Rigoni et al. report that hypomorphic Rag2R229Q mutation is associated with altered microbiota composition and defects in the gut–blood barrier and suggest that intestinal microbes may play a critical role in the distinctive immune dysregulation of Omenn syndrome.
Abstract
Omenn syndrome (OS) is caused by hypomorphic Rag mutations and characterized by a profound immunodeficiency associated with autoimmune-like manifestations. Both in humans and mice, OS is mediated by oligoclonal activated T and B cells. The role of microbial signals in disease pathogenesis is debated. Here, we show that Rag2R229Q knock-in mice developed an inflammatory bowel disease affecting both the small bowel and colon. Lymphocytes were sufficient for disease induction, as intestinal CD4 T cells with a Th1/Th17 phenotype reproduced the pathological picture when transplanted into immunocompromised hosts. Moreover, oral tolerance was impaired in Rag2R229Q mice, and transfer of wild-type (WT) regulatory T cells ameliorated bowel inflammation. Mucosal immunoglobulin A (IgA) deficiency in the gut resulted in enhanced absorption of microbial products and altered composition of commensal communities. The Rag2R229Q microbiota further contributed to the immunopathology because its transplant into WT recipients promoted Th1/Th17 immune response. Consistently, long-term dosing of broad-spectrum antibiotics (ABXs) in Rag2R229Q mice ameliorated intestinal and systemic autoimmunity by diminishing the frequency of mucosal and circulating gut-tropic CCR9+ Th1 and Th17 T cells. Remarkably, serum hyper-IgE, a hallmark of the disease, was also normalized by ABX treatment. These results indicate that intestinal microbes may play a critical role in the distinctive immune dysregulation of OS.
The immune system plays a fundamental role in the maintenance of a mutualistic relationship between host and intestinal microbiota (Hooper and Macpherson, 2010). The development and maturation of the gut immune system depends on these microorganisms (Smith et al., 2007), and the composition of microbiota, in turn, plays a critical role in the regulation of immune system activation in the gut. For example, a lack of regulatory T (T reg) cell induction results in excessive adaptive immune responses to gut microbial antigens and intestinal inflammation (Cong et al., 2002; Lodes et al., 2004). Moreover, intestinal bacteria shape host systemic immune responses by conditioning both pro- and antiinflammatory T cell populations (Gaboriau-Routhiau et al., 2009; Ivanov et al., 2009; Atarashi et al., 2011; Round et al., 2011). Homeostatic T cell proliferation is driven by the microbial flora or their penetrant molecules (Kieper et al., 2005), and this expansion of the T cell compartment can be important in the pathogenesis of autoimmune diseases (King et al., 2004; Milner et al., 2007; Chang et al., 2008).
Hypomorphic mutations in Rag genes result in immunodeficiency associated with autoimmune-like manifestations in both humans and mice (Villa et al., 1998; Khiong et al., 2007; Marrella et al., 2007). The disease, known as Omenn syndrome (OS), is characterized by homeostatically proliferating self-reactive T and B cells with a limited receptor repertoire generated by the residual recombination activity (Rieux-Laucat et al., 1998; Signorini et al., 1999). Moreover, poor generation of thymic Foxp3+ cells and functional impairments in the peripheral T regulatory compartment have been reported in OS patients (Poliani et al., 2009; Cassani et al., 2010b) and in the murine model (Marrella et al., 2007), indicating that a break in immune tolerance contributes to the development of autoimmunity in OS. The symptoms are very similar to graft-versus-host disease, as inflammatory reactions particularly involve the environmental interfaces such as the skin and gut, leading to distinctive early onset erythroderma and protracted diarrhea. Infiltration in other organs such as the kidney and liver is also reported, and other features include eosinophilia, extremely elevated serum IgE levels and hypogammaglobulinaemia, susceptibility to infections, and failure to thrive (Omenn, 1965; Ochs et al., 1974). The disease is rapidly fatal unless treated by allogeneic bone marrow transplantation (de la Morena and Nelson, 2014). Interestingly, the clinical and immunological spectrum of OS presentation is extremely broad. In fact, the same mutation or different mutations affecting the same codon can manifest with different phenotypes, ranging from leaky to full-blown forms of severe combined immunodeficiency with severe autoimmunity, even in the same family (Marrella et al., 2011). The underlying causes are largely unknown, but epigenetic and environmental factors have been considered. A role for microbial flora in the disease pathogenesis is suggested by the peculiar pathological involvement of the mucosal interfaces. However, whether chronic immune inflammation and autoimmune-like disease in OS is mediated by faults in the establishment of intestinal tolerance is unknown.
We found that hypomorphic Rag2R229Q mutation is associated with altered microbiota composition and defects in the gut–blood barrier, leading to enhanced systemic translocation of microbial products. Decreasing bacterial load in Rag2R229Q mice with long-term dosing of antibiotics (ABXs) reduced local and circulating proinflammatory Th1 and Th17 T cell populations, visibly ameliorated both intestinal and systemic autoimmunity, and normalized serum hyper-IgE. Our results suggest that gut microbial flora play a crucial role in the pathogenesis of OS.
RESULTS
Rag2R229Q mice develop an inflammatory bowel disease(IBD) affecting both small and large intestines
Analysis of intestinal pathology in Rag2R229Q/R229Q (herein referred to as Rag2R229Q) mice revealed different degrees of spontaneous colitis, wasting diarrhea, and rectal prolapse. The overall incidence of rectal prolapse in the Rag2R229Q colony was 5% in mice by 24 wk of age. No rectal prolapse was detected in Rag2+/+ littermates. Nonetheless, 70–80% of all mutant mice showed substantial thickening of the colon throughout their entire length and microscopic evidence of inflammation (Fig. 1 A). Histologically, colonic inflammation is characterized by crypt elongation, epithelial hyperplasia, and a large inflammatory cell infiltrate extending to the colonic lamina propria (LP), with occasional crypt abscesses. Remarkably, such morphological changes also affected the small intestine (SI) of the mutant mice with increased mucosal thickness and histological score and lower villus/crypt ratio (Fig. 1, B and C).
Figure 1.
Characterization of intestinal inflammation in Rag2R229Q mice. (A and B) Representative colon (A) and SI (B) sections from Rag2+/+ and Rag2R229Q mice (8–12 wk old) stained with H&E and CD3 immunostaining. Histograms show the inflammation score in the colon (A) and SI (B) of n = 14–17 mice/group from four experiments. (C) Quantification of villous/crypt ratio of ileum from Rag2+/+ and Rag2R229Q mice (n = 10 from three experiments). (D) Representative FACS plots showing the CD4+ and CD8+ T subsets within the CD3+ population from SI LP of Rag2+/+ and Rag2R229Q mice. (E and F) Representative FACS plots, cumulative frequencies, and absolute numbers of SI (E) and colonic (F) LP CD4+IL-17+ (Th17), CD4+IFN-γ+ (Th1), and CD4+IL-17+IFN-γ+ (Th17/Th1) T cells from Rag2+/+ and Rag2R229Q mice. The numbers in the plots indicate the frequency of cells in each quadrant. Results are mean ± SEM. Cumulative results of three independent experiments are shown (n = 11–20 mice/group). (G) Gene expression analysis of cytokines in the ileal and colonic tissues of mice. Target mRNA was normalized to Actb mRNA. RNA contents are shown as arbitrary units (A.U.). Data are representative results of three independent experiments with at least five mice per group. Bars, 100 µm. Values are mean ± SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
LP infiltrate consisted mainly of T lymphocytes (Fig. 1, A and B; and Fig. S1), particularly CD4+, with very few CD8+ T cells (Fig. 1 D). Correlating with mucosal thickening, absolute counts were three- to sixfold higher in the mutant mice than controls, in sharp contrast with the lymphoid depletion observed in the peripheral lymphoid organs. The proportion of intestinal Th17 cells was significantly higher in Rag2R229Q mice than Rag2+/+ mice, and mutants also had more IFN-γ–producing LP T cells compared with controls (Fig. 1, E and F). Furthermore, IFN-γ IL-17 double-producing T cells, frequently identified in the inflamed LP of patients and murine models of IBD (Annunziato et al., 2007; Ahern et al., 2010), were abundant in Rag2R229Q mice (Fig. 1, E and F). To comprehensively characterize the gut inflammatory environment in the Rag2R229Q mice, we measured the expression of several cytokines. Transcript levels of IFN-γ, IL-17A, IL-6, IL-2, IL-1β, IL-22, and TNF were markedly up-regulated in the Rag2R229Q ileum and colonic tissues (Fig. 1 G). In contrast, IL-10 and IL-12p35 levels were significantly reduced, whereas levels of IL-4, TGF-β, and IL-23p19 were not markedly different (Fig. 1 G). Overall, these results indicate that in Rag2R229Q mice, gut inflammation is mediated by Th1 and Th17 immune responses.
Hypomorphic CD4 T cells are sufficient to induce colitis
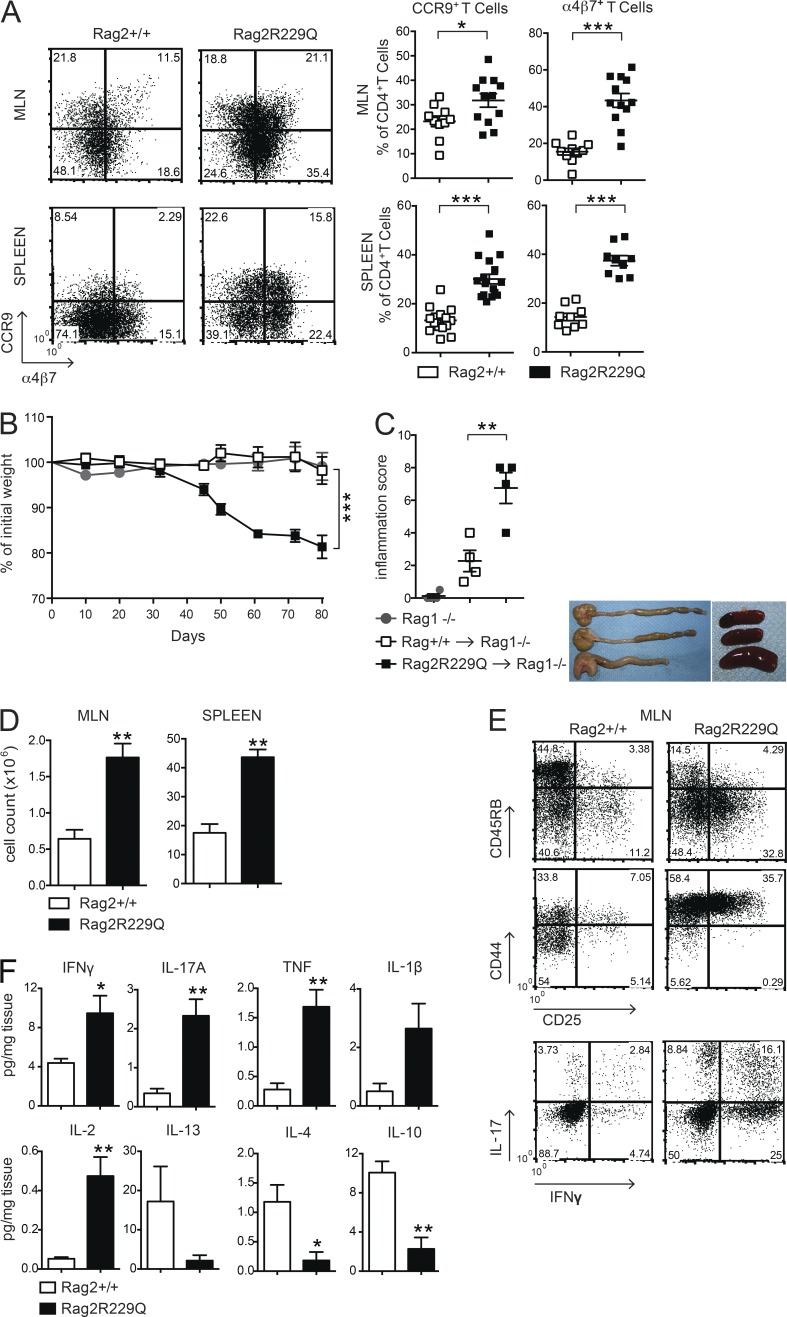
CD4 T cells from mesenteric LNs (MLNs) of Rag2R229Q mice showed an activated memory phenotype (CD44hiCD62L−) and expressed the gut-homing receptors CCR9 and α4β7 (Mora and Von Andrian, 2006) at higher frequency compared with the same T cell population from Rag2+/+ mice (Fig. 2 A). Of note, we detected CCR9 and α4β7 expression in the splenic T effector cell compartment (Fig. 2 A), suggesting that gut inflammation resulted in the circulation of cells activated within the intestine of Rag2R229Q mice. In this line, CCR9+ T lymphocytes are increased in the circulation of IBD patients (Papadakis et al., 2001).
Figure 2.
Rag2R229Q CD4 T cells have enhanced gut tropism and cause colitis. (A) Representative FACS plots showing the expression of gut homing receptors, CCR9 and α4β7, on gated CD44hiCD62L−CD4+ T cells from MLNs and spleen of Rag2+/+ and Rag2R229Q mice. Numbers in quadrants indicate the percentage of cells in each. Graphs show cumulative frequencies from one representative experiment out of nine independent with n = 9–16 mice/group. (B) Body weight as a percentage of starting weight of Rag1−/− recipient mice after transfer of total MLNs CD4+ T cells from Rag2+/+ or Rag2R229Q mice. (C, left) Graph shows the colon inflammation scores of Rag1−/− mice receiving Rag2+/+ or Rag2R229Q MLNs CD4+ T cells. Right: Representative picture of macroscopic observation in colons and spleens. (D) Total cellularity of MLNs and spleens from Rag1−/− mice receiving Rag2+/+ or Rag2R229Q MLN CD4+ T cells. (E) Representative FACS plots show immunophenotype and cytokine production by CD4+ T cells recovered from MLNs of recipient mice. (F) Cytokine profile in the supernatants from cultured colonic tissue of Rag1−/− mice adoptively transferred with Rag2+/+ or Rag2R229Q CD4+ T cells. Data from B–F are representative of two independent experiments with at least n = 4 animals. Values are mean ± SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
To determine whether Rag2R229Q CD4 T cells were sufficient to cause intestinal inflammation, total CD4+ T cells derived from either Rag2+/+ or Rag2R229Q MLNs were transferred into Rag1−/− recipient mice. Analysis 10 wk afterward revealed signs of colitis in all recipients of mutant CD4+ T cells. Rag2R229Q chimeric Rag1−/− mice showed significantly greater weight loss, colon shortening, and inflammatory scores than Rag1−/− mice receiving Rag2+/+ CD4+ T cells (Fig. 2, B and C). Furthermore, MLNs and spleens in recipients of Rag2R229Q T cells were enlarged (Fig. 2, C and D). Immunophenotype analysis revealed a marked expansion of activated T cells (CD44hi, CD45RBlow, and CD25+) in the MLN (Fig. 2 E) with a Th1/Th17 phenotype, similar to that observed in the Rag2R229Q mice at steady state (Fig. 2 E). When we examined the cytokine profile in the supernatants from cultured colonic tissue of transplanted mice, we found that the intestinal inflammatory response in the Rag2R229Q chimeric Rag1−/− mice is an exacerbated form of the disease described in Rag2R229Q mice. Indeed, culture supernatants from the colon of Rag2R229Q chimeras contained two- to fivefold increased levels of IL-2, IFN-γ, IL-17, and TNF than supernatants of corresponding cultures from Rag1−/− mice transferred with Rag2+/+ cells (Fig. 2 F). On the contrary, IL-4 and IL-10 levels were significantly reduced (Fig. 2 F). Overall, these results indicate that Rag2R229Q CD4 T cells are able to transfer the disease into immunodeficient hosts.
Oral tolerance is impaired in Rag2R229Q mice
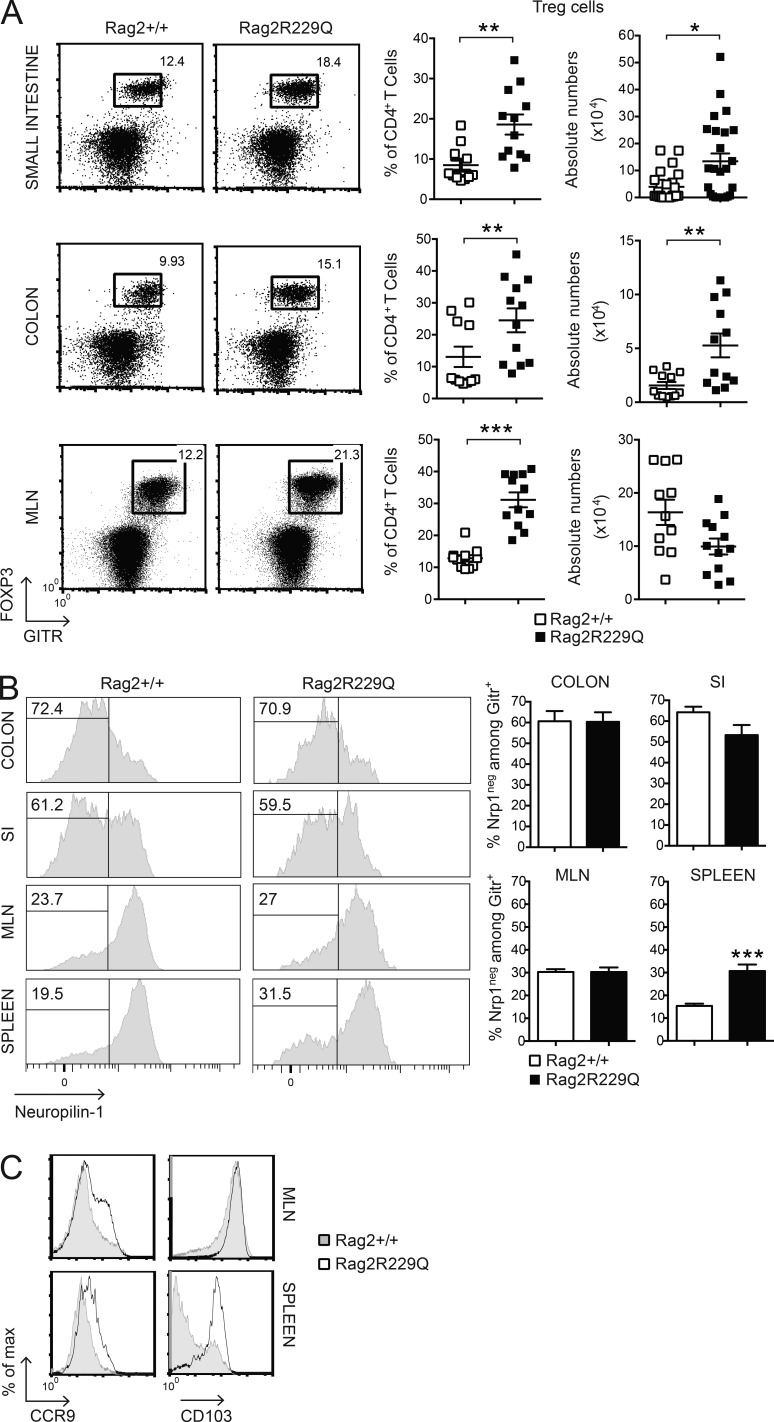
We found that Foxp3+GITR+ T reg cells were abundantly present in the inflamed intestine of Rag2R229Q mice. In fact, a significant increase in the frequency and number of T reg cells was detected in the SI and colonic LP (Fig. 3 A). In the MLNs, T reg cells were augmented only in frequency (Fig. 3 A), suggesting that T reg cell expansion is unique to the LP compartment. The gut mucosa is considered a primary site for extrathymic T reg cell generation. We therefore analyzed the expression of Neuropilin-1 (Nrp-1) as a marker of inducible T reg (iT reg) cells (Weiss et al., 2012; Yadav et al., 2012) in the gut and in secondary lymphoid organs of Rag2R229Q mice.
Figure 3.
Phenotype of T reg cells in Rag2R229Q mice. (A) Frequency and absolute number of CD4+Foxp3+GITR+ (T reg) T cells infiltrating the SI, colonic LP, and MLNs of Rag2+/+ and Rag2R229Q mice (n = 11–25 from three independent experiments). FACS plots show the coexpression of GITR+ and FOXP3+ gated on CD4+ T cells. (B) Representative FACS analysis of Nrp-1 expression by CD4+GITR+ T cells in the intestines and secondary lymphoid organs of Rag2+/+ and Rag2R229Q mice. Histograms show cumulative data from two experiments (n = 7–19). The numbers in the histograms indicate the frequency of Nrp-1− cells (iT reg cells). (C) Representative histogram plots depicting the expression of CCR9 and CD103 markers on gated CD4+GITR+FOXP3+ from MLNs and spleens of Rag2+/+ and Rag2R229Q mice. Values are mean ± SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
The percentages of Nrp-1low (iT reg) cells among GITR+ T reg cells were comparable between mutant and WT mice in both small and large intestines and higher than in MLNs (Fig. 3 B). The analysis of the spleens revealed that 30% of peripheral T reg cells in Rag2R229Q mice were constituted by iT reg cells, a significantly higher fraction than in control mice (Fig. 3 B). This result could reflect the poor generation of natural T reg cells in the thymus of mutant mice (Marrella et al., 2007). Alternatively, iT reg cell maintenance could be favored in the periphery under homeostatic conditions.
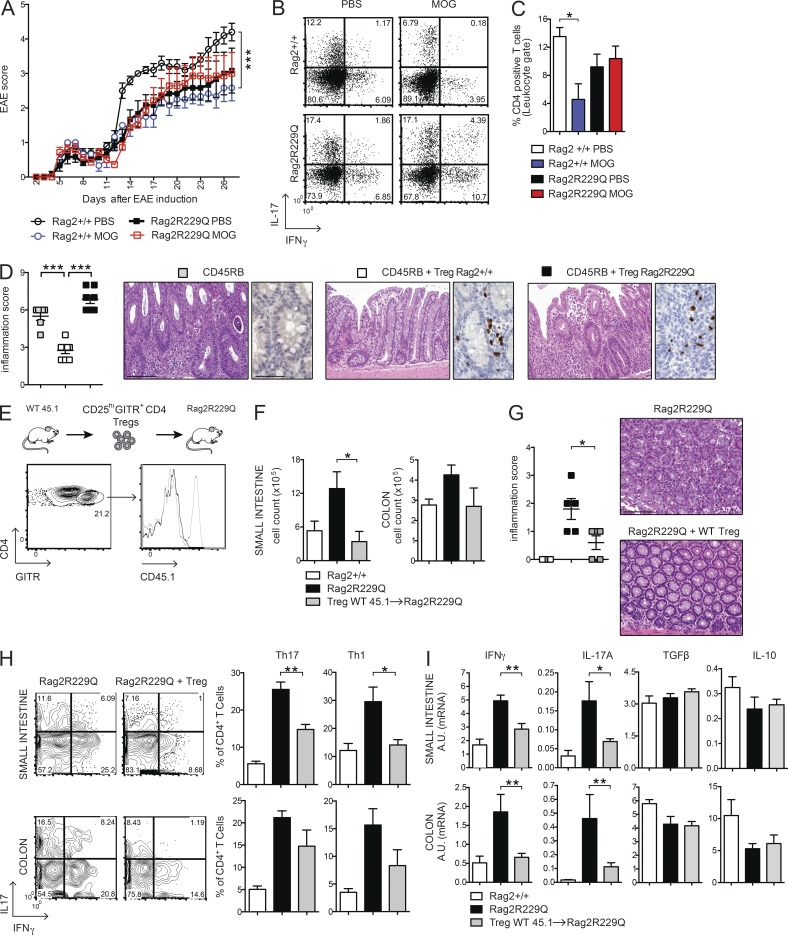
Mutant T reg cells from MLNs expressed the CD103 marker, indicative of antigen-specific expansion and differentiation, similar to control cells, but displayed enhanced expression of the CCR9 receptor (Fig. 3 C). The analogous effector/memory–like phenotype was observed in the peripheral T reg cell pool of Rag2R229Q but not of Rag2+/+ mice, and a major fraction (30–55% vs. 9–15% in the Rag2+/+ mice; P < 0.05) of these cells retained the CCR9 expression (Fig. 3 C), indicative of their gut tropism. Hence, T reg cells accumulate at the intestinal mucosal areas, although they fail to control the disease in Rag2R229Q mice. Thus, we assessed the capacity of Rag2R229Q mice to generate functionally competent T reg cells by the induction of oral tolerance (Dubois et al., 2009; Cassani et al., 2011). We used the well-established model of prevention of experimental allergic encephalomyelitis (EAE) that can be generated by orally supplementing mice with a peptide derived from myelin oligodendrocyte glycoprotein (MOG35–55; Chen et al., 1994). EAE developed in the Rag2R229Q mice supplemented orally with PBS, albeit at a lower extent than Rag2+/+ mice (Fig. 4 A). However, orally administered MOG35–55 prevented EAE only in Rag2+/+ but not in Rag2R229Q mice (Fig. 4 A). Th17 and Th1 cells are involved in EAE pathogenesis, and oral tolerance induction abrogates the generation of these pro-inflammatory T cells (Peron et al., 2010). Consistently, MOG-tolerized Rag2+/+ mice showed lower frequencies of IL-17– and IFN-γ–producing cells compared with nontolerized controls (Fig. 4 B). On the contrary, mutant mice did not show a decrease in the proportions of Th17 or Th1 cells upon oral MOG35–55 administration. In agreement, brain lymphocyte infiltration was reduced in the tolerized Rag2+/+ but not in the mutant animals (Fig. 4 C). These results suggested that Rag2R229Q iT reg cells could not control autoimmunity in this setting. To substantiate the hypothesis of functional impairment of Rag2R229Q T reg cells, we assessed their immunosuppressive potential in the severe combined immunodeficiency model of colitis induced by CD4+ CD45RBhi T cells (Powrie et al., 1994). Mice were sacrificed at week four after cell transfer, and IBD was assessed by weight loss (IBD mice: 21.1% ± 4.5; Rag2R229Q mice: 21.1% ± 2.6; WT mice: 12.6% ± 4.6; n = 6–8) and histological examination of the colon. The disease severity was significantly attenuated when naive CD4 T cells were cotransferred with Rag2+/+ T reg cells (Fig. 4 D). On the contrary, Rag2R229Q T reg cells failed to protect lymphopenic mice from colitis development, despite the fact that they were readily detected in the gut tissues as in control mice (Fig. 4 D). In summary, these results demonstrate that T reg cell functionality is compromised in Rag2R229Q mice.
Figure 4.
Oral tolerance is impaired in Rag2R229Q mice, and transfer of WT T reg cells attenuates gut inflammation. (A) EAE model of oral tolerance in Rag2R229Q mice. EAE induction and progression was scored from days 0 to 26 in mice orally administered with MOG35–55 or PBS. (B) Cytokine production profile of CD4+ T cells. At day 26, the mice were sacrificed, and total splenocytes were stimulated ex vivo with MOG35–55. The numbers in the plots indicate the frequency of cells in each quadrant. (C) Frequency of CD4+ T cells infiltrating the brain of Rag2+/+ and Rag2R229Q mice. Data are shown from one representative experiment out of two (n = 6–7 mice/group). (D) Colitis score and representative colonic sections from Rag1−/− transferred with CD45RB alone or with T reg cells from Rag2+/+ and Rag2R229Q mice stained with H&E and CD3 immunostaining. Cumulative score results of two independent experiments are shown (n = 6–8 mice/group). (E) WT CD45.1+CD25hiGITR+CD4+ T reg cells were adoptively transferred into Rag2R229Q mice. Histogram plot show the CD45.1 expression on GITR+CD4+ T cells in the LP of recipient Rag2R229Q mice. (F) Total cell counts from SI and colonic LP. (G) Representative colonic sections stained with H&E and colitis score in Rag2R229Q mice recipient mice and controls (n = 5 from two experiments). (H) Representative FACS plots and frequency of LP Th17 and Th1 cells from SI and colon. (I) SI and colonic tissue expression of cytokines. RNA contents are shown as arbitrary units (A.U.). Data shown are cumulative results from two independent experiments with nine mice per group. Bars, 100 µm. Values are mean ± SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Adoptive transfer of WT T reg cells attenuates gut inflammation in Rag2R229Q mice
To address whether the intestinal inflammation in Rag2R229Q is a consequence of T reg cell functional impairment, we transferred sorted splenic CD25hiGITR+CD4+ T reg cells from Rag2+/+ (CD45.1) donors into Rag2R229Q (CD45.2) recipients and followed them for 4 wk. Transferred T reg cells expanded well and were readily detectable in the gut LP of Rag2R229Q mice (Fig. 4 E), indicating that T reg cells efficiently colonized the inflamed intestine. In the T reg cells transfer group, lymphocytic infiltration decreased both in the SI and colon (Fig. 4 F), resulting in a reduced inflammatory score (Fig. 4 G). Accordingly, Th1 and Th17 cell populations lowered in the gut of Rag2R229Q mice receiving Rag2+/+ T reg cells (Fig. 4, H and I). However, production of IL-10 and TGF-β did not change (Fig. 4 I). Overall, these data confirm that impaired T reg cell function plays a role in the development of intestinal inflammation in Rag2R229Q mice.
Mucosal B cell deficiency in Rag2R229Q mice causes enhanced bacterial translocation and altered microbiota composition
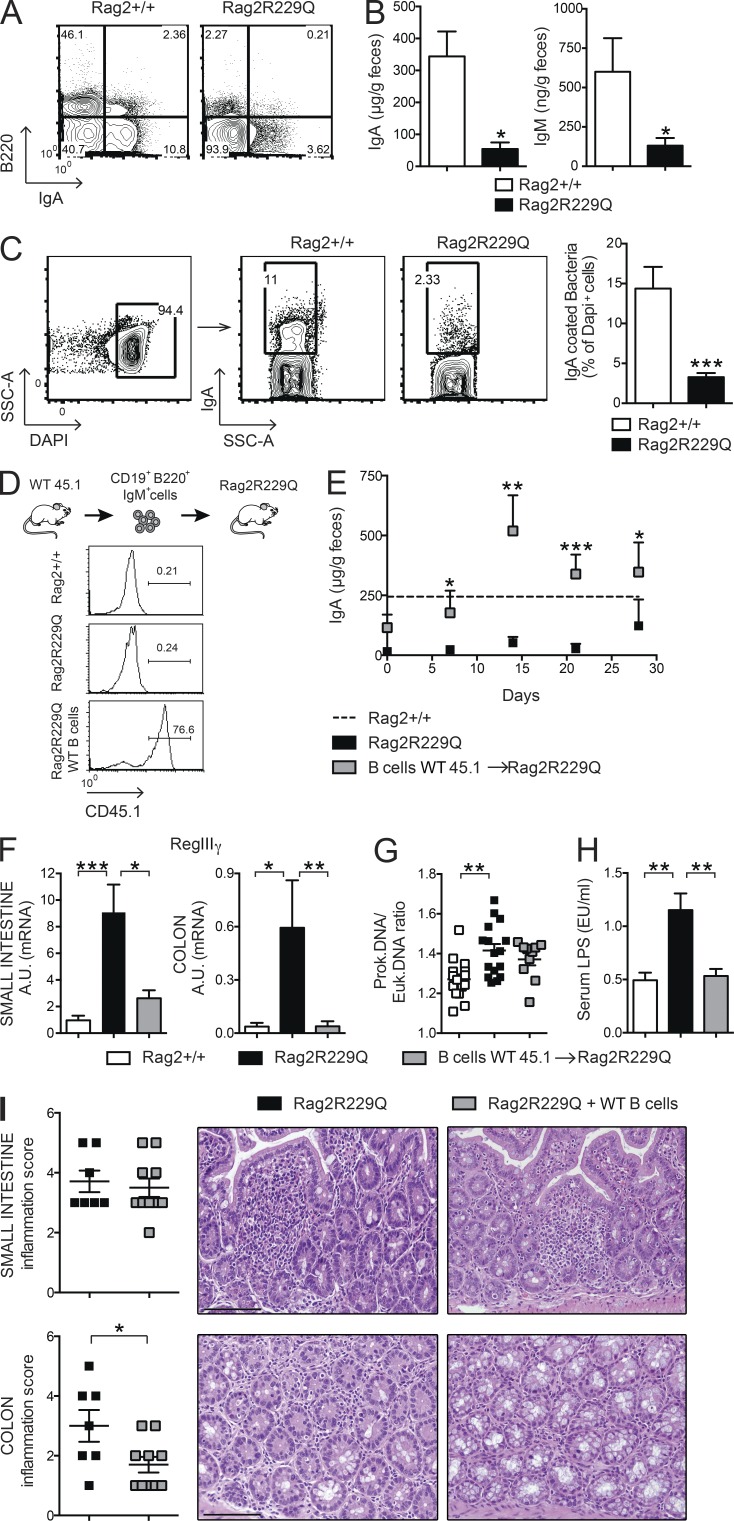
Intestinal B cells are poorly represented in Rag2R229Q mice. Peyer’s patches (PPs) are rudimentary, and few LP B220+ cells and IgA+ plasma cells could be detected (Fig. 5 A). In agreement, fecal IgA and IgM levels were markedly diminished (Fig. 5 B), indicating a general deficiency in B cell function at the intestinal mucosal interface. As a consequence, a significant decrease of IgA-coated bacteria was detected in the large intestine of mutant mice (Fig. 5 C). Mucosal IgA operates immune exclusion of potentially harmful antigens and pathogens and contains the gut microbiota (Macpherson et al., 2000; Macpherson and Uhr, 2004; Palm et al., 2014). Lack of IgA results in augmented serum LPS levels (Shulzhenko et al., 2011). Quantification of bacteria adherent to the ileal mucosa by real-time PCR (RT-PCR; Proietti et al., 2014) showed a higher prokaryotic/eukaryotic DNA ratio in Rag2R229Q mice (Fig. 5 G) associated with increased serum LPS concentrations (Fig. 5 H), indicating that mucosal B cell deficiency in mutant mice resulted in enhanced mucosal colonization and peripheral translocation of bacterial products. To better evaluate the link between B cell deficiency and gut inflammation, we adoptively transferred naive CD45.1+B220+IgM+ WT B cells into adult Rag2R229Q recipients (Fig. 5 D). As shown in Fig. 5 E, transferred B cells generated IgA-producing plasma cells rapidly, reconstituting the secretory IgA compartment in recipient mice to the control level. Bacterial translocation and intestinal inflammation were assessed 4 wk after the transfer. In agreement with a previous study reporting the expression of epithelial antimicrobial peptide RegIIIγ in response to mucosal IgA deficiency (Shulzhenko et al., 2011), we found that correction of IgA defect substantially reduced RegIIIγ mRNA level in the gut of Rag2R229Q recipient mice, suggesting diminished microbial stimulation of the intestinal epithelium (Fig. 5 F). Accordingly, bacterial adherence to ileal mucosa was diminished, although not significantly (Fig. 5 G). These results suggest that low-affinity T cell–independent IgA responses, such as those generated in mutant mice in the absence of T cell help (Cassani et al., 2010a), are not sufficient to contain more invasive bacteria, which tightly adhere to epithelial cells in terminal ileum. Nonetheless, LPS translocation was visibly reduced in the recipients of WT B cells (Fig. 5 H). Interestingly, we found that decreased stimulation of the epithelium attenuated the inflammation in the colonic but not in the SI tissue (Fig. 5 I). Overall, these data suggest that B cells are required but not sufficient to maintain gut homeostasis.
Figure 5.
In Rag2R229Q mice, mucosal B cell deficiency correlates with enhanced bacterial translocation. (A) Representative FACS plots of B220+ B cells and IgA+ plasma cells infiltrating the SI LP of Rag2+/+ and Rag2R229Q mice. Numbers indicate the frequency of cells in each quadrant. (B) Fecal IgA and IgM were further analyzed by ELISA. Cumulative results from three different experiments with at least n = 12 mice are shown. (C) FACS staining of IgA-coated bacteria in fecal pellets of Rag2+/+ and Rag2R229Q mice. Numbers in quadrants indicate the percentage of cells in each. Cumulative results from analysis of n = 10 mice from two experiments are reported in the bar graph. (D) Expression of CD45.1 on LP B220+ cells in recipient Rag2R229Q mice. WT CD45.1+CD19+ B220+IgM+ cells were adoptively transferred into Rag2R229Q mice. (E) Fecal IgA levels in recipient Rag2R229Q mice and controls at the indicated time points after the cell transfer. The dashed line indicates the mean level of IgA in Rag2+/+ mice. Cumulative results from two different experiments with n = 5–10 mice are shown. (F) Gene expression analysis of antimicrobial RegIIIγ in the ileal and colonic tissues of mice. RNA contents are shown as arbitrary units (A.U.). Cumulative results from analysis of n = 8–20 mice in two experiments are reported in the graphs. (G) Quantitative PCR analysis of ileal adherent bacteria in Rag2+/+, Rag2R229Q, and Rag2R229Q mice adoptively transferred with WT B cells expressed as prokaryotic/eukaryotic DNA ratios. Cumulative results from two independent experiments with n = 10–16 mice/group are shown. (H) Serum LPS concentrations. Data shown are cumulative of two experiments with at least 10 mice/group. (I) Representative SI and colon sections from Rag2R229Q mice and Rag2R229Q mice adoptively transferred with WT B cells stained with H&E. Bars, 100 µm. Histograms show the SI and colonic inflammation score. Cumulative results of two experiments with n = 7–10 mice/group are reported. Values from E–I are mean ± SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
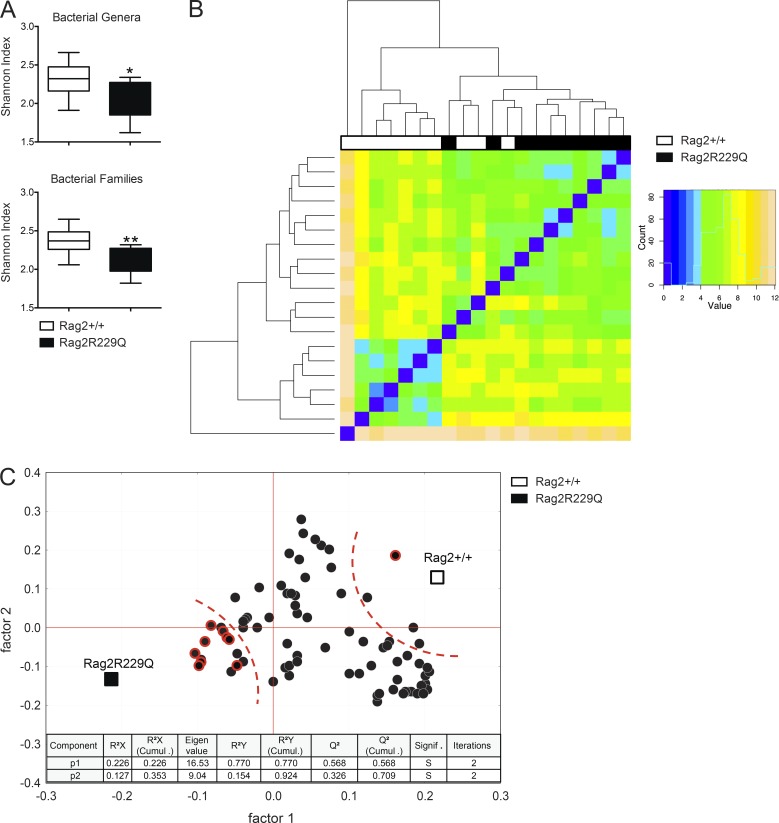
Defects in the adaptive immune compartment have profound impact on gut microbial ecology (Fagarasan et al., 2002; Kawamoto et al., 2014). We therefore evaluated the microbiota composition in Rag2R229Q mice and their cohoused WT littermates by 16S rRNA gene profiling. Overall, no marked variation was observed in the relative abundance of the dominant intestinal bacterial phyla (Firmicutes, Bacterioidetes, Actinobacteria, and Proteobacteria) between the two sets of mice (Fig. S2 A). However, we found that mutant mice had considerably less diverse bacterial communities at the family and genus levels compared with controls, as determined by the Shannon and Chao1 indexes of α-diversity (Fig. 6 A and not depicted). Moreover, hierarchical cluster analysis based on data of relative abundance of the bacterial families revealed that Rag2R229Q and Rag2+/+ mice formed approximate clustering according to their genotype (Fig. 6 B). Then, we observed that most of the differences in the relative abundances of single bacterial genera between Rag2R229Q and Rag2+/+ mice was due to a higher abundance in the group of WT mice (Fig. S2 B); in addition, we found that modified taxa belonged most frequently to the phylum Proteobacteria (Fig. S2 B). Finally, to identify the bacterial genera that better describe the difference between genotypes, we performed a partial least squares (PLS1) regression analysis, which takes into consideration the overall composition of the microbiota, using genotype as categorical responses and the relative abundance of bacterial genera as continuous predictors. In accordance to α-diversity analysis, PLS1 results confirmed the overall dearth of the microbiota richness in the Rag2R229Q mice compared with Rag2+/+ mice (Fig. 6 C). Furthermore, PLS1 analysis revealed that Rag2R229Q microbiota is mainly described by several genera of the phylum Proteobacteria (Fig. 6 C), previously associated with chronic inflammatory condition in humans (Manichanh et al., 2012). Of note, segmented filamentous bacteria (Ivanov et al., 2009) were not present in our colonies, thus excluding their role in the Th17 polarizing gut environment of mutant mice (data not shown). Overall, these data indicate that intestinal inflammation in mutant mice is associated with important alterations of the microbial communities in the gut.
Figure 6.
Rag2R229Q mice show altered microbiota composition. Adults Rag2+/+ and Rag2R229Q littermates were cohoused for 2 mo after the weaning. Cecum fecal content was analyzed for microbiota profiling with 16s sequencing. (A) Box and whisker plots of the Shannon diversity index calculated at the bacterial genera and family level. (B) Heat map visualization at the family level, which was obtained applying the DESeq package on the taxonomic data produced by BioMaS. (C) Correlation between mouse genotype and bacterial genera in fecal samples. X and Y loading plots (factors 1 and 2) were obtained by partial least squares analysis performed by using the genotype as categorical response and relative abundance of bacterial genera as continuous predictors. Taxa belonging to the phylum Proteobacteria are shown with a red circle. 92% of sum of squares is explained by the first two extracted components. *, P < 0.05; **, P < 0.01.
Reducing intestinal bacterial load ameliorates gut inflammation in Rag2R229Q mice
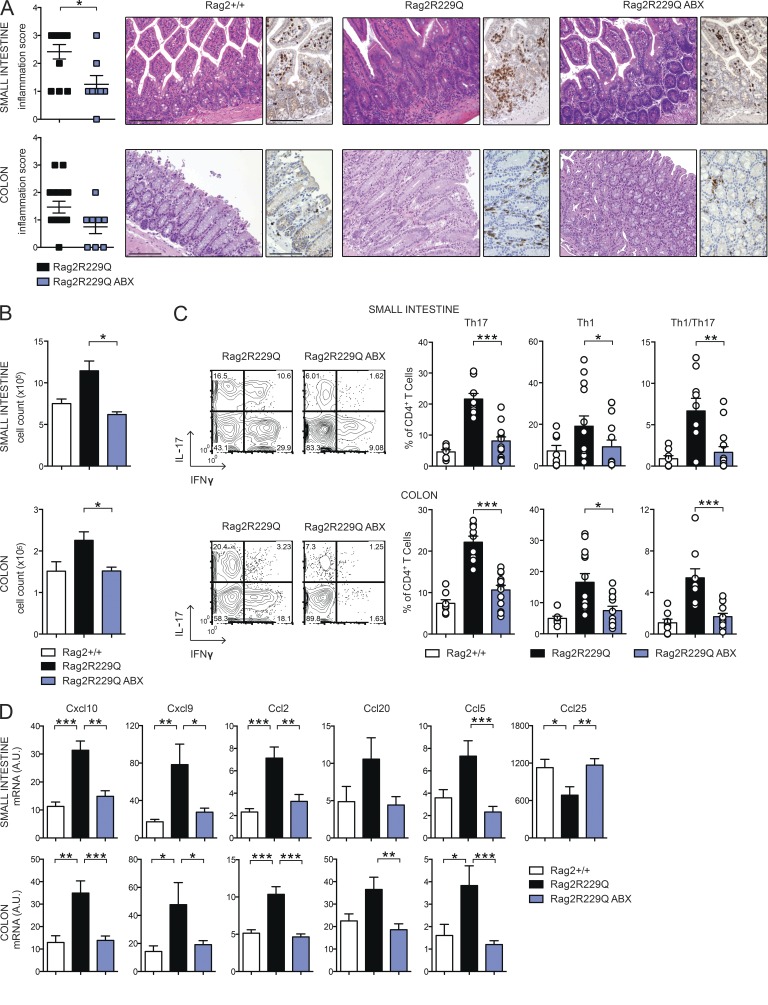
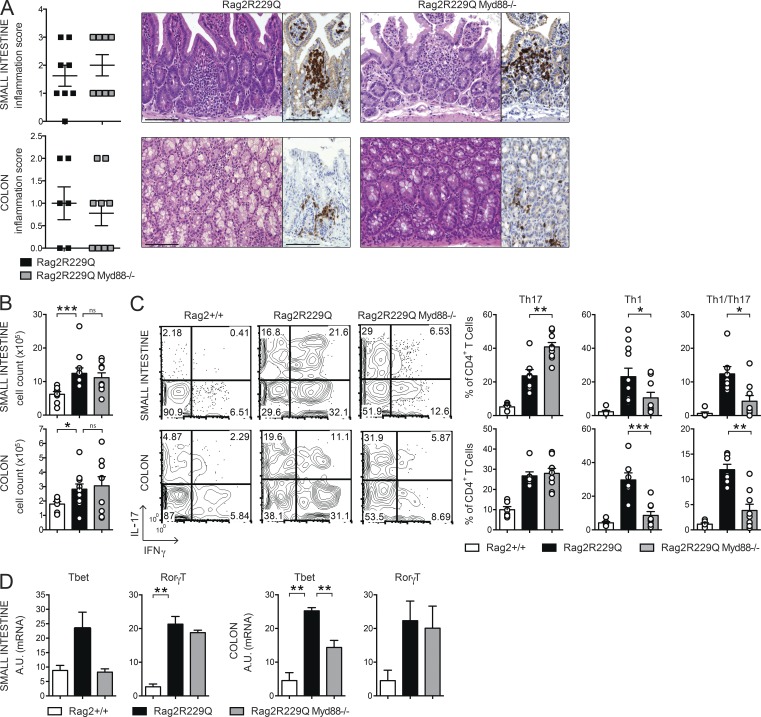
To directly explore the role of microbial flora in gut inflammation in Rag2R229Q mice, we treated mice with broad-spectrum ABXs. A regimen of three ABXs was administered to 8–12-wk-old mice for 4 wk. We did not observe substantial changes in body weight between treated and untreated animals, thus ruling out potential toxicity related to the pharmacological treatment. In ABX-treated Rag2R229Q mice, we found amelioration of gut inflammation associated with a resolution of lymphocytic mucosal infiltrates (Fig. 7 A) and diminished percentages and absolute numbers of IFN-γ– and IL-17–expressing CD4+ T cells in the LP and MLNs (Fig. 7, B and C; and not depicted). On the contrary, a minor reduction of these cell types was observed in the Rag2+/+-treated group (SI Rag2+/+ vs. Rag2+/+ ABX: Th17, 4.6 ± 2% vs. 3.5 ± 2.18%; Th1, 1.01 ± 1.06% vs. 1.27 ± 0.52%; Colon: Th17, 7.4 ± 2.46% vs. 3.1 ± 1.89%; Th1, 4.9 ± 1.99% vs. 3.85 ± 2.06%). Of note, concurrent deficiency of MyD88 (and consequent lack of most TLR signaling) did not protect from bowel inflammation. Rag2R229Q × MyD88−/− mice showed waste disease and analogous accumulation of infiltrating inflammatory T cells to Rag2R229Q littermates (Fig. 8, A and B). Of note, lack of MyD88 abolished Th1 but not Th17 intestinal immune responses in Rag2R229Q mice, indicating that Th17 cell generation in mutants occurs independently from the TLR-MyD88–dependent sensing of commensal microbiota (Fig. 8, C and D).
Figure 7.
ABX treatment dampens gut inflammation in Rag2R229Q mice. (A) Representative SI and colonic sections from controls and ABX-treated Rag2R229Q stained with H&E and CD3 immunostaining. Bars, 100 µm. Histograms show the inflammation score in gut tissues. Data are cumulative results of three independent experiments (n = 8–15). (B) Total cell counts from SI and colonic LP of ABX-treated Rag2R229Q and controls. (C) Representative FACS plots and frequency of SI and colonic LP IL-17+, IFN-γ+, and IL-17+IFN-γ+ CD4 T cells. Numbers in the plots indicate the frequency of cells in each quadrant. (D) Expression of the indicated chemokines in the ileal and colonic tissues assessed by quantitative RT-PCR. Normalized values are indicated as arbitrary units (A.U.). Data from B–D are representative results of four independent experiments (n = 8–13 animals/group). Values are mean ± SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Figure 8.
Myd88 deficiency limits Th1 but not Th17 responses in the gut of Rag2R229Q mice. (A) Representative SI and colonic section from control and Rag2R229QMyd88−/− stained with H&E and CD3 immunostaining. Bars, 100 µm. Histograms show the inflammation score in the SI and colon. Data are the cumulative results of two independent experiments with n = 7–9. (B) Total cell counts from SI and colonic LP. (C) Representative FACS plots and frequencies of CD4+IL-17+, CD4+IFN-γ+, and CD4+IL-17+IFN-γ+ T cells from SI and colon LP. Numbers in the plots indicate the frequency of cells in each quadrant. Data represent one of two independent experiments with 6–11 mice/group. (D) Expression of Tbet (Tbx21) and RorγT (Rorc) on sorted CD4+ T cells from SI and colonic LP. RNA contents are shown as arbitrary units (A.U.). Data represent three independent experiments each using pooled cells from at least five mice per group. Values are mean ± SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Chemokines play a major role in recruiting immune cells to sites of injury. Consistent with the increased tissue infiltration and cytokine skewing, transcript levels of Th1 (Cxcl10 and Cxcl9)- and Th17 (Ccl20)-associated chemokines, as well as those of Ccl2 and Ccl5, were up-regulated in the bowels of Rag2R229Q mice (Fig. 7 D). However, upon ABX treatment, their expression returned to control values (Fig. 7 D). Notably, analogously to Crohn’s disease (Papadakis et al., 2001), Ccl25 expression was lower in the Rag2R229Q ileum with respect to control mice (Fig. 7 D), whereas it reached normal levels in ABX Rag2R229Q mice (Fig. 7 D). Collectively, these results show that reducing microbial load beneficially affects Rag2R229Q mice.
Reducing intestinal bacterial load ameliorates systemic autoimmunity in Rag2R229Q mice
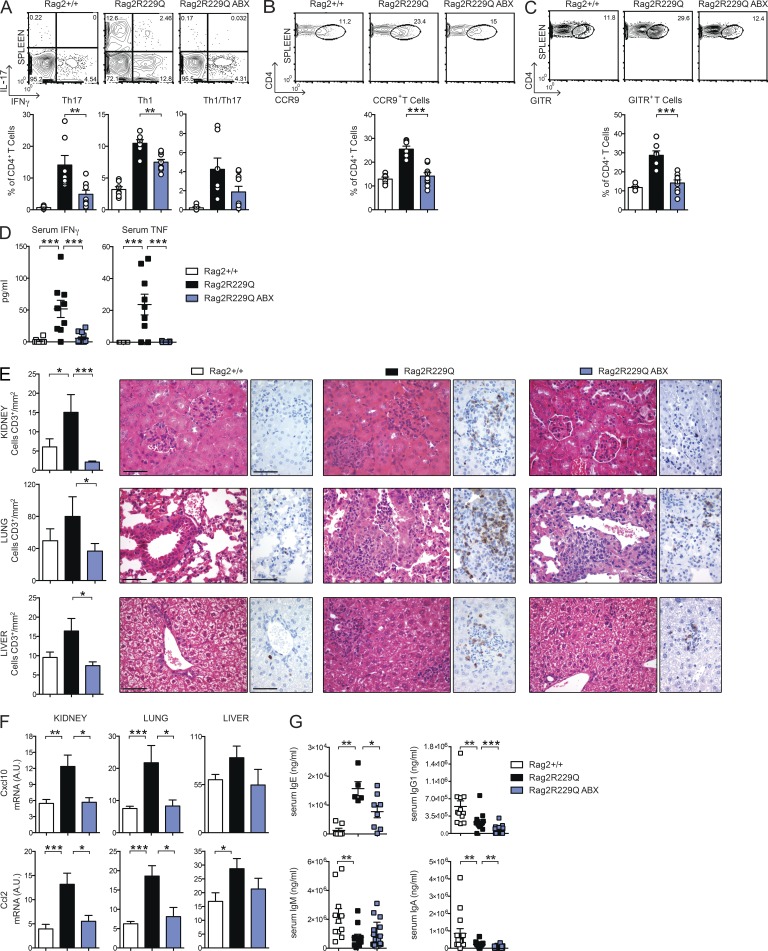
The same Th cells polarization dominating the intestinal immune response was evident in the spleens of mutant mice (Fig. 9 A). Hence, we examined the possibility that the antiinflammatory effect of ABXs extended beyond the intestinal mucosa. We found that frequencies of Th1 and Th17 cells were considerably reduced in the spleens of ABX Rag2R229Q mice (Fig. 9 A), whereas their proportion was low and unaffected in ABX Rag2+/+ mice, indicating a role for microbial flora in the maintenance of circulating, other than intestinal, effector Rag2R229Q T cell populations. Interestingly, ABX Rag2R229Q mice also showed significantly decreased frequencies of systemic CCR9+ CD4 T cells (Fig. 9 B), indicating that resolution of gut inflammation normalized the circulating pool of these cells.
Figure 9.
ABX treatment ameliorates systemic autoimmunity in Rag2R229Q mice. (A) Representative FACS plots and frequency of IL-17+–, IFN-γ+–, and IL-17+IFN-γ+–producing CD4 T cells in the spleens of Rag2+/+, Rag2R229Q, and ABX-treated Rag2R229Q mice. (B) Representative FACS plots and frequency of splenic CCR9+ T cells within the CD44+CD62L− CD4+ T cell gate. (C) Representative FACS plots and frequency of splenic CD4+GITR+ T reg cells. Data are representative results of four independent experiments with at least six animals per group. (D) Levels of IFN-γ and TNF in the serum of ABX-treated Rag2R229Q mice and controls were analyzed in three independent experiments (n = 8–13). Each serum sample was run in duplicate. (E) Representative kidney, lung, and liver sections from controls and ABX Rag2R229Q stained with H&E and CD3 immunostaining. Bars, 50 µm. Histograms show quantification of infiltrating CD3+ T cells in the kidney, lung, and liver sections. (F) Quantitative PCR analysis of chemokine expression in the kidney, liver, and lung of Rag2+/+, Rag2R229Q, and ABX-treated Rag2R229Q mice. RNA contents are shown as arbitrary units (A.U.). Results from two experiments with n = 4–9 mice analyzed are shown. (G) Serum levels of IgE, IgG1, IgM, and IgA were analyzed by ELISA in Rag2+/+, Rag2R229Q, and ABX-treated Rag2R229Q mice. Data shown are cumulative results from four independent experiments with at least 11 mice/group. For IgE analysis, data are representative results of two independent experiments with five to eight mice/group. All values are mean ± SEM *, P < 0.05; **, P < 0.01; ***, P < 0.001.
To confirm the influence of microbial flora on the inflammatory T cell phenotype of Rag2R229Q mice, independently from the host genetic susceptibility, we recolonized ABX-treated adult Rag2+/+ mice with fecal materials obtained from mutant mice (Rag2R229Q → Rag2+/+). Fecal transplant was repeated twice 1 wk apart, and mice were sacrificed 3 wk later. Recolonized ABX Rag2+/+ mice showed augmented frequencies of Th1 and Th17 cells in the gut LP as well as spleens with respect to ABX Rag2+/+ mice receiving feces from Rag2+/+ mice (Rag2+/+→ Rag2+/+; Fig. 10). On the contrary, ABX Rag2R229Q mice recolonized with Rag2+/+ feces displayed considerably fewer intestinal and peripheral Th1/Th17 cells compared with controls (Fig. 10). Like effector T cells, intestinal microbiota contributes to local induction of T reg cells (Round and Mazmanian, 2010; Atarashi et al., 2011), and ABX-treated mice had fewer intestinal T reg cells (Rosser et al., 2014). In agreement with these published data, we found that the proportion of GITR+ T reg cells was reduced in the LP and MLNs of ABX mice (not depicted). However, a drop of their splenic frequency was observed in the ABX Rag2R229Q but not Rag2+/+ mice (Fig. 9 C; Rag2+/+ vs. Rag2+/+ ABX: 11.7 ± 1.43% vs. 12.2 ± 5.36%), suggesting that most T reg cells in Rag2R229Q mice are microbiota dependent. Overall, these findings point to a substantial role of gut microenvironment and commensal bacteria in the OS pathogenesis. Next, we assessed whether the ABX regimen protected against the multisystemic autoimmune-like manifestations of mutant mice. Indeed, Rag2R229Q mice show important infiltrates of effector T cells in the skin, kidney, lung, and liver, which correlate with abnormal concentration of IFN-γ–induced CXCL10 in the serum (Cassani et al., 2010a; Marrella et al., 2012). Importantly, the decline of circulating inflammatory T cells in ABX Rag2R229Q mice lowered IFN-γ and TNF serum concentrations (Fig. 9 D). We then examined the CD3+ T cell infiltrates in target organs. A decreased number of T cells was observed in the kidney, lung, and liver of ABX Rag2R229Q mice compared with untreated controls (Fig. 9 E). In line with these results, tissue expression of inflammatory chemokines (Ccl2 and Cxcl10) was lower in ABX mutant mice (Fig. 9 F). On the contrary, skin inflammation was not affected by the ABX treatment (not depicted), suggesting that this district did not benefit from the reduced gut bacterial load and/or that an independent pathogenetic mechanism sustained tissue inflammation.
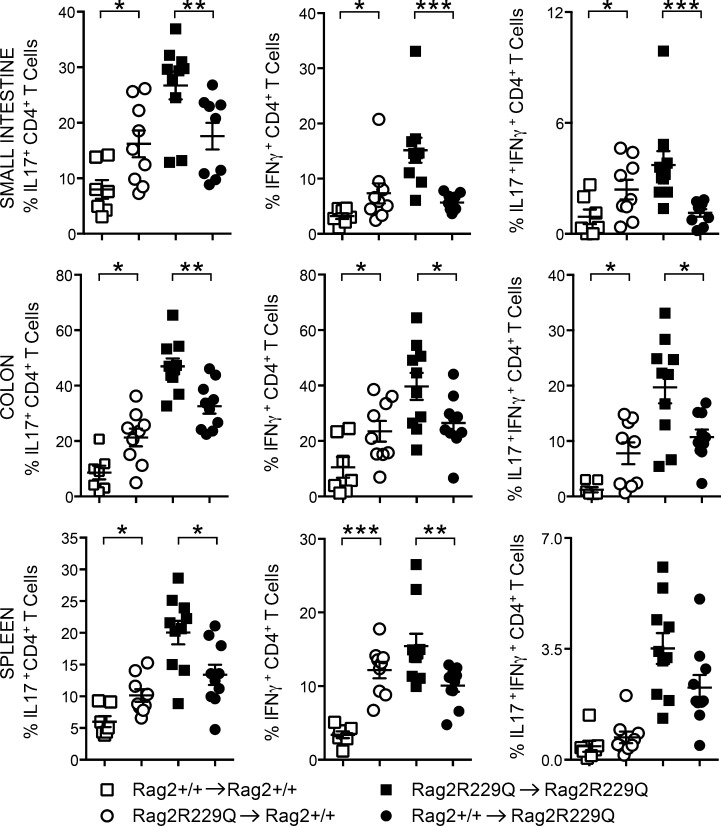
Figure 10.
Rag2R229Q gut microbiota promotes a pro-inflammatory Th1-Th17 skewing. ABX-treated adult mice were recolonized with fecal matters from Rag2+/+ or Rag2R229Q donors. Graphs show the percentages of intestinal and splenic CD4 T cells producing IL-17 and/or IFN-γ cytokines after fecal transplantation. Data are representative results of two independent experiments with 7–10 mice/group. Values are mean ± SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
ABX treatment reduces hyper-IgE in Rag2R229Q mice
CD4 T cells have a pivotal role in B cell abnormalities, including plasma cell expansion and high IgE serum levels (Cassani et al., 2010a). Given that reducing microbial load in mutant mice had a clear impact on peripheral T cell phenotype, we asked whether ABX administration could impinge on B cell defects. In Rag2+/+ mice, microbiota depletion reduced serum IgA levels, whereas other Ig isotypes remained unchanged (mean values of Rag2+/+ vs. Rag2+/+ ABX: IgA, 8.5 × 105 vs. 2.2 × 105 ng/ml; IgG1, 5.4 × 105 vs. 3.6 × 105 ng/ml; IgM, 2.2 × 106 vs. 2.6 × 106 ng/ml). Remarkably, in Rag2R229Q mice, the long-term treatment resulted in a significantly reduced serum IgE (Fig. 9 G). A similar trend was observed for IgA and IgG, but not for IgM (Fig. 9 G). Of note, frequencies and numbers of peripheral B cells were not affected by ABX treatment. Because hyper-IgE constitutes a hallmark of OS, these results indicate that microbiota can constitute a pharmacological target for this pathological trait of the disease.
DISCUSSION
Here, we investigated the role of intestinal immune responses and gut microbiota in the pathogenesis of OS. A large fraction of Rag2R229Q mice developed bowel inflammation characterized by marked infiltration of T cells in the LP. We found that lymphocytes are sufficient for disease initiation. In fact, CD4 T cells, either isolated from colitic mice or not, were able to transfer the disease into immunodeficient hosts. Cytokine production profiling of gut infiltrating lymphocytes revealed a dominant mixed Th1/Th17 skewing, frequently implicated in the pathogenesis of IBD and experimental colitis (Powrie et al., 1994; Berg et al., 1996; Fuss et al., 1996; Ahern et al., 2010). An abundant presence of Th17 cells, also producing the IL-22 cytokine, might also contribute to the aberrant expression of antimicrobial peptide RegIIIγ (Liang et al., 2006).
Multiple studies revealed the importance of T reg cells for the maintenance of immune tolerance in the gastrointestinal tract owing to the constant immune stimulation by commensal flora and food antigens (Izcue et al., 2009; Pedros et al., 2016). Impaired intestinal tolerance is believed to contribute to inflammation in response to microflora in IBD (Xavier and Podolsky, 2007). Several experiments in this study addressed the role of T reg cells in the gut homeostasis of Rag2R229Q mice. We showed that T reg cells accumulate in the intestine but fail to control disease in mutant mice. In the adoptive transfer model of colitis, Rag2R229Q T reg cells cotransferred with WT naive CD45RBhi CD4 T cells did not prevent disease development in Rag1−/− mice. Interestingly, similar failure in controlling ongoing autoimmune responses has been described for Nrp-1low iT reg cells generated in inflammatory and lymphopenic environment (Yadav et al., 2012). On the contrary, the transfer of a limited number of Rag2+/+ T reg cells significantly ameliorated intestinal inflammation by dampening Th1 and Th17 immune responses in Rag2R229Q hosts. Thus, it is conceivable that the absence of functional T reg cells plays a major role in the pathogenesis of intestinal inflammation in Rag2R229Q mice.
Distinct effector mechanisms from those operating in the Rag2R229Q T reg cells may be required to control mucosal inflammation. These may include the expression of CD62L and CCR7 (Fu et al., 2004; Schneider et al., 2007), which are necessary for homing to the inductive sites, and production of IL-10, which is essential to limiting the induction of pro-inflammatory Th1 and Th17 cells at the intestinal mucosa interface (Geuking et al., 2011). IL-10 mRNA levels were significantly reduced in the intestines of Rag2R229Q mice, and IL-10 is also required for oral tolerance induction in different models (Cong et al., 2004; Navarro et al., 2011). Finally, iT reg cells derived from an oligoclonal population of CD4+ T cells may be restricted in their ability to effectively suppress autoimmune disease and maintain tolerogenicity to microbiota (Huter et al., 2008; Nishio et al., 2015).
Mucosal T reg cells, together with secretory IgAs, critically preserve the gut barrier and control diversification of commensal species, thus ensuring host–bacterial mutualism (Kawamoto et al., 2014). Conversely, biased expansion of certain bacterial species impairs epithelial integrity and induces hyperactivation of the immune system, also promoting the generation of pro-inflammatory T cell subsets (Kawamoto et al., 2012; Kamada et al., 2013). We demonstrated that the hypomorphic Rag defect in B cells favors microbial access to the LP and circulation and considerably impacts on the diversity of the microbial communities in the gut. Our findings correlate with previous observations in mice harboring immune deficiencies affecting inflammatory responses in adaptive immune cells (Garrett et al., 2007; Kawamoto et al., 2014). We also showed that such altered microbiota from Rag2R229Q mice was sufficient to promote a skewed Th1/Th17 pro-inflammatory phenotype into Rag2+/+ mice. This observation indicates that Rag2R229Q microbiota has a pathogenic potential.
The clinical benefits deriving from long-term ABX dosing formally implicates intestinal microbes in the disease pathogenesis. We showed that both mucosal and circulating pro-inflammatory Th1 and Th17 cell subsets were significantly reduced in ABX-treated mice, establishing that skewed polarization of adaptive immunity in Rag2R229Q mice was microbiota dependent. Of note, CCR9-expressing T cells were similarly affected by the treatment, consistent with resolution of gut inflammation. However, because CCR9 expression is acquired by T cells upon priming by GALT-derived dendritic cells (Mora, 2008), we propose that its presence on the vast majority of circulating T cells in Rag2R229Q mice might reflect their original intestinal differentiation.
Although CD4+ T cells from Rag2R229Q mice are able to transfer colitis to immunodeficient hosts, it still remains to be determined whether an aberrant non–T cell population is required for the priming of dysregulated T cells. On this point, it has been shown that both innate and adaptive immune system stimulation by microbiota are required for driving spontaneous T cell proliferation and induction of colitis in immunodeficient mice (Feng et al., 2011). Furthermore, we previously reported perturbed cell trafficking and altered lymphoid distribution of dendritic cells in Rag2R229Q mice, which tend instead to ectopically accumulate and halt at the environmental interfaces (Maina et al., 2013). It seems therefore logical to envisage a crucial role for innate immune cells in colitis development in mutant mice.
In contrast to previous publications describing its protective effects in different models of IBD induced by immune deficiencies (Rakoff-Nahoum et al., 2006; Rivas et al., 2012), we found that inhibition of MyD88-dependent pathways did not restrain intestinal inflammation in Rag2R229Q mice. Rather, Rag2R229Q × MyD88−/− mice displayed worsening of wasting syndrome and unaffected accumulation of mucosal lymphocytic infiltrates. These results, however, are consistent with the intricate effect of MyD88 deficiency on host–microbial mutualism (Slack et al., 2009; Vaishnava et al., 2011). Interestingly, the absence of MyD88 signaling uncouples, in our model, the molecular requirements for generation of Th1 and Th17 cells in the gut. Despite observations that MyD88 ablation is required for the induction of both Th1 and Th17 cell responses (Fukata et al., 2008; Schenten et al., 2014), our data showed that signaling through MyD88 is crucial for Th1 immune responses yet has a nonessential role for generation of Th17 cells. Therefore, multiple sensors may be involved in the development of these cells in the gut. On this line, it has been suggested that bacterially produced ATP can directly augment intestinal Th17 cell differentiation (Atarashi et al., 2008).
In addition to gut inflammation, reducing intestinal microbial load visibly ameliorated multisystem autoimmunity in Rag2R229Q mice, confirming the critical role of gut flora in sustaining chronic immune inflammation and autoimmune disease onset/progression in genetically susceptible hosts. The evidence that this inflammatory immune phenotype is transmissible to an intact immune system in a microbiota-dependent manner leads us to speculate that pharmacological gut decontamination might constitute a valid strategy to limit side effects of hematopoietic stem cell transplantation in these patients (Vossen et al., 2014). In contrast, given the importance of the intestinal microbiota, interventions aimed at promoting intestinal diversity may improve immunological outcomes in OS transplant settings. Replenishment of commensal bacterial populations through fecal microbiota transplantation or targeted flora reintroduction may theoretically be effective, although these procedures still need to be carefully evaluated in a transplant context. Alternatively, the use of dietary supplements, which may help to maintain beneficial bacteria (Gerbitz et al., 2004), might be considered to avoid episodes of dominance of harmful bacterial species during posttransplant recovery (Lane et al., 2015) and to promote healthy mucosal responses.
Importantly, our results showed that reducing the microbial load is beneficial for normalizing IgE levels in Rag2R229Q mice. Because our previously published data indicated that T cells in mutant mice do have a crucial role in B cell abnormalities (Cassani et al., 2010a), diminished IgE levels after ABXs might reflect the lower serum concentration of cytokines promoting class switch recombination. Interestingly, it has been recently proposed that IgE production is strongly dependent on microbial exposure during early life and that a critical level of microbial diversity is required to establish the proper immunoregulatory networks to inhibit IgE production (Cahenzli et al., 2013). Thus, it is conceivable that reduced microbiota diversity in Rag2R229Q mice also impinges the immune regulation that functions to maintain IgE levels at the baseline.
In summary, our findings might provide novel insights into the pathogenesis of OS. We showed in a murine model that hypomorphic Rag mutation compromises gut barrier integrity and alters microbiota composition. Furthermore, the loss of T cell tolerance to commensal microbiota leads to gut Th17/Th1 inflammation which, in turn, sustains dysbiosis in a well-recognized regulatory loop.
Rag2R229Q mice represent an important tool to investigate whether intestinal microbiota can be manipulated therapeutically to dampen immune activation and perpetuation of inflammation. Furthermore, they may help to determine the extent to which a limited adaptive immune system also controls microbial community composition in other organ systems, such as the skin, and to clarify whether these other resident communities of indigenous microorganisms shape local immune cell differentiation and similarly influence the host health.
MATERIALS AND METHODS
Mice and treatments
129Sv/C57BL/6 knock-in Rag2R229Q mice were backcrossed to C57BL/6 strain for four generations. The colony was maintained onsite with heterozygous breeders, and littermates were kept in the same cages until weaning at 4 wk of age. Rag2R229QMyd88−/− double-mutant mice and littermates controls were generated from heterozygous–heterozygous crossing and maintained onsite as described for single mutants. All mouse strains were used for experiments between 8 and 13 wk of age, and, unless indicated otherwise, adult littermates were raised in separate cages according to the genotype after weaning. Rag1−/− (Rag1tm1Mom/J) mice were obtained from The Jackson Laboratory and C57BL/6 Ly5.1 mice from Charles River. Mice were maintained under specific pathogen–free conditions at the Istituto Clinico Humanitas. To deplete gut flora, adult Rag2+/+ and Rag2R229Q mice, housed in separate cages, were treated daily with an ABX cocktail containing 2.5 mg metronidazole, 2.5 mg ampicillin, and 1.25 mg vancomycin in 200-µl/mouse doses by oral gavage for 4 wk. All animal procedures were performed according to protocols approved by the Istituto Clinico Humanitas and the Italian Institutional Animal Care and Use Committee.
Induction of oral tolerance and EAE
Mice were fed daily 0.25 mg MOG peptide (MOG35–55) via oral gavage for 7 d as described previously (Cassani et al., 2011). Mice were then injected s.c. in the flank with 100 µg MOG35–55 in 0.15 ml PBS emulsified in an equal volume of CFA containing 4 mg Mycobacterium tuberculosis H37 RA/ml (Difco). Pertussis toxin (200 ng/mouse/injection; List Biological Laboratories) was administered i.p. at the time of immunization and 48 h later. Animals were scored for EAE as follows: 0, no disease; 1, tail paralysis; 2, hind limb weakness; 3, hind limb paralysis; 4, hind limb plus forelimb paralysis; 5, moribund.
Induction of colitis by adoptive T cell transfer
CD4+ T cells were isolated by pooled spleens and MLNs of 10-wk-old Rag2+/+ or Rag2R229Q mice using CD4-specific magnetic beads via negative selection according to the manufacturer’s instructions (Miltenyi Biotec). Enriched CD4 T cells were subsequently sorted into naive CD4+CD25−CD45RBhi and regulatory CD4+CD45RB−CD25hi populations (>98% purity) using a cell sorter (FACSAria; BD). For colitis induction, 4 × 105 WT CD4+CD25−CD45RBhi naive T cells were i.p. transferred into Rag1−/− mice alone or with 1.5 × 105 CD4+CD25hi T reg cells from Rag2+/+ or Rag2R229Q mice. Recipient mice were weighted twice a week and sacrificed at week four after the transfer, when IBD was diagnosed by severe weight loss.
Evaluation of IgA-coated bacteria by FACS
Fecal pellets were suspended in filtered PBS, homogenized, and centrifuged at 700 g for 5 min to remove large particles from bacteria. Then, supernatants were collected and centrifuged at 8,000 g for 10 min to remove unbound Ig’s. The bacterial pellet was suspended in PBS 1% BSA and stained with PE-labeled anti–mouse IgA on ice for 30 min. Bacteria was suspended in 4% formalin overnight at 4°C for fixation and resuspended in DAPI/PBS for acquisition at a cell analyzer (FACSCanto II; BD) using forward scatter (FSC) and side scatter parameters in logarithmic mode. Data were analyzed with FlowJo software (version 7.6.5; Tree Star).
ELISA assay
For evaluation of fecal Ig’s, feces were collected, weighed, and resuspended in 1 ml PBS containing 0.1% sodium azide and protein inhibitors. After overnight incubation, fecal pellets were centrifuged at 10,000 g for 15 min, and supernatants were collected. Fecal suspensions were stored at −20°C. Levels of fecal IgA and IgM were measured by a mouse IgA/IgM ELISA quantitation set (Bethyl Laboratories, Inc.) according to the manufacturer’s instructions. Serum levels of IgG1, IgA, and IgM were measured by a multiplex assay kit (Beadlyte Mouse Immunoglobulin Isotyping kit; EMD Millipore) according to the manufacturer’s instructions and run using a Bio-Plex reader (Bio-Rad Laboratories). Levels of IgE were measured in sera by ELISA assay (BD). Cytokine levels in sera were measured using a specific ELISA kit for mouse IFN-γ and TNF (R&D Systems). For cytokine determination, colonic tissues were incubated at 37°C for 48 h. Cultured supernatants were collected after 48 h, and cytokine concentrations were determined by ELISA (R&D Systems) and normalized for tissue weight.
RT-PCR
Total RNA was extracted from 1 cm of ileal and colonic tissues. Tissues from intestines, kidney, lung, and liver were homogenized in PureZOL reagent (Bio-Rad Laboratories) using TissueLyser II (QIAGEN). RNA was extracted using the RNeasy Lipid Tissue kit (QIAGEN), and cDNA was synthetized using the High-Capacity cDNA Reverse Transcription kit (Thermo Fisher Scientific). Quantitative RT-PCR was performed on a CFX384 system (Bio-Rad Laboratories) with the SYBR green master mix (Thermo Fisher Scientific). For sorted cells, total RNA was extracted using the RNeasy Plus Micro kit (QIAGEN). RNA was retrotranscribed using the High-Capacity cDNA Reverse Transcription kit followed by a preamplification PCR using TaqMan PreAmp Master Mix (Applied Biosystems). Quantitative RT-PCR was performed using a RT-PCR system (ViiA 7; Thermo Fisher Scientific) with TaqMan probes (Table S1). The relative amounts of mRNAs were calculated as 2−ΔCT and expressed as arbitrary units. Each sample was analyzed in duplicate, and the relative level of expression was determined by normalization to β-actin (Actb) ribosomal RNA.
Quantification of mucosal adherent bacteria by RT-PCR
The quantification of prokaryotic/eukaryotic ratios was performed as described previously (Proietti et al., 2014). In brief, the latest segment of terminal ileum was collected and digested in lysis buffer (10 mM Tris, 100 mM NaCl, 10 mM EDTA, 0.5% SDS, and 0.4 mg/ml proteinase K). Total DNA was extracted with the isopropanol-ethanol method. The microbial load was determined by RT-PCR (SYBR green) using specific primers for microbial 16s and genomic 18s.
LPS measurement
Sera were collected from Rag2+/+ and Rag2R229Q mice in endotoxin-free conditions. LPS concentration was determined by Limulus amebocyte lysate assay (Lonza) according to the manufacturer’s instructions.
LP preparation
SIs and colons were flushed to remove contents, opened longitudinally, and cut into 3–5-mm pieces. PPs were removed from SIs. Tissues were washed in HBSS buffer (0.5 M EDTA, 1 mM DTT, 15 mM Hepes, and 10% FCS) under agitation for 20 min at 37°C to remove the epithelial layer. Intestine pieces were then Liberase-digested (0.15 mg/ml; Roche) for 30 min at 37°C. Cells were washed and passed through a 100-µm cell strainer. The cell suspension obtained was loaded into a 44/67% Percoll gradient (GE Healthcare) and centrifuged at 600 g for 20 min at room temperature. Lymphoid fractions were collected at the interphase of the Percoll gradient and used for flow cytometry stainings.
Cell isolation and adoptive transfer
Single-cell suspensions were prepared from pooled MLNs harvested from Rag2R229Q and Rag2+/+ mice. Total CD4 T cells were negatively selected using the CD4 T Cell Isolation kit (Miltenyi Biotec) according to the manufacturer’s instructions (purity > 95%). Purified CD4+ T cells (4 × 106) were then i.v. injected into Rag1−/− mice. CD25hiGITR+CD4+ T reg cells were sorted from splenic cell suspension derived from WT CD45.1 mice using the FACSAria cell sorter. Sorted cells (5 × 105) were then injected i.v. into Rag2R229Q mice. The recipient mice were analyzed 4 wk after the transfer. For B cell adoptive transfer, total CD19+ B cells were isolated from pooled spleens, MLNs, and PPs derived from WT CD45.1 mice with anti-CD19 magnetic beads via positive selection (Miltenyi Biotec). B220+IgM+IgA− B cells were then sorted from the enriched CD19+ fraction (purity > 98%) using the FACSAria cell sorter. Rag2R229Q mice were i.v. transferred with 30 × 106 sorted B cells. Recipient mice were monitored for B cell reconstitution and were sacrificed 4 wk after the transfer.
Histological analysis
2-µm-thick formalin-fixed and paraffin-embedded tissue sections from liver, lung, kidney, and gut were used for routine hematoxylin and eosin (H&E) staining and immunostains. In brief, sections were dewaxed and rehydrated, endogenous peroxidase activity was blocked by 0.3% H2O2/methanol for 20 min, and heat-induced antigen retrieval was obtained by microwave or thermostatic bath treatment using EDTA buffer, pH 8.0, followed by 1-h incubation with primary antibody rabbit anti-CD3 (1:100; Dako) or rat anti-FoxP3 (1:100; eBioscience). Signal was revealed by using Real EnVision Rabbit-HRP (Dako) or Rat-on-Mouse HRP-Polymer (Biocare Medical) detection system followed by diaminobenzidine and nuclei counterstained with hematoxylin. Digital imagines were acquired by a camera (DP70; Olympus) mounted on a microscope (Bx60; Olympus) using CellF Imaging software (Soft Imaging System GmbH). Quantification of CD3+ cells was performed on representative sections of liver, lung, and kidney acquired by Aperio ScanScope CS Slide Scanner (Aperio Technologies). Total area of the sections and number of CD3+ cells were obtained by using Aperio ImageScope Software, and the absolute number of CD3+ cells was expressed as the number of CD3+ cells/mm2. The degree of inflammation of the gut (colon and SI) was double-blind graded using combined scores, including the grade of inflammation (grade 0, no evidence of inflammation; grade 1, low; grade 2, moderate; grade 3, high; grade 4, intense and diffuse level of inflammation), the structural changes of the glands (grade 0, absence; grade 1, focal; grade 2, partial; grade 3, diffuse level of structural changes), and the goblet cell depletion (grade 0, no loss; grade 1, low; grade 2, moderate; grade 3, diffuse level of goblet cell depletion). The cumulative total scores ranged from 0 to 10.
Villous/crypt ratio
Villous height and crypt depth were randomly measured for 16 well-oriented villi of distal ileum per mouse and averaged. Data were analyzed using commercial image analysis software (ImageJ; National Institutes of Health).
Flow cytometry and cell sorting
Cell suspensions were obtained from intestines, spleens, and MLNs and stained in PBS 2% FCS with conjugated antibodies to the following markers: TCR-β (eBioscience), TCR-γδ (eBioscience), CD3 (eBioscience), CD45 (eBioscience), CD4 (eBioscience), CD8a (eBioscience), CD62L (eBioscience), CD44 (BD), CD45R (B220; eBioscience), CD357 (GITR; eBioscience), CCR9 (eBioscience), α4β7 (BioLegend), Foxp3 (eBioscience), IgA (eBioscience), IgM (eBioscience), CD25 (eBioscience), CD103 (eBioscience), CD45RB (BioLegend), and Nrp-1 (R&D Systems). Live/dead cell discrimination was obtained using the Aqua Dead Cell Stain kit (Invitrogen). For cytokine production, cells were stimulated for 4 h with 20 ng/ml PMA (Sigma-Aldrich) and 0.5 µg/ml ionomycin (Sigma-Aldrich). Golgi Stop (1,000×; BD) was added during the last 3 h of stimulation. Cells were fixed and permeabilized using intracellular fixation and permeabilization buffer kit (eBioscience) and stained for conjugated anti–IL-17A (eBioscience) and anti–IFN-γ (eBioscience). Flow cytometry data were acquired at FACSCanto II. Data were analyzed with FlowJo software (version 7.6.5).
Microbiota analysis
For the evaluation of intestinal microbiota, weaned Rag2R229Q and Rag2+/+ littermates were cohoused in the same cages until analysis. The bacterial microbiota of 20 fecal samples from 10 Rag2+/+ and 10 Rag2R229Q mice was investigated by sequencing the V5-V6 hypervariable regions of 16S rDNA gene by using the Illumina MiSeq platform as described previously (Manzari et al., 2015). The obtained 250 × 2 paired-end reads were taxonomically profiled by applying the BioMaS pipeline (Fosso et al., 2015). Paired-end reads of each sample were merged into consensus sequences using Flash (Adobe; Magoč and Salzberg, 2011) and then dereplicated by Usearch (drive5; Edgar, 2010). The unmerged reads were trimmed of low-quality regions (Phred score cutoff of 25), and paired ends containing reads shorter than 50 nt were removed. Both the merged sequences and the unmerged reads were mapped against the RDP II database (release 10.32; Cole et al., 2009) by using Bowtie 2 (Langmead and Salzberg, 2012). The mapping data were filtered according to query coverage (≥70%) and similarity percentage (≥97%) and were stored in a file format suitable for the taxonomic assignment steps. Finally, the Tango tool processed the similarity analysis data to assign the sequences to a taxonomic clade in the NCBI taxonomy (Kim et al., 2013; Alonso-Alemany et al., 2014). Statistically significant differences of the bacterial population in Rag+/+ and Rag2R229Q mice were determined at family, genus, and species levels by using the R/Bioconductor DESeq package (Anders et al., 2012).
Fecal transplantation
10–12-wk-old adult male Rag2R229Q and Rag2+/+ mice, housed in separated cages since weaning, received long-term (30 d) dosing of ABXs. For the fecal suspension preparation, pellets were collected from adult female Rag2+/+ and Rag2R229Q donors, suspended in sterile PBS (three to four pellets in 2 ml PBS), and homogenized. Then, homogenized pooled fecal samples were subsequently introduced by gavage with a flexible plastic tube into the stomachs of each ABX-treated mouse recipient. The fecal suspension was administered twice, 1 wk apart, in 200-µl doses. Transplant recipients were maintained in cages dedicated to mice colonized with the same donor microbiota, Rag2R229Q or WT, until the sacrifice (n = 5–6 mice per cage per strain per donor microbiota sample per experiment). Recipient mice were analyzed 3 wk after the second microbiota transfer.
Statistical analysis
Results were analyzed using the nonparametric Mann-Whitney test, Student’s unpaired t test, and two-way ANOVA with Bonferroni post-test analysis. Results are presented as mean ± SEM. Values of P < 0.05 were considered statistically significant.
Online supplemental material
Fig. S1 shows the experimental strategy for FACS staining of intestinal LP cells. Fig. S2 includes graphs showing distribution of bacterial phyla and classes and heat map showing the relative abundance of gut bacterial genera discriminating Rag2+/+ and Rag2R229Q littermates. Table S1 lists PCR primers used for the experiments in this study. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20151116/DC1.
Supplementary Material
ACKNOWLEDGMENTS
The technical assistance of Enrica Mira Catò is acknowledged. We also thank Teresa De Filippis (University of Bari, Bari, Italy) and Caterina Manzari (Consiglio Nazionale delle Ricerche-Istituto di Biomembrane e Bioenergetica and Molecular Biodiversity Lab, Lifewatch, Bari, Italy) for high-throughput sequencing of metagenomic samples.
This work was supported by Ministry of Education, University, and Research grant MIUR-FIR RBFR12I3UB and Ministry of Health Giovani Ricercatori grant GR-2011-02349759 to B. Cassani, Ministry of Education, University, and Research grant MIUR-FIRB RBAP11H2R9-004 to P. Vezzoni, Ministry of Education, University, and Research grant MIUR PON01_02589 and Consiglio Nazionale delle Ricerche Medicina Personalizzata to G. Pesole and A.M. D’Erchia, San Raffaele Telethon Institute for Gene Therapy (TIGET) core grant A3 from the Italian Telethon Foundation to A. Villa and Fondazione Cariplo grant N-2012-0519 to A. Villa, P.L. Poliani, and F. Grassi, and Ministry of Health Giovani Ricercatori grant GR-2009-1607206 to R. Pedotti. B. Cassani was the recipient of a Gerry Scotti fellowship, and R. Rigoni was the recipient of a short-term European Federation of Immunological Societies fellowship.
The authors declare no competing financial interests.
Author contributions: B. Cassani and A. Villa designed research experiments. R. Rigoni performed most experiments. E. Fontana, V. Maina, M.C. Castiello, G. Pacchiana, and S. Musio performed experiments. E. Fontana and S. Mantero contributed histological analysis. B. Fosso, A.M. D’Erchia, G. Pesole, V. Taverniti, and S. Guglielmetti contributed sequencing and bioinformatic analysis of intestinal microbiota. C. Selmi, J.R. Mora, and R. Pedotti contributed reagents and intellectual input. P. Vezzoni, S. Guglielmetti, P.L. Poliani, and F. Grassi contributed intellectual input and data analysis and revised the paper. R. Rigoni, A. Villa, and B. Cassani analyzed data and wrote the paper.
Footnotes
Abbreviations used:
- ABX
- antibiotic
- EAE
- experimental allergic encephalomyelitis
- IBD
- inflammatory bowel disease
- LP
- lamina propria
- MLN
- mesenteric LN
- MOG
- myelin oligodendrocyte glycoprotein
- OS
- Omenn syndrome
- PP
- Peyer’s patch
- RT-PCR
- real-time PCR
- SI
- small intestine
References
- Ahern P.P., Schiering C., Buonocore S., McGeachy M.J., Cua D.J., Maloy K.J., and Powrie F.. 2010. Interleukin-23 drives intestinal inflammation through direct activity on T cells. Immunity. 33:279–288. 10.1016/j.immuni.2010.08.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso-Alemany D., Barré A., Beretta S., Bonizzoni P., Nikolski M., and Valiente G.. 2014. Further steps in TANGO: improved taxonomic assignment in metagenomics. Bioinformatics. 30:17–23. 10.1093/bioinformatics/btt256 [DOI] [PubMed] [Google Scholar]
- Anders S., Reyes A., and Huber W.. 2012. Detecting differential usage of exons from RNA-seq data. Genome Res. 22:2008–2017. 10.1101/gr.133744.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Annunziato F., Cosmi L., Santarlasci V., Maggi L., Liotta F., Mazzinghi B., Parente E., Filì L., Ferri S., Frosali F., et al. . 2007. Phenotypic and functional features of human Th17 cells. J. Exp. Med. 204:1849–1861. 10.1084/jem.20070663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atarashi K., Nishimura J., Shima T., Umesaki Y., Yamamoto M., Onoue M., Yagita H., Ishii N., Evans R., Honda K., and Takeda K.. 2008. ATP drives lamina propria TH17 cell differentiation. Nature. 455:808–812. 10.1038/nature07240 [DOI] [PubMed] [Google Scholar]
- Atarashi K., Tanoue T., Shima T., Imaoka A., Kuwahara T., Momose Y., Cheng G., Yamasaki S., Saito T., Ohba Y., et al. . 2011. Induction of colonic regulatory T cells by indigenous Clostridium species. Science. 331:337–341. 10.1126/science.1198469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg D.J., Davidson N., Kühn R., Müller W., Menon S., Holland G., Thompson-Snipes L., Leach M.W., and Rennick D.. 1996. Enterocolitis and colon cancer in interleukin-10-deficient mice are associated with aberrant cytokine production and CD4(+) TH1-like responses. J. Clin. Invest. 98:1010–1020. 10.1172/JCI118861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahenzli J., Köller Y., Wyss M., Geuking M.B., and McCoy K.D.. 2013. Intestinal microbial diversity during early-life colonization shapes long-term IgE levels. Cell Host Microbe. 14:559–570. 10.1016/j.chom.2013.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassani B., Poliani P.L., Marrella V., Schena F., Sauer A.V., Ravanini M., Strina D., Busse C.E., Regenass S., Wardemann H., et al. . 2010a Homeostatic expansion of autoreactive immunoglobulin-secreting cells in the Rag2 mouse model of Omenn syndrome. J. Exp. Med. 207:1525–1540. 10.1084/jem.20091928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassani B., Poliani P.L., Moratto D., Sobacchi C., Marrella V., Imperatori L., Vairo D., Plebani A., Giliani S., Vezzoni P., et al. . 2010b Defect of regulatory T cells in patients with Omenn syndrome. J. Allergy Clin. Immunol. 125:209–216. 10.1016/j.jaci.2009.10.023 [DOI] [PubMed] [Google Scholar]
- Cassani B., Villablanca E.J., Quintana F.J., Love P.E., Lacy-Hulbert A., Blaner W.S., Sparwasser T., Snapper S.B., Weiner H.L., and Mora J.R.. 2011. Gut-tropic T cells that express integrin α4β7 and CCR9 are required for induction of oral immune tolerance in mice. Gastroenterology. 141:2109–2118. 10.1053/j.gastro.2011.09.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang X., Zheng P., and Liu Y.. 2008. Homeostatic proliferation in the mice with germline FoxP3 mutation and its contribution to fatal autoimmunity. J. Immunol. 181:2399–2406. 10.4049/jimmunol.181.4.2399 [DOI] [PubMed] [Google Scholar]
- Chen Y., Kuchroo V.K., Inobe J., Hafler D.A., and Weiner H.L.. 1994. Regulatory T cell clones induced by oral tolerance: suppression of autoimmune encephalomyelitis. Science. 265:1237–1240. 10.1126/science.7520605 [DOI] [PubMed] [Google Scholar]
- Cole J.R., Wang Q., Cardenas E., Fish J., Chai B., Farris R.J., Kulam-Syed-Mohideen A.S., McGarrell D.M., Marsh T., Garrity G.M., and Tiedje J.M.. 2009. The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 37:D141–D145. 10.1093/nar/gkn879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong Y., Weaver C.T., Lazenby A., and Elson C.O.. 2002. Bacterial-reactive T regulatory cells inhibit pathogenic immune responses to the enteric flora. J. Immunol. 169:6112–6119. 10.4049/jimmunol.169.11.6112 [DOI] [PubMed] [Google Scholar]
- Cong Y., Liu C., Weaver C.T., and Elson C.O.. 2004. Early upregulation of T cell IL-10 production plays an important role in oral tolerance induction. Ann. N. Y. Acad. Sci. 1029:319–320. 10.1196/annals.1309.037 [DOI] [PubMed] [Google Scholar]
- de la Morena M.T., and Nelson R.P. Jr. 2014. Recent advances in transplantation for primary immune deficiency diseases: a comprehensive review. Clin. Rev. Allergy Immunol. 46:131–144. 10.1007/s12016-013-8379-6 [DOI] [PubMed] [Google Scholar]
- Dubois B., Joubert G., Gomez de Agüero M., Gouanvic M., Goubier A., and Kaiserlian D.. 2009. Sequential role of plasmacytoid dendritic cells and regulatory T cells in oral tolerance. Gastroenterology. 137:1019–1028. 10.1053/j.gastro.2009.03.055 [DOI] [PubMed] [Google Scholar]
- Edgar R.C. 2010. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 26:2460–2461. 10.1093/bioinformatics/btq461 [DOI] [PubMed] [Google Scholar]
- Fagarasan S., Muramatsu M., Suzuki K., Nagaoka H., Hiai H., and Honjo T.. 2002. Critical roles of activation-induced cytidine deaminase in the homeostasis of gut flora. Science. 298:1424–1427. 10.1126/science.1077336 [DOI] [PubMed] [Google Scholar]
- Feng T., Qin H., Wang L., Benveniste E.N., Elson C.O., and Cong Y.. 2011. Th17 cells induce colitis and promote Th1 cell responses through IL-17 induction of innate IL-12 and IL-23 production. J. Immunol. 186:6313–6318. 10.4049/jimmunol.1001454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fosso B., Santamaria M., Marzano M., Alonso-Alemany D., Valiente G., Donvito G., Monaco A., Notarangelo P., and Pesole G.. 2015. BioMaS: a modular pipeline for Bioinformatic analysis of Metagenomic AmpliconS. BMC Bioinformatics. 16:203 10.1186/s12859-015-0595-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu S., Yopp A.C., Mao X., Chen D., Zhang N., Chen D., Mao M., Ding Y., and Bromberg J.S.. 2004. CD4+ CD25+ CD62+ T-regulatory cell subset has optimal suppressive and proliferative potential. Am. J. Transplant. 4:65–78. 10.1046/j.1600-6143.2003.00293.x [DOI] [PubMed] [Google Scholar]
- Fukata M., Breglio K., Chen A., Vamadevan A.S., Goo T., Hsu D., Conduah D., Xu R., and Abreu M.T.. 2008. The myeloid differentiation factor 88 (MyD88) is required for CD4+ T cell effector function in a murine model of inflammatory bowel disease. J. Immunol. 180:1886–1894. 10.4049/jimmunol.180.3.1886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuss I.J., Neurath M., Boirivant M., Klein J.S., de la Motte C., Strong S.A., Fiocchi C., and Strober W.. 1996. Disparate CD4+ lamina propria (LP) lymphokine secretion profiles in inflammatory bowel disease. Crohn’s disease LP cells manifest increased secretion of IFN-γ, whereas ulcerative colitis LP cells manifest increased secretion of IL-5. J. Immunol. 157:1261–1270. [PubMed] [Google Scholar]
- Gaboriau-Routhiau V., Rakotobe S., Lécuyer E., Mulder I., Lan A., Bridonneau C., Rochet V., Pisi A., De Paepe M., Brandi G., et al. . 2009. The key role of segmented filamentous bacteria in the coordinated maturation of gut helper T cell responses. Immunity. 31:677–689. 10.1016/j.immuni.2009.08.020 [DOI] [PubMed] [Google Scholar]
- Garrett W.S., Lord G.M., Punit S., Lugo-Villarino G., Mazmanian S.K., Ito S., Glickman J.N., and Glimcher L.H.. 2007. Communicable ulcerative colitis induced by T-bet deficiency in the innate immune system. Cell. 131:33–45. 10.1016/j.cell.2007.08.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerbitz A., Schultz M., Wilke A., Linde H.J., Schölmerich J., Andreesen R., and Holler E.. 2004. Probiotic effects on experimental graft-versus-host disease: let them eat yogurt. Blood. 103:4365–4367. 10.1182/blood-2003-11-3769 [DOI] [PubMed] [Google Scholar]
- Geuking M.B., Cahenzli J., Lawson M.A., Ng D.C., Slack E., Hapfelmeier S., McCoy K.D., and Macpherson A.J.. 2011. Intestinal bacterial colonization induces mutualistic regulatory T cell responses. Immunity. 34:794–806. 10.1016/j.immuni.2011.03.021 [DOI] [PubMed] [Google Scholar]
- Hooper L.V., and Macpherson A.J.. 2010. Immune adaptations that maintain homeostasis with the intestinal microbiota. Nat. Rev. Immunol. 10:159–169. 10.1038/nri2710 [DOI] [PubMed] [Google Scholar]
- Huter E.N., Punkosdy G.A., Glass D.D., Cheng L.I., Ward J.M., and Shevach E.M.. 2008. TGF-β-induced Foxp3+ regulatory T cells rescue scurfy mice. Eur. J. Immunol. 38:1814–1821. 10.1002/eji.200838346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov I.I., Atarashi K., Manel N., Brodie E.L., Shima T., Karaoz U., Wei D., Goldfarb K.C., Santee C.A., Lynch S.V., et al. . 2009. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 139:485–498. 10.1016/j.cell.2009.09.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izcue A., Coombes J.L., and Powrie F.. 2009. Regulatory lymphocytes and intestinal inflammation. Annu. Rev. Immunol. 27:313–338. 10.1146/annurev.immunol.021908.132657 [DOI] [PubMed] [Google Scholar]
- Kamada N., Seo S.U., Chen G.Y., and Núñez G.. 2013. Role of the gut microbiota in immunity and inflammatory disease. Nat. Rev. Immunol. 13:321–335. 10.1038/nri3430 [DOI] [PubMed] [Google Scholar]
- Kawamoto S., Tran T.H., Maruya M., Suzuki K., Doi Y., Tsutsui Y., Kato L.M., and Fagarasan S.. 2012. The inhibitory receptor PD-1 regulates IgA selection and bacterial composition in the gut. Science. 336:485–489. 10.1126/science.1217718 [DOI] [PubMed] [Google Scholar]
- Kawamoto S., Maruya M., Kato L.M., Suda W., Atarashi K., Doi Y., Tsutsui Y., Qin H., Honda K., Okada T., et al. . 2014. Foxp3+ T cells regulate immunoglobulin A selection and facilitate diversification of bacterial species responsible for immune homeostasis. Immunity. 41:152–165. 10.1016/j.immuni.2014.05.016 [DOI] [PubMed] [Google Scholar]
- Khiong K., Murakami M., Kitabayashi C., Ueda N., Sawa S., Sakamoto A., Kotzin B.L., Rozzo S.J., Ishihara K., Verella-Garcia M., et al. . 2007. Homeostatically proliferating CD4 T cells are involved in the pathogenesis of an Omenn syndrome murine model. J. Clin. Invest. 117:1270–1281. 10.1172/JCI30513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieper W.C., Troy A., Burghardt J.T., Ramsey C., Lee J.Y., Jiang H.Q., Dummer W., Shen H., Cebra J.J., and Surh C.D.. 2005. Recent immune status determines the source of antigens that drive homeostatic T cell expansion. J. Immunol. 174:3158–3163. 10.4049/jimmunol.174.6.3158 [DOI] [PubMed] [Google Scholar]
- Kim M., Lee K.H., Yoon S.W., Kim B.S., Chun J., and Yi H.. 2013. Analytical tools and databases for metagenomics in the next-generation sequencing era. Genomics Inform. 11:102–113. 10.5808/GI.2013.11.3.102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King C., Ilic A., Koelsch K., and Sarvetnick N.. 2004. Homeostatic expansion of T cells during immune insufficiency generates autoimmunity. Cell. 117:265–277. 10.1016/S0092-8674(04)00335-6 [DOI] [PubMed] [Google Scholar]
- Lane J.P., Stewart C.J., Cummings S.P., and Gennery A.R.. 2015. Gut microbiome variations during hematopoietic stem cell transplant in severe combined immunodeficiency. J. Allergy Clin. Immunol. 135:1654–1656. 10.1016/j.jaci.2015.01.024 [DOI] [PubMed] [Google Scholar]
- Langmead B., and Salzberg S.L.. 2012. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 9:357–359. 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang S.C., Tan X.Y., Luxenberg D.P., Karim R., Dunussi-Joannopoulos K., Collins M., and Fouser L.A.. 2006. Interleukin (IL)-22 and IL-17 are coexpressed by Th17 cells and cooperatively enhance expression of antimicrobial peptides. J. Exp. Med. 203:2271–2279. 10.1084/jem.20061308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodes M.J., Cong Y., Elson C.O., Mohamath R., Landers C.J., Targan S.R., Fort M., and Hershberg R.M.. 2004. Bacterial flagellin is a dominant antigen in Crohn disease. J. Clin. Invest. 113:1296–1306. 10.1172/JCI200420295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macpherson A.J., and Uhr T.. 2004. Induction of protective IgA by intestinal dendritic cells carrying commensal bacteria. Science. 303:1662–1665. 10.1126/science.1091334 [DOI] [PubMed] [Google Scholar]
- Macpherson A.J., Gatto D., Sainsbury E., Harriman G.R., Hengartner H., and Zinkernagel R.M.. 2000. A primitive T cell-independent mechanism of intestinal mucosal IgA responses to commensal bacteria. Science. 288:2222–2226. 10.1126/science.288.5474.2222 [DOI] [PubMed] [Google Scholar]
- Magoč T., and Salzberg S.L.. 2011. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics. 27:2957–2963. 10.1093/bioinformatics/btr507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maina V., Marrella V., Mantero S., Cassani B., Fontana E., Anselmo A., Del Prete A., Sozzani S., Vezzoni P., Poliani P.L., and Villa A.. 2013. Hypomorphic mutation in the RAG2 gene affects dendritic cell distribution and migration. J. Leukoc. Biol. 94:1221–1230. 10.1189/jlb.0713365 [DOI] [PubMed] [Google Scholar]
- Manichanh C., Borruel N., Casellas F., and Guarner F.. 2012. The gut microbiota in IBD. Nat. Rev. Gastroenterol. Hepatol. 9:599–608. 10.1038/nrgastro.2012.152 [DOI] [PubMed] [Google Scholar]
- Manzari C., Fosso B., Marzano M., Annese A., Caprioli R., D’Erchia A., Gissi C., Intranuovo M., Picardi E., Santamaria M., et al. . 2015. The influence of invasive jellyfish blooms on the aquatic microbiome in a coastal lagoon (Varano, SE Italy) detected by an Illumina-based deep sequencing strategy. Biol. Invasions. 17:923–940. 10.1007/s10530-014-0810-2 [DOI] [Google Scholar]
- Marrella V., Poliani P.L., Casati A., Rucci F., Frascoli L., Gougeon M.L., Lemercier B., Bosticardo M., Ravanini M., Battaglia M., et al. . 2007. A hypomorphic R229Q Rag2 mouse mutant recapitulates human Omenn syndrome. J. Clin. Invest. 117:1260–1269. 10.1172/JCI30928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marrella V., Maina V., and Villa A.. 2011. Omenn syndrome does not live by V(D)J recombination alone. Curr. Opin. Allergy Clin. Immunol. 11:525–531. 10.1097/ACI.0b013e32834c311a [DOI] [PubMed] [Google Scholar]
- Marrella V., Poliani P.L., Fontana E., Casati A., Maina V., Cassani B., Ficara F., Cominelli M., Schena F., Paulis M., et al. . 2012. Anti-CD3ε mAb improves thymic architecture and prevents autoimmune manifestations in a mouse model of Omenn syndrome: therapeutic implications. Blood. 120:1005–1014. 10.1182/blood-2012-01-406827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milner J.D., Ward J.M., Keane-Myers A., and Paul W.E.. 2007. Lymphopenic mice reconstituted with limited repertoire T cells develop severe, multiorgan, Th2-associated inflammatory disease. Proc. Natl. Acad. Sci. USA. 104:576–581. 10.1073/pnas.0610289104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mora J.R. 2008. Homing imprinting and immunomodulation in the gut: role of dendritic cells and retinoids. Inflamm. Bowel Dis. 14:275–289. 10.1002/ibd.20280 [DOI] [PubMed] [Google Scholar]
- Mora J.R., and Von Andrian U.H.. 2006. Specificity and plasticity of memory lymphocyte migration. Curr. Top. Microbiol. Immunol. 308:83–116. [PubMed] [Google Scholar]
- Navarro S., Cossalter G., Chiavaroli C., Kanda A., Fleury S., Lazzari A., Cazareth J., Sparwasser T., Dombrowicz D., Glaichenhaus N., and Julia V.. 2011. The oral administration of bacterial extracts prevents asthma via the recruitment of regulatory T cells to the airways. Mucosal Immunol. 4:53–65. 10.1038/mi.2010.51 [DOI] [PubMed] [Google Scholar]
- Nishio J., Baba M., Atarashi K., Tanoue T., Negishi H., Yanai H., Habu S., Hori S., Honda K., and Taniguchi T.. 2015. Requirement of full TCR repertoire for regulatory T cells to maintain intestinal homeostasis. Proc. Natl. Acad. Sci. USA. 112:12770–12775. 10.1073/pnas.1516617112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochs H.D., Davis S.D., Mickelson E., Lerner K.G., and Wedgwood R.J.. 1974. Combined immunodeficiency and reticuloendotheliosis with eosinophilia. J. Pediatr. 85:463–465. 10.1016/S0022-3476(74)80445-2 [DOI] [PubMed] [Google Scholar]
- Omenn G.S. 1965. Familial reticuloendotheliosis with eosinophilia. N. Engl. J. Med. 273:427–432. 10.1056/NEJM196508192730806 [DOI] [PubMed] [Google Scholar]
- Palm N.W., de Zoete M.R., Cullen T.W., Barry N.A., Stefanowski J., Hao L., Degnan P.H., Hu J., Peter I., Zhang W., et al. . 2014. Immunoglobulin A coating identifies colitogenic bacteria in inflammatory bowel disease. Cell. 158:1000–1010. 10.1016/j.cell.2014.08.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papadakis K.A., Prehn J., Moreno S.T., Cheng L., Kouroumalis E.A., Deem R., Breaverman T., Ponath P.D., Andrew D.P., Green P.H., et al. . 2001. CCR9-positive lymphocytes and thymus-expressed chemokine distinguish small bowel from colonic Crohn’s disease. Gastroenterology. 121:246–254. 10.1053/gast.2001.27154 [DOI] [PubMed] [Google Scholar]
- Pedros C., Duguet F., Saoudi A., and Chabod M.. 2016. Distrupted regulatory T cell homeostasis in inflammatory bowel diseases. World J. Gastroenterol. 22:974–995. 10.3748/wjg.v22.i3.974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peron J.P., Yang K., Chen M.L., Brandao W.N., Basso A.S., Commodaro A.G., Weiner H.L., and Rizzo L.V.. 2010. Oral tolerance reduces Th17 cells as well as the overall inflammation in the central nervous system of EAE mice. J. Neuroimmunol. 227:10–17. 10.1016/j.jneuroim.2010.06.002 [DOI] [PubMed] [Google Scholar]
- Poliani P.L., Facchetti F., Ravanini M., Gennery A.R., Villa A., Roifman C.M., and Notarangelo L.D.. 2009. Early defects in human T-cell development severely affect distribution and maturation of thymic stromal cells: possible implications for the pathophysiology of Omenn syndrome. Blood. 114:105–108. 10.1182/blood-2009-03-211029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powrie F., Correa-Oliveira R., Mauze S., and Coffman R.L.. 1994. Regulatory interactions between CD45RBhigh and CD45RBlow CD4+ T cells are important for the balance between protective and pathogenic cell-mediated immunity. J. Exp. Med. 179:589–600. 10.1084/jem.179.2.589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proietti M., Cornacchione V., Rezzonico Jost T., Romagnani A., Faliti C.E., Perruzza L., Rigoni R., Radaelli E., Caprioli F., Preziuso S., et al. . 2014. ATP-gated ionotropic P2X7 receptor controls follicular T helper cell numbers in Peyer’s patches to promote host-microbiota mutualism. Immunity. 41:789–801. 10.1016/j.immuni.2014.10.010 [DOI] [PubMed] [Google Scholar]
- Rakoff-Nahoum S., Hao L., and Medzhitov R.. 2006. Role of toll-like receptors in spontaneous commensal-dependent colitis. Immunity. 25:319–329. 10.1016/j.immuni.2006.06.010 [DOI] [PubMed] [Google Scholar]
- Rieux-Laucat F., Bahadoran P., Brousse N., Selz F., Fischer A., Le Deist F., and De Villartay J.P.. 1998. Highly restricted human T cell repertoire in peripheral blood and tissue-infiltrating lymphocytes in Omenn’s syndrome. J. Clin. Invest. 102:312–321. 10.1172/JCI332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivas M.N., Koh Y.T., Chen A., Nguyen A., Lee Y.H., Lawson G., and Chatila T.A.. 2012. MyD88 is critically involved in immune tolerance breakdown at environmental interfaces of Foxp3-deficient mice. J. Clin. Invest. 122:1933–1947. 10.1172/JCI40591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosser E.C., Oleinika K., Tonon S., Doyle R., Bosma A., Carter N.A., Harris K.A., Jones S.A., Klein N., and Mauri C.. 2014. Regulatory B cells are induced by gut microbiota-driven interleukin-1β and interleukin-6 production. Nat. Med. 20:1334–1339. 10.1038/nm.3680 [DOI] [PubMed] [Google Scholar]
- Round J.L., and Mazmanian S.K.. 2010. Inducible Foxp3+ regulatory T-cell development by a commensal bacterium of the intestinal microbiota. Proc. Natl. Acad. Sci. USA. 107:12204–12209. 10.1073/pnas.0909122107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Round J.L., Lee S.M., Li J., Tran G., Jabri B., Chatila T.A., and Mazmanian S.K.. 2011. The Toll-like receptor 2 pathway establishes colonization by a commensal of the human microbiota. Science. 332:974–977. 10.1126/science.1206095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schenten D., Nish S.A., Yu S., Yan X., Lee H.K., Brodsky I., Pasman L., Yordy B., Wunderlich F.T., Brüning J.C., et al. . 2014. Signaling through the adaptor molecule MyD88 in CD4+ T cells is required to overcome suppression by regulatory T cells. Immunity. 40:78–90. 10.1016/j.immuni.2013.10.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider M.A., Meingassner J.G., Lipp M., Moore H.D., and Rot A.. 2007. CCR7 is required for the in vivo function of CD4+ CD25+ regulatory T cells. J. Exp. Med. 204:735–745. 10.1084/jem.20061405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shulzhenko N., Morgun A., Hsiao W., Battle M., Yao M., Gavrilova O., Orandle M., Mayer L., Macpherson A.J., McCoy K.D., et al. . 2011. Crosstalk between B lymphocytes, microbiota and the intestinal epithelium governs immunity versus metabolism in the gut. Nat. Med. 17:1585–1593. 10.1038/nm.2505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Signorini S., Imberti L., Pirovano S., Villa A., Facchetti F., Ungari M., Bozzi F., Albertini A., Ugazio A.G., Vezzoni P., and Notarangelo L.D.. 1999. Intrathymic restriction and peripheral expansion of the T-cell repertoire in Omenn syndrome. Blood. 94:3468–3478. [PubMed] [Google Scholar]
- Slack E., Hapfelmeier S., Stecher B., Velykoredko Y., Stoel M., Lawson M.A., Geuking M.B., Beutler B., Tedder T.F., Hardt W.D., et al. . 2009. Innate and adaptive immunity cooperate flexibly to maintain host-microbiota mutualism. Science. 325:617–620. 10.1126/science.1172747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith K., McCoy K.D., and Macpherson A.J.. 2007. Use of axenic animals in studying the adaptation of mammals to their commensal intestinal microbiota. Semin. Immunol. 19:59–69. 10.1016/j.smim.2006.10.002 [DOI] [PubMed] [Google Scholar]
- Vaishnava S., Yamamoto M., Severson K.M., Ruhn K.A., Yu X., Koren O., Ley R., Wakeland E.K., and Hooper L.V.. 2011. The antibacterial lectin RegIIIγ promotes the spatial segregation of microbiota and host in the intestine. Science. 334:255–258. 10.1126/science.1209791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villa A., Santagata S., Bozzi F., Giliani S., Frattini A., Imberti L., Gatta L.B., Ochs H.D., Schwarz K., Notarangelo L.D., et al. . 1998. Partial V(D)J recombination activity leads to Omenn syndrome. Cell. 93:885–896. 10.1016/S0092-8674(00)81448-8 [DOI] [PubMed] [Google Scholar]
- Vossen J.M., Guiot H.F., Lankester A.C., Vossen A.C., Bredius R.G., Wolterbeek R., Bakker H.D., and Heidt P.J.. 2014. Complete suppression of the gut microbiome prevents acute graft-versus-host disease following allogeneic bone marrow transplantation. PLoS One. 9:e105706 10.1371/journal.pone.0105706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss J.M., Bilate A.M., Gobert M., Ding Y., Curotto de Lafaille M.A., Parkhurst C.N., Xiong H., Dolpady J., Frey A.B., Ruocco M.G., et al. . 2012. Neuropilin 1 is expressed on thymus-derived natural regulatory T cells, but not mucosa-generated induced Foxp3+ T reg cells. J. Exp. Med. 209:1723–1742. 10.1084/jem.20120914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xavier R.J., and Podolsky D.K.. 2007. Unravelling the pathogenesis of inflammatory bowel disease. Nature. 448:427–434. 10.1038/nature06005 [DOI] [PubMed] [Google Scholar]
- Yadav M., Louvet C., Davini D., Gardner J.M., Martinez-Llordella M., Bailey-Bucktrout S., Anthony B.A., Sverdrup F.M., Head R., Kuster D.J., et al. . 2012. Neuropilin-1 distinguishes natural and inducible regulatory T cells among regulatory T cell subsets in vivo. J. Exp. Med. 209:1713–1722. 10.1084/jem.20120822 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.