Mice with a mutated form of RNase H2 found in patients with the neuroinflammatory Aicardi-Goutières Syndrome develop a lethal, cGAS–STING–dependent disease.
Abstract
The neuroinflammatory autoimmune disease Aicardi-Goutières syndrome (AGS) develops from mutations in genes encoding several nucleotide-processing proteins, including RNase H2. Defective RNase H2 may induce accumulation of self-nucleic acid species that trigger chronic type I interferon and inflammatory responses, leading to AGS pathology. We created a knock-in mouse model with an RNase H2 AGS mutation in a highly conserved residue of the catalytic subunit, Rnaseh2aG37S/G37S (G37S), to understand disease pathology. G37S homozygotes are perinatal lethal, in contrast to the early embryonic lethality previously reported for Rnaseh2b- or Rnaseh2c-null mice. Importantly, we found that the G37S mutation led to increased expression of interferon-stimulated genes dependent on the cGAS–STING signaling pathway. Ablation of STING in the G37S mice results in partial rescue of the perinatal lethality, with viable mice exhibiting white spotting on their ventral surface. We believe that the G37S knock-in mouse provides an excellent animal model for studying RNASEH2-associated autoimmune diseases.
Aicardi-Goutières syndrome (AGS) is a rare neuroinflammatory disorder (Crow et al., 2015). In most cases, it is present at birth with symptoms indistinguishable from those associated with congenital viral infection, such as elevated levels of type I IFN in the serum and cerebrospinal fluid. AGS arises from mutations in seven different genes: RNASEH2A, RNASEH2B, RNASEH2C, TREX1, SAMHD1, ADAR1, and IFIH1, all of which are nucleic acid–transacting enzymes. AGS is believed to result from activation of the innate immune pathway by nucleic acids accumulating in the cytosol when an AGS-associated gene is defective. AGS shares some disease hallmarks with systemic lupus erythematosus (Günther et al., 2015). Over 50% of AGS patients carry biallelic mutations in the genes encoding the three subunits of the heterotrimeric RNase H2 complex (RNase H2A, RNase H2B, and RNase 2C; Crow et al., 2015).
RNase H2 provides the main RNase H activity in humans (Cerritelli and Crouch, 2009) and is essential for removing ribonucleotides incorporated in genomic DNA during replication, as well as for resolving R-loops formed during transcription (Nick McElhinny et al., 2010; Reijns et al., 2012; Chon et al., 2013). The crystal structures of human and mouse RNase H2 revealed the interactions of the subunits and the positions of the more than 50 known AGS-related mutations in the three subunits (Figiel et al., 2011; Reijns et al., 2011; Reijns and Jackson, 2014). Some mutations are located near the catalytic center and affect catalysis, whereas others affect stability or alter protein interactions. The most common mutations reported in AGS patients are found in the B subunit (Crow et al., 2015) and are associated with less severe disease phenotype than mutations in the catalytic A subunit. A mutation in a highly conserved glycine (G37S) near the catalytic center on the RNase H2A subunit causes a severe early onset presentation of AGS, likely as a result of a substantial loss of RNase H activity (Crow et al., 2006). Many in vitro studies showed that Serine substitution for Glycine 37 in RNases H2 of eukaryotes reduces RNase activity (Crow et al., 2006; Rohman et al., 2008; Chon et al., 2009; Coffin et al., 2011). Mouse models using deletions of RNASE2B and RNASEH2C exhibit significant DNA damage, resulting in embryonic lethality at E9.5 (Hiller et al., 2012; Reijns et al., 2012). These mice have elucidated important information on the role of RNase H2 in genome stability, but because of their early death, have not yielded insight into the innate immune pathways responsible for disease manifestation. Likewise, neither do mice with residual levels of RNase H2B (R2B KOF; Hiller et al., 2012), or the viable and asymptomatic Rnaseh2bA174T/A174T mouse model (Reijns and Jackson, 2014). Therefore, it remains unclear how RNASEH2 mutations lead to the development of AGS.
RESULTS AND DISCUSSION
G37S homozygous mice are perinatal lethal, and G37S embryos show increased expression in IFN-stimulated genes
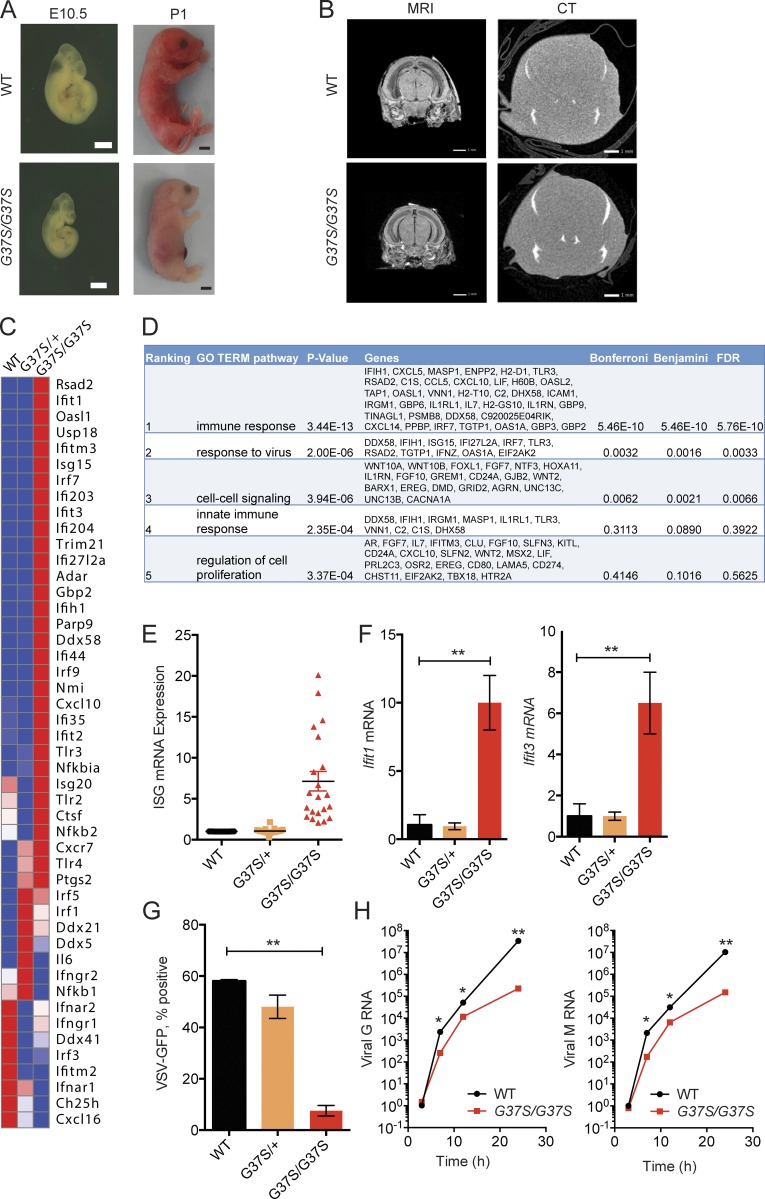
We generated Rnaseh2a-G37S knock-in mice to mimic the exact mutation present in AGS patients. Rnaseh2aG37S/+ mice were viable, with no evident deleterious phenotype. Breeding of Rnaseh2aG37S/+ mice generate still-born pups that were homozygotes for the mutation (Fig. 1 A). No viable G37S homozygotes were observed (dead at or within hours after birth). The G37S homozygote embryos were smaller from an early embryogenesis period of E10.5, present at expected Mendelian ratio, and were ∼20% smaller in size compared with their uterine mates (Fig. 1 A). Magnetic resonance imaging and micro-computed tomography scans did not reveal any additional phenotypic abnormalities in Rnaseh2aG37S/G37S embryos in the brain (Fig. 1 B) or elsewhere (not depicted). We also did not observe any inflammation in histology staining of tissues of Rnaseh2aG37S/G37S embryos, including the brain (unpublished data). The lack of neuroinflammation could be caused by early death of the animal, or by different disease presentation in mouse versus human. Other AGS mouse models such as Trex1−/− or Samhd1−/− also lack evidence of neuroinflammation (Gall et al., 2012; Rehwinkel et al., 2013).
Figure 1.
Primary cells from Rnaseh2aG37S/G37S embryos show increased expression in ISGs. (A) Images of WT and Rnaseh2aG37S/G37S embryos at indicated stage (bottom). Bars, 2 mm. (B) Transcranial images of ex vivo mouse E18.5 embryos WT and Rnaseh2aG37S/G37S in MRI gradient echo image and micro computed tomography scans. No appreciable calcium, other than in the bones of the cranium (top arrow) and jaw (bottom arrow), which are still under formation. Bars, 1 mm. (C) A heat map of immune gene expression in WT, Rnaseh2aG37S/+ and Rnaseh2aG37S/G37S primary MEFs. Data from RNA-seq (Table S1). (D) Gene ontology analysis of 388 genes that are increased by twofold or more in Rnaseh2aG37S/G37S compared with WT MEFs. Top five enriched pathways are shown. (E) Expression of ISGs in WT, Rnaseh2aG37S/+, and Rnaseh2aG37S/G37S primary MEFs. Each dot represents a different ISG. Data from RNA-seq. (F) Quantitative RT-PCR analysis of Ifit1 and Ifit3 mRNA (ISGs) in WT, Rnaseh2aG37S/+, and Rnaseh2aG37S/G37S primary MEFs. (G and H) VSV-GFP replication in WT, Rnaseh2aG37S/+, and Rnaseh2aG37S/G37S primary MEFs. FACS analysis measures VSV-GFP signal at 24 h after infection (G). Quantitative RT-PCR analysis of VSV G and M RNA measure viral RNA replication at indicated time after infection (H). *, P < 0.05; **, P < 0.01. Mice were compared with littermate controls. Data are representative of at least three independent experiments. Error bars represent the SEM. Unpaired Student’s t test (F–H).
We next examined where we can detect a molecular signature of immune activation in Rnaseh2aG37S/G37S embryos, as it would be expected from its association with AGS. We performed RNA-seq analysis comparing gene expression profiles of WT, Rnaseh2aG37S/+, and Rnaseh2aG37S/G37S primary MEFs isolated from E13.5 embryos. We found that 388 genes were up-regulated twofold or more in Rnaseh2aG37S/G37S MEFs compared with WT; of those, the most enriched gene network was “immune response” (DAVID GO term analysis; Fig. 1, C and D). Many of the highly up-regulated genes in Rnaseh2aG37S/G37S cells were IFN-stimulated genes (ISGs), such as Ifit44, Usp18, Ifit1, Rsad2, Isg15, Irf7, and Cxcl10 (Fig. 1 D). We validated increased expression of Ifit1, Ifit3, Rsad2, and Cxcl10 by quantitative RT-PCR (Figs. 1 E and 2). As ISGs provide defense mechanisms against viral infection, we infected WT, Rnaseh2aG37S/+, and Rnaseh2aG37S/G37S primary MEFs with vesicular stomatitis virus (VSV)-PeGFP, to assess their functionality. We found that Rnaseh2aG37S/G37S MEFs were highly refractory to VSV infection, as measured by reduced GFP fluorescence at 24 h or by reduced VSV G and M RNA from 6 to 24 h after infection (Fig. 1 F). Collectively, our data showed that the homozygous G37S mutation in mice invokes innate immune activation of ISGs, similar to that of AGS patients (Crow et al., 2015).
Figure 2.
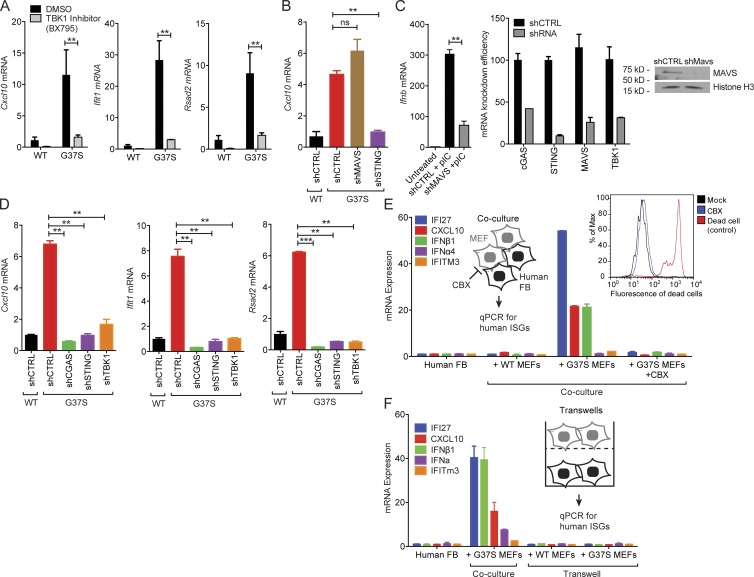
Immune activation in Rnaseh2aG37S/G37S primary MEFs requires the cGAS–STING innate immune pathway. (A) Quantitative RT-PCR analysis of Cxcl10, Ifit1 and Rsad2 mRNA (all ISGs) in WT and Rnaseh2aG37S/G37S (G37S, same below) MEFs treated with DMSO or TBK1 inhibitor BX795 (10 µM) for 6 h. (B) Quantitative RT-PCR analysis of Cxcl10 mRNA in WT and G37S MEFs treated with shRNA against indicated genes involved in cytosolic nucleic acid-sensing. (C) shMAVS knockdown reduces poly(I:C)-induced IFN response. Knockdown efficiency is shown on the right. (D) Quantitative RT-PCR analysis of Cxcl10, Ifit1, and Rsad2 mRNA in WT and G37S MEFs treated with shRNA against indicated genes involved in DNA-sensing pathway. (E) Quantitative RT-PCR analysis of a panel of human ISGs and IFN genes in human fibroblasts (BJ-1 cells) co-cultured with WT or G37S MEFs for 18 h, with or without CBX treatment (inhibits gap junction). Left inset shows a schematic diagram of the gap junction assay. Right inset shows FACS analysis of cell death in mock- and CBX-treated cells. (F) Quantitative RT-PCR analysis of human ISGs in human fibroblasts in a trans-well assay co-cultured with WT or G37S MEFs for 18 h. Mice were compared with littermate controls. **, P < 0.01; ***, P < 0.001. ns, not significant. Data are representative of at least three independent experiments. Error bars represent the SEM. Unpaired Student’s t test (A–D).
Immune activation in Rnaseh2aG37S/G37S primary MEFs requires the cGAS–STING innate immune pathway
We next aimed to determine the signaling pathways responsible for the innate immune activation in Rnaseh2aG37S/G37S cells. Many of the up-regulated ISGs we observed are direct targets of the transcription factor IRF3 (Diamond and Farzan, 2013; Lazear et al., 2013), which is activated by phosphorylation by TBK1. We treated Rnaseh2aG37S/G37S cells with a TBK1 inhibitor BX795, and observed reduced expression of activated genes, Cxcl10, Ifit1, and Rsad2 in Rnaseh2aG37S/G37S cells (Fig. 2 A), suggesting the involvement of a cytosolic immune-sensing pathway. Using shRNA directed against Mavs or Sting (adaptor proteins for cytosolic RNA or DNA sensing, respectively) in Rnaseh2aG37S/G37S cells, we found that shRNA against Sting restored the low expression levels of Cxcl10 mRNA to WT levels. In contrast, Mavs knockdown significantly reduced poly(I:C)-induced IFN response, but had no effect on the G37S-induced ISG expression (Fig. 2, B and C), suggesting that the G37S mutation leads to activation of a DNA- but not RNA-sensing pathway.
We next used shRNA to knock down components of the cytosolic DNA-sensing pathway in Rnaseh2aG37S/G37S cells, and then examined expression of ISGs. shSting and shTbk1 effectively restored mRNA levels to those seen in WT cells (Fig. 2 D). A prominent driver of cytosolic DNA detection, cGAS, responds to microbial or self-DNA and long, homopolymeric RNA/DNA hybrids (Ablasser et al., 2014; Mankan et al., 2014; Gao et al., 2015; Gray et al., 2015). shRNA-mediated knockdown of cGAS in Rnaseh2aG37S/G37S cells also returned ISG expression to low WT levels (Fig. 2 D). As the production of cGAMP by cGAS leads to activation of STING-mediated signaling, we next examined the presence of cGAMP in Rnaseh2aG37S/G37S cells using the gap junction transfer cGAMP bioassay (Ablasser et al., 2013). cGAMP can be transferred from producing cells to neighboring cells through gap junctions, thereby enabling a co-culture–based transactivation assay for cGAMP detection (Ablasser et al., 2013). We co-cultured Rnaseh2aG37S/G37S MEFs (producing cells) with human fibroblasts (target cells), and then used human-specific primers to probe the immune activation status of the human cells. Rnaseh2aG37S/G37S MEFs induced strong immune activation of the human ISGs tested, whereas WT MEFs had no effect (Fig. 2 E). Importantly, we also found that inhibiting gap junctions with carbenoxolone (CBX), a nontoxic gap junction inhibitor (Fig. 2 E), or trans-well separation completely abolished Rnaseh2aG37S/G37S MEF’s transactivation activity (Fig. 2 F). These data suggest that Rnaseh2aG37S/G37S primary MEFs produce cGAMP that activates the STING pathway.
Sting−/− partially rescues perinatal lethality of G37S mice
In the AGS mouse model of Trex1 gene deletion, viability of Trex1−/− mice is extended dramatically when the response to secreted type I IFN is ablated by deleting the IFN receptor, Infar1, or by eliminating the adaptive immune response through Rag2 gene deletion (Stetson et al., 2008). We thus bred G37S to Infar1−/− or Rag2−/− backgrounds and found that neither of these genetic knockouts was able to rescue the perinatal lethal phenotype (Table 1). These findings are consistent with lack of inflammation in the Rnaseh2aG37S/G37S embryos, further suggesting cell-intrinsic defects may be responsible. Partial rescue of the embryonic development defect seen in Rnaseh2b-null mice was obtained by deleting the p53−/− gene (Reijns et al., 2012). However, we found Rnaseh2aG37S/G37S p53−/− mice also exhibited perinatal lethality indistinguishable from Rnaseh2aG37S/G37S alone. We also did not observe elevated expression of p53 pathway genes from our RNA-seq analysis (unpublished data).
Table 1. Genetic crosses of G37S mice.
| Mice | p53−/− | INFAR−/− | Rag2−/− | Mavs−/− | Sting−/− | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Neonates (no. embryos) | Weaned pups (no. mice) | Neonates (no. embryos) | Weaned pups (no. mice) | Neonates (no. embryos) | Weaned pups (no. mice) | Neonates (no. embryos) | Weaned pups (no. mice) | Neonates (no. embryos) | Weaned pups (no. mice) | |||||
| WT | 23% (7) | 33% (17) | 24% (8) | 44% (67) | 26% (10) | 32% (30) | 22% (4) | 41% (52) | 23% (7) | 32% (95) | ||||
| G37S/+ | 58% (18) | 67% (34) | 52% (17) | 56% (87) | 53% (20) | 68% (65) | 56% (10) | 59% (74) | 60% (18) | 66% (196) | ||||
| G37S/ G37S | 19% (6) | 0% (0) | 24% (8) | 0% (0) | 21% (8) | 0% (0) | 22% (4) | 0% (0) | 17% (5) | 2% (6) | ||||
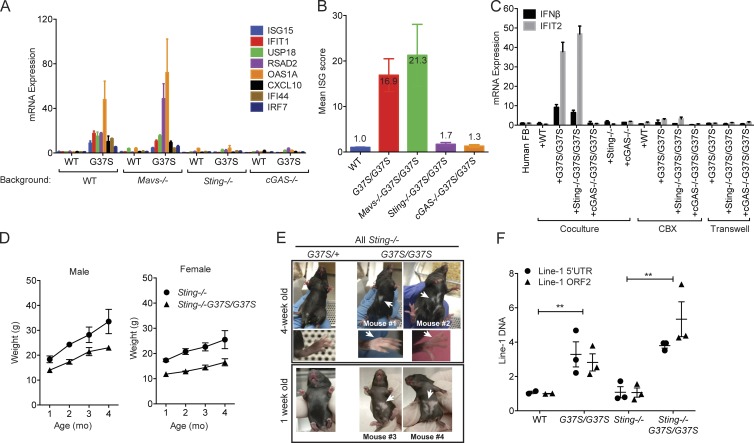
We next bred Rnaseh2aG37S/G37S onto Mavs−/− or Sting−/− background. Rnaseh2aG37S/G37S remains perinatal lethal on Mavs−/− background (Table 1), consistent with RNA-sensing pathways being uninvolved (Fig. 2). Remarkably, we obtained viable pups of Rnaseh2aG37S/G37SSting−/− genotype, albeit at 6% of expected frequency (or 2% of weaned pups from heterozygous crosses on the Sting−/− background; Table 1). Most of the Rnaseh2aG37S/G37SSting−/− still exhibited perinatal lethality, similar to Rnaseh2aG37S/G37S Sting+/− or Sting+/+ (Table 1). Mice were either perinatal lethal or survived after birth. The failure to rescue any lethality in G37S−/− Ifnar1−/− could reflect the limited number of offspring examined. We compared ISG expression in primary E14.5 MEFs, and found that Rnaseh2aG37S/G37SSting−/− completely returned ISG expression to the low level in Rnaseh2a+/+Sting−/−, whereas Mavs−/− had no effect (Fig. 3 A). We bred Rnaseh2aG37S/G37S onto cGAS−/− background and found that Rnaseh2aG37S/G37ScGAS−/− embryos also restored ISG expression to WT levels (Fig. 3 A). Moreover, the mean ISG score is highly elevated in Rnaseh2aG37S/G37S and Rnaseh2aG37S/G37SMavs−/− MEFs, whereas both Rnaseh2aG37S/G37SSting−/− and Rnaseh2aG37S/G37ScGAS−/− MEFs show similar baseline values as in WT (Fig. 3 B). We also measured cGAMP in both rescued MEFs using the gap junction bioassay. Both Rnaseh2aG37S/G37S and Rnaseh2aG37S/G37SSting−/− produce cGAMP, whereas Rnaseh2aG37S/G37ScGAS−/− MEFs did not (Fig. 3 C). These data further demonstrate that the cGAS–cGAMP–STING pathway is mediating the immune activation in G37S mice. Because only a small fraction of the progeny with Rnaseh2aG37S/G37SSting−/− genotype is viable, our data also suggest that innate immune activation through the cGAS–cGAMP–STING pathway only partially contributed to the lethality of G37S mice.
Figure 3.
Sting−/− partially rescues perinatal lethality of G37S mice. (A) Quantitative RT-PCR analysis of a panel of mouse ISGs in WT or G37S embryos on Mavs−/− or Sting−/− or cGAS−/− background. Total RNA was isolated from primary MEFs (E13.5) of indicated genotype. (B) Mean ISG score of indicated genotypes. Data from A. (C) Gap junction cGAMP bioassay. As in Fig. 2 E, MEFs of indicated genotype were co-cultured with human BJ-1 cells for 18 h, with or without CBX or direct contact (indicated on the bottom). Quantitative RT-PCR analysis of IFN-β and IFIT1 indicates cGAMP activity in MEFs. (D) Mouse body weights. n = 4. (E) White-spotting phenotype in Rnaseh2aG37S/G37S Sting−/− viable adults. (F) Quantitative PCR analysis of mouse Line-1 5′ UTR and ORF2 DNA in WT or G37S embryos (isolated from E13.5 or E15.5; n = 3). Each dot represents an individual embryo. Mice were compared with littermate controls and with age-matched knock-out mice **, P < 0.01. Data are representative of at least two independent experiments (A–C), or pooled data from multiple animals (D–F). Error bars represent the SEM. Unpaired Student’s t test (F).
White-spotting phenotype and increased LINE-1 expression
We also observed several interesting phenotypes in the viable Rnaseh2aG37S/G37SSting−/− mice. These mice are ∼70% in body size and weight compared with WT or heterozygous controls (Fig. 3 D). We have observed both male and female progeny for Rnaseh2aG37S/G37SSting−/−, and all have so far failed to produce offspring, whereas littermate controls are fertile. These rescued Rnaseh2aG37S/G37SSting−/− mice are grossly healthy, with the oldest animal approaching 1 yr of age. Intriguingly, all of the viable Rnaseh2aG37S/G37SSting−/− mice presented a ventral white spotting phenotype, as well as white hind- and forepaws that are not observed in WT or heterozygous littermates (Fig. 3 E). This phenotype was consistent from birth and remained throughout the lifespan of the mice. Histopathology analysis did not find any abnormalities or inflammation in internal organs, including the brain, of Rnaseh2aG37S/G37SSting−/− mice (not depicted). Skin histology from the white patches of Rnaseh2aG37S/G37SSting−/− mice is structurally normal, although lack of melanin in hair shafts is evident (unpublished data). As endogenous retroelements have been implicated in the pathogenesis of AGS (Volkman and Stetson, 2014) and the RNA/DNA hybrids or DNA of murine endogenous retroviruses can function as a ligand of the cGAS–STING pathway (Mankan et al., 2014), we measured LINE-1 element in WT, Rnaseh2aG37S/G37S, and Rnaseh2aG37S/G37SSting−/− E13.5 and E15.5 embryos. Indeed, we found that LINE-1 DNA level from cytosolic extract is increased in both Rnaseh2aG37S/G37S and Rnaseh2aG37S/G37SSting−/− embryos to similar levels compared with littermate WT embryos, suggesting that it is independent of immune activation (Fig. 3 F). However, we failed to observe elevation of L1 ORF1 protein by Western blot (not depicted). It remains unclear whether the increase in LINE-1 DNA is a result of an increase in LINE element activity, or because of defects in genomic structures where LINE elements are enriched as was recently suggested (Lim et al., 2015). Together, our data suggest that the G37S mutation causes white-spotting phenotype in Rnaseh2aG37S/G37SSting−/− mice, likely resulting from defects in melanocyte development or migration from progenitors at the neural crest. G37S mutation also causes increased level of LINE-1 DNA, which may contribute to the activation of the cGAS–STING pathway.
In summary, the G37S mouse represents the first RNase H2 mouse model with a clear immune activation phenotype, making it uniquely useful for understanding the associated human disease. Embryonic development until birth allowed the expression of innate immune signaling proteins or immune ligands, which uncovered the active cGAS–STING innate immune pathway in G37S mice. RNA/DNA hybrids or rNMPs in DNA could be a direct source of nucleic acids activating the DNA-sensing pathway. Alternatively, specific nucleic acids, such as LINE-1–derived nucleic acids, could elicit the innate immune response. The perinatal lethality of the G37S mice is likely caused in large part by a yet-to-be-identified biochemical defect associated with the mutation. Sting−/− only partially rescued the lethality, despite complete suppression of ISG expression, and p53−/− failed to rescue the lethality. Similarly, another AGS mouse model, the Adar1−/− mouse, exhibits early embryonic lethality, and it can be partially rescued to birth by Mavs−/− and to adult by Ifih1−/− (Mannion et al., 2014; Liddicoat et al., 2015). These findings indicate that Rnaseh2 and Adar1 genes associated with AGS have important functions that are critical for embryonic development in mice, beyond that of prohibiting formation of aberrant nucleic acids that activate innate immunity (Pestal et al., 2015). Further biochemical analysis comparing G37S and other existing Rnaseh2 knockout mouse models are necessary to elucidate the differences in biochemical defects and to shed light on the possible source of nucleic acids that trigger the cGAS–STING pathway in the G37S mice. Recently studies also showed that cGAS−/− can rescue the inflammation and mortality of Trex1−/− mice (another AGS mouse model; Gao et al., 2015; Gray et al., 2015). Therefore, our genetic and immunological analysis of the G37S mice further establish a critical role of the cGAS–STING pathway in G37S-induced immune activation, and suggest that therapeutic intervention of this pathway may be beneficial for treating AGS patients.
MATERIALS AND METHODS
Mice, cells, and viruses
G37S mice were generated by introducing the human disease associated point mutation into the conserved residue of mouse Rnaseh2a gene. Infar1−/− and Rag2−/− mice were obtained from Taconic Biosciences. p53−/− mice were obtained from The Jackson Laboratory. Mavs−/− and cGAS−/− mice were obtained from Z. Chen (University of Texas Southwestern Medical Center, Dallas, TX) and Sting−/− mice were obtained from G. Barber (University of Miami, Miami, FL). Primary MEFs were isolated from embryos of indicated embryonic dates. These cells were maintained in DMEM with 20% (vol/vol) heat-inactivated FCS, 2 mM l-glutamine, 10 mM Hepes, and 1 mM sodium pyruvate (complete DMEM) with the addition of 100 U/ml penicillin and 100 mg/ml streptomycin and were cultured at 37°C with 5% CO2. VSV-PeGFP is a gift from A. Pattnaik (University of Nebraska, Lincoln, NE; Das et al., 2014). Cells were plated overnight and, the next day, infected overnight with VSV-GFP at a multiplicity of infection of 1. Cells were washed with PBS before standard fixation with 4% paraformaldehyde in PBS (Affymetrix). Percentage of infectivity was assessed with FACSCalibur (BD). For viral RNA measurement, total RNA was extracted at various time points after infection, and VSV G and M RNA were measured with specific primers (Hasan et al., 2013). Experiments performed in BSL2 conditions were approved by the Environmental Health and Safety Committee at University of Texas Southwestern Medical Center. Experiments involving mouse materials were approved by the Institutional Animal Care and Use Committees of the University of Texas Southwestern Medical Center and the National Institute for Child Health and Human Development (Bethesda, MD).
RNA isolation and quantitative RT-PCR
Total RNA was isolated with TRI reagent according to the manufacturer’s protocol (Sigma-Aldrich), and cDNA was synthesized with iScript cDNA synthesis kit (Bio-Rad Laboratories). iTaq Universal SYBR Green Supermix (Bio-Rad Laboratories) and an ABI-7500 Fast Real-Time PCR system (Applied Biosystems) were used for quantitative RT-PCR analysis (primer sequences listed in Table S2). Hprt and Gapdh were used as housekeeping genes for data normalization. RNA-seq was performed as previously described (Hasan et al., 2013).
cGAMP activity bioassay
cGAMP activity in MEFs was measured by a co-culture bioassay as previously described (Ablasser et al., 2014). In brief, 2 × 105 human fibroblasts/ml were plated overnight. After attachment, 4 × 105 primary murine fibroblasts/ml were plated onto the human cells, with or without 200 µM CBX (Sigma-Aldrich) treatment or with separation by 0.4-µm polycarbonate Trans-well inserts (Corning) for 18 h. Subsequent analysis was performed using a human-specific PrimePCR Array plate (Bio-Rad Laboratories).
shRNA knockdown and TBK inhibitor
shRNA oligos were synthesized (Sigma-Aldrich) and cloned into a pLKO.1-TRC cloning vector following Addgene’s protocol. Lentiviral particles were packaged in HEK-293 T cells and filtered with Amicon Ultra-15 centrifugal filters (EMD Millipore). shRNA-harboring lentiviruses were allowed to infect cells overnight with polybrene (10 µg/ml) treatment, and cells expressing the shRNA were selected with a puromycin concentration (Life Technologies) for several days. Puromycin selection was removed several days before subsequent analysis. shRNA oligo sequences are listed in Table S1. For TBK1 inhibitor experiments, cells were treated with DMSO (% vol/vol) or 10 µM BX795 (InvivoGen) for 6 h. Cells were then washed with PBS and grown in DMEM with 20% FCS alone for several hours before subsequent analysis.
Line-1 retroelements
E13.5 or E15.5 embryos were homogenized into single-cell suspensions and cytosolic lysates obtained using a cytosolic extraction buffer as previously described (Yang et al., 2007). In brief, PBS-washed cell pellets were lysed in 10 mM Hepes, pH 7.9, 10 mM KCl, 1.5 mM MgCl2, 0.34 M sucrose, 10% glycerol, and 0.1% Triton X-100 for 5 min on ice. Cell lysates were treated with Proteinase K at 55°C for 1 h and with RNase I (Life Technologies) before phenol/chloroform extraction and ethanol precipitation overnight. Isolated DNA was resuspended in nuclease free H2O and directly subjected to quantitative PCR analysis using Line-1 primers (Table S1).
Statistical methods
Data are presented as the mean ± SEM. Prism 6 (GraphPad) was used for statistical analysis. Statistical tests performed are indicated in figure legends. *, P < 0.05; **, P < 0.01; ***, P < 0.001; and ****, P < 0.0001.
Online supplemental material
Table S1 lists RNA-seq data. Table S2 lists oligonucleotides used in this study. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20151464/DC1.
Supplementary Material
ACKNOWLEDGMENTS
We thank Zhijian James Chen for Mavs−/− and cGAS−/− mice, Glen Barber for Sting−/− mice, and members of the Yan and Crouch laboratories for helpful discussions.
This work is supported by grants from the National Institutes of Health (AI098569 and AR067135 to N. Yan), Alliance for Lupus Research (329774 N. Yan), UT Southwestern Immunology graduate program training grant (2T32AI005284 to V. Pokatayev), and in part by the Intramural Research Program of the National Institutes of Health.
The authors declare no competing financial interests.
Author contributions: V. Pokatayev performed immunology related experiments, including crosses between G37S mice and Mavs−/− or Sting−/− or cGAS−/− mice. S.M. Ceritelli was responsible for production of Rnaseh2aG37S ES cells. N. Hasin, H. Chon, and K. Sakhuja generated and characterized the initial phenotypes of the G37S mouse, including crosses between G37S mice and Infar−/−, Rag2−/−, or P53−/− mice. J.M. Ward performed pathology observations, H. Douglas Morris. performed MRI, CT and analyzed the images. N. Yan and R.J. Crouch jointly supervised the study. N. Yan, R.J. Crouch, V. Pokatayev, N. Hasin, and S.M. Cerritelli wrote the paper.
Footnotes
Abbreviations used:
- AGS
- Aicardi-Goutières syndrome
- CBX
- carbenoxolone
- ISG
- IFN-stimulated gene
- VSV
- vesicular stomatitis virus
References
- Ablasser A., Schmid-Burgk J.L., Hemmerling I., Horvath G.L., Schmidt T., Latz E., and Hornung V.. 2013. Cell intrinsic immunity spreads to bystander cells via the intercellular transfer of cGAMP. Nature. 503:530–534. 10.1038/nature12640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ablasser A., Hemmerling I., Schmid-Burgk J.L., Behrendt R., Roers A., and Hornung V.. 2014. TREX1 deficiency triggers cell-autonomous immunity in a cGAS-dependent manner. J. Immunol. 192:5993–5997. 10.4049/jimmunol.1400737 [DOI] [PubMed] [Google Scholar]
- Cerritelli S.M., and Crouch R.J.. 2009. Ribonuclease H: the enzymes in eukaryotes. FEBS J. 276:1494–1505. 10.1111/j.1742-4658.2009.06908.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chon H., Vassilev A., DePamphilis M.L., Zhao Y., Zhang J., Burgers P.M., Crouch R.J., and Cerritelli S.M.. 2009. Contributions of the two accessory subunits, RNASEH2B and RNASEH2C, to the activity and properties of the human RNase H2 complex. Nucleic Acids Res. 37:96–110. 10.1093/nar/gkn913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chon H., Sparks J.L., Rychlik M., Nowotny M., Burgers P.M., Crouch R.J., and Cerritelli S.M.. 2013. RNase H2 roles in genome integrity revealed by unlinking its activities. Nucleic Acids Res. 41:3130–3143. 10.1093/nar/gkt027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffin S.R., Hollis T., and Perrino F.W.. 2011. Functional consequences of the RNase H2A subunit mutations that cause Aicardi-Goutieres syndrome. J. Biol. Chem. 286:16984–16991. 10.1074/jbc.M111.228833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crow Y.J., Leitch A., Hayward B.E., Garner A., Parmar R., Griffith E., Ali M., Semple C., Aicardi J., Babul-Hirji R., et al. 2006. Mutations in genes encoding ribonuclease H2 subunits cause Aicardi-Goutières syndrome and mimic congenital viral brain infection. Nat. Genet. 38:910–916. 10.1038/ng1842 [DOI] [PubMed] [Google Scholar]
- Crow Y.J., Chase D.S., Lowenstein Schmidt J., Szynkiewicz M., Forte G.M., Gornall H.L., Oojageer A., Anderson B., Pizzino A., Helman G., et al. 2015. Characterization of human disease phenotypes associated with mutations in TREX1, RNASEH2A, RNASEH2B, RNASEH2C, SAMHD1, ADAR, and IFIH1. Am. J. Med. Genet. A. 167A:296–312. 10.1002/ajmg.a.36887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das A., Dinh P.X., Panda D., and Pattnaik A.K.. 2014. Interferon-inducible protein IFI35 negatively regulates RIG-I antiviral signaling and supports vesicular stomatitis virus replication. J. Virol. 88:3103–3113. 10.1128/JVI.03202-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diamond M.S., and Farzan M.. 2013. The broad-spectrum antiviral functions of IFIT and IFITM proteins. Nat. Rev. Immunol. 13:46–57. 10.1038/nri3344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figiel M., Chon H., Cerritelli S.M., Cybulska M., Crouch R.J., and Nowotny M.. 2011. The structural and biochemical characterization of human RNase H2 complex reveals the molecular basis for substrate recognition and Aicardi-Goutières syndrome defects. J. Biol. Chem. 286:10540–10550. 10.1074/jbc.M110.181974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gall A., Treuting P., Elkon K.B., Loo Y.-M., Gale M. Jr., Barber G.N., and Stetson D.B.. 2012. Autoimmunity initiates in nonhematopoietic cells and progresses via lymphocytes in an interferon-dependent autoimmune disease. Immunity. 36:120–131. 10.1016/j.immuni.2011.11.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao D., Li T., Li X.D., Chen X., Li Q.Z., Wight-Carter M., and Chen Z.J.. 2015. Activation of cyclic GMP-AMP synthase by self-DNA causes autoimmune diseases. Proc. Natl. Acad. Sci. USA. 112:E5699–E5705. 10.1073/pnas.1516465112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray E.E., Treuting P.M., Woodward J.J., and Stetson D.B.. 2015. Cutting Edge: cGAS Is Required for Lethal Autoimmune Disease in the Trex1-Deficient Mouse Model of Aicardi-Goutières Syndrome. J. Immunol. 195:1939–1943. 10.4049/jimmunol.1500969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Günther C., Kind B., Reijns M.A., Berndt N., Martinez-Bueno M., Wolf C., Tüngler V., Chara O., Lee Y.A., Hübner N., et al. 2015. Defective removal of ribonucleotides from DNA promotes systemic autoimmunity. J. Clin. Invest. 125:413–424. 10.1172/JCI78001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasan M., Koch J., Rakheja D., Pattnaik A.K., Brugarolas J., Dozmorov I., Levine B., Wakeland E.K., Lee-Kirsch M.A., and Yan N.. 2013. Trex1 regulates lysosomal biogenesis and interferon-independent activation of antiviral genes. Nat. Immunol. 14:61–71. 10.1038/ni.2475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiller B., Achleitner M., Glage S., Naumann R., Behrendt R., and Roers A.. 2012. Mammalian RNase H2 removes ribonucleotides from DNA to maintain genome integrity. J. Exp. Med. 209:1419–1426. 10.1084/jem.20120876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazear H.M., Lancaster A., Wilkins C., Suthar M.S., Huang A., Vick S.C., Clepper L., Thackray L., Brassil M.M., Virgin H.W., et al. 2013. IRF-3, IRF-5, and IRF-7 coordinately regulate the type I IFN response in myeloid dendritic cells downstream of MAVS signaling. PLoS Pathog. 9:e1003118 10.1371/journal.ppat.1003118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liddicoat B.J., Piskol R., Chalk A.M., Ramaswami G., Higuchi M., Hartner J.C., Li J.B., Seeburg P.H., and Walkley C.R.. 2015. RNA editing by ADAR1 prevents MDA5 sensing of endogenous dsRNA as nonself. Science. 349:1115–1120. 10.1126/science.aac7049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim Y.W., Sanz L.A., Xu X., Hartono S.R., and Chédin F.. 2015. Genome-wide DNA hypomethylation and RNA:DNA hybrid accumulation in Aicardi-Goutières syndrome. eLife. 4:111–112. 10.7554/eLife.08007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mankan A.K., Schmidt T., Chauhan D., Goldeck M., Höning K., Gaidt M., Kubarenko A.V., Andreeva L., Hopfner K.-P., and Hornung V.. 2014. Cytosolic RNA:DNA hybrids activate the cGAS-STING axis. EMBO J. 33:2937–2946. 10.15252/embj.201488726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mannion N.M., Greenwood S.M., Young R., Cox S., Brindle J., Read D., Nellåker C., Vesely C., Ponting C.P., McLaughlin P.J., et al. 2014. The RNA-editing enzyme ADAR1 controls innate immune responses to RNA. Cell Reports. 9:1482–1494. 10.1016/j.celrep.2014.10.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nick McElhinny S.A., Watts B.E., Kumar D., Watt D.L., Burgers P.M., Johansson E., Chabes A., and Kunkel T.A.. 2010. Abundant ribonucleotide incorporation into DNA by yeast replicative polymerases. Proc. Natl. Acad. Sci. USA. 107:4949–4954. 10.1073/pnas.0914857107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestal K., Funk C.C., Snyder J.M., Price N.D., Treuting P.M., and Stetson D.B.. 2015. Isoforms of RNA-Editing Enzyme ADAR1 Independently Control Nucleic Acid Sensor MDA5-Driven Autoimmunity and Multi-organ Development. Immunity. 43:933–944. 10.1016/j.immuni.2015.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehwinkel J., Maelfait J., Bridgeman A., Rigby R., Hayward B., Liberatore R.A., Bieniasz P.D., Towers G.J., Moita L.F., Crow Y.J., et al. 2013. SAMHD1-dependent retroviral control and escape in mice. EMBO J. 32:2454–2462. 10.1038/emboj.2013.163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reijns M.A., and Jackson A.P.. 2014. Ribonuclease H2 in health and disease. Biochem. Soc. Trans. 42:717–725. 10.1042/BST20140079 [DOI] [PubMed] [Google Scholar]
- Reijns M.A., Bubeck D., Gibson L.C., Graham S.C., Baillie G.S., Jones E.Y., and Jackson A.P.. 2011. The structure of the human RNase H2 complex defines key interaction interfaces relevant to enzyme function and human disease. J. Biol. Chem. 286:10530–10539. 10.1074/jbc.M110.177394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reijns M.A., Rabe B., Rigby R.E., Mill P., Astell K.R., Lettice L.A., Boyle S., Leitch A., Keighren M., Kilanowski F., et al. 2012. Enzymatic removal of ribonucleotides from DNA is essential for mammalian genome integrity and development. Cell. 149:1008–1022. 10.1016/j.cell.2012.04.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohman M.S., Koga Y., Takano K., Chon H., Crouch R.J., and Kanaya S.. 2008. Effect of the disease-causing mutations identified in human ribonuclease (RNase) H2 on the activities and stabilities of yeast RNase H2 and archaeal RNase HII. FEBS J. 275:4836–4849. 10.1111/j.1742-4658.2008.06622.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stetson D.B., Ko J.S., Heidmann T., and Medzhitov R.. 2008. Trex1 prevents cell-intrinsic initiation of autoimmunity. Cell. 134:587–598. 10.1016/j.cell.2008.06.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkman H.E., and Stetson D.B.. 2014. The enemy within: endogenous retroelements and autoimmune disease. Nat. Immunol. 15:415–422. 10.1038/ni.2872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y.G., Lindahl T., and Barnes D.E.. 2007. Trex1 exonuclease degrades ssDNA to prevent chronic checkpoint activation and autoimmune disease. Cell. 131:873–886. 10.1016/j.cell.2007.10.017 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.