressively during translation from codon 40 to 45. At the take-off site, the KKYK portion of the nascent peptide is ~30 amino acids from the P-site tRNA (as opposed to interaction of SecM, which is 17 amino acids (Nakatogawa and Ito, 2002; Tsai et al., 2014). Thus, even though the dynamic signatures are similar to other stalling sequences, the interaction in bypassing is different from that of SecM; the SecM stalling mechanism does not promote bypassing. We recently showed that co-translational folding of a short peptide sequence upstream of the SecM sequence in the exit tunnel beyond the constriction point, “pulls” on the peptide releaving the stall (Nilsson et al., 2015). Since the nascent peptide signal sequence from codons 14 – 29 in bypassing has been predicted to fold into a α-helical structure (Bhushan et al., 2010; Samatova et al., 2014) (see Figure S7), it may play a similar role in “pulling” on the peptidyl-tRNA to cause the disruption of anticodon-codon interactions necessary for take-off. This suggests that for efficient bypassing, stall is insufficient; the specific interaction and force direction from the traditional SecM stall may not be conducive for bypassing (Goldman et al., 2015). The precise interactions of the nascent peptide with the tunnel will require further study using structural methods. Nonetheless, this nascent peptide interaction is a prerequisite to the ribosome pausing in the rotated state at codon Gly45.
Ribosomes at the bypass site stochastically continue translating or bypass. We propose that the – UUCG– hairpin is the origin for this branchpoint of pathways, similar to the role played by a helical stem-loop for −1 frameshifting (Chen et al., 2014b). The role of the bypass hairpin, however, is puzzling, since at the take-off site, the hairpin has been melted by the ribosome. However, the stability of the UUCG tetraloop (Ennifar et al., 2000; Todd and Walter, 2013), which has a propensity to form a compact structure may cause the apical portion of the hairpin stem loop to refold. In addition, the recent work by Samatova et al. and our work have identified a previously uncharacterized 5’ stem loop that also contributes to bypassing (Samatova et al., 2014). When tRNAGly at codon 45 accommodates into the ribosome and the ribosome rotates, the A site is over codon 45, which places the ribosome such that the UUCG tetraloop is just within the 3’ mRNA channel and the 5’ stem-loop is mostly folded except for the bottom base pairs (Figure 7). The tendency for the 5’ stem-loop and the UUCG tetraloop to re-fold, in addition to the “pull” on the tRNA through the cascade of nascent peptide interactions, likely creates a tension in the system. Thus, the re-folding of the 5’ stem-loop and the ribosome stochastically encountering a folded or unfolded UUCG tetraloop may cause the initial branchpoint.
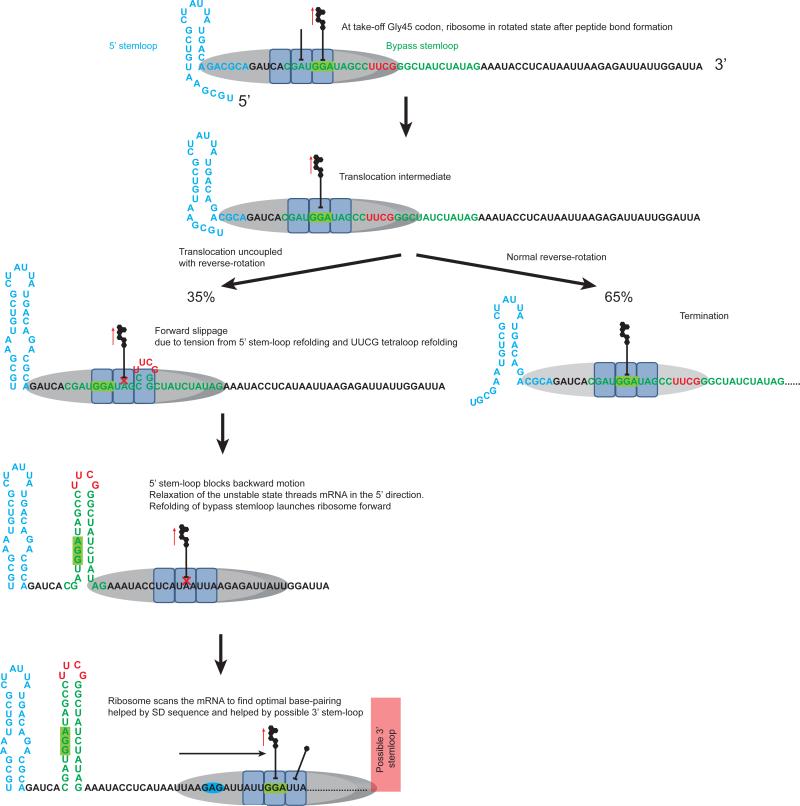
Figure 7. Model of translational bypassing.
At the take-off Gly45 (GGA) codon, after the arrival of tRNAGly to the A site and peptidyl transfer, the ribosome rotates. The nascent peptide signal interaction pulls on the peptidyl-tRNA, as indicated by the red arrow. The 5’ stem-loop is shown in blue, the bypass hairpin is shown in green, and the UUCG tetraloop is shown in red. EF-G catalyzes translocation, moving the GGA codon to the P site. Combined with the propensity of the UUCG tetraloop to re-fold, the ribosome slips forward and leads to uncoupled translocation, allowing the UUCG tetraloop and a few base pairs to re-fold within the A site and the 5’ stem-loop to completely refold. Since the 5’ stem-loop blocks backwards movement, relaxation of the unstable state threads the mRNA in the 5’ direction. Refolding of the bypass hairpin launches ribosome forward. The ribosome then scans the mRNA to find the optimal base pairing, assisted by the GAG Shine-Dalgarno-like sequence and a possible 3’ stem-loop. Upon landing at the landing-site, the next tRNA accommodates to the rotated ribosome to help re-define the reading frame, resuming translation. See also Figure S7.
EF-G catalyzed translocation occurs, and combined with the 5’ stem-loop refolding, we propose that the ribosome slips forward in the 3’ direction, allowing for the 5’ stem-loop to completely refold. This is consistent with the observation that the lower part of the secondary structure is important for bypassing (Nosek et al., 2015; Samatova et al., 2014). Simultaneously, the –UUCG– tetraloop becomes positioned such that it is able to refold. In one model this is within the A site of the ribosome. The folding of a tetraloop or hairpin within the A site is not without precedent: a crystal structure of the 70S ribosome showed the mRNA forming a hairpin with a 4 base pair stem and a tetraloop in the A-site, overlapping the natural codon-anticodon interaction region (Yusupova et al., 2001). Along similar lines, a previous model of bypassing suggested that the hairpin re-folds within the A site of the ribosome (Wills et al., 2008). Alternatively, the mRNA may be forced a short distance in the forward direction before the tetraloop hairpin forms, perhaps even in the ribosomal E site, with formation of the stem loop it nucleates enhancing forward mRNA to position the ribosome to a more 3’ position on the mRNA. Here, we propose that the tetraloop hairpin forms within the A site, though only the top base pairs of the stem are formed.
We further propose that the slip caused by the refolding of the 5’ stem-loop and tetraloop uncouples anticodon-codon interactions and translocation from ribosome reverse-rotation. This non-canonical conformation may be hyper-rotated (Qin et al., 2014) or represent a translocation intermediate (Tourigny et al., 2013). The rotated state, with its weakened ribosome-tRNA-mRNA interactions, is key to allowing the mRNA rearrangements that promote bypassing. This ribosome state with a hairpin within the A site may be unstable, and relaxation of this unstable state threads the mRNA in the 5’ direction. This forward bias is due to the 5’ stem-loop preventing backward movement (Figure 7).
The bypass begins and ends during the long rotated state pause, with the movement occurring in two steps. First, as soon as the tetraloop clears the ribosome on the 5’ side, the high tendency for the hairpin to refold may cause the mRNA to fold directionally in the 5’ direction and the hairpin to fold 5’ of the ribosome. This launches the ribosome forward towards the landing site. However, even with hairpin folding 5’-to the ribosome, the distance threaded is not sufficient to place the ribosome over the landing codon. Thus in the second step, as we have demonstrated, the ribosome scans a short distance to find the optimal landing codon, possibly with the aid of the internal Shine-Dalgarno sequence. This is consistent with the delay between mRNA rearrangement and resumption of translation as measured here.
A combination of mRNA rearrangement-induced movement with processive scanning builds upon and reconciles inconsistencies in earlier models of bypassing (Samatova et al., 2014; Wills et al., 2008). The model proposed here, although still speculative in some aspects, explains many outstanding questions and provides a testable model for future studies. In our model, the re-pairing of the peptidyl-tRNA to the correct position on the mRNA during scanning may be stabilized by the SD-like sequence or a possible downstream 3’ stem-loop (Samatova et al., 2014); the SD-like sequence has a moderate effect on bypassing but may be important for the fidelity of landing site selection (Herr et al., 2004; Wills et al., 2008). All of these events happen during the rotated state pause; the majority of the pause is the ribosome sampling and exploring the reading frame widely, with movements possibly similar to the excursions and sliding behaviors observed previously (Koutmou et al., 2015; Yan et al., 2015). In the mechanism proposed here, bypassing is not induced by A-site (UAG stop codon) starvation, explaining why the absence of RF1 did not significantly affect the bypassing efficiency (Herr et al., 2000b). Bypassing induced by A-site starvation may follow a different mechanism (Lindsley et al., 2005a; Lindsley et al., 2005b).
How does the ribosome resume translation? After successful landing and initial contact of the peptidyl-tRNA in the P site with the mRNA codon, the ribosome remains in a non-canonical rotated state with an exposed A site, similar to what was observed for frameshifting. The subsequent tRNA can bind to the ribosomal A site, which may help the ribosome re-define the correct reading frame. Peptidyl-transfer in this state is slow, since the rotated ribosome may not position the A and P-site tRNAs correctly for peptidyl transfer to occur efficiently. Subsequent to peptide bond formation, EF-G can then act on the ribosome and translocate the tRNAs, allowing for normal translation to resume. However, normal rates are not immediately resumed. The nascent peptide is major contributor to this slow down, suggesting that it still inhibiting subsequent peptidyl transfer and slowing non-rotated state lifetimes until the key sequences leave the ribosomal exit tunnel. This is consistent with the inference from mutagenesis experiments that the nascent peptide signal also has affects at the completion stage (Herr et al., 2000b).
Our results provide a glimpse of an unexpectedly versatile translation scheme with widespread implications. Bypassing may be more widespread than previously thought, suggesting that this phenomenon is not limited to gene 60 (Lang et al., 2014). Furthermore, the issue of a fidelity check may be significant for bypassing. Any mismatches upon codon-anticodon re-pairing during reading frame sampling before landing would not be susceptible to the fidelity controls governing proper mRNA decoding (Yan et al., 2015). Lastly, the mechanisms presented here may have parallel in eukaryotic scanning during initiation or other recoding events.
Here, we present a general mechanistic and conformational framework for ribosomal bypassing that may be applicable to different recoding signals. Many aspects of the framework are speculative and still require further investigation, especially the high-resolution structures of the many bypassing intermediates. Nonetheless, a long-lived, non-canonical translational state is the centerpiece of this mechanism, and provides a window for reading-frame reset through mRNA structure rearrangement. This state, whose formation is driven by mRNA and nascent chain energy barriers in bypassing, may be universal for many recoding events and possibly a central feature of translational control.
EXPERIMENTAL PROCEDURES
Reagents and buffers for translation experiments
Escherichia coli translation factors (IF2, EF-Tu, EF-G, EF-Ts) and initiator fMet-tRNA for the single-molecule experiments were prepared and purified as described before (Blanchard et al., 2004b; Marshall et al., 2008). Ribosomes purification, tRNA aminoacylation, preparation of biotinylated mRNA and in vivo bypass assays are described in Supplemental Experimental Procedures.
All experiments were conducted in a Tris-based polymix buffer consisting of 50 mM Tris-acetate (pH 7.5), 100 mM KCl, 5 mM ammonium acetate, 0.5 mM calcium acetate, 5 mM magnesium acetate, 0.5 mM EDTA, 5 mM putrescine-HCl and 1 mM spermidine. All single-molecule experiments had 4 mM GTP and were performed at 20°C.
Single-molecule profiling experiments
Translation experiments with ribosome Cy3B/BHQ conformational FRET are performed as described (Chen et al., 2014b). Before each experiment 30S (helix 44 mutant) and 50S (helix 101 mutant) ribosomal subunits (at 1 μM) were mixed in 1:1 ratio with the 3′ dye-labeled oligonucleotides specific for the hairpin extensions in each subunit for 37°C for 10 min and then at 30°C for 20 min in a polymix buffer system. 30S pre-initiation complexes (PICs) were formed as described (Marshall et al., 2008) by incubating the following at 37 °C for 5 minutes: 0.25 μM Cy3B-30S, pre-incubated with stoichiometric S1, 1 μM IF2, 1 μM fMet-tRNAfMet, 1 μM mRNA, and 4 mM GTP to form 30S PICs in the polymix buffer. Before use, the mRNA is heated to 90°C for 1 minute and then snap cooled to 4°C for 20 minutes to promote mRNA folding.
Before use, we pre-incubate a SMRT Cell V3 (Pacific Biosciences), a zero-mode waveguide (ZMW) chip, with a 1 mg/ml Neutravidin solution in 50 mM Tris-acetate pH 7.5 and 50 mM KCl at room temperature for 5 minutes. The cell is then washed with the Tris-based polymix buffer. After washing, 40 μl of the buffer is left in the cell to keep the cell surface wet. We then dilute the 30S PICs with polymix buffer containing 1 μM IF2 and 4 mM GTP down to 100 nM PIC concentration. A higher immobilization concentration compared to previous reports was used since not all the PICs will have an mRNA with a biotin (Chen et al., 2014b). The diluted PICs are loaded into the SMRT cell at room temperature for 3 min to immobilize the 30S PICs into the ZMW wells. We wash away unbound material with polymix buffer containing 1 μM IF2, 4 mM GTP, 2.5 mM Trolox, and a PCA/PCD oxygen scavenging system (2.5 mM 3,4-dihydroxybenzoic acid and 250 nM protocatechuate deoxygenase (Aitken et al., 2008)). After washing, 20 μl of the washing mix were added to the cell to keep the surface wet.
We formed ternary complexes (TCs) between total charged E. coli tRNAs and EF-Tu(GTP) as described (Marshall et al., 2008). Total or Δ(Phe) aminoacyl-tRNA·EF-Tu·GTP ternary complexes were pre-formed by incubating (2 min at 37C) the aa-tRNAs with five-fold excess of EF-Tu, GTP (1 mM), PEP (3 mM) and EF-Ts (40 μM) in polymix. The ternary complexes (3 ~ 6 μM) were added to BHQ-50S (200 nM), EF-G (240 ~ 480 nM), IF2 (1 μM), GTP (4 mM), 2.5 mM Trolox, and the oxygen scavenging system to form a delivery mix in polymix buffer. Experiments are done at 3 μM ternary complexes and 240 nM EF-G (chosen to have well-defined, detectable FRET transition signals), unless indicated otherwise. Before an experiment, the SMRT Cell is loaded into a modified PacBio RS sequencer. At the start of the elongation experiment, the instrument illuminates the SMRT cell with a green laser and then automatically delivers 20 μl of a delivery mixture onto the cell surface at t ~ 10 sec. Experiments involving labeled tRNAs and ribosome-mRNA FRET were performed similarly. See Supplemental Experimental Procedures.
ZMW instrumentation and data analysis
All single-molecule fluorescence experiments were conducted using a modified PacBio RS sequencer that allows the collection of single-molecule fluorescence from individual ZMW wells in 4 dye channels corresponding to Cy3, Cy3.5, Cy5, and Cy5.5(Chen et al., 2014a). The RS sequencer has 532 nm and 632 nm excitation lasers. In all experiments, data was collected at 10 frames per second (100 ms exposure time) for 10 minutes. The energy flux of the green laser is 0.32 μW/μm2 and the red laser is at 0.14 μW/μm2.
Data analyses for all experiments are conducted with MATLAB (MathWorks) scripts written in-house, see Supplemental Experimental Procedures. All error bars are standard error or measurement (s.e.m.).
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Gary Loughran for support. This work was supported by US National Institutes of Health (NIH) grant GM51266 to J.C. and J.D.P., by NIH grant GM099687 to A.P., S.E.O'L, and J.D.P., by NIH grant GM111858 to S.E.O'L, by a Stanford Interdisciplinary Graduate Fellowship to J.C. and by Science Foundation Ireland grants (SFI grant codes are 12/IP1492 and 13/1A/1853) to J.F.A.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS
J.C., J.F.A., and J.D.P. conceived and designed the experiments. J.C. performed and analyzed the single-molecule experiments. A.C. and M.O'C performed the in vivo experiments. J.C. wrote the manuscript with input from all the authors.
References
- Aitken CE, Marshall RA, Puglisi JD. An oxygen scavenging system for improvement of dye stability in single-molecule fluorescence experiments. Biophysical journal. 2008;94:1826–1835. doi: 10.1529/biophysj.107.117689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aitken CE, Puglisi JD. Following the intersubunit conformation of the ribosome during translation in real time. Nature structural & molecular biology. 2010;17:793–800. doi: 10.1038/nsmb.1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhushan S, Gartmann M, Halic M, Armache JP, Jarasch A, Mielke T, Berninghausen O, Wilson DN, Beckmann R. alpha-Helical nascent polypeptide chains visualized within distinct regions of the ribosomal exit tunnel. Nature structural & molecular biology. 2010;17:313–317. doi: 10.1038/nsmb.1756. [DOI] [PubMed] [Google Scholar]
- Blanchard SC, Gonzalez RL, Kim HD, Chu S, Puglisi JD. tRNA selection and kinetic proofreading in translation. Nature structural & molecular biology. 2004a;11:1008–1014. doi: 10.1038/nsmb831. [DOI] [PubMed] [Google Scholar]
- Blanchard SC, Kim HD, Gonzalez RL, Jr., Puglisi JD, Chu S. tRNA dynamics on the ribosome during translation. Proceedings of the National Academy of Sciences of the United States of America. 2004b;101:12893–12898. doi: 10.1073/pnas.0403884101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucklin DJ, Wills NM, Gesteland RF, Atkins JF. P-site pairing subtleties revealed by the effects of different tRNAs on programmed translational bypassing where anticodon re-pairing to mRNA is separated from dissociation. Journal of molecular biology. 2005;345:39–49. doi: 10.1016/j.jmb.2004.10.037. [DOI] [PubMed] [Google Scholar]
- Caliskan N, Katunin VI, Belardinelli R, Peske F, Rodnina MV. Programmed -1 frameshifting by kinetic partitioning during impeded translocation. Cell. 2014;157:1619–1631. doi: 10.1016/j.cell.2014.04.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Dalal RV, Petrov AN, Tsai A, O'Leary SE, Chapin K, Cheng J, Ewan M, Hsiung PL, Lundquist P, et al. High-throughput platform for real-time monitoring of biological processes by multicolor single-molecule fluorescence. Proceedings of the National Academy of Sciences of the United States of America. 2014a;111:664–669. doi: 10.1073/pnas.1315735111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Petrov A, Johansson M, Tsai A, O'Leary SE, Puglisi JD. Dynamic pathways of -1 translational frameshifting. Nature. 2014b;512:328–332. doi: 10.1038/nature13428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Petrov A, Tsai A, O'Leary SE, Puglisi JD. Coordinated conformational and compositional dynamics drive ribosome translocation. Nature structural & molecular biology. 2013;20:718–727. doi: 10.1038/nsmb.2567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Tsai A, O'Leary SE, Petrov A, Puglisi JD. Unraveling the dynamics of ribosome translocation. Curr Opin Struct Biol. 2012a;22:804–814. doi: 10.1016/j.sbi.2012.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Tsai A, Petrov A, Puglisi JD. Nonfluorescent quenchers to correlate single-molecule conformational and compositional dynamics. Journal of the American Chemical Society. 2012b;134:5734–5737. doi: 10.1021/ja2119964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornish PV, Ermolenko DN, Noller HF, Ha T. Spontaneous intersubunit rotation in single ribosomes. Molecular cell. 2008;30:578–588. doi: 10.1016/j.molcel.2008.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunkle JA, Dunham CM. Mechanisms of mRNA frame maintenance and its subversion during translation of the genetic code. Biochimie. 2015 doi: 10.1016/j.biochi.2015.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ennifar E, Nikulin A, Tishchenko S, Serganov A, Nevskaya N, Garber M, Ehresmann B, Ehresmann C, Nikonov S, Dumas P. The crystal structure of UUCG tetraloop. Journal of molecular biology. 2000;304:35–42. doi: 10.1006/jmbi.2000.4204. [DOI] [PubMed] [Google Scholar]
- Goldman DH, Kaiser CM, Milin A, Righini M, Tinoco I, Jr., Bustamante C. Ribosome. Mechanical force releases nascent chain-mediated ribosome arrest in vitro and in vivo. Science. 2015;348:457–460. doi: 10.1126/science.1261909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen TM, Baranov PV, Ivanov IP, Gesteland RF, Atkins JF. Maintenance of the correct open reading frame by the ribosome. EMBO reports. 2003;4:499–504. doi: 10.1038/sj.embor.embor825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herr AJ, Atkins JF, Gesteland RF. Mutations which alter the elbow region of tRNA2Gly reduce T4 gene 60 translational bypassing efficiency. The EMBO journal. 1999;18:2886–2896. doi: 10.1093/emboj/18.10.2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herr AJ, Atkins JF, Gesteland RF. Coupling of open reading frames by translational bypassing. Annual review of biochemistry. 2000a;69:343–372. doi: 10.1146/annurev.biochem.69.1.343. [DOI] [PubMed] [Google Scholar]
- Herr AJ, Gesteland RF, Atkins JF. One protein from two open reading frames: mechanism of a 50 nt translational bypass. The EMBO journal. 2000b;19:2671–2680. doi: 10.1093/emboj/19.11.2671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herr AJ, Wills NM, Nelson CC, Gesteland RF, Atkins JF. Drop-off during ribosome hopping. Journal of molecular biology. 2001;311:445–452. doi: 10.1006/jmbi.2001.4899. [DOI] [PubMed] [Google Scholar]
- Herr AJ, Wills NM, Nelson CC, Gesteland RF, Atkins JF. Factors that influence selection of coding resumption sites in translational bypassing: minimal conventional peptidyl-tRNA:mRNA pairing can suffice. The Journal of biological chemistry. 2004;279:11081–11087. doi: 10.1074/jbc.M311491200. [DOI] [PubMed] [Google Scholar]
- Huang WM, Ao SZ, Casjens S, Orlandi R, Zeikus R, Weiss R, Winge D, Fang M. A persistent untranslated sequence within bacteriophage T4 DNA topoisomerase gene 60. Science. 1988;239:1005–1012. doi: 10.1126/science.2830666. [DOI] [PubMed] [Google Scholar]
- Jenner LB, Demeshkina N, Yusupova G, Yusupov M. Structural aspects of messenger RNA reading frame maintenance by the ribosome. Nature structural & molecular biology. 2010;17:555–560. doi: 10.1038/nsmb.1790. [DOI] [PubMed] [Google Scholar]
- Johansson M, Chen J, Tsai A, Kornberg G, Puglisi JD. Sequence-dependent elongation dynamics on macrolide-bound ribosomes. Cell Rep. 2014;7:1534–1546. doi: 10.1016/j.celrep.2014.04.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koutmou KS, Schuller AP, Brunelle JL, Radhakrishnan A, Djuranovic S, Green R. Ribosomes slide on lysine-encoding homopolymeric A stretches. eLife. 2015;4 doi: 10.7554/eLife.05534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang BF, Jakubkova M, Hegedusova E, Daoud R, Forget L, Brejova B, Vinar T, Kosa P, Fricova D, Nebohacova M, et al. Massive programmed translational jumping in mitochondria. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:5926–5931. doi: 10.1073/pnas.1322190111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsley D, Bonthuis P, Gallant J, Tofoleanu T, Elf J, Ehrenberg M. Ribosome bypassing at serine codons as a test of the model of selective transfer RNA charging. EMBO reports. 2005a;6:147–150. doi: 10.1038/sj.embor.7400332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsley D, Gallant J, Doneanu C, Bonthuis P, Caldwell S, Fontelera A. Spontaneous ribosome bypassing in growing cells. Journal of molecular biology. 2005b;349:261–272. doi: 10.1016/j.jmb.2005.03.031. [DOI] [PubMed] [Google Scholar]
- Liu CY, Qureshi MT, Lee TH. Interaction strengths between the ribosome and tRNA at various steps of translocation. Biophysical journal. 2011;100:2201–2208. doi: 10.1016/j.bpj.2011.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maldonado R, Herr AJ. Efficiency of T4 gene 60 translational bypassing. Journal of bacteriology. 1998;180:1822–1830. doi: 10.1128/jb.180.7.1822-1830.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marquez V, Wilson DN, Tate WP, Triana-Alonso F, Nierhaus KH. Maintaining the ribosomal reading frame: the influence of the E site during translational regulation of release factor 2. Cell. 2004;118:45–55. doi: 10.1016/j.cell.2004.06.012. [DOI] [PubMed] [Google Scholar]
- Marshall RA, Dorywalska M, Puglisi JD. Irreversible chemical steps control intersubunit dynamics during translation. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:15364–15369. doi: 10.1073/pnas.0805299105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatogawa H, Ito K. The ribosomal exit tunnel functions as a discriminating gate. Cell. 2002;108:629–636. doi: 10.1016/s0092-8674(02)00649-9. [DOI] [PubMed] [Google Scholar]
- Nilsson OB, Hedman R, Marino J, Wickles S, Bischoff L, Johansson M, Muller-Lucks A, Trovato F, Puglisi JD, O'Brien EP, et al. Cotranslational Protein Folding inside the Ribosome Exit Tunnel. Cell Rep. 2015 doi: 10.1016/j.celrep.2015.07.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nosek J, Tomaska L, Burger G, Lang BF. Programmed translational bypassing elements in mitochondria: structure, mobility, and evolutionary origin. Trends in genetics : TIG. 2015;31:187–194. doi: 10.1016/j.tig.2015.02.010. [DOI] [PubMed] [Google Scholar]
- Qin P, Yu D, Zuo X, Cornish PV. Structured mRNA induces the ribosome into a hyper-rotated state. EMBO reports. 2014;15:185–190. doi: 10.1002/embr.201337762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu X, Wen JD, Lancaster L, Noller HF, Bustamante C, Tinoco I., Jr. The ribosome uses two active mechanisms to unwind messenger RNA during translation. Nature. 2011;475:118–121. doi: 10.1038/nature10126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samatova E, Konevega AL, Wills NM, Atkins JF, Rodnina MV. High-efficiency translational bypassing of non-coding nucleotides specified by mRNA structure and nascent peptide. Nature communications. 2014;5:4459. doi: 10.1038/ncomms5459. [DOI] [PubMed] [Google Scholar]
- Tinoco I, Jr., Kim HK, Yan S. Frameshifting dynamics. Biopolymers. 2013;99:1147–1166. doi: 10.1002/bip.22293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todd GC, Walter NG. Secondary structure of bacteriophage T4 gene 60 mRNA: implications for translational bypassing. RNA. 2013;19:685–700. doi: 10.1261/rna.037291.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tourigny DS, Fernandez IS, Kelley AC, Ramakrishnan V. Elongation factor G bound to the ribosome in an intermediate state of translocation. Science. 2013;340:1235490. doi: 10.1126/science.1235490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai A, Kornberg G, Johansson M, Chen J, Puglisi JD. The dynamics of SecM-induced translational stalling. Cell Rep. 2014;7:1521–1533. doi: 10.1016/j.celrep.2014.04.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valle M, Zavialov A, Sengupta J, Rawat U, Ehrenberg M, Frank J. Locking and unlocking of ribosomal motions. Cell. 2003;114:123–134. doi: 10.1016/s0092-8674(03)00476-8. [DOI] [PubMed] [Google Scholar]
- Weiss RB, Huang WM, Dunn DM. A nascent peptide is required for ribosomal bypass of the coding gap in bacteriophage T4 gene 60. Cell. 1990;62:117–126. doi: 10.1016/0092-8674(90)90245-A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wills NM. Translational Bypassing - Peptidyl-tRNA Re-pairing at Non-overlapping Sites. Nucleic Acids Mol Bi. 2010;24:365–381. [Google Scholar]
- Wills NM, O'Connor M, Nelson CC, Rettberg CC, Huang WM, Gesteland RF, Atkins JF. Translational bypassing without peptidyl-tRNA anticodon scanning of coding gap mRNA. The EMBO journal. 2008;27:2533–2544. doi: 10.1038/emboj.2008.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan S, Wen JD, Bustamante C, Tinoco I., Jr. Ribosome Excursions during mRNA Translocation Mediate Broad Branching of Frameshift Pathways. Cell. 2015;160:870–881. doi: 10.1016/j.cell.2015.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yusupova GZ, Yusupov MM, Cate JH, Noller HF. The path of messenger RNA through the ribosome. Cell. 2001;106:233–241. doi: 10.1016/s0092-8674(01)00435-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.